Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Regulation of Arabidopsis gene expression by low fluence rate UV-B independently of UVR8 and stress signaling†
Andrew
O'Hara‡
 ab,
Lauren R.
Headland§‡
ab,
Lauren R.
Headland§‡
 b,
L. Aranzazú
Díaz-Ramos
b,
L. Aranzazú
Díaz-Ramos
 b,
Luis O.
Morales
b,
Luis O.
Morales
 a,
Åke
Strid
a,
Åke
Strid
 a and
Gareth I.
Jenkins
a and
Gareth I.
Jenkins
 *b
*b
aÖrebro Life Science Center, School of Science and Technology, Örebro University, SE-70182 Örebro, Sweden
bInstitute of Molecular, Cell and Systems Biology, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow G12 8QQ, UK. E-mail: Gareth.Jenkins@Glasgow.ac.uk
First published on 20th June 2019
Abstract
UV-B exposure of plants regulates expression of numerous genes concerned with various responses. Sudden exposure of non-acclimated plants to high fluence rate, short wavelength UV-B induces expression via stress-related signaling pathways that are not specific to the UV-B stimulus, whereas low fluence rates of UV-B can regulate expression via the UV-B photoreceptor UV RESISTANCE LOCUS 8 (UVR8). However, there is little information about whether non-stressful, low fluence rate UV-B treatments can activate gene expression independently of UVR8. Here, transcriptomic analysis of wild-type and uvr8 mutant Arabidopsis exposed to low fluence rate UV-B showed that numerous genes were regulated independently of UVR8. Moreover, nearly all of these genes were distinct to those induced by stress treatments. A small number of genes were expressed at all UV-B fluence rates employed and may be concerned with activation of eustress responses that facilitate acclimation to changing conditions. Expression of the gene encoding the transcription factor ARABIDOPSIS NAC DOMAIN CONTAINING PROTEIN 13 (ANAC13) was studied to characterise a low fluence rate, UVR8-independent response. ANAC13 is induced by as little as 0.1 μmol m−2 s−1 UV-B and its regulation is independent of components of the canonical UVR8 signaling pathway COP1 and HY5/HYH. Furthermore, UV-B induced expression of ANAC13 is independent of the photoreceptors CRY1, CRY2, PHOT1 and PHOT2 and phytochromes A, B, D and E. ANAC13 expression is induced over a range of UV-B wavelengths at low doses, with maximum response at 310 nm. This study provides a basis for further investigation of UVR8 and stress independent, low fluence rate UV-B signaling pathway(s).
Introduction
Ultraviolet-B radiation (UV-B; 280–315 nm) makes up less than 0.5% of the total solar spectrum but can have a major impact on all organisms. Plants, in particular, are dependent on sunlight as a source of energy and have evolved elaborate mechanisms to utilize solar energy in a positive manner and overcome the potentially negative effects of its most energetic wavelengths. Perception of specific wavelengths of light is carried out by photoreceptors, and the first UV-B photoreceptor identified in plants was the UV RESISTANCE LOCUS 8 (UVR8) protein.1–3 Since its discovery, a growing body of research has revealed UVR8's function, and its relationship with other abiotic and biotic pathways.4–10Additionally, progress has been made in determining the crystal structure and mechanistic action of UVR8 in UV-B perception, including its intrinsic tryptophan-based chromophore.2,3,11–13 In essence, UVR8 senses UV-B photons via specific tryptophan residues within its structure, and this brings about monomerization of the homodimer. UVR8 monomers bind to CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1), ultimately triggering a network of transcriptional responses.7,10 The E3 ubiquitin ligase component COP1 is a master regulator of numerous proteins involved in photomorphogenesis and acts as a negative regulator in darkness. However, COP1 has a positive role in the UV-B signaling pathway and is required for expression of UVR8-regulated genes.14–17 ELONGATED HYPOCOTYL5 (HY5) and HY5 HOMOLOG (HYH) are both transcription factors that act downstream of UVR8 and COP1 to regulate expression of many UVR8 target genes.15,18 In addition, UVR8 has recently been found to interact directly with specific transcription factors to mediate responses to UV-B.19,20
Overall, the UVR8 pathway gives the plant protection against potentially harmful UV-B wavelengths and initiates other processes, including morphogenic and physiological responses, entrainment of the circadian clock and protection against specific pathogens.5,9,10,15,18,21–23 Nevertheless, it has become apparent that other pathways mediate responses to UV-B that are independent of UVR8. Several studies, mainly involving microarrays, have shown that numerous genes are regulated by UV-B exposure, in some cases through the activation of stress pathways by relatively short wavelengths and high fluence rates of UV-B.14,18,24–27 However, UVR8-independent UV-B pathways remain poorly characterised, and the possible existence of additional UV-B photoreceptor(s) cannot be excluded.28
The aim of this study was, firstly, to determine whether low, photomorphogenic exposures to UV-B initiate gene expression responses independently of UVR8 and to identify genes regulated by low fluence UV-B via UVR8-independent pathway(s), using transcriptomic analysis in Arabidopsis thaliana. Secondly, the UVR8-independent pathway regulating expression of a gene selected from the transcriptomic data was characterised. This gene encodes NAC DOMAIN CONTAINING PROTEIN 13 (ANAC13), a putative transcription factor.
Materials and methods
Plant material and light treatments
Seeds of wild-type Arabidopsis thaliana ecotypes Landsberg erecta (Ler), Columbia (Col-0), Wassilewskija (Ws) and the mutants uvr8-1,4uvr8-6,15cry1cry2uvr8-2,29phyAphyBphyDphyE,30cry1cry2,31phot1-5phot2-1,32hy5-ks50hyh33 and cop1-434 were sown on compost, stratified at 4 °C for 48 h, and then grown in continuous white light of 20 μmol m−2 s−1 (warm white fluorescent tubes L36W/30, Osram) at 20 °C for 21 days. Plants were exposed to UV-B using either a broadband UVB-313 fluorescent tube (Q-Panel Co., USA) covered by cellulose acetate film (FLM400110/2925, West Design Products) or a narrowband tube (Philips TL20 W/01RS, λmax 312 nm) at the fluence rates indicated in the figure legends. Control plants were kept in 20 μmol m−2 s−1 white light. The spectra of the UV-B sources, measured with a Macam Photometrics spectroradiometer model SR9910 are shown in Fig. S1.† Assays of expression at different UV wavelengths were undertaken using a pulsed Opolette 355 + UV II tuneable laser (Opotek Inc., USA) with a half bandwidth of 0.4 nm as described in ref. 35. Following UV exposure with the tuneable laser, plant material was left in darkness for 1 hour to allow transcripts to accumulate and then harvested and snap frozen in liquid nitrogen. All the data presented were obtained from 3 independent experiments with different sets of plants.Transcript measurements
RNA extraction was performed using the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer's instructions. cDNA synthesis was performed as described in ref. 18. Quantitative PCR was performed using the MX4000 Stratagene real-time PCR system and a Brilliant III SYBR Green qPCR kit (Stratagene) following the manufacturer's instructions. A master mix was prepared of 1× SYBR Green Master Mix (Stratagene), 0.2 M of each primer, and appropriate volumes of cDNA and DEPC treated water. The PCR conditions were as follows: 3 min at 95 °C, 40 cycles of 10 s at 95 °C, 20 s at 60 °C, followed by a 60 to 95 °C dissociation protocol. Stratagene MX software was used to automatically calculate the cycle threshold (Ct) value for each reaction. Each reaction was performed in duplicate in three independent experiments. As a control for variation in RNA quantification, reverse transcription efficiency, and template preparation, the expression of HY5, CHS and ANAC13 (At1g32870) transcripts was normalized to the mean of either 18S rRNA or ACTIN2 (as indicated in the figure legends). The relative levels of transcripts were calculated following the ΔΔCt method. The primers used for HY5 were: 5′-CTGAAGAGGTTGTTGAGGAAC-3′ and 5′-AGCATCTGGTT CTCGTTCTGAAGA-3′ (or 5′-GGCTGAAGAGGTTGTTGAGG-3′ and 5′-CAGCATTAGAACCACCACCA-3′ for the data in Fig. 4); for ANAC13: 5′-AAGAAAGATCCGTCGGAAAAA-3′ and 5′-CCAATAGCCACGTTCAGTAGC-3′; for CHS: 5′-CTACTTCCGCATCACCAACA-3′ and 5′-TTAGGGACTTCGACCACCAC-3′; for ACTIN2: 5′-ACTAAAACGCAAAACGAAAGCGGTT-3′ and 5′-CTAAGCTCTCAAGATCAAAGGCTTA-3′; and for 18S rRNA: 5′-AAACGGCTACCACATCCAAG-3′ and 5′-CCTCCAATGGATCCTCGTTA-3′.For semi-quantitative PCR, to the appropriate volume of cDNA, a master mix was added consisting of 1× PCR Buffer (New England Biolabs), 0.2 mM dNTPs, 0.5 μM of each primer, 0.625 Units of Taq DNA Polymerase (New England Biolabs) and RNase free water to a final volume of 25 μl. The PCR cycle used was (step 1) incubation for 2 min 30 s at 95 °C, (step 2) a further 45 s at 95 °C, (step 3) incubation at 55–59 °C for 1 min, (step 4) elongation at 72 °C for 1 min and a final step of a further elongation at 72 °C for 5 min (step 5). Steps 2–4 were repeated 24–28 times depending on the primers used; the cycle number was selected to ensure that PCR product was quantitatively related to transcript level over a linear range of amplification. The primers used were as follows, ACTIN2: 5′-CTTACAATTTCCCGCTCTGC-3′ and 5′-GTTGGGATGAACCAGAAGGA-3′; ANAC13: 5′-AGCTCGTTGTTTCGGCTAGT-3′and 5′-TCAGGAGACCAGAACCATCC-3′; CHS: 5′-ATCTTTGAGATGGTGTCTGC-3′ and 5′-CGTCTAGTATGAAGAGAACG-3′; At5g51440: 5′-GCGGAAATGAAGAATGGTGT-3′ and 5′-AAGTCAAAATCCCGAACACA-3′; At2g41730: 5′-GTCACCAAGGCATCGTAAGG-3′ and 5′-ACTTGATAGCTGGCGACACG-3′; At3g22060: 5′-ACAATGCGTTTCTCTTCCACA-3′ and 5′-GCGAGTTGAATGTTGATGGAT-3′.
Transcriptomics
Three independent RNA samples were extracted as described above and transcriptomic analysis was undertaken by the Sir Henry Wellcome Functional Genomics Facility (SHWFGF, University of Glasgow). RNA quality was checked using an Agilent RNA BioAnalyzer 2100 (Austin, TX). The samples were then reverse transcribed and biotinylated cRNA hybridised to Affymetrix Arabidopsis ATH1 GeneChips (High Wycombe, UK) as per the manufacturer's protocols. Subsequent washes and staining were performed using a Fluidics Station 400 (Affymetrix) according to manufacturer's instructions. The chips were scanned on a GeneArray Scanner 2500 (Affymetrix) and analysed by the SHWFGF using FUNALYSE version 2.0 (University of Glasgow, UK). Analysis involved normalization using Robust Multi Chip Average36 and differentially expressed genes were determined using the Rank Products method.37Venn diagrams were constructed using GeneVenn software (http://mcbc.usm.edu/genevenn/genevenn.htm) and jvenn.38 Microarray comparisons across studies were performed by incorporating all data into a FileMaker Pro 10® (Filemaker Inc., CA, USA) database. Data was then exported to Excel® (Microsoft, USA) and sorted according to frequency of occurrence for each gene. Gene Ontology (GO) term (biological processes) enrichment was performed using the R package clusterProfiler 3.10.1.39 For the Venn analysis in Fig. 2, the gene lists were from Kilian et al. (2007)26 Supplemental Table 2 (genes induced by multiple stresses) and Supplemental Tables 3 and 4 (genes induced by UV-B stress at different time points).
Results
Transcriptome analysis identifies genes regulated by low UV-B fluence rates independently of UVR8 and stress signaling
Several studies have been carried out previously with Arabidopsis to unravel UV-B regulated and UVR8 dependent pathways at the transcriptional level using microarrays and other techniques.6,15,18,24–26,40 Our initial research18 identified 639 genes induced in response to UV-B, of which 72 are dependent on UVR8. The implication is that many genes are induced by UVR8-independent pathway(s). However, the UV-B treatment used by Brown et al. (2005)18 (3 μmol m−2 s−1 broadband UV-B for 4 hours) is sufficient to trigger several stress related genes in non-acclimated plants,25 which could explain the preponderance of UVR8-independent genes in the microarray data. Hence, in this study we wanted to determine whether UVR8-independent pathways regulate gene expression at low fluence rates of UV-B. We therefore gave WT plants UV-B treatments (0.3 and 1 μmol m−2 s−1 for 4 hours) that have been shown to initiate photomorphogenic gene expression rather than non-specific stress responses.25 Microarray analyses were carried out similarly to Brown et al. (2005),18 comparing transcript profiles for each UV-B treatment to a non-UV-B-treated control. The gene lists are shown in Tables S1 (0.3 μmol m−2 s−1) and S2 (1 μmol m−2 s−1).† The data are presented with a false discovery rate (FDR), which represents the probability that a given gene in a list is a false positive. We use 5% FDR to facilitate comparison with previous published data, but are aware from transcript measurements that at this % FDR some genes in the lists may be only weakly differential. Comparisons at 2% or 1% FDR reduce the likelihood of false positives.Fig. 1 shows Venn diagrams constructed from the microarray data at different false discovery rates (FDR) and depicts the overlap between the microarrays. Intuitively one might expect that lower fluence rates of UV-B would up-regulate many fewer genes than higher fluence rates because the latter are likely to induce more stress related, non-specific genes. Conversely, this was not the case. At a FDR of 5%, 549 and 572 genes were up-regulated in the 0.3 and 1 μmol m−2 s−1 microarrays respectively, which is comparable to the 639 genes induced at 3 μmol m−2 s−1 in ref. 18. A similar outcome was obtained at FDRs of 2% and 1% (Fig. 1).
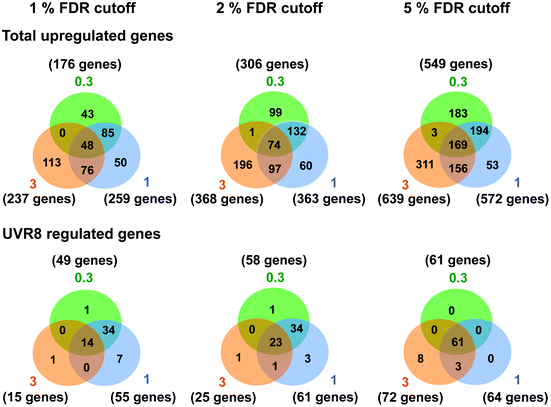 | ||
| Fig. 1 Venn diagrams depicting the overlap in gene expression at different fluence rates of UV-B. Three-weeks-old Arabidopsis plants grown in a low fluence rate of white light (20 μmol m−2 s−1) were treated with either 4 hours of 0.3, 1 or 3 μmol m−2 s−1 broadband UV-B or were left in low white light as a control. This corresponds to a plant weighted UV51 of 0.08, 0.27, and 0.82 kJ m−2, respectively, which compares with 4.8 kJ m−2 day−1 in Lund, Sweden, at midsummer under a cloudless sky.51 The numbers of genes that showed an increase in transcript level were calculated for each of three False Discovery Rates (FDR). Gene lists were then compared to those published by Brown et al. (2005)18 to determine overlap and dependence on UVR8. Numbers in orange circles denote transcripts identified in the Brown et al. (2005)18 3 μmol m−2 s−1 UV-B microarray, those in blue and green are those found in this study to be induced by 1 and 0.3 μmol m−2 s−1 UV-B respectively. | ||
There was a degree of overlap between the genes induced by each fluence rate, with 169 genes (at 5% FDR) in common between the 3 treatments. However, there was a greater degree of overlap when comparing 0.3 and 1 μmol m−2 s−1 (approximately 65% genes in common) and 1 and 3 μmol m−2 s−1 (approximately 55% genes in common) than when comparing 0.3 and 3 μmol m−2 s−1 (approximately 30% genes in common), which highlights the differences in signaling processes regulating gene expression at the lowest and highest fluence rates. The intermediate 1 μmol m−2 s−1 UV-B treatment initiates expression of both ‘low’ and ‘high’ fluence rate genes and consequently less than 10% of the genes induced are specific to that treatment, whereas 33% and 49% of genes are specific to the 0.3 and 3 μmol m−2 s−1 treatments, respectively.
The requirement for UVR8 for expression of specific genes was determined from previous analyses using a uvr8 mutant.18 Most of the 72 UVR8-dependent genes induced by 3 μmol m−2 s−1 UV-B (at 5% FDR18) were also detected in the present microarrays (64 and 61 genes for 0.3 and 1 μmol m−2 s−1 respectively). Interestingly, UVR8-regulated genes show a greater representation at lower fluence rates than at 3 μmol m−2 s−1 when the stringency is increased to 1% FDR. At 0.3 μmol m−2 s−1 28% of genes are UVR8-dependent at 1% FDR, compared to 11% at 5% FDR. Conversely, at 3 μmol m−2 s−1 only 6% of genes are UVR8-dependent at 1% FDR, in contrast to 11% at 5% FDR. The above analysis indicates that a substantial number of genes induced by low fluence rates of UV-B are not regulated by UVR8. To extend the analysis, we compared the genes induced at low fluence rate UV-B with the set of UVR8 regulated genes identified by Favory et al. (2009),15 who listed a total of over 700 genes potentially regulated by UVR8 in the different UV-B exposures in their study. As shown in Fig. 2A, there is extensive overlap between the sets of UVR8 regulated genes identified in ref. 18 and 15. In addition, the Favory15 list increases the number of genes identified as UVR8 regulated at low fluence rate UV-B. Nevertheless, 44% of the 306 genes (at 2% FDR) induced by 0.3 μmol m−2 s−1 UV-B are not dependent on UVR8 (Fig. 2A).
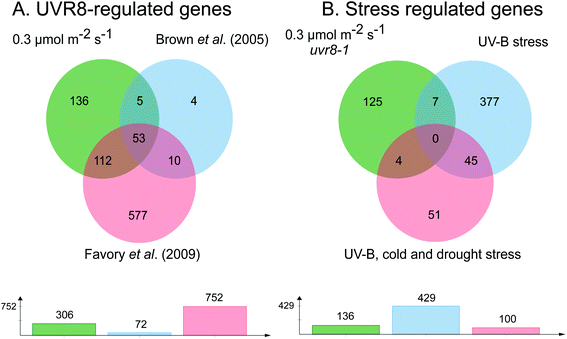 | ||
| Fig. 2 Venn diagrams showing genes induced by low fluence rate UV-B independently of UVR8 and stress signalling pathways. (A) Genes induced by exposure of Arabidopsis to 0.3 μmol m−2 s−1 broadband UV-B for 4 hours (corresponding to a plant weighted UV51 of 0.08 kJ m−2), cut at 2% FDR (as shown in Fig. 1) compared to genes shown to be regulated by UVR8 in microarray analyses of Brown et al. (2005)18 and Favory et al. (2009).15 (B) UVR8-independent genes identified in A compared to genes shown to be induced either by UV-B stress or in common by UV-B, cold and drought stresses in the microarray study of Kilian et al. (2007).26 Venn diagrams are shown above with numbers of genes in each set below. The diagrams were constructed using jvenn.38 | ||
Since UVR8 independent UV-B signaling pathways are known to overlap with stress-related signaling pathways, at least at relatively high UV-B fluence rates,41,42 we examined whether any of the UVR8-independent genes induced by 0.3 μmol m−2 s−1 UV-B are known targets of stress signaling pathways. We tested whether any of these genes were induced by UV-B stress in the microarray study of Kilian et al. (2007).26 Of the 429 UV-B stress-induced genes,26 only 11 were among the UVR8-independent genes induced by 0.3 μmol m−2 s−1 UV-B (Fig. 2B). Four of these 11 genes were among a set of 100 genes induced in common by UV-B, cold and drought stresses26 (Fig. 2B). Interestingly, 9 of the above 11 genes are among 52 UVR8-independent genes expressed at all 3 fluence rates employed in the present study, demonstrating that expression of this small set of putative ‘stress-related’ genes is not confined to stress conditions; it may be that these genes are expressed as a result of a change in environment rather than ‘damage stress’ per se.
According to the above analysis, 41% of the genes induced at 0.3 μmol m−2 s−1 UV-B (125 genes in total at 2% FDR; listed in Table S3†) are not known targets of either UVR8 or stress signaling pathways, indicating that a distinct type of UV-B signaling operates at low UV-B fluence rates. To gain insights into the potential functions of genes induced via this UVR8 and stress independent pathway(s) we used GO enrichment analysis (Fig. S2†). This revealed that a substantial number of the genes are concerned with carbohydrate metabolism.
ANAC13 expression is induced by low fluence rate UV-B independently of UVR8
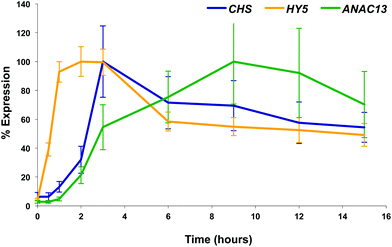
We examined a number of genes identified as UVR8-independent in the microarray analysis to test the validity of the results with respect to UV-B fluence rate: ANAC13, At5g51440 (a putative HSP20-like chaperone), At3g22060 (a gene expressed in response to abscisic acid) and At2g41730 (a gene expressed in rosette leaves and responsive to high concentrations of Boron). As shown in Fig. 3, semi-quantitative RT-PCR confirmed the microarray result in that all four genes were induced by low fluence rate UV-B: ANAC13, At2g41730, and At3g22060 were induced at 0.1 μmol m−2 s−1, similar to CHS, whereas At5g51440 was induced at 0.2 μmol m−2 s−1. To further explore the potential low fluence rate UV-B, UVR8-independent pathway we decided to focus on one gene and selected ANAC13. Quantitative PCR analysis of the kinetics of ANAC13 expression (Fig. 4) revealed that transcripts peak at around 6–12 hours after the start of UV-B exposure. This increase in expression is slower than that of the UVR8 dependent transcription factor HY5 and CHS, which is downstream of the UVR8/HY5 UV-B pathway. In addition, we wanted to test whether ANAC13 is in fact UVR8-independent, as suggested by the microarray results and previously reported by Safrany et al. (2008).27Fig. 5 confirms that ANAC13 is induced at low fluence rates (0.5 μmol m−2 s−1) and, unlike HY5, this induction still occurs in the uvr8 mutant. Fig. 5 further shows that HY5 and ANAC13 transcript levels are similar after 2 hours UV-B in WT, but at 6 hours ANAC13 expression continues to rise with HY5 starting to decline, consistent with the kinetics in Fig. 4. Overall, these results confirm that ANAC13 expression is UVR8-independent and induced at low UV-B doses.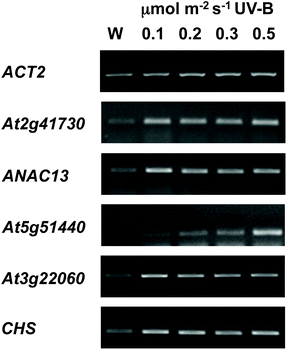 | ||
| Fig. 3 Expression of UVR8-independent genes under very low fluence rates of UV-B. Three-weeks-old wild type plants grown in a low fluence rate of white light (20 μmol m−2 s−1) were treated for either 4 hours with 0.1, 0.2, 0.3, or 0.5 μmol m−2 s−1 broadband UV-B (corresponding to plant weighted UV51 of 0.03, 0.05, 0.08, and 0.14 kJ m−2) or were left in low white light (W) as a control. Transcript levels were assayed using RT-PCR and compared with control ACT2 transcripts. | ||
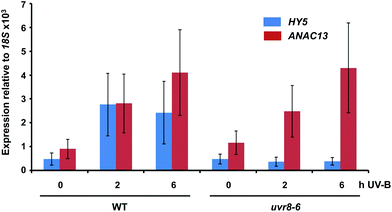 | ||
| Fig. 5 ANAC13 is UV-B induced and UVR8 independent. Quantitative RT-PCR assays of HY5 and ANAC13 transcripts, normalized to control 18S transcript levels, in wild-type Col-0 and uvr8-6. Plants were exposed (+) or not (−) to 0.5 μmol m−2 s−1 of narrowband UV-B for 2 or 6 h. The plant weighted UV51 was 0.005 kJ m−2 for the 2 h exposure and 0.015 kJ m−2 for the 6 h exposure, which compares with 4.8 kJ m−2 day−1 in Lund, Sweden, at midsummer with a cloudless sky.51 Data are means of three experiments ±S.E. | ||
ANAC13 UV-B induction is independent of COP1 and HY5/HYH
To further examine whether ANAC13 is induced by the low fluence rate UV-B, UVR8-independent pathway, we tested ANAC13 expression in cop1 and hy5hyh mutants. Since both COP1 and the transcription factors HY5/HYH are associated with UVR8 signaling, one would expect ANAC13 expression to be unaffected in such mutants. In agreement with this notion ANAC13 expression was induced by UV-B in cop1-4 (Fig. 6A), as reported previously.27 Furthermore, expression was similar in hy5hyh and WT plants (Fig. 6B), indicating that the UVR8 independent pathway regulating ANAC13 expression is independent of HY5/HYH as well as COP1.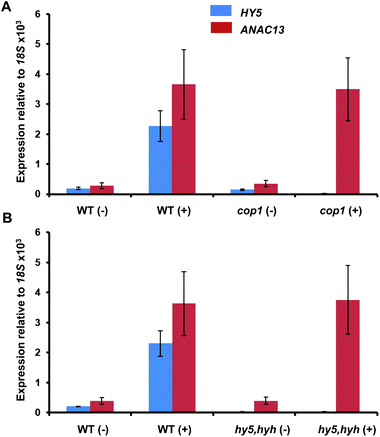 | ||
| Fig. 6 UV-B induction of ANAC13 is independent of COP1, HY5 and HYH. Quantitative RT-PCR assays of HY5 and ANAC13 transcripts, normalized to control 18S transcript levels, in wild-type Ws, cop1-4 (A) and hy5/hyh (B). 21-day old wild type, cop1-4 or hy5/hyh mutant plants were grown in a low fluence rate of fluorescent white light (20 μmol m−2 s−1) and then exposed (+) or not (−) to 0.5 μmol m−2 s−1 of narrowband UV-B light for 2 h. The plant weighted UV51 was 0.005 kJ m−2, which compares with 4.8 kJ m−2 day−1 in Lund, Sweden, at midsummer with a cloudless sky.51 Data are means of three experiments ±S.E. | ||
ANAC13 UV-B induced expression is independent of several other known photoreceptors
To test the possibility that a photoreceptor other than UVR8 is able to detect low fluence UV-B and regulate ANAC13, or perhaps act redundantly with UVR8, we tested several photoreceptor mutants for their ability to induce ANAC13 in response to UV-B. To do this we used the cry1cry2 mutant which lacks both cryptochromes, phot1phot2 which lacks both phototropins, and phyabde which is deficient in all the phytochromes except PHYC. We also utilized the triple mutant, cry1cry2uvr8-2 to rule out the possibility of redundancy between the cryptochromes and UVR8. Fig. 7 shows that UV-B induction of ANAC13 is similar to WT in all the mutants tested. Furthermore, in the triple mutant cry1cry2uvr8-2, HY5 expression is not induced upon UV-B exposure, in contrast to ANAC13 expression (Fig. 7D). Overall, these data indicate that ANAC13 UV-B up-regulation is not activated via the known photoreceptors tested here and in addition ANAC13 expression is not induced via an overlapping, redundant pathway operating between UVR8 and CRY1/CRY2.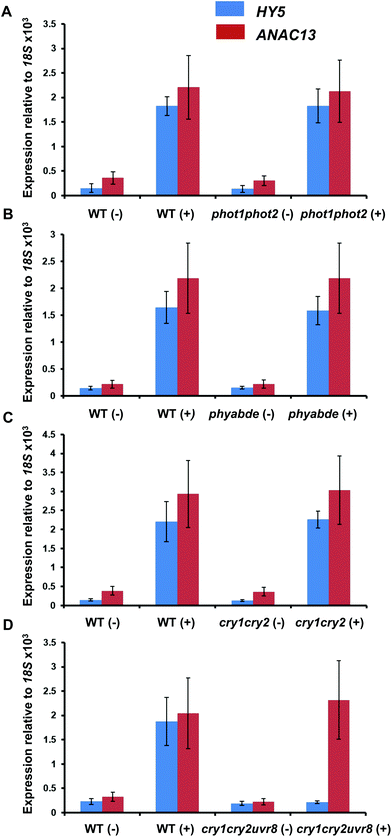 | ||
| Fig. 7 UV-B induction of ANAC13 is independent of multiple photoreceptors. Quantitative RT-PCR assays of HY5 and ANAC13 transcripts, normalized to control 18S transcript levels, in wild-type Ler (or Col-0 for phot1phot2 comparison), phot1phot2 (A), phyABDE (B), cry1cry2 (C) and cry1cry2uvr8-2 (D). Plants were exposed (+) or not (−) to 0.5 μmol m−2 s−1 of narrowband UV-B light for 2 h. The plant weighted UV51 was 0.005 kJ m−2, which compares with 4.8 kJ m−2 day−1 in Lund, Sweden, at midsummer with a cloudless sky.51 Data are means of three experiments ±S.E. | ||
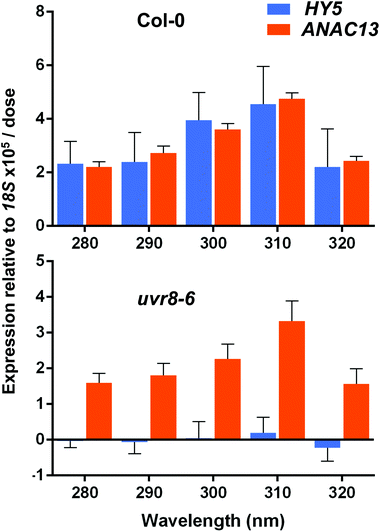
ANAC13 shows maximum expression at longer wavelengths of UV-B in WT and uvr8-6 plants
Finally, we wanted to determine which UV-B wavelengths produce the maximal expression of ANAC13. To do this we exposed both WT and uvr8-6 plants to a range of UV-B wavelengths in increments of 10 nm from 280 to 320 nm for 1 hour, followed by 1 hour in darkness to allow transcripts to accumulate, then carried out qPCR to examine ANAC13 and HY5 transcript levels. Fig. 8 shows that both ANAC13 and HY5 expression are induced over the range of UV-B wavelengths in WT and only ANAC13 expression is induced in the uvr8-6 mutant. Fig. 8 also demonstrates that both ANAC13 and HY5 are maximally expressed at 310 nm and that the expression levels descend accordingly 300 > 290 > 280 > 320. The p-values (Table 1) show that ANAC13 expression in WT and uvr8-6 were not significantly different (>0.05) over the range of wavelengths tested. Overall, these data further support the claim that ANAC13 is UVR8-independent and regulated by lower energy, longer wavelength UV-B.| Col-0 | uvr8-6 | HY5 | ANAC13 | |
|---|---|---|---|---|
| Comparison | HY5/ANAC13 | HY5/ANAC13 | Col-0/uvr8-6 | Col-0/uvr8-6 |
| 280 | 0.991 | 0.015 | 0.002 | 0.491 |
| 290 | 0.926 | 0.046 | 0.011 | 0.402 |
| 300 | 0.888 | 0.012 | 0.0001 | 0.097 |
| 310 | 0.988 | 0.005 | 0.001 | 0.179 |
| 320 | 0.981 | 0.046 | 0.011 | 0.5 |
Discussion
It is well established that UV-B exposure of plants regulates expression of a large number of genes.15,18,24,26 Such reprogramming of the transcriptome enables plants to re-adjust their metabolism, morphology and physiology to acclimate to changes in their radiation environment. While UVR8 regulates a substantial, functionally important subset of UV-B-regulated genes, it is evident that other processes can also mediate UV-B-induced gene expression. A number of studies have shown that sudden exposure of non-acclimated plants to high fluence rate, short wavelength UV-B induces expression of numerous genes that are not specific to the UV-B stimulus and are also induced by various stress treatments.24,26,41 The signal transduction processes initiated by UV-B stress treatments are reported to include DNA damage signaling, ROS signaling, MAPK kinase activity, and wound/defence signaling molecules.42–45 However, there is very little information about whether non-stressful, low fluence rate, longer wavelength UV-B treatments can regulate gene expression independently of UVR8, and that is the question addressed in this study.The UV-B exposures chosen for the microarray analysis were based on previous research that identified conditions for the induction of UVR8-dependent and UVR8-independent gene expression in Arabidopsis.25 Plants were grown under identical conditions and given the same spectral quality and duration of UV-B exposure as previously,25 however, the 0.3 μmol m−2 s−1 treatment used here is below the threshold of 1 μmol m−2 s−1 found to be required for UVR8-independent induction of stress related genes.25 In the present study the UVR8-dependent genes were defined by both the Brown et al. (2005)18 and Favory et al. (2009)15 microarray analyses, which involved quite different plant growth and UV-B exposure conditions. It is therefore interesting that a substantial number of genes were induced by 0.3 μmol m−2 s−1 UV-B independently of UVR8 (Fig. 1 and 2). Moreover, there is very little overlap between these UVR8-independent genes and genes reported to be induced by UV-B stress by Kilian et al. (2007).26 This is not surprising because 0.3 μmol m−2 s−1 UV-B is very unlikely to induce any of the known UV-B stress signaling pathways. Overall, the data indicate that a significant proportion of UV-B-responsive genes are regulated independently of both UVR8 and stress signaling.
The sets of genes induced by the 0.3 and 3.0 μmol m−2 s−1 UV-B treatments had little overlap, but a small number of the UV-B regulated UVR8-independent genes were induced at all 3 UV-B fluence rates employed and several were also found to be induced under stress conditions.26 At least some of these genes may be activated by ‘change’ rather than ‘stress’ per se. Hideg et al. (2013)45 highlight the distinction between constructive stress, termed eustress, which promotes acclimation, and destructive stress in UV-B responses. It may be that transfer of plants that have not previously been exposed to UV-B to even a very low fluence rate of UV-B is sufficient to activate ‘eustress’ as opposed to ‘damage stress’ (distress), and that this UV-B induced eustress response is at least partly independent of UVR8.
It is not known how many different signaling pathways mediate the response to low fluence rate UV-B, and it is not clear why multiple mechanisms are employed. Redundancy is common in plant photoreception and signaling as it ensures that key stimuli, such as UV-B, will be perceived, and introduces flexibility in response. Characterisation of ANAC13 gene expression provided information on one low fluence UV-B, UVR8-independent signaling pathway. ANAC13 encodes a putative NAC domain containing transcription factor. The NAC domain (NAM, ATAF1/2 and CUC2) proteins are unique to plants and are thought to be involved in a wide range of processes including stress responses, development and growth.46–50 A previous study27 reported that ANAC13 could also be induced by short wavelength, relatively high fluence rate UV-B, but this is likely via a different signaling pathway. The kinetics of ANAC13 induction by low fluence rate UV-B are different to those of the classic UVR8-regulated HY5 gene and its downstream target CHS, supporting the notion that it is regulated via a distinct pathway. In addition, ANAC13 is regulated by UV-B independently of COP1 and HY5/HYH, which are associated with UVR8 signaling. Moreover, at low fluence rates ANAC13 is regulated by longer wavelength UV-B, maximally at 310 nm, further suggesting that it is not activated by stress signaling under these conditions. No detailed information is yet available on the nature of the low fluence pathway that regulates ANAC13 expression. It does not appear to be regulated by the known photoreceptors, at least based on our experiments with a range of photoreceptor mutants described above. However, the possibility of a novel photoreceptor cannot be excluded. There has been speculation about the existence of UV-B photoreceptors in addition to UVR8,28 although no such molecule has yet been identified.
An important, but largely unanswered question is to what extent UVR8-independent pathways regulate gene expression in plants growing in natural environments where plants are not usually subject to UV-B stress. There is evidence that UVR8-independent pathways do operate,6 but they are not well defined. The present study highlights the potential for low, non-damaging fluence rates of UV-B, well within the wavelength range experienced by plants in nature, to regulate a substantial number of genes. Moreover, these pathways should not be considered as stress-related as they are evidently independent of classic stress signaling pathways. We therefore refer to these pathways as UVR8 and Stress-Independent (UASI) UV-B signaling pathways. Further research is now required to define the molecular mechanisms involved in these pathways and to assess the functional significance of gene expression responses that they mediate. The GO enrichment analysis suggests that carbohydrate metabolism may be an important function of genes regulated by UASI UV-B signaling, but no further insights are available at present.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
AO and LRH were supported by UK Biotechnology and Biological Sciences research council PhD studentships at the University of Glasgow. GIJ thanks Dr Pawel Herzyk and staff of the Sir Henry Wellcome Functional Genomics Facility (University of Glasgow) for producing the microarray data. GIJ thanks the University of Glasgow for the support of his research. ÅS was supported by grants from the Knowledge Foundation (kks.se; contract no. 20130164) and The Swedish Research Council Formas (formas.se/en; Contract no. 942-2015-516). ÅS and AO were also supported by the Faculty for Business, Science, and Technology at Örebro University, and LOM was supported by the Strategic Young Researchers Recruitment Programme at Örebro University.References
- L. Rizzini, J.-J. Favory, C. Cloix, D. Faggionato, A. O'Hara, E. Kaiserli, R. Baumeister, E. Schäfer, F. Nagy, G. I. Jenkins and R. Ulm, Science, 2011, 332, 103–106 CrossRef CAS PubMed.
- J. M. Christie, A. S. Arvai, K. J. Baxter, M. Heilmann, A. J. Pratt, A. O'Hara, S. M. Kelly, M. Hothorn, B. O. Smith, K. Hitomi, G. I. Jenkins and E. D. Getzoff, Science, 2012, 335, 1492–1496 CrossRef CAS PubMed.
- D. Wu, Q. Hu, Z. Yan, W. Chen, C. Yan, X. Huang, J. Zhang, P. Yang, H. Deng, J. Wang, X. W. Deng and Y. Shi, Nature, 2012, 484, 214–219 CrossRef PubMed.
- D. J. Kliebenstein, J. E. Lim, L. G. Landry and R. L. Last, Plant Physiol., 2002, 130, 234–243 CrossRef CAS PubMed.
- P. V. Demkura and C. L. Ballaré, Mol. Plant, 2002, 5, 642–652 CrossRef PubMed.
- L. O. Morales, M. Brosché, J. Vainonen, G. I. Jenkins, J. J. Wargent, N. Sipari, Å. Strid, A. V. Lindfors, R. Tegelberg and P. J. Aphalo, Plant Physiol., 2013, 161, 744–7597 CrossRef CAS PubMed.
- G. I. Jenkins, Plant Cell, 2014, 26, 21–37 CrossRef CAS PubMed.
- S. Hayes, A. Sharma, D. P. Fraser, M. Trevisan, C. K. Cragg-Barber, E. Tavridou, C. Fankhauser, G. I. Jenkins and K. A. Franklin, Curr. Biol., 2016, 27, 120–127 CrossRef PubMed.
- G. I. Jenkins, Plant, Cell Environ., 2017, 40, 2544–2557 CrossRef CAS PubMed.
- R. Yin and R. Ulm, Curr. Opin. Plant Biol., 2017, 37, 42–48 CrossRef CAS PubMed.
- M. Wu, Å. Strid and L. A. Eriksson, J. Phys. Chem. B, 2014, 118, 951–965 CrossRef CAS PubMed.
- T. Mathes, M. Heilmann, A. Pandit, J. Zhu, J. Ravensbergen, M. Klos, Y. Fu, B. O. Smith, J. M. Christie, G. I. Jenkins and J. T. M. Kennis, J. Am. Chem. Soc., 2015, 137, 8113–8120 CrossRef CAS PubMed.
- X. Zeng, Z. Ren, Q. Wu, J. Fan, P. Peng, K. Tang, R. Zhang, K.-H. Zhao and X. Yang, Nat. Plants, 2015, 1, 14006 CrossRef CAS PubMed.
- A. Oravecz, A. Baumann, Z. Máté, A. Brzezinska, J. Molinier, E. J. Oakeley, É. Adám, E. Schäfer, F. Nagy and R. Ulm, Plant Cell, 2006, 18, 1975–1990 CrossRef CAS PubMed.
- J. J. Favory, A. Stec, H. Gruber, L. Rizzini, A. Oravecz, M. Funk, A. Albert, C. Cloix, G. I. Jenkins, E. J. Oakeley, H. K. Seidlitz, F. Nagy and R. Ulm, EMBO J., 2009, 28, 591–601 CrossRef CAS PubMed.
- C. Cloix, E. Kaiserli, M. Heilmann, K. J. Baxter, B. A. Brown, A. O'Hara, B. O. Smith, J. M. Christie and G. I. Jenkins, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 16366–16370 CrossRef CAS PubMed.
- R. Yin, M. Y. Skvortsova, S. Loubéry and R. Ulm, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E4415–E4422 CrossRef CAS PubMed.
- B. A. Brown, C. Cloix, G. H. Jiang, E. Kaiserli, P. Herzyk, D. J. Kliebenstein and G. I. Jenkins, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 18225–18230 CrossRef CAS PubMed.
- Y. Yang, T. Liang, L. Zhang, K. Shao, X. Gu, R. Shang, N. Shi, X. Li, P. Zhang and H. Liu, Nat. Plants, 2018, 4, 980–107 CrossRef PubMed.
- T. Liang, S. Mei, C. Shi, Y. Yang, Y. Peng, L. Ma, F. Wang, X. Li, X. Huang, Y. Yin and H. Liu, Dev. Cell, 2018, 44, 1–12 CrossRef PubMed.
- B. Fehér, L. Kozma-Bognár, E. Kevei, A. Hajdu, M. Binkert, S. J. Davis, E. Schäfer, R. Ulm and F. Nagy, Plant J., 2011, 67, 37–48 CrossRef PubMed.
- A. O'Hara and G. I. Jenkins, Plant Cell, 2012, 24, 3755–3766 CrossRef PubMed.
- J. J. Wargent, V. C. Gegas, G. I. Jenkins, J. H. Doonan and N. D. Paul, New Phytol., 2009, 183, 315–326 CrossRef CAS PubMed.
- R. Ulm, A. Baumann, A. Oravecz, Z. Mate, E. Adam, E. J. Oakeley, E. Schäfer and F. Nagy, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 1397–1402 CrossRef CAS PubMed.
- B. A. Brown and G. I. Jenkins, Plant Physiol., 2008, 146, 576–588 CrossRef CAS PubMed.
- J. Kilian, D. Whitehead, J. Horak, D. Wanke, S. Weinl, O. Batistic, C. D'Angelo, E. Bornberg-Bauer, J. Kudla and K. Harter, Plant J., 2007, 50, 347–363 CrossRef CAS PubMed.
- J. Safrany, V. Haasz, Z. Mate, A. Ciolfi, B. Feher, A. Oravecz, A. Stec, G. Dallmann, G. Morelli, R. Ulm and F. Nagy, Plant J., 2008, 54, 402–414 CrossRef CAS PubMed.
- J. Takeda, R. Nakata, H. Ueno, A. Murakami, M. Iseki and M. Watanabe, Photochem. Photobiol., 2014, 90, 1043–1049 CAS.
- N. Rai, S. Neugart, Y. Yan, F. Wang, S. M. Siipola, A. V. Lindfors, J. B. Winkler, A. Albert, M. Brosché, T. Lehto, L. O. Morales and P. J. Aphalo, J. Exp. Bot., 2019 DOI:10.1093/jxb/erz236.
- K. A. Franklin, U. Praekelt, W. M. Stoddart, O. E. Billingham, K. J. Halliday and G. C. Whitelam, Plant Physiol., 2003, 131, 1340–1346 CrossRef CAS PubMed.
- H. K. Wade, A. K. Sohal and G. I. Jenkins, Plant Physiol., 2003, 131, 707–715 CrossRef CAS PubMed.
- T. Kagawa, T. Sakai, N. Suetsugu, K. Oikawa, S. Ishiguro, T. Kato, S. Tabata, K. Okada and M. Wada, Science, 2001, 291, 2138–2141 CrossRef CAS PubMed.
- M. Holm, C. Hardtke, R. Gaudet and X. W. Deng, EMBO J., 2001, 20, 118–127 CrossRef CAS PubMed.
- X. W. Deng and P. H. Quail, Plant J., 1992, 2, 83–95 CrossRef CAS.
- L. A. Díaz-Ramos, A. O'Hara, S. Kanagarajan, D. Farkas, Å. Strid and G. I. Jenkins, Photochem. Photobiol. Sci., 2018, 17, 1108–1117 RSC.
- R. A. Irizarry, B. M. Bolstad, F. Collin, L. M. Cope, B. Hobbs and T. P. Speed, Nucleic Acids Res., 2003, 31, e15 CrossRef PubMed.
- R. Breitling, P. Armengaud, A. Amtmann and P. Herzyk, FEBS Lett., 2004, 573, 83–92 CrossRef CAS PubMed.
- P. Bardou, J. Mariette, F. Escudié, C. Djemiel and C. Klopp, BMC Bioinf., 2014, 15, 293 CrossRef PubMed.
- G. Yu, L.-G. Wang, Y. Han and Q.-Y. He, OMICS, 2012, 16, 284–287 CrossRef CAS PubMed.
- K. Hectors, E. Prinsen, W. De Coen, M. A. K. Jansen and Y. Guisez, New Phytol., 2007, 175, 255–270 CrossRef CAS PubMed.
- M. Brosché and Å. Strid, Physiol. Plant., 2003, 117, 1–10 CrossRef.
- G. I. Jenkins, Annu. Rev. Plant Biol., 2009, 60, 407–431 CrossRef CAS PubMed.
- R. Ulm and F. Nagy, Curr. Opin. Plant Biol., 2005, 8, 477–482 CrossRef CAS PubMed.
- M. A. González-Besteiro, S. Bartels, A. Albert and R. Ulm, Plant J., 2011, 68, 727–737 CrossRef PubMed.
- É. Hideg, M. A. K. Jansen and Å. Strid, Trends Plant Sci., 2013, 18, 107–115 CrossRef PubMed.
- A. N. Olsen, H. A. Ernst, L. L. Leggio and K. Skriver, Trends Plant Sci., 2005, 10, 79–87 CrossRef CAS PubMed.
- H. Ooka, K. Satoh, K. Doi, T. Nagata, Y. Otomo, K. Murakami, K. Matsubara, N. Osato, J. Kawai, P. Carninci, Y. Hayashizaki, K. Suzuki, K. Kojima, Y. Takahara, K. Yamamoto and S. Kikuchi, DNA Res., 2003, 10, 239–247 CrossRef CAS PubMed.
- K. Nakashima, H. Takasaki, J. Mizoi, K. Shinozaki and K. Yamaguchi-Shinozaki, Biochim. Biophys. Acta, 2012, 1819, 97–10 CrossRef CAS PubMed.
- I. De Clercq, V. Vermeirssen, O. Van Aken, K. Vandepoele, M. W. Murcha, S. R. Law, A. Inzé, S. Ng, A. Ivanova, D. Rombaut, B. van de Cotte, P. Jaspers, Y. Van de Peer, J. Kangasjärvi, J. Whelan and F. Van Breusegem, Plant Cell, 2013, 25, 3472–3490 CrossRef CAS PubMed.
- C. O'Shea, L. Staby, S. K. Bendsen, F. G. Tidemand, A. Redsted, M. Willemoës, B. B. Kragelund and K. Skriver, Biochem. J., 2015, 465, 281–294 CrossRef PubMed.
- S. G. Yu and L. O. Björn, J. Photochem. Photobiol., B, 1997, 37, 212–218 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9pp00151d |
| ‡ These authors contributed equally to the work. |
| § Present address: Essex Pathways, University of Essex, Wivenhoe Park, Colchester, CO4 3SQ, UK. |
| This journal is © The Royal Society of Chemistry and Owner Societies 2019 |