Different platinum crystal surfaces show very distinct protein denaturation capabilities†
Abstract
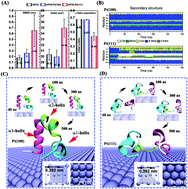
Different platinum (Pt) surfaces of nanocrystals usually exhibit significant distinctions with regard to various biological, physical, and chemical characteristics, such as bio-recognition, surface wetting, and catalytic activities. In this study, we report for the first time that two shape-controlled Pt nanocrystals with the most common low-index surfaces, Pt(100) and Pt(111), show very dissimilar protein denaturation capabilities based on all-atom molecular dynamics simulations employing the widely used model protein, villin headpiece (HP35). We demonstrate that HP35 is well preserved on the Pt(100) crystal surface, whereas it is severely disrupted on the Pt(111) crystal surface. This surprising difference originates from the distinct water behavior in the first solvation shell (FSS) of the two Pt crystal surfaces. Within the FSS of the Pt(100) crystal surface, water molecules form a very compact and stable monolayer through a highly uniform rhombic hydrogen-bond network. This water monolayer prefers the adsorption of acidic residues (such as Glu and Asp) and acts as a shield to prevent other residues from directly coming into contact with the metal surface. On the other hand, the hydrogen bond network in the water monolayer in the FSS of the Pt(111) crystal surface is very sparse and quite defective, which makes it more vulnerable to the penetration of various residues, particularly those with planar side chains such as Phe, Trp and Arg due to strong dispersion interactions, leading to subsequent protein unfolding. The binding free energy calculations for some key amino acids on the two different crystal surfaces further uncover the molecular origin behind their distinct protein denaturation capability. Our study reveals the vital importance of interfacial water in determining the structure of proteins when binding to different metal crystal surfaces. The discovered molecular mechanisms may be helpful for the future development of a bio-assisted programmable synthetic strategy of sophisticated Pt nanostructures for biomedical applications.
- This article is part of the themed collection: Nanoscale 10th Anniversary Special Issue


 Please wait while we load your content...
Please wait while we load your content...