Open Access Article
Open Access ArticleLipid-based colloidal nanoparticles for applications in targeted vaccine delivery
Muhammad Saad
Khan
ab,
Sila Appak
Baskoy
bc,
Celina
Yang
ab,
Joohye
Hong
d,
Jayoung
Chae
d,
Heejin
Ha
d,
Sungjun
Lee
de,
Masayoshi
Tanaka
 f,
Yonghyun
Choi
*de and
Jonghoon
Choi
f,
Yonghyun
Choi
*de and
Jonghoon
Choi
 *de
*de
aDepartment of Physics, Toronto Metropolitan University, 350 Victoria Street, Toronto, M5B2K3, Canada
bInstitute for Biomedical Engineering, Science and Technology (iBEST), St. Michael's Hospital, 209 Victoria Street, Toronto, M5B1W8, Canada
cDepartment of Chemistry and Biology, Toronto Metropolitan University, Faculty of Science, 350 Victoria Street, Toronto, M5B2K3, ON, Canada
dSchool of Integrative Engineering, Chung-Ang University, Seoul 06974, Republic of Korea. E-mail: dydgus5057@cau.ac.kr; nanomed@cau.ac.kr
eFeynman Institute of Technology, Nanomedicine Corporation, Seoul 06974, Republic of Korea
fDepartment of Chemical Science and Engineering, Tokyo Institute of Technology, 4259 Nagatsuta-cho, Midori-ku, Yokohama-shi 226-8503, Kanagawa, Japan
First published on 17th February 2023
Abstract
Bioactive molecules and their effects have been influenced by their solubility and administration route. In many therapeutic reagents, the performance of therapeutics is dependent on physiological barriers in the human body and delivery efficacy. Therefore, an effective and stable therapeutic delivery promotes pharmaceutical advancement and suitable biological usage of drugs. In the biological and pharmacological industries, lipid nanoparticles (LNPs) have emerged as a potential carrier to deliver therapeutics. Since studies reported doxorubicin-loaded liposomes (Doxil®), LNPs have been applied to numerous clinical trials. Lipid-based nanoparticles, including liposomes, solid lipid nanoparticles (SLNs), and nanostructured lipid nanoparticles, have also been developed to deliver active ingredients in vaccines. In this review, we present the type of LNPs used to develop vaccines with attractive advantages. We then discuss messenger RNA (mRNA) delivery for the clinical application of mRNA therapeutic-loaded LNPs and recent research trend of LNP-based vaccine development.
1. Introduction
Vaccination is one of the most significant and effective strategies to control the spread of infectious diseases. The first introduced human vaccines against viruses were attenuated live viruses to generate immunity. According to the National Center for Immunization and Respiratory Diseases (NCIRD), many diseases, including smallpox, polio, diphtheria, mumps, measles, and rubella, have been eradicated or nearly eliminated through widespread vaccination efforts. Over the last several decades, various types of vaccines, such as subunit and conjugate vaccines which contain only parts of their target pathogens, have been developed.DNA and RNA have been widely investigated in vaccine technologies since their nucleotide sequence-independent characteristics allow cost- and time-effective development of new vaccines that target new diseases or new strains of diseases. However, these therapeutic methods require suitable delivery systems because DNA requires nuclear delivery and RNA requires cytoplasmic delivery.1 Nanomedicine is an emerging branch of medicine that uses nanotechnology that can be tailored to specific diseases and target sites depending on the diagnosis, treatment, and prevention.2–7 Nanotechnology research has gained popularity, and billions of dollars are spent on the development of goods incorporating nanotechnology each year.8–11 Nanoparticles are advantageous because they can be tailored according to specific diagnostic or therapeutic applications due to their physiochemical properties and nanosize.12 In diagnostic and bioimaging techniques, nanoparticles can be engineered to target specific cells or biomarkers. In therapeutic systems, specific biomolecules, such as antibodies, proteins, and therapeutics, can be incorporated into nanoparticles for drug delivery applications. Therefore, nanoparticles possess unique advantages over traditional medicine because of drug loading capabilities, cellular uptake, and targeting abilities8,13–15 Nanoparticles can be used to solve key antiviral drug-related challenges, including low solubility and low bioavailability, by enhancing their pharmacokinetic properties to reduce dose and toxicity, improve drug bioavailability, and suppress viral spread.16
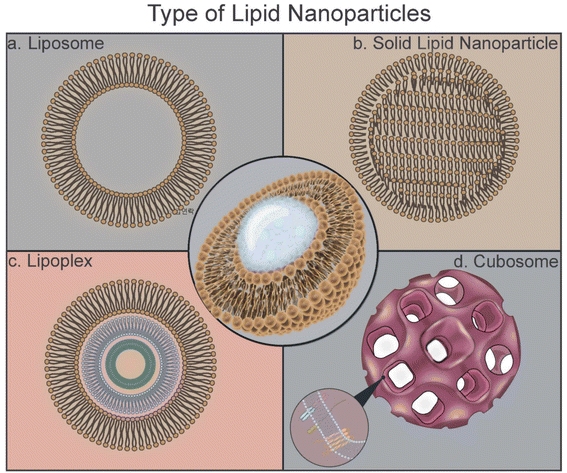
Natural and synthetic lipids are conventional materials that can be tailored to form nanoparticles via varied conjugation processes, such as sonication, extrusion, or the reverse-phase evaporation method.17 Lipid nanoparticles (LNPs) include liposomes, lipoplexes, solid lipid nanoparticles, lipid-based nanoemulsions, and nanostructured lipid carriers. The characteristics of LNPs are suitable for drug and vaccine delivery applications because of their enhanced biodistribution, biocompatibility, biodegradability, high cellular uptake, long half-life, and ease of synthesis. Phospholipids enhance the ability of LNPs in endosomal escape. Polyethylene glycol (PEG) and cholesterols improve stability and shelf-life. Therefore, LNPs are promising nonviral delivery carriers.
