Open Access Article
Open Access ArticleLabel-free detection of MiRNA biomarkers using broadband multi-resonant infrared metasurfaces for early breast cancer diagnosis†
Shuyan
Zhang
 a,
Qing Yang Steve
Wu
a,
Yi Fan
Chen
a,
Melissa
Hum
b,
Dave Chi Lok
Wong
c,
Ern Yu
Tan
de,
Ann Siew Gek
Lee
*b,
Jinghua
Teng
a,
Qing Yang Steve
Wu
a,
Yi Fan
Chen
a,
Melissa
Hum
b,
Dave Chi Lok
Wong
c,
Ern Yu
Tan
de,
Ann Siew Gek
Lee
*b,
Jinghua
Teng
 *a,
Dinish
U.S
*a and
Malini
Olivo
*a
*a,
Dinish
U.S
*a and
Malini
Olivo
*a
aInstitute of Materials Research and Engineering (IMRE), Agency for Science, Technology and Research (A*STAR), 2 Fusionopolis Way, Innovis #08-03, Singapore, 138634, Republic of Singapore. E-mail: dinish@imre.a-star.edu.sg; malini_olivo@imre.a-star.edu.sg; jh-teng@imre.a-star.edu.sg
bDivision of Cellular and Molecular Research, Humphrey Oei Institute of Cancer Research, National Cancer Centre (NCC), 30 Hospital Boulevard, Singapore, 169610, Republic of Singapore. E-mail: dmslsg@nccs.com.sg
cChang Gung University, No. 259, Wenhua 1 st Rd, Guishan District, Taoyuan City, 333, Taiwan, Republic of China
dBreast & Endocrine Surgery, Tan Tock Seng Hospital (TTSH), 11 Jln Tan Tock Seng, Singapore, 308433, Republic of Singapore
eLee Kong Chian School of Medicine, Nanyang Technological University, 50 Nanyang Avenue, Singapore, 639798, Republic of Singapore
First published on 24th May 2023
Abstract
Breast cancer is the most prevalent cancer globally. Early detection is crucial and can be achieved by detecting cancer biomarkers in blood, such as circulating miRNAs (microRNAs). In this study, we present a label-free detection method based on broadband multi-resonant infrared metasurface for surface-enhanced infrared absorption (SEIRA) spectroscopy to detect miRNAs. The SEIRA resonances were optimized to match the miRNA biomarker fingerprint regions in the range of 800 to 2000 cm−1 and 2800 to 3500 cm−1, resulting in a simulated resonance enhancement of up to 103 times. Nine patient samples (six cancerous and three non-cancerous) were measured using SEIRA multi-well sensor chips. A novel analysis method, SEIRA-AR, was also developed to benchmark the results against industry standards, such as quantitative reverse transcription polymerase chain reaction (RT-qPCR) and next-generation sequencing (NGS). Our results showed an excellent linear correlation with a Pearson's r value of up to 0.99 and an R Squared value of up to 0.98. This study represents the first use of a SEIRA sensor for biomarker detection on clinical breast cancer samples and introduces an analysis method that produces results comparable to industry standards. Our findings pave the way for routine cancer diagnosis in the future. Additionally, the method discussed can be generalized to other biosensing activities involving two-step binding processes with complementary molecule-capturing agents.
Introduction
Breast cancer is the most commonly diagnosed cancer globally, with about 2.3 million newly diagnosed cases and 685![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths in 2020.1 The current approach for screening breast cancer in asymptomatic individuals is mammography. Although mammography is deemed the gold standard for breast cancer screening, it has many well-recognized shortcomings, such as high false-positive results, overdiagnosis, and poor participation rates.2,3 False-positive mammograms are also more common among younger women or women of Asian ethnicities due to the presence of denser breast tissue, affecting the sensitivity and specificity of mammography.4,5 A study in the U.S. has shown that 11–13% of all screening mammograms are abnormal, with true-positive rates standing below 0.5% of all screening mammograms. Hence, a person with an abnormal mammogram has a ∼96% chance of having a false-positive result.6 Furthermore, women with false-positive mammograms are subjected to unnecessary additional diagnostic imaging tests and tissue biopsies, which are stressful, invasive, expensive, and laborious. In addition, it has been reported that the national cost of false-positive mammograms and breast cancer overdiagnoses is $4 billion annually, based on a study conducted in 2015.6 Hence, there is a need for a less-invasive, quick, and reliable test to discriminate breast cancers and non-breast cancers in women with abnormal screening mammograms.
000 deaths in 2020.1 The current approach for screening breast cancer in asymptomatic individuals is mammography. Although mammography is deemed the gold standard for breast cancer screening, it has many well-recognized shortcomings, such as high false-positive results, overdiagnosis, and poor participation rates.2,3 False-positive mammograms are also more common among younger women or women of Asian ethnicities due to the presence of denser breast tissue, affecting the sensitivity and specificity of mammography.4,5 A study in the U.S. has shown that 11–13% of all screening mammograms are abnormal, with true-positive rates standing below 0.5% of all screening mammograms. Hence, a person with an abnormal mammogram has a ∼96% chance of having a false-positive result.6 Furthermore, women with false-positive mammograms are subjected to unnecessary additional diagnostic imaging tests and tissue biopsies, which are stressful, invasive, expensive, and laborious. In addition, it has been reported that the national cost of false-positive mammograms and breast cancer overdiagnoses is $4 billion annually, based on a study conducted in 2015.6 Hence, there is a need for a less-invasive, quick, and reliable test to discriminate breast cancers and non-breast cancers in women with abnormal screening mammograms.
Various blood-based biomarkers have been reviewed for the early detection of breast cancer.7 Circulating microRNAs (miRNAs) have emerged as a promising non-invasive biomarker for cancer diagnosis and prognosis because of their remarkable stability in the bloodstream and their involvement in cancer development and progression.8,9 They are a class of small non-coding RNAs with 19 to 25 nucleotides that play important roles in regulating gene expression. Conventional methods such as northern blotting, microarray-based hybridization, quantitative reverse transcription polymerase chain reaction (RT-qPCR), and next-generation sequencing (NGS) have been widely used to detect miRNAs.10 However, some of these methods are costly, and they involve complex and time-consuming procedures. Hence, we propose an alternative label-free approach based on surface-enhanced infrared absorption (SEIRA) to measure the target miRNA molecule contents in total RNA solutions extracted from the patient's blood serum samples. The infrared spectroscopy technique detects the molecular vibrations that result in a change of the dipole moment of the molecules, and it measures the absolute frequency (molecular fingerprints) where the molecules absorb infrared light in a label-free manner. SEIRA is a technique based on the resonant effect of the metasurface nanostructures at the substrate surface, where the molecules interact with the incident light to improve the sensitivity of infrared spectroscopy measurements.11,12 The resonance wavelength(s) can be tuned by the material, shape, and size of the nanostructures. Many nanostructures have been proposed, including zig-zag nanotips, nanorods, nanodiscs, nanoislands, etc.13–21
Here, we demonstrate a multi-well SEIRA sensor chip based on metallic nanorod structures featuring a broad-band multiband resonance. The structures are specifically designed so that the resonances coincide with the absorption fingerprint regions of the target miRNA biomarkers, hence maximizing the light interaction with the biomarker molecules and collected SEIRA signals. In addition, we demonstrate the possibility of multiplexed detection. The sensor chip comprises 15 millimeter-sized sensor wells capable of measuring multiple biomarkers and patient samples on a single platform. The SEIRA sensor structures are repeated in each well, and each well is isolated from its neighbors so that it can be treated as a separate SEIRA measurement site. We have earlier performed extensive studies on establishing a miRNA biomarker panel for early breast cancer diagnosis.22–24 Here, we detected miRNAs hsa-miR-let-7a-5p, hsa-miR-451a, hsa-miR126-5p, and hsa-miR-195-5p, for patients who were clinically diagnosed with breast cancer by mammography and histopathology. The detection process involves binding thiol group-modified complementary single-stranded DNA (ssDNA) molecules to the SEIRA sensor and subsequent selective binding of target miRNA molecules to the complementary ssDNA molecules. We then propose to use the area ratio of the change in the area under the curve of the SEIRA signal at each step to represent the concentration of miR-NAs in the sample. The analysis method is termed “SEIRAAR”. It takes into account both the shift in the absorption peak position and the change in the intensity values across the entire infrared absorption spectrum. An excellent correlation has been established between the SEIRA-AR index values and the industry-standard RT-qPCR and NGS read counts. This demonstrates that our proposed approach could accurately detect specific target miRNA molecules in a mixed environment. Moreover, our approach is label-free without the need to use fluorescent dyes, and it is relatively fast. It is one of the first successful demonstrations of a SEIRA sensor being used for biomarker detection in a clinical setting and compared with industry standards. The study paves the way for routine cancer diagnosis in the future. It can also be used for real-time and in situ label-free measurements of any biomarker, which can be captured and immobilized using a complementary targeting moiety.
Materials and methedology
Breast cancer sample preparation
The authors extracted the microRNAs from serum samples, which were prepared from the collected blood samples. Peripheral blood samples were collected at the National Cancer Centre Singapore (Singapore) and Tan Tock Seng Hospital (Singapore) prior to biopsy or surgery. Serum samples were also obtained from the SingHealth Tissue Repository (Singapore). The materials were not purchased/donated. There were five women with invasive breast carcinomas, one with ductal carcinoma in situ (DCIS), and three with benign fibroadenoma. Women with invasive carcinomas or DCIS were defined as cancerous cases, and others were non-cancerous (control) cases. This study was conducted in accordance with the Declaration of Helsinki, and approved by the Centralized Institutional Review Board of SingHealth (CIRB Ref: 2018/2874). Written informed consent was obtained from all study participants.Total RNA was isolated from 200 μl of serum using the miRNeasy Serum/Plasma Advanced Kit (Qiagen N. V.). This was performed according to the manufacturer's protocol, except for an additional step where bacteriophage MS2 RNA was added into the sample lysis buffer (1 μg mL−1 of QIAzol) to improve the RNA yield.
SEIRA sensor fabrication
The SEIRA sensor consists of rod-shaped nanostructures fabricated using electron beam lithography (ELS-7000, Elionix Inc.). In this process, two layers of photoresists were spin-coated (PMMA 495 A3, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with IPA, 4000 rpm 90 seconds and hot plate cured at 180 °C for 10 minutes, followed by PMMA 950 A5, 1
1 with IPA, 4000 rpm 90 seconds and hot plate cured at 180 °C for 10 minutes, followed by PMMA 950 A5, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 with IPA, 4000 rpm 90 seconds and hot plate cured at 180 °C for 10 minutes) on a clean CaF2 substrate. The electron beam current setting was 500 pA, 100 kV, and dosage 0.6. After the electron beam exposure, the sample was dipped into MIBK
2 with IPA, 4000 rpm 90 seconds and hot plate cured at 180 °C for 10 minutes) on a clean CaF2 substrate. The electron beam current setting was 500 pA, 100 kV, and dosage 0.6. After the electron beam exposure, the sample was dipped into MIBK![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IPA 1
IPA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 solution for 1 minute, followed by rinsing and N2 air gun drying. Electron beam evaporation (Explorer, Denton Vacuum) was performed subsequently to deposit 10 nm of Cr adhesion layer and 100 nm of Au layer. Lift off process was then carried out using acetone solution to remove the extra film of PMMA, followed by washing in the IPA solution and DI water and then drying.
