Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Quaternary ammonia compounds in disinfectant products: evaluating the potential for promoting antibiotic resistance and disrupting wastewater treatment plant performance
Zihao
Lu
 a,
Anna K.
Mahony
a,
Anna K.
Mahony
 b,
William A.
Arnold
b,
William A.
Arnold
 b,
Christopher W.
Marshall
c and
Patrick J.
McNamara
b,
Christopher W.
Marshall
c and
Patrick J.
McNamara
 *a
*a
aDept. of Civil, Construction, Environmental Engineering, Marquette University, USA. E-mail: patrick.mcnamara@marquette.edu
bDept. of Civil, Environmental, and Geo-Engineering, University of Minnesota-Twin Cities, 500 Pillsbury Dr SE, Minneapolis, MN 55455, USA
cDept. of Biological Sciences, Marquette University, USA
First published on 28th November 2023
Abstract
Quaternary ammonium compounds (QACs) are a class of compounds that were widely used as disinfectants during the COVID-19 pandemic and continue to be used as disinfecting agents. After consumer usage, QAC concentrations are diluted in wastewater as they enter wastewater treatment plants. At sub-inhibitory concentrations, QACs may have unintended repercussions, including increased antibiotic resistance and inhibition of process performance in wastewater treatment plants. This review first summarizes how QACs inhibit bacteria and then highlights the mechanisms by which QACs can promote antibiotic resistance in general. Reported environmental concentrations of QACs are compared to concentrations that are suspected to impact antibiotic resistance, and the role QACs may have on antibiotic resistance proliferation in wastewater treatment is addressed. Finally, the specific impacts that QACs can have on biological wastewater processes (activated sludge and anaerobic digestion) are reviewed. We highlight key research gaps along with recommendations for future research. Of particular interest, research is needed to elucidate the relationship between the chemical structure of QACs and impacts on antibiotic resistance as well as process performance in wastewater treatment plants. Finally, the ability to mitigate (reverse) these impacts if QACs are removed needs to be determined.
Environmental significanceConsidering the expectation of increased loading of QACs due to the COVID-19 pandemic and long-term changes in consumer/business purchasing of products containing QACs, it is necessary to evaluate the potential effects of QACs on microbial communities, antibiotic resistance, and the performance of wastewater treatment. Understanding the impacts of specific QACs on environmental processes and antibiotic resistance can help protect public health while minimizing environmental impacts. |
1. Introduction
Antibiotic resistance is a critical global health problem.1,2 A report from the Organization for Economic Co-operation and Development (OECD) predicts that approximately 2.4 million people will die in Europe, North America, and Australia between 2015–2050 from infections by antibiotic resistant bacteria, costing up to USD 3.5 billion per year.3 The actual situation, however, may be worse than estimated in the report. A recent predictive model revealed that a median of 1.27 million deaths in 2019 was directly attributable to antibiotic resistance, which is only less than the number of deaths due to SARS-CoV-2 (COVID-19) and tuberculosis.1 Additionally, the Centers for Disease Control and Prevention (CDC) estimated over 2.8 million people in the United States are infected with antibiotic-resistant bacteria each year, which carries an economic burden of $20 billion.4 In Europe, antibiotic resistance is estimated to be the annual cause of 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 fatalities and cost EU €1.5 billion for healthcare expenditures and lost productivity.5 In spite of different actions taken to tackle this issue, the threat of antibiotic resistance is not only still present, but also has been exacerbated by the SARS-CoV-2 pandemic.6,7 CDC data showed that antibiotic resistant infections and deaths both increased at least 15% from 2019 to 2020.8 The reasons for the increasing prevalence of antibiotic resistance are complex. In addition to the overuse and misuse of antibiotics, the widespread use of biocides and disinfectants could be a significant driver of antibiotic resistance, particularly during the pandemic.9–18
000 fatalities and cost EU €1.5 billion for healthcare expenditures and lost productivity.5 In spite of different actions taken to tackle this issue, the threat of antibiotic resistance is not only still present, but also has been exacerbated by the SARS-CoV-2 pandemic.6,7 CDC data showed that antibiotic resistant infections and deaths both increased at least 15% from 2019 to 2020.8 The reasons for the increasing prevalence of antibiotic resistance are complex. In addition to the overuse and misuse of antibiotics, the widespread use of biocides and disinfectants could be a significant driver of antibiotic resistance, particularly during the pandemic.9–18
The use of disinfectants has risen dramatically in various settings as a result of the COVID-19 outbreak.19 Among disinfecting products, quaternary ammonium compounds (QACs) are predominant in industrial, commercial, hospital, and consumer products due to their ability to inactivate enveloped viruses.20–22 QACs are a class of organic, cationic chemicals that contain one or more positively charged quaternary amine groups and at least one hydrophobic alkyl chain.23 The cationic charge is usually balanced by a chloride ion or bromide ion. The most commonly used and studied groups of QACs are mono-cationic compounds including benzyl alkyl dimethyl ammonium compounds (BACs, also known as benzalkonium compounds), dialkyl dimethyl ammonium compounds (DADMACs), and alkyl trimethyl ammonium compounds (ATMACs) (Fig. 1). Additionally, ethyl benzyl ammonium compounds (EtBACs) usage has been increasing in recent years.24–26 Typically, these compounds are used as mixtures with varying chain lengths ranging from 8–20 carbon atoms. QACs are amphiphilic, due to their hydrophobic alkyl groups and hydrophilic positively charged N. In general, QACs are water soluble and stable, but their solubility diminishes as chain length increases.23,27 Within the same class of QACs, as the alkyl chain length increases the octanol–water partition coefficient (Kow) increases. QACs with longer alkyl chains are more readily sorbed by solid phases via electrostatic and hydrophobic interactions and are more likely to be found in biosolids, soil, and sediments.28–32 This characteristic of QACs not only impacts the distribution and bioavailability of QAC in environments, but also restricts the biodegradation efficiency and toxicity of QACs.
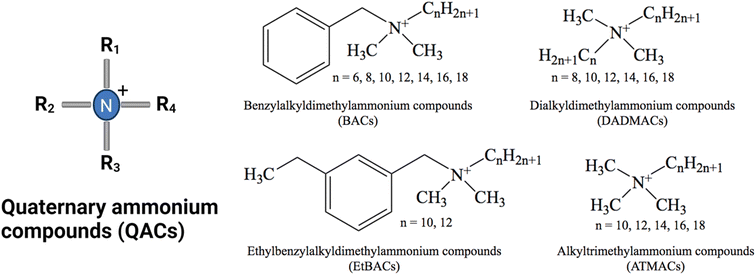 | ||
| Fig. 1 General structure of the four main QAC chemical groups. The most commonly used QACs are C8–C18 BAC, C12–C14 EtBAC and C8, C8/10 and C10 DADMAC.33 | ||
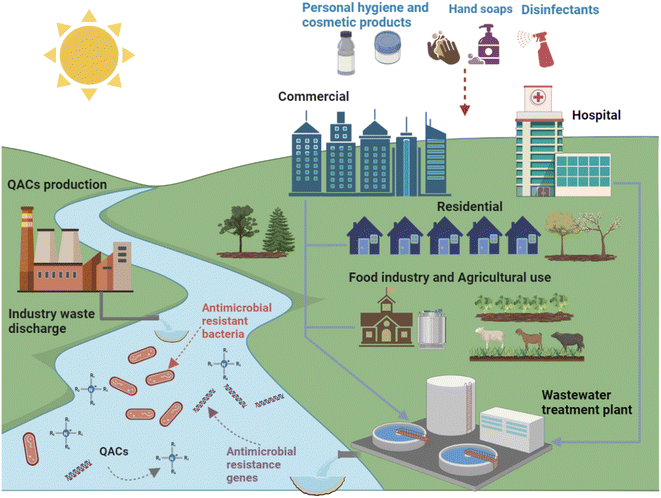
Since the invention of BAC in 1935, QACs have been extensively used for a wide variety of purposes (Fig. 2).34 In addition to being the main active ingredient in disinfectants, QACs can also be used as fabric softeners, surfactants, antistatic agents and wood preservatives. In the United States, BACs are used in applications on indoor and outdoor surfaces, humidifiers, decorative ponds, fountains, and more.35 The global QACs market was valued at $963.7 million in 2019, and it will continue to grow at a rate of 6.8% after the COVID-19 pandemic to reach $1.63 billion by 2027 due to increased demand for disinfectant products from hospitals, clinics, the food industry, and households.36 Currently over 200 disinfectants recommended by the US Environmental Protection Agency for use against COVID-19 contain QACs.33 Thus, it should be anticipated that the amounts of QACs used and released to the environment will increase. Previous studies showed that QACs have been detected in a variety of natural and engineered environments, including drinking water, surface water, wastewater, biosolids, soil, house dust, and sediments, due to their widespread use.37–42 It is estimated that approximately 75% of QACs used end up in sewers and wastewater treatment plants (WWTPs).43 The majority of QACs (>90%) in influent wastewater can be removed via aerobic biodegradation and sorption to the activated sludge. The sludge is then transferred to solids handling processes such as anaerobic digestion where the solids are stabilized and then, in some cases, land applied.28,44–46
Pre-pandemic concentrations of QACs in WWTPs were typically hundreds μg L−1 in influents and ng L−1 in effluents, and mid to high μg g−1 range for sewage sludge.34,40,47–50 While degradation of QACs could occur via biodegradation and photolysis, both degradation pathways are slow and thus QACs are persistent in various environments.51 QACs tend to accumulate in anaerobic media such as anaerobic digesters and sediments.28,43,52 Unfortunately, long-term QAC exposure not only negatively impacts the performance of microorganisms in industrial processes, but also may alter environmental microorganism community structure via enrichment of microorganisms with QAC and antibiotic resistance, leading to the dissemination of antibiotic resistance.53–56 However, the impact of chronic QAC exposure on WWTP performance, i.e., biological reactor treatment efficiency, and the development of antibiotic resistance are still major research gaps. Specifically, there is still a lack of sufficient direct evidence to elucidate whether, and how, environmental levels of QACs affect the activity and resistance of microbial communities in complex engineering environments. Thus, the broad objective of this review is to summarize the state of knowledge regarding QAC impacts on antibiotic resistance and process performance in WWTPs. Specifically, this review covers the general antimicrobial activity of QACs as well as the mechanisms by which QACs impact antibiotic resistance, and then highlights how the environmental concentrations of QACs could impact antibiotic resistance and biological processes in WWTPs. Critical research gaps and proposed research plans to fill these gaps are presented. In general, this paper provides a comprehensive discussion on QACs as environmental stressors that could impact treatment process performance and antibiotic resistance.
2. Antimicrobial activity of QACs and resistance mechanisms of bacteria
2.1 Mode of action
QACs have antimicrobial properties that inhibit bacteria, yeasts, molds, and viruses, among other microorganisms.35,57,58 The antimicrobial activity of individual QACs varies based on chemical properties including hydrophobicity, charge, and chemical functional groups.23,32,59,60 In contrast to antibiotics which work on specific target sites, QACs exert their microbial inactivation by interacting with multiple target sites. The known modes of antimicrobial action include disruption of the cell membrane, inhibition of microbial aerobic respiratory system, precipitation of cytoplasmic material, and deactivation of the protective lipid coating of viruses.9,61–63The interaction between QACs and the membranes of the target cells is the essential component of their antibacterial activity, which means QACs are broad-spectrum antimicrobials.64–66 The positively charged quaternary nitrogen of QACs interacts with the negatively charged head groups of phospholipids in bacterial membranes via electrostatic interactions. The hydrophobic chain of QACs enters the bacterial lipid bilayer of the cell membrane.67–69 In these processes, antimicrobial effects of QACs occur by disrupting membrane integrity, leading to leakage of cellular content.70,71 Potassium ions, phosphates, and larger molecular weight molecules are released. Depending on the chain length, the effectiveness of QACs against bacteria varies, and this is one reason many commercial disinfectants contain multiple QACs with varying chain lengths. In general, alkyl chain lengths of C10 to C16 have higher antibacterial activity, while twin-chained compounds like DADMACs have superior bioactivity against Gram-positive bacteria compared to BACs.68,72 When the concentration is below the minimum inhibitory concentration (MIC), QACs bind to anionic sites found on the membrane surface, which results in loss of membrane osmoregulation, inhibition of respiratory enzymes, dissipation of the proton motive force (which leads to decreased ATP production), and/or oxidative stress.43,66,73
QACs also act on intracellular targets such as bacterial DNA. A study indicated that nostocarboline (a bis-cationic QAC) exposure induced a SOS response following DNA damage in Escherichia coli, which may suggest some specific interaction with DNA.74 In another study, a mono-cationic QAC with aromatic rings displayed considerable inhibitory potencies to Escherichia coli DNA gyrase supercoiling assays.75 Meanwhile, a newly synthesized mono-cationic QAC has been found to bind with DNA to block the synthesis of enzyme and receptor, and inhibit the migration of genomic DNA.76,77
2.2 Tolerance and resistance to QACs
Generally speaking, tolerance is the capacity of microorganisms to survive a transient exposure to antibiotics or disinfectants without a change in the mini MIC.78 Understanding when microorganisms are more tolerant to QACs could mean that a greater exposure time to QACs, rather than a greater concentration of QACs, is required to produce the same extent of disinfection.78 Mechanisms of tolerance to QACs is generally correlated with decreased growth rates, transcriptional modification of the density and structure of porins, and expression of efflux pumps.79–81 In contrast to tolerance, resistance is the heritable ability of microorganisms to maintain biological activity at QAC concentrations above the MIC. QAC resistance is typically associated with modification of porins, regulatory hyperexpression of efflux pumps, enzymatic degradation, and acquisition of QAC resistance genes through horizontal genes transfer (HGT) (see Fig. 3).43,45,73,82–88 It is important to highlight that effectiveness of QACs is decreased by mechanisms of bacterial resistance; specifically, a larger dose of the QACs is needed to have the same impact on a resistant strain as it does on a susceptible strain.89,90 Unfortunately, current studies of QAC susceptibility do not distinguish between tolerance and resistance, which may result in the misclassification of tolerant microorganisms as resistant.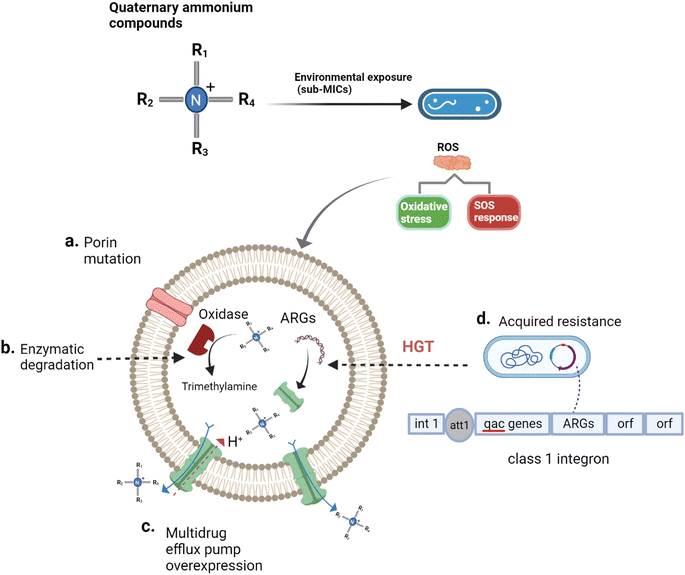 | ||
| Fig. 3 Mechanisms of QACs tolerance and resistance.35,66,86–88,91–94 QACs stimulate microorganisms to produce reactive oxygen species (ROS). ROS impact the expression of efflux pumps, promote porin mutation, and frequency of horizontal gene transfer (HGT) via oxidative stress and SOS response. (a) QACs exposure reduces the number of porins on the cell surface and alters the porin structure to decrease the permeability of cell surface which could prevent the passage of QACs into the cell, (b) the hydroxylation and oxidation of QACs via enzymes, (c) QACs activate the regulatory system of multidrug efflux pumps leading to their overexpression and QAC removal using proton gradients, (d) QACs promote HGT via an increase in the conjugation rate. | ||
The structure of the cell wall and cell membrane directly influence the tolerance and resistance of microorganisms to QACs.80 For instance, QACs are more effective at inhibiting Gram-positive bacteria and algae compared with Gram-negative bacteria and molds because the outer membrane of Gram-negative bacteria act as a permeability barrier to limit the entry of QACs into a cell.73,95 For example, Gram-negative bacteria including Escherichia coli, Salmonella typhimurium, and Proteus mirabilis exhibit less susceptibility to QACs or other disinfectants.63,91 Less acidic outer membrane lipopolysaccharides (LPS), small porins caused by strong LPS–LPS linkages, fewer porins, and a slime layer are physiological features that provide QAC tolerance.73,87,91,96 In addition, mycobacteria have significant resistance to QACs due to their unusual cell wall made up of a hydrophobic mycolate layer and a peptidoglycan layer connected by the polysaccharide arabinogalactan.97 Thus, the affinity of the outer cellular structure is one of the key determinants of QAC resistance.
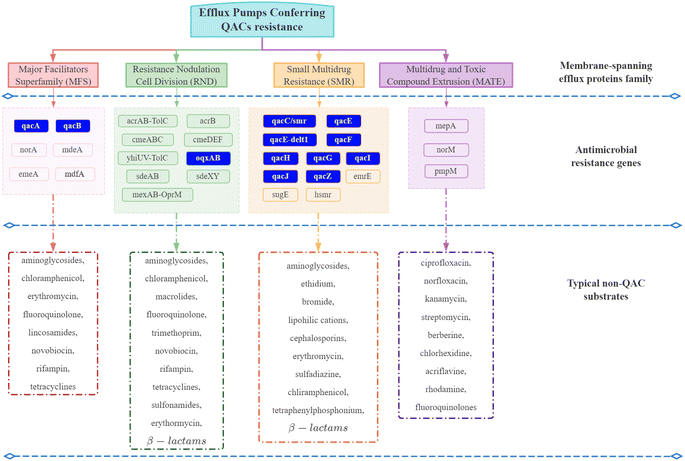 | ||
| Fig. 4 Efflux pumps conferring QAC resistance. The efflux pumps in the blue box are often plasmid-encoded. Others are primarily chromosome-encoded.92–94,99–101,103–108,119,126–128 | ||
Microorganisms metabolize QACs under aerobic conditions via three pathways. The main difference between these pathways lies in the carbon location where the hydroxylation takes place. In the first pathway, hydroxylation of the alkyl chain at the terminal carbon takes place, followed by oxidation of the aldehyde group and then the acid group.130 Subsequently, the compounds are processed by β-oxidation-generating acetyl-CoA. This strategy was proposed by Dean-Raymond and Aleksander in a study of the decyltrimethylammonium salt (C10 ATMAC) biodegradation carried out by Xanthomonas.84 The results showed that decyl- and hexadecyltrimethylammonium bromides were decomposed by organisms derived from sewage and soil. A mixture consisting of individual strains of Pseudomonas and Xanthomonas grew in solutions containing C10 ATMAC as the sole carbon source.84 The second mechanism is the hydroxylation at the carbon adjacent to the central nitrogen followed by the central fission of the molecule resulting in separation of the hydrophobic and hydrophilic moieties.131 It was proposed that the hexadecyltrimethylammonium chloride (C16 ATMAC) degradation carried out by Pseudomonas began with N-dealkylation catalyzed by an oxygen/NADH-dependent monooxygenase and yielded trimethylamine.131 The first intermediate in the biodegradation pathway of C16 ATMAC was hexadecanol, which was oxidized to hexadecenoic acid by an alkanol dehydrogenase. The acid was further metabolized through β-oxidation. The third aerobic QAC degradation pathway involves hydroxylation at methyl C attached to the central N, followed by demethylation. This scheme was proposed for the degradation of dodecyltrimethyl ammonium chloride (C12 ATMAC) by Pseudomonas sp. strain 7-6 isolated from a WWTP.85
Aerobic biodegradation of QACs has been attributed mainly to bacterial species in the genera of Xanthomonas, Aeromonas, and Pseudomonas. Several microorganisms have been identified that are resistant to QACs and capable of QACs degradation including Xanthomonas,84Pseudomonas B1,131Pseudomonas Fluorescens TN4,132Pseudomonas spp. Strain 7-6,85Pseudomonas putida A ATCC 12633,129Pseudomonas nitroreducens B and DB,83 and Pseudomonas sp. BIOMIG1.133 The ability to aerobically degrade QACs depends largely on the presence of monooxygenases, dioxygenases, amine dehydrogenases, and/or amine oxidases. An amine oxidase can reduce the BAC toxicity to a bacteria 500 fold.83 In addition, Ertekin et al. characterized a gene cluster encoding for transporters; an integrase and a dioxygenase were involved in BAC biotransformation under aerobic conditions.133 Multi-drug efflux pump genes such as sugE, PmpM, mexAB-oprM and mexEF-oprN were enriched in a BAC-degrading community, which indicates that the efflux pumps and oxidase reinforce microbial resistance to QACs.38,83,134 The impact of BAC degradation products should also be investigated to determine if degradation is beneficial, or if potentially the degradation products impact antibiotic resistance as well.
3. How QACs can promote the spread of antibiotic resistance
Considering their biological activity and amphiphilic properties, QACs may play multiple roles in the development of antibiotic resistance. First, as biocides, QACs have potential to directly select for antibiotic resistant microorganisms. Secondly, QACs can stimulate horizontal gene transfer of antibiotic resistance genes (ARGs). Lastly, QACs could indirectly promote the transmission of ARGs by increasing the bioavailability of other stressors, thereby reducing the actual concentration of a stressor (e.g., metal or antibiotic) required to trigger a response. Additionally, QACs could increase cell membrane permeability which would allow stressors to more readily have an effect.3.1 Selection of antibiotic resistant microorganisms
Although QACs and antibiotics have different modes of action against microorganisms,70,87,98 QAC exposure can promote the development of intrinsic non-specific cross-resistance to QACs and antibiotics through expression of multi-drug resistant efflux pump genes and a reduction in surface permeability.54,105,119,135 For example, MFS and RND family efflux proteins have a broad range of substrates including nine kinds of antibiotics (Fig. 4).98,103Pseudomonas exposed to sub-inhibitory concentrations of mixed BACs (60% C12 BAC & 40% C14 BAC) had high levels of mexCD-oprJ multi-drug efflux pump gene expression, which could lead to resistance to BACs, chlorhexidine, quinolones, macrolides and six other antibiotics classes.9,53 Another study also confirmed that the expression of the multi-drug efflux pumps sugE-A, sugE-B, and acrB, increased when BAC was added to the growth media.136 Thus, QAC exposure can directly increase the expression of multi-drug resistant efflux pump genes. In addition, QAC resistance genes and ARGs are frequently located together (co-resistance) on mobile genetic elements, which could be another important reason for the development of antibiotic resistance due to QAC exposure.137–140 The first identified qac gene (qacA) was found on a transmissible plasmid that also contained β-lactam and heavy metal resistance genes.141 In that case, all three resistance genes may be co-selected and transmitted if the microorganism is exposed to one of these stressors. Co-selection caused by QAC exposure is usually associated with class I integrons. Approximately 1–5% of bacterial cells in soil, freshwater, and biofilms contain Class I integrons.117 Class I integrons can acquire diverse gene cassettes that confer adaptive phenotypes and move between chromosomal locations, species, and lineages.142 The qac cassette, which has potential to provide a survival advantage under the selective stress of QACs, is carried by approximately 50% of environmental class I integrons.118 Ghaly et al. proposed a modified hypothesis for integron assembly based on the structure of class I integrons,139 where one chromosomal class I integron carrying a qac gene cassette was captured by a Tn5090-like transposon to form Tn402. The position of this complex mosaic component on a broad host range IncP plasmid would then have allowed its conjugative transfer into a diversity of bacterial hosts when the selective stress caused by QACs was present.139 In Enterobacteriaceae, the qac genes have been frequently found in combination with ARGs to aminoglycosides, chloramphenicol, sulfonamides, trimethoprim, and β-lactams on large, transmissible plasmids (resistance plasmids) that carry integrons.143 Thus, co-selection and transfer of QAC resistance genes and ARGs has the ability to occur from QAC exposure.It is important to note, however, that even carrying QAC resistance genes does not necessarily confer a corresponding survival advantage to the microorganism. Cervinkova et al. investigated the adaptation of Staphylococcus aureus (SK982) carrying the QAC efflux pumps (qacA and norA) when exposed to BAC under various circumstances.144 The results showed that the expression of norA, one of the efflux pumps conferring QAC resistance belonging to MFS family, was relatively low regardless of the BAC concentration. The qacA gene was only overexpressed in the exponential phase of growth. Several studies also proposed that the role of QAC resistance genes may be overstated.89,145–147 It is crucial, therefore, to understand the practical significance of QAC resistance genes, while monitoring the residual concentration and persistence of QACs in environments to predict the fate of ARGs.