In this review, the types of LNPs, their synthesis techniques, formulations, triggered releases, and potential in vaccine applications are summarized (Fig. 1).13
2. Nucleic acids for therapeutics and vaccine applications
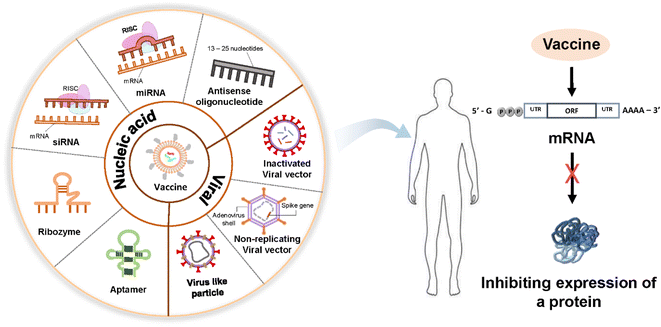
DNA and RNA are two linear polymers responsible for carrying all the genetic information of organisms. DNA is a double-stranded molecule that is much larger than a single-stranded RNA molecule. Their structures have similarities: they have a sugar backbone, phosphates, and nucleobases. However, the sugar in a DNA molecule is deoxyribose, whose number of hydroxyl groups is one less than that of ribose in RNA. DNA is more stable than RNA because of the lack of one oxygen-containing hydroxyl group, double-stranded nature, possession of a damage response and repair mechanisms, and presence of histone proteins.18 DNA stability is necessary to maintain all the genetic information of organisms through evolution. Although DNA and RNA have a phosphate group and a sugar molecule, RNA carries the nucleobase uracil instead of the nitrogenous base thymine in DNA.Targeted DNA or RNA inhibition, which consequently inhibits the expression of a protein responsible for a certain disease, underlines the common aim of all nucleic acid-based therapeutics except gene therapy (Fig. 2).19,20 The common mechanism in nucleic acid-based therapies is mediated by the sequence-specific recognition of endogenous nucleic acids through Watson–Crick base pairing. Some examples of nucleic acid-based drugs are plasmids, oligonucleotides, aptamers, ribozymes, and small-interfering ribonucleic acids, which can be delivered through nonviral nanocarriers, such as liposomes and cationic polymers. Viral vectors can be used for direct nucleic acid transfer.21
In DNA therapeutics, antisense oligonucleotides, DNA aptamers, and gene therapy are used. In RNA therapeutics, antisense oligonucleotides, such as microRNAs (mRNA) and agents of RNA interference, including short-interfering RNAs (siRNA) and short hairpin RNA (shRNA), are used to block mRNA synthesis. In addition, ligand RNAs (such as aptamers and decoys) and catalytic RNAs (such as ribozymes) are important components of RNA therapeutics.