3 solution for 1 minute, followed by rinsing and N2 air gun drying. Electron beam evaporation (Explorer, Denton Vacuum) was performed subsequently to deposit 10 nm of Cr adhesion layer and 100 nm of Au layer. Lift off process was then carried out using acetone solution to remove the extra film of PMMA, followed by washing in the IPA solution and DI water and then drying.
Multi-well SEIRA sensor chip
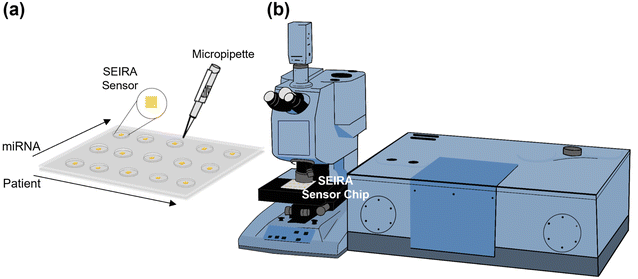
Fig. 1(a) shows a multi-well sensor chip consisting of 5 rows and 3 columns with a total of 15 individual wells. Firstly, the SEIRA sensor structures were repeated 15 times, each with a dimension of 500 μm × 500 μm and a center-to-center distance of 7.5 mm. Next, a multi-well mask was created by a polydimethylsiloxane (PDMS) layer of thickness 1 mm with the well positions aligned with the patterned SEIRA structures. Each well was made by a pen puncher with a tip size of 1.5 mm. The PDMS layer was then placed on top of the SEIRA structures and adhered to by van der Waals force. The final sensor chip allows detection at multiple sites to be performed independently. The spacing between the cells ensures that the solutions will not cross over to neighboring wells. The size of each well ensures that SEIRA reflectance signals are collected with a reasonable sample area. For one of the experiments, we have arranged the wells so that the rows measure different patients (i.e., patient sample 1–5) and the columns measure different target miRNA types (i.e., miRNA 1–3). Finally, a micropipette was used to dispense the sample solutions precisely.Fig. 1(b) is a schematic of the Fourier transform infrared (FTIR) system attached with a microscope accessory (Vertex 80v with Hyperion 2000, Bruker Corporation). A 15× reflective microscope objective was used to focus the incident light onto the SEIRA structure and collect the reflected signal. A region of interest of 100 μm × 100 μm was selected for each measurement. Three measurements were performed at different locations for each SEIRA structure to minimize the effect of fabrication non-uniformity and ensure measurement repeatability. Each FTIR measurement was taken at the wavenumber range of 800–5000 cm−1 with a resolution of 4 cm−1 and 64 scans.
Design of experiments
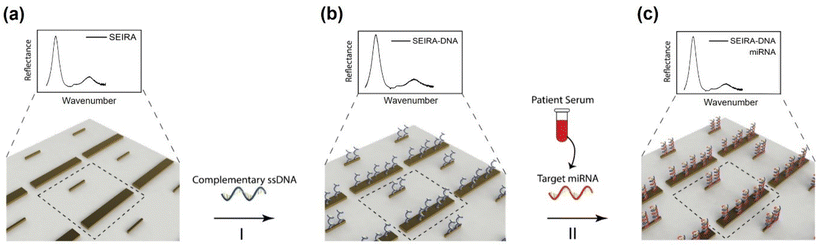
There were four experiments performed. The first was a baseline measurement using synthetic thiol group-modified ssDNA and miRNA samples (Integrated DNA Technologies, Inc.). The measurements were performed using an FTIR spectroscopy instrument with the attenuated total reflection (ATR) accessory (VERTEX 70v, Bruker Corporation) under the vacuum condition. The results were used to guide the SEIRA sensor nanostructures design and simulation. After the SEIRA sensor was designed and fabricated, the second to fourth experiments were conducted using the FTIR microscope system, as shown in Fig. 1(b). The second experiment was to quantify the SEIRA enhancement by comparing the measured FTIR signal of a bare area (without SEIRA structures) and with SEIRA structures. The third experiment sought to validate the proposed SEIRA-AR analysis method with both RT-qPCR and NGS results by measuring the miRNA concentration in five clinical samples. Finally, the fourth experiment demonstrated the capability of using the SEIRA sensor for multiplexed detection by measuring three breast cancer miRNA biomarkers from another set of five clinical samples and establishing a linear relationship between SEIRA-AR index values and RT-qPCR results.The experimental workflow using the SEIRA sensor is as follows. Firstly, FTIR signals were collected from a clean SEIRA sensor. Fig. 2(a) shows a zoom-in view of the rod-shaped nanostructures of the SEIRA sensor with the dashed square highlighting the design unit cell. Secondly, solutions containing thiol group-modified complementary ssDNA molecules (2 μL, 50 μM) were immobilized to the SEIRA substrate (Fig. 2(b)). The thiol group will react to Au, so the ssDNA molecules will bind to the SEIRA substrate. After 1 hour, the substrate was rinsed thoroughly with RNase-free water to remove the excess unbound ssDNA molecules. Thirdly, solutions containing target miRNA molecules (2 μL) extracted from breast cancer patients’ serums were immobilized on the SEIRA-DNA substrate. Only target miRNA molecules will bind to the ss-DNA molecules because of the peptide bonds. After 1 hour, the sensor was rinsed thoroughly with RNase-free water to remove the excess unbound miRNA molecules. The specificity of this two-step binding process was previously studied for similar devices in our earlier papers.25–27 FTIR microscope measurements were taken at each step, i.e., SEIRA, SEIRA-DNA, and SEIRA-DNA-miRNA. The same workflow was applied to each well on the sensor.
Data analysis workflow
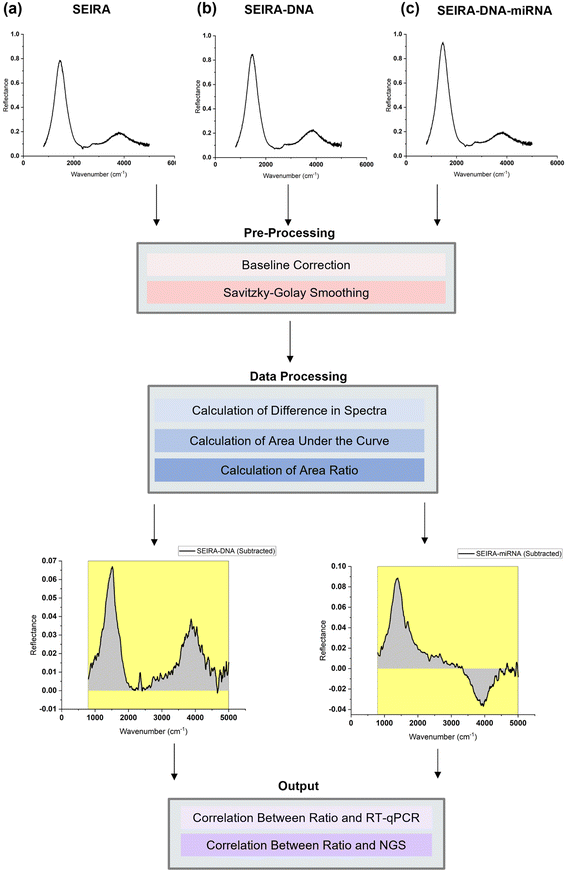
To best quantify the change in the optical signals during the two-step binding process, a method based on calculating the area ratio under the curve of the difference spectrum was proposed, as shown in Fig. 3. Firstly, the reflectance spectrum of SEIRA (RSEIRA(λ)), SEIRA-DNA (RSEIRA-DNA(λ)), and SEIRA-DNA-miRNA (RSEIRA-DNA-miRNA(λ)) were measured three times at different locations. Pre-processing steps were then carried out, including baseline correction and Savitzky–Golay smoothing. Data processing steps involve firstly the calculation of the differences between the two spectra, which are eqn (1) and (2), and secondly the calculation of the area under the curves, which are eqn (3) and (4), and finally the calculation of the ratio between the two areas, which is eqn (5).| ΔDNA(λ) = RSEIRA-DNA(λ) − RSEIRA-DNA(λ) | (1) |
| ΔmiRNA(λ) = RSEIRA-DNA-miRNA(λ) − RSEIRA-DNA(λ) | (2) |
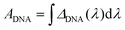 | (3) |
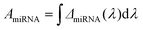 | (4) |
| Ratio = |AmiRNA/ADNA| | (5) |
The method is termed “SEIRA-AR” in short, which stands for SEIRA Area Ratio. The ratio between the two areas under the curve represents the miRNA-DNA binding events as a percentage of the DNA-SEIRA binding events so that unsuccessful DNA binding events are not reflected. The result is a SEIRA-AR index calculated for each target miRNA type. It is worth mentioning that this index accounts for both the shift in the peak position and the change in the intensity values across the entire spectrum. Hence, it allows us to capture the total change in the optical signals due to the DNA and miRNA binding events accurately. Finally, the SEIRA-AR index values are compared with the RT-qPCR and NGS read counts for a miRNA biomarker, and a linear correlation is established to validate the method.
Results and discussion
Simulation results of SEIRA sensor
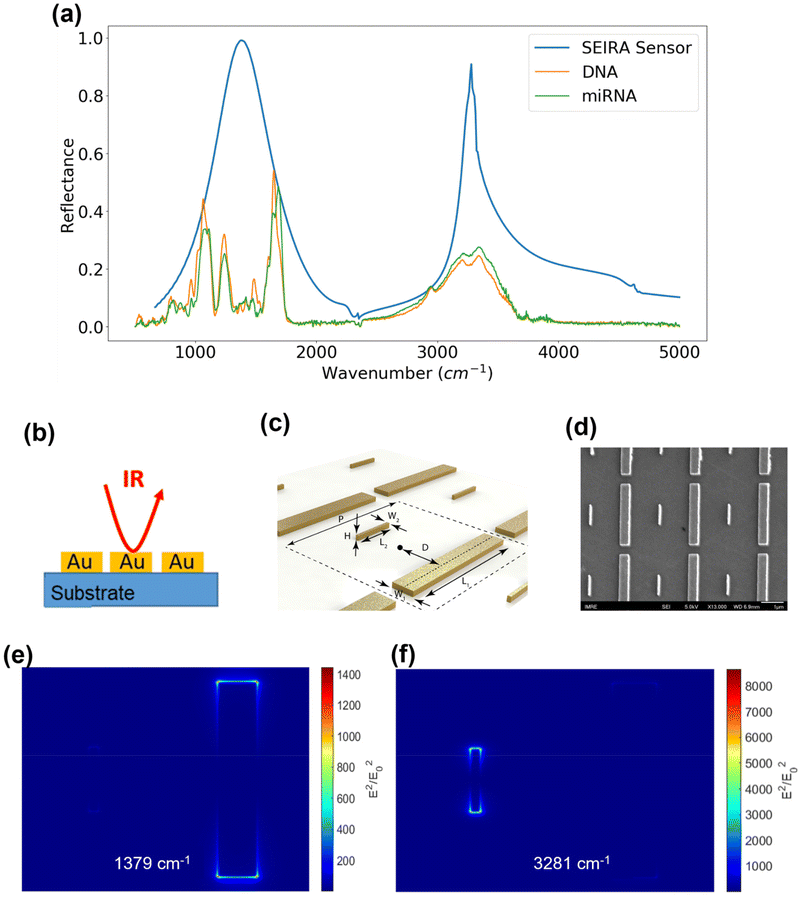
The SEIRA sensor design and simulation were performed using the Lumerical FDTD software (Ansys, Inc.). It was guided by the infrared absorption features of the DNA and miRNA molecules because the purpose of SEIRA is to enhance their interaction with the incident light. Hence, a first experiment was performed on a variety of synthetic thiol group-modified ssDNA and miRNA samples to obtain their infrared spectrum. As mentioned in the methodology section, this experiment was conducted using an ATR-FTIR system under a vacuum environment. The ATR accessory improves the measurement sensitivity of a conventional FTIR instrument. In addition, water has strong absorption features in the infrared region, so the vacuum condition is necessary to remove the interference from water. We have measured five types of synthetic thiolated ssDNA and miRNA solution samples under vacuum and atmospheric conditions. The results are provided in Fig. S1 and S2 in the ESI.† We note that the absorption peaks, i.e., the molecular fingerprints of the ss-DNAs and miRNAs could be visualized only under the vacuum condition. Selected spectra of the synthetic hsa-DNA-let-7a-5p and hsa-miR-let-7a-5p were shown in Fig. 4(a) as the orange and green curves, respectively. The identified fingerprints are shown in Table 1, where each fingerprint is represented by a peak in the spectrum corresponding to a type of molecular vibration or chemical bond.28–31 It was found that although there are slight variations in the spectra of different kinds of DNA and miRNA samples, there exist two prominent fingerprint regions, 800–2000 cm−1 and 2800–3500 cm−1. Hence, SEIRA nanostructures with broadband multi-resonances that match these two finger-print regions are ideal. A metallic nanorod structure was applied and optimized so that the resonance peaks coincide with the centers of the fingerprint regions. Fig. 4(b) is a schematic showing the infrared light interacting with the SEIRA structures that consist of a CaF2 substrate and gold (Au) nanorods. Fig. 4(c) is a schematic of the unit cell of the SEIRA structure with a long and wide nanorod and a short and narrow nanorod. The unit cell repeats in a period P = 3 μm. The spacing between the two nanorods is 2D = 1.5 μm. The length, width, and height of the long and wide nanorod are: L1 = 2.6 μm, W1 = 0.4 μm, H1 = H = 0.1 μm. The length, width, and height of the short and narrow nanorod are: L2 = 0.85 μm, W2 = 0.08 μm, H2 = H = 0.1 μm. Fig. 4(d) is a scanning electron microscope (SEM) image of the fabricated structures. Fig. 4(e) and (f) show the electric field intensity distributions of the SEIRA structures at the two resonance peaks at 1379 cm−1 and 3281 cm−1, respectively. Specifically, the long nanorod has a resonance at 1379 cm−1 with an intensity enhancement of 1.4 × 103 times at the edges of the rod. The short nanorod has a resonance at 3281 cm−1 with an intensity enhancement of more than 8 × 103 times at the edges of the rod. The resonance peak and width can be tuned by changing the length, width, and height of the nanorods. For example, an increase in the width will increase the wavenumber of the resonance peak and width. We have performed extensive optimization of the SEIRA structures using Lumerical software simulations. The optimized parameters are shown in Fig. 4(c). To determine the optimal shape and size of the Au nanorods, we selectively presented the optimization results for W2 and h variations in Fig. S3.† The selection criteria included maximizing the overlapping between the SEIRA spectrum peak wavenumbers and the DNA and miRNA reflectance peak wavenumber ranges, i.e., around 1379 cm−1 and 3281 cm−1; maximizing the reflectance intensity of the SEIRA spectrum peaks; and maximizing the bandwidth of the SEIRA spectrum so all DNA and miRNA peaks could be enhanced. The final optimized result of the SEIRA structure is shown as the blue curve in Fig. 4(a).| Peak (cm−1) | Chemical bond |
|---|---|
| 788 | Free cytosine vibration |
| 798 | Free adenine vibration |
| 813, 866 | Main N-type sugar marker |
| 915 | Ribose ring vibration |
| 966 | C–C and C–O ribose stretching |
| 995 | Ribose-phosphate main chain vibration |
| 1082 | PO2− symmetric stretching |
| 1110 | C–O ribose ring stretching |
| 1240 | PO2− anti-symmetric stretching |
| 1475 | Adenine and guanine ring vibration |
| 1492 | Cytosine in-plane vibration |
| 1600 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N ring vibration of guanine N ring vibration of guanine |
| 1650 | C2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O2 stretching in cytosine O2 stretching in cytosine |
| 1685 | C2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O2 stretching in guanine O2 stretching in guanine |
| 2950 | C–H stretching |
| 3140, 3210 | O–H stretching |
| 3330, 3340 | N–H stretching |
Experimental results of SEIRA enhancement
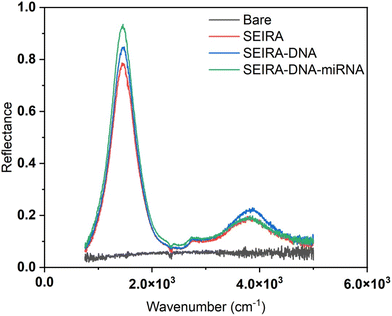
Fig. 5 shows a representative measurement result of a bare substrate area without SEIRA structures and an area with SEIRA structures. The reflectance with SEIRA was enhanced 20 times compared to a bare area. The biological reaction after each step, SEIRA, SEIRA-DNA, and SEIRA-DNA-miRNA was clearly captured in the optical measurements, which involves changes in the reflectance spectrum over the entire wavenumber range, not only a shift in the peak position.Experimental validation of SEIRA-AR index
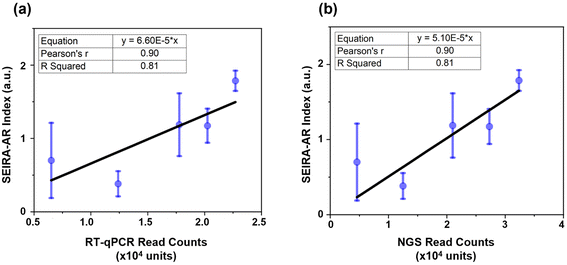
Firstly, the SEIRA-AR analysis method was compared with industry standards. In our previous studies, hsa-miR-let-7a-5p (5′-UGAGGUAGUAGGUUGUAUAGUU-3′) was detected through RT-qPCR and NGS methods, and a good correlation was observed between these detection methods.22,23 For the SEIRA sensor experiment, the complementary ssDNA with thiol group modification was 5′-AACTATACAACCTACTACCTCA/3ThioMC3-D/-3′. Five patients (three cancerous and two non-cancerous) were measured. Their hsa-miR-let-7a-5p SEIRA-AR index values were calculated and plotted against the RT-qPCR and NGS read counts, as shown in Fig. 6(a) and (b), respectively. The mean values and the standard deviations were shown in blue circles and error bars. A linear curve fitting was performed with an equation y = a × x, with a being the fitting parameter. The constraint that the curve intercepts at the origin reflects the physical meaning that when the read count is zero, the SEIRA-AR index value should be zero. The fitting parameters are shown in the top left corner with a = 6.60 × 10−5 for RT-qPCR and a = 5.10 × 10−5 for NGS.Pearson's r, also known as Pearson's correlation coefficient, measures the strength of the linear relationship. A value closer to 1 indicates a strong positive linear correlation between the paired data. R Squared value, also known as the coefficient of determination, measures the quality of the linear relationship. A value closer to 1 indicates a better-fitted line that explains the variability of the response variable. It is seen that Pearson's r and R Squared values are 0.90 and 0.81 in both cases, indicating that there exists a good correlation between SEIRA-AR index values and RT-qPCR and NGS read counts. The error bars appear larger for the data at lower miRNA read counts, indicating a potential limitation of the sensor at low concentrations. This could be due to the reduced sensitivity of the sensor at low miRNA concentrations, which could result in higher measurement variability. While we have attempted to optimize the sensor design and data analysis methods to mitigate this effect, future studies may explore more complex structures to improve the accuracy and sensitivity of the sensor at low miRNA concentrations, for example, using a tetrahedral DNA nanostructure.16 Nonetheless, we believe that our findings demonstrate the potential of the sensor as a sensitive and accurate platform for miRNA biomarker detection.
SEIRA measurements of three breast cancer biomarkers
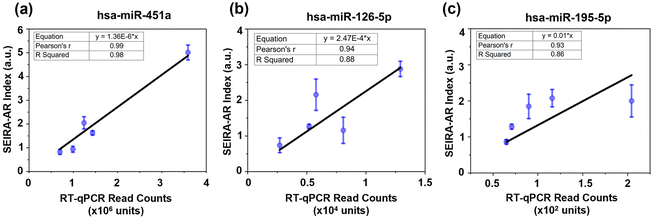
Next, established breast cancer miRNA biomarkers were measured. An optimal six-miRNA panel was reported for differentiating malignant and benign breast cancer samples based on a study of 638 patients with abnormal mammograms and 100 healthy controls.22 Here, three miRNAs from the panel were selected for detection, namely, hsa-miR-451a (5′-AAACCGUUACCAUUACUGAGUU-3′), hsa-miR-126-5p (5′-CAUUAUUACUUUUGGUACGCG-3′), and hsa-miR-195-5p (5′-UAGCAGCACAGAAAUAUUGGC-3′). The corresponding ssDNA capturing molecules with thiol group modifications were 5′-TTTGGCAATGGTAATGACTCAA/3ThioMC3-D/-3′, 5′-GTAATAATGTTTTCCATGCGC/3ThioMC3-D/-3′, 5′-ATCGTCGTGTCTTTATAACCG/3ThioMC3-D/-3′, respectively. There were five patients (four cancerous and one non-cancerous) measured. The SEIRA-AR index values obtained were plotted against known RT-qPCR read counts, as shown in Fig. 7. A linear correlation was established for each miRNA biomarker, i.e., y = 1.36 × 10−6x (for hsa-miR-451a), y = 2.47 × 10−4x (for hsa-miR-126-5p), and y = 1.00 × 10−2x (for hsa-miR-195-5p). The Pearson's r values are 0.99, 0.94, 0.93, and R Squared values are 0.98, 0.88, 0.86, which indicate strong correlations. This shows that the SEIRA sensor and the SEIRA-AR analysis method are capable of multiplexed measuring of biomarker molecules of clinical samples accurately. To improve the accuracy and reliability of the linear relationship between the new index value and the industry standards, future studies may consider increasing the sample size by reliable sensor platform for miRNA biomarker detection.Conclusions
This paper presents a multi-well SEIRA sensor chip capable of label-free, multiplexed sensing of DNA and RNA molecules based on their infrared absorption molecular fingerprints. The broadband multi-resonant SEIRA structures were designed to match the resonances with the absorption peaks of the target molecules, ensuring maximum light-matter interaction at the SEIRA/molecule interface. Simulations showed that the resonance enhancement was on the order of 103 times. The sensor can measure multiple biomarkers and samples on a single platform. We also developed a new parameter, the SEIRA-AR index, to measure specific target miRNA biomarkers in a patient's serum solution containing a mixture of RNAs, using complementary ssDNA with thiol group modifications as capturing molecules. We proposed using the ratio of the change in the area under the curve of the SEIRA spectra at each binding step as a reflection of the concentration of the target miRNA biomarker in the sample. The SEIRA-AR index values were compared with RT-qPCR and NGS read counts collecting additional data points. This could be achieved by increasing the number of measurements per well or increasing the number of samples per biomarker. The larger sample size would enable a more comprehensive evaluation of the sensor's performance and improve the statistical significance of the results. However, we recognize that increasing the sample size may require additional resources and time, and thus it may not be feasible in all cases. Nonetheless, we believe that this is an important direction for future research in developing a robust and for breast cancer biomarkers using clinical samples. An excellent correlation was established, with a Pearson's r value up to 0.99 and an R Squared value up to 0.98 for detecting hsa-miR-let-7a-5p, hsa-miR-451a, hsa-miR-126-5p, and hsa-miR-195-5p. Compared to RT-qPCR and NGS methods, the proposed SEIRA sensor offers significant advantages: it is label-free, relatively fast (less than 2 hours), and cost-effective. In summary, we have demonstrated, for the first time, the use of a SEIRA sensor on clinical breast cancer samples for miRNA biomarker detection, and developed an analysis method that produces results that can be benchmarked with industry standards. The sensor and analysis approach could also be applied to other biomarker detection and biosensing activities that involve the use of a complementary molecule as the capturing agent. An area for improvement of the current device is to increase detection speed, such as developing a read-out system to capture spectral information at all detection sites simultaneously. With necessary refinement and optimization, we envision that this sensor can be used for routine medical diagnosis in the future.Author contributions
Conception: S. Zhang, D. C. L. Wong, A. S. G. Lee, J. Teng, D. U. S., and M. Olivo. Simulation: S. Zhang and D. C. L. Wong. Clinical samples: M. Hum, E. Y. Tan, and A. S. G. Lee. Experiment: S. Zhang, Q. Y. S. Wu, and Y. F. Chen, Manuscript preparation: S. Zhang, Q. Y. S. Wu, Y. F. Chen, M. Hum, and D. U. S. Supervision: A. S. G. Lee, J. Teng, D. U. S., and M. Olivo.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank Jayakumar Perumal for helping with the PDMS-made wells; Ghayathri Balasundaram for insightful discussions; Grace Chong, Sahad Zamir, and Casey Ang for helping with the data collection; Valerie Teo and Sonia Tan for helping with data analysis and figure preparation. Agency for Science, Technology and Research, Singapore: IAF-PP Grant H19H6a0025 and BMRC UIBR Grant; National Medical Research Council, Singapore: NMRC/CBRG/0087/2015.References
- M. Arnold, E. Morgan, H. Rumgay, A. Mafra, D. Singh, M. Laversanne, J. Vignat, J. R. Gralow, F. Cardoso, S. Siesling and I. Soerjomataram, Breast, 2000, 66, 15–23 CrossRef PubMed.
- H. D. Nelson, M. Pappas, A. Cantor, J. Griffin, M. Daeges and L. Humphrey, Ann. Intern. Med., 2016, 164, 256–256 CrossRef PubMed.
- C. V. D. Ende, A. M. Oordt-Speets, H. Vroling and H. M. E. V. Agt, Int. J. Cancer, 2017, 141, 1295–1306 CrossRef PubMed.
- H. D. Nelson, E. S. O'meara, K. Kerlikowske, S. Balch and D. Miglioretti, Ann. Intern. Med., 2016, 164, 226–226 CrossRef PubMed.
- T. Li, J. Li, R. Heard, Z. Gandomkar, J. Ren, M. Dai and P. Brennan, Asia. Pac. J. Clin. Oncol., 2022, 696–705 CrossRef PubMed.
- M. S. Ong and K. D. Mandl, Health Aff., 2015, 34, 576–583 CrossRef PubMed.
- S. Y. Loke and A. S. G. Lee, Eur. J. Cancer, 2018, 92, 54–68 CrossRef CAS PubMed.
- L. Sempere, C. Graveel, H. Calderone, J. Westerhuis and M. Winn, Breast Cancer: Targets Ther., 2015, 7, 59–79 Search PubMed.
- H. M. Heneghan, N. Miller, A. J. Lowery, K. J. Sweeney and M. J. Kerin, J. Oncol., 2010, 950201–950201 Search PubMed.
- T. Ouyang, Z. Liu, Z. Han and Q. Ge, Anal. Chem., 2019, 91, 3179–3186 CrossRef CAS PubMed.
- M. Janneh, Results Opt., 2022, 6, 100201 CrossRef.
- F. Neubrech, C. Huck, K. Weber, A. Pucci and H. Giessen, Chem. Rev., 2017, 117, 5110–5145 CrossRef CAS PubMed.
- H. Aouani, M. Rahmani, H. Šípová, V. Torres, K. Hegnerová, M. Beruete, J. Homola, M. Hong, M. Navarro-Cía and S. A. Maier, J. Phys. Chem. C, 2013, 117, 18620–18626 CrossRef CAS.
- D. Rodrigo, A. Tittl, N. Ait-Bouziad, A. John-Herpin, O. Limaj, C. Kelly, D. Yoo, N. J. Wittenberg, S. H. Oh, H. A. Lashuel and H. Altug, Nat. Commun., 2018, 9, 2160–2160 CrossRef PubMed.
- T. D. Dao, K. Chen and T. Nagao, Nanoscale, 2019, 11, 9508–9517 RSC.
- X. Hui, C. Yang, D. Li, X. He, H. Huang, H. Zhou, M. Chen, C. Lee and X. Mu, Adv. Sci., 2021, 8, 2100583 CrossRef CAS PubMed.
- M. Zhao, J. Li, M. Sebek, L. Yang, Y. J. Liu, M. Bosman, Q. Wang, X. Zheng, J. Lu and J. Teng, Adv. Mater., 2021, 2101950–2101950 CrossRef CAS PubMed.
- Z. Yao, Q. Zhang, W. Zhu, M. Galluzzi, W. Zhou, J. Li, A. V. Zayats and X. F. Yu, Nanoscale, 2021, 13, 10133–10142 RSC.
- Y. Bian, Y. Li, K. Liu, H. He, Y. Feng and X. Yu, J. Biophotonics, 2022, e202200277 Search PubMed.
- L. Dong, X. Yang, C. Zhang, B. Cerjan, L. Zhou, M. L. Tseng, Y. Zhang, A. Alabastri, P. Nordlander and N. J. Halas, Nano Lett., 2017, 17, 5768–5774 CrossRef CAS PubMed.
- S. Zhang, C. L. Wong, S. Zeng, R. Bi, K. Tai, K. Dholakia and M. Olivo, Nanophotonics, 2020, 10, 259–293 Search PubMed.
- R. Zou, S. Y. Loke, V. K. Tan, S. T. Quek, P. Jagmohan, Y. C. Tang, P. Madhukumar, B. K. Tan, W. S. Yong, Y. Sim, S. Z. Lim, E. Png, S. Y. S. Lee, M. Y. P. Chan, T. S. J. Ho, B. K. J. Khoo, S. L. J. Wong, C. H. Thng, B. K. Chong, Y. Y. Teo, H. P. Too, M. Hartman, N. C. Tan, E. Y. Tan, S. C. Lee, L. Zhou and A. S. G. Lee, Cancers, 2021, 13, 2130–2130 CrossRef CAS PubMed.
- S. Y. Loke, P. Munusamy, G. L. Koh, C. H. T. Chan, P. Madhukumar, J. L. Thung, K. T. B. Tan, K. W. Ong, W. S. Yong, Y. Sim, C. L. Oey, S. Z. Lim, M. Y. P. Chan, T. S. J. Ho, B. K. J. Khoo, S. L. J. Wong, C. H. Thng, B. K. Chong, E. Y. Tan, V. K. M. Tan and A. S. G. Lee, Cancers, 2019, 11, 1872 CrossRef CAS PubMed.
- R. Zou, S. Y. Loke, Y. C. Tang, H. P. Too, L. Zhou, A. S. G. Lee and M. Hartman, Br. J. Cancer, 2022, 126, 472–481 CrossRef CAS PubMed.
- C. L. Wong, S. Y. Loke, H. Q. Lim, G. Balasun-daram, P. Chan, B. K. Chong, E. Y. Tan, A. S. G. Lee and M. Olivo, J. Biophotonics, 2021, 14, e202100153 CrossRef CAS PubMed.
- K. K. Maiti, U. S. Dinish, A. Samanta, M. Vendrell, K. S. Soh, S. J. Park, M. Olivo and Y. T. Chang, Nano Today, 2012, 7, 85–93 CrossRef CAS.
- D. U.S, C. Y. Fu, K. S. Soh, B. Ramaswamy, A. Kumar and M. Olivo, Biosens. Bioelectron., 2012, 33, 293–298 CrossRef CAS PubMed.
- F. Geinguenaud, V. Militello and V. Arluison, RNA Spectroscopy Methods and Protocols, 2020, pp. 119–133 Search PubMed.
- M. Banyay, M. Sarkar and A. Gräslund, Biophys. Chem., 2003, 104, 477–488 CrossRef CAS PubMed.
- M. Tsuboi, Appl. Spectrosc. Rev., 1970, 3, 45–90 CrossRef.
- I.R. Spectrum Table & Chart. Retrieved from: https://www.sigmaaldrich.com/SG/en/technical-documents/technical-article/analytical-chemistry/photometry-and-reflectometry/ir-spectrum-table.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3nr01369c |
| This journal is © The Royal Society of Chemistry 2023 |