3.2 Promotion of horizontal gene transfer (HGT)
HGT is an important aspect affecting how microorganisms acquire antibiotic resistance and can be achieved through three pathways: conjugation, transformation, and transduction.148–152 HGT can be affected by various environmental factors, such as antibiotics, disinfectants, oxidants, nanomaterials, metals, and microplastics.153,154 QACs have also been confirmed to be HGT-promoting stressors. The low concentration exposure of different QACs (C10ATMAC, C12DADMAC, C12–16BAC) promoted conjugation frequency of the RP4-resistant plasmids in E. coli.54 C12BAC had the most obvious effect on the transconjugative efficiency, with a 15-fold increase at a concentration of 0.1 mg L−1. In other studies, the promotional effect of different QACs on conjugative transfer at sub-MIC concentrations exposure has also been demonstrated, and the facilitating ability is concentration and core structure dependent.155,156 QACs facilitate plasmid conjugation transfer via multiple pathways including increasing cell membrane permeability, altering the composition and content of extracellular polymeric substances (EPS), and regulating gene expression associated with plasmid conjugation.54,155,156 A recent study indicated that the ratio of extracellular polysaccharides and proteins increased with the exposure of QACs (C10 DADMAC and C12 BAC), which caused an increase in cellular hydrophobicity, thereby contributing to the intercellular contact between donors and recipients.156 Additionally, QACs regulate the transcriptional expression levels of the genes that code for quorum sensing regulator (luxS), global regulators (korA, korB, trbA), DNA replication and translocation (trbB) and mating pairing formation (trbB).156 Besides conjugation, QACs also have potential to facilitate the transformation of exogenous ARGs via multiple pathways. Jia et al. studied the effect of benzalkonium bromide (BB) and benzalkonium chloride (BC) on bacterial transformation at environmentally relevant concentrations (0–0.1 μg ml−1).157 BB and BC significantly enhanced the transformation of ARGs in E. coli via increased membrane permeability, improved bacterial flagellum motility and enhanced expression of genes related to secretion systems. These studies illustrate the potential risk associated with QAC exposure at environmental residual concentrations on the spread of antibiotic resistance. However, these findings are based on simple, artificially constructed HGT models. It is important to determine if QACs are equally effective in complex engineered environments, especially for antibiotic resistance development in WWTPs.3.3 Indirect promotion of antibiotic resistance through altered bioavailability
QACs as cationic surfactants have the potential to indirectly affect the transmission of ARGs via altering the bioavailability of poorly soluble antibiotics or other hydrophobic stressors in water.158 Previous studies showed that the dissolution of hydrophobic substances (i.e., antibiotics) in suspension requires a significant amount of energy to counteract the effect of increased area of liquid–solid interface and interfacial surface tension.159–161 QACs can lower the interfacial tension to reduce surface energy and alter the zeta potential of particles in the suspension system due to their amphiphilic property from hydrophobic alkyl groups and hydrophilic positively charged N atom.68,161–163 For instance, the solubility of erythromycin increased linearly with increasing ATMAC concentration.158 Due to electrostatic attractions, C16 ATMAC showed the highest molar solubilization capacity for erythromycin. Therefore, QAC exposure can theoretically facilitate the dispersion of the hydrophobic antibiotics/stressors in an aqueous solution and increase the thermodynamic stability of the system, which enhances mass transfer of antibiotics and increases toxicity, thereby subsequently increasing the selective pressure. A recent study by Wang et al. revealed the roles and critical mechanisms of anionic surfactants (sodium dodecylbenzenesulfonate, SDBS) on sulfadiazine (SDZ) stressing for ARGs during anaerobic fermentation.164 Evidently, the exogenous SDBS sped up the propagation of ARGs stressed by SDZ. In addition to enhancing SDZ solubility in the waste activated sludge fermentation system, the presence of SDBS also increased cell permeability and destroyed EPS, which hastened the SDZ interaction with ARG hosts and aided in the spread of ARGs. Although this is about the role of anionic surfactants (SDBS), previous studies have demonstrated that QAC cationic surfactants (ATMAC) had a higher solubilization capability in comparison to nonionic (Brij) and anionic surfactants (SDBS).158 Thus, QAC exposure may have a similar or stronger effect, especially in some important wastewater treatment processes where various contaminants co-occur, such as aerobic biological oxidation, nitrification/denitrification, and anaerobic digestion. However, there is still a lack of relevant studies about the role of QACs on antibiotic selective pressure in WWTPs, making it difficult to estimate the selection pressure of QACs in real-world settings.4. Linking QACs to antibiotic resistance in WWTPs
WWTPs are reservoirs for three key factors in antibiotic resistance development: selective stressors (such as QACs and antibiotics), resistance donors, and resistance recipients.40,165–168 While concentrations of QACs in consumer and industrial products are typically several orders of magnitude greater than necessary to kill bacteria, when QACs enter WWTPs their concentrations are diluted, exposing bacteria to sub-inhibitory concentrations of QACs (Fig. 5).19,40,50 Levels of QACs were reported in mg L−1 range in influent wastewater and μg L−1 range in effluent.34 As mentioned in Section 3, these sub-inhibitory concentrations of QACs can still lead to the spread of resistance. Some unit operations may have elevated QAC levels compared to other unit operations, particularly in solid treatment process like anaerobic digestion where QACs levels are as high as 500 mg kg−1.22 Due to the high densities and metabolic activities of microbes in biological processes (activated sludge tanks or anaerobic digesters), QACs may play a role in the spread of antibiotic resistance during wastewater treatment.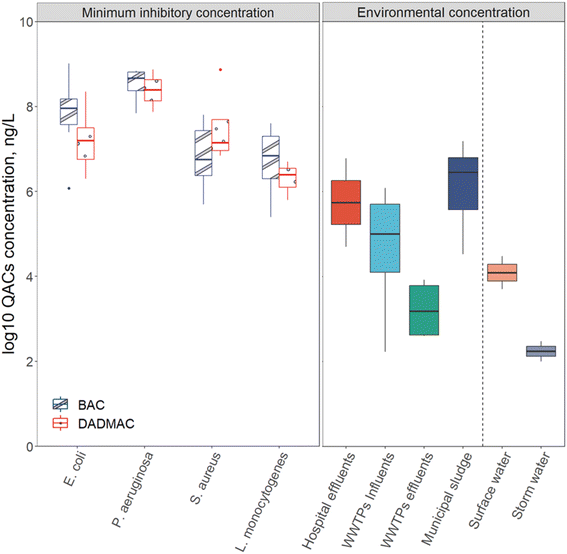 | ||
| Fig. 5 The QACs MIC of various isolates and total concentration of QACs in various environments. Left boxes are the MIC of BAC and DADMAC for different sensitive strains. Right boxes are range of total QACs concentrations found in each environmental setting. Biosolids (municipal sludge) concentrations were converted from mg kg−1 to mg L−1 by assuming 3% total solids in reactors that produce biosolids. The line extending above the box is the maximum and the line extending from the bottom of the box is the minimum. The box's up side represents the first quartile of the data, the line in the middle of the box represents the median of the data, and the box's bottom side represents the third quartile.46–50,73,106,169–185 | ||
In recent years, genes that encode for multi-drug efflux pumps and ARGs were frequently detected in WWTPs.186–190 Pärnänen et al. investigated the prevalence of ARGs in 12 WWTPs in seven European countries. Results showed that qacEΔ1 and sul1 (sulfonamide resistance gene) were core wastewater ARGs that were present in all influent and effluent samples. Another QAC resistance gene, qacH, persisted after treatment in 92.1% of the analyzed samples.191 Meanwhile, qacH was also detected in foam and activated sludge in wastewater treatment plants.192 In a recent study, Tan et al. investigated the abundance of ARGs and mobile genetic elements (e.g., integrases) in aerobic composting systems for sewage sludge, finding a high relative abundance of QACs resistance genes (qacEΔ1, qacH, sugE, emrE).165 The co-occurrence of QAC resistance genes, ARGs and MGEs was also detected in industrial wastewater.94,193,194 However, in most studies, no statistically significant correlation could be established between antibiotic consumption, antibiotic residues, and abundance of ARGs.195,196 This raised the speculation that other potential stressors in wastewater, as well as conditions and processes of wastewater treatment, may play an important role in the fate of ARGs. Of all the possible interfering factors, QAC concentration is one that cannot be ignored considering that residual concentrations are generally higher than those of antibiotics.96
Moreover, QAC resistance genes, specifically qacF, qacEΔ1, and qacH, were identified on the resistance plasmid pSp1 isolated from activated sludge bacteria residing in a municipal wastewater treatment plant and linked with other ARGs.197 As noted in Fig. 4, these genes confer resistance to QACs and antibiotics. IncP-1 resistance plasmids isolated from WWTP bacteria also have been found to encode QAC efflux pumps linked to ARGs such as streptomycin, β-lactam, and sulfonamide resistance genes.113 Similar associations were confirmed in three Swedish sewage treatment plants;198qacEΔ1, sul1, and the aminoglycoside resistance gene ant(3′′)-la were located with an int1 integrase gene in microorganisms in the influent and primary sludge. The most recent studies showed chronic exposure to QACs (C16 ATMAC, benzethonium chloride) significantly enriched QAC resistance genes (qacEΔ1 and qacH) and ARGs (aadA, aac(6′)-Ib and blaTEM) in a partial nitrification/anammox system.199–201 Thus, WWTPs may be a hotspot for the development and spread of QAC resistance genes and ARGs.166,202
Genes encoding efflux-driven QAC resistance are also abundant in natural aquatic environments, especially in rivers or lakes located downstream of WWTPs and livestock farms.203–207 A study by Chen investigated the prevalence, abundance, and distribution of the RND family efflux pump genes (acrA/B), which is responsible for resistance to QACs, and 20 other ARGs conferring resistance to five major classes of antibiotics including tetracycline, sulfonamide, quinolone, aminoglycoside, and macrolide in wastewater from 12 livestock farms and their downstream surface water.208 The results showed that the detected frequency of QAC resistance genes (arcA and arcB) were above 90%, although livestock farms are not an application scenario for QACs, indicating promiscuity of the enzymes or selection on other traits in the genome where the QAC resistance genes reside. The relative abundance of these QAC resistance genes is less than tetracycline and sulfonamide resistance genes, which are the two primary veterinary antibiotics used in animal husbandry. Moreover, arcA and arcB were also detected in drinking water sources with presumably no or little exposure to QACs.205 Canonical correlation analysis between antibiotic concentration and ARG subtype abundance revealed that ARG abundance was closely related with the total concentration of quinolones. Thus, exposure to antibiotics may lead to QAC resistance through efflux and, similarly, exposure to QACs could provide resistance to antibiotics. Likewise, a large abundance of QAC resistance genes (qacEΔ1, qacH) was reported in an estuary from China,155 indicating that estuary sediments are a reservoir of QAC resistance genes. The detection frequencies of QAC resistance genes (qacEΔ1 and qacH) were 100% in estuary sediments. Meanwhile, qacEΔ1, qacH-01, and oprD were the top three most abundant genes among the fifteen detected multidrug resistance genes in the studied estuary area. In addition to the above-mentioned environments that are highly disturbed by anthropogenic activities, 250 ARGs were subtypes found in both remote lakes (7.25 × 10−2 copies/16S rRNA) and marine environments (2.70 × 10−2 copies/16S rRNA), four multi-drug resistance genes (qacA, acrA, acrB, emrE) that confer QAC resistance were found,209 which may indirectly confirm that ARGs and QAC resistance genes are naturally present.
5. The potential impact of QACs on process performance in wastewater treatment plants
Due to the extensive use of QACs in medical, industrial, and personal consumer products, most of these compounds are released into WWTPs and then are dispersed into natural environments via liquid effluent discharges and land application of biosolids.34,96 Both the activated sludge process and anaerobic digestion serve as core technologies to treat wastewater, have a high solids content, and are therefore potentially impacted by QACs that enter the reactor sorbed to solids. A previous study showed that mixed BACs (40% C12 BAC, 50% C14 BAC, 10% C16 BAC) inhibited oxygen uptake and utilization in the activated sludge process, which hindered COD removal.45 Chen and Zhang also found that exposure to high BAC concentration affected aerobic respiratory behaviors and the tricarboxylic acid (TCA) cycle of microorganisms in membrane bioreactors, resulting in decreased ATP.210 Zhang proposed that the decrease in ATP production may be caused by damage to energy-production enzymes (e.g., adenylate kinase, ADK).79 These studies indicate that BACs can impact bulk carbon removal (BOD5), but they can also impact nutrient removal.Nutrient (nitrogen and phosphorus) removal is a key goal for wastwater treatment. Different biological processes can be employed to remove nitrogen from wastewater including nitrification, denitrification, and anammox.211–213 Nitrification is a two-step process. Ammonia oxidizing bacteria (AOB) convert ammonia to nitrite. Then, nitrite oxidizing bacteria (NOB) convert nitrite to nitrate. Denitrification occurs in the absence of oxygen under anoxic conditions using heterotrophic bacteria (usually Pseudomonas) to reduce nitrate to nitric oxide, nitrous oxide and nitrogen gas.214 Anammox is another nitrogen-removal technique for treating nitrogen-rich wastewater.215 Autotrophic anammox bacteria get energy from converting ammonium and nitrite simultaneously into nitrogen gas in the absence of oxygen. A recent study indicated that chronic exposure of benzethonium chloride (BEC) at low concentration (1–2 mg L−1) significantly inhibited the nitrification capacity of a lab-scale reactor.200 During the initial phase of exposure, the ammonia oxidation was inhibited, but recovered completely within 3 days with nitrite accumulation, which means ammonia oxidizing bacteria have excellent adaptability to BEC. In contrast, the activity of nitrite oxidizing bacteria (NOB) severely decreased with BEC exposure. BEC exposure converted nitrification to partial nitrification, but this effect was reversible. Other studies also showed BEC can selectively eliminate NOB in activated sludge at high levels (70 mg L−1).199,216 Meanwhile, BEC has potential to increase EPS secretion and alter protein secondary structure, while inhibiting the dominant NOB-Nitrospira.199 C16 ATMAC has also been found to affect the nitrogen removal performance via inhibiting the anammox (hzsB) gene at environmental concentrations (0.5 mg L−1).201 C16 ATMAC exposure enriched two types of anammox bacteria (Candidatus Jettenia and Candidatus Kuenenia) to maintain a good nitrogen removal performance in a partial nitrification/anammox system. In addition, QACs (C12 BAC, C12 ATMAC, C12 DADMAC) inhibited denitrification.217 With the increase of QACs concentration, the abundance of Thauera, a denitrifying genera, decreased significantly. At the time of this review, no papers were found that discussed how QACs impact biological phosphorus removal treatment processes. This lack of knowledge represents a key research gap that would be important to WWTPs that employ biological phosphorus removal.
The strong sorption capacity of activated sludge for QACs (C12 & C16 ATMAC, and C12 & C16 BAC) results in QACs entering solids handling processes such as anaerobic digestion.28 During anaerobic digestion, complex organic matter is converted to methane and carbon dioxide through hydrolysis, acidogenesis, acetogenesis, and methanogenesis.218 Under the stress of C12–C16 BAC and C8 and C10 DADMAC, both methanogenesis and acidogenesis were inhibited at concentrations ≥25 mg L−1 meaning that the overall anaerobic digestion process was inhibited and methane production was decreased.219 Another study indicated that BACs (C12, C14, and C16) caused release of soluble polysaccharides and proteins, which were positively related to the residual levels of BACs in an anaerobic digester.220 Higher concentrations or shorter alkyl chain lengths caused more inhibition of methane production and the accumulation of short chain fatty acids.221 The long-term stress of BACs (C12 BAC, C14 BAC, and C16 BAC) reduced microbial diversity and decreased the relative abundance of aceticlastic methanogens and increased the relative abundance of hydrogenotrophic methanogens.90,221 C12 BAC, with a shorter alkyl chain length than the other BACs in this study, had more significant effects on microbial diversity and shifted the main acid-producing bacteria from Proteiniclasticum and Intestinibacter to Candidatus Microthrix and Lutispora. Meanwhile, the proportion of Firmicutes, Bacteroidetes and Chloroflexi in the community decreased significantly under the impact of mixed BACs (69% C12, 23% C14, 3% C16), while Proteobacteria and Actinobacteria were increased. The effect of BAC (C12–C16) on Archaea in anaerobic digestion is mainly reflected in the screening of methanogenic trophic types.220 BACs led to a shift in methanogens from Methanosaeta, a strictly aceticlastic methanogen group, to hydrogenotrophic methanogens, such as Methanobacterium and Methanobrevibacter.220,221 Overall more volatile fatty acids were formed and less methane was produced. It is still unclear, however, which microorganisms responsible for anaerobic digestion are most significantly affected by QACs. Minor impacts could include changes to the microbial community structure with no loss in treatment efficiency. If there is functional redundancy between QAC-sensitive microorganisms and QAC-tolerant microorganisms, then QAC impacts would be expected to be minor. Conversely, major impacts could include loss of process function at concentrations not-yet-seen in an anaerobic digester. Furthermore, it is important to determine if and how microbial communities respond after QAC stressors are removed to know if the impacts are reversible.222
6. Conclusions and research gaps
QACs are commonly used cationic disinfectants and surfactants, and a vast amount of QACs enter WWTPs following usage by consumers. QACs disrupt microbial cell membranes leading to gradual leakage of cytoplasmic components out of the cell, thereby inhibiting microbial growth. As a result, QAC resistant microorganisms and QAC resistance genes may be selected and enriched under specific conditions in WWTPs. Due to the mechanism of QAC resistance and the fact that QAC resistance genes are often encoded on mobile genetic elements with other ARGs, persistent residues of QACs may lead to co-transformation and co-accumulation of QAC resistance genes and ARGs in WWTPs, further exacerbating the growing risk of antibiotic resistance. Meanwhile, microbial community structure and process performance of WWTPs could be affected by long-term exposure to QACs.Against the backdrop of increased use of disinfectants such as QACs due to the COVID-19 pandemic, there are still many knowledge gaps that need to be filled to clarify the role of QACs on the development of antibiotic resistance. First, it is not known if exposure to QACs contributes to the rise of antibiotic resistant pathogens in complex engineered environments. Although previous studies revealed that QAC exposure can promote the development of antibiotic resistance,155,156,223,224 evidence is lacking about how QACs impact clinical antibiotic resistance. The impact of the coexistence of QACs and other stressors on antibiotic resistance is also an important knowledge gap. Antibiotics, disinfectants, and heavy metals are common selective pressures leading to antibiotic resistance in wastewater. Although they have different modes of action on microorganisms, these chemicals may trigger the same resistance mechanisms, such as upregulation of multi-drug efflux pumps. In addition, QACs as surfactants have potential to alter the bioavailability of other organic compounds. Thus, the simultaneous presence of multiple stressors may significantly reduce the threshold of effect concentration of a single stressor, leading to an easier transfer of resistance genes. Additionally, the synergistic effect of multiple contaminants may create oxidative stress, thereby enhancing the capture efficiency of mobile genetic elements to antibiotic resistance cassettes, and the frequency of HGT. Studies pursuing these issues could further elucidate the molecular mechanisms of cross-resistance and co-resistance between QACs and other stressors.
Complex microbial environments may be conducive to the spread and dissemination of antibiotic resistance. The microbial community structure in wastewater and different unit operations such as aerobic nitrification reactors or anaerobic digesters can vary widely. The sensitivity of different microbial communities to QACs also varies. It is important, therefore, to understand which microbial communities are most susceptible to changes induced by QACs. The conventional physicochemical characteristics of environmental systems, such as temperature, ionic strength, pH, and organic matter content may also play a role in QACs affecting the susceptibility of specific microbial communities to antibiotic resistance.
In addition, it is not known how the core chemical structure of QACs is linked to impacts on biological wastewater treatment process performance, including nitrification, denitrification, annamox, biological phosphorus removal, and anaerobic digestion. Although previous studies have revealed the acute impacts of select BACs, ATMACs and DADMACs on the activated sludge process and anaerobic digestion, structural differences in QACs may significantly affect bioavailability and toxicity. Furthermore, the impacts of EtBACs on any of these biological wastewater treatment processes are yet to be studied. Overall, as the amount, and types, of QACs in consumer usage continues to grow, so too must our understanding of environmental impacts. Only then can we make the best-informed decisions to protect public health and ensure stability of environmental engineering treatment systems.
Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This work was funded by the National Science Foundation through projects CBET 2051336 and 2051313.References
- C. J. Murray, K. S. Ikuta, F. Sharara, L. Swetschinski, G. Robles Aguilar and A. Gray, et al., Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis, Lancet, 2022, 399(10325), 629–655 CrossRef CAS PubMed.
- P. Dadgostar, Antimicrobial Resistance: Implications and Costs, Infect. Drug Resist., 2019, 12, 3903–3910 CrossRef CAS PubMed.
- OECD, Stemming the Superbug Tide: Just A Few Dollars More, OECD, 2018, [cited 2023 Feb 20]. (OECD Health Policy Studies), available from: https://www.oecd-ilibrary.org/social-issues-migration-health/stemming-the-superbug-tide_9789264307599-en.
- Centers for Disease Control and Prevention (U.S.), Antibiotic resistance threats in the United States, Centers for Disease Control and Prevention (U.S.), 2019 Nov, [cited 2023 Feb 20], available from: https://stacks.cdc.gov/view/cdc/82532.
- European Centre for Disease Prevention and Control, Antimicrobial resistance in the EU/EEA (EARS-Net): annual epidemiological report for 2019, LU: Publications Office, 2020, [cited 2023 Feb 20], available from: https://data.europa.eu/doi/10.2900/10103.
- G. M. Knight, R. E. Glover, C. F. McQuaid, I. D. Olaru, K. Gallandat and Q. J. Leclerc, et al., Antimicrobial resistance and COVID-19: Intersections and implications, eLife, 2021, 10, e64139 CrossRef CAS PubMed.
- C. Wang, D. Mantilla-Calderon, Y. Xiong, M. Alkahtani, Y. M. Bashawri and H. Al Qarni, et al., Investigation of Antibiotic Resistome in Hospital Wastewater during the COVID-19 Pandemic: Is the Initial Phase of the Pandemic Contributing to Antimicrobial Resistance?, Environ. Sci. Technol., 2022, 56(21), 15007–15018 CrossRef CAS PubMed.
- COVID-19: U.S. Impact on Antimicrobial Resistance, Special Report 2022, National Center for Emerging and Zoonotic Infectious Diseases, 2022 Jun, [cited 2023 Feb 20], available from: https://stacks.cdc.gov/view/cdc/117915.
- K. Hegstad, S. Langsrud, B. T. Lunestad, A. A. Scheie, M. Sunde and S. P. Yazdankhah, Does the Wide Use of Quaternary Ammonium Compounds Enhance the Selection and Spread of Antimicrobial Resistance and Thus Threaten Our Health?, Microb. Drug Resist., 2010, 16(2), 91–104 CrossRef CAS PubMed.
- R. Laxminarayan, The overlooked pandemic of antimicrobial resistance, Lancet, 2022, 399(10325), 606–607 CrossRef CAS PubMed.
- S. Ansari, J. P. Hays, A. Kemp, R. Okechukwu, J. Murugaiyan and M. D. Ekwanzala, et al., The potential impact of the COVID-19 pandemic on global antimicrobial and biocide resistance: an AMR Insights global perspective, JAC-Antimicrob. Resist., 2021, 3(2), dlab038 CrossRef PubMed.
- J. Y. Maillard, S. Bloomfield, J. R. Coelho, P. Collier, B. Cookson and S. Fanning, et al., Does Microbicide Use in Consumer Products Promote Antimicrobial Resistance? A Critical Review and Recommendations for a Cohesive Approach to Risk Assessment, Microb. Drug Resist., 2013, 19(5), 344–354 CrossRef PubMed.
- D. E. Carey and P. J. McNamara, The impact of triclosan on the spread of antibiotic resistance in the environment, Front Microbiol., 2015, 5, [cited 2023 Oct 18], available from: https://www.frontiersin.org/articles/10.3389/fmicb.2014.00780.
- D. E. Carey, D. H. Zitomer, K. R. Hristova, A. D. Kappell and P. J. McNamara, Triclocarban Influences Antibiotic Resistance and Alters Anaerobic Digester Microbial Community Structure, Environ. Sci. Technol., 2016, 50(1), 126–134 CrossRef CAS PubMed.
- P. J. McNamara, T. M. LaPara and P. J. Novak, The Impacts of Triclosan on Anaerobic Community Structures, Function, and Antimicrobial Resistance, Environ. Sci. Technol., 2014, 48(13), 7393–7400 CrossRef CAS PubMed.
- D. E. Carey, D. H. Zitomer, A. D. Kappell, M. J. Choi, K. R. Hristova and P. J. McNamara, Chronic exposure to triclosan sustains microbial community shifts and alters antibiotic resistance gene levels in anaerobic digesters, Environ. Sci.: Processes Impacts, 2016, 18(8), 1060–1067 RSC.
- K. R. Harrison, A. D. Kappell and P. J. McNamara, Benzalkonium chloride alters phenotypic and genotypic antibiotic resistance profiles in a source water used for drinking water treatment, Environ. Pollut., 2020, 257, 113472 CrossRef CAS PubMed.
- D. E. Carey and P. J. McNamara, Altered antibiotic tolerance in anaerobic digesters acclimated to triclosan or triclocarban, Chemosphere, 2016, 163, 22–26 CrossRef CAS PubMed.
- P. I. Hora, S. G. Pati, P. J. McNamara and W. A. Arnold, Increased Use of Quaternary Ammonium Compounds during the SARS-CoV-2 Pandemic and Beyond: Consideration of Environmental Implications, Environ. Sci. Technol. Lett., 2020, 7(9), 622–631 CrossRef CAS PubMed.
- E. Tuladhar, M. C. de Koning, I. Fundeanu, R. Beumer and E. Duizer, Different Virucidal Activities of Hyperbranched Quaternary Ammonium Coatings on Poliovirus and Influenza Virus, Appl. Environ. Microbiol., 2012, 78(7), 2456–2458 CrossRef CAS PubMed.
- N. Baker, A. J. Williams, A. Tropsha and S. Ekins, Repurposing Quaternary Ammonium Compounds as Potential Treatments for COVID-19, Pharm. Res., 2020, 37(6), 104 CrossRef CAS PubMed.
- W. A. Arnold, A. Blum, J. Branyan, T. A. Bruton, C. C. Carignan and G. Cortopassi, et al., Quaternary Ammonium Compounds: A Chemical Class of Emerging Concern, Environ. Sci. Technol., 2023, 57(20), 7645–7665 CrossRef CAS PubMed.
- F. Bureš, Quaternary Ammonium Compounds: Simple in Structure, Complex in Application, Top. Curr. Chem., 2019, 377(3), 14 CrossRef PubMed.
- S. C. Marteinson, M. J. Lawrence, Z. E. Taranu, K. Kosziwka, J. J. Taylor and A. Green, et al., Increased use of sanitizers and disinfectants during the COVID-19 pandemic: identification of antimicrobial chemicals and considerations for aquatic environmental contamination, Environ. Rev., 2022, 31(1), 76–94 CrossRef.
- H. S. J. Al-Maliki, R. A. F. Al-jaberi, A. A. H. Kadhum, M. E. Al-Gazally, M. Rahem and S. H. A. Al-Rekabi, The Role of Immune System and Sterilization on the Covid-19 Spread Control, Syst. Rev. Pharm., 2021, 12(1), 579–592 CAS.
- K. A. Nguyen, H. Förster and J. E. Adaskaveg, Quaternary Ammonium Compounds as New Sanitizers for Reducing the Spread of the Olive Knot Pathogen on Orchard Equipment, Plant Dis., 2017, 101(7), 1188–1193 CrossRef CAS PubMed.
- R. S. Boethling and D. G. Lynch, Quaternary Ammonium Surfactants, in Detergents, Anthropogenic Compounds, ed. N. T. de Oude, Springer, Berlin, Heidelberg, 1992, pp. 145–77, [cited 2023 Feb 20], available from: DOI:10.1007/978-3-540-47108-0_3.
- Z. Z. Ismail, U. Tezel and S. G. Pavlostathis, Sorption of quaternary ammonium compounds to municipal sludge, Water Res., 2010, 44(7), 2303–2313 CrossRef CAS PubMed.
- G. P. Zanini, R. G. Ovesen, H. C. B. Hansen and B. W. Strobel, Adsorption of the disinfectant benzalkonium chloride on montmorillonite. Synergistic effect in mixture of molecules with different chain lengths, J. Environ. Manage., 2013, 128, 100–105 CrossRef CAS PubMed.
- L. Xiang, T. F. Sun, M. J. Zheng, Y. W. Li, H. Li and M. H. Wong, et al., Sorption of dodecyltrimethylammonium chloride (DTAC) to agricultural soils, Sci. Total Environ., 2016, 560–561, 197–203 CrossRef CAS PubMed.
- A. H. Khan, S. M. Macfie and M. B. Ray, Sorption and leaching of benzalkonium chlorides in agricultural soils, J. Environ. Manage., 2017, 196, 26–35 CrossRef CAS PubMed.
- M. N. Nadagouda, P. Vijayasarathy, A. Sin, H. Nam, S. Khan and J. B. M. Parambath, et al., Antimicrobial activity of quaternary ammonium salts: structure-activity relationship, Med. Chem. Res., 2022, 31(10), 1663–1678 CrossRef CAS.
- US EPA O, About List N: Disinfectants for Coronavirus (COVID-19), 2020, [cited 2023 Feb 20], available from: https://www.epa.gov/coronavirus/about-list-n-disinfectants-coronavirus-covid-19-0.
- C. Zhang, F. Cui, G. M. Zeng, M. Jiang, Y. Z. zhu and Z. gang Yu, et al., Quaternary ammonium compounds (QACs): A review on occurrence, fate and toxicity in the environment, Sci. Total Environ., 2015, 518–519, 352–362 CrossRef CAS PubMed.
- B. Merchel Piovesan Pereira and I. Tagkopoulos, Benzalkonium Chlorides: Uses, Regulatory Status, and Microbial Resistance, Appl. Environ. Microbiol., 2019, 85(13), e00377 CrossRef PubMed.
- Fact.MR – Quaternary Ammonium Salts Market Analysis By Product Type (Benzyldimethyldecyl Ammonium Chloride, Benzyldimethyldodecyl Ammonium Chloride), By Function (Flocculant, Drilling, Fluids, Antistatic Agents, Disinfectants), By End Use & Regional Forecast, 2022–2032, [cited 2023 Feb 14], available from: https://www.factmr.com/report/quaternary-ammonium-salts-market.
- I. Ferrer and E. T. Furlong, Identification of Alkyl Dimethylbenzylammonium Surfactants in Water Samples by Solid-Phase Extraction Followed by Ion Trap LC/MS and LC/MS/MS, Environ. Sci. Technol., 2001, 35(12), 2583–2588 CrossRef CAS PubMed.
- A. S. Liffourrena and G. I. Lucchesi, Identification, Cloning and Biochemical Characterization of Pseudomonas putida A (ATCC 12633) Monooxygenase Enzyme necessary for the Metabolism of Tetradecyltrimethylammonium Bromide, Appl. Biochem. Biotechnol., 2014, 173(2), 552–561 CrossRef CAS PubMed.
- A. K. Mahony, P. J. McNamara and W. A. Arnold, Quaternary Ammonium Compounds (QACs) in Wastewater Influent and Effluent Throughout the COVID-19 Pandemic, Environ. Sci. Technol., 2023 DOI:10.1021/acs.est.3c04413.
- M. Östman, R. H. Lindberg, J. Fick, E. Björn and M. Tysklind, Screening of biocides, metals and antibiotics in Swedish sewage sludge and wastewater, Water Res., 2017, 115, 318–328 CrossRef PubMed.
- W. L. Li, Z. F. Zhang, C. Sparham and Y. F. Li, Validation of sampling techniques and SPE-UPLC/MS/MS for home and personal care chemicals in the Songhua Catchment, Northeast China, Sci. Total Environ., 2020, 707, 136038 CrossRef CAS PubMed.
- G. Zheng, G. M. Filippelli and A. Salamova, Increased Indoor Exposure to Commonly Used Disinfectants during the COVID-19 Pandemic, Environ. Sci. Technol. Lett., 2020, 7(10), 760–765 CrossRef CAS PubMed.
- U. Tezel and S. G. Pavlostathis, Quaternary ammonium disinfectants: microbial adaptation, degradation and ecology, Curr. Opin. Biotechnol., 2015, 33, 296–304 CrossRef CAS PubMed.
- A. H. Khan, E. Topp, A. Scott, M. Sumarah, S. M. Macfie and M. B. Ray, Biodegradation of benzalkonium chlorides singly and in mixtures by a Pseudomonas sp. isolated from returned activated sludge, J. Hazard. Mater., 2015, 299, 595–602 CrossRef CAS PubMed.
- C. Zhang, U. Tezel, K. Li, D. Liu, R. Ren and J. Du, et al., Evaluation and modeling of benzalkonium chloride inhibition and biodegradation in activated sludge, Water Res., 2011, 45(3), 1238–1246 CrossRef CAS PubMed.
- G. G. Ying, Fate, behavior and effects of surfactants and their degradation products in the environment, Environ. Int., 2006, 32(3), 417–431 CrossRef CAS PubMed.
- T. Ruan, S. Song, T. Wang, R. Liu, Y. Lin and G. Jiang, Identification and Composition of Emerging Quaternary Ammonium Compounds in Municipal Sewage Sludge in China, Environ. Sci. Technol., 2014, 48(8), 4289–4297 CrossRef CAS PubMed.
- E. Martínez-Carballo, A. Sitka, C. González-Barreiro, N. Kreuzinger, M. Fürhacker and S. Scharf, et al., Determination of selected quaternary ammonium compounds by liquid chromatography with mass spectrometry. Part I. Application to surface, waste and indirect discharge water samples in Austria, Environ. Pollut., 2007, 145(2), 489–496 CrossRef PubMed.
- E. Martínez-Carballo, C. González-Barreiro, A. Sitka, N. Kreuzinger, S. Scharf and O. Gans, Determination of selected quaternary ammonium compounds by liquid chromatography with mass spectrometry. Part II. Application to sediment and sludge samples in Austria, Environ. Pollut., 2007, 146(2), 543–547 CrossRef PubMed.
- S. G. Pati and W. A. Arnold, Comprehensive screening of quaternary ammonium surfactants and ionic liquids in wastewater effluents and lake sediments, Environ. Sci.: Processes Impacts, 2020, 22(2), 430–441 RSC.
- P. I. Hora and W. A. Arnold, Photochemical fate of quaternary ammonium compounds in river water, Environ. Sci.: Processes Impacts, 2020, 22(6), 1368–1381 RSC.
- R. J. T. Kleijwegt, W. Winkenwerder, W. Baan and J. van der Schaaf, Degradation kinetics and solvent effects of various long-chain quaternary ammonium salts, Int. J. Chem. Kinet., 2022, 54(1), 16–27 CrossRef CAS.
- M. Kim, M. R. Weigand, S. Oh, J. K. Hatt, R. Krishnan and U. Tezel, et al., Widely Used Benzalkonium Chloride Disinfectants Can Promote Antibiotic Resistance. Dozois CM, Appl. Environ. Microbiol., 2018, 84(17), e01201–e01218 CAS.
- Y. Han, Z. C. Zhou, L. Zhu, Y. Y. Wei, W. Q. Feng and L. Xu, et al., The impact and mechanism of quaternary ammonium compounds on the transmission of antibiotic resistance genes, Environ. Sci. Pollut. Res., 2019, 26(27), 28352–28360 CrossRef CAS PubMed.
- M. Zhao, J. Gao, H. Zhang, Y. Cui, Z. Wang and Y. Zhao, et al., Quaternary ammonium compounds promoted anoxic sludge granulation and altered propagation risk of intracellular and extracellular antibiotic resistance genes, J. Hazard. Mater., 2023, 445, 130464 CrossRef CAS PubMed.
- K. Yang, M. L. Chen and D. Zhu, Exposure to benzalkonium chloride disinfectants promotes antibiotic resistance in sewage sludge microbiomes, Sci. Total Environ., 2023, 867, 161527 CrossRef CAS PubMed.
- D. L. Fredell, Biological Properties and Applications of Cationic Surfactants, in Cationic Surfactants, CRC Press, 1994 Search PubMed.
- C. L. Schrank, K. P. C. Minbiole and W. M. Wuest, Are Quaternary Ammonium Compounds, the Workhorse Disinfectants, Effective against Severe Acute Respiratory Syndrome-Coronavirus-2?, ACS Infect. Dis., 2020, 6(7), 1553–1557 CrossRef CAS PubMed.
- T. G. Osimitz and W. Droege, Quaternary ammonium compounds: perspectives on benefits, hazards, and risk, Toxicol. Res. Appl., 2021, 5, 239784732110490 Search PubMed.
- W. Dan, J. Gao, X. Qi, J. Wang and J. Dai, Antibacterial quaternary ammonium agents: Chemical diversity and biological mechanism, Eur. J. Med. Chem., 2022, 243, 114765 CrossRef CAS PubMed.
- T. G. Osimitz and W. Droege, Adverse Outcome Pathway for Antimicrobial Quaternary Ammonium Compounds, J. Toxicol. Environ. Health, Part A, 2022, 85(12), 494–510 CrossRef CAS PubMed.
- Â. S. Inácio, G. N. Costa, N. S. Domingues, M. S. Santos, A. J. M. Moreno and W. L. C. Vaz, et al., Mitochondrial Dysfunction Is the Focus of Quaternary Ammonium Surfactant Toxicity to Mammalian Epithelial Cells, Antimicrob. Agents Chemother., 2013, 57(6), 2631–2639 CrossRef PubMed.
- M. Tischer, G. Pradel, K. Ohlsen and U. Holzgrabe, Quaternary Ammonium Salts and Their Antimicrobial Potential: Targets or Nonspecific Interactions?, ChemMedChem, 2012, 7(1), 22–31 CrossRef CAS PubMed.
- D. B. Vieira, Cationic lipids and surfactants as antifungal agents: mode of action, J. Antimicrob. Chemother., 2006, 58(4), 760–767 CrossRef CAS PubMed.
- C. J. Ioannou, G. W. Hanlon and S. P. Denyer, Action of Disinfectant Quaternary Ammonium Compounds against Staphylococcus aureus, Antimicrob. Agents Chemother., 2007, 51(1), 296–306 CrossRef CAS PubMed.
- J. Y. Maillard, Bacterial target sites for biocide action: Antibacterial Mechanisms Of Action Of Biocides, J. Appl. Microbiol., 2002, 92, 16S–27S CrossRef PubMed.
- S. P. Denyer, Mechanisms of action of antibacterial biocides, Int. Biodeterior. Biodegrad., 1995, 36(3–4), 227–245 CrossRef CAS.
- C. P. Gerba, Quaternary Ammonium Biocides: Efficacy in Application, Appl. Environ. Microbiol., 2015, 81(2), 464–469 CrossRef PubMed.
- Z. Zhou, S. Zhou, X. Zhang, S. Zeng, Y. Xu and W. Nie, et al., Quaternary Ammonium Salts: Insights into Synthesis and New Directions in Antibacterial Applications, Bioconjugate Chem., 2023, 34(2), 302–325 CrossRef CAS PubMed.
- P. Gilbert and L. E. Moore, Cationic antiseptics: diversity of action under a common epithet, J. Appl. Microbiol., 2005, 99(4), 703–715 CrossRef CAS PubMed.
- S. Wessels and H. Ingmer, Modes of action of three disinfectant active substances: A review, Regul. Toxicol. Pharmacol., 2013, 67(3), 456–467 CrossRef CAS PubMed.
- M. C. Jennings, K. P. C. Minbiole and W. M. Wuest, Quaternary Ammonium Compounds: An Antimicrobial Mainstay and Platform for Innovation to Address Bacterial Resistance, ACS Infect. Dis., 2015, 1(7), 288–303 CrossRef CAS PubMed.
- G. McDonnell and A. D. Russell, Antiseptics and Disinfectants: Activity, Action, and Resistance, Clin. Microbiol. Rev., 1999, 12(1), 147–179 CrossRef CAS PubMed.
- H. H. Locher, D. Ritz, P. Pfaff, M. Gaertner, A. Knezevic and D. Sabato, et al., Dimers of Nostocarboline with Potent Antibacterial Activity, Chemotherapy, 2010, 56(4), 318–324 CrossRef CAS PubMed.
- J. Fedorowicz, J. Sączewski, A. Konopacka, K. Waleron, D. Lejnowski and K. Ciura, et al., Synthesis and biological evaluation of hybrid quinolone-based quaternary ammonium antibacterial agents, Eur. J. Med. Chem., 2019, 179, 576–590 CrossRef CAS PubMed.
- S. V. Sapozhnikov, A. E. Sabirova, N. V. Shtyrlin, A. Y. Druk, M. N. Agafonova and M. N. Chirkova, et al., Design, synthesis, antibacterial activity and toxicity of novel quaternary ammonium compounds based on pyridoxine and fatty acids, Eur. J. Med. Chem., 2021, 211, 113100 CrossRef CAS PubMed.
- X. Wang, H. Qiu, N. Yang, H. Xie, W. Liang and J. Lin, et al., Fascaplysin derivatives binding to DNA via unique cationic five-ring coplanar backbone showed potent antimicrobial/antibiofilm activity against MRSA in vitro and in vivo, Eur. J. Med. Chem., 2022, 230, 114099 CrossRef CAS PubMed.
- A. Brauner, O. Fridman, O. Gefen and N. Q. Balaban, Distinguishing between resistance, tolerance and persistence to antibiotic treatment, Nat. Rev. Microbiol., 2016, 14(5), 320–330 CrossRef CAS PubMed.
- X. Zhang, J. Ma, M. Chen, Z. Wu and Z. Wang, Microbial responses to transient shock loads of quaternary ammonium compounds with different length of alkyl chain in a membrane bioreactor, AMB Express, 2018, 8(1), 118 CrossRef PubMed.
- B. Chen, J. Han, H. Dai and P. Jia, Biocide-tolerance and antibiotic-resistance in community environments and risk of direct transfers to humans: Unintended consequences of community-wide surface disinfecting during COVID-19?, Environ. Pollut., 2021, 283, 117074 CrossRef CAS PubMed.
- B. Moen, K. Rudi, E. Bore and S. Langsrud, Subminimal Inhibitory Concentrations of the Disinfectant Benzalkonium Chloride Select for a Tolerant Subpopulation of Escherichia coli with Inheritable Characteristics, Int. J. Mol. Sci., 2012, 13(4), 4101–4123 CrossRef CAS PubMed.
- E. Obłąk, B. Futoma-Kołoch and A. Wieczyńska, Biological activity of quaternary ammonium salts and resistance of microorganisms to these compounds, World J. Microbiol. Biotechnol., 2021, 37(2), 22 CrossRef PubMed.
- S. Oh, Z. Kurt, D. Tsementzi, M. R. Weigand, M. Kim and J. K. Hatt, et al., Microbial Community Degradation of Widely Used Quaternary Ammonium Disinfectants, Appl. Environ. Microbiol., 2014, 80(19), 5892–5900 CrossRef PubMed.
- D. Dean-Raymond and M. Alexander, Bacterial metabolism of quaternary ammonium compounds, Appl. Environ. Microbiol., 1977, 33(5), 1037–1041 CrossRef CAS PubMed.
- S. Takenaka, T. Tonoki, K. Taira, S. Murakami and K. Aoki, Adaptation of Pseudomonas sp. Strain 7-6 to Quaternary Ammonium Compounds and Their Degradation via Dual Pathways, Appl. Environ. Microbiol., 2007, 73(6), 1797–1802 CrossRef CAS PubMed.
- Antibiotic Resistance: Mechanisms and New Antimicrobial Approaches, ed. K. V. Kon and M. Rai, Elsevier, Academic Press, Amsterdam, 2016, p. 413 Search PubMed.
- E. Christaki, M. Marcou and A. Tofarides, Antimicrobial Resistance in Bacteria: Mechanisms, Evolution, and Persistence, J. Mol. Evol., 2020, 88(1), 26–40 CrossRef CAS PubMed.
- M. N. Alekshun and S. B. Levy, Molecular Mechanisms of Antibacterial Multidrug Resistance, Cell, 2007, 128(6), 1037–1050 CrossRef CAS PubMed.
- M. C. Jennings, M. E. Forman, S. M. Duggan, K. P. C. Minbiole and W. M. Wuest, Efflux Pumps Might Not Be the Major Drivers of QAC Resistance in Methicillin-Resistant Staphylococcus aureus, ChemBioChem, 2017, 18(16), 1573–1577 CrossRef CAS PubMed.
- M. Tandukar, S. Oh, U. Tezel, K. T. Konstantinidis and S. G. Pavlostathis, Long-Term Exposure to Benzalkonium Chloride Disinfectants Results in Change of Microbial Community Structure and Increased Antimicrobial Resistance, Environ. Sci. Technol., 2013, 47(17), 9730–9738 CrossRef CAS PubMed.
- U. Tezel and S. G. Pavlostathis, Role of Quaternary Ammonium Compounds on Antimicrobial Resistance in the Environment, in Antimicrobial Resistance in the Environment, ed. P. L. Keen and M. H. M. M. Montforts, John Wiley & Sons, Inc., Hoboken, NJ, USA, 2011, pp. 349–87, [cited 2023 Feb 14], available from: https://onlinelibrary.wiley.com/doi/10.1002/9781118156247.ch20 Search PubMed.
- F. Van Bambeke, E. Balzi and P. M. Tulkens, Antibiotic efflux pumps, Biochem. Pharmacol., 2000, 60(4), 457–470 CrossRef CAS PubMed.
- J. Bjorland, T. Steinum, M. Sunde, S. Waage and E. Heir, Novel Plasmid-Borne Gene qacJ Mediates Resistance to Quaternary Ammonium Compounds in Equine Staphylococcus aureus , Staphylococcus simulans , and Staphylococcus intermedius, Antimicrob. Agents Chemother., 2003, 47(10), 3046–3052 CrossRef CAS PubMed.
- W. H. Gaze, N. Abdouslam, P. M. Hawkey and E. M. H. Wellington, Incidence of Class 1 Integrons in a Quaternary Ammonium Compound-Polluted Environment, Antimicrob. Agents Chemother., 2005, 49(5), 1802–1807 CrossRef CAS PubMed.
- E. Ortega Morente, M. A. Fernández-Fuentes, M. J. Grande Burgos, H. Abriouel, R. Pérez Pulido and A. Gálvez, Biocide tolerance in bacteria, Int. J. Food Microbiol., 2013, 162(1), 13–25 CrossRef PubMed.
- S. Mohapatra, L. Yutao, S. G. Goh, C. Ng, Y. Luhua and N. H. Tran, et al., Quaternary ammonium compounds of emerging concern: Classification, occurrence, fate, toxicity and antimicrobial resistance, J. Hazard. Mater., 2023, 445, 130393 CrossRef CAS PubMed.
- S. j. Broadley, P. a. Jenkins, J. r. Furr and A. d. Russell, Anti-mycobacterial activity of biocides, Lett. Appl. Microbiol., 1991, 13(3), 118–122 CrossRef CAS.
- J. M. A. Blair, M. A. Webber, A. J. Baylay, D. O. Ogbolu and L. J. V. Piddock, Molecular mechanisms of antibiotic resistance, Nat. Rev. Microbiol., 2015, 13(1), 42–51 CrossRef CAS PubMed.
- N. Sigal, O. Lewinson, S. G. Wolf and E. Bibi, E. coli Multidrug Transporter MdfA Is a Monomer, Biochemistry, 2007, 46(17), 5200–5208 CrossRef CAS PubMed.
- D. Patel, C. Kosmidis, S. M. Seo and G. W. Kaatz, Ethidium Bromide MIC Screening for Enhanced Efflux Pump Gene Expression or Efflux Activity in Staphylococcus aureus, Antimicrob. Agents Chemother., 2010, 54(12), 5070–5073 CrossRef CAS PubMed.
- M. Braoudaki and A. C. Hilton, Mechanisms of resistance in Salmonella enterica adapted to erythromycin, benzalkonium chloride and triclosan, Int. J. Antimicrob. Agents, 2005, 25(1), 31–37 CrossRef CAS PubMed.
- P. Nordmann, L. Poirel, J. K. Mak, P. A. White, C. J. McIver and P. Taylor, Multidrug-Resistant Salmonella Strains Expressing Emerging Antibiotic Resistance Determinants, Clin. Infect. Dis., 2008, 46(2), 324–325 CrossRef PubMed.
- K. Poole, Efflux pumps as antimicrobial resistance mechanisms, Ann. Med., 2007, 39(3), 162–176 CrossRef CAS PubMed.
- L. Pumbwe, L. P. Randall, M. J. Woodward and L. J. V. Piddock, Evidence for Multiple-Antibiotic Resistance in Campylobacter jejuni Not Mediated by CmeB or CmeF, Antimicrob. Agents Chemother., 2005, 49(4), 1289–1293 CrossRef CAS PubMed.
- P. Wieczorek, P. Sacha, T. Hauschild, M. Zórawski, M. Krawczyk and E. Tryniszewska, Multidrug resistant Acinetobacter baumannii—the role of AdeABC (RND family) efflux pump in resistance to antibiotics, Folia Histochem. Cytobiol., 2008, 46(3), 257–267 CAS.
- G. X. He, T. Kuroda, T. Mima, Y. Morita, T. Mizushima and T. Tsuchiya, An H+ -Coupled Multidrug Efflux Pump, PmpM, a Member of the MATE Family of Transporters, from Pseudomonas aeruginosa, J. Bacteriol., 2004, 186(1), 262–265 CrossRef CAS PubMed.
- Y. J. Chung and M. H. Saier, Overexpression of the Escherichia coli sugE Gene Confers Resistance to a Narrow Range of Quaternary Ammonium Compounds, J. Bacteriol., 2002, 184(9), 2543–2545 CrossRef CAS PubMed.
- M. E. Enany, A. M. Algammal, S. A. Nasef, S. A. M. Abo-Eillil, M. Bin-Jumah and A. E. Taha, et al., The occurrence of the multidrug resistance (MDR) and the prevalence of virulence genes and QACs resistance genes in E. coli isolated from environmental and avian sources, AMB Express, 2019, 9(1), 192 CrossRef CAS PubMed.
- A. Norman, L. H. Hansen and S. J. Sørensen, Conjugative plasmids: vessels of the communal gene pool, Philos. Trans. R. Soc., B, 2009, 364(1527), 2275–2289 CrossRef CAS PubMed.
- A. J. Lopatkin, T. A. Sysoeva and L. You, Dissecting the effects of antibiotics on horizontal gene transfer: Analysis suggests a critical role of selection dynamics, BioEssays, 2016, 38(12), 1283–1292 CrossRef CAS PubMed.
- J. Bengtsson-Palme, E. Kristiansson and D. G. J. Larsson, Environmental factors influencing the development and spread of antibiotic resistance, FEMS Microbiol Rev, 2018 Jan 1;42(1), [cited 2023 Feb 22], available from: https://academic.oup.com/femsre/article/doi/10.1093/femsre/fux053/4563583.
- D. C. Bay, K. L. Rommens and R. J. Turner, Small multidrug resistance proteins: A multidrug transporter family that continues to grow, Biochim. Biophys. Acta, Biomembr., 2008, 1778(9), 1814–1838 CrossRef CAS PubMed.
- A. Schlüter, R. Szczepanowski, A. Pühler and E. M. Top, Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool, FEMS Microbiol. Rev., 2007, 31(4), 449–477 CrossRef PubMed.
- M. R. Gillings, D. Xuejun, S. A. Hardwick, M. P. Holley and H. W. Stokes, Gene cassettes encoding resistance to quaternary ammonium compounds: a role in the origin of clinical class 1 integrons?, ISME J., 2009, 3(2), 209–215 CrossRef CAS PubMed.
- T. M. Braga, P. E. Marujo, C. Pomba and M. F. S. Lopes, Involvement, and dissemination, of the enterococcal small multidrug resistance transporter QacZ in resistance to quaternary ammonium compounds, J. Antimicrob. Chemother., 2011, 66(2), 283–286 CrossRef CAS PubMed.
- I. T. Paulsen, T. G. Littlejohn, P. Rådström, L. Sundström, O. Sköld and G. Swedberg, et al., The 3′ conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants, Antimicrob. Agents Chemother., 1993, 37(4), 761–768 CrossRef CAS PubMed.
- M. R. Gillings, Class 1 integrons as invasive species, Curr. Opin. Microbiol., 2017, 38, 10–15 CrossRef CAS PubMed.
- M. R. Gillings, I. T. Paulsen and S. G. Tetu, Genomics and the evolution of antibiotic resistance: Genomics and antibiotic resistance, Ann. N. Y. Acad. Sci., 2017, 1388(1), 92–107 CrossRef PubMed.
- P. J. F. Henderson, C. Maher, L. D. H. Elbourne, B. A. Eijkelkamp, I. T. Paulsen and K. A. Hassan, Physiological Functions of Bacterial “Multidrug” Efflux Pumps, Chem. Rev., 2021, 121(9), 5417–5478 CrossRef CAS PubMed.
- K. Poole, Efflux-mediated antimicrobial resistance, J. Antimicrob. Chemother., 2005, 56(1), 20–51 CrossRef CAS PubMed.
- R. Vijayakumar and T. Sandle, A review on biocide reduced susceptibility due to plasmid-borne antiseptic-resistant genes—special notes on pharmaceutical environmental isolates, J. Appl. Microbiol., 2019, 126(4), 1011–1022 CrossRef CAS PubMed.
- X. Li, A. C. Doherty, B. Brownawell and P. A. Lara-Martin, Distribution and diagenetic fate of synthetic surfactants and their metabolites in sewage-impacted estuarine sediments, Environ. Pollut., 2018, 242, 209–218 CrossRef CAS PubMed.
- M. Saidijam, G. Benedetti, Q. Ren, Z. Xu, C. Hoyle and S. Palmer, et al., Microbial Drug Efflux Proteins of the Major Facilitator Superfamily, Curr. Drug Targets, 2006, 7(7), 793–811 CrossRef CAS PubMed.
- M. A. Schumacher and R. G. Brennan, Structural mechanisms of multidrug recognition and regulation by bacterial multidrug transcription factors, Mol. Microbiol., 2002, 45(4), 885–893 CrossRef CAS PubMed.
- M. A. Schumacher, M. C. Miller, S. Grkovic, M. H. Brown, R. A. Skurray and R. G. Brennan, Structural Mechanisms of QacR Induction and Multidrug Recognition, Science, 2001, 294(5549), 2158–2163 CrossRef CAS PubMed.
- W. H. Gaze, L. Zhang, N. A. Abdouslam, P. M. Hawkey, L. Calvo-Bado and J. Royle, et al., Impacts of anthropogenic activity on the ecology of class 1 integrons and integron-associated genes in the environment, ISME J., 2011, 5(8), 1253–1261 CrossRef CAS PubMed.
- H. Maseda, Y. Hashida, R. Konaka, A. Shirai and H. Kourai, Mutational Upregulation of a Resistance-Nodulation-Cell Division-Type Multidrug Efflux Pump, SdeAB, upon Exposure to a Biocide, Cetylpyridinium Chloride, and Antibiotic Resistance in Serratia marcescens, Antimicrob. Agents Chemother., 2009, 53(12), 5230–5235 CrossRef CAS PubMed.
- T. M. Wassenaar, D. W. Ussery and H. Ingmer, The qacC Gene Has Recently Spread between Rolling Circle Plasmids of Staphylococcus, Indicative of a Novel Gene Transfer Mechanism, Front. Microbiol., 2016, 7, 1528 Search PubMed.
- A. S. Liffourrena, F. G. López, M. A. Salvano, C. E. Domenech and G. I. Lucchesi, Degradation of tetradecyltrimethylammonium by Pseudomonas putida A ATCC 12633 restricted by accumulation of trimethylamine is alleviated by addition of Al 3+ ions, J. Appl. Microbiol., 2008, 104(2), 396–402 CAS.
- B. Brycki, M. Waligórska and A. Szulc, The biodegradation of monomeric and dimeric alkylammonium surfactants, J. Hazard. Mater., 2014, 280, 797–815 CrossRef CAS PubMed.
- C. G. V. Ginkel, J. B. V. Dijk and A. G. M. Kroon’, Metabolism of Hexadecyltrimethylammonium Chloride in Pseudomonas Strain Bi, Appl. Environ. Microbiol., 1992, 58(9), 3083–3087 CrossRef PubMed.
- T. Nishihara, T. Okamoto and N. Nishiyama, Biodegradation of didecyldimethylammonium chloride by Pseudomonas fluorescens TN4 isolated from activated sludge, J. Appl. Microbiol., 2000, 88(4), 641–647 CrossRef CAS PubMed.
- E. Ertekin, J. K. Hatt, K. T. Konstantinidis and U. Tezel, Similar Microbial Consortia and Genes Are Involved in the Biodegradation of Benzalkonium Chlorides in Different Environments, Environ. Sci. Technol., 2016, 50(8), 4304–4313 CrossRef CAS PubMed.
- U. Tezel and S. G. Pavlostathis, Fate and Biotransformation of Quaternary Ammonium Compounds in Biological Treatment Processes, Proc. Water. Environ. Fed., 2010, 2010(17), 641 CrossRef.
- Y. Takatsuka, C. Chen and H. Nikaido, Mechanism of recognition of compounds of diverse structures by the multidrug efflux pump AcrB of Escherichia coli, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(15), 6559–6565 CrossRef CAS PubMed.
- M. Kim, J. K. Hatt, M. R. Weigand, R. Krishnan, S. G. Pavlostathis and K. T. Konstantinidis, Genomic and Transcriptomic Insights into How Bacteria Withstand High Concentrations of Benzalkonium Chloride Biocides, Appl. Environ. Microbiol., 2018, 84(12), e00197 CrossRef CAS PubMed.
- Z. Han, Y. Zhang, W. An, J. Lu, J. Hu and M. Yang, Antibiotic resistomes in drinking water sources across a large geographical scale: Multiple drivers and co-occurrence with opportunistic bacterial pathogens, Water Res., 2020, 183, 116088 CrossRef CAS PubMed.
- M. Nowrotek, E. Kotlarska, A. Łuczkiewicz, E. Felis, A. Sochacki and K. Miksch, The treatment of wastewater containing pharmaceuticals in microcosm constructed wetlands: the occurrence of integrons (int1–2) and associated resistance genes (sul1–3, qacEΔ1), Environ. Sci. Pollut. Res., 2017, 24(17), 15055–15066 CrossRef CAS PubMed.
- T. M. Ghaly, L. Chow, A. J. Asher, L. S. Waldron and M. R. Gillings, Evolution of class 1 integrons: Mobilization and dispersal via food-borne bacteria, PLoS One, 2017, 12(6), e0179169 CrossRef PubMed.
- X. L. An, Q. L. Chen, D. Zhu, Y. G. Zhu, M. R. Gillings and J. Q. Su, Impact of Wastewater Treatment on the Prevalence of Integrons and the Genetic Diversity of Integron Gene Cassettes, Appl. Environ. Microbiol., 2018, 84(9), e02766 CrossRef CAS PubMed.
- B. R. Lyon and R. Skurray, Antimicrobial resistance of Staphylococcus aureus: genetic basis, Microbiol. Rev., 1987, 51 Search PubMed.
- M. R. Gillings, Integrons: Past, Present, and Future, Microbiol. Mol. Biol. Rev., 2014, 78(2), 257–277 CrossRef PubMed.
- W. H. Zhao, G. Chen, R. Ito, S. Kimura and Z. Q. Hu, Identification of a plasmid-borne bla IMP-11 gene in clinical isolates of Escherichia coli and Klebsiella pneumoniae, J. Med. Microbiol., 2012, 61(2), 246–251 CrossRef CAS PubMed.
- D. Cervinkova, V. Babak, D. Marosevic, I. Kubikova and Z. Jaglic, The Role of the qacA Gene in Mediating Resistance to Quaternary Ammonium Compounds, Microb. Drug Resist., 2013, 19(3), 160–167 CrossRef CAS PubMed.
- K. Smith, C. G. Gemmell and I. S. Hunter, The association between biocide tolerance and the presence or absence of qac genes among hospital-acquired and community-acquired MRSA isolates, J. Antimicrob. Chemother., 2007, 61(1), 78–84 CrossRef PubMed.
- Z. Jaglic, D. Červinková, H. Vlková, E. Michu, G. Kunová and V. Babák, Bacterial biofilms resist oxidising agents due to the presence of organic matter, Czech J. Food Sci., 2012, 30(2), 178–187 CrossRef CAS.
- C. Romão, C. A. Miranda, J. Silva, M. Mandetta Clementino, I. de Filippis and M. Asensi, Presence of qacEΔ1 Gene and Susceptibility to a Hospital Biocide in Clinical Isolates of Pseudomonas aeruginosa Resistant to Antibiotics, Curr. Microbiol., 2011, 63(1), 16–21 CrossRef PubMed.
- A. S. Lang, O. Zhaxybayeva and J. T. Beatty, Gene transfer agents: phage-like elements of genetic exchange, Nat. Rev. Microbiol., 2012, 10(7), 472–482 CrossRef CAS PubMed.
- A. S. Lang, A. B. Westbye and J. T. Beatty, The Distribution, Evolution, and Roles of Gene Transfer Agents in Prokaryotic Genetic Exchange, Annu. Rev. Virol., 2017, 4(1), 87–104 CrossRef CAS PubMed.
- M. Gaudin, E. Gauliard, S. Schouten, L. Houel-Renault, P. Lenormand and E. Marguet, et al., Hyperthermophilic archaea produce membrane vesicles that can transfer DNA, Environ. Microbiol. Rep., 2013, 5(1), 109–116 CrossRef CAS PubMed.
- A. Wagner, R. J. Whitaker, D. J. Krause, J. H. Heilers, M. Van Wolferen and C. Van Der Does, et al., Mechanisms of gene flow in archaea, Nat. Rev. Microbiol., 2017, 15(8), 492–501 CrossRef CAS PubMed.
- S. M. Soucy, J. Huang and J. P. Gogarten, Horizontal gene transfer: building the web of life, Nat. Rev. Genet., 2015, 16(8), 472–482 CrossRef CAS PubMed.
- S. Zhu, B. Yang, Z. Wang and Y. Liu, Augmented dissemination of antibiotic resistance elicited by non-antibiotic factors, Ecotoxicol. Environ. Saf., 2023, 262, 115124 CrossRef CAS PubMed.
- W. Li and G. Zhang, Detection and various environmental factors of antibiotic resistance gene horizontal transfer, Environ. Res., 2022, 212, 113267 CrossRef CAS PubMed.
- S. Zhu, B. Yang, Y. Jia, F. Yu, Z. Wang and Y. Liu, Comprehensive analysis of disinfectants on the horizontal transfer of antibiotic resistance genes, J. Hazard. Mater., 2023, 453, 131428 CrossRef CAS PubMed.
- C. Liu, S. G. Goh, L. You, Q. Yuan, S. Mohapatra and K. Y. H. Gin, et al., Low concentration quaternary ammonium compounds promoted antibiotic resistance gene transfer via plasmid conjugation, Sci. Total Environ., 2023, 887, 163781 CrossRef CAS PubMed.
- Y. Jia, Z. Wang, S. Zhu, Z. Wang and Y. Liu, Disinfectants facilitate the transformation of exogenous antibiotic resistance genes via multiple pathways, Ecotoxicol. Environ. Saf., 2023, 253, 114678 CrossRef CAS PubMed.
- P. A. Bhat, A. A. Dar and G. M. Rather, Solubilization Capabilities of Some Cationic, Anionic, and Nonionic Surfactants toward the Poorly Water-Soluble Antibiotic Drug Erythromycin, J. Chem. Eng. Data, 2008, 53(6), 1271–1277 CrossRef CAS.
- A. D. Brunaugh, H. D. C. Smyth and R. O. Williams III, Essential Pharmaceutics, AAPS Introductions in the Pharmaceutical Sciences, Springer International Publishing, Cham, 2019, [cited 2023 Feb 24], available from: http://link.springer.com/10.1007/978-3-030-31745-4 Search PubMed.
- J. Duffy, M. Larsson and A. Hill, Suspension Stability; Why Particle Size, Zeta Potential and Rheology are Important, Annu. Trans. Nord. Rheol. Soc., 2012, 20 Search PubMed.
- W. Smułek and E. Kaczorek, Factors Influencing the Bioavailability of Organic Molecules to Bacterial Cells—A Mini-Review, Molecules, 2022, 27(19), 6579 CrossRef PubMed.
- H. Katepalli and A. Bose, Response of Surfactant Stabilized Oil-in-Water Emulsions to the Addition of Particles in an Aqueous Suspension, Langmuir, 2014, 30(43), 12736–12742 CrossRef CAS PubMed.
- M. Zembyla, B. S. Murray and A. Sarkar, Water-in-oil emulsions stabilized by surfactants, biopolymers and/or particles: a review, Trends Food Sci. Technol., 2020, 104, 49–59 CrossRef CAS.
- F. Wang, L. Zhang, Y. Luo, Y. Li, X. Cheng and J. Cao, et al., Surfactant aggravated the antibiotic's stress on antibiotic resistance genes proliferation by altering antibiotic solubilization and microbial traits in sludge anaerobic fermentation, Sci. Total Environ., 2023, 873, 162440 CrossRef CAS PubMed.
- Y. Tan, X. Cao, S. Chen, X. Ao, J. Li and K. Hu, et al., Antibiotic and heavy metal resistance genes in sewage sludge survive during aerobic composting, Sci. Total Environ., 2023, 866, 161386 CrossRef CAS PubMed.
- A. Karkman, T. T. Do, F. Walsh and M. P. J. Virta, Antibiotic-Resistance Genes in Waste Water, Trends Microbiol., 2018, 26(3), 220–228 CrossRef CAS PubMed.
- R. Pallares-Vega, H. Blaak, R. van der Plaats, A. M. de Roda Husman, L. Hernandez Leal and M. C. M. van Loosdrecht, et al., Determinants of presence and removal of antibiotic resistance genes during WWTP treatment: A cross-sectional study, Water Res., 2019, 161, 319–328 CrossRef CAS PubMed.
- W. Yang, C. Cai, R. Wang and X. Dai, Insights into the impact of quaternary ammonium disinfectant on sewage sludge anaerobic digestion: Dose-response, performance variation, and potential mechanisms, J. Hazard. Mater., 2023, 444, 130341 CrossRef CAS PubMed.
- K. Juksu, J. L. Zhao, Y. S. Liu, L. Yao, C. Sarin and S. Sreesai, et al., Occurrence, fate and risk assessment of biocides in wastewater treatment plants and aquatic environments in Thailand, Sci. Total Environ., 2019, 690, 1110–1119 CrossRef CAS PubMed.
- A. Van de Voorde, C. Lorgeoux, M. C. Gromaire and G. Chebbo, Analysis of quaternary ammonium compounds in urban stormwater samples, Environ. Pollut., 2012, 164, 150–157 CrossRef CAS PubMed.
- S. Kim, K. Ji, H. Shin, S. Park, Y. Kho and K. Park, et al., Occurrences of benzalkonium chloride in streams near a pharmaceutical manufacturing complex in Korea and associated ecological risk, Chemosphere, 2020, 256, 127084 CrossRef CAS PubMed.
- B. J. Heyde, A. Barthel, J. Siemens and I. Mulder, A fast and robust method for the extraction and analysis of quaternary alkyl ammonium compounds from soil and sewage sludge, PLoS One, 2020, 15(8), e0237020 CrossRef CAS PubMed.
- E. Bore, M. Hébraud, I. Chafsey, C. Chambon, C. Skjæret and B. Moen, et al., Adapted tolerance to benzalkonium chloride in Escherichia coli K-12 studied by transcriptome and proteome analyses, Microbiology, 2007, 153(4), 935–946 CrossRef CAS PubMed.
- M. Braoudaki and A. C. Hilton, Adaptive Resistance to Biocides in Salmonella enterica and Escherichia coli O157 and Cross-Resistance to Antimicrobial Agents, J. Clin. Microbiol., 2004, 42(1), 73–78 CrossRef CAS PubMed.
- V. Dutta, D. Elhanafi and S. Kathariou, Conservation and Distribution of the Benzalkonium Chloride Resistance Cassette bcrABC in Listeria monocytogenes, Appl. Environ. Microbiol., 2013, 79(19), 6067–6074 CrossRef CAS PubMed.
- S. Katharios-Lanwermeyer, M. Rakic-Martinez, D. Elhanafi, S. Ratani, J. M. Tiedje and S. Kathariou, Coselection of Cadmium and Benzalkonium Chloride Resistance in Conjugative Transfers from Nonpathogenic Listeria spp. to Other Listeriae, Appl. Environ. Microbiol., 2012, 78(21), 7549–7556 CrossRef CAS PubMed.
- S. Langsrud, G. Sundheim and A. L. Holck, Cross-resistance to antibiotics of Escherichia coli adapted to benzalkonium chloride or exposed to stress-inducers, J. Appl. Microbiol., 2004, 96(1), 201–208 CrossRef CAS PubMed.
- R. Edgar and E. Bibi, MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition, J. Bacteriol., 1997, 179(7), 2274–2280 CrossRef CAS PubMed.
- B. Aase, G. Sundheim, S. Langsrud and L. M. Rørvik, Occurrence of and a possible mechanism for resistance to a quaternary ammonium compound in Listeria monocytogenes, Int. J. Food Microbiol., 2000, 62(1–2), 57–63 CrossRef CAS PubMed.
- R. J. W. Lambert, J. Joynson and B. Forbes, The relationships and susceptibilities of some industrial, laboratory and clinical isolates of Pseudomonas aeruginosa to some antibiotics and biocides, J. Appl. Microbiol., 2001, 91(6), 972–984 CrossRef CAS PubMed.
- J. Kovacevic, J. Ziegler, E. Wałecka-Zacharska, A. Reimer, D. D. Kitts and M. W. Gilmour, Tolerance of Listeria monocytogenes to Quaternary Ammonium Sanitizers Is Mediated by a Novel Efflux Pump Encoded by emrE. Drake HL, Appl. Environ. Microbiol., 2016, 82(3), 939–953 CrossRef CAS PubMed.
- C. Rodríguez-Melcón, C. Alonso-Calleja, C. García-Fernández, J. Carballo and R. Capita, Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) for Twelve Antimicrobials (Biocides and Antibiotics) in Eight Strains of Listeria monocytogenes, Biology, 2021, 11(1), 46 CrossRef PubMed.
- N. Taheri, A. Ardebili, A. Amouzandeh-Nobaveh and E. Ghaznavi-Rad, Frequency of Antiseptic Resistance Among Staphylococcus aureus and Coagulase-Negative Staphylococci Isolated From a University Hospital in Central Iran, Oman Med. J., 2016, 31(6), 426–432 CrossRef CAS PubMed.
- S. Buffet-Bataillon, B. Branger, M. Cormier, M. Bonnaure-Mallet and A. Jolivet-Gougeon, Effect of higher minimum inhibitory concentrations of quaternary ammonium compounds in clinical E. coli isolates on antibiotic susceptibilities and clinical outcomes, J. Hosp. Infect., 2011, 79(2), 141–146 CrossRef CAS PubMed.
- P. M. Davidson, J. N. Sofos and A. L. Branen, Antimicrobials in food, CRC press, 2005 Search PubMed.
- J. Zhang, J. Liu, T. Lu, P. Shen, H. Zhong and J. Tong, et al., Fate of antibiotic resistance genes during anaerobic digestion of sewage sludge: Role of solids retention times in different configurations, Bioresour. Technol., 2019, 274, 488–495 CrossRef CAS PubMed.
- M. Mukherjee, E. Laird, T. J. Gentry, J. P. Brooks and R. Karthikeyan, Increased Antimicrobial and Multidrug Resistance Downstream of Wastewater Treatment Plants in an Urban Watershed, Front. Microbiol., 2021, 12, 657353 CrossRef PubMed.
- B. Gurmessa, E. F. Pedretti, S. Cocco, V. Cardelli and G. Corti, Manure anaerobic digestion effects and the role of pre- and post-treatments on veterinary antibiotics and antibiotic resistance genes removal efficiency, Sci. Total Environ., 2020, 721, 137532 CrossRef CAS PubMed.
- K. Yoo, H. Yoo, J. Lee, E. J. Choi and J. Park, Exploring the antibiotic resistome in activated sludge and anaerobic digestion sludge in an urban wastewater treatment plant via metagenomic analysis, J. Microbiol., 2020, 58(2), 123–130 CrossRef CAS PubMed.
- Y. N. Jiao, Z. C. Zhou, T. Chen, Y. Y. Wei, J. Zheng and R. X. Gao, et al., Biomarkers of antibiotic resistance genes during seasonal changes in wastewater treatment systems, Environ. Pollut., 2018, 234, 79–87 CrossRef CAS PubMed.
- K. M. M. Pärnänen, C. Narciso-da-Rocha, D. Kneis, T. U. Berendonk, D. Cacace and T. T. Do, et al., Antibiotic resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence, Sci. Adv., 2019, 5(3), eaau9124 CrossRef PubMed.
- H. Zhang, Z. Zhang, J. Song, L. Cai, Y. Yu and H. Fang, Foam shares antibiotic resistomes and bacterial pathogens with activated sludge in wastewater treatment plants, J. Hazard. Mater., 2021, 408, 124855 CrossRef CAS PubMed.
- J. Bengtsson-Palme, M. Milakovic, H. Švecová, M. Ganjto, V. Jonsson and R. Grabic, et al., Industrial wastewater treatment plant enriches antibiotic resistance genes and alters the structure of microbial communities, Water Res., 2019, 162, 437–445 CrossRef CAS PubMed.
- Y. Tang, Z. Liang, G. Li, H. Zhao and T. An, Metagenomic profiles and health risks of pathogens and antibiotic resistance genes in various industrial wastewaters and the associated receiving surface water, Chemosphere, 2021, 283, 131224 CrossRef CAS PubMed.
- S. Rodriguez-Mozaz, I. Vaz-Moreira, S. Varela Della Giustina, M. Llorca, D. Barceló and S. Schubert, et al., Antibiotic residues in final effluents of European wastewater treatment plants and their impact on the aquatic environment, Environ. Int., 2020, 140, 105733 CrossRef CAS PubMed.
- I. Michael, L. Rizzo, C. S. McArdell, C. M. Manaia, C. Merlin and T. Schwartz, et al., Urban wastewater treatment plants as hotspots for the release of antibiotics in the environment: A review, Water Res., 2013, 47(3), 957–995 CrossRef CAS PubMed.
- T. Tennstedt, R. Szczepanowski, S. Braun, A. Pühler and A. Schlüter, Occurrence of integron-associated resistance gene cassettes located on antibiotic resistance plasmids isolated from a wastewater treatment plant, FEMS Microbiol. Ecol., 2003, 45(3), 239–252 CrossRef CAS PubMed.
- J. Bengtsson-Palme, R. Hammarén, C. Pal, M. Östman, B. Björlenius and C. F. Flach, et al., Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics, Sci. Total Environ., 2016, 572, 697–712 CrossRef CAS PubMed.
- Z. Wu, J. Gao, Y. Cui, Z. Wang, Y. Zhao and H. Zhang, et al., Feeding low-level benzethonium chloride can promote the start-up, fast recovery and long-term stable maintenance of partial nitrification for low-ammonium wastewater, Bioresour. Technol., 2022, 353, 127152 CrossRef CAS PubMed.
- Y. Cui, J. Gao, M. Zhao, Y. Guo, Y. Zhao and Z. Wang, Deciphering the interaction impacts between antiseptic benzethonium chloride and biofilm nitrification system: Performance, resistance mechanisms and biodegradation, Water Res., 2023, 240, 120062 CrossRef CAS PubMed.
- D. Li, J. Gao, H. Dai, Z. Wang, Y. Cui and Y. Zhao, et al., Fates of quaternary ammonium compound resistance genes and the corresponding resistant strain in partial nitrification/anammox system under pressure of hexadecyl trimethyl ammonium chloride, Water Res., 2022, 217, 118395 CrossRef CAS PubMed.
- H. Zhang, J. Gao, M. Zhao, Z. Wang, D. Li and Z. Wu, et al., The spread of different resistance genes fractions in nitrification system under chronic exposure to varying alkyl chain length benzalkyl dimethylammonium compounds, Bioresour. Technol., 2023, 371, 128588 CrossRef CAS PubMed.
- O. Thakali, J. P. Brooks, S. Shahin, S. P. Sherchan and E. Haramoto, Removal of Antibiotic Resistance Genes at Two Conventional Wastewater Treatment Plants of Louisiana, Water, 2020, 12(6), 1729 CrossRef CAS.
- X. Pan, L. Lin, W. Zhang, L. Dong and Y. Yang, Metagenome sequencing to unveil the resistome in a deep subtropical lake on the Yunnan-Guizhou Plateau, China, Environ. Pollut., 2020, 263, 114470 CrossRef CAS PubMed.
- Y. Bai, X. Ruan, X. Xie and Z. Yan, Antibiotic resistome profile based on metagenomics in raw surface drinking water source and the influence of environmental factor: A case study in Huaihe River Basin, China, Environ. Pollut., 2019, 248, 438–447 CrossRef CAS PubMed.
- G. Reichert, S. Hilgert, J. Alexander, J. C. Rodrigues de Azevedo, T. Morck and S. Fuchs, et al., Determination of antibiotic resistance genes in a WWTP-impacted river in surface water, sediment, and biofilm: Influence of seasonality and water quality, Sci. Total Environ., 2021, 768, 144526 CrossRef CAS PubMed.
- I. Buriánková, P. Kuchta, A. Molíková, K. Sovová, D. Výravský and M. Rulík, et al., Antibiotic Resistance in Wastewater and Its Impact on a Receiving River: A Case Study of WWTP Brno-Modřice, Czech Republic, Water, 2021, 13(16), 2309 CrossRef.
- B. Chen, L. Hao, X. Guo, N. Wang and B. Ye, Prevalence of antibiotic resistance genes of wastewater and surface water in livestock farms of Jiangsu Province, China, Environ. Sci. Pollut. Res., 2015, 22(18), 13950–13959 CrossRef CAS PubMed.
- Y. Yang, Z. Li, W. Song, L. Du, C. Ye and B. Zhao, et al., Metagenomic insights into the abundance and composition of resistance genes in aquatic environments: Influence of stratification and geography, Environ. Int., 2019, 127, 371–380 CrossRef CAS PubMed.
- M. Chen, X. Zhang, Z. Wang, M. Liu, L. Wang and Z. Wu, Impacts of quaternary ammonium compounds on membrane bioreactor performance: Acute and chronic responses of microorganisms, Water Res., 2018, 134, 153–161 CrossRef CAS PubMed.
- S. Mishra, V. Singh, L. Cheng, A. Hussain and B. Ormeci, Nitrogen removal from wastewater: A comprehensive review of biological nitrogen removal processes, critical operation parameters and bioreactor design, J. Environ. Chem. Eng., 2022, 10(3), 107387 CrossRef CAS.
- H. Chandel, K. Shyam, N. Kumar, G. Sharma, M. Yadav, S. Murugesan, et al., Chapter 2 – Anaerobic ammonium oxidation (anammox) technology for nitrogen removal from wastewater: Recent advances and challenges, in Integrated Environmental Technologies for Wastewater Treatment and Sustainable Development, ed. V. Kumar and M. Kumar, Elsevier, 2022, pp. 23–48, [cited 2023 Oct 25], available from: https://www.sciencedirect.com/science/article/pii/B9780323911801000090 Search PubMed.
- J. G. Kuenen, Anammox bacteria: from discovery to application, Nat. Rev. Microbiol., 2008, 6(4), 320–326 CrossRef CAS PubMed.
- M. Hayatsu, K. Tago and M. Saito, Various players in the nitrogen cycle: Diversity and functions of the microorganisms involved in nitrification and denitrification, Soil Sci. Plant Nutr., 2008, 54(1), 33–45 CrossRef CAS.
- S. Cho, C. Kambey and V. K. Nguyen, Performance of Anammox Processes for Wastewater Treatment: A Critical Review on Effects of Operational Conditions and Environmental Stresses, Water, 2020, 12(1), 20 CrossRef CAS.
- Y. Cui, J. Gao, D. Zhang, Y. Zhao and Y. Wang, Rapid start-up of partial nitrification process using benzethonium chloride—a novel nitrite oxidation inhibitor, Bioresour. Technol., 2020, 315, 123860 CrossRef CAS PubMed.
- M. Zhao, J. Gao, Y. Liu, Z. Wang, Z. Wu and H. Zhang, et al., Short-term stress of quaternary ammonium compounds on intracellular and extracellular resistance genes in denitrification systems, Chem. Eng. J., 2023, 452, 139166 CrossRef CAS.
- S. P. Lohani and J. Havukainen, Anaerobic Digestion: Factors Affecting Anaerobic Digestion Process, in Waste Bioremediation, Energy, Environment, and Sustainability, ed. S. J. Varjani, E. Gnansounou, B. Gurunathan, D. Pant and Z. A. Zakaria, Springer Singapore, Singapore, 2018, pp. 343–59, [cited 2023 Feb 23], available from: http://link.springer.com/10.1007/978-981-10-7413-4_18 Search PubMed.
- U. Tezel, J. A. Pierson and S. G. Pavlostathis, Fate and effect of quaternary ammonium compounds on a mixed methanogenic culture, Water Res., 2006, 40(19), 3660–3668 CrossRef CAS PubMed.
- Z. W. He, W. Z. Liu, C. C. Tang, B. Liang, Z. C. Guo and L. Wang, et al., Performance and microbial community responses of anaerobic digestion of waste activated sludge to residual benzalkonium chlorides, Energy Convers. Manage., 2019, 202, 112211 CrossRef.
- Z. W. He, W. Z. Liu, C. C. Tang, B. Liang, A. J. Zhou and F. Chen, et al., Responses of anaerobic digestion of waste activated sludge to long-term stress of benzalkonium chlorides: Insights to extracellular polymeric substances and microbial communities, Sci. Total Environ., 2021, 796, 148957 CrossRef CAS PubMed.
- A. D. Kappell, D. E. Carey, D. H. Zitomer and P. J. McNamara, Effect of antimicrobial washout from anaerobic digesters on microbial community composition, Environ. Sci.: Water Res. Technol., 2020, 6(6), 1658–1671 RSC.
- A. J. McBain, R. G. Ledder, L. E. Moore, C. E. Catrenich and P. Gilbert, Effects of Quaternary-Ammonium-Based Formulations on Bacterial Community Dynamics and Antimicrobial Susceptibility, Appl. Environ. Microbiol., 2004, 70(6), 3449–3456 CrossRef CAS PubMed.
- Y. Wang, J. Lu, J. Engelstädter, S. Zhang, P. Ding and L. Mao, et al., Non-antibiotic pharmaceuticals enhance the transmission of exogenous antibiotic resistance genes through bacterial transformation, ISME J., 2020, 14(8), 2179–2196 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |