Hydrogel 3D in vitro tumor models for screening cell aggregation mediated drug response†
Maria V.
Monteiro
,
Vítor M.
Gaspar
 *,
Luís P.
Ferreira
and
João F.
Mano
*,
Luís P.
Ferreira
and
João F.
Mano
 *
*
Department of Chemistry, CICECO – Aveiro Institute of Materials, University of Aveiro, Campus Universitário de Santiago, 3810-193, Aveiro, Portugal. E-mail: vm.gaspar@ua.pt; jmano@ua.pt
First published on 17th February 2020
Abstract
Hydrogel-based 3D in vitro models comprising tumor ECM-mimetic biomaterials exhibit superlative potential as preclinical testing platforms for drug discovery and bioperformance screening. However, during hydrogel design and testing stages, the ideal selection between cancer cell laden 3D models or spheroid embedded hydrogel platforms remains to be elucidated. Selecting a disease-mimicking cellular arrangement within ECM hydrogels is paramount for anti-cancer therapeutics performance evaluation and may lead to differential outcomes. To investigate the effects assigned to varying cellular-arrangement, we developed dense 3D spheroid microtumors and cell-laden MG-63 osteosarcoma platforms embedded in GelMA and Matrigel ECM-mimetic scaffolds. These platforms enabled cancer cells/3D microtissues maturation and lorlatinib drug performance screening. Initial 3D spheroids assembly via the liquid overlay technique, resulted in the fabrication of dense cellular aggregates with reproducible size, morphology and necrotic core formation, thus mimicking the native tumor. Upon in vitro maturation, MG-63 spheroids encapsulated in hydrogel scaffolds exhibited significantly higher invasion and drug resistance than their cell laden hydrogel counterparts. Such data reveals inherent physiological and drug response variances among randomly distributed osteosarcoma cells and 3D spheroid-laden hydrogels. Overall, this highlights the importance of evaluating different cellular aggregation states when designing ECM-mimetic hydrogels for in vitro tumor modeling and high-throughput screening of anti-cancer therapeutics.
Introduction
In the last decades, researchers endeavored to capture the complexity of the tumor microenvironment (TME) in a laboratory setting by developing various types of in vitro and in vivo models. However, despite significant advances in the available preclinical testing platforms, the attrition rates in pharmaceutical development and drug discovery pipeline remain remarkably high.During in vitro preclinical testing stages, bidimensional (2D) cell cultures still remain the gold standard models for benchmarking cancer research and for rapidly screening candidate anti-cancer therapeutics.1,2 Nevertheless, researchers are increasingly recognizing that traditional 2D in vitro models established in tissue culture treated polystyrene flat substrates are in essence monolayers and overly simplistic. Monolayer culture models ultimately fail to recapitulate key human tumor hallmarks including the complex three-dimensional (3D) architecture of solid tumors, the pH/nutrient/metabolite gradients, necrotic regions, as well as the 3D cell–cell and cell–matrix interactions, all of which are instrumental for mimicking cancer complexity.3,4 In the long run, conventional 2D cultures have been unable to provide robust data that can predict the performance of new anti-cancer compounds with proper correlation with the in vivo scenario.5–7 On the other hand, despite xenograft in vivo models reveal higher physiological relevance in the study of malignant diseases, they are complex, low-throughput, often involve immunosuppressed animals and do not recapitulate the human anatomic scale.8,9 These fundamental limitations have encouraged the development of more physiomimetic 3D microtumors by using advanced in vitro laboratory setups.2
3D in vitro microphysiological tumor models outperform standard 2D platforms by recapitulating solid human tumors complexity and key disease hallmarks including gene expression patterns.5,6,10,11 In a laboratory setting such tumor-mimicking 3D cell cultures have been generated through two different approaches: (i) scaffold-free 3D tumor spheroid models comprising cell-rich aggregates assembled by self-aggregation,12–14 as well as (ii) scaffold-based platforms combining cancer cells and ECM-mimetic biomaterials.7,15 Although scaffold-free, cell-rich 3D spheroid platforms closely mimic oxygen/nutrient/pH gradients of human solid tumors, these models do not recapitulate tumor-ECM components at initial development stages.14,16,17 Such limitations are overcome in scaffold-based models comprising cancer cells laden in a ECM-mimetic hydrogel to form a 3D in vitro model that more closely resembles the in vivo set up. To date, biomaterials such as Matrigel™, collagen, gelatin and hyaluronic acid have been explored to formulate hydrogel platforms for 3D in vitro tumor modeling since they more closely recreate native ECM cues than heir scaffold-free counterparts.1,18 However, to date few fundamental aspects regarding the development of cell-laden hydrogel models for 3D in vitro disease modeling have been addressed. Particularly, the influence of cancer cells arrangement upon incorporation in scaffold-based ECM-mimetic hydrogels during their design and testing stages is a key parameter to be evaluated. At this stage, differential and random cellular arrangements in hydrogels matrix may impact cell–cell adhesion, physiology and drug resistance, therefore influencing therapeutics bioactivity screening. Addressing this parameter may provide new insights into the development of cell-rich ECM-mimetic hydrogels that better recapitulate in vitro the physiology and drug response of realistic in vivo tumors.
Herein, we fabricated osteosarcoma tumor models comprised either by cell-laden or 3D-spheroids embedded in ECM-mimetic hydrogel to evaluate the influence of cellular aggregation state on cellular metabolic activity, invasive potential and response to a standard-of-care chemotherapeutic drug. Overall, the results demonstrated that MG-63 osteosarcoma spheroids embedding in tumor-ECM mimetic scaffolds provided a better platform for the recapitulation of solid tumor invasive potential and resistance to anti-cancer therapeutics in comparison to their single cell laden hydrogels.
Results and discussion
During the design stages of 3D in vitro models the influence of differential cellular aggregation states i.e. (i) cell-laden hydrogels with dispersed cells versus (ii) preformed 3D spheroids and post-embedded in hydrogels – should therefore be further investigated. In this study, such effects were evaluated by seeding pre-established MG-63 osteosarcoma spheroids in ECM-mimetic scaffolds comprised of GelMA or Matrigel hydrogels. The interest in these two biomaterials is related to fact that GelMA and Matrigel have both shown significant promise as tumor ECM-mimetic platforms in several studies.19,20 Matrigel™ is a commercially available ECM21,22 a natural basement membrane matrix extracted from Engelbreth–Holm–Swarm (EHS) mouse sarcoma and comprises a mixture of several ECM proteins (collagen, fibronectin, laminin, filamin, actin, etc.) and growth factors (EGF, TGF-β, NGF, PDGF, IGF, FGF, etc.).23 This biologically active platform has been useful as support matrix of 3D cancer models that aim to study cancer progression, angiogenesis, metastasis process and drug-screening.24 In addition, gelatin was selected because it is a biomaterial derived from collagen type I, the major component of tumor-ECM and of the bone microenvironment. It retains natural cell-binding sequences (e.g. Arg-Gly-Asp-RGD) and protease sensitive sites for matrix metalloproteinases (MMPs), being therefore useful for modeling cell attachment, motility, survival and differentiation.25,26 Commonly, gelatin chemical versatility allows its modification (e.g. reaction with methacrylic anhydride to yield GelMA) in order to produce photocrosslinkable gelatin hydrogels which has been widely used for mimicking native ECM in in vitro disease models.27,28 According to previous reports, GelMA 10% upholds cell viability, stimulates proliferation and yields culture stable gels, showing a higher proliferation rate compared to other GelMA concentrations.29,30Therefore, three distinct osteosarcoma in vitro models comprised by (i) osteosarcoma 3D spheroids (scaffold-free control), (ii) 3D spheroids embedded in ECM-mimetic hydrogels and (iii) cancer cells laden in ECM-mimetic hydrogel were established and characterized regarding their metabolic activity, growth dynamics, invasive potential and susceptibility to lorlatinib, a potent chemotherapeutic.
ECM-biomimetic matrix formulation and characterization
To provide a better in vitro/in vivo correlation hydrogel-based disease models must provide biomimetic cues to support cell adhesion and hence recapitulate the tumor microenvironment in an in vitro setting.16 Previous studies reported that both GelMA and Matrigel hydrogels provide a suitable environment for cellular proliferation in 3D and allow nutrients diffusion through their network during culture at 37 °C, 5% CO2.31 Adding to this, hydrogels physicochemical properties play a key role in in vitro cancer cells spreading, migration, and differentiation. Rheological analysis was performed to characterize GelMA 10% and Matrigel photo- and thermal crosslinking process, respectively, as well as the resulting viscoelastic properties of these ECM-mimetic hydrogels (Fig. S1†). The obtained results indicate that GelMA 10% scaffold has the highest storage modulus of the tested hydrogel formulations. The obtained Matrigel's storage modulus is similar to that reported in the literature, with this supporting matrix behaving mostly as a soft biomaterial.32,33Having characterized the supporting matrixes for establishing the scaffold-based models we then optimized the formulation of the control scaffold-free osteosarcoma 3D spheroids by using the liquid overlay technique.
3D MG-63 spheroids assembly and characterization
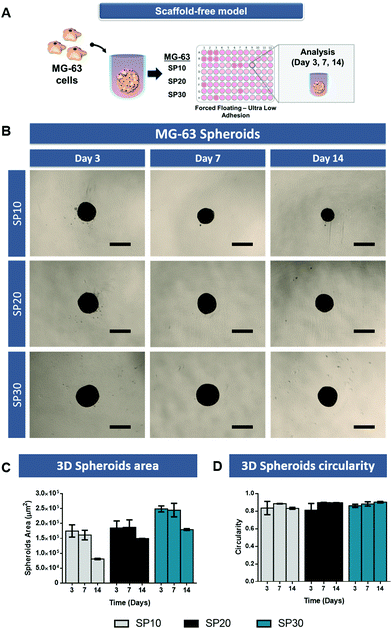
3D tumor spheroids are generally established via a streamlined cell-mediated self-assembly in vitro through different techniques.34 These models retain 3D cell–cell adhesion promoting a more physiologically accurate response to the surrounding microenvironment and prolongs cell survival.8 Moreover, 3D spheroids better recapitulate dense solid human tumors hallmarks including the establishment of nutrient, oxygen and pH gradients from the proliferative outer rim to the tumor core.Prior to the assembly of 3D osteosarcoma spheroids-laden hydrogels, a preliminary optimization of MG-63 spheroid cultures with different cell densities (e.g. 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, 20
000, 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 30
000 to 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells – SP10, SP20, SP30) was performed. Image analysis over time allowed to access 3D spheroids growth, compaction along time and circularity (Fig. 1). Such variations in cell densities were explored to establish 3D osteosarcoma models with sizes that are reported to promote the establishment of biomimetic nutrient, pH and hypoxic gradients (spheroids size >400–600 μm in diameter).17,35 Alongside, we also aimed to verify the establishment of the characteristic necrotic core and maintenance of spheroids shape uniformity over culture period, so as to assure batch-to-batch reproducibility.
000 cells – SP10, SP20, SP30) was performed. Image analysis over time allowed to access 3D spheroids growth, compaction along time and circularity (Fig. 1). Such variations in cell densities were explored to establish 3D osteosarcoma models with sizes that are reported to promote the establishment of biomimetic nutrient, pH and hypoxic gradients (spheroids size >400–600 μm in diameter).17,35 Alongside, we also aimed to verify the establishment of the characteristic necrotic core and maintenance of spheroids shape uniformity over culture period, so as to assure batch-to-batch reproducibility.
The MG-63 3D spheroids micrographs (Fig. 1B) demonstrate that over the period of 14 days, the formed spheroids had a gradual process of aggregation, becoming a more compact and circular 3D microtissue along time (Fig. 1C and D). MG-63 spheroids with cellular density of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells (SP-30) presented the largest areas, with associated diameters of approximately 470–500 μm. During the culture period all scaffold-free models maintained their morphological features.
000 cells (SP-30) presented the largest areas, with associated diameters of approximately 470–500 μm. During the culture period all scaffold-free models maintained their morphological features.
Characterization of osteosarcoma 3D spheroids
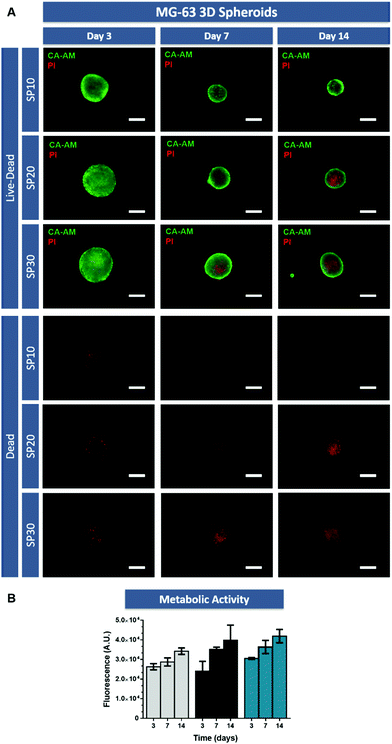
The ability of 3D tumor spheroids to recapitulate the characteristic necrotic core similar to the in vivo solid tumors was then evaluated by using Calcein-AM and PI (live/dead assay). As evidenced by fluorescence microscopy, the necrotic core formation was visible after 14 days of culture (Fig. 2A) in models with 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (SP-20) and 30
000 (SP-20) and 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells (SP-30), but not with lower cell density.
000 cells (SP-30), but not with lower cell density.
This is an important finding since the establishment of a necrotic core in the osteosarcoma microenvironment promotes tumorigenesis and chemoresistance.36 Factors associated with necrotic core formation could promote pro-tumoral interactions between mesenchymal stem cells and breast cancer cells as shown by Chaturvedi and coworkers, in which hypoxia induced interactions promoted metastasis.37 Additionally, the existence of a necrotic core where nutrient and medium penetration into the spheroid is reduced, establishes a region where therapeutic agents penetration is hindered by physical and biological barriers.38 Therefore, recapitulating these hallmarks of in vivo tumors with the established 3D in vitro tumor models is highly desirable.
Furthermore, to complement live/dead assays, cell metabolic activity was analyzed in 3D MG-63 spheroids by using the AlamarBlue® assay (Fig. 2B). These assays demonstrated that in the tested culture conditions, cellular metabolic activity increased from day 3 to day 14 in all cellular conditions.
Taking into consideration the former results regarding area, circularity, necrotic core formation and metabolic activity over time, osteosarcoma 3D spheroids formed with an initial number of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells (SP-30) were selected for subsequent studies focusing on investigating the cellular distribution influence in the design and predictive potential of scaffold-based hydrogel 3D in vitro tumor models.
000 cells (SP-30) were selected for subsequent studies focusing on investigating the cellular distribution influence in the design and predictive potential of scaffold-based hydrogel 3D in vitro tumor models.
Comparison of 3D osteosarcoma models established in ECM-mimetic hydrogels
The majority of solid 3D tumor models currently employed for cancer research and drug screening are based on scaffold-free 3D tumor spheroids or hydrogel-based models (cell-laden hydrogels). However, scaffold-free 3D tumor spheroids utterly lack pre-existent tumor-ECM components, being a rather simplistic representation of this disease. Conversely, a random dispersion of cancer cells within hydrogels matrix does not necessarily lead to the establishment of dense cellular aggregates capable of recapitulating solid tumors cellular macrostructure that generally occurs in in vivo solid tumors. These cellular arrangements present in in vivo tumors result both from the physical pressures exerted by the growing cell-mass on surrounding tissues, and from a complex set of cell–cell and cell–ECM interactions, that combined with micro-evolutive drifts culminate in cancer cells invasion and metastasis to healthy organs. Recognizing these important aspects and the influence of cellular aggregation state is key when developing new ECM-mimetic hydrogels to support tumor models establishment in vitro.To evaluate possible differences arising from differential cellular aggregation states different osteosarcoma models comprising MG-63 cell laden hydrogels and 3D spheroid-embedded hydrogels were established.
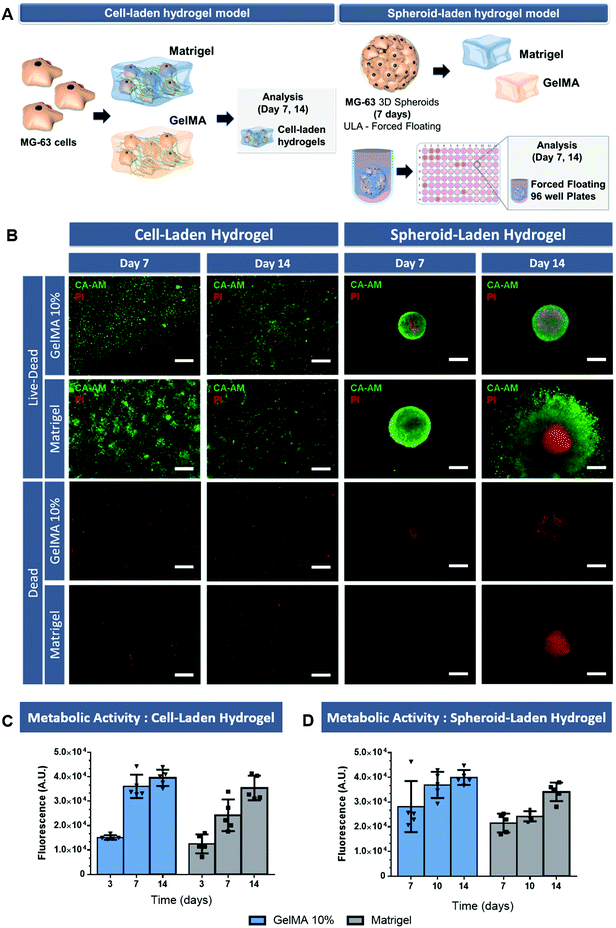
Live/dead analysis demonstrated that randomly distributed MG-63 cells remain viable up to 14 days of culture within the hydrogels (Fig. 3B, cell-laden hydrogel). In fact, the high density of viable MG-63 cells in the GelMA 10% and Matrigel supporting matrix indicate the favorable microenvironment that these ECM-biomimetic matrices provide for these cells. However, it is important to emphasize that unlike 3D spheroids, cell laden hydrogels did not allow the formation of a tumor macrostructure with a necrotic core, similar to that of in vivo solid tumors. Therefore, depriving cancer cells of priming factors that are generally present in osteosarcoma microenvironment and that are key for cell invasion and resistance to therapeutics.
The complementary AlamarBlue® assay allowed to evaluate the metabolic activity of both cell laden hydrogels (Fig. 3C). The obtained results indicate that the metabolic activity increases up to 14 days, with a slightly decreased rate at the later stage of culture probably due to increased oxygen and nutrient consumption. Interestingly, the metabolic activity was higher in the gelatin-based hydrogels compared to that obtained in Matrigel scaffold. Such findings were also reported in the literature by Jiang and co-workers which discovered that MG-63 cells activity was higher in collagen hydrogels than in Matrigel.39
Regarding 3D spheroid-embedded hydrogels these were established by using spheroids pre-formed for 7 days in ULA 96-well plates and then embedded in the tested hydrogel matrixes for additional 7 days following photo- or thermal-based crosslinking.
The live/dead analysis of these platforms revealed the existence of 3D spheroids with an outer rim of viable cells combined with a necrotic core comprised of non-viable cells, after 7 days of culture within both ECM-mimetic hydrogels (Fig. 3B, spheroid-embedded hydrogel models). In addition, the metabolic activity of spheroids embedded within both hydrogels at the same timepoints was evaluated (Fig. 3D). The metabolic activity increased up to 14 days and was higher in gelatin-based models when compared to that obtained in Matrigel scaffolds. These findings that indicate the tumor-supporting role of these matrixes, but also the feasibility of using photo- or thermally-induced crosslinking methodologies to establish such models.
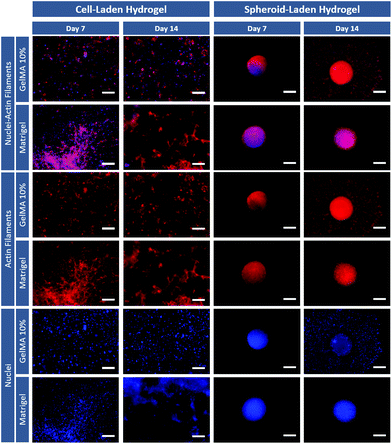
Immunofluorescent staining of F-actin was then performed at different time points to evaluate cell spreading and cytoskeletal organization of the different osteosarcoma models (Fig. 4). Fluorescence microscopy images show that individual cancer cells laden in both GelMA and Matrigel hydrogels, present significant spreading at 7 and 14 days. Also, it is important to emphasize that very few 3D cellular agglomerates are observed and that cells exhibit a more layered type structure than that of a 3D mass. Modeling the formation of compact cancer cells aggregates is key when bioengineering in vitro tumor models of solid tumors as previously mentioned.
Interestingly, the 3D spheroids embedded in hydrogels maintained their compact morphology 1 day after removal from the ULA plate and embedding in the hydrogel matrix (day 7–3D spheroids). The models demonstrate a compact cytoskeleton with well-established cell–cell interactions even after incorporation into the hydrogel matrix. Also, at day 14 (which correspond to day 7 after embedding) cancers cell from the 3D spheroids clearly invaded the surrounding Matrigel and GelMA matrix, recapitulating a key characteristic of late-stage tumors. These differential results clearly indicate the influence of cancer cells arrangement within hydrogels and emphasizes the importance of not only exploring cell-laden hydrogels, but also of adding 3D spheroid-hydrogel models during the design stages of ECM-mimetic scaffolds for in vitro disease modeling. Aiming to further evaluate the influence of hydrogels physicochemical properties on embedded 3D spheroids invasive potential and their ability to model tumor metastasis, invasion assays were performed.
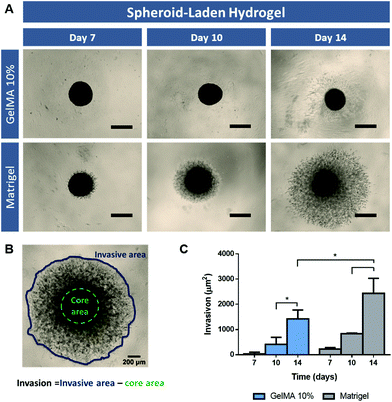
3D spheroid-embedded hydrogel models invasive potential
Tumor metastasis is a highly complex process which represents the main cause of cancer-associated mortality. Despite some advances, osteosarcoma still presents poor prognosis when metastasis naturally occur, commonly, to organs such as the lung and other bones. The invasion of surrounding tissues and ultimately the migration of tumor cells into secondary metastatic sites is directly correlated with alterations in tumor ECM composition and organization, which allow cancer cells migration away from the primary tumor.40The influence of the supporting matrix (i.e. GelMA 10% or Matrigel) in the invasion capacity of embedded 3D spheroids was evaluated to address the ability of these models to model the natural metastasis process occurring in vivo. Similar to the previous assays, the pre-formed 3D spheroids were transferred to gelatin and Matrigel scaffolds on day 6 of culture, and their invasion capacity in the surrounding matrix was evaluated on days 7, 10 and 14 after incorporation in hydrogels (Spheroid-laden hydrogel, Fig. 5A). The results demonstrate a clear cell migration from 3D spheroids in Matrigel on the first day after embedding, whereas this only occurred after day 3 for GelMA 10% scaffolds (Fig. 5C). Since Matrigel presents a softer matrix than GelMA 10% hydrogel, the obtained results are in accordance with the study of Lam and co-workers which report that increased stiffness of collagen gels significantly impeded the invasion of cancer cells from 3D spheroids.41 Most importantly, these findings indicate that cell-laden hydrogel models may present limitations when evaluating anti-metastatic compounds bioactivity in the context of in vitro disease models development.
Influence of the cellular aggregation state on drug response
Having confirmed fundamental phenotypic and bioactivity differences of cell-laden and 3D spheroid-hydrogel models the effect of their differential cellular aggregation states regarding the ultimate response to anti-cancer therapeutics was investigated. For this purpose, we screened lorlatinib, a potent third-generation anaplastic lymphoma kinase (ALK) and ROS1 tyrosine kinase inhibitor. This compound was selected since several ALK domains have been reported to be mutated in pediatric tumors including osteosarcoma.42,43 Moreover, lorlatinib has shown superior progression-free survival in phase 3 clinical trials for ALK responsive tumors in comparison to the widely used crizotinib (ClinicalTrials.gov Identifier: NCT03052608). This compound was administered to in vitro models exhibiting differential cellular aggregation states as follows.The first model – 3D scaffold-free spheroids – closely mimics specific features of the in vivo tumor, namely the 3D cell–cell agglomeration and necrotic core but fails to recapitulate pre-existing ECM cues. The second model – cell-laden hydrogels – recapitulates the 3D tumor microenvironment by including a 3D biomimetic matrix comprised of either GelMA 10% or Matrigel. It is important to emphasize that embedding dispersed cells in a hydrogel matrix fails to recapitulate the compact and cell-rich macrostructure that most human solid tumors exhibit in their native microenvironment. The third model – comprises an ECM-mimetic matrix (i.e. GelMA 10% or Matrigel hydrogels) where an osteosarcoma 3D spheroid is embedded. This platform allows the reproduction of cell–cell interactions existing in solid tumors and the characteristic necrotic core, while also incorporating the supporting ECM, therefore providing the key architectural and microenvironmental cues found in vivo.
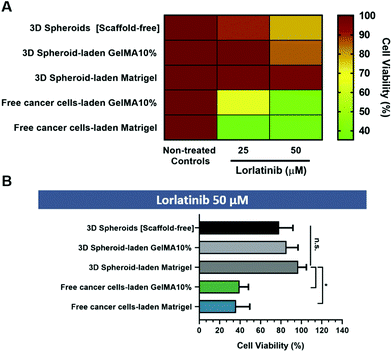
The abovementioned 3D in vitro osteosarcoma models were incubated with different lorlatinib concentrations (25 μM and 50 μM) and cell viability was accessed after 24 h by using a luminescence-based assay optimized for 3D cell cultures (CellTiter-Glo®) (Fig. 6A and B).
The obtained results indicate that randomly dispersed cells in GelMA 10% and Matrigel hydrogels (cell-laden hydrogel models) exhibited the highest sensitivity to lorlatinib in comparison to scaffold-free and scaffold-based 3D spheroid models. This finding may be correlated with the absence of a dense cellular agglomerate which is known to play an important role in drug resistance due to hindered drugs diffusion across microtumors volume. Such results may also be related with the fact that drugs diffusion does not occur evenly neither on dense 3D spheroids nor on the entire hydrogel matrix, thus leading to the establishment of concentration gradients. The physical barrier imposed by the hydrogel matrix hinders drugs availability in deeper regions, similarly to what occurs in in vivo solid tumors due to their ECM. The existence of such barrier is increasingly recognized as one of the major hallmarks that can be targeted to improve chemotherapeutics bioavailability in deeper regions of the tumor. Recent studies have therefore focused on modulating ECM density either by reducing type I collagen synthesis via Halofunginone drug administration, or by targeting lysyl oxidase activity and consequently collagen ECM-component crosslinking.44
On the other hand, although the drug cytotoxicity assay (Fig. 6) does not show significant differences between scaffold-free 3D spheroids and hydrogel embedded 3D spheroids, the later provides an ECM-mimetic scaffold that allows to better recapitulate the native tumor microenvironment providing both biophysical cues to 3D spheroids and recapitulating the diffusional barrier provided by native in vivo ECM. Importantly, 3D spheroid embedded hydrogel models provide a unique platform to evaluate cancer cells fundamental invasion mechanisms and to screen anti-metastatic drugs as well as therapeutics specifically targeting cancer cells–ECM interactions in vitro, constituting a more realistic model in comparison to scaffold-free 3D spheroids.
Overall these findings emphasize the necessity to establish both cell-laden hydrogels and 3D spheroid-embedded hydrogel platforms during the development of new biomaterials designed to mimic native tumor-ECM. Designing different 3D hydrogel-based cancer models is important to reflect the distinct stages of cancer progression. Therefore, cell-laden models better recapitulate early stage disease and 3D spheroids microtumors mimic later stages that include the acquisition of solid macrostructures and invasive phenotypes which contributes to the disease progression.
Experimental
Gelatin methacrylate synthesis and characterization
Gelatin methacrylate (GelMA) was synthesized and characterized as previously described, with minor modifications.45 In brief, gelatin was dissolved in PBS (pH 7.4), at 50 °C overnight, to yield a 10% (w/v) working solution. Thereafter, 0.6 g of methacrylic anhydride/gram of gelatin was dropwise added under mild magnetic stirring. The methacrylation reaction was then allowed to proceed for 5 h, at RT, under mild magnetic stirring. After this period the mixture was centrifuged (3 min, 3500g, RT) to allow for phase separation between gelatin solution and the denser methacrylic anhydride pellet. The remaining methacrylated gelatin (GelMA) was then diluted and dialyzed (MWCO 6–8 kDa), at 50 °C, by using double distilled deionized and filtered water (ddH2O, 17.9 mΩ) as the dialysant. The purified GelMA was then recovered by freeze drying (−86 °C, Telstar LyoQuest) for 6 days, in the dark. The determined gelatin degree of methacrylation was ∼73.2 ± 0.36%, n = 3, as determined by the fluoraldehyde assay.2D in vitro cell culture
The MG-63 human osteosarcoma cell line (ATCC® CRL-1427™) was cultured in vitro in tissue culture treated polystyrene 2D substrates by using α-Minimum Essential Medium (α-MEM), supplemented with 10% (v/v) Fetal Bovine Serum (FBS), 1% (v/v) ATB, as recommended by the provider. Cells were maintained in culture under a temperature-controlled environment (37 °C) and an atmosphere containing 5% CO2, as recommended by the manufacturer. MG-63 cells were expanded until reaching 70–80% confluency and routinely passed by using TrypLE Express detachment reagent.Generation of osteosarcoma 3D spheroids by liquid overlay technique
Monocultured 3D tumor spheroids comprised by MG-63 cells were fabricated by using the liquid-overlay technique where cells self-aggregated into spheroidal structures during culture in ULA 96-well plates. To achieve the ideal cellular density to produce dense 3D in vitro models that would better recapitulate osteosarcoma tumor microenvironment and necrotic core formation, MG-63 cell suspensions comprised by different cellular densities (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000; 20
000; 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 30
000 and 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells – SP10; SP20; SP30, respectively) were established. During 3D spheroids in vitro maturation, the culture medium was completely removed and replaced with fresh medium every 3 days. Cellular aggregation, size and 3D microtissues morphological features was monitored along time by using an inverted optical contrast microscope equiped with a 3MPix color camera and a Plan-Achromat 4×/0.10 objective (Zeiss Primovert, Carl Zeiss Microscopy GmbH, Germany). Optical contrast images were acquired at specific timepoints during MG-63 3D spheroids culture (3, 7 and 14 days).
000 cells – SP10; SP20; SP30, respectively) were established. During 3D spheroids in vitro maturation, the culture medium was completely removed and replaced with fresh medium every 3 days. Cellular aggregation, size and 3D microtissues morphological features was monitored along time by using an inverted optical contrast microscope equiped with a 3MPix color camera and a Plan-Achromat 4×/0.10 objective (Zeiss Primovert, Carl Zeiss Microscopy GmbH, Germany). Optical contrast images were acquired at specific timepoints during MG-63 3D spheroids culture (3, 7 and 14 days).
Cell-laden hydrogel models fabrication
For establishing cell laden hydrogel models MG-63 osteosarcoma cells were initially mixed with GelMA (10 mg mL−1, at 37 °C) containing Irgacure 2959 (0.5% w/v) and injected into flat-bottom μ-Slide Angiogenesis multi-well plates. GelMA-cancer cells mixture was then exposed to collimated U.V. light for 60 s (OmniCure S2000, 2.23 W cm−2). Following UV crosslinking, the resulting MG-63 3D hydrogels were cultured in complete culture medium at 37 °C in an atmosphere with 5% CO2. For Matrigel models, MG-63 were initially detached, centrifuged and resuspended in cell culture medium supplemented with 10% FBS. Afterwards, cells were bioencapsulated in ECM-mimetic matrix pre-cooled at 4 °C and injected into μ-Slide Angiogenesis multi-well plates, at RT. Incubation of cells-Matrigel mixtures at 37 °C promoted basal membrane extract crosslinking and the establishment of osteosarcoma MG-63 cell laden hydrogels.Hydrogel-3D spheroid models fabrication
For generating hydrogel-spheroid models, MG-63 3D spheroids were initially pre-matured for 7 days in scaffold-free ULA plates and then bioencapsulated in GelMA (10 mg mL−1, at 37 °C) or Matrigel (4 °C) ECM-mimetic platforms. For this purpose, before embedding, 3D tumor spheroids were gently washed in dPBS, followed by the addition of 20 μL GelMA or Matrigel hydrogel precursors. Upon UV irradiation (60 s, GelMA) or incubation at physiological temperature (Matrigel) the precursors were crosslinked. This approach allowed to maintain cellular aggregation and sphericity, while also enabled to embed a single 3D spheroid in a hydrogel matrix formed in situ in each ULA plate well. The 3D MG-63 spheroids loaded in the different hydrogels were cultured in complete culture medium, at 37 °C, 5% CO2, for up to 14 days. The cellular viability, cellular morphology, metabolic activity and cell invasion were then analyzed at various timepoints (day 1, 3 and 7, after spheroids embedding in hydrogels, in total corresponding to day 7, 10 and 14 of MG-63 cells 3D culture).Cell viability analysis
The cell viability of MG-63 cell laden hydrogels and 3D spheroid-hydrogel hybrids was accessed by using the AlamarBlue® Cell Viability assay. In brief, MG-63 spheroids and hydrogel-3D spheroid models were incubated with cell culture medium containing 10% (v/v) AlamarBlue® reagent, at specific timepoints according to the manufacturer's instructions. After incubation, the fluorescence of the resorufin product was recorded in a Synergy HTX multi-modal microplate reader (λex: 540 nm, λem: 600 nm), by using a black-clear bottom 96-well plate (Corning, NY, US). Both assays were performed in accordance with the manufacturer's instructions. To further access viability, cell laden hydrogels and 3D spheroid-embedded hydrogels were incubated with Calcein AM and PI (Live/Dead assay), for imaging live and dead cells, respectively. Briefly, samples were labelled with Calcein-AM (5 μg mL−1, in dPBS) and Propidium Iodide (PI) (5 μg mL−1 in dPBS) for 1 h, at 37 °C. Following the incubation period, the samples were washed three times with dPBS. Bioimaging of Live/Dead labelled 3D spheroids, cell laden hydrogels and hydrogel-3D spheroid models was performed in a Zeiss Imager M2 upright widefield fluorescence microscope equiped with a Plan-Apochromat 10×/0.45 objective. All fluorescence images were acquired and post-processed in Zeiss Zen Software SP 2.1 (Carl Zeiss, Germany).Cytoskeletal proteins Labelling
The cellular distribution and cytoskeletal spatial arrangement in different osteosarcoma in vitro models were evaluated via F-actin/nuclei staining. Briefly, MG-63 cell laden GelMA and Matrigel hydrogels were fixed with formaldehyde 4% (v/v) solution, at different time points. After washing three times with dPBS and cells membrane was permeabilized with 0.5% Triton X-100 then, cells were stained with Flash Phalloidin™ Red 594 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 (v/v), in dPBS) in a humidified atmosphere, for 24 h, at RT. After extensive washing with dPBS, samples were stained with DAPI (4′,6-diamidino-2-phenylindole, dihydrochloride) (1
40 (v/v), in dPBS) in a humidified atmosphere, for 24 h, at RT. After extensive washing with dPBS, samples were stained with DAPI (4′,6-diamidino-2-phenylindole, dihydrochloride) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250 (v/v)) and incubated for 30 min, at RT. MG-63 spheroids embedded into GelMA and Matrigel 3D matrix were stained for F-actin/nuclei using the above described procedure. Widefield fluorescence bioimaging was then performed as described above.
250 (v/v)) and incubated for 30 min, at RT. MG-63 spheroids embedded into GelMA and Matrigel 3D matrix were stained for F-actin/nuclei using the above described procedure. Widefield fluorescence bioimaging was then performed as described above.
3D tumor spheroids invasion analysis
Cellular protrusions from GelMA and Matrigel hydrogel-3D spheroid platforms was evaluated along time to determine the invasive/metastatic potential of these in vitro osteosarcoma models. The cellular movement and invasion were evaluated along time by using an inverted optical contrast microscope (Primovert, Carl Zeiss, Germany) equiped with a 3MPix color camera and a 4×/0.5 objective. Data from at least three spheroids was obtained at 7, 10 and 14 days after embedding in the ECM-mimetic hydrogel matrix. Image analysis and invasion quantification was performed in Image J software (ImageJ, NIH, USA) by using image segmentation and area analysis algorithms. Invasion quantification was determined for cells protruding from each spheroid and was defined as the difference between the invasive area and core area.46Chemotherapeutic drug performance screening
The developed 3D osteosarcoma in vitro models were then used as testing platforms for the evaluation of lorlatinib anti-cancer activity (Lorviqua®, Pfizer). On 14 day of culture 3D in vitro models were incubated with the drug at different concentrations (25 μM and 50 μM), for 24 h. Cell viability was assessed by quantifying cellular ATP through a luminescence-based assay specific for 3D cell culture (3D Cell Titer Glo Luminescent viability assay, Promega, Madison, WI, USA). Luminescence analysis was performed according to the manufacturer's instructions. After lorlatinib incubation, the medium was removed, and 3D tumor models were incubated with a mixture of α-MEM complete medium and CellTiter-Glo® reagent (ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The mixture was homogenized for 5 min and incubated for 25 min, in the dark, at RT. Luminescence was measured on flat bottom 96-well white plates by using a Synergy HTX microplate reader. Acquisition settings were according to the manufacturer's instructions.
1). The mixture was homogenized for 5 min and incubated for 25 min, in the dark, at RT. Luminescence was measured on flat bottom 96-well white plates by using a Synergy HTX microplate reader. Acquisition settings were according to the manufacturer's instructions.
Statistical analysis
Statistical analysis was performed in GraphPad Prism 8 Software (Prism 8™, trial version). One-way analysis of variance (One-ANOVA) with Holm–Sidak post-hoc test was generally used for data analysis. A value of p < 0.05 was considered to be statistically significant.Conclusions
In this study the effect of osteosarcoma cells agglomeration state in in vitro disease models’ ability to recapitulate key hallmarks of in vivo tumors was evaluated. Three distinct platforms – cell-laden hydrogels, 3D spheroid-embedded hydrogels and scaffold-free 3D spheroids – were established by using different methodologies including the liquid overlay technique and photo/thermally crosslinked hydrogel platforms. The morphological and phenotypical features, as well as drug response were evaluated in the fabricated models.Our findings indicate that 3D spheroids embedding in ECM-mimetic hydrogels provided a better platform for recapitulating the invasive potential and resistance to therapeutics in comparison to their single cell laden hydrogels. More importantly, these findings emphasize the importance of evaluating multiple aggregation states during the development of tumor mimetic models to leverage their full predictive potential for drug development, screening and also for streamlining fundamental biology studies. We envision that 3D tumor modelling in hydrogel platforms will evolve to consider cellular agglomeration states and that the field will adapt cell-laden and 3D spheroids-embedded models in vitro screening platforms according to the tumor stage (i.e. early or late stage) that is envisioned to be modelled according. This is particularly relevant considering that such platforms can be in the foreseeable future be explored for generating patient-personalized microtumors that recapitulate the disease state of specific individuals.
Conflicts of interest
The authors declare that there are no conflicts of interest.Acknowledgements
The authors would like to acknowledge the support of the European Research Council grant agreement ERC-2014-ADG-669858 for project “ATLAS” and for the proof-of-concept project Microbone (PoC-2017-7897602017). This work was also developed within the scope of the project CICECO-Aveiro Institute of Materials, UIDB/50011/2020 & UIDP/50011/2020, financed by national funds through the FCT/MEC and when appropriate co-financed by FEDER under the PT2020 Partnership Agreement. Vítor Gaspar acknowledges funding in the form of a Junior Researcher Contract under the scope of the project PANGEIA (PTDC/BTM-SAL/30503/2017), supported by the Programa Operacional Competitividade.References
- P. Saglam-metiner, S. Gulce-iz and C. Biray-avci, Gene, 2018, 686, 203–212 CrossRef PubMed
.
- S. Breslin and L. O'Driscoll, Drug Discovery Today, 2013, 18, 240–249 CrossRef CAS PubMed
.
- A. Nyga, U. Cheema and M. Loizidou, J. Cell Commun. Signal., 2011, 5, 239–248 CrossRef PubMed
.
- F. Pampaloni, E. G. Reynaud and E. H. K. Stelzer, Nat. Rev. Mol. Cell Biol., 2007, 8, 839–845 CrossRef CAS PubMed
.
- K. Stock, M. F. Estrada, S. Vidic, K. Gjerde, A. Rudisch, V. E. Santo, M. Barbier, S. Blom, S. C. Arundkar, I. Selvam, A. Osswald, Y. Stein, S. Gruenewald, C. Brito, W. Van Weerden, V. Rotter, E. Boghaert, M. Oren, W. Sommergruber, Y. Chong, R. De Hoogt and R. Graeser, Sci. Rep., 2016, 6, 1–16 CrossRef PubMed
.
- N. Charbe, P. A. McCarron and M. M. Tambuwala, World J. Clin. Oncol., 2017, 8, 21–36 CrossRef PubMed
.
- K. A. Fitzgerald, M. Malhotra, C. M. Curtin, J. O. Brien and C. M. O. Driscoll, J. Controlled Release, 2015, 215, 39–54 CrossRef CAS PubMed
.
- P. Mcgonigle and B. Ruggeri, Biochem. Pharmacol., 2013, 87, 162–171 CrossRef PubMed
.
- I. W. Y. Mak, N. Evaniew and M. Ghert, Am. J. Transl. Res., 2014, 6, 114–118 Search PubMed
.
- Y. Fang and R. M. Eglen, SLAS Discovery, 2017, 22, 456–472 CAS
.
- L. P. Ferreira, V. M. Gaspar and J. F. Mano, Acta Biomater., 2018, 75, 11–34 CrossRef CAS PubMed
.
- L. P. Ferreira, V. M. Gaspar and J. F. Mano, Biomaterials, 2018, 185, 155–173 CrossRef CAS PubMed
.
- A. S. Nunes, I. J. Correia and E. C. Costa, Biotechnol. Bioeng., 2019, 116, 206–226 CrossRef CAS PubMed
.
- A. I. Neto, C. R. Correia, M. B. Oliveira, M. I. Rial-Hermida, C. Alvarez-Lorenzo, R. L. Reis and J. F. Mano, Biomater. Sci., 2015, 3, 581–585 RSC
.
- B. K. Jong, R. Stein and M. J. O'Hare, Breast Cancer Res. Treat., 2004, 85, 281–291 CrossRef PubMed
.
- P. Kaphle, Y. Li and L. Yao, J. Cell. Physiol., 2018, 234, 3948–3960 CrossRef PubMed
.
- S. Nath and G. R. Devi, Pharmacol. Ther., 2016, 163, 94–108 CrossRef CAS PubMed
.
- J. A. Belgodere, C. T. King, J. B. Bursavich, M. E. Burow, E. C. Martin and J. P. Jung, Front. Bioeng. Biotechnol., 2018, 6, 1–27 CrossRef PubMed
.
- H. Y. Liu, M. Korc and C. C. Lin, Biomaterials, 2018, 160, 24–36 CrossRef CAS PubMed
.
- S. Pradhan, A. M. Smith, C. J. Garson, I. Hassani, W. J. Seeto, K. Pant, R. D. Arnold, B. Prabhakarpandian and E. A. Lipke, Sci. Rep., 2018, 8, 1–15 CrossRef CAS PubMed
.
- M. Zanoni, S. Pignatta, C. Arienti, M. Bonafè, M. Zanoni, S. Pignatta, C. Arienti, M. Bonafè and A. Tesei, Expert Opin. Drug Discovery, 2019, 14, 289–301 CrossRef CAS PubMed
.
- G. Benton, I. Arnaoutova, J. George, H. K. Kleinman and J. Koblinski, Adv. Drug Delivery Rev., 2014, 79–80, 3–18 CrossRef CAS PubMed
.
- C. S. Hughes, L. M. Postovit and G. A. Lajoie, Proteomics, 2010, 10, 1886–1890 CrossRef CAS PubMed
.
- F. Monteiro, C. A. Cust and F. Mano, Adv. Ther., 2019, 2, 1–18 Search PubMed
.
- K. S. Kopanska, Y. Alcheikh, R. Staneva and D. Vignjevic, PLoS, 2016, 11, 1–23 Search PubMed
.
- T. Jiang, J. G. Munguia-lopez, S. Flores-torres, J. Grant, A. De Leon-rodriguez and J. M. Kinsella, Sci. Rep., 2017, 7, 1–9 CrossRef PubMed
.
- A. R. Donaldson, C. E. Tanase, D. Awuah, P. Vasanthi Bathrinarayanan, L. Hall, M. Nikkhah, A. Khademhosseini, F. Rose, C. Alexander and A. M. Ghaemmaghami, Front. Bioeng. Biotechnol., 2018, 6, 1–11 CrossRef PubMed
.
- D. Loessner, C. Meinert, E. Kaemmerer, L. C. Martine, K. Yue, P. A. Levett, T. J. Klein, F. P. W. Melchels, A. Khademhosseini and D. W. Hutmacher, Nat. Protoc., 2016, 11, 727–746 CrossRef CAS PubMed
.
- A. D. Arya, P. M. Hallur, A. G. Karkisaval, A. Gudipati, S. Rajendiran, V. Dhavale, B. Ramachandran, A. Jayaprakash, N. Gundiah and A. Chaubey, ACS Appl. Mater. Interfaces, 2016, 8, 22005–22017 CrossRef CAS PubMed
.
- J. W. Nichol, S. Koshy, H. Bae, C. M. Hwang and A. Khademhosseini, Biomaterials, 2011, 31, 5536–5544 CrossRef PubMed
.
- A. De Luca, L. Raimondi, F. Salamanna, V. Carina, V. Costa, D. Bellavia, R. Alessandro, M. Fini and G. Giavaresi, J. Exp. Clin. Cancer Res., 2018, 37, 2–16 CrossRef PubMed
.
- M. H. Zaman, L. M. Trapani, A. L. Sieminski, D. Mackellar, H. Gong, R. D. Kamm, A. Wells, D. A. Lauffenburger and P. Matsudaira, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 10889–10894 CrossRef CAS PubMed
.
- X. Hu, L. Ma, C. Wang and C. Gao, Macromol. Biosci., 2009, 9, 1194–1201 CrossRef CAS PubMed
.
- J. A. Hickman, R. Graeser, R. de Hoogt, S. Vidic, C. Brito, M. Gutekunst, H. van der Kuip and Imi Predect consortium, Biotechnol. J., 2014, 9, 1115–1128 CrossRef CAS PubMed
.
- L. A. Kunz-schughart, M. Kreutz and R. Knuechel, Int. J. Exp. Pathol., 1998, 79, 1–23 CrossRef CAS PubMed
.
-
Y. Zhang, Q. Mai, X. Chunyuan, X. Zhang and Y. Zhang, in Osteosarcoma - Biology, Behavior and Mechanisms, 2017, pp. 73–102 Search PubMed
.
- P. Chaturvedi, D. M. Gilkes, C. Chak, L. Wong, W. Luo, H. Zhang, H. Wei, N. Takano, L. Schito, A. Levchenko and G. L. Semenza, J. Clin. Invest., 2013, 123, 189–205 CrossRef CAS PubMed
.
- W. Asghar, R. El Assal, H. Shafiee, S. Pitteri, R. Paulmurugan and U. Demirci, Biochem.
Pharmacol., 2015, 18, 539–553 CAS
.
- T. Jiang, G. Xu, X. Chen, X. Huang, J. Zhao and L. Zheng, Adv. Healthcare Mater., 2019, 8, 1–11 Search PubMed
.
- C. Liu, D. Lewin Mejia, B. Chiang, K. E. Luker and G. D. Luker, Acta Biomater., 2018, 75, 213–225 CrossRef CAS PubMed
.
- I. Lam, H. K. Wong, S. Nai, C. K. Chua, N. S. Tan and L. P. Tan, Mol. Pharm., 2016, 11, 2016–2021 CrossRef PubMed
.
- J. Takita, Cancer Sci., 2017, 108, 1913–1920 CrossRef CAS PubMed
.
- X. Chen, A. Pappo and M. A. Dyer, Oncogene, 2015, 34, 5207–5215 CrossRef CAS PubMed
.
- K. C. Valkenburg, A. E. De Groot and K. J. Pienta, Nat. Rev. Clin. Oncol., 2018, 15, 366–381 CrossRef PubMed
.
- J. Antunes, V. M. Gaspar, L. Ferreira, M. Monteiro, R. Henrique, C. Jerónimo and J. F. Mano, Acta Biomater., 2019, 94, 392–409 CrossRef CAS PubMed
.
- E. B. Berens, J. M. Holy, A. T. Riegel and A. Wellstein, J. Visualized Exp., 2015, 105, 1–6 Search PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9bm02075f |
| This journal is © The Royal Society of Chemistry 2020 |