2.1. DNA therapeutics
2.2. RNA-based therapeutics
The exploration of the RNA moiety of ribonuclease P as the catalytic subunit of this enzyme in 1983 (ref. 28) and other important discoveries in RNA biology such as RNA interference have further elucidated the function of RNA in gene regulation and protein synthesis that enables the utility of RNA as a therapeutic tool or a target. Generally, RNA-based therapies are mostly targeted against proteins or nucleic acids; however, new approaches that can target both proteins and nucleic acids have been developed.29RNA-based therapeutics have different mechanisms of activities. These mechanisms include mRNA translation inhibitors (antisense), RNA interference (RNAi) agents, catalytically active RNA molecules (ribozymes), and RNAs that bind to proteins and other molecular ligands (aptamers).30
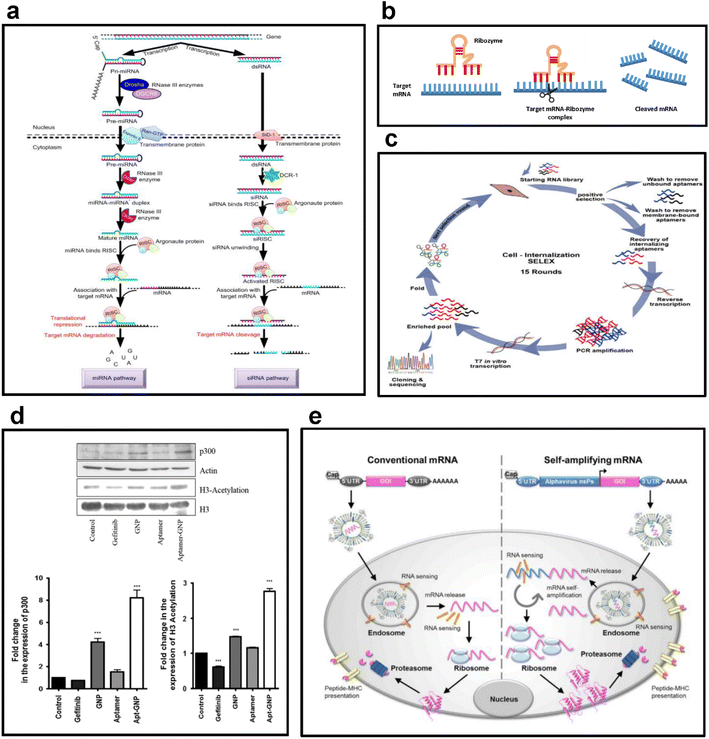 | ||
| Fig. 3 (a) Examples of antisense RNA. This figure shows the formation and regulatory mechanisms of miRNA and siRNA, the most used forms of antisense RNA. Reproduced with permission from ref. 56 (Copyright (2018) by Springer Nature). (b) Mechanism of action of ribozymes. This figure shows the sequence of processes through which a ribozyme binds to a target mRNA to form an mRNA-ribozyme complex and cleaves the target mRNA to suppress gene expression. Replicated with permission from ref. 57 (Copyright (2019) by MDPI) and made by the author. (c) Selection strategy for RNA aptamers. This figure shows the cycle of cell internalization SELEX for internalizing-RNA aptamer isolation for skeletal muscles. Reproduced with permission from ref. 58 (Copyright (2018) by Elsevier B.V.). (d) Selective targeting ability of RNA aptamers. This figure shows the results of evaluating the changes in the expression of p300 and H3 acetylation after the treatment of H1975 cells with 8 μM gefitinib, GNP, aptamer, and apt-GNP. Reproduced with permission from ref. 59 (Copyright (2015) by Springer Nature). (e) Antigen expression mechanism of mRNA vaccines. This figure schematically shows the structures of conventional mRNA and self-amplifying mRNA vaccines and exhibits their overall antigen-generating process. Reproduced with permission from ref. 60 (Copyright (2019) by Elsevier Inc.). | ||
Some miRNAs, such as let-7 (the first miRNA discovered), act as tumor suppressors by regulating the expression of Ras.35 In the analysis of the let-7 expression in normal and tumor lung tissues, the let-7 expression of normal tissues is significantly higher than that in tumor tissues, and let-7 regulates cellular proliferation.36 The expression of miRNAs in certain tumors and liquid biopsies is also correlated with survival and treatment response in cancer. Some miRNAs are important mediators of the development of the heart, muscle, liver, pancreas, central nervous system, and hematopoiesis, in addition to their role in cancer progression, diagnosis, and therapy.37–40
The role of miRNAs in several pathological and physiological processes can be investigated with the gain of function and loss of function studies, and silencing of miRNAs is maintained with specific oligonucleotides known as antagomirs that inhibit miRNA binding to its target.41 Antagomirs can be considered important therapeutic agents in disease treatment and enhanced delivery systems, which can deliver small RNAs that help their targeted delivery to the correct cell types without enhancing immune responses; therefore, they will be useful to ameliorate various diseases.
In recent years, siRNAs have been attractive therapeutic tools for the treatment of genetic diseases, metabolic diseases, and cancer. The first RNAi-based therapeutic tool is Patisiran, ALN-TTR02, which was approved to be used in adults in 2018 for the treatment of hereditary amyloidogenic transthyretin amyloidosis with polyneuropathy.
RNA aptamers have specific advantages over other therapeutic RNA molecules since aptamers not only enter cells but also act on extracellular targets by directly binding to them to inhibit or activate target functions (Fig. 3d).49 With the ability of RNA aptamers to bind to extracellular targets, they are considered a versatile tool to deliver RNA-based therapeutics to combat various human diseases. In 2004, the first RNA aptamer drug Macugen (pegaptanib sodium) that blocks the activity of vascular endothelial growth factor was approved for the treatment of neovascular age-related macular degeneration to inhibit angiogenesis. Since then, many RNA aptamers have been used in clinical trials. RNA aptamers have also been used for imaging and diagnostic purposes.50
The process of mRNA-based vaccine development is rather simple. A target pathogen is identified, and its gene is sequenced. Then, the mRNA is loaded into a nanoparticle and injected into the subject. Host cells translate these mRNAs into antigens to trigger the immune response of hosts. The mechanism is similar to viral infections that activate immune responses (Fig. 3e). Incorporating mRNA into a suitable nanoparticle is critical for its protection from extracellular degradation and enhancement of cellular uptake.55 Since the procedure is synthetic, a sequence can be designed and delivered as an mRNA vaccine. mRNA vaccines can be rapidly produced according to a target pathogen, which makes them suitable for a rapid response. mRNA can either be encapsulated in a delivery vesicle or bound to a surface through electrostatic interaction. In both cases, the vesicle must be taken up by the cell most likely through endocytosis pathways. mRNA is unloaded in the cytosol after particle degradation, and the translation process begins.
Moderna, Inc. and Pfizer-BioNTech developed mRNA vaccines with nucleoside-modified mRNA formulated in lipid nanoparticles. After the intramuscular injection of these vaccines, cells take up lipid nanoparticles, effectively delivering the mRNA sequence into host cells for the expression of the SARS-CoV-2 S antigen, which induces neutralizing antibodies and cellular immune responses to the spike (S) antigen. These responses in turn help protect against COVID-19.
2.3. Gene therapy and genomic-based vaccine development
In 2001, the results of the Human Genome Project (HGP) that aimed at sequencing the whole human DNA were published by two research groups in Nature and Science.61,62 HGP has been considered the most important project in biology and biotechnology;63 the data acquired from the project enabled researchers to analyze genomic data at various levels and develop big-data high-throughput technologies that uncover information at genomic, transcriptomic, metabolomic, and epigenomic levels. Although the mortality rate and life expectancy of many diseases have not changed since the results of the HGP were conveyed, they helped researchers gain information as a basis for the development of genomic-based therapeutics.64 The first nucleic acid vaccine was formulated using naked or non-formulated plasmid DNA obtained via bacteria and it generated a potent immune response against influenza virus. However, since plasmid DNA is restricted to reach the nuclear membrane after the injection, there was a weak and non-sustaining immune response.65 Although device-mediated delivery such as electroporation and jet injectors have been able to improve delivery, equipment requirements and high working volume have limited the widespread use of plasmid DNA vaccines.66 Numerous clinical trials have therefore focused on appropriate routes of delivery and optimization strategies for various DNA vaccines to enhance their immunogenicity.67 Moreover, the use of mRNA as a recent vaccine platform has attracted considerable interest. Although, mRNA vaccines had many limitations, such as nuclease degradation, instability, and unwanted innate immune system stimulation, advances in mRNA delivery and immunology over the past decade have led to the emergence of several mRNA vaccines that are in clinical trials today.68 In particular, the Pfizer/BioNTech (BNT162b2)69 and Moderna (mRNA1273)70 vaccines developed against COVID-19 have made the greatest contribution to the widespread acceptance of the use of mRNA vaccines. This effectively showed that mRNA vaccines are rapidly customizable for any disease.713. Composition and characteristics of LNPs
Lipid nanoparticles have been actively studied for decades as an effective carrier capable of delivering various therapeutic agents. Lipid-based nanoparticles include liposomes, solid lipid nanoparticles (SLN), and nanostructured lipid carriers (NLCs). Many clinical trials have shown that they can deliver both hydrophilic and hydrophobic therapeutics.72,73 In addition, LNPs have recently been attracting great attention as an essential component of COVID-19 mRNA vaccines by effectively protecting and delivering mRNA to cells. As such, since LNPs exhibit characteristics such as high biocompatibility and targeting efficiency, they are attracting great attention as nucleic acid delivery systems such as DNA, mRNA, and siRNA.74Most DNA vaccines using lipid-based delivery systems are in the development and clinical stages, and only a small number have been approved by the FDA. Mosquirix (RTS,S/AS01), which is currently in phase 3 clinical trials, is a vaccine against malaria caused by Plasmodium falciparum.75,76 It consists of RTS,S using recombinant DNA technology and AS01, a liposome-based adjuvant. Shingrix is an FDA-approved vaccine for the prevention of shingles in adults over the age of 50. Shingrix is also a vaccine using recombinant DNA technology with the composition of AS01, a liposome-based adjuvant.77
Many RNA vaccines using lipid formulations have been approved. Representatively, BNT162b2 (Pfizer-BioNTech) and mRNA-1273 vaccine (Moderna), which have been clinically approved and approved for COVID-19, are mRNA vaccines using LNP formulations. A number of LNP-formulated mRNA vaccines are in clinical and approval stages, with Moderna's mRNA vaccines against influenza virus mRNA-1440 (H10N8) and mRNA-1851 (H7N9) completing phase 1 clinical trials being another example.78 More specifically, Patisiran (ONPATTRO), is a therapeutic agent that delivers siRNA using a liposome formulation and was approved by the FDA in 2018.79 Another lipid formulation is cationic nanoemulsion (CNE). The CNE formulation was used as a vaccine adjuvant in studies to deliver self-amplifying mRNA expressing H1 haemagglutinin of influenza virus.80–82 In this section, the characteristics and compositions of various LNPs are discussed by focusing on their applications in vaccines (Fig. 4a).
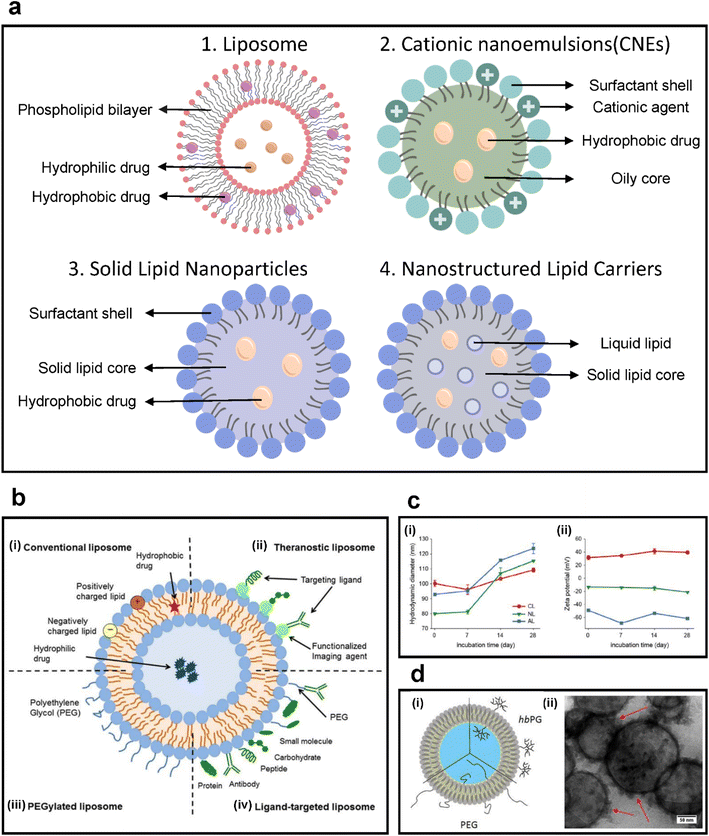 | ||
| Fig. 4 (a) Types of lipid-based colloidal nanoparticles. (b) Scheme of various modifications on liposomal drug delivery systems. (i) Conventional liposomes, (ii) theragnostic liposomes, (iii) PEGylated liposomes, and (iv) ligand-targeted liposomes. Reproduced with permission from ref. 83 (Copyright (2018) by Frontiers Media S.A.). (c) Characterization of three liposomes: cationic liposomes (CL), neutral liposomes (NL), and anionic liposomes (AL). (i) Hydrodynamic size and (ii) zeta potential of each liposome measured for 28 days. Reproduced with permission from ref. 84 (Copyright (2021) by Wiley). (d) (i) Schematic of the structure of PEG- or hbPG-functionalized liposomes. (ii) TEM image of liposomes. The spherical liposome structure was preserved during separation. The red arrows indicate protein corona formation after incubation with plasma. More proteins were observed around functionalized liposomes, causing a higher cellular uptake. Reproduced with permission from ref. 85 (Copyright (2019) by ACS). | ||
3.1. Liposomes
Liposomes are lipid bilayer colloidal structures with a central aqueous core and are one of the most common lipid-based carriers, with sizes generally in the range of 100–300 nm.86–88 Liposomes are ideal delivery platforms for various types of drugs, antibodies, nucleic acids, and other molecules because hydrophobic and lipophilic substances can be loaded within the lipid bilayer while hydrophilic components can be loaded in the aqueous core.89,90 Liposomes are generally biocompatible, and liposome-based drug delivery has several advantages to the administration of free drugs as liposomes improve uptake by tumors due to long circulation half-life.86–88 The composition of liposomes determines the size, stability, surface charge, and membrane fluidity. These properties are determined by the choice of phospholipids and incorporation of cholesterol or PEGs.89,91 Liposomes are promising agents for particulates with low aqueous solubility and poor stability (Fig. 4b).83,87,90–95Several FDA-approved liposomal drugs are used in tumor therapy, including DaunoXome (liposomal daunorubicin), DepoCyt (liposomal cytarabine), and Myocet (liposomal doxorubicin).96,97 Doxil, a PEG-liposome containing doxorubicin, is approved for AIDS-related Kaposi's sarcoma, ovarian cancer, and multiple myeloma. The half-life of Doxil is significantly longer and its cardiotoxicity is reduced compared with that in free doxorubicin, but skin toxicity that does not occur with doxorubicin has been discovered.98–100 Many studies focus on the encapsulation of loaded multiple drugs by using liposomes. Multi-drug loading efficiency is effectively demonstrated by exploiting the structural properties of liposomes (Fig. 5a). Through the development of multilayered liposomes and the development of long-circulating liposomes, drug delivery efficacy has been greatly improved (Fig. 5b).101,102
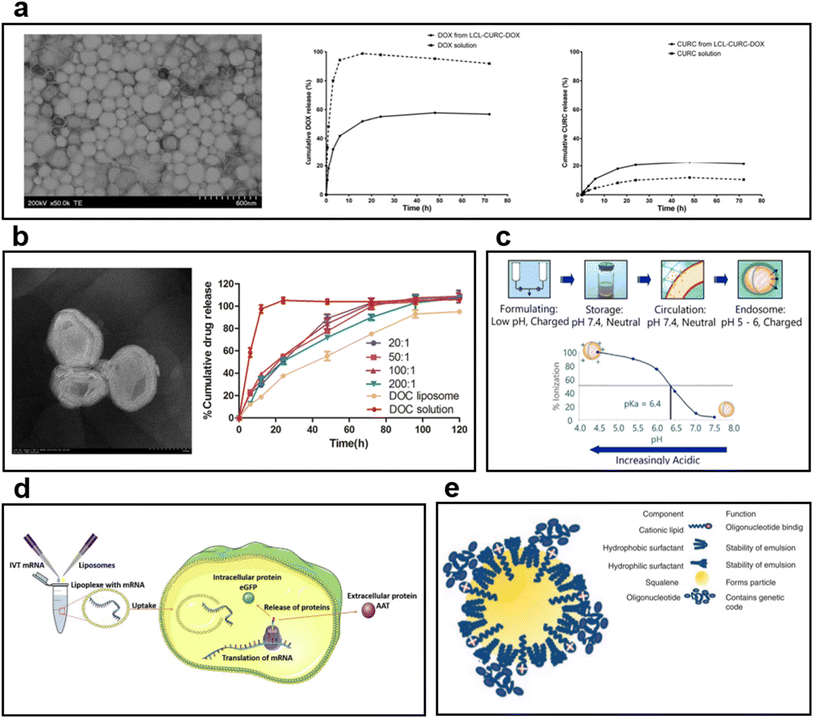 | ||
| Fig. 5 (a) TEM image of long-circulating liposome (LCL) co-encapsulating doxorubicin (DOX) and curcumin (CURC).101 In the case of DOX, 55% of the release was observed in liposome formulation after 24 h. In the case of CURC, the release from the liposome was faster than the solution, and 20% was released after 24 h. (b) TEM images and drug release behavior graphs of multilayered liposomes co-encapsulating docetaxel (DOC) and palmitoyl ascorbate (PA).102 The DOC solution showed a rapid release, but DOC liposome and DOC-PA-liposome exhibited sustained release behavior. The results confirmed that the drug release rate can be controlled by PA addition. (c) Mechanism of ionizing cationic liposomes at different pH levels.110 In physiological pH, it is neutral; in acidic environments, an ionizing lipid becomes a cationic liposome. (d) The process by which mRNA is encapsulated into liposomes produces a protein.111 After entering the cytoplasm by endocytosis, mRNA is released to the outside of the liposome; it undergoes transcription and translation to produce target proteins. (e) Cationic nanoemulsion (CNE) complexed with self-amplifying mRNA (SAM).112 Binding of SAM to CNE maintained the long loading of mRNA by cationic lipids and improved the delivery capacity. | ||
Liposomes have also shown promising results for the delivery of mRNA-based vaccines in infectious diseases and cancer immunotherapy.98,99,103 The intratumoral injection of mRNA-liposomal complexes is highly effective in achieving in situ tumor transfection.104 Zhou et al.105 evaluated neutral liposomes for mRNA vaccine encoded with human melanoma antigen glycoprotein 100 (gp100), which showed evidence of tumor suppression and enhanced survival. Allison et al.106 in 1974 first described the potential use of liposomes as a vaccine vehicle, where the intramuscular injection of diphtheria toxoid (DT) in negatively charged liposomes elicits significantly higher antibody levels than free DT in in vivo models. Since then, liposomal vaccine formulations have been studied, developed, and approved for marketing. Epaxal®, a hepatitis A vaccine, and Inflexal V®, an influenza vaccine, are examples of liposomal vaccines developed by Crucell (formerly Berna Biotech Ltd).107,108 Liposomes can be used for imaging applications as well as drug delivery. Representatively, radiolabeled liposomes are being actively studied in the preclinical and clinical fields as well as in vitro study. Wong et al. compared the imaging efficiencies of Cu-labelled liposomes, Cu-labelled albumin, and FDG. In particular, when imaging was performed after 18–24 h, liposomes were superior to FDG in detecting small tumors and revealing heterogeneity within tumors.109
DOTAP has been proposed as an immunomodulator to stimulate the immune response of the host in a pathogen-independent manner.116 This characteristic of immune stimulation has been linked with the type of amine headgroups. Therefore, lipids possessing quaternary amine headgroups are better in immune stimulation than lipids with tertiary amine headgroups. Additionally, lipids with unsaturated acyl chains stimulate cytokine release. Hence, the potential immune response of cationic liposomes must be carefully evaluated before designing a delivery system.72,117,118
3.2. Cationic nanoemulsions (CNEs)
Oil-in-water nanoemulsions can be used as drug and vaccine delivery systems. CNEs are proposed as a potential alternative to nucleic acid therapeutics to overcome liposome-associated problems, such as particle aggregation in biological fluids. CNEs contain an oil core stabilized by either cationic lipids or a combination of phospholipids, PEG-lipids, and/or nonionic surfactants. The incorporation of cationic lipids is crucial for nucleic acid complexation through electrostatic interactions.124,125 Brito et al.112 developed CNEs using cationic lipid DOTAP to deliver self-amplifying RNA-based vaccines for HIV, RSV, and human cytomegalovirus (Fig. 5e). Nanoemulsions offer the advantages of high-loading capacity, ease of large-scale manufacturing, and toxicological safety.3.3. Solid lipid nanoparticles (SLNs)
SLNs have emerged as an alternative to liposomes. Unlike liposomes, SLNs have a biodegradable solid core and are dispersed in an aqueous solution as colloidal particles with the help of a surfactant layer.126 They provide higher drug stability and can carry a higher payload than conventional liposomes. They also offer the opportunity of controlled drug release because of the solid core. Solid lipids have been used to reduce the drug release rate in pellets.126–128 SLNs have excellent stability and intracellular uptake efficiency as mRNA and DNA carriers; they also have an excellent drug release control ability. SLNs, developed as a combination of cationic and ionizing lipids, remain stable for a long time as an mRNA delivery vector; the binding ability of SLN to mRNA is maintained (Fig. 6a).129 In another study using SLN as a DNA vector, confocal image analysis confirmed the results widely observed in the nucleus and cytoplasm of cells following SLN transfection. Furthermore, the transfection efficiency of SLN is higher than that of Lipofectin®, a commercial transfection reagent (Fig. 6b).130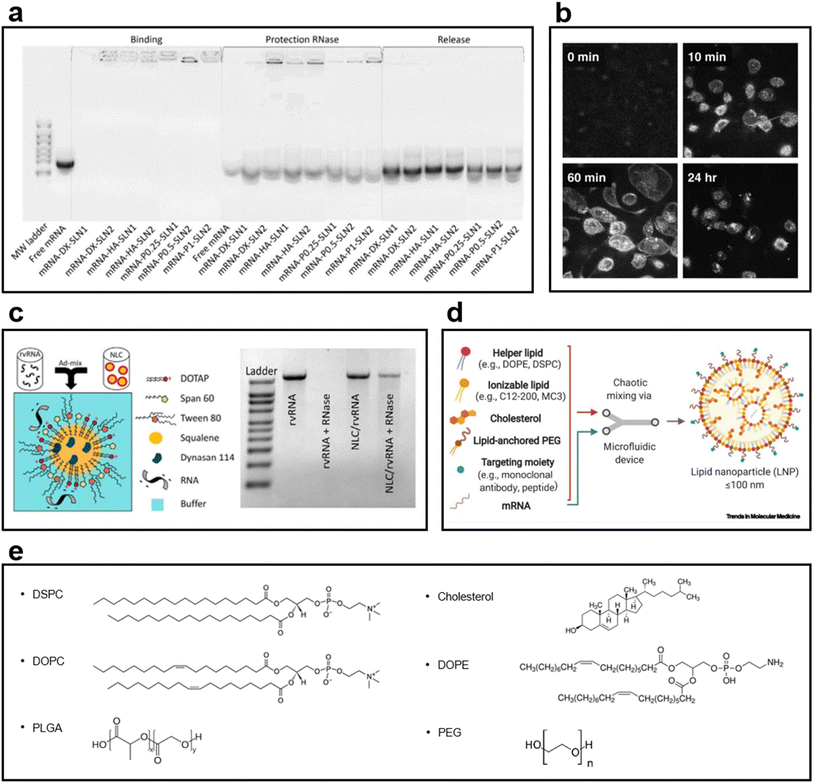 | ||
| Fig. 6 (a) Gel electrophoresis of particles loaded with mRNA into SLN1 composed of the cationic lipid DOTAP and SLN2 composed of DOTAP and the ionizing lipid DODAP129 (Copyright (2020) by MDPI). After 3 months of loading, SLNs maintained their ability to bind to mRNA, exhibited stability, and provided mRNA protection for a long period. (b) Confocal image of the endocytosis of fluorescently labeled SLN into H1299 cells130 (Copyright (2008) by Elsevier). The SLN synthesized using a fluorescence-conjugated DOPE was confirmed to enter the cytosol after 60 min of treatment and verified in the nucleus after 24 h. (c) NLC structure image in which a replicating viral RNA (rvRNA) binds to its surface1 (Copyright (2018) by ASGCT). A relatively low amount of Tween 80, a surfactant, was used, i.e., 35% of the lipid mass, to prevent rvRNA degradation; the ladder of gel electrophoresis revealed that rvRNA did not appear when RNase was used. (d) Components used in the LNP formulations131 (Copyright (2021) by Elsevier). A helper lipid for encapsulating mRNA, an ionizing lipid for improved delivery and loading stability, and cholesterol to improve integrity are used. In addition, PEGylated lipids for enhancing biocompatibility and reducing cytotoxicity and various types of targeting moieties for delivering to more precise targets are used. (e) Chemical structures of phospholipids, cholesterol, and emulsifiers used in LNP formulations. | ||
3.4. Nanostructured lipid carriers (NLCs)
NLCs are the advanced stage of SLNs, with a hybrid formulation of SLNs and nanoemulsions. A semicrystalline matrix is formed in NLCs with a controlled mixture of solid and liquid lipids, improving colloidal stability, drug loading, and drug release. NLCs have been reported for the delivery of mRNA-based vaccines for Zika virus, providing 100% protection from death in a small single-dose intramuscular injection in mice (Fig. 6c).13.5. Role of helper lipids in the formulation of LNPs
The composition plays a critical role in the characteristics of LNPs. The type of phospholipids and their molar ratio determine the size, spherical charge, drug loading capabilities, cellular uptake, longevity, and stability of particles (Fig. 6d).131 The attributes of helper lipids, such as PEGylated lipids, cholesterol, and phospholipids, have been described below.Some concerns regarding anti-PEG antibodies developed in people who received PEGylated drugs or PEG-containing products have been reported. Hypersensitivity reactions have been described in some patients but their occurrence is low. The modification of PEG molecules to reduce immunogenic reactions may provide optimal solutions to overcome adverse reactivity toward PEG. Moderna, Inc. and Pfizer-BioNTech's lipid nanoparticle-encapsulated mRNA vaccines contain PEG in their vaccine ingredients, and the Centers for Disease Control and Prevention (CDC) stated that those with known allergies to PEG should not receive these mRNA COVID-19 vaccines and opt for vaccines that do not contain PEG.
DOPE is a typical phosphoethanlamine and has a conical shape. The two unsaturated tails of DOPE form this structure and have a smaller head group than the tertiary amine group of PC. DOPE adopts an inverted hexagonal H(II) phase that promotes endosomal escape of lipid nanoparticles by destabilizing the endosomal membrane and improves intracellular delivery of nucleic acids. For example, when synthesizing LNP by adding DOPE to cationic lipid, mRNA vaccine delivery was improved by targeting splenic antigen-presenting cells.141
4. Mechanism of the cellular uptake of LNPs and targeting strategies
4.1. Cellular uptake
LNPs undergo cellular uptake via the endocytic pathway. Endocytosis pathways can be broadly divided into phagocytosis and pinocytosis. Phagocytosis is a triggered process by which cells such as monocytes/macrophages, neutrophils, and dendritic cells engulf large particles. Pinocytosis is a continuous process and is associated with the internalization of body fluids and molecules by small vesicles, such as NPs.144Pinocytosis is further divided into four courses: clathrin-mediated, caveolin-mediated, clathrin/caveolin-independent endocytosis, and micropinocytosis. The particular pathway that particles enter depends on several characteristics such as size, shape, particle surface, and rigidity of NPs.145 Clathrin-mediated endocytosis relies on receptor-mediated, hydrophobic, or electrostatic interactions between ligands and extracellular receptors in the region of clathrin expression to internalize specific molecules into cells. All lipid-based NPs use this pathway. After entering cells, internalized nanoparticles are usually trapped inside endosomal/lysosomal vesicles, where a cargo material is degraded by lysosomal enzymes.146 Caveolin-mediated endocytosis can occur in molecules smaller than 60 nm and is a cell entry pathway involving flask-shaped membrane inclusions called “caveolae”. They form special vesicles after they are swallowed using lipid rafts.147 Clathrin- and caveolin-independent endocytosis occurs in cells deficient in clathrin and caveolin, internalizing cargos such as growth hormone, cellular fluid, interleukin-2, and folic acid.148 Macropinocytosis is a pathway by which extracellular fluid and its debris are absorbed by forming large heterogeneous vesicles called “macropinosomes”. It mainly internalizes apoptotic cells, extracellular fluid, bacteria, and viruses and can internalize micron-sized NPs that cannot enter through other pathways.149
4.2. Targeting
Cellular uptake and cargo release in the cytosol are critical steps and important areas of research. However, researchers have reported that less than 2% of the genetic material is released into the cytosol. Nucleic acid therapeutics have been clinically approved to treat a small number of diseases, but their potential has been limited due to inefficient delivery. Nucleic acids can be activated until they are delivered to specific cellular compartments (cytoplasm, nucleus, and mitochondria).71 Trafficking to the appropriate site is therefore one of the challenges to be solved and numerous strategies have been implemented to improve cellular uptake. Among them, this aim is achieved by attaching target ligands such as specific ligand molecules of various receptors such as amino acids, peptide proteins, receptors, antibodies, and growth factor receptors that are functionalized to the nanoparticle surface.150,151 Such surface-modified delivery systems can also be applied to a variety of nucleic acid therapeutic delivery from short siRNAs to large plasmid DNAs.71 A recent study investigated the cyclic peptide iRGD (CCRGDKGPDC)-conjugated solid lipid nanoparticles for the delivery of siRNA for the epidermal growth factor receptor (EGFR) and programmed cell death ligand-1 (PD-L1). As a result, the effectiveness of targeting and immunotherapy against glioblastoma is improved to treat gliomas by bypassing the blood–brain barrier.152 Cationic liposomes or polymers may protect mRNA from degradation by RNAse, aid internalization by DCs, and allow intracellular delivery. In particular, DOTAP:DOPE liposomes resulted in enhanced vaccination against HIV by delivering mRNA encoding the HIV gag antigen.153 In addition, a lectin subtype with high specificity for a particular carbohydrate residue (α-L-fucose) expressed on the cell surface bound to liposomes, leading to oral vaccine delivery for intestinal immunization.154 Lipid-based delivery systems can play a flexible and important role in designing effective vaccines and therapeutic strategies against infectious and non-infectious diseases. Therefore, recent advances in preclinical and clinical trials of lipid-based delivery systems that enhance the efficiency and