Open Access Article
Open Access ArticlePerspectives in growth production trade-off in microbial bioproduction
Deepanwita
Banerjee
 ab and
Aindrila
Mukhopadhyay
ab and
Aindrila
Mukhopadhyay
 *ab
*ab
aBiological Systems and Engineering, Lawrence Berkeley National Laboratory, 1 Cyclotron Road, Berkeley, CA 94720, USA. E-mail: amukhopadhyay@lbl.gov
bJoint BioEnergy Institute, Emeryville, CA 94608, USA
First published on 26th November 2022
Abstract
Metabolic engineering of microbial systems and conversion routes can provide robust platforms for the production of bulk commodities for food, materials and fuel targets. For products in this range, the maximum conversion of starting materials and stable phenotypes in a bioreactor are vital for an economically viable process. Strain engineering approaches to improve the conversion efficiency and reduce the phenotypic variability have witnessed significant development in the past decade. Herein, we review several of the main categories of these approaches including growth coupling, growth decoupling, regulatory control and use of non-metabolic cellular functions. We discuss these topics in the context of microbial host physiology and its impact in the selection of the most effective approach. We also discuss the importance of growth medium optimization and studies using bioreactors in delineating a bioproduction system that is most likely to provide stable conversion over a longer period.
Sustainability spotlightIn 2019, the renewable energy consumption increased but its share in the total energy consumption was 17.7%, which is only 1.6% higher than that in 2010. In 2020, governments spent $375 billion on subsidies for fossil fuels. Subsequently, in 2021, the fuel-related emissions were at their highest and eliminated the pandemic-related reduction seen in 2020. These challenges can be addressed through the faster scale-up of renewable fuels and commodity chemicals. The sustainable production of many previously petrochemically derived chemicals can be realized via microbial production using renewable starting materials, but for success, this requires the maximum conversion and balancing cultivation with final product formation in the process. This is in alignment with the UN Sustainable Development Goals including affordable and clean energy, responsible consumption and production, and climate action. |
Introduction
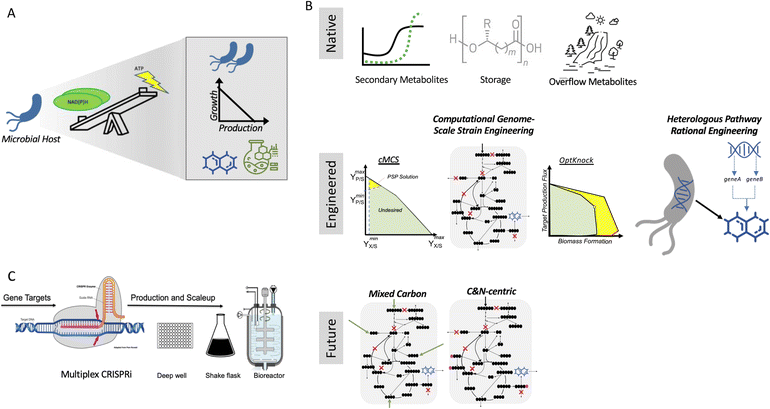
Biotechnological application is no longer limited to the pharmaceutical industry and fine chemicals but is also widely applied for the production of food, fuel and other commodity chemicals. After optimization of the biosynthetic pathway, the production yield required for these commodities remains high. and therefore maximum conversion of the starting materials is necessary1 for economical, sustainable production of these bulk chemicals. To achieve this, several engineering strategies can be employed, which are both rational and computationally driven. In principle, all these approaches can be implemented for all microbial systems and target products, but in practice, their experimental implementation is conducted in a small set of model hosts (e.g., E. coli and S. cerevisiae) and targets. Furthermore, in the case of systems that have been experimentally vetted, few have been scaled up. Consequently, these microbial bioconversion methods for commodity chemical production are still far from being host-, product-, substrate-, and scale-agnostic.A key parameter to consider in de-risking microbial production is that growth and bioproduction are linked. Although robust growth is required for efficient production, both growth and production also utilize the same pool of starting materials, creating a trade-off (Fig. 1A). Bioproduction mainly falls into three categories (Fig. 1B) that utilize a natural phenotype for accumulating a bioproduct. The first is secondary metabolism, rerouting a small amount of cellular intermediates to make a final product that has specialized use. Most engineering pathways (including heterologous pathways) fall in this category. The second is in the form of storage molecules such as polyhydroxyalkanoates (PHA), lipids, and fatty acids. The third is redox balancing by-product accumulation such as CO2, ethanol, and organic acids. Ethanol is one of the best bioconversion examples that works in exactly the desired conditions, i.e., high sugar load and low O2. Moving forward, mixed carbon co-feed and carbon (C) and nitrogen (N)-centric genome-scale metabolic representations hold immense potential in addressing the trade-off for the bulk production of commodity chemicals.
For the maximum conversion and ideal hosts, products, substrates, and scale agnostic bioconversion success of any type (Fig. 1C), the trade-off introduced through strain/host engineering can be addressed with the right combination of host and product, and thus these methods need to be more widely applied.
