Open Access Article
Open Access ArticleInteraction of graphene and WS2 with neutrophils and mesenchymal stem cells: implications for peripheral nerve regeneration†
Domenica
Convertino‡
 *a,
Martina
Nencioni‡
b,
Lara
Russo‡
b,
Neeraj
Mishra
ac,
Vesa-Matti
Hiltunen
ac,
Maria Sofia
Bertilacchi
b,
Laura
Marchetti
ab,
Chiara
Giacomelli
*a,
Martina
Nencioni‡
b,
Lara
Russo‡
b,
Neeraj
Mishra
ac,
Vesa-Matti
Hiltunen
ac,
Maria Sofia
Bertilacchi
b,
Laura
Marchetti
ab,
Chiara
Giacomelli
 *b,
Maria Letizia
Trincavelli§
b and
Camilla
Coletti§
*b,
Maria Letizia
Trincavelli§
b and
Camilla
Coletti§
 *ac
*ac
aCenter for Nanotechnology Innovation @ NEST, Istituto Italiano di Tecnologia, Piazza San Silvestro 12, Pisa, Italy. E-mail: domenica.convertino@iit.it; camilla.coletti@iit.it
bDepartment of Pharmacy, University of Pisa, Via Bonanno 6, Pisa, Italy. E-mail: chiara.giacomelli@unipi.it
cGraphene Labs, Istituto Italiano di Tecnologia, Via Morego 30, Genova, Italy
First published on 13th December 2023
Abstract
Graphene and bidimensional (2D) materials have been widely used in nerve conduits to boost peripheral nerve regeneration. Nevertheless, the experimental and commercial variability in graphene-based materials generates graphene forms with different structures and properties that can trigger entirely diverse biological responses from all the players involved in nerve repair. Herein, we focus on the graphene and tungsten disulfide (WS2) interaction with non-neuronal cell types involved in nerve tissue regeneration. We synthesize highly crystalline graphene and WS2 with scalable techniques such as thermal decomposition and chemical vapor deposition. The materials were able to trigger the activation of a neutrophil human model promoting Neutrophil Extracellular Traps (NETs) production, particularly under basal conditions, although neutrophils were not able to degrade graphene. Of note is that pristine graphene acts as a repellent for the NET adhesion, a beneficial property for nerve conduit long-term applications. Mesenchymal stem cells (MSCs) have been proposed as a promising strategy for nerve regeneration in combination with a conduit. Thus, the interaction of graphene with MSCs was also investigated, and reduced viability was observed only on specific graphene substrates. Overall, the results confirm the possibility of regulating the cell response by varying graphene properties and selecting the most suitable graphene forms.
1 Introduction
In the last few years of studies on nerve injury, biocompatible scaffolds have increasingly been investigated for their capability of supporting the regeneration of the damaged nerve. Many works have reported on how the use of conduits could beneficially affect the healing process and ameliorate the outcome of nerve regeneration, thanks to an optimal combination of specific physiochemical, mechanical, and electrical properties.1–3 Also, these structures can be functionalized with biological materials such as growth factors, extracellular vesicles and cells, to further support regeneration.4,5Regeneration conduits are mainly composed of biocompatible natural and synthetic materials, endowed with mechanical resistance, electrical conductivity, and topographical guiding features.1,6 The interactions of these materials with neuronal cells as well as Schwann cells, i.e. the main cells that build up nerves, are nowadays well documented;7–10 the effects that they exert on neuronal differentiation and neurite outgrowth have been studied down to the molecular detail.11–13 Nevertheless, injured nerve regions typically include several other cell types playing a role in the regeneration pathways.4,14–18 An example is constituted by macrophages and other immune system cells like neutrophils.19 Indeed, almost immediately after the damage, Schwann cells dissociate from axons, dedifferentiate, and secrete cytokines and chemokines,2 prompting the recruitment of neutrophils, the first immune cell type that arrives at the site of inflammation from the bloodstream. They infiltrate the tissue within 8 hours after damage and carry out their immune action via three main mechanisms: phagocytosis, degranulation and NETosis.20,21 NETosis is a process of programmed cell death characterized by the release of a network of chromatin, histones and proteins from all types of neutrophil granules to the extracellular space. Initially reported for their ability to form a physical trap for pathogens (from which the name neutrophil extracellular traps is derived), NETs have also been causally linked to the propagation of inflammation.22 Neutrophils are well-known to interact with the other immune cells to direct adaptive immune responses.23 They contribute to recruiting and activating monocytes,24 mediate immune modulation of dendritic cells,25 affect the phenotype and function of T cells,26 and are probably involved in macrophage polarization, regulating their pro- or anti-inflammatory profile switching.27 Although neutrophil action is limited to the first few hours of the immune response, numerous studies show how excessive activation of neutrophils or their dysregulated deletion in an injured site may amplify the inflammatory response and tissue damage.28
Other cell types involved in nerve regeneration are constituted by stem cells, which either infiltrate the injury site or are administered locally as cell therapy or in a combinatorial tissue engineering/cell therapy approach. Stem cells constitute a promising approach to promote nerve healing and damage resolution, due to their capacity to differentiate into neuronal cells or Schwann cells, in the latter case promoting the secretion of neurotrophic factors to prompt regeneration.4,29–31 In particular, both bone marrow and adipose-derived mesenchymal stem cells (MSCs) have been widely investigated for their pro-regenerative properties for nerve repair.29,31 The main advantages brought by MSCs are their easy isolation, differentiation potential, and immunomodulation properties.32 MSCs are known for their immunomodulatory activity, which mainly relies on the release of several growth factors (i.e. brain-derived growth factor (BDNF), vascular endothelial growth factor (VEGF), glia-derived nerve growth factor (GDNF), nerve growth factor (NGF), and neurotrophic 3 (NT-3)) and cytokines that modulate the surrounding tissue.4,33,34 MSCs spontaneously migrate from the bloodstream to the site of the lesion, in response to the several cytokines released by demyelinated Schwann cells, and contribute to wound healing and tissue regeneration by reducing inflammation, promoting angiogenesis and sustaining neuritogenesis, axonal growth and re-myelination.4,35,36 In particular, MSCs produce several anti-inflammatory factors, such as IL-10 and IL-4, and decrease the release of inflammatory factors such as IL-1, IL-6 and TNF-α, to attenuate inflammation and thus accelerate regeneration.34
In recent years, graphene-based materials (GBMs) have attracted the attention of the scientific community in the field of tissue engineering as promising electroactive conductive materials for nerve tissue regeneration.18,37–39 Graphene has been extensively demonstrated to be an excellent platform for cell culture, promoting adhesion, growth, proliferation and differentiation of a variety of cells, including neural and Schwann cells.7,12,40–43 The peculiar electrical properties of graphene are key parameters that could be implemented in nerve conduits to enhance nerve regeneration and restore proper signal transmission.5,13,44–47 In fact, graphene-based conduits have been used in combination with exogenous electrical stimulation.46,48,49 In particular graphene-based materials combined with electrical stimulation showed improved cell differentiation and proliferation,50,51 neurite extension46 and Schwann cell migration, proliferation and myelination,49,52 and functional recovery comparable to the gold standard autograft.48 However, there is a great experimental and commercial variability in GBMs that leads to materials with different compositions and structures that may elicit completely different biological responses.53,54 The most common production methods include “top down” approaches that start from a layered precursor that can be exfoliated in a liquid medium (i.e., liquid phase exfoliation), or use graphite oxide to produce graphene oxide (GO), eventually turned into reduced graphene oxide (rGO).54 The size, shape and surface functionalization of the resulting flakes strongly influence their biocompatibility, often showing time- and dose-dependent cytotoxicity that can be improved by increasing the hydrophilicity, reducing the flakes’ dimension and modifying their surface.54 In addition, graphene can be grown on various substrates ranging from semiconductors (e.g., silicon carbide (SiC)) to transition metals (e.g., Ni and Cu) and insulators (e.g., sapphire (Al2O3)) with different techniques such as thermal decomposition and chemical vapour deposition (CVD).55–59 These techniques allow one to obtain a continuous planar layer of high quality and crystallinity. In addition, CVD graphene can be easily transferred and integrated to different target substrates, while preserving its properties. This versatility is advantageous for neural interfaces and nervous system regeneration.7,60 Along with graphene, transition metal dichalcogenides (TMDs) have emerged as functional materials for neuroscience applications, as they could be adopted to fabricate sensors and neural implants.61,62 Interestingly, bidimensional (2D) materials also display quantum properties that could be used to modulate selective biological functions and to build up advanced, tunable therapeutic or drug delivery tools.63 The cytocompatibility of TMD flakes has been explored, revealing that the choice of the chalcogen atom plays a significant role in determining the toxicological response of cells, being the telluride and selenide forms more toxic than the sulfide forms for the same transition metal.64,65 As regards CVD WS2, a pioneering study by Palumbo and coworkers examined the morphology and adhesion of fibroblast cells on MoS2 and WS2 islands, directly grown on SiO2 and transferred onto PDMS.66 They reported an enhanced morphology on TMDs, explained by their large surface area, the absence of dangling bonds, and van der Waals affinity, which favored a higher physical adsorption of protein on TMDs. Indeed, this might favor the cell binding to the protein adsorbed on the TMD surface from culture media.67 Recently, our group assessed the impact of WS2 on SH-SY5Y human neuroblastoma cell culture, using a uniformly CVD-grown WS2. The cells uniformly adhered to the substrates and were characterized by neural morphology with significant neurite outgrowth.62 TMDs have also been combined with a non-invasive external magnetic field to stimulate mechanosensitive proteins and signaling pathways to remotely regulate cell behavior.68
Despite the significant interest in using graphene and TMDs for neural interfaces,69 so far their interaction with cells different from neurons and Schwann cells in the context of peripheral nerve regeneration has been scarcely unravelled, especially concerning the one synthesized by CVD. Yet, to achieve an optimal pro-regenerative performance of 2D material-based conduits, the interaction of the 2D material with the different cell types involved in peripheral nerve injury needs to be understood. Indeed, the implantation of foreign materials may reinforce the establishment of a local inflammatory microenvironment, which leads to the recruitment of even more cells like neutrophils or MSCs, influencing the correct activation of the resulting immune cascade.70 Moreover, this microenvironment could lead to a non-specific destruction of the adjacent healthy tissue or of the biomaterial and an improper integration between them.71 Furthermore, both graphene and carbon nanotubes may undergo neutrophil-mediated degradation.72,73
In this study, we prepared different graphene substrates, namely CVD graphene on sapphire, epitaxial graphene on SiC via thermal decomposition, both as-grown and hydrogen intercalated (H-intercalated), and CVD graphene grown on copper and transferred on glass, along with CVD monolayer tungsten disulfide (WS2) on sapphire, as an alternative to graphene. Subsequently, we tested them as culture substrates for human neutrophils and human MSCs to shed light on the immune and pro-regenerative responses they could stimulate when adopted as nerve conduit materials in the context of peripheral nerve regeneration (Fig. 1). No external stimulation was applied, since we were mostly interested in the intrinsic properties of the materials, to assess how surface chemistry and electrical conductivity per se could affect the cell behaviour.
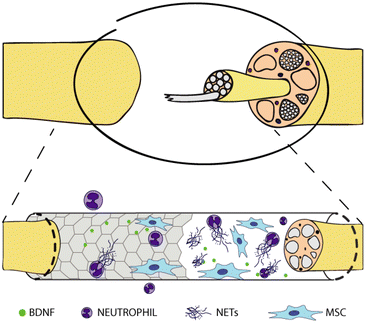 | ||
| Fig. 1 Graphene-coated nerve conduit for nerve regeneration. Interaction with neutrophils and mesenchymal stem cells. | ||
2 Materials and methods
2.1 Substrate preparation and characterization
Graphene on glass (G glass) was obtained by transferring on glass coverslips graphene grown on copper (Cu). Polycrystalline monolayer graphene was synthesized on electropolished Cu foil (purity 99.8%, 46365, Alfa-Aesar) using a 4′′ Aixtron BM Pro cold-wall reactor as previously described.11,55 Graphene was transferred on glass coverslips using a standard wet etching process with ammonium persulfate (APS, 248614, Sigma Aldrich) as copper etchant.11Graphene was grown on the silicon face (Si-face) of hydrogen etched silicon carbide (6H n-type SiC, TankeBlue Semiconductor, China) in an Aixtron HT-BM reactor via thermal decomposition (G SiC). Quasi-free standing monolayer graphene (G SiC Hint) was obtained starting from buffer layer graphene obtained via thermal decomposition and subsequently intercalated under a hydrogen-rich atmosphere.7,74
Monolayer graphene on sapphire (Hangzhou Silan Microelectronics, China) with bilayer patches (G sapp) was synthesized via CVD in the same Aixtron HT-BM reactor adopted to grow graphene on SiC.62,75
Monolayer tungsten disulphide (WS2) was grown directly on 2-inch c-plane sapphire wafer (Hangzhou Silan Microelectronics, China) via a low-pressure CVD process using tungsten trioxide (WO3, 204781, Sigma Aldrich) and sulphur (S, 213292, Sigma Aldrich) as solid precursors in a horizontal hot-wall furnace (Carbolite Gero). The growth was performed at 950 °C for 10 minutes under a pressure of 6 mbar and 420 sccm flow of a gas mixture of 3% H2 in argon.62,76
The controls adopted in the experiments were: (i) bare glass coverslips (Glass, Corning, USA); (ii) hydrogen etched Si-face SiC dices (the same substrates where graphene was grown) (SiC);7,77 (iii) sapphire dices (Sapp).
When graphene and WS2 samples needed to be cut to fit the culture well plates, the surfaces were protected with a polymer (PMMA, 679.04, All-resist) deposited by spin-coating to reduce the scratch formation. The polymer was then removed by immersing the samples in acetone for at least 2 h and rinsing them with isopropanol. The size of all the samples was about 4 × 4 mm2.
Before cell culture, all substrates were sterilized by 30 minutes immersion in 96% ethanol and then rinsed several times with deionized (DI) water.
The topography, quality and number of layers of graphene and WS2 were assessed by both atomic force microscopy (AFM) and Raman spectroscopy.
2.2 HL60 cell culture and differentiation
The human promyelocytic leukaemia (HL60) cell line was used as a cellular model to study NETosis on the 2D material surfaces.78 HL60 cells were obtained from the European Collection of Authenticated Cell Cultures (ECACC, UK Health Security Agency, UK; 98070106). Cells were cultured in RPMI-1640 culture medium (Corning™ RPMI 1640 medium without L-glutamine; 15-040-CV) supplemented with 10% fetal bovine serum (Corning™ 35-016-CV), 2 mM L-glutamine (Euroclone ECB 3000D), 1% penicillin/streptomycin solution (Corning™ 30-002-CI) and incubated at 37 °C in 5% CO2. The cells were maintained in suspension at 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–1
000–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per ml confluency. The differentiation of HL60 to neutrophil-like phenotype was promoted through a 3 day treatment with a 1 μM solution of retinoic acid (RA) in DMSO for 3 days in complete medium.79 The solutions were diluted 1000-fold in the growth medium such that the final DMSO concentration was not higher than 0.1%.
000 cells per ml confluency. The differentiation of HL60 to neutrophil-like phenotype was promoted through a 3 day treatment with a 1 μM solution of retinoic acid (RA) in DMSO for 3 days in complete medium.79 The solutions were diluted 1000-fold in the growth medium such that the final DMSO concentration was not higher than 0.1%.
2.3 NETosis induction and quantification
HL60 differentiated into neutrophils were counted and seeded into a black 96-well plate containing sterilized 2D materials, at a density of 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in a 200 μl total volume. Basal NETosis production was compared to that induced by stimulation with 100 nM phorbol 12-myristate 13-acetate (PMA) in DMSO, for 4 h at 37 °C.80 In both cases NETs were quantified by adding 220 nM Sytox™ Green nucleic acid stain (Invitrogen™, Thermo Fisher Scientific; S7020) in DMSO during the same 4 h incubation.81 After the incubation, the fluorescence intensity was read using a spectrophotometer (excitation/emission: 504/523 nm; EnSight plate reader, PerkinElmer). NETs produced by neutrophils seeded on 2D materials were analysed and normalized to the material areas and to the respective value obtained for the control (glass). Following quantification, the materials were washed in PBS and fixed with PFA 4% in PBS for 20 minutes, to evaluate the adhesion of NETs. The samples were mounted between a glass support and a coverslip and imaged with a laser scanning confocal microscope (Nikon Eclipse Ti-A1), using a 20× air objective (NA 0.45) and the pinhole was set to 1 Airy Unit. Images were acquired at 1024 × 1024 pixel resolution using a 488 nm laser (500–600 emission window). NETs area was analysed with FIJI (ImageJ software, ImageJ 1.53q, Java 1.8.0_322 64 bit). Briefly, the background was subtracted from all the fields and a threshold was applied to select the NET signal. The thresholded image was converted into a binary 0–1 mask. For each binary image, the ratio between the integrated density and the image area was calculated, to get the NETs covered surface fraction.
000 cells per well in a 200 μl total volume. Basal NETosis production was compared to that induced by stimulation with 100 nM phorbol 12-myristate 13-acetate (PMA) in DMSO, for 4 h at 37 °C.80 In both cases NETs were quantified by adding 220 nM Sytox™ Green nucleic acid stain (Invitrogen™, Thermo Fisher Scientific; S7020) in DMSO during the same 4 h incubation.81 After the incubation, the fluorescence intensity was read using a spectrophotometer (excitation/emission: 504/523 nm; EnSight plate reader, PerkinElmer). NETs produced by neutrophils seeded on 2D materials were analysed and normalized to the material areas and to the respective value obtained for the control (glass). Following quantification, the materials were washed in PBS and fixed with PFA 4% in PBS for 20 minutes, to evaluate the adhesion of NETs. The samples were mounted between a glass support and a coverslip and imaged with a laser scanning confocal microscope (Nikon Eclipse Ti-A1), using a 20× air objective (NA 0.45) and the pinhole was set to 1 Airy Unit. Images were acquired at 1024 × 1024 pixel resolution using a 488 nm laser (500–600 emission window). NETs area was analysed with FIJI (ImageJ software, ImageJ 1.53q, Java 1.8.0_322 64 bit). Briefly, the background was subtracted from all the fields and a threshold was applied to select the NET signal. The thresholded image was converted into a binary 0–1 mask. For each binary image, the ratio between the integrated density and the image area was calculated, to get the NETs covered surface fraction.
2.4 Analysis of the 2D material degradation by neutrophils
The stability of graphene in the presence of activated neutrophils was assessed by Raman spectroscopy. Differentiated HL60 were seeded on different materials in a 48 well multiplate (400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well) and incubated with PMA 100 nM for 24 h or 96 h at 37 °C. Then the materials were washed in PBS and fixed with PFA 4% for 15 minutes. The possible degradation of the 2D materials caused by NETosis was evaluated by Raman spectroscopy analysis by analysing the graphene Raman spectra of each substrate before and after incubation with differentiated HL60. Three different areas per sample (bigger than 15 × 15 μm2) were mapped with a Renishaw inVia microRaman system, equipped with a 532 nm (green) laser. All the measurements were performed for 1 s, at 5% laser power, using a 100× (0.89 NA) objective with a spot size of ≈1 μm. From each sample an average of 675 spectra were recorded. The reported distributions obtained from the Raman maps of graphene on glass have been taken on more than 1250 spectra.
000 cells per well) and incubated with PMA 100 nM for 24 h or 96 h at 37 °C. Then the materials were washed in PBS and fixed with PFA 4% for 15 minutes. The possible degradation of the 2D materials caused by NETosis was evaluated by Raman spectroscopy analysis by analysing the graphene Raman spectra of each substrate before and after incubation with differentiated HL60. Three different areas per sample (bigger than 15 × 15 μm2) were mapped with a Renishaw inVia microRaman system, equipped with a 532 nm (green) laser. All the measurements were performed for 1 s, at 5% laser power, using a 100× (0.89 NA) objective with a spot size of ≈1 μm. From each sample an average of 675 spectra were recorded. The reported distributions obtained from the Raman maps of graphene on glass have been taken on more than 1250 spectra.
2.5 Mesenchymal stem cell culture and viability
Human, bone marrow derived, mesenchymal stem cells (MSCs) were obtained from Cell Lines Service (CLS, Heidelberg, Germany) and maintained in a xeno-free medium (XF MSC expansion medium, Sigma Aldrich) at 37 °C in 5% CO2. Upon reaching 70% confluence, the cells were harvested, counted, and seeded on the 2D materials for the experiments as described. The cell viability was measured by the CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega). MSCs were seeded on different materials into a 96 well multiplate at 3000 cells per well density. After 72 hours of incubation in DMEM-F12 (Corning™ 10-092 CV), supplied with 10% FBS and 1% P/S, the MTS assay was performed according to the manufacturer's instruction. Briefly, 100 μL of fresh medium were supplemented with 15 μl of the MTS reagent per well. The multiplate was then incubated for 2 hours at 37 °C in 5% CO2. The supernatant from each well was transferred to a new multiplate to prevent the interference of the materials with the measurements, and the absorbance of the formazan product was read at 490 nm using a multiplate reader (absorbance: 490 nm; EnSight plate reader, PerkinElmer).2.6 AFM analysis on MSCs
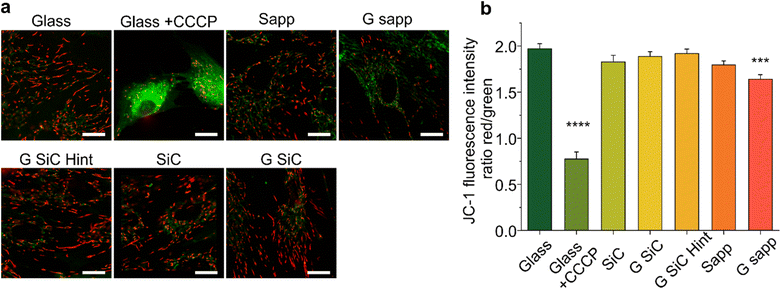
To elucidate the MSCs’ morphology, atomic force microscopy (AFM) was performed with a Dimension ICON (Bruker). Briefly, MSCs were incubated with the material as described in the MSCs’ viability assay. After 72 hours, cells were fixed with PFA 4% in PBS for 20 minutes, washed in PBS and finally washed in DI water to remove salt residuals. After drying, the topography of the cells on different 2D materials was imaged in the peak force tapping mode (Bruker Scan-Asyst). Gwyddion software (Gwyddion 2.58) was used to process the AFM images and extract the surface profiles.2.7 Evaluation of the mitochondrial membrane potential of MSCs on graphene using JC-1 dye
The mitochondrial activity of MSCs was checked by measuring the mitochondrial membrane potential marker 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide (JC-1 iodide salt mitochondria dye, 70014, Biotium).82,83 MSCs were seeded on SiC or sapphire, with or without graphene. After 72 h, the cells were washed in warm dye-free culture medium and incubated with 5 μg ml−1 JC-1 probe for 10 minutes followed by washing two times with fresh medium. Carbonyl cyanide m-chlorophenyl hydrazone (1 mM, CCCP, 21855, Sigma Aldrich) was added for 5 minutes before JC-1 incubation, as a positive control to confirm the probe sensitivity to changes in membrane potential.83 The MSCs on different materials were imaged, by placing the substrate face down in a 22 mm glass-bottom dish, at an inverted Zeiss LSM 880 confocal microscope, equipped with a 63×/1.47 oil objective and 488 and 568 nm lasers to acquire fluorescent images. The JC-1 monomers were excited with a 488 nm laser, while the JC-1 aggregates using a 568 nm laser. The fluorescence emission of JC-1 monomers was collected in the 490–550 nm interval, while the 575–630 nm interval was selected for the aggregates. Typically, 10 fields per sample were acquired with an average cell number of 13. Variations in the mitochondrial potential of MSCs seeded on different materials were evaluated by quantifying the green/red fluorescence intensity ratio.842.8 Determination of BDNF released by MSCs on 2D materials
The amount of produced brain-derived neurotrophic factor (BDNF) was evaluated with an enzyme-linked immunosorbent assay (RayBio, cod. ELH-BDNF) according to the manufacturer's instructions. 3000 cells per well were seeded on each material in a 96 well multiplate and maintained in DMEM-F12, 10% FBS, 1% P/S for 48 h. The medium was then replaced with unsupplemented DMEM-F12 and incubated for a further 24 h. The supernatants were collected, centrifuged, and used to evaluate the amount of BDNF using a multiplate reader (EnSight Plate Reader, PerkinElmer).3 Results and discussion
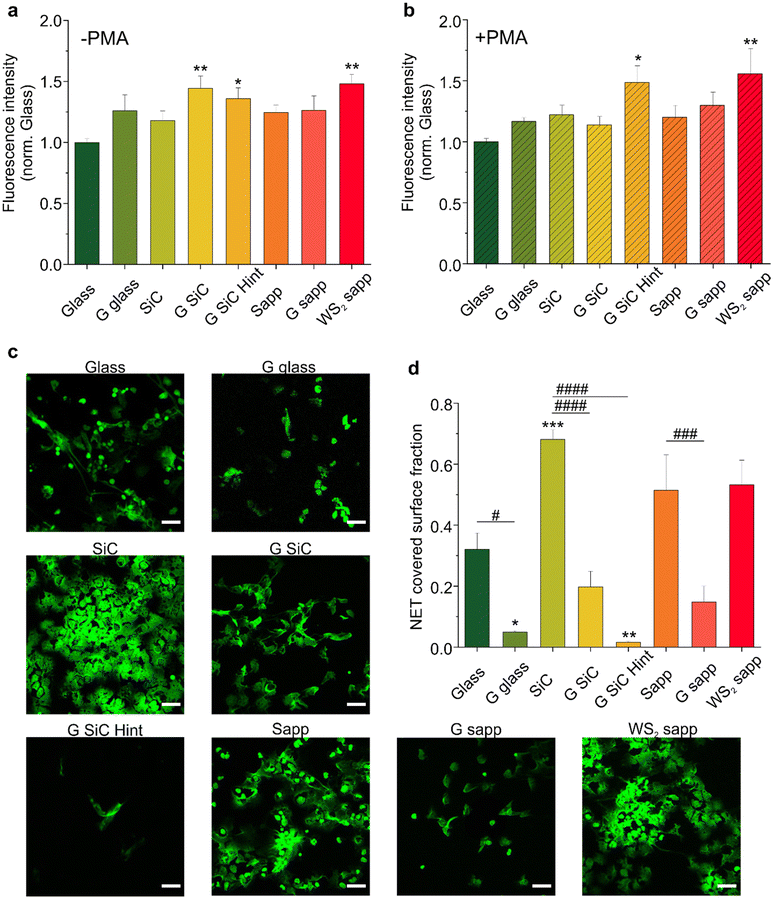
3.1 Fluorimetric quantification of NETosis on graphene and WS2
High-quality fully covered graphene and WS2 samples were grown using different growth techniques, to investigate how the production method influences the immune response and the regenerative process following a peripheral nerve injury. Graphene was grown via thermal decomposition of SiC (G SiC) as an example of a clean and transfer free approach. H-intercalated graphene on SiC (G SiC Hint) was also tested to have a quasi-free-standing material, with reduced SiC substrate interactions.74 CVD growth on copper was chosen as the most common technique to obtain large-scale monolayer graphene that can be easily transferred; such graphene was transferred on glass (G glass). CVD on sapphire (G sapp) was selected to have graphene with reduced metal contaminations.58 CVD WS2 on sapphire (WS2 sapp) was also tested as an additional interesting 2D material that could be adopted in neural interfaces.62 Bare substrates where 2D materials were grown or transferred (i.e., SiC, sapphire and glass) were used as controls. The quality and thickness of each sample were assessed by Raman spectroscopy. As reported in Fig. S1a,† we observed typical monolayer Raman spectra for CVD graphene on glass and H-intercalated graphene, while graphene on sapphire and on SiC presented measurable bilayer contributions as indicated by an enlarged full-width-at-half-maximum (FHWM) of the 2D peak. The Raman spectra of the synthesized WS2 confirmed a monolayer thickness (Fig. S1b†).To evaluate the ability of graphene and WS2 to promote NETosis, neutrophil-like differentiated HL60 cells (dHL60) were seeded on all of them, and NETosis was evaluated either under basal conditions or in response to 4 hours of stimulation with phorbol myristate acetate (PMA), a well-known NETosis stimulus.85 To quantify NET production, a plasma membrane-impermeable DNA-binding dye (Sytox™ Green) was used, and the resulting fluorescence was quantified. No fluorescence was observed for the materials in the absence of the cells, which excludes the contribution of the substrates to the measured fluorescence. All materials showed the ability to slightly increase basal NETosis after 4 hours of incubation compared with the control (glass); however only graphene on SiC (**p < 0.01), graphene on SiC H-intercalated (*p < 0.05) and WS2 on sapphire (**p < 0.01) showed a significant difference compared to the glass control (Fig. 2a). In contrast graphene transferred on glass and graphene on sapphire per se did not significantly increase NETosis. We exclude that the difference observed between the graphene samples was due to topography, since all the graphene samples had comparable roughness, regardless of the growth substrates (Fig. S2†).7,62 The RMS variations at the nanometer scale between the different graphenes are far away from the surface roughness that could influence neurons’ biological response.86
To determine whether the effects of graphene and WS2 on NETosis were quantitatively higher upon stimulation, we analyzed the NET production following PMA addition (Fig. 2b). As expected, PMA triggered the formation of NETs in all the substrates, evidenced by the doubling of fluorescence values with respect to the basal NETosis (Fig. S3†). Interestingly, the PMA-activated NETosis induced on both H-intercalated graphene on SiC and WS2 was significantly higher than the other substrates, confirming the trend previously observed in the not-treated samples. Conversely, we did not observe a significant increase in NETosis on graphene on SiC (G SiC) which instead increased the basal NETosis (Fig. 2a). The reason for this difference may not be attributed to the material per se, as it could be that the PMA activation hampers the possibility to appreciate significant differences in NETosis due to the material. Recently, it has been demonstrated that a polyacrylamide (PAA) gel substrate does not provide a suitable surface for neutrophil adhesion in the absence of specific functionalization.87 In accordance, we did not evidence a specific adhesion of neutrophils on the 2D materials. The same authors reported that PMA-induced NETosis is independent of cell adhesion and scaffold elasticity. Thus, the increased NETosis promoted by G SiC Hint and WS2 sapp may as well not depend on a direct cell–material contact. However, we could hypothesize that the increase of NET release could be related to a transient interaction of neutrophil suspension with the 2D material surface even in the absence of cell adhesion. Of note, WS2 has comparable roughness to graphene7,62 and is more hydrophilic than G SiC Hint (Fig. S4†) thus, other chemical–physical properties of 2D materials may be involved.
3.2 Adhesion of NETs on graphene and WS2
NETs have a known sticky and highly charged nature,88 and constitute a platform for pathological thrombosis onset89 and for the integrin-dependent adhesion of several other inflammatory cell types.90 Thus, materials adopted in conduits and implants shall not unspecifically adsorb NETs on their surface as this could lead to undesired effects. This prompted us to quantify by confocal microscopy how much of the NETs (stained with Sytox green) induced by PMA-stimulated neutrophils remained adhered to each tested substrate after 4 h incubation and subsequent removal of the cells (Fig. 2c). The analysis of NET covered surface fraction reported in Fig. 2d showed that all graphene substrates reduced the adhesion of NETs with respect to the glass control. In particular, a significant reduction, with respect the glass control, was observed for graphene transferred on glass (*p < 0.01) and H-intercalated graphene on SiC (**p < 0.01). Among the bare substrates, SiC significantly increased the adhesion of NETs over glass, indicating a substantially higher adhesion relative to graphene and H-intercalated graphene on SiC (####p < 0.0001). Finally, WS2 did not reduce the adhesion of NETs like graphene, resulting in an area occupation similar to the respective sapphire control. Overall, all the graphene samples reduce the adhesion of NETs, possibly as a result of the high hydrophobicity of graphene compared to WS2 and the control substrates (Fig. S4†). Interestingly WS2 significantly attracted NETs (Fig. 2d). Yet, in addition to hydrophilicity (Fig. S4†), different surface chemistry and the presence of substitutional oxygen atoms (well known to take the place of sulfur in WS2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 91) may significantly affect NET adhesion. The mechanism behind the NET formation on graphene and WS2 needs to be further investigated. Additional studies could aim at exploring the intrinsic material properties that are involved in the NET formation and adhesion. All in all, these experiments convey an important message: graphene promotes NETosis while acting as a repellent for NETs, an important set of properties for the realization of nerve conduits.
91) may significantly affect NET adhesion. The mechanism behind the NET formation on graphene and WS2 needs to be further investigated. Additional studies could aim at exploring the intrinsic material properties that are involved in the NET formation and adhesion. All in all, these experiments convey an important message: graphene promotes NETosis while acting as a repellent for NETs, an important set of properties for the realization of nerve conduits.
3.3 Graphene and WS2 resistance to NET-induced degradation
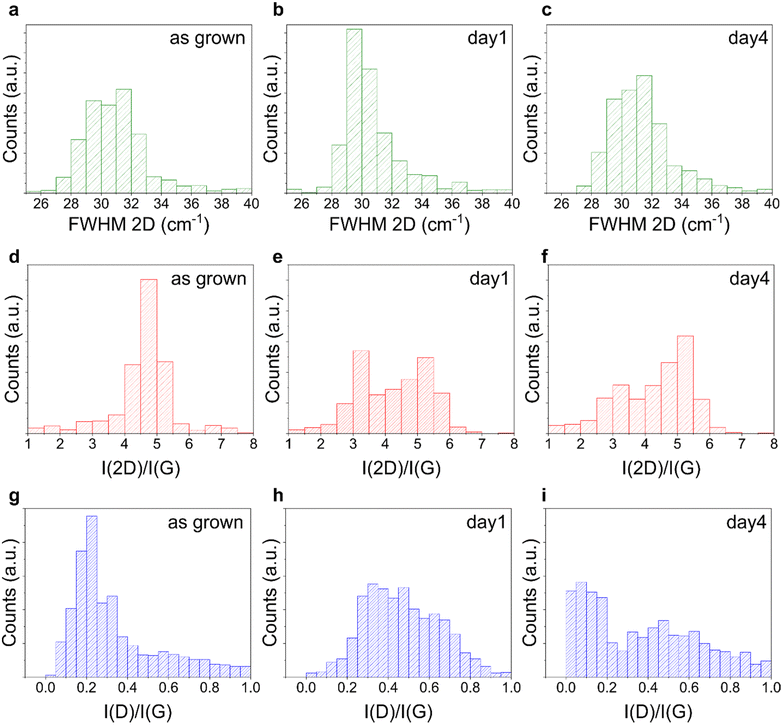
Recent results have shown that factors secreted by neutrophils are responsible for the degradation of carbon based materials, such as graphene and carbon nanotubes (CNTs).72,73,92,93 CNTs were shown to undergo oxidative biodegradation when exposed to primary human neutrophils or to isolated NETs that released myeloperoxidase (MPO). MPO was also shown to degrade highly dispersed GO flakes.94,95 Also single-layer and few-layer graphene flakes were not exempt from degradation,72 although at a slower speed, due to the reduced number of defects and functional groups, that are determinant factors for the enzymatic degradation process.95In light of these considerations, we investigated whether our graphene was subjected to degradation. As previously reported before for graphene flakes,72 we used Raman spectroscopy to assess structural integrity and defects in graphene and thus evaluate possible alterations in the material induced by dHL60. First, we characterized the as-grown graphene samples, and all displayed the characteristic Raman spectrum, with low disorder-induced D peak (∼1340–1350 cm−1) indicating high crystallinity of the grown graphene (Fig. S1†).
Each graphene sample was then incubated with dHL60 and analyzed using Raman spectroscopy to identify possible neutrophil-induced graphene alterations. We compared the representative Raman spectra of graphene on glass, with the characteristic D, G and 2D bands, before and after 1 and 4 days of incubation with dHL60 and we did not observe significant alterations in graphene Raman spectra in the presence of PMA-stimulated neutrophils (Fig. S5†). To verify the hypothesis that NETs do not introduce defects in planar graphene, we performed the Raman analysis of distinct areas of the graphene substrates. Fig. 3 plots the histograms of the FWHM of the graphene 2D Raman peak (FWHM 2D) and of the ratio of the intensities of the 2D and G peaks (I(2D)/I(G)) and the D and G peaks I(D)/I(G) for six selected areas (15 × 15 μm2) of graphene on glass, before and after the incubation. By analyzing FWHM(2D), I(2D)/I(G) and I(D)/I(G) relevant information about graphene thickness, doping level and defect density can be extracted.96,97 We found that their distributions after the incubation overlapped with the one retrieved for the bare substrate, with a difference that can be ascribed to the intrinsic variations between samples of distinct growths, indicating the absence of degradation in both conditions. We performed the Raman mapping also for graphene on sapphire (Fig. S6†) and graphene on SiC, confirming no alteration in the Raman peaks after the treatment (Fig. S7†). Similar to graphene WS2 also did not show alteration following 4 days of incubation with dHL60, as confirmed by the histograms of the ratio of the intensity of the 2LA + E2g/A1g Raman modes reported in Fig. S8.†
Overall, the collective evidence suggests that CVD and epitaxial graphene are not degraded by activated neutrophils after 1 and 4 days of culture, in contrast to what was previously reported for GO. Notably, our graphene presented a negligible number of defects, as clearly visible by their characteristic Raman spectra with a low or null D-peak (Fig. S1a†). In addition they are planar and not suspended, a crucial aspect to be considered knowing how differences in shape and lateral size can produce dramatically different biological results.98
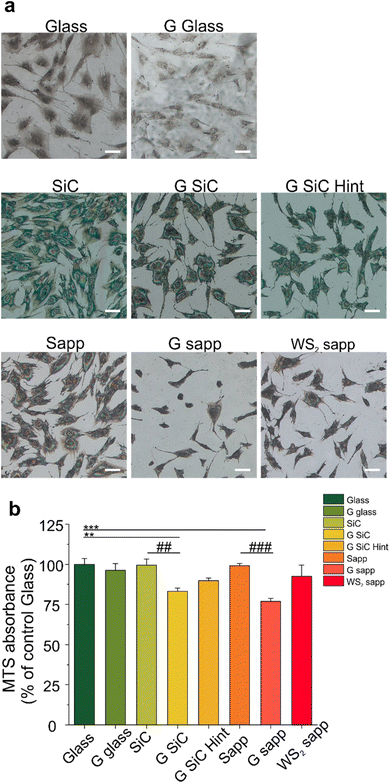
3.4 Viability and morphology of MSCs on 2D materials
We then focused on another player involved in nerve regeneration, MSCs. First of all, we investigated whether MSC adhesion would be affected by 2D materials. We cultured MSCs on all the substrates for 72 hours and quantified the cell number. As visible in Fig. 4a and Fig. S9,† 2D materials and their growth substrates differently affected cell adhesion. Interestingly the number of cells was higher on SiC and sapphire compared to the bare glass control. All the graphene substrates showed a reduced number of cells compared to the growth substrate, with a significant decrease for the H-intercalated graphene on SiC (31.3%) and for graphene on sapphire (29.9%) (####p < 0.0001) (Fig. S9†). A similar result was previously observed by Kim and coworkers for hMSC cultured on CVD graphene transferred on glass.99Then, the viability of MSCs was quantified by tetrazolium MTS reduction assay, that is based on the quantification of intracellular NADH. The reduced adhesion on graphene, seemed not to have a significant impact on cell viability, apart from graphene on SiC and sapphire (Fig. 4b). We observed a similar cell viability for cells cultured on the bare substrates used to grow graphene (SiC and sapphire) and glass control. Also, WS2 displays a similar cell viability to both glass and sapphire, probably helped by good hydrophilic surfaces that could enhance the early stages of cell adhesion and proliferation, in comparison to hydrophobic surfaces.67 Among the graphene-coated samples, those on SiC and on sapphire showed a significant decrease of 16.2 and 23.1% in comparison to the glass control group (100 ± 8.9%) (**p < 0.01 and ***p < 0.001, respectively) and a decrease in the percentage of viability of 19.6 and 22.4% when compared to the bare substrates on which graphene was grown, namely SiC and sapphire (##p < 0.01 and ###p < 0.001, respectively). These variations in cell viability were, however, not high enough to promote significant variations in the BDNF released by MSCs when incubated with the different substrates (Fig. S10†). A previous study showed a PC12 cell viability on graphene on SiC comparable to that obtained for other controls (glass and SiC).7 However, in that work a polymeric coating was used to improve the cell adhesion on the substrates. When an uncoated G SiC Hint was used, cell bodies aggregated and neurite bundles were reported, a clear sign of reduced adhesion.7,41 Interestingly, the performance of graphene on SiC was different from that of H-intercalated graphene, our results show a higher viability despite the reduced number of cells (Fig. 4b, G SiC Hint). The reduced MSCs viability on graphene on sapphire confirmed what was previously observed for SH-SY5Y on the same graphene.62 Something that these two substrates (i.e., graphene on SiC and graphene on sapphire) have in common, and that differentiates them from the other graphene substrates, is the higher percentage of areas with bilayer thickness. In addition, we noticed that the graphene film on sapphire can be easily scratched forming graphene debris, resembling the condition of suspended graphene flakes in the cell medium.
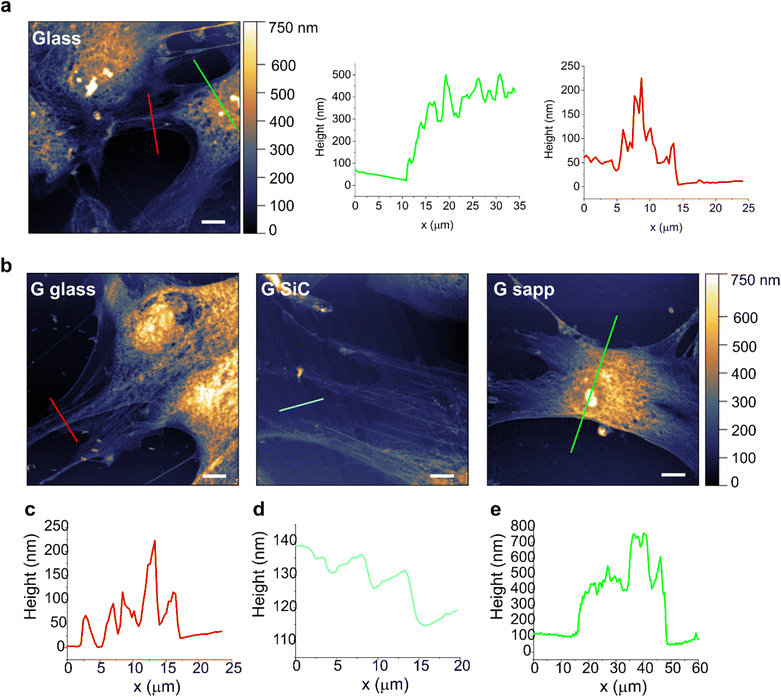
In order to study if a loose adhesion to the substrate could affect MSC viability on graphene, cells grown on each sample were then assessed by AFM, to evaluate possible alterations in cell morphologies and topographies (Fig. 5). AFM topographies of fixed MSCs revealed the height of contours of the cell surface. The bright areas identified the elevated parts of the cell, where the nuclei are located. Besides the intrinsic heterogeneity in cell morphology, results evidenced no clear alterations in the shape and morphology of MSCs grown on most of the graphene substrates compared to the glass control. Only the shape of some MSCs on graphene on sapphire seemed more rounded, with shorter protrusions, pointing to a possible reduced cell adhesion (Fig. 5b, G sapp), confirming what was previously observed in Fig. 4a. On the other substrates the cells were mainly spread, showing a healthy and normal shape (Fig. 5b and Fig. S11†). The high cell body is clearly visible (green line profiles in Fig. 6), as well as the cell protrusions that extend toward the cells for more than 100 μm (Fig. 5b, G SiC). A similar morphology was also observed in MSCs grown on the other substrates (Fig. S11†). Interestingly, AFM of G on SiC revealed the nanometric terraces of the SiC substrates (Fig. 5b and c, G SiC), which do not influence the cell polarity, with the cell protrusions aligned independently from the direction of the terraces.
Overall, the AFM micrographs revealed a comparable behaviour of MSCs on graphene, with spreading adherent cells. Despite the reduced viability on G SiC, we did not observe considerable cell shape alterations. The cells expressed variable extensions on the substrate, independent of the substrate topography.
Concerning material stability issues, it should be noted that both graphene and WS2 showed good stability and remained intact during the entire culture period. No visible alterations were observed (Fig. 4a and Fig. S11a, S12†), resembling the results previously reported for PC12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 and SH-SY5Y62 cells.
7 and SH-SY5Y62 cells.
3.5 Assessment of mitochondrial function on graphene
To explore whether the reduced viability measured by the MTS assay for MSC grown on graphene on SiC and graphene on sapphire is mediated by a mitochondrial instability (which may justify a reduced NADH measured by MTS), we also determined the mitochondrial potential (Δψm) of MSCs grown on graphene using a fluorescent cationic dye JC-1 (Fig. 6). Indeed, the MTS assay results depend on the mitochondrial electron transport chain; thus, a decreased formazan production could be ascribed to a reduced metabolic activity due to mitochondrial damage. Furthermore, Δψm is considered a valuable indicator of cell functional status that helps to evaluate their physicopathological conditions,83 with a decrease in Δψ usually being associated with reduced cell viability and apoptosis.84The mitochondrial activity was evaluated by detecting the red/green fluorescence intensity ratio of JC-1. As shown in Fig. 6a, in the glass control JC-1 emits red fluorescence. After MSC cells were exposed to CCCP (positive control of depolarization), a significant increase in green fluorescence intensity was observed, indicating, as expected, a reduction in Δψm (****p < 0.0001, Fig. 6b).84 Then, the effect of the materials on Δψm was investigated. As reported in Fig. 6b, in the control the ratio was 1.97 ± 0.23. After treatment with CCCP the ratio drastically decreased (0.77 ± 0.26). A significant decrease was also observed in graphene on sapphire (***p < 0.001), while the other substrates did not affect the mitochondrial activity.
Overall, the variation in Δψm for graphene on sapphire (G sapp, Fig. 6) confirmed what was previously reported in the MTS results. In contrast, no alterations were observed on graphene on SiC, even though this substrate showed a reduced viability similar to graphene on sapphire (Fig. 4). Graphene-based materials have been shown to alter the stability of mitochondria in different cell models.100–102 Xiaoli and coworkers reported an increased mitochondrial stress with a reduction in the membrane potential (Δψm).101 A positively charged polyethylenimine (PEI)-functionalized GO was also shown to promote mitochondrial fragmentation and cell apoptosis.100 However, these studies were conducted by incubating the cells with GO or graphene flakes, a completely different condition compared to cells seeded on planar GBMs. As a matter of fact, studies showed the downregulation of neuronal signalling of graphene-exposed neurons, in contrast to the unaltered neuronal activity when seeded on planar graphene, clear proof of the influence of the materials’ physical–chemical features on their interaction with the cells.103,104
In conclusion, only graphene on sapphire shows poorer cell morphology, reduced cell viability and mitochondrial activity. One could speculate that the lower viability might be induced by small fragments of graphene that could be released by the samples and could be internalized by the cells, with harmful effects. Bilayer graphene is loosely bonded to the substrate and in the presence of a significant number of small bilayer islands, a detachment and internalization of these flakes could indeed negatively affect cell viability. In addition, the debris formed following graphene scratching could release similar flakes with negative effects. Unfortunately, following 24 h incubation with the cell medium, the morphological variations due to medium residuals were comparable to a single layer of graphene thickness and did not allow to appreciate a potential bilayer patch detachment (Fig. S2†) and also micro-Raman spectroscopy did not allow us to appreciate variations at the submicrometric scale. Therefore, further studies are necessary to evaluate if other morphological, electrical and/or chemical properties could have impacted the cell behaviour.
4 Conclusion
In order to fully understand the potential of 2D materials for tissue engineering applications, it is critical to explore their interaction with all the cells involved in the process of interest. This is particularly true for the implementation of graphene and the other 2D materials in nerve conduits for peripheral nerve regeneration, where highly different cells (including neurons, Schwann cells, immune cells, stem cells, and fibroblasts) play a role. While the interaction of graphene with nerve cells has been widely explored, less is known about other cells in the context of peripheral nerve injury.19Here, first, we assessed the interaction of graphene and WS2 with neutrophils. Phagocytic cells, such as neutrophils and monocytes, represent the first line of defence against a foreign material and play a critical role in immune response.70,105 Among the strategies through which neutrophils eliminate the pathogenic insult, recently it has been reported that neutrophils can extrude NETs, which are mainly composed of DNA and granule proteins.20 The effects of neutrophils on implanted biomaterials vary considerably based on the material used to make implants and the anatomical site of the implant.105 Neutrophil activation in the first phase of biomaterial implant is pivotal to guarantee the lack of bacterial infection.71 Notably, excessive neutrophil activation can lead to negative effects. Therefore, their modulation is a key factor to improve tissue-material integration.105,106 Keshavan et al. discussed the impact of engineered nanomaterials on neutrophils and how neutrophils, in turn, may digest carbon nanotubes and graphene oxide.19 They also observed that micrometric GO sheets disrupt the lipid rafts in neutrophils and induce the formation of NETs, associated with calcium influx and the production of ROS.73 A similar size-dependency was observed also by Huang and coworkers, who showed how neutrophils gradually degraded GO following two distinct defensive pathways dependent on the dimension of the sheets: NETosis for micrometer-sized flakes or degranulation for nanometer-sized flakes.107 The degradation mechanism was probably triggered by defects and oxygenated functions on the sheet surfaces, since GO showed faster degradability, when compared to pristine graphene sheets produced by mechanochemical exfoliation.72 In view of these considerations, here, we explored if our graphene substrates triggered the release of NETs. We found that graphene and WS2 were both able to activate NETosis, although the effect was more significant under basal conditions than under conditions in which neutrophils are activated. However, it is worth noting that unlike GO and pristine graphene sheets, our materials were planar and continuous. Only a few nanometric features were present, namely the ridges on graphene on sapphire, formed as a consequence of the different thermal expansion coefficients of graphene and sapphire,58 and the atomically flat terraces separated by nanometric steps on graphene on SiC, formed after hydrogen etching of the SiC substrate.7,108 However, these features are different from the sharp corners and asperities that characterize the dispersed graphene flakes. Indeed, the flakes have been shown to elicit neutrophil's membrane stripping73 and easily pierce cell membranes, allowing graphene to penetrate the lipid bilayers and interfere with the normal cell function.109 In agreement with these observations, we found that differently from other graphene forms, our materials were not degraded up to 4 days of incubation with neutrophils. Also, we highlight here the unprecedented observation that pristine graphene may act as a repelling agent for NETs persistence in the injury site, which may be a useful property to exploit for the design of new nerve conduits.
Furthermore, herein, we enriched the details of graphene interactions with MSCs. To date, many studies have reported encouraging results regarding adhesion, proliferation and neuronal differentiation of MSCs on GO, rGO and other graphene-based hybrid materials.110,111 All these graphene forms have been extensively used to realize scaffolds, showing improved nerve regeneration and neurite sprouting and outgrowth.43,112–116 In contrast, the effect of monolayer CVD graphene on MSCs is still little explored. CVD graphene has been demonstrated to promote the neurogenesis of hMSCs as well as the neurite outgrowths by enhancing MSC–substrate and cell–cell interactions.99 However, the majority of the studies focus on the application of CVD graphene in bone tissue engineering for osteogenic differentiation.117–119 We focus on CVD and epitaxial graphene, in a relatively pure and flat form, and demonstrate as substrates that are nominally the same material, graphene, but have different effects on MSCs due to their intrinsic properties, a key point in perspective to use graphene in peripheral nerve regeneration. Despite many studies proving that graphene is highly biocompatible,7,62,120,121 here graphene on SiC and on sapphire was shown to induce a significant decrease in the viability of MSCs. This, at least for the graphene on sapphire, seems to correlate with a lower mitochondrial activity of the cell and a looser adhesion when they grow on this substrate. Further studies that deepen the knowledge of the surface characteristics and homogeneity of graphene will certainly aid in understanding better the cell–graphene interface that strongly influences the resulting interaction.
Overall, these findings emphasize the importance of choosing the right graphene form to adequately regulate the cell response. The possibility to change and tune graphene properties (i.e. via intercalation or functionalization) can provide the possibility to avoid undesirable characteristics and explore the use of graphene as a peripheral neural interface. Indeed, once the effect of 2D materials on the players involved in nerve regeneration is unravelled, it would be possible to integrate the materials onto a biopolymer support to realize a planar polymeric structure rolled to form tubular nerve conduits. In the literature there are many examples of 3D tubes prepared from a planar scaffold that can be simply rolled along the lengthwise edge and taped,122 implanted and wrapped directly around the nerve stumps123 or prepared using self-rolling films.124 Similar examples are also between FDA-approved nerve grafts, such as NeuraWrap™ (Integra LifeSciences Co.) that is used by wrapping around the injured nerves, or NeuroMend™ (Collagen Matric, Inc.) that can be unrolled and self-curled to match the dimensions of the injured tissue.
Author contributions
D. C.: conceptualization, investigation, writing – original draft, and visualization; M. N.: conceptualization, investigation, writing – original draft, and visualization; L. R.: conceptualization, investigation, writing – original draft, and visualization; N. M.: investigation and writing – reviewing and editing; V. H.: investigation and writing – reviewing and editing; M. S. B.: investigation and writing – reviewing and editing; L. M.: conceptualization, writing – reviewing and editing, and supervision; C. G.: conceptualization, writing – reviewing and editing, and supervision; M. L. T.: writing – reviewing and editing, and supervision; C. C.: conceptualization, writing – reviewing and editing, and supervision.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge the support from the University of Pisa under the “PRA – Progetti di Ricerca di Ateneo” (Institutional Research Grants) – Project No. PRA 2020–2021 92 “Quantum Computing, Technologies and Applications”. This work has received funding from the European Union's Horizon 2020 Research and Innovation Programme under grant agreement 881603 and by the Next Generation EU project ECS00000017 ‘Ecosistema dell'Innovazione’ Tuscany Health Ecosystem (THE, PNRR, Spoke 4: Nanotechnologies for Diagnosis and Therapy). This research work is supported also by the “TRITONE project” founded by “Regione Toscana” with “BANDO RICERCA SALUTE 2018”. We thank CISUP at the University of Pisa for the access to the confocal microscopy laboratory facility.References
- A. Faroni, S. A. Mobasseri, P. J. Kingham and A. J. Reid, Adv. Drug Delivery Rev., 2015, 82, 160–167 CrossRef PubMed.
- R. R. López-Cebral, J. Silva-Correia, R. L. Reis, T. H. Silva and J. M. Oliveira, ACS Biomater. Sci. Eng., 2017, 3, 3098–3122 CrossRef PubMed.
- S. Vijayavenkataraman, Acta Biomater., 2020, 106, 54–69 CrossRef CAS PubMed.
- A. Lavorato, S. Raimondo, M. Boido, L. Muratori, G. Durante, F. Cofano, F. Vincitorio, S. Petrone, P. Titolo, F. Tartara, A. Vercelli and D. Garbossa, Int. J. Mol. Sci., 2021, 22(2), 572 CrossRef CAS PubMed.
- Q. Huang, Y. Cai, X. Zhang, J. Liu, Z. Liu, B. Li, H. Wong, F. Xu, L. Sheng, D. Sun, J. Qin, Z. Luo and X. Lu, ACS Appl. Mater. Interfaces, 2021, 13, 112–122 CrossRef CAS PubMed.
- A. C. Pinho, A. C. Fonseca, A. C. Serra, J. D. Santos and J. F. J. Coelho, Adv. Healthcare Mater., 2016, 5, 2732–2744 CrossRef CAS PubMed.
- D. Convertino, S. Luin, L. Marchetti and C. Coletti, Front. Neurosci., 2018, 12, 1–8 CrossRef PubMed.
- L. Scaccini, R. Mezzena, A. De Masi, M. Gagliardi, G. Gambarotta, M. Cecchini and I. Tonazzini, Int. J. Mol. Sci., 2021, 22, 7901 CrossRef CAS PubMed.
- B. E. Fornasari, G. Carta, G. Gambarotta and S. Raimondo, Front. Bioeng. Biotechnol., 2020, 8, 554257 CrossRef PubMed.
- A. J. Petty, R. L. Keate, B. Jiang, G. A. Ameer and J. Rivnay, Chem. Mater., 2020, 32, 4095–4115 CrossRef CAS.
- D. Convertino, F. Fabbri, N. Mishra, M. Mainardi, V. Cappello, G. Testa, S. Capsoni, L. Albertazzi, S. Luin, L. Marchetti and C. Coletti, Nano Lett., 2020, 20, 3633–3641 CrossRef CAS PubMed.
- N. Li, X. Zhang, Q. Song, R. Su, Q. Zhang, T. Kong, L. Liu, G. Jin, M. Tang and G. Cheng, Biomaterials, 2011, 32, 9374–9382 CrossRef CAS PubMed.
- L. Wang, D. Song, X. Zhang, Z. Ding, X. Kong, Q. Lu and D. L. Kaplan, ACS Biomater. Sci. Eng., 2019, 5, 613–622 CrossRef CAS PubMed.
- R. C. Rennert, M. Sorkin, R. K. Garg and G. C. Gurtner, Regener. Med., 2012, 7, 833–850 CrossRef CAS PubMed.
- P. Liu, J. Peng, G. H. Han, X. Ding, S. Wei, G. Gao, K. Huang, F. Chang and Y. Wang, Neural Regener. Res., 2018, 1335–1342 Search PubMed.
- A. R. Sas, K. S. Carbajal, A. D. Jerome, R. Menon, C. Yoon, A. L. Kalinski, R. J. Giger and B. M. Segal, Nat. Immunol., 2020, 21, 1496–1505 CrossRef PubMed.
- B. E. Fornasari, M. El Soury, G. Nato, A. Fucini, G. Carta, G. Ronchi, A. Crosio, I. Perroteau, S. Geuna, S. Raimondo and G. Gambarotta, Cells, 2020, 9, 1–19 CrossRef PubMed.
- D. Convertino, M.L. Trincavelli, C. Giacomelli, L. Marchetti and C. Coletti, Front. Bioeng. Biotechnol., 2023, 11, 1–15, DOI:10.3389/fbioe.2023.1306184.
- S. Keshavan, P. Calligari, L. Stella, L. Fusco, L. G. Delogu and B. Fadeel, Cell Death Dis., 2019, 10, 569 CrossRef PubMed.
- V. Brinkmann, U. Reichard, C. Goosmann, B. Fauler, Y. Uhlemann, D. S. Weiss, Y. Weinrauch and A. Zychlinsky, Science, 2004, 303, 1532–1536 CrossRef CAS PubMed.
- J. Wang and H. Arase, Ann. N. Y. Acad. Sci., 2014, 1319, 66–81 CrossRef CAS PubMed.
- N. V. Vorobjeva and B. V. Chernyak, Biochemistry, 2020, 85, 1178–1190 CAS.
- E. Kolaczkowska and P. Kubes, Nat. Rev. Immunol., 2013, 13, 159–175 CrossRef CAS PubMed.
- O. Soehnlein and L. Lindbom, Nat. Rev. Immunol., 2010, 10, 427–439 CrossRef CAS PubMed.
- K. P. J. M. Van Gisbergen, M. Sanchez-hernandez, T. B. H. Geijtenbeek and Y. Van Kooyk, J. Exp. Med., 2005, 201, 1281–1292 CrossRef CAS PubMed.
- D. Minns, K. J. Smith, G. Hardisty, A. G. Rossi and E. G. Findlay, Front. Immunol., 2021, 12, 15–17 Search PubMed.
- J. A. Marwick, R. Mills, O. Kay, K. Michail, J. Stephen, A. G. Rossi, I. Dransfield and N. Hirani, Cell Death Dis., 2018, 9, 665 CrossRef PubMed.
- E. Frangou, D. Vassilopoulos, J. Boletis and D. T. Boumpas, Autoimmun. Rev., 2019, 18, 751–760 CrossRef CAS PubMed.
- L. Jiang, S. Jones and X. Jia, Int. J. Mol. Sci., 2017, 18, 1–17 Search PubMed.
- C. A. Kubiak, J. Grochmal, T. A. Kung, P. S. Cederna, R. Midha and S. W. P. Kemp, Muscle Nerve, 2020, 449–459 CrossRef PubMed.
- S. Blando, I. Anchesi, E. Mazzon and A. Gugliandolo, Int. J. Mol. Sci., 2022, 23(14), 7545 CrossRef CAS PubMed.
- M. F. Pittenger, A. M. Mackay, S. C. Beck, R. K. Jaiswal, R. Douglas, J. D. Mosca, M. A. Moorman, D. W. Simonetti, S. Craig and D. R. Marshak, Science, 1999, 284, 143–148 CrossRef CAS PubMed.
- T. Sumarwoto, H. Suroto, F. Mahyudin, D. Novembri, D. Tinduh, H. Basuki, C. Rosita, S. Prakoeswa, F. Abdul and S. Rhatomy, Ann. Med. Surg. (Lond), 2021, 67, 102482 Search PubMed.
- X. Li, Y. Guan, C. Li, T. Zhang, F. Meng, J. Zhang, J. Li, S. Chen, Q. Wang, Y. Wang, J. Peng and J. Tang, Stem Cell Res. Ther., 2022, 1–13 Search PubMed.
- Y. Chen, L. Xiang, J. Shao and R. Pan, J. Cell. Mol. Med., 2010, 14, 1494–1508 CrossRef PubMed.
- L. Crigler, R. C. Robey, A. Asawachaicharn, D. Gaupp and D. G. Phinney, Exp. Neurol., 2006, 198, 54–64 CrossRef CAS PubMed.
- S. Grijalvo and D. Díaz, Neurochem. Int., 2021, 147, 105005 CrossRef CAS PubMed.
- T. Aydin, C. Gurcan, H. Taheri and Y. Açelya, Cell Biol. Transl. Med., 2018, 3, 129–142 Search PubMed.
- D. Convertino, L. Marchetti and C. Coletti, in Silicon Carbide Technology for Advanced Human Healthcare Applications, ed. S. E. Saddow, Elsevier, 2022, pp. 65–98 Search PubMed.
- J. S. Lee, A. Lipatov, L. Ha, M. Shekhirev, M. N. Andalib, A. Sinitskii and J. Y. Lim, Biochem. Biophys. Res. Commun., 2015, 460, 267–273 CrossRef CAS PubMed.
- A. Bendali, L. H. Hess, M. Seifert, V. Forster, A. F. Stephan, J. A. Garrido and S. Picaud, Adv. Healthcare Mater., 2013, 2, 929–933 CrossRef CAS PubMed.
- Z. Huang, M. Sun, Y. Li, Z. Guo and H. Li, J. Mater. Chem. B, 2021, 9, 2656–2665 RSC.
- J. Wang, W. Zheng, L. Chen, T. Zhu, W. Shen, C. Fan, H. Wang and X. Mo, ACS Biomater. Sci. Eng., 2019, 5, 2444–2456 CrossRef CAS PubMed.
- X. Song, T. Gao, Y. Nie, J. Zhuang, J. Sun, D. Ma, J. Shi, Y. Lin, F. Ding, Y. Zhang and Z. Liu, Nano Lett., 2016, 16, 6109–6116 CrossRef CAS PubMed.
- N. B. Tolou, H. Salimijazi, T. Dikonimos and G. Faggio, J. Mater. Sci., 2021, 56, 5581–5594 CrossRef.
- Y. Sun, X. Liu, M. N. George, S. Park, B. Gaihre, A. Terzic and L. Lu, J. Biomed. Mater. Res., 2021, 193–206 CrossRef.
- X. Fang, J. Deng, W. Zhang, H. Guo, F. Yu, F. Rao, Q. Li, P. Zhang, S. Bai and B. Jiang, RSC Adv., 2020, 10, 16769–16775 RSC.
- C. Dong, F. Qiao, W. Hou, L. Yang and Y. Lv, Appl. Mater. Today, 2020, 21, 100870 CrossRef.
- S. Lu, W. Chen, J. Wang, Z. Guo, L. Xiao, L. Wei, J. Yu, Y. Yuan, W. Chen, M. Bian, L. Huang, Y. Liu, J. Zhang, Y. Li and L. Jiang, Small Methods, 2023, 2200883 CrossRef CAS PubMed.
- S. Meng, Tissue Eng. Regener. Med., 2014, 11, 274–283 CrossRef CAS.
- C. Fu, S. Pan, Y. Ma, W. Kong, Z. Qi and X. Yang, Artif. Cells, Nanomed., Biotechnol., 2019, 47, 1867–1876 CrossRef CAS PubMed.
- J. Wang, Y. Cheng, L. Chen, T. Zhu, K. Ye, C. Jia, H. Wang, M. Zhu, C. Fan and X. Mo, Acta Biomater., 2019, 84, 98–113 CrossRef CAS PubMed.
- M. Bramini, G. Alberini, E. Colombo, M. Chiacchiaretta, M. L. DiFrancesco, J. F. Maya-Vetencourt, L. Maragliano, F. Benfenati and F. Cesca, Front. Syst. Neurosci., 2018, 12, 1–22 Search PubMed.
- J. Li, H. Zeng, Z. Zeng, Y. Zeng and T. Xie, ACS Biomater. Sci. Eng., 2021, 7, 5363–5396 CrossRef CAS PubMed.
- V. Miseikis, D. Convertino, N. Mishra, M. Gemmi, T. Mashoff, S. Heun, N. Haghighian, F. Bisio, M. Canepa, V. Piazza and C. Coletti, 2D Mater., 2015, 2, 014006 CrossRef.
- X. Li, W. Cai, J. An, S. Kim, J. Nah, D. Yang, R. Piner, A. Velamakanni, I. Jung, E. Tutuc, S. K. Banerjee, L. Colombo and R. S. Ruoff, Science, 2009, 324, 1312–1314 CrossRef CAS.
- M. A. Fanton, J. A. Robinson, C. Puls, Y. Liu, M. J. Hollander, B. E. Weiland, M. Labella, K. Trumbull, R. Kasarda, C. Howsare, J. Stitt and D. W. Snyder, ACS Nano, 2011, 5, 8062–8069 CrossRef CAS.
- N. Mishra, S. Forti, F. Fabbri, L. Martini, C. McAleese, B. R. Conran, P. R. Whelan, A. Shivayogimath, B. S. Jessen, L. Buß, J. Falta, I. Aliaj, S. Roddaro, J. I. Flege, P. Bøggild, K. B. K. Teo and C. Coletti, Small, 2019, 1904906 CrossRef CAS PubMed.
- K. V. Emtsev, A. Bostwick, K. Horn, J. Jobst, G. L. Kellogg, L. Ley, J. L. McChesney, T. Ohta, S. A. Reshanov, J. Röhrl, E. Rotenberg, A. K. Schmid, D. Waldmann, H. B. Weber and T. Seyller, Nat. Mater., 2009, 8, 203–207 CrossRef CAS PubMed.
- F. Veliev, A. Briançon-Marjollet, V. Bouchiat and C. Delacour, Biomaterials, 2016, 86, 33–41 CrossRef CAS PubMed.
- S. N. Faisal and F. Iacopi, ACS Appl. Nano Mater., 2022, 5, 10137–10150 CrossRef CAS.
- D. Convertino, N. Mishra, L. Marchetti, M. Calvello, A. Viegi, A. Cattaneo, F. Fabbri and C. Coletti, Front. Neurosci., 2020, 14, 1–10 Search PubMed.
- A. Qu, M. Sun, J. Kim, L. Xu, C. Hao, W. Ma, X. Wu, X. Liu, H. Kuang, N. A. Kotov and C. Xu, Nat. Biomed. Eng., 2021, 5, 103–113 CrossRef CAS.
- W. Z. Teo, E. L. K. Chng, Z. Sofer and M. Pumera, Chem. – Eur. J., 2014, 20, 9627–9632 CrossRef CAS PubMed.
- A. Murali, G. Lokhande, K. A. Deo, A. Brokesh and A. K. Gaharwar, Mater. Today, 2021, 50, 276–302 CrossRef CAS PubMed.
- A. Palumbo, F. Tourlomousis, R. C. Chang and E. H. Yang, ACS Appl. Bio Mater., 2018, 1, 1448–1457 CrossRef CAS PubMed.
- L. Bačáková, E. Filová, F. Rypáček, V. Švorčík and V. Starý, Physiol. Res., 2004, 53(Suppl. 1), S35–S45 Search PubMed.
- J. Luo, Q. Zhou, X. Hu, X. Hou, D. Li, S. Guo and S. Sun, ACS Appl. Nano Mater., 2021, 4, 10828–10835 CrossRef CAS.
- K. Kostarelos, M. Vincent, C. Hebert and J. A. Garrido, Adv. Mater., 2017, 29, 1–7 CrossRef PubMed.
- S. Jhunjhunwala, ACS Biomater. Sci. Eng., 2018, 4, 1128–1136 CrossRef CAS PubMed.
- S. A. Yavari, S. M. Castenmiller, J. A. G. Van Strijp and M. Croes, Adv. Mater., 2020, 2002962 CrossRef PubMed.
- R. Kurapati, S. P. Mukherjee, C. Martín, G. Bepete, E. Vázquez, A. Pénicaud, B. Fadeel and A. Bianco, Angew. Chem., Int. Ed., 2018, 57, 11722–11727 CrossRef CAS.
- S. P. Mukherjee, B. Lazzaretto, K. Hultenby, L. Newman, A. F. Rodrigues, N. Lozano, K. Kostarelos, P. Malmberg and B. Fadeel, Chem, 2017, 4, 334–358 Search PubMed.
- C. Riedl, C. Coletti, T. Iwasaki, A. A. Zakharov and U. Starke, Phys. Rev. Lett., 2009, 103, 1–4 CrossRef PubMed.
- N. Mishra, V. Miseikis, D. Convertino, M. Gemmi, V. Piazza and C. Coletti, Carbon, 2016, 96, 497–502 CrossRef CAS.
- A. Rossi, H. Büch, C. Di Rienzi, V. Miseikis, D. Convertino, A. Al-Temimy, V. Volani, M. Gemmi, V. Piazza and C. Coletti, 2D Mater., 2016, 3, 031013 CrossRef.
- C. L. Frewin, C. Coletti, C. Riedl, U. Starke and S. E. Saddow, Mater. Sci. Forum, 2009, 615–617, 589–592 CAS.
- S. J. Martin, J. G. Bradley and T. G. Cotter, Clin. Exp. Immunol., 1990, 79, 448–453 CrossRef CAS PubMed.
- C. Muller, S. Monferran, A. Gamp, P. Calsou and B. Salles, Oncogene, 2001, 20, 4373–4382 CrossRef CAS PubMed.
- C. Carmona-Rivera and M. J. Kaplan, Curr. Protoc. Immunol., 2016, 1–14 Search PubMed.
- G. Sollberger, B. Amulic and A. Zychlinsky, PLoS One, 2016, 11, 1–10 CrossRef PubMed.
- S. T. Smiley, M. Reerstt, C. Mottola-hartshorn, M. Lin, A. Chen, T. W. Smithtt, G. D. Steele and L. B. Chen, Proc. Natl. Acad. Sci. U. S. A., 1991, 88, 3671–3675 CrossRef CAS PubMed.
- A. Perelman, C. Wachtel, M. Cohen, S. Haupt, H. Shapiro and A. Tzur, Cell Death Dis., 2012, 6, 1–7 Search PubMed.
- F. Sivandzade, A. Bhalerao and L. Cucullo, Bio-Protoc., 2019, 9, 1–13 Search PubMed.
- D. Hylane Luiz, A. W. Silveira, M. S. Castro and W. Fontes, Cells, 2022, 11, 2889 CrossRef PubMed.
- V. Brunetti, G. Maiorano, L. Rizzello, B. Sorce, S. Sabella, R. Cingolani and P. P. Pompa, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 6264–6269 CrossRef CAS PubMed.
- L. Erpenbeck, A. L. Gruhn, G. Kudryasheva, G. Günay, D. Meyer, J. Busse, E. Neubert, M. P. Schön, F. Rehfeldt and S. Kruss, Front. Immunol., 2019, 10, 1–12 CrossRef.
- G. Schönrich and M. J. Raftery, Front. Immunol., 2016, 7, 11–14 Search PubMed.
- K. Martinod and D. D. Wagner, Blood, 2014, 123, 2768–2776 CrossRef CAS PubMed.
- E. J. Folco, T. L. Mawson, A. Vromman, B. Bernardes-souza, G. Franck, O. Persson, M. Nakamura, G. Newton, F. W. Luscinskas and P. Libby, Arterioscler. Thromb. Vasc. Biol., 2018, 38, 1901–1912 CrossRef CAS PubMed.
- H. Büch, A. Rossi, S. Forti, D. Convertino, V. Tozzini and C. Coletti, Nano Res., 2018, 11, 5946–5956 CrossRef.
- C. Farrera, K. Bhattacharya, B. Lazzaretto, F. T. Andòn, K. Hultenby, G. P. Kotchey, A. Star and B. Fadeel, Nanoscale, 2014, 6, 6974–6983 RSC.
- V. E. Kagan, N. V. Konduru, W. Feng, B. L. Allen, J. Conroy, Y. Volkov, I. I. Vlasova, N. A. Belikova, N. Yanamala, A. Kapralov, Y. Y. Tyurina, J. Shi, E. R. Kisin, A. R. Murray, J. Franks, D. Stolz, P. Gou, J. Klein-seetharaman, B. Fadeel, A. Star and A. A. Shvedova, Nat. Nanotechnol., 2010, 5, 1–6 CrossRef PubMed.
- S. P. Mukherjee, A. R. Gliga, B. Lazzaretto, B. Brandner, M. Fielden, C. Vogt, L. Newman, A. F. Rodrigues, W. Shao, P. M. Fournier, M. S. Toprak, S. Alexander, K. Kostarelos, B. Kunal and B. Fadeel, Nanoscale, 2018, 10, 1180–1188 RSC.
- R. Kurapati, J. Russier, M. A. Squillaci, E. Treossi, C. Ménard-Moyon, A. E. Del Rio-Castillo, E. Vazquez, P. Samorì, V. Palermo and A. Bianco, Small, 2015, 11, 3985–3994 CrossRef CAS.
- A. C. Ferrari and D. M. Basko, Nat. Nanotechnol., 2013, 8, 235–246 CrossRef CAS.
- M. A. Giambra, V. Miseikis, S. Pezzini, S. Marconi, A. Montanaro, F. Fabbri, V. Sorianello, A. C. Ferrari, C. Coletti and M. Romagnoli, ACS Nano, 2021, 15, 3171–3187 CrossRef CAS PubMed.
- K. Kostarelos and K. S. Novoselov, Science, 2014, 344, 261–263 CrossRef CAS PubMed.
- J. Kim, S. Park, Y. J. Kim, C. S. Jeon, K. T. Lim, H. Seonwoo, S.-P. Cho, T. D. Chung, P.-H. Choung, Y.-H. Choung, B. H. Hong and J. H. Chung, J. Biomed. Nanotechnol., 2015, 11, 2024–2033 CrossRef CAS PubMed.
- S. Ye, P. Yang, K. Cheng, T. Zhou, Y. Wang, Z. Hou, Y. Jiang and L. Ren, ACS Biomater. Sci. Eng., 2016, 2, 722–733 CrossRef CAS PubMed.
- F. Xiaoli, Z. Yaqing, L. Ruhui, L. Xuan, C. Aijie, Z. Yanli, H. Chen, C. Lili and S. Longquan, J. Hazard. Mater., 2021, 416, 126158 CrossRef PubMed.
- S. Jaworski, B. Strojny, E. Sawosz, M. Wierzbicki, M. Grodzik, M. Kutwin, K. Daniluk and A. Chwalibog, Int. J. Mol. Sci., 2019, 20(3), 650 CrossRef CAS PubMed.
- A. Fabbro, D. Scaini, V. León, E. Vázquez, G. Cellot, G. Privitera, L. Lombardi, F. Torrisi, F. Tomarchio, F. Bonaccorso, S. Bosi, A. C. Ferrari, L. Ballerini and M. Prato, ACS Nano, 2016, 10, 615–623 CrossRef CAS PubMed.
- R. Rauti, N. Lozano, V. León, D. Scaini, M. Musto, I. Rago, F. P. Ulloa Severino, A. Fabbro, L. Casalis, E. Vázquez, K. Kostarelos, M. Prato and L. Ballerini, ACS Nano, 2016, 10, 4459–4471 CrossRef CAS PubMed.
- G. S. Selders, A. E. Fetz, M. Z. Radic and G. L. Bowlin, Regener. Biomater., 2017, 55–68 CrossRef CAS PubMed.
- W. G. Brodbeck, G. Voskerician, N. P. Ziats, Y. Nakayama, T. Matsuda and J. M. Anderson, J. Biomed. Mater. Res., Part A, 2003, 64A, 320–329 CrossRef CAS PubMed.
- S. Huang, S. Li, Y. Liu, B. Ghalandari, L. Hao, C. Huang, W. Su, Y. Ke, D. Cui, X. Zhi and X. Ding, Adv. Healthcare Mater., 2022, 11, 2102439 CrossRef CAS PubMed.
- V. Ramachandran, M. F. Brady, A. R. Smith, R. M. Feenstra and D. W. Greve, J. Electron. Mater., 1998, 27, 308–312 CrossRef CAS.
- Y. Li, H. Yuan, A. von dem Bussche, M. Creighton, R. H. Hurt, A. B. Kane and H. Gao, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 12295–12300 CrossRef CAS PubMed.
- X. Yao, Z. Yan, X. Wang, H. Jiang, Y. Qian and C. Fan, Regener. Biomater., 2021, 1–9 Search PubMed.
- Y. Hui, Z. Yan, H. Yang, X. Xu, W. Yuan and Y. Qian, ACS Appl. Bio Mater., 2022, 5, 4741–4759 CrossRef CAS PubMed.
- A. E. Jakus, E. B. Secor, A. L. Rutz, S. W. Jordan, M. C. Hersam and R. N. Shah, ACS Nano, 2015, 9, 4636–4648 CrossRef CAS PubMed.
- Y. Qian, X. Zhao, Q. Han, W. Chen, H. Li and W. Yuan, Nat. Commun., 2018, 9, 323 CrossRef PubMed.
- W. Zhang, X. Fang, Q. Li, W. Pi and N. Han, Neural Regener. Res., 2023, 18, 200–206 CrossRef CAS PubMed.
- X. Liu, A. L. Miller, S. Park, B. E. Waletzki, Z. Zhou, A. Terzic and L. Lu, ACS Appl. Mater. Interfaces, 2017, 9, 14677–14690 CrossRef CAS PubMed.
- Y. Qian, X. Wang, J. Song, W. Chen, S. Chen, Y. Jin, Y. Ouyang, Y. Wei-En and C. Fan, npj Regener. Med., 2021, 6, 31 CrossRef CAS PubMed.
- Y. Liu, T. Chen, F. Du, M. Gu, P. Zhang, X. Zhang, J. Liu, L. Lv, C. Xiong and Y. Zhou, J. Biomed. Nanotechnol., 2016, 12, 1270–1284 CrossRef CAS PubMed.
- J. Li, G. Wang, H. Geng, H. Zhu, M. Zhang, Z. Di, X. Liu, P. K. Chu and X. Wang, ACS Appl. Mater. Interfaces, 2015, 7, 19876–19881 CrossRef CAS PubMed.
- S. C. Park, W. S. Yang, J. Y. Ahn, J. B. Park, J. Lee, Y. Jung, H. R. Kim, J. Y. Kim, J. M. Lim and B. H. Hong, 2D Mater., 2021, 8, 035012 CrossRef CAS.
- Kenry, W. C. Lee, K. P. Loh and C. T. Lim, Biomaterials, 2018, 155, 236–250 CrossRef CAS PubMed.
- I. Lasocka, E. Jastrzębska, L. Szulc-Dąbrowska, M. Skibniewski, I. Pasternak, M. Hubalek Kalbacova and E. M. Skibniewska, Int. J. Nanomed., 2019, 14, 2281–2299 CrossRef CAS PubMed.
- J. Litowczenko, J. K. Wychowaniec, K. Załęski, Ł. Marczak, C. J. C. Edwards-Gayle, K. Tadyszak and B. M. Maciejewska, Biomater. Adv., 2023, 154, 213653 CrossRef CAS PubMed.
- L. Cicero, R. Puleio, G. Cassata, R. Cirincione, L. Camarda, D. Caracappa, L. D'Itri, M. Licciardi and G. E. Vigni, Polymers, 2023, 15, 1–13 Search PubMed.
- T. B. Aigner, C. Haynl, S. Salehi, A. O'Connor and T. Scheibel, Mater. Today Bio., 2020, 5, 100042 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3nr04927b |
| ‡ These authors equally contributed to the work. |
| § These authors share last authorship. |
| This journal is © The Royal Society of Chemistry 2024 |