Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Bucking the trend: understanding lipopolysaccharide structure and outer membrane dynamics in cold-adapted Pseudomonas isolated from Enigma Lake, Antarctica†
Marcello
Mercogliano
a,
Stefania
De Chiara
a,
Antonio
De Nicola
*bc,
Jacopo
Cardellini
de,
Costanza
Montis
*de,
Mikhail M.
Yakimov
f,
Violetta
La Cono
f,
Francesca
Crisafi
f,
Alba
Silipo
 ag,
Debora
Berti
ag,
Debora
Berti
 de,
Giuseppe
Milano
h,
Antonio
Molinaro
agi and
Flaviana
Di Lorenzo
de,
Giuseppe
Milano
h,
Antonio
Molinaro
agi and
Flaviana
Di Lorenzo
 *ag
*ag
aDepartment of Chemical Science, University of Naples Federico II, Via Cinthia, 4, Naples 80126, Italy. E-mail: flaviana.dilorenzov@unina.it
bCineca, Casalecchio di Reno (BO) 40033, Italy. E-mail: adenicola.chem@gmail.com
cResearch Center for Organic Electronics (ROEL), Yamagata University, 4-3-16 Jonan, Yonezawa, Yamagata 992-8510, Japan
dConsorzio Interuniversitario Per Lo Sviluppo Dei Sistemi A Grande Interfase, Via Della Lastruccia 3, Firenze 50019, Italy. E-mail: costanza.montis@unifi.it
eDipartimento Di Chimica “Ugo Schiff”, Università Degli Studi di Firenze, Via Della Lastruccia 3, Firenze 50019, Italy
fInstitute of Polar Sciences, National Research Council of Italy (ISP-CNR), Via S. Raineri 4, Messina 98122, Italy
gCEINGE-Biotecnologie Avanzate Franco Salvatore, Via Gaetano Salvatore 486, Napoli 80145, Italy
hDepartment of Chemical, Material and Production Engineering, University of Naples Federico II, Piazzale V. Tecchio, 80, Naples 80125, Italy
iDepartment of Chemistry, School of Science, Osaka University 1-1 Osaka University Machikaneyama, Toyonaka, Osaka 560-0043, Japan
First published on 18th September 2024
Abstract
Cold environments are predominant over the Earth and are inhabited by bacteria able to cope with a series of simultaneous environmental pressures. Gram-negative species of the Pseudomonas genus are the predominant ones isolated from cold habitats, making them an excellent model for studying the mechanisms of bacterial adaptation to the most extreme habitats on our planet. Here we focused on the lipopolysaccharide (LPS) structure and the outer membrane dynamics of Pseudomonas sp. EW#7, a strain isolated from Enigma Lake in Antarctica where, among other extreme characteristics, water temperature can reach 0.4 °C. We show that near-zero growth temperature mostly affects the LPS lipid A component. An uncommon tendency of decreasing lipid A secondary hydroxylation while increasing its phosphorylation degree was observed. This resulted in a faster lateral diffusion of lipid chains in the membrane and therefore in an enhancement of its fluctuations that guarantee membrane integrity and flexibility.
Introduction
Pseudomonas, which currently is the genus of Gram-negative bacteria with the largest number of species, is an excellent example of widespread microorganisms able to adapt to various growth conditions and complex environments, comprising terrestrial, freshwater, marine ecosystems as well as the human body.1 This remarkable versatility makes Pseudomonas a perfect research model to explore the influence of environmental factors on chemical and physical properties of the bacterial outer membrane. Since growth temperature significantly impacts both on structural and biological properties, seminal studies focused on the analysis of Pseudomonas strains grown at different temperatures revealing an optimal growth temperature in the range 25–30 °C.2–5 Nevertheless, low-temperature habitats are ideal sites for the colonization of several Pseudomonas species, which make them among the predominant ones isolated from cold environments.6 Considerable interest has been developed and progress has been made to understand how Pseudomonas spp. acclimatize to low temperature, thus disclosing a number of survival strategies based on bacteria physiological changes.7 These include the production and release of anti-freeze proteins and cryoprotectants, alteration of metabolic processes and acquisition of energy via alternate routes. Certainly, one key tactic for survival at low temperatures is to ensure membrane integrity and functionality by controlling the fluidity of the membrane itself.7 As a matter of fact, Pseudomonas typically reshapes membrane fluidity by altering the lipid polar head, charge, location and size, length and distribution of fatty acid chains.8 For example, it has been extensively reviewed that an increased amount of low-melting-point fatty acids, which comprises monounsaturated, polyunsaturated and branched-chain fatty acids, results in an improved membrane fluidity compared to membranes constituted by their saturated straight analogs.9–11 Of note, transcriptome analyses have demonstrated that exposure to cold temperatures induces up-regulation of specific genes involved in outer membrane biogenesis, such as fatty acid and lipopolysaccharide (LPS) biosynthesis, whereas the expression of genes encoding other outer membrane structures, like flagella, is mostly suppressed.12As the main constituent of the Pseudomonas outer membrane, LPS heavily contributes to the structural integrity and fluidity of the cell envelope, thus providing resilience against external stressors.13 The dynamic properties of LPSs are strictly related to their chemical structure, which includes a glycolipid anchor to the membrane, the lipid A, a core oligosaccharide (core OS), and a polysaccharide chain, termed O-antigen.13 An LPS made up of these three components is defined smooth-type LPS (or S-LPS), whereas when the polysaccharide moiety is absent the terminology employed is rough-type LPS (or R-LPS).14 Contextually, bacteria typically modify the inner core OS-lipid A area to enhance the integrity of the outer membrane. This is achieved through changes in the composition of acyl chains, phosphate groups, and other functional groups.15 Likewise, modulation of the strength of membrane bilayers through O-antigen expression was also investigated by molecular coarse-grained Molecular Dynamics (MD) simulations, which evidenced how the lamellar packing of O-antigen chains affects lipid mobility within the membrane and confers an increased capacity to withstand significant surface tension.16,17
In this work we focused on a specific cold adapted strain Pseudomonas sp. EW#7, which was isolated from water column of the permanently ice-covered Enigma Lake (Antarctica).18,19 Of note, the main water masses of the Antarctica Enigma Lake present unique extreme conditions characterized by strong alkalinity, oxygen oversaturation, and a temperature that does not exceed 0.6 °C.20 The uniqueness of this microbial isolation boosted our interest in investigating the structure and molecular characteristics of the outer membrane and its LPS constituent from such an intriguing polyextremophile. Specifically, we have isolated LPS from Pseudomonas sp. EW#7 grown at two different temperatures, i.e. 0.4 °C (in situ temperature of the collected water of Enigma Lake) and 20 °C (referred to hereafter as LPS0.4C and LPS20C). We have defined their chemical structures, the morphology of their assemblies in water, the viscoelastic properties of LPS membranes, and the effects of their lipid A in membrane rigidity. An uncommon tendency of decreasing lipid A hydroxylation while increasing its phosphorylation was observed for the bacterium grown at 0.4 °C, which resulted in a faster lateral diffusion of lipid chains and therefore in an enhanced membrane fluctuation responsible for membrane flexibility and integrity preservation.
Results and discussion
LPS isolation and structural characterization
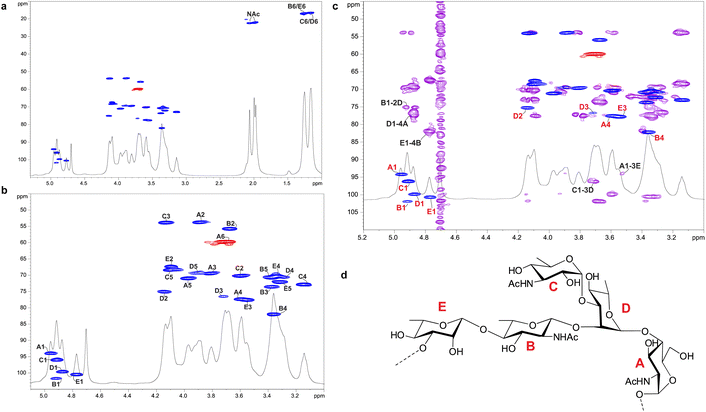
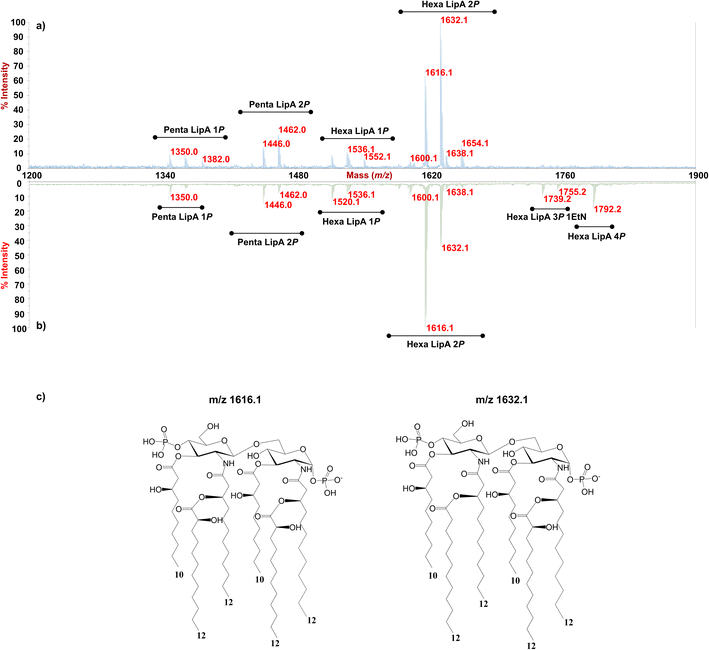
Enigma Lake is the largest and deepest perennially frozen lake occurring in the Northern Foothills, Northern Victoria Land in Antarctica.18,19,21 This peculiar lake, whose name is due to the presence of an unusual debris-laden area in its middle, hosts a large variety of microorganisms, like Pseudomonas species, able to withstand harsh conditions. As a matter of fact, Pseudomonas species are widely recognized as one of the most metabolically diversified bacterial colonizers of cold habitats.6,22 Therefore, we have isolated Pseudomonas sp. EW#7 grown at 0.4 °C and 20 °C and characterized the structure and morphology of its membrane. LPS0.4C and LPS20C were isolated from lyophilized bacterial pellets through the hot phenol-water extraction method23 and then extensively purified by means of enzymatic digestion and ultracentrifugation. Purified LPS fractions were both inspected via SDS-PAGE followed by silver nitrate staining, which revealed that both Pseudomonas produce a smooth-type LPS. Chemical analyses24–26 highlighted an identical sugar and lipid composition. In detail, monosaccharide analyses revealed the presence of L-rhamnose (L-Rha), 2,6-dideoxy-2-amino-L-glucose (L-quinovosamine, L-QuiN) and 3,6-dideoxy-3-amino-D-glucose (D-Qui3N), 2-amino-2-deoxy-D-glucose (D-GlcN), and in minor amount D-glucose (D-Glc), 2-amino-2-deoxy-D-galactose (D-GalN), L-glycero-D-manno-heptose (L,D-Hep), and 3-deoxy-D-manno-oct-2-ulopyranosonicacid (Kdo). Linkage analysis revealed the occurrence mainly of 2,3-disubstituted L-Rha and 3-substituted L-Rha, 4-substituted-D-GlcN, 4-substituted-L-QuiN, terminal D-Qui3N. As stated above, fatty acids composition, which was obtained by GC-MS analysis of the related methyl ester derivatives, showed the same composition for both LPSs. However, a higher abundance of 2-hydroxylated acyl moieties was noticed for LPS20C compared to LPS0.4C. In detail, in both LPSs it was highlighted the presence of R-3-hydroxydodecanoic acid [12:0(3-OH)] and S-2-hydroxydodecanoic acid [12:0(2-OH)], R-3-hydroxydecanoic [10:0(3-OH)] and dodecanoic acid (12:0), which was in agreement with lipid A fatty acid composition found for other Pseudomonas strains.27,28 To define the structure of the carbohydrate and lipid portion, a mild acid hydrolysis was performed on an aliquot of both purified LPS0.4C and LPS20C to selectively cleave the linkage between the non-reducing GlcN of the lipid A and the Kdo. The lipid A fractions were then inspected by MALDI-TOF MS and MS/MS while the carbohydrate portions were further purified by size-exclusion chromatography and then analyzed via 1D and 2D NMR. The NMR investigation revealed that bacteria grown at different temperatures express the same O-antigen repeating unit that was fully characterized. In detail, the 1H, 1H DOSY and 1H, 13C HSQC NMR spectra of the carbohydrate portion of both LPS0.4C and LPS20C (Fig. 1a, b and S1†) highlighted the presence of five anomeric signals, diagnostic for five spin systems (A–E, Table S1†). The structural assignment of each spin system was performed by tracing the spin connectivity visible in the COSY and TOCSY spectra, while the assignment of carbon atoms was deduced by analyzing the HSQC spectrum. The anomeric configuration of each monosaccharide unit was defined tracing the intra-residue NOE correlations observable in the NOESY spectrum.29 In detail, residues A, B and C were identified as gluco-configured sugar units as inferred by the efficient propagation of magnetization in the TOCSY spectrum and by their 13C chemical shift values, with C-4 of A and B (77.2 and 82.0 ppm, respectively, Table S1†) shifted at low field by glycosylation. An α-anomeric configuration was deduced for residues A and C, while B was a β-configured sugar unit as proven by the evident intra-residue NOE correlation between its H-1, H-3 and H-5. The observation that from the anomeric proton of B and C it was possible to identify connections up to the methyl groups (H-6) at 1.27 ppm (B) and 1.14 ppm (C), along with the correlation of the H-2 of B and H-3 of C with two nitrogen-bearing carbon atoms (55.8 and 53.8 ppm), led to their identification as QuiN and Qui3N, respectively. Likewise, residue A was a terminal GlcN assigned on the basis of its H-2 proton that correlated with a nitrogen-bearing carbon atom at 53.8 ppm. All the three amino sugar residues (A-C) also bear N-acetyl groups (Table S1†). Spin systems D and E were assigned to rhamnoses, as indicated in the TOCSY spectrum by the scalar correlations from H-2 to H-6 methyl protons (1.16 and 1.26 ppm). D and E were α- and β-configured respectively at the anomeric center on the basis of the agreement between their C-5 values (69.3 and 72.1 ppm) and the value reported for the related O-methyl rhamnoside (69.4 ppm for α anomer, 73.6 for β anomer).29 Downfield displacement of carbon signals revealed substitution at O-2 and O-3 of D and at O-3 of E. The primary sequence of the O-antigen repeating unit was then established by tracing both inter-residue NOE contacts and the long–range correlations visible in the 1H,13C HMBC (Fig. 1c and S2†). Briefly, β-L-QuiNAc residue B was found to be substituted at its position O-4 by β -L-Rha E, in turn substituted at its position O-3 by α-D-GlcNAc A, as proven by the related NOE contact between H-1 E and H-4 B and H-1 A and H-3 E (Fig. S2†). Residue A was in turn substituted at position O-4 by α-L-Rha D that carried the terminal α-D-Qui3NAc C at position O-3 and β-L-QuiNAc B at position O-2, as demonstrated by the detection of the related HMBC correlations (Fig. 1c). Therefore, the O-antigen repeating unit was defined as the branched pentasaccharide reported in Fig. 1d. These results led to the conclusion that the growth temperature did not affect the carbohydrate portion of LPS, as both Pseudomonas sp. EW#7 produced the same O-antigen repeating unit.As for the lipid A moiety, negative-ion MALDI-TOF MS spectra were recorded for both LPSs highlighting key structural differences. As shown in Fig. 2, both Pseudomonas produced a mixture of mono and bis-phosphorylated penta- and hexa-acylated lipid A species. For both lipid A variants, MALDI-TOF MS spectra displayed three distinct peaks with differences of 16 amu, due to the presence or absence of a hydroxyl group in the secondary C12 acyl substituents, although different relative abundances were observable between the two spectra. In fact, the main lipid A species identified for the bacterium grown at 20 °C (Fig. 2a) was detected at m/z 1632.1 and assigned to bis-phosphorylated hexa-acyl lipid A bearing four 12:0(OH) and two 10:0(OH), whereas the main species for the Pseudomonas sp. EW#7 grown at 0.4 °C (Fig. 2b) was detected at m/z 1616.2 and matched with bis-phosphorylated hexa-acyl lipid A bearing three 12:0(OH), two 10:0(OH), and one 12:0. Minor peaks at −16 amu (m/z 1600.1), detected in both spectra, indicated the absence of a hydroxyl group on the acyl chains. Therefore, a reduction in the lipid A hydroxylation state was observed when growth temperature was dramatically decreased to 0.4 °C. Mono- and bis-phosphorylated penta-acylated forms were also identified and were assigned to species devoid of a C10:0(OH) moiety. Besides the hydroxylation pattern, a slight decrease in the relative intensity of peaks corresponding to penta-acylated lipid A species was also noticed when Pseudomonas sp. EW#7 was grown at low temperature. Notably, only when Pseudomonas sp. EW#7 was grown at 0.4 °C it was possible to identify an additional cluster of peaks that matched with bis-phosphorylated hexa-acylated species further decorated by a 2-aminoethyl phosphate group (at m/z 1755.2) and by two additional phosphate groups (m/z 1792.2). Negative-ion MALDI-TOF MS/MS analysis of the main species provided specific fragmentation patterns that guided the structural characterization of lipid A structures (Fig S3 and Table S2†), which agreed with previous structures from Pseudomonas sp., and confirmed a higher degree of secondary hydroxylation of fatty acid chains for LPS20C compared to LPS0.4C. According to chemical analyses, it is plausible to conclude that LPS20C and LPS0.4C are built with the same core OS structure, although it remains to be defined. Therefore, the growth temperature seems to exclusively affect the lipid A portion in Pseudomonas sp. EW#7, producing at 0.4 °C lipid A species with a lower degree of secondary 2-hydroxylation than at 20 °C, but with a higher degree of phosphorylation. These results were extremely interesting as previous studies have highlighted an increment of hydroxylated fatty acids in LPS from another cold-adapted Pseudomonas, i.e. P. syringae, at low temperature compared to 22 °C temperature-grown cells, with differences affecting only primary acyl chains and not secondary 12:0 (2-OH).30 In addition, Pseudomonas sp. EW#7 did not produce lipid A species carrying unsaturated or branched acyl chains, which are well known strategies employed by bacteria to provide an adequate membrane fluidity in harsh and highly variable habitats.9,31–35 As it has been reviewed by Hassan et al.,9Pseudomonas is the most dominant genus isolated from cold environments, followed by Arthrobacter, Brevundimonas, and Massilia, whose cell membrane fatty acids, in 24 out of 42 strains, were mainly straight-chain monounsaturated, whereas 18 species were found to have branched acyl chains, both saturated and monounsaturated.9 Among these, P. mandelii and P. veronii displayed the highest percentage of unsaturated acyl chains and P. frederiksbergensis the highest percentage of branched fatty acids.9 In addition, the alteration of LPS phosphorylation in response to high and low temperatures is acknowledged as aiding modulation of membrane function and consequently help in cold-temperature adaptation. On the contrary, previous studies on P. syringae showed that LPS was phosphorylated more at 22 °C and less at 4 °C, although most of this increment in phosphorylation was associated with the core OS region.36
Cryo-EM and SAXS characterization of Pseudomonas sp. EW#7 LPS0.4C and LPS20C
To glean molecular information about LPS0.4C and LPS20C, we first investigated structure and morphology of both LPS assemblies in water via Cryo-EM images and Small-Angle X-ray Scattering (SAXS) experiments, which are complementary techniques (in the direct and reciprocal space, respectively) enabling structural characterization at the sub-micron scale.Both LPS0.4C and LPS20C were dispersed in water at a concentration of 1 mg mL−1 (see experimental section). As shown, in Fig. 3a, LPS0.4C and LPS20C dispersion assembled into vesicle-like structures, featuring a well-defined bilayer that separates an inner aqueous pool from the external medium. It is worth noting that a minority of spherical and rod-like structures without the bilayer were also visible (Fig. S4†). However, although being very informative on the shape and size of aggregates, Cryo-EM may be not fully representative of the whole sample, since only a few assemblies are imaged. To fill this gap and to identify possible structural differences between the LPS0.4C and LPS20C at the ensemble-average level, SAXS was used to assess the overall shape of LPS assemblies. The scattering intensity I(Q) as a function of the scattering vector (Q) is reported in Fig. 3b. SAXS profiles originating from the scattering of LPS0.4C and LPS20C (red circles and blue circles, respectively) were almost superimposable. This experimental result clearly indicated that the differences in the LPS molecular structures (i.e. in the lipid A moiety) did not significantly affect the overall structures of the aggregates. To get additional molecular insights, the SAXS curves were analyzed through a bilayered vesicle model with the SASfit software package.37 According to this model, the inner and outer layers of hydrophilic heads (th) are considered as two concentric shells with identical scattering length density (SLD), separated by an additional shell of hydrophobic tails (tt) with a lower SLD with respect to the solvent. The SLD values inserted in the model fitting were consistent with recent literature reports.38 Fitting parameters (Table S3†) and further details about SAXS measurements are reported in the ESI.† This analysis confirmed the locally bilayered shape of LPS0.4C and LPS20C assemblies. The total bilayer thickness (tt + 2 th) for LPS0.4C and LPS20C was in agreement with literature values evaluated for several LPS membranes.39–41 Overall, SAXS and Cryo-EM results highlighted that the molecular differences in the LPS molecular structures do not affect the overall morphology of their assemblies in water and slightly affect the bilayer thickness.
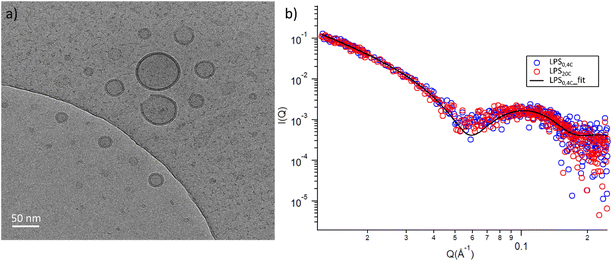 | ||
| Fig. 3 (a) Cryo-EM micrographs of 1 mg mL−1 LPS water solution; (b) log–log SAXS profiles of 1 mg mL−1 water dispersion of LPS0.4C (red circles) and LPS20C (blue circles). | ||
To shed light on the viscoelastic properties of LPS membranes, we collected lateral–pressure isotherms of the LPS films at the air water-interface, as LPS monolayers have proven to be accurate, albeit simple model interfaces to simulate the behavior of the outer membranes of Gram-negative bacteria.42,43
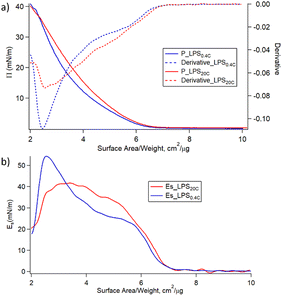
Fig. 4a shows the variation of the surface pressure (π) as a function of the air–water interfacial area for LPS0.4C and LPS20C. Further compression leads to the increase of π until the film collapses occurring at 35–40 mN m−1 for both LPS molecules. Such low lateral pressure values of the collapse of the monolayers highlights the high water solubility of LPS monomers. When the pressure increases, the LPS monomers desorb from the air–water interface, eventually forming the spontaneous assemblies observed in SAXS and Cryo-EM measurements. Additionally, the monolayer compressibility modulus (Es) was evaluated as a function of the area per unit weight and is reported in Fig. 4b. Initially, Es increases similarly for the two LPSs. Then, for higher values of lateral pressure (5 mN m−1 < π < 25 mN m−1) the compressibility, and in turn, the rigidity of the LPS20C monolayer, overcomes the one of LPS0.4C. However, when approaching pressure values relevant for membrane conditions (25 mN m−1 < π < 30 mN m−1), the compressibility moduli of the monolayers assume similar values for the two LPS. Reasonably, this evidence suggests that, in these experimental conditions, the compositional differences between the two LPSs slightly affect the rigidity of the monolayer. The same trends of isotherms and, in turn, similar behavior of compressive moduli were obtained during the compression of heterogeneous monolayers composed of LPS, DOPE, and DSPE, selected as more realistic compositional mimics of Gram-negative outer membranes (Fig. S5†). As a more general consideration, the compressibility values obtained at membrane pressures relevant for membrane conditions (Es = 40 mN m−1) correspond to the formation of liquid-expanded (LE) films,43 suggesting the formation of soft LPSs monolayers. Accordingly, as shown in Table 1, the compressibility of LPS0.4C and LPS20C are significantly lower compared to compressibility moduli obtained for E. coli LPS monolayers and literature values obtained for different LPSs (Fig. S5†).44,45
Molecular dynamics simulations
Intrigued by the diverse degree of secondary 2-hydroxylation observed for LPS0.4C and LPS20C, we next used MD computational technique to evaluate whether the structural differences in the glycolipid anchor of the two LPSs impact on membrane properties. For simplicity, we chose to investigate the most representative bis-phosphorylated hexa-acyl lipid A structures, as experimentally identified, adopting a full atom representation of the molecules. Computational details and atomistic model description are reported in the ESI section.† To understand the effects of hydroxylation on membrane properties, in terms of the lipid A chemical distinctions, three different lipid bilayer systems were constructed. The lower layers of the three systems were composed exclusively by DOPE and DSPE phospholipids, while for the upper layers we set: one having exclusively a single secondary 2-hydroxylated acyl substitution (referred to as LipAmono-hydrated), one presenting two 2-hydroxylated acyl substitutions (LipAdi-hydrated) and one comprising a mixture of both types (LipAmix). The choice to have different lipid compositions of the simulated systems has been taken to closely match the estimated distribution of mono and bis-phosphorylated hexa-acylated lipid A produced by Pseudomonas in low temperature conditions. In Table 2, the composition of simulated system is reported, while in Fig. S6† a pictorial view of the difference in the lipid A composition at low and higher temperature is shown. From MD simulations, we extracted structural information about distribution of molecules in the cross-section direction of the bacteria membrane plane, enabling us to compute the thickness of the hydrophobic core of and compared it with experimental estimations made by SAXS measurements. Specifically, simulations yielded a hydrophobic tail thickness of approximately 2.2 nm at both temperatures, which is close to the experimentally measured value of 2.8 nm. The overall membrane thickness, calculated as the peak-to-peak distance of PO4 groups, was found to be 4.1 nm for the system simulated at 20 °C, and 4.2 nm for the system at 0 °C. The results from MD simulations match the experimental indication about the little structural differences between the two bilayers growth at low and higher temperature. Calculated mass density profiles are reported in Fig. 5. Visual inspection of the MD runs revealed a common behavior at the simulated temperatures regarding the interactions between mono- and di-hydrated LipA in the mixed systems. Starting from a random initial distribution of LipAdi-hydrated in the upper layer of the bacterial membrane, we observed the formation of small aggregates of LipAdi-hydrated as the simulation progressed. Notably, once these small aggregates (comprising 2 or 3 LipA molecules) are formed, they remained stable longer and diffused as larger single units. The picture of this behavior can be gained looking at the time sequence of LipA molecules reported in the snapshots of Fig. 6. The snapshots sequence of the upper-layer shows the formation of stable LipAdi-hydrated clusters (top-view of LipAs). Above 300 ns, clusters of LipAdi-hydrated (depicted in light blue) are formed and remain stable for the whole simulated time. The presence of these stable clusters can potentially affect the behavior of the lateral diffusion of both mono- and di-hydrated LipA, as well as membrane area fluctuations, thereby altering membrane fluidity. To explore this phenomenon, we have exploited the concept that rigidity or fluidity of a membrane is linked to the amount of energy required to deform it. This can be expressed as the work needed to compress or expand a membrane (referred to as the Helmholtz free energy change) and from this relationship we could calculate the isothermal area compressibility (see the Area Compressibility Calculation section in the ESI†). Through MD simulations, it is possible to directly compute the isothermal area compressibility, providing insights into membrane rigidity. Through this approach, we have reported in Fig. 7a the membrane area fluctuations calculated from MD runs. As observed in Fig. 7a, the isothermal area compressibility remained almost unchanged for systems with a higher percentage of LipAmono-hydrated (Table 2), but slightly higher than systems comprised solely of LipAdi-hydrated. This observation suggested an increased flexibility for membranes containing a higher proportion of LipAmono-hydrated, in alignment with experimental observations and monolayer compressibility modulus measurements.| System | Lipid A* | DOPE | DSPE | Water | Box [nm] | Temp. [°C] | |
|---|---|---|---|---|---|---|---|
| Mono-hydrated | Di-hydrated | ||||||
| LipA0.4mix | 52 | 23 | 21 (upper) | 4 (upper) | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 107 107 |
9.64804 | 0 |
| LipA0.4mono-hydrated | 75 | — | 84 (lower) | 16 (lower) | 9.64804 | ||
| LipA0.4di-hydrated | — | 75 | 8.79987 | ||||
| LipA20mix | 45 | 30 | 21 (upper) | 4 (upper) | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 107 107 |
9.77029 | 20 |
| LipA20mono-hydrated | 75 | — | 84 (lower) | 16 (lower) | 9.77029 | ||
| LipA20di-hydrated | — | 75 | 8.45202 | ||||
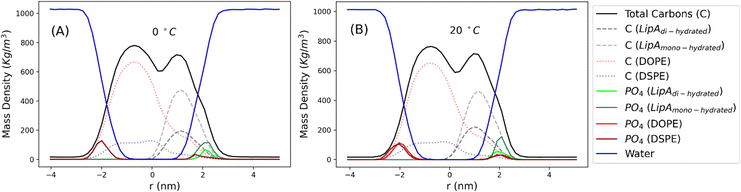 | ||
| Fig. 5 Mass density profiles calculated for the mix systems (see Table 2 for composition details) simulated at 0 and 20 °C. Profiles have been calculated averaging over the last 250 ns of production runs. | ||
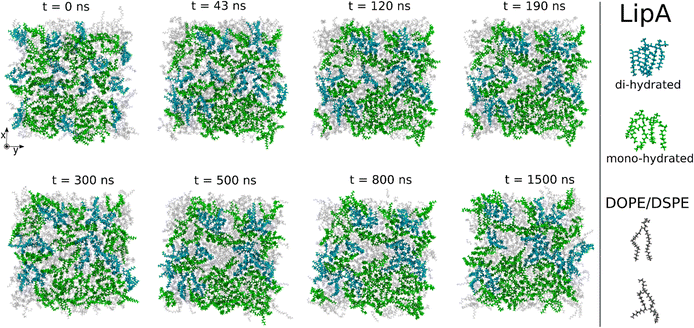 | ||
| Fig. 6 Sequence of top-view snapshots of the mixed system (see Table 2) simulated at 20 °C. All lipids are shown only in the upper layer of the membrane. LipAmono-hydrated molecules are depicted in green, LipAdi-hydrated molecules are shown in blue, and DSPE and DOPE phospholipids are both represented in light gray. A similar behavior is observed in the simulations of the mixed system at 0 °C. | ||
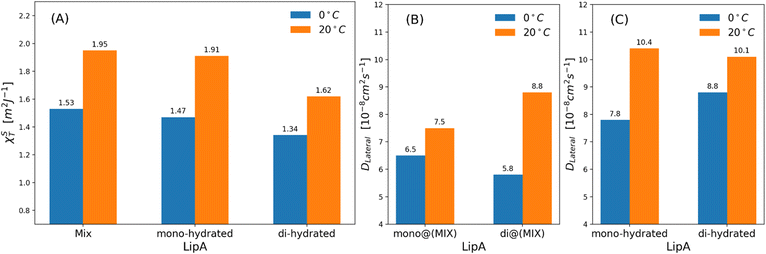 | ||
| Fig. 7 (a) Isothermal area compressibility calculated for lipid membranes as function of lipid A composition and temperatures. The calculations have been made considering the last 500 ns of production runs. Lateral diffusion DLateral calculated at two different temperatures and for: (b) mixed systems (see Table 2), (c) systems in which Lipids A are exclusively mono-hydrated or di-hydrated. The time behavior of area per lipid is reported, for all systems, in Fig. S7. | ||
Interestingly, the decrease of the isothermal area compressibility (a reduction of about 17–26%) due to the reduction of temperature from 20 °C to 0 °C was modest. For the sake of comparison, the isothermal area compressibility of a DPPC membrane decreases from to 6.9–7.5 m2 J−1 (at 25 °C) to 1.1 m2 J−1 (at 8 °C), indicating, in this case, a strong temperature-dependence and a drastic change of membrane fluidity.46,47 The results here reported clearly show how membrane fluidity of LipAmix is preserved, thus allowing for membrane integrity at low temperature by means of modulation of lipid A chemistry. Finally, we calculated lateral self-diffusion coefficient DL from MD simulations because DL is correlated to the translational membrane fluidity (further details in the ESI†). In Fig. 7b and c, DL is computed and averaged over all LipA molecules belonging to the upper leaflet of the membrane. DL coefficients were calculated based on 2 μs-long trajectories. The bar plot in Fig. 7b, in the case of the mixtures, revealed an inversion in lateral diffusion for LipAmono-hydrated, which exhibited faster diffusion (mono@(MIX) in Fig. 7bDL = 6.5 × 10−8 cm2 s−1) than LipAdi-hydrated di@(MIX) in Fig. 7b.
D L = 5.8 × 10−8 cm2 s−1) at low temperature (0 °C), but a slightly slower diffusion (mono@(MIX) in Fig. 7bDL = 7.5 × 10−8 cm2 s−1), compared to LipAdi-hydrated (di@(MIX) in Fig. 7bDL = 8.8 × 10−8 cm2 s−1) at higher temperature (20 °C). Differently, in systems where the Lipid A composition is exclusively LipAmono-hydrated or LipAdi-hydrated, the lateral diffusion LipAmono-hydrated at 0 °C (DL = 7.8 × 10−8 cm2 s−1) is lower than that of LipAdi-hydrated (DL = 8.8 × 10−8 cm2 s−1), while at 20 °C, both Lipid As show similar DL values (see Fig. 7c). These results indicate that the significant decrease in lateral diffusion at 0 °C and 20 °C for LipAdi-hydrated in the mixed systems is somehow mitigated by the DL of LipAmono-hydrated, which is less affected by temperature changes. Additionally, given that the composition of the mixed system at lower temperatures is richer in LipAmono-hydrated compared to the composition at 20 °C, we can speculate that this contributes to preserve the membrane area fluctuations, thereby maintaining membrane fluidity.
Conclusions
Enigma Lake in Antarctica is home to a unique microbial community able to face the strongly alkalinity, oxygen oversaturation and near-zero temperatures of this habitat.20 Among these microorganisms, we have isolated Pseudomonas sp. EW#7. Structural and morphological characterization of membrane of bacteria inhabiting Enigma Lake is crucial for understanding the essential components that contribute to their innate adaptive capacity to cope with cold and its associated stresses. In this perspective, we have focused on the main constituent of Pseudomonas sp. EW#7 outer membrane, the LPS, and have defined its chemical structure, the morphology of LPS assemblies in water, the viscoelastic properties of LPS membranes, as well as the impact of its lipid A component in membrane rigidity. To do this, we have isolated the LPS from Pseudomonas sp. EW#7 grown at 0.4 °C (LPS0.4C) and compared to the LPS from the same bacterium grown at 20 °C (LPS20C). We have disclosed that the growth temperature did not affect the carbohydrate portion of LPS, as both Pseudomonas sp. EW#7 produced the same O-antigen repeating unit, which was identified as a novel pentasaccharide made up of Rha, QuiNAc, Qui3NAc and GlcNAc (Fig. 1d). Indeed, also our SAXS and Cryo-EM results highlighted a similar morphology of LPS0.4C and LPS20C assemblies in water, likewise the mechanical properties of the LPS monolayer at the air–water interface resulted similar. On the contrary, structural differences were identified in the lipid A portion, surprisingly showing that Pseudomonas sp. EW#7 tends to decrease the degree of lipid A secondary hydroxylation to grow at 0.4 °C. Our molecular dynamics studies have evidenced that, at low temperature, lipid As carrying only one secondary hydroxylation (LipAmono-hydrated) exhibit faster lateral diffusion than lipid As with two secondary 12:0 (2-OH) (LipAdi-hydrated), thus facilitating membrane fluctuations and consequently preserving membrane flexibility and integrity. Of note, and still against the trend, lipid A from Pseudomonas sp. EW#7 grown at 0.4 °C also displayed a more heterogenous phosphorylation pattern, with minor species carrying four phosphate groups or three phosphates and one EtN group, which were not produced by the bacterium when grown at 20 °C. Therefore, although only present as minor species, our data demonstrates that Pseudomonas sp. EW#7 may adopt lipid A hyperphosphorylation as a strategy to adapt to the extremely cold environmental temperature.Overall, the combination of experimental and computational approaches enabled us to investigate the systems at multiple levels of resolution, providing a more comprehensive understanding of the structural and dynamic properties of the LPS membranes. However, more in-depth studies will be conducted to understand the full significance of Pseudomonas sp. EW#7 lipid A phosphorylation in the physiology of its outer membrane. In addition, whether these structural differences are species-dependent or are due to the growth temperature of 0.4 °C, i.e. lower than the typical 4 °C used in other studies, remains to be defined.
Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
A. M. conceived the study. A. M., A. S. and F. D. L. designed the research. M. M. and S. D. C. extracted, purified the LPS and analyzed the data. F. D. L. run MS and NMR studies. A. D. N. and G. M. run computational studies. J. C., C. M. and D. B. performed SAXS and Cryo-EM analyses. M. M. Y., V. L. C and F. C. provided bacterial cells. All the authors contributed to analyzing the data and writing the manuscript and have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
AM acknowledges National Recovery and Resilience Plan (NRRP), Mission 4 Component 2 Investment 1.3 – Call for tender No. 341, Project code PE00000003, ON Foods; CN2 Agritech National Research Center and the European Union Next-Generation EU Missione 4, Componente 2, Investimento 1.4 D.D. 1032 CN00000022; POS5 Italian Ministry of Health – Health Operational Plan Trajectory 5 – Mediterranean Diet for Human Health Lab “MeDiHealthLab”; code T5-AN-07. FDL acknowledges European Research Council (ERC) under the Horizon Europe program under grant agreement no 101039841 (DEBUGGING LPS). This work was also supported in part by the Italian Ministry of Foreign Affairs and International Cooperation (Italy-Germany Science and Technology cooperation–Call for joint research proposals for the years 2023–2025) to FDL. FDL also acknowledges Ministry of Education, Universities and Research, PRIN MUR 2022 (2022SHW3KY) and PRIN MUR PNRR 2022 (P202293ZMC). AS acknowledges Ministry of Education, Universities and Research, PRIN MUR 2022 (2022ZEZS45), PRIN MUR PNRR 2022 (P2022M457Z), and PNRR, Missione 4 – Componente 2 – NextGenerationEU – Partenariato Esteso INF-ACT – One Health Basic and Translational Research Actions Addressing Unmet Needs on Emerging Infectious Diseases MUR: PE00000007. AS also acknowledges the National Antarctic Research Program (PNRA), Italian Ministry of Education and Research (Research project PNRA16_00089) and MMY, VLC and FC acknowledge PNRA, Italian Ministry of Education and Research (Research Projects PNRA 16_00121).References
- M. Gomila, A. Peña, M. Mulet, J. Lalucat and E. García-Valdés, Front. Microbiol., 2015, 6, 214 CrossRef.
- D. Sengupta and M. K. Chattopadhyay, J. Biosci., 2013, 38, 409–412 CrossRef CAS.
- P. De Maayer, D. Anderson, C. Cary and D. A. Cowan, EMBO Rep., 2014, 15, 508–517 CrossRef CAS PubMed.
- W. Schumann, Cell Stress Chaperones, 2016, 21, 959–968 CrossRef CAS PubMed.
- P. M. Tribelli and N. I. López, Appl. Microbiol. Biotechnol., 2022, 106, 7699–7709 CrossRef CAS PubMed.
- M. Chauhan, A. Kimothi, A. Sharma and A. Pandey, Front. Microbiol., 2023, 14, 1218708 CrossRef PubMed.
- P. M. Tribelli, E. C. Solar Venero, M. M. Ricardi, M. Gómez-Lozano, L. J. Raiger Iustman, S. Molin and N. I. López, PLoS One, 2015, 10, e0145353 CrossRef.
- A. Tamby, J. S. Sinninghe Damsté and L. Villanueva, Front. Mol. Biosci., 2023, 9, 1058381 CrossRef PubMed.
- N. Hassan, A. M. Anesio, M. Rafiq, J. Holtvoeth, I. Bull, A. Haleem, A. A. Shah and F. Hasan, Front. Microbiol., 2020, 11, 824 CrossRef PubMed.
- S. D'Amico, T. Collins, J.-C. Marx, G. Feller and C. Gerday, EMBO Rep., 2006, 7, 385–389 CrossRef PubMed.
- J. W. Deming, Curr. Opin. Microbiol., 2002, 5, 301–309 CrossRef CAS.
- S. Frank, F. Schmidt, J. Klockgether, C. F. Davenport, M. Gesell Salazar, U. Völker and B. Tümmler, FEMS Microbiol. Lett., 2011, 318, 47–54 CrossRef CAS PubMed.
- F. Di Lorenzo, K. A. Duda, R. Lanzetta, A. Silipo, C. De Castro and A. Molinaro, Chem. Rev., 2022, 122, 15767–15821 CrossRef CAS PubMed.
- M. Pallach, F. Di Lorenzo, F. A. Facchini, D. Gully, E. Giraud, F. Peri, K. A. Duda, A. Molinaro and A. Silipo, Int. J. Biol. Macromol., 2018, 119, 1027–1035 CrossRef CAS.
- B. D. Needham and M. S. Trent, Nat. Rev. Microbiol., 2013, 11, 467–481 CrossRef CAS.
- D. Jefferies, J. Shearer and S. Khalid, J. Phys. Chem. B, 2019, 123, 3567–3575 CrossRef CAS PubMed.
- M. D. Tomasini, Exp. Biol. Med., 2010, 235, 181–188 CrossRef CAS.
- T. J. Chinn, Glacial history and glaciology of Terra Nova Bay area. Logistic report for K 162, Christchurch, NZ, 1985 Search PubMed.
- H. M. French and M. Guglielmin, Geogr. Ann. Ser. A Phys. Geogr., 2000, 82, 513–526 CrossRef.
- F. Smedile, V. La Cono, S. Urbini, G. Benedetti, G. La Spada, F. Crisafi, M. Azzaro, N. Porcino, S. Fazi, S. Amalfitano, F. Tassi, O. Vaselli, S. Venturi, M. T. Madigan, J. E. Hallsworth and M. M. Yakimov, Nat. Comm. Earth Environ., 2024, in press Search PubMed.
- M. French and M. Guglielmin, Phys. Geogr., 2000, 82, 513–526 Search PubMed.
- F. Di Lorenzo, J. M. Billod, S. Martín-Santamaría, A. Silipo and A. Molinaro, Eur. J. Org Chem., 2017, 2017, 4055–4073 CrossRef CAS.
- O. Westphal, O. Lüderitz, F. Bister and B. Naturforsch, J. Chem. Sci., 1952, 7, 148–155 Search PubMed.
- M. D. Pither, A. Silipo, A. Molinaro and F. Di Lorenzo, Methods Mol. Biol., 2023, 2613, 153–179 CrossRef CAS.
- P. Garcia-Vello, I. Speciale, F. Di Lorenzo, A. Molinaro and C. De Castro, Methods Mol. Biol., 2022, 2548, 181–209H CrossRef CAS PubMed.
- F. D. Lorenzo, L. Sturiale, A. Palmigiano, L. Lembo-Fazio, I. Paciello, C. P. Coutinho, I. Sá-Correia, M. L. Bernardini, R. Lanzetta, D. Garozzo, A. Silipo and A. Molinaro, ChemBioChem., 2013, 14(9), 1105–1115 CrossRef.
- F. Di Lorenzo, A. Silipo, I. Bianconi, N. I. Lore', A. Scamporrino, L. Sturiale, D. Garozzo, R. Lanzetta, M. Parrilli, A. Bragonzi and A. Molinaro, Mol. Immunol., 2015, 63, 166–175 CrossRef CAS.
- C. E. Hofstaedter, C. E. Chandler, C. M. Met, J. J. Gillespie, J. M. Harro, D. R. Goodlett, D. A. Rasko and R. K. Ernst, mBio, 2024, 15, e0282323 CrossRef PubMed.
- I. Speciale, A. Notaro, P. Garcia-Vello, F. Di Lorenzo, S. Armiento, A. Molinaro, R. Marchetti, A. Silipo and C. De Castro, Carbohydr. Polym., 2022, 277, 118885 CrossRef CAS.
- G. S. Kumar, M. V. Jagannadham and M. K. Ray, J. Bacteriol., 2002, 184, 6746–6749 CrossRef CAS.
- R. Ernst, C. S. Ejsing and B. Antonny, J. Mol. Biol., 2016, 428, 4776–4791 CrossRef CAS.
- R. Cirella, C. Pagliuca, M. D. Pither, E. Scaglione, O. I. Nedashkovskaya, O. V. Chernikov, K. F. Hua, R. Colicchio, M. Vitiello, M. S. Kokoulin, A. Silipo, P. Salvatore, A. Molinaro and F. Di Lorenzo, ChemBioChem, 2023, 24, e202300183 CrossRef CAS PubMed.
- F. Di Lorenzo, F. Crisafi, V. La Cono, M. M. Yakimov, A. Molinaro and A. Silipo, Mar. Drugs, 2020, 18, 592 CrossRef CAS.
- F. Di Lorenzo, A. Palmigiano, I. Paciello, M. Pallach, D. Garozzo, M. L. Bernardini, V. Cono, M. M. Yakimov, A. Molinaro and A. Silipo, Mar. Drugs, 2017, 15, 201 CrossRef.
- F. Di Lorenzo, I. Paciello, L. L. Fazio, L. Albuquerque, L. Sturiale, M. S. da Costa, R. Lanzetta, M. Parrilli, D. Garozzo, M. L. Bernardini, A. Silipo and A. Molinaro, ChemBioChem, 2014, 15, 2146–2155 CrossRef CAS PubMed.
- M. K. Ray, G. S. Kumar and S. Shivaji, J. Bacteriol., 1994, 176, 4243–4249 CrossRef CAS PubMed.
- I. Breßler, J. Kohlbrecher and A. F. Thünemann, J. Appl. Crystallogr., 2015, 48(Pt 5), 1587–1598 CrossRef.
- E. F. Semeraro, L. Marx, J. Mandl, M. P. K. Frewein, H. L. Scott, S. Prévost, H. Bergler, K. Lohner and G. Pabst, J. Appl. Crystallogr., 2021, 54(Pt 2), 473–485 CrossRef CAS PubMed.
- J. Tong and T. J. McIntosh, Biophys. J., 2004, 86, 3759–3771 CrossRef CAS PubMed.
- T. Abraham, S. R. Schooling, M. P. Nieh, N. Kucerka, T. J. Beveridge and J. Katsaras, J. Phys. Chem. B, 2007, 111, 2477–2483 CrossRef CAS.
- M. Kastowsky, T. Gutberlet and H. Bradaczek, Eur. J. Biochem., 1993, 217, 771–779 CrossRef CAS PubMed.
- P. Dynarowicz-Łatka and K. Hac-Wydro, Colloids Surf., B, 2004, 37, 21–25 CrossRef.
- J. Hu, H. Liu, P. Xu, Y. Shang and H. Liu, Langmuir, 2019, 35, 13452–13460 CrossRef CAS.
- T. Abraham, S. R. Schooling, T. J. Beveridge and J. Katsaras, Biomacromolecules, 2008, 9, 2799–2804 CrossRef CAS.
- S. Micciulla, Y. Gerelli and E. Schneck, Biophys. J., 2019, 116, 1259–1269 CrossRef CAS.
- E. A. Evans and R. Kwok, Biochem., 1982, 21, 4874–4879 CrossRef CAS.
- D. Needhamn and E. Evans, Biochem., 1988, 27, 8261–8269 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc05116e |
| This journal is © The Royal Society of Chemistry 2024 |