Growth coupling (GC)
The translation of the majority of proof of principles tested on the lab scale to industrial production platforms is very unpredictable, and thus most systems have not been tested.2 Among the various challenges, one major bottleneck is that if the production is independent of microbial growth, growth inhibition due to the expression of a burdensome pathway or production of an inhibitory compound can cause engineered strains to drift away from their optimal production phenotype due to its selective advantage. Thus, to address this challenge, coupling growth with production is a powerful strategy for evolving strains to maintain high titers, rates, and yields (TRY). Three main definitions of growth coupling have been widely used in the metabolic engineering for microbial conversion. First, the most popular approach is optimal production at the optimal growth rates (weak coupling), where the earliest example includes the use of Optknock for the production of lactate in E. coli3 and one popular example is Optknock-based strain engineering for the production of 1,4 BDO in E. coli4. Second, when production is coupled with a driving force such as ATP production or NADPH or NADH cofactor balancing. These coupling strategies have been used for the production of short chain primary alcohols (1-butanol, isobutanol, and 2-methylbutanol) in Escherichia coli5 and Synechococcus sp.6,7 as well as for 3-hydroxypropionic acid production in Saccharomyces cerevisiae.8 Third, when metabolite production is essential for growth, which is also termed strong coupling or obligatory production. This type of strong growth coupling is found in nature, where examples include ethanol under fermentative state (e.g., S. cerevisiae), carbon dioxide during microbial aerobic growth and acetate in acetogenic bacteria. An iterative implementation of computed constrained Minimal Cut Sets (cMCSs) using a modified central metabolic model of E. coli for itaconate production9 resulted in significantly high titers (32 g L−1) and itaconate yield (0.77 mol mol−1 glucose) with negligible by-product (pyruvate) although it required glutamate supplementation in the fed batch cultivation. Recent advancement in cMCS computation at the genome-scale10,11 led to the engineering of Pseudomonas putida for the production of indigoidine, a bipyridyl molecule, using a product-substrate-pairing (PSP) approach (Fig. 2).12 This study reported the highest indigoidine titer (26 g L−1) to date at that time, which remains the highest using glucose minimal media and made more of indigoidine during glucose feeding, i.e., the indigoidine production was in the growth phase. In general, these GC prediction strategies assume the best or optimal growth conditions for the host, and therefore GC engineering when followed by adaptive laboratory evolution (ALE) should improve the production phenotype given that growth is hard-wired to production and ALE will trace the path for better growth in the fitness landscape. In the case of non-conventional microbial systems, it may be a challenge to determine the best growth cultivation condition (discussed later in this review). This bottleneck can be addressed using medium optimization methods such as design of experiment (DOE) or high throughput (HT) phenotypic profiling in combination with machine learning.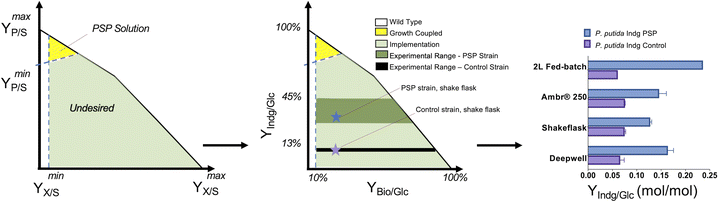 | ||
| Fig. 2 Product substrate pairing (PSP) approach for the growth-coupled microbial conversion of glucose (Glc) to indigoidine (Indg), adapted from Banerjee et al. | ||
Although growth-coupled strategies are promising, sometimes their direct implementation may result in auxotrophy. For example, OptGene-based S. cerevisiae strain engineering for the production of succinic acid resulted in glycine auxotrophy.13 Upon ALE the resulting strain had significant improvement in succinate titre (0.9 g L−1) and growth rates, but the yields were still low (0.05 g g−1 of glucose or 0.43 versus 0.69 g g−1 of biomass) due to the trade-off between growth and production. This trade-off due to competition for the same resources also resulted in significant dependence on available oxygen when growth-coupled strain engineering in E. coli was performed using elementary mode analysis for ethanol production.14 Recently, growth-coupled production examples have been comprehensively reviewed.15,16 Growth-coupled schemes are also being investigated for in vivo biohalogenation17 as well as protein production.18
Decoupling growth and production (DC)
To address the trade-off between growth and production, the separation of the two phases during any microbial process, i.e., growth phase and production phase, has been shown to be promising. This avoids the metabolic burden of the production pathway during the growth phase and helps in the allocation of all metabolic resources to the synthesis of the product during the production phase. These approaches for microbial conversion have been widely used for the utilization of non-canonical carbon sources such as glycerol,19 xylose,20 protocatechuate,21 hydrocinnamic acids,22 and toxic products such as 1,2-indandiol production in Rhodococcus sp.23 The transition from one phase to another is induced by several different strategies. Examples include glucose starvation and carbon source switching for the production of PHB in E. coli in rich and minimal media,24 nitrogen starvation for fatty alcohol production in E. coli,25 phosphate limitation for linalool production in Pantoea ananatis,26 oxygen supply limitation,27 biotin limitation for proline production in Corynebacterium glutamicum,28 pH control for propionic acid production in Propionibacterium jensenii19 and temperature shift29 to regulate isocitrate dehydrogenase activity to increase itaconate production in E. coli.30 Extreme examples are growth in one carbon source and conversion of a different source to the product21,23 or use of whole cells as biocatalysts for the production of acetoin, gamma-amino-butyric acid (GABA) and ε-caprolactone using engineered E. coli.31–33 The earliest example of growth decoupling to separate growth from production using stationary phase production was the production of trans-(1R,2R)-indandiol in Rhodococcus sp. I24.23 Examples of co-production include a growth-decoupled fed-batch production process, which resulted in the formation of about 11 g L−1 α-ketoglutarate and succinate from xylose in C. glutamicum.20 Advancement in computational implementations for identifying a range of operating points within a feasible solution space has also been reported.34A major challenge of decoupled microbial conversions that use constitutive production pathways is that during the growth phase, there may be a phenotypic drift due to suboptimal growth or toxicity and low-producing variants with fitness advantages may overtake the population in the bioconversion process. One approach to address this challenge is the native auto-induction method for bioconversion. Auto-induction is based on the lac operon regulatory function during diauxic growth in the presence of multiple carbon sources such as glucose, glycerol, and lactose.35 Although the control of these natural circuits is well characterized in E. coli, catabolite repression and preferential carbon source utilization in “rising star” hosts still need to be elucidated for bioproduction purposes. In this case, tighter control through synthetic feedback or feed-forward loops may help devise better solutions to overcome these trade-offs.
Synthetic circuits for dynamic control (feed-back/feed-forward loops)
Advances in gene editing technologies36,37 have made the design of gene circuits easier for accurate dynamic control to balance the existing trade-off between growth and bioconversion. One of the earliest examples is a two-layered circuit design that decoupled growth and production and utilized product-addicted promoters to amplify production in E. coli.22 This lowered the metabolic stress and increased the growth rate and vanillic acid productivity. An example of GC together with dynamic control resulted in a high naringenin-producing E. coli strain (523.7 ± 51.8 mg L−1) with 20% increase in cell growth compared to the control strain.38 An example of this dynamic control is the switch between growth and production phase for GABA production in C. glutamicum, which produced 45.6 g L−1 of GABA with a yield of 0.4 g g−1 glycerol.39 Cell density based quorum-sensing circuits in E. coli resulted in 520 ± 7 mg L−1 salicylic acid, which is a 1.8-fold improvement compared to the control strain.40 Nitrogen limitation-driven dynamic control for the production of itaconate41 was extended to produce itaconate at 1.4 g L−1 in P. putida from deconstructed lignin. Another example of dynamic control of pH has been reported for the production of lycopene in E. coli.42 On–off switches, which have been also used by genetic engineers, are inversion promoter regions based on recombination events.Gene circuits for synthetic addiction have been successfully developed in model organisms but can be context specific. Metabolite-responsive transcription factor-based biosensors have been the most successful thus far and hold immense potential but their dynamic range may be growth dependent and vary under different media conditions and growth environments. Recently, it was reported that the dynamic range of aTc-TetR and IPTG-LacI sensors has positive correlations with the cell growth rate, whereas the FA-FadR biosensor has a negative correlation with the cell growth rates when tested for several carbon sources in minimal media condition, confirming the trade-off between the dynamic range and growth condition.43 However, optimization is challenging when using a synthetic feed, given that only narrow sweet spots exist. These gene circuits are still not scalable across products and formats due to several challenges including their narrow dynamic range, linearity, and signal to noise ratio. A successful alternative was the design of genetically stable circuits or “landing pads” in the E. coli genome, where the expression level is high,44 and creation of insulators using the NOR gate logic. Recently, this has been extended to S. cerevisiae and Bacteroides thetaiotaomicron.45 Computational workflows are also being developed for multi-objective optimization for the trade-offs associated with biosensor development.46,47 Another challenge that remains is the enrichment of producers versus escapers or non-producers, as reported for baker's yeast.48 Recent examples of biosensor-based dynamic regulation have been reviewed elsewhere.49
Recently, optogenetic tools have also been utilized for the production of various value-added products such as lactic acid, isobutanol, and shikimic acid. Zhao et al.50 produced 8.49 g L−1 of isobutanol in S. cerevisiae using OptoEXP- and OptoINVRT-based control of EL222 transcriptional activator for metabolic switching to the production phase. Subsequently, a further optimized version, i.e., OptoAMP-based control of LDH, was reported for the production of lactic acid 6 g L−1 in S. cerevisiae even at low light intensities.51 In another study, optogenetic tools were developed using the TetR system together with the tobacco etch virus protease (TEVp) for the production of shikimic acid in E. coli at 35 g L−1 using glucose minimal media.52 The major bottlenecks with optogenetic tools include the limited number of photo-switchable proteins, restricted implementation in popular industrial hosts, insufficient and heterogeneous lighting at high cell density in large-scale bioreactors and cost associated with specifically designed industrial-scale light-bioreactors (up to 5000–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 L) for the production of commodity chemicals. Another major challenge is the metabolic burden associated with the cellular resource allocation to build the multiple proteins and cofactors (FMN, NADH, and NADPH) required for the functioning of these optogenetic systems. Examples of various optogenetic tools have been recently reviewed elsewhere.53
000 L) for the production of commodity chemicals. Another major challenge is the metabolic burden associated with the cellular resource allocation to build the multiple proteins and cofactors (FMN, NADH, and NADPH) required for the functioning of these optogenetic systems. Examples of various optogenetic tools have been recently reviewed elsewhere.53
Multi-substrate-based synthetic circuits for metabolic control of bioconversion processes have been reported54 but are still to be routinely implemented for production control. These synthetic genetic control systems should at least include substrate-induced transitions from the growth to production phase (decoupled production). Thus far, some progress in this direction has been reported for S. cerevisiae55 and E. coli24 but there has not been much progress in growth-coupled bioproduction strategies to the best of our knowledge. An aspect to consider in dynamic control circuits is the maintenance of the phenotype in a variable or heterogeneous environment. Specifically, for use in larger-scale production, the circuit must retain a high signal to noise ratio and have minimal interference from any crosstalk. Given that it is challenging to predict these issues a priori, implementation and examination of these systems in larger-scale fed-batch mode with industrially relevant feed sources are necessary.
Medium optimization for scalable bioconversion systems
Another critical factor in the growth-production trade-off, and consequently in the advancement and scaleup of bioconversion systems is the medium composition and its role in engineered strain cultivation during the bioconversion process. Examples of medium optimization to improve productivity are both essential and well-reported. In this section, we review medium optimization in the context of growth-production trade-off, such as the use of mixed carbon and the impact of carbon![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nitrogen (C/N) ratios.
nitrogen (C/N) ratios.
Well-defined media with minimal nutrient supplementation are ideal from a techno-economic perspective; however, the use of resources coming from poorly utilized but abundant feedstocks is also necessary, both aiming towards a carbon-negative bioconversion process. Furthermore, successful bioconversion for commodity chemicals is measured by titer, rates, and yields (TRY), which is challenging to quantify in rich media or in undefined media such as plant biomass-derived feedstocks. These factors play a role in the use of growth-pairing strategies. To date, growth-coupled strategies have been examined for only a single C source. Mixed C-sources have been shown with DC strategies but it is necessary to further engineer strategies that account for more than one carbon source to address the growth productivity trade-off. An example towards advanced bioconversion systems using carbon negative feedstocks reported that a cofeed of CO2 and glucose enhanced acetogenesis in Moorella thermoacetica, whereas gluconate and acetate cofeed enhanced fatty acid production in Yarrowia lipolytica under fed-batch condition.56 Another example involved DC engineering of a heterologous Weimberg pathway bypass in the itaconate producer E. coli Δicd strain, which led to a high titer of 20 g L−1 itaconate and ∼65% of maximum theoretical yield (glycerol) using a cofeed of glycerol and xylose.57 Given that GC strategies are hard wired to the carbon source, it will be interesting to see how the production phenotype performs when multiple carbon sources or a nutrient-rich medium such as plant biomass-derived hydrolysate is used as the carbon stream for bioconversion to commodity chemicals using such engineered strains. Carbon sources that are incompatible with growth-coupling engineering may reduce the TRY, whereas compatible carbon sources may have a synergistic effect on the product yield (for example, growth coupling with glucose is synergistic with galactose but may be incompatible with aromatic substrates).12 The GC algorithm used in the PSP pipeline is customizable for co-feed substrate utilization but it has not been experimentally implemented thus far. To truly understand the growth versus production Pareto front, accurate measurements of TRY are required, but remain a challenge to obtain routinely. In the case of DC strategies, glucose is a well-studied and preferred carbon source, whereas the bioconversion medium components such as nitrogen, oxygen, phosphate, temperature and other limitations may have far more complex relationships to have tight transitions between the two phases and may not scale successfully to industry relevant bioreactor cultivation conditions. For example, biotin limitation under the fed-batch regime induced glutamate secretion as a by-product in C. glutamicum.28 In another extreme example, only 4 out of 16 phosphate-regulated promoters tested in E. coli scaled successfully to the bioreactor condition (ugpB, yibD, phoA, and phoB promoters).58
Finally, instead of implementing GC with multiple substrates in the same strain, a viable near-future alternative is a one-pot synthetic microbial community of GC-engineered strains for the division of labor for the bioconversion of specific substrates efficiently. A similar division of labor has been used to reduce the metabolic burden in a P. putida and S. cerevisiae consortium to produce 295.7 mg L−1 mcl-PHA titer.59 Recently, a synthetic microbial consortium of P. putida and E. coli was reported, where substrate utilization and production were decoupled in the two strains, which resulted in 1.32 g L−1 of mcl-PHA from 20 g L−1 of a glucose–xylose mixture (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1).60 Computational dynamic modeling has also been used to study the trade-off between productivity and efficiency of substrate utilization in a synthetic consortium of E. coli strains, with one producing a heterologous protein together with a second E. coli strain engineered to scavenge acetate.61
1).60 Computational dynamic modeling has also been used to study the trade-off between productivity and efficiency of substrate utilization in a synthetic consortium of E. coli strains, with one producing a heterologous protein together with a second E. coli strain engineered to scavenge acetate.61
Nitrogen is another major constraint that is usually overlooked in various engineering strategies. The optimization of the C/N ratio in bioprocess optimization is required to identify the best bioconversion cultivation condition62–64 or supplementation of large amounts of nitrogen sources (e.g., 1 g L−1 or higher supplementation in production medium during scale-up). Firstly, nitrogen has been extensively used for metabolic engineering purposes to address the growth versus production trade-off of storage metabolites that are triggered by N starvation such as PHA production in P. putida strains.65,66 Secondly, research is shifting towards “greener” non-conventional carbon feedstocks containing aromatics, furfurals, etc., which follow different catabolic routes and regulatory mechanisms to generate energy and maintain cellular biomass (not the conventional glycolysis → TCA → oxidative phosphorylation). Aromatic catabolism is very different from traditional sugar glycolytic pathways. Conventional knowledge is that when glucose, fatty acids, or some amino acids are utilized as a carbon source under aerobic conditions, the C/N ratio sensing metabolic node is alpha-ketoglutarate (AKG).67 However, this may not be true for aromatics or other carbon sources. C and N-centric context-specific genome-scale metabolic models (GSM) and 13C- and 15N-labeled metabolic flux analysis (MFA) under a range of different conditions will advance the GSM closer to C- and N-relevant experimental conditions. Further, as is often the case with the carbon source, a certain N source can also be more suitable for a host or a conversion process, and a given medium formulation may not hold true if the host, pathway or culture format is changed for the same bioconversion system.63,68
Given that the final products span a greater range of targets, we now encounter products that are metabolized or degraded by the host microbe. Product degradation or catabolism also indirectly related to growth-production pairing and medium amendments has been used to prevent product catabolism. For example, in isoprenol production in P. putida, it was found that the natural catabolism of isoprenol occurs only after the consumption of glucose (Xi et al.,82 in their review). In general, metabolically versatile hosts such as P. putida have catabolic pathways for many final products or precursors. This can also be addressed via the deletion of the catabolic route when known.69,70 However, to the best of our knowledge, there are not enough systematic studies on the effect of media components on highly engineered strains that are tailored for bioconversion on a large scale.
Medium optimization is an integral part of process optimization for any bioconversion process.71–73 From a synthetic biology perspective, metabolic pathways and host engineering research must also incorporate the knowledge of the medium components on the metabolism being engineered. It is understood that during scale-up from the lab to industrial level, medium and process optimization plays a significant role in research and development. However, even at the lab-scale design, requirements of media formulations can be incorporated into strain design. In the case of strains devoid of hierarchical substrate uptake and utilization (using modification in master regulators, e.g., delta crc), accessory machinery (e.g., EM42 strain) allows rewiring the host metabolism as well as cell physiology, and thus the trade-off is now substantially altered relative to the basal strain. The C/N ratio for the new strain designs will be different, and therefore requires reintegration in the form of a medium optimization module. However, despite the essentiality of medium optimization, it remains ad hoc in the literature73 and few examples of high throughput data-driven medium optimization exist.74,75 One can anticipate that with improvements in automation and data-driven approaches and the ability to examine configurations in high throughput (e.g., via microfluidics76), this aspect will also witness a lot of improvements. The consilience of metabolic and host engineering approaches with the optimization of production medium will enable ideal growth-production ratios that are stable (and thus predictable) across scales.
Beyond metabolism
For sustainable industrial scale-up, microbial bioconversion goes beyond metabolic engineering. Computational fluid dynamic (CFD)-based models have been used to understand the trade-off in industrial-scale bioreactors (https://www.nrel.gov/docs/fy21osti/78334.pdf) together with several other bioprocess models, which have been recently reviewed,77 to assess the aeration and other parameters that are currently bottlenecks for these bioconversion processes. Heavily engineered strains designed for high TRY may not achieve this due to the regulatory mechanisms involved with nutrient starvation, stress and tolerance response, which are not well understood in non-conventional, fairly new microbial hosts. The engineering process itself may alter or change the tolerance, e.g. ref. 78. Non-traditional strategies have been used for the strain engineering of ethanol-producing S. cerevisiae by the overexpression of key protein kinases associated with flocculation-associated stress tolerance.79 Alternatively, the intrinsic heterogeneity of phenotypes caused by small molecule messengers has been leveraged to generate strains that segregate the population into stem cells and production cells.80 For cholesterol-like molecule production, the membrane physiology and trafficking were engineered in E. coli.81 Currently, many of these approaches remain disconnected and are implemented in isolation from each other. Thus, combining these approaches with mainstream metabolic engineering and medium optimization will allow us to develop sophisticated strains with potentially highly predictable performance in high-scale production environments.Conclusion and future scope
In this review, we discussed relevant examples of GC and DC, synthetic control for dynamic control, impact of the medium formulations, and research advances that go beyond metabolism (Table 1). Although examples of multiple carbon substrates exist for DC approaches, thus far, GC approaches have been only tested for a single carbon source. Thus, it will be interesting to see how GC-engineered strains perform with a mixed carbon source or when cultivated using rich media such as plant biomass hydrolysate or other industrial waste to make the bioconversion more sustainable and carbon neutral. In addition to designing GC strategies for the co-utilization of substrates, mixed community culturing of GC strains that are hard wired for each of the carbon sources that exist in the rich medium stream is necessary. Another challenge is the real-time unbiased comparison of different types of computational algorithms used for strain optimization, where the question remains, do they converge onto similar solutions or do they favor certain beachhead metabolites? A broader suite of methods should be tested rather than cherry picking the familiar ones. Common platform and digital resources are available, which provide details on the different methods that have been tested thus far and the respective success (TRY fold improvement) and failures to identify gaps that need to be tested further. Advancement in genome-wide editing approaches, miniaturization of production systems and combinatorial methods that integrate designs for pathway, host and production conditions is increasing to test and reach the ideal growth-production trade-off.| Product | TRYa | Host | Media | Reference | |
|---|---|---|---|---|---|
| a TRY – titer (T), rate (R) and yield (Y). b Commodity chemical has been commercialized. c AA – amino acid; CDW – cell dry weight; Gly – glycine; His – histidine; MAD – modified AD7; MOPS – 3-(N-morpholino)propanesulfonic acid; PHA – polyhydroxyalkanoate; SD – synthetic defined; Thr – threonine; TSB – tryptic soy broth; YE – yeast extract; YPD – yeast extract peptone dextrose. | |||||
| Growth coupled | |||||
| 1 | Lactic acidb | 1.75 g L−1 | E. coli | M9 glucose | Fong et al., 2005 (ref. 3) |
| 2 | 1,4 BDOb | 18 g L−1 | E. coli | M9 glucose | Yim et al., 2011 (ref. 4) |
| 3 | 1-Butanolb | 30 g L−1, 70% yield, 0.18 g L−1 h−1 | E. coli | Glucose | Shen et al., 2011 (ref. 5) |
| 4 | 1-Butanolb | 29.9 mg L−1 | Synechococcus sp. | BG-11 | Lan and Liao, 2012 (ref. 6) |
| 5 | 2-Methyl-1-butanol, isobutanol | 171 mg L−1 and 181 mg L−1 | Synechococcus sp. | MAD media | Purdy et al., 2022 (ref. 7) |
| 6 | 3-Hydroxypropionic acidb | 463 mg L−1 | S. cerevisiae | Glucose | Chen et al., 2014 (ref. 8) |
| 7 | Itaconateb | 32 g L−1, 0.68 mol mol−1 and 0.45 g L−1 h−1 | E. coli | Glucose + glutamic acid | Harder et al., 2016 (ref. 9) |
| 8 | Indigoidine | 26 g L−1 | P. putida | M9 glucose | Banerjee et al., 2020 (ref. 12) |
| 9 | Succinic acidb | 0.9 g L−1, 0.05 g g−1 | S. cerevisiae | Glucose + Gly/Thr | Otero et al., 2013 (ref. 13) |
| 10 | Ethanolb | 0.44 g g−1, 0.39 g L−1 h−1 | E. coli | Glycerol | Trinh and Srienc, 2009 (ref. 14) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Growth decoupled | |||||
| 1 | Propionic acid | 34.62 g L−1 | P. jensenii | Glycerol | Zhuge et al., 2014 (ref. 19) |
| 2 | Alpha-ketoglutarate, succinate | 11 g L−1 | C. glutamicum | Xylose | Tenhaef et al., 2021 (ref. 20) |
| 3 | Protocatechuate | 9.51 g L−1 | C. glutamicum | Glucose then xylose | Labib et al., 2021 (ref. 21) |
| 4 | Vanillin | 900 mg L−1 | E. coli | Ferulic acid | Lo et al., 2016 (ref. 22) |
| 5 | 1,2-Indandiol | 262 mg L−1 | Rhodococcus sp. | Glucose | Stafford et al., 2002 (ref. 23) |
| 6 | Polyhydroxybutyrate | 1.4 g L−1 | E. coli | Rich and minimal media glucose | Bothfeld et al., 2017 (ref. 24) |
| 7 | Fatty alcohol | 150 mg L−1, 0.03 g g−1 | E. coli | Glucose | Chubukov et al., 2017 (ref. 25) |
| 8 | Linalool | 10.9 g L−1, 5.1% yield | Pantoea ananatis | Glucose + YE | Nitta et al., 2021 (ref. 26) |
| 9 | Isobutanolb | 0.53 g L−1, 0.31 C-mol C-mol−1 | C. glutamicum | Hemicellulose fraction + YE | Lange et al., 2018 (ref. 27) |
| 10 | Proline | 142.4 g L−1, 2.9 g L−1 h−1, 0.31 g g−1 | C. glutamicum | Rich TSB media | Liu et al., 2022 (ref. 28) |
| 11 | Itaconateb | 47 g L−1, 0.86 g L−1 h−1 | E. coli | Glucose | Harder et al., 2018 (ref. 29) |
| 12 | Gamma-aminobutyric acid | 614 g L−1, 40.94 g L−1 h−1 | E. coli | Glucose + YE | Ke et al., 2016 (ref. 30) |
| 13 | Alpha-hydroxy ketones | 62–84% yield | E. coli | Pyruvate and benzaldehyde | Liang et al., 2020 (ref. 31) |
| 14 | Caprolactone | 126 mM, 0.78 mol mol−1 | E. coli | Cyclohexanol | Xiong et al., 2021 (ref. 32) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Feedback loops | |||||
| 1 | Vanillin | 900 mg L−1 | E. coli | Ferulic acid | Lo et al., 2016 (ref. 22) |
| 2 | Naringenin | 523.7 mg L−1 | E. coli | MOPS glucose, glycerol, potassium acetate, palmitic acid, stearic acid | Zhou et al., 2021 (ref. 38) |
| 3 | Gamma-aminobutyric acid | 45.6 g L−1, 0.4 g g−1 | C. glutamicum | Glycerol | Wei et al., 2022 (ref. 39) |
| 4 | Salicylic acid | 520 mg L−1 | E. coli | M9 glucose + glycerol | Dinh and Prather, 2019 (ref. 40) |
| 5 | Itaconateb | 1.4 g L−1 | P. putida | Deconstructed lignin | Elmore et al., 2021 (ref. 41) |
| 6 | Lycopene | 150.9 mg L−1 | E. coli | Glucose + YE + peptone | Li et al., 2020 (ref. 42) |
| 7 | Insulated genetic landing pads with logic gate circuits | — | E. coli | M9 glucose + thiamine and casamino acids | Park et al., 2020 (ref. 44) |
| 8 | Insulated genetic landing pads with logic gate circuits | — | E. coli, S. cerevisiae, B. thetaiotaomicron | — | Jones et al., 2022 (ref. 45) |
| 9 | N-Acetylglucosamine | 0.2 g L−1 | S. cerevisiae | Glucose + SD-Leu + yeast nitrogen base without AA | Lee et al., 2021 (ref. 48) |
| 10 | Isobutanol | 8.49 g L−1, 53.5 mg g−1 | S. cerevisiae | YPD or SC-dropout medium with 2% glucose and branched AA | Zhao et al., 2018 (ref. 50) |
| 11 | Lactic acid | 6 g L−1 | S. cerevisiae | YPD or SC-His with 2% glucose | Zhao et al., 2021 (ref. 51) |
| 12 | Shikimic acid | 35 g L−1, 0.43 g g−1, 0.48 g L−1 h−1 | E. coli | Glucose minimal media | Komera et al., 2022 (ref. 52) |
| 13 | Nerolidol | 4 g L−1, 3.8–4.5% yield | S. cerevisiae | Glucose, sucrose, glucose + ethanol | Peng et al., 2017 (ref. 55) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Media optimization | |||||
| 1 | Fatty acids | 38% energy yield | M. thermoacetica and Y. lipolytica | CO2, glucose and gluconate, acetate | Park et al., 2019 (ref. 56) |
| 2 | Itaconate | 20 g L−1 | E. coli | Glycerol and xylose | Lu et al., 2021 (ref. 57) |
| 3 | — | — | E. coli | Rich and minimal media | Moreb et al., 2020 (ref. 58) |
| 4 | PHA | 295.7 mg L−1, 19.3% yield per CDW | P. putida and S. cerevisiae | Xylose and octanoate | Wei et al., 2022 (ref. 59) |
| 5 | PHA | 1.32 g L−1 | P. putida and E. coli | Glucose or xylose minimal media | Zhu et al., 2021 (ref. 60) |
| 4 | PHA | 48.5% yield per CDW | P. putida | E2 minimal media with glucose, fructose, glycerol, octanoate or decanoate | Huijberts et al., 1992 (ref. 65) |
| 5 | PHA | 41.5 g L−1, 67% yield per CDW, 0.83 g L−1h− 1 | P. putida | Glucose minimal media | Poblete-Castro et al., 2014 (ref. 66) |
Author contributions
DB and AM conceived the study. DB wrote the first draft of the manuscript and prepared figures. AM reviewed and edited the final manuscript. All authors have read and approved the final version of this manuscript for publication.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This perspective is dedicated to celebrating Prof. David G. Lynn’s 70th birthday. Both DB and AM are funded by the Joint BioEnergy Institute (https://www.jbei.org) supported by the US Department of Energy, Office of Science, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory (LBNL) and the US Department of Energy.References
- N. R. Baral, O. Kavvada, D. Mendez-Perez, A. Mukhopadhyay, T. S. Lee, B. A. Simmons and C. D. Scown, Energy Environ. Sci., 2019, 12, 807–824 RSC.
- M. Wehrs, D. Tanjore, T. Eng, J. Lievense, T. R. Pray and A. Mukhopadhyay, Trends Microbiol., 2019, 27, 524–537 CrossRef CAS PubMed.
- S. S. Fong, A. P. Burgard, C. D. Herring, E. M. Knight, F. R. Blattner, C. D. Maranas and B. O. Palsson, Biotechnol. Bioeng., 2005, 91, 643–648 CrossRef CAS PubMed.
- H. Yim, R. Haselbeck, W. Niu, C. Pujol-Baxley, A. Burgard, J. Boldt, J. Khandurina, J. D. Trawick, R. E. Osterhout, R. Stephen, J. Estadilla, S. Teisan, H. B. Schreyer, S. Andrae, T. H. Yang, S. Y. Lee, M. J. Burk and S. Van Dien, Nat. Chem. Biol., 2011, 7, 445–452 CrossRef CAS PubMed.
- C. R. Shen, E. I. Lan, Y. Dekishima, A. Baez, K. M. Cho and J. C. Liao, Appl. Environ. Microbiol., 2011, 77, 2905–2915 CrossRef CAS PubMed.
- E. I. Lan and J. C. Liao, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 6018–6023 CrossRef CAS PubMed.
- H. M. Purdy, B. F. Pfleger and J. L. Reed, Metab. Eng., 2022, 69, 87–97 CrossRef CAS PubMed.
- Y. Chen, J. Bao, I.-K. Kim, V. Siewers and J. Nielsen, Metab. Eng., 2014, 22, 104–109 CrossRef CAS PubMed.
- B.-J. Harder, K. Bettenbrock and S. Klamt, Metab. Eng., 2016, 38, 29–37 CrossRef CAS PubMed.
- A. von Kamp and S. Klamt, Nat. Commun., 2017, 8, 15956 CrossRef CAS PubMed.
- P. Schneider, A. von Kamp and S. Klamt, PLoS Comput. Biol., 2020, 16, e1008110 CrossRef CAS PubMed.
- D. Banerjee, T. Eng, A. K. Lau, Y. Sasaki, B. Wang, Y. Chen, J.-P. Prahl, V. R. Singan, R. A. Herbert, Y. Liu, D. Tanjore, C. J. Petzold, J. D. Keasling and A. Mukhopadhyay, Nat. Commun., 2020, 11, 5385 CrossRef CAS PubMed.
- J. M. Otero, D. Cimini, K. R. Patil, S. G. Poulsen, L. Olsson and J. Nielsen, PLoS One, 2013, 8, e54144 CrossRef CAS PubMed.
- C. T. Trinh and F. Srienc, Appl. Environ. Microbiol., 2009, 75, 6696–6705 CrossRef CAS PubMed.
- E. Orsi, N. J. Claassens, P. I. Nikel and S. N. Lindner, Nat. Commun., 2021, 12, 5295 CrossRef CAS PubMed.
- P. Schneider, R. Mahadevan and S. Klamt, Biotechnol. J., 2021, 16, e2100236 CrossRef PubMed.
- A. Cros, G. Alfaro-Espinoza, A. De Maria, N. T. Wirth and P. I. Nikel, Curr. Opin. Biotechnol., 2022, 74, 180–193 CrossRef CAS PubMed.
- J. Chen, Y. Wang, P. Zheng and J. Sun, Trends Biotechnol., 2022, 40, 773–776 CrossRef CAS PubMed.
- X. Zhuge, L. Liu, H. Shin, J. Li, G. Du and J. Chen, Bioresour. Technol., 2014, 152, 519–525 CrossRef CAS PubMed.
- N. Tenhaef, J. Kappelmann, A. Eich, M. Weiske, L. Brieß, C. Brüsseler, J. Marienhagen, W. Wiechert and S. Noack, Biotechnol. J., 2021, 16, e2100043 CrossRef PubMed.
- M. Labib, J. Görtz, C. Brüsseler, N. Kallscheuer, J. Gätgens, A. Jupke, J. Marienhagen and S. Noack, Biotechnol. Bioeng., 2021, 118, 4414–4427 CrossRef CAS PubMed.
- T.-M. Lo, S. H. Chng, W. S. Teo, H.-S. Cho and M. W. Chang, Cell Syst., 2016, 3, 133–143 CrossRef CAS PubMed.
- D. E. Stafford, K. S. Yanagimachi, P. A. Lessard, S. K. Rijhwani, A. J. Sinskey and G. Stephanopoulos, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 1801–1806 CrossRef CAS PubMed.
- W. Bothfeld, G. Kapov and K. E. J. Tyo, ACS Synth. Biol., 2017, 6, 1296–1304 CrossRef CAS PubMed.
- V. Chubukov, J. J. Desmarais, G. Wang, L. J. G. Chan, E. E. Baidoo, C. J. Petzold, J. D. Keasling and A. Mukhopadhyay, npj Syst. Biol. Appl., 2017, 3, 16035 CrossRef PubMed.
- N. Nitta, Y. Tajima, Y. Yamamoto, M. Moriya, A. Matsudaira, Y. Hoshino, Y. Nishio and Y. Usuda, Microb. Cell Fact., 2021, 20, 54 CrossRef CAS PubMed.
- J. Lange, F. Müller, R. Takors and B. Blombach, Microb. Biotechnol., 2018, 11, 257–263 CrossRef CAS PubMed.
- J. Liu, M. Liu, T. Shi, G. Sun, N. Gao, X. Zhao, X. Guo, X. Ni, Q. Yuan, J. Feng, Z. Liu, Y. Guo, J. Chen, Y. Wang, P. Zheng and J. Sun, Nat. Commun., 2022, 13, 891 CrossRef CAS PubMed.
- S. Zhang, Y. Fang, L. Zhu, H. Li, Z. Wang, Y. Li and X. Wang, Syst. Microbiol. Biomanuf., 2021, 1, 444–458 CrossRef CAS.
- B.-J. Harder, K. Bettenbrock and S. Klamt, Biotechnol. Bioeng., 2018, 115, 156–164 CrossRef CAS PubMed.
- C. Ke, X. Yang, H. Rao, W. Zeng, M. Hu, Y. Tao and J. Huang, SpringerPlus, 2016, 5, 591 CrossRef PubMed.
- Y.-F. Liang, L.-T. Yan, Q. Yue, J.-K. Zhao, C.-Y. Luo, F. Gao, H. Li and W.-Y. Gao, Sci. Rep., 2020, 10, 15404 CrossRef CAS PubMed.
- J. Xiong, H. Chen, R. Liu, H. Yu, M. Zhuo, T. Zhou and S. Li, Bioresour. Bioprocess., 2021, 8, 32 CrossRef.
- K. Raj, N. Venayak and R. Mahadevan, Metab. Eng., 2020, 62, 186–197 CrossRef CAS PubMed.
- F. Jacob and J. Monod, J. Mol. Biol., 1961, 3, 318–356 CrossRef CAS PubMed.
- A. Pickar-Oliver and C. A. Gersbach, Nat. Rev. Mol. Cell Biol., 2019, 20, 490–507 CrossRef CAS PubMed.
- A. V. Anzalone, L. W. Koblan and D. R. Liu, Nat. Biotechnol., 2020, 38, 824–844 CrossRef CAS PubMed.
- S. Zhou, S.-F. Yuan, P. H. Nair, H. S. Alper, Y. Deng and J. Zhou, Metab. Eng., 2021, 67, 41–52 CrossRef CAS PubMed.
- L. Wei, J. Zhao, Y. Wang, J. Gao, M. Du, Y. Zhang, N. Xu, H. Du, J. Ju, Q. Liu and J. Liu, Metab. Eng., 2022, 69, 134–146 CrossRef CAS PubMed.
- C. V. Dinh and K. L. J. Prather, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 25562–25568 CrossRef CAS PubMed.
- J. R. Elmore, G. N. Dexter, D. Salvachúa, J. Martinez-Baird, E. A. Hatmaker, J. D. Huenemann, D. M. Klingeman, G. L. Peabody, D. J. Peterson, C. Singer, G. T. Beckham and A. M. Guss, Nat. Commun., 2021, 12, 2261 CrossRef CAS PubMed.
- C. Li, X. Gao, X. Peng, J. Li, W. Bai, J. Zhong, M. He, K. Xu, Y. Wang and C. Li, Microb. Cell Fact., 2020, 19, 202 CrossRef CAS PubMed.
- C. J. Hartline and F. Zhang, ACS Synth. Biol., 2022, 11, 2247–2258 CrossRef CAS PubMed.
- Y. Park, A. Espah Borujeni, T. E. Gorochowski, J. Shin and C. A. Voigt, Mol. Syst. Biol., 2020, 16, e9584 CrossRef CAS PubMed.
- T. S. Jones, S. M. D. Oliveira, C. J. Myers, C. A. Voigt and D. Densmore, Nat. Protoc., 2022, 17, 1097–1113 CrossRef CAS PubMed.
- B. K. Verma, A. A. Mannan, F. Zhang and D. A. Oyarzún, ACS Synth. Biol., 2022, 11, 228–240 CrossRef CAS PubMed.
- Y. Boada, F. N. Santos-Navarro, J. Picó and A. Vignoni, Front. Mol. Biosci., 2022, 9, 801032 CrossRef CAS PubMed.
- S.-W. Lee, P. Rugbjerg and M. O. A. Sommer, ACS Synth. Biol., 2021, 10, 2842–2849 CrossRef CAS PubMed.
- Y. Teng, J. Zhang, T. Jiang, Y. Zou, X. Gong and Y. Yan, Curr. Opin. Biotechnol., 2022, 75, 102696 CrossRef CAS PubMed.
- E. M. Zhao, Y. Zhang, J. Mehl, H. Park, M. A. Lalwani, J. E. Toettcher and J. L. Avalos, Nature, 2018, 555, 683–687 CrossRef CAS PubMed.
- E. M. Zhao, M. A. Lalwani, J.-M. Chen, P. Orillac, J. E. Toettcher and J. L. Avalos, ACS Synth. Biol., 2021, 10, 1143–1154 CrossRef CAS PubMed.
- I. Komera, C. Gao, L. Guo, G. Hu, X. Chen and L. Liu, Biotechnol. Biofuels Bioprod., 2022, 15, 13 CrossRef CAS PubMed.
- V. V. Reshetnikov, S. V. Smolskaya, S. G. Feoktistova and V. V. Verkhusha, Trends Biotechnol., 2022, 40, 858–874 CrossRef CAS PubMed.
- T. S. Moon, C. Lou, A. Tamsir, B. C. Stanton and C. A. Voigt, Nature, 2012, 491, 249–253 CrossRef CAS PubMed.
- B. Peng, M. R. Plan, A. Carpenter, L. K. Nielsen and C. E. Vickers, Biotechnol. Biofuels, 2017, 10, 43 CrossRef PubMed.
- J. O. Park, N. Liu, K. M. Holinski, D. F. Emerson, K. Qiao, B. M. Woolston, J. Xu, Z. Lazar, M. A. Islam, C. Vidoudez, P. R. Girguis and G. Stephanopoulos, Nat. Metab., 2019, 1, 643–651 CrossRef CAS PubMed.
- K. W. Lu, C. T. Wang, H. Chang, R. S. Wang and C. R. Shen, Metab. Eng. Commun., 2021, 13, e00190 CrossRef CAS PubMed.
- E. A. Moreb, Z. Ye, J. P. Efromson, J. N. Hennigan, R. Menacho-Melgar and M. D. Lynch, ACS Synth. Biol., 2020, 9, 1483–1486 CrossRef CAS PubMed.
- Z. Wei, Y. Zhu, M. Ai, C. Liu and X. Jia, Can. J. Chem. Eng., 2022 DOI:10.1002/cjce.24751 , article ASAP.
- Y. Zhu, M. Ai and X. Jia, Front. Bioeng. Biotechnol., 2021, 9, 794331 CrossRef PubMed.
- M. Mauri, J.-L. Gouzé, H. de Jong and E. Cinquemani, PLoS Comput. Biol., 2020, 16, e1007795 CrossRef CAS PubMed.
- H. Joshi, J. Isar, D. A. Jain, S. S. Badle, S. B. Dhoot and V. Rangaswamy, Appl. Biochem. Biotechnol., 2021, 193, 2403–2419 CrossRef CAS PubMed.
- M. Wehrs, J. M. Gladden, Y. Liu, L. Platz, J.-P. Prahl, J. Moon, G. Papa, E. Sundstrom, G. M. Geiselman, D. Tanjore, J. D. Keasling, T. R. Pray, B. A. Simmons and A. Mukhopadhyay, Green Chem., 2019, 21, 3394–3406 RSC.
- Y. Sasaki, T. Eng, R. A. Herbert, J. Trinh, Y. Chen, A. Rodriguez, J. Gladden, B. A. Simmons, C. J. Petzold and A. Mukhopadhyay, Biotechnol. Biofuels, 2019, 12, 41 CrossRef PubMed.
- G. N. Huijberts, G. Eggink, P. de Waard, G. W. Huisman and B. Witholt, Appl. Environ. Microbiol., 1992, 58, 536–544 CrossRef CAS PubMed.
- I. Poblete-Castro, A. L. Rodriguez, C. M. C. Lam and W. Kessler, J. Microbiol. Biotechnol., 2014, 24, 59–69 CrossRef CAS PubMed.
- L. F. Huergo and R. Dixon, Microbiol. Mol. Biol. Rev., 2015, 79, 419–435 CrossRef CAS PubMed.
- M. Wehrs, J.-P. Prahl, J. Moon, Y. Li, D. Tanjore, J. D. Keasling, T. Pray and A. Mukhopadhyay, Microb. Cell Fact., 2018, 17, 193 CrossRef CAS PubMed.
- M. G. Thompson, L. E. Valencia, J. M. Blake-Hedges, P. Cruz-Morales, A. E. Velasquez, A. N. Pearson, L. N. Sermeno, W. A. Sharpless, V. T. Benites, Y. Chen, E. E. K. Baidoo, C. J. Petzold, A. M. Deutschbauer and J. D. Keasling, Metab. Eng. Commun., 2019, 9, e00098 CrossRef PubMed.
- M. G. Thompson, M. R. Incha, A. N. Pearson, M. Schmidt, W. A. Sharpless, C. B. Eiben, P. Cruz-Morales, J. M. Blake-Hedges, Y. Liu, C. A. Adams, R. W. Haushalter, R. N. Krishna, P. Lichtner, L. M. Blank, A. Mukhopadhyay, A. M. Deutschbauer, P. M. Shih and J. D. Keasling, Appl. Environ. Microbiol., 2020, 86, 21 CrossRef PubMed.
- S. D. Doig, F. Baganz and G. J. Lye, in Basic Biotechnology, ed. C. Ratledge and B. Kristiansen, Cambridge University Press, 2006, pp. 289–306 Search PubMed.
- P. Rohe, D. Venkanna, B. Kleine, R. Freudl and M. Oldiges, Microb. Cell Fact., 2012, 11, 144 CrossRef CAS PubMed.
- V. Singh, S. Haque, R. Niwas, A. Srivastava, M. Pasupuleti and C. K. M. Tripathi, Front. Microb., 2016, 7, 2087 Search PubMed.
- L. F. Motta Dos Santos, F. Coutte, R. Ravallec, P. Dhulster, L. Tournier-Couturier and P. Jacques, Bioresour. Technol., 2016, 218, 944–952 CrossRef CAS PubMed.
- P. Biniarz, F. Coutte, F. Gancel and M. Łukaszewicz, Microb. Cell Fact., 2018, 17, 121 CrossRef PubMed.
- K. Iwai, M. Wehrs, M. Garber, J. Sustarich, L. Washburn, Z. Costello, P. W. Kim, D. Ando, W. R. Gaillard, N. J. Hillson, P. D. Adams, A. Mukhopadhyay, H. Garcia Martin and A. K. Singh, Microsyst. Nanoeng., 2022, 8, 31 CrossRef CAS PubMed.
- G. Wang, C. Haringa, H. Noorman, J. Chu and Y. Zhuang, Trends Biotechnol., 2020, 38, 846–856 CrossRef CAS PubMed.
- J. Gao, Y. Li, W. Yu and Y. J. Zhou, Nat. Metab., 2022, 4, 932–943 CrossRef CAS PubMed.
- P.-L. Ye, X.-Q. Wang, B. Yuan, C.-G. Liu and X.-Q. Zhao, Bioresour. Technol., 2022, 348, 126758 CrossRef CAS PubMed.
- G. Bowman, M. Gomelsky and N. Mushnikov, Microbial Stem Cell Technology, US pat., 20190322980 A1, 2019.
- M. C. Santoscoy and L. R. Jarboe, Metab. Eng., 2022, 73, 134–143 CrossRef CAS PubMed.
- X. Wang, E. E. K. Baidoo, R. Kakumanu, S. Xie, A. Mukhopadhyay and T. S. Lee, Biotechnol. Biofuels, 2022, 15, 137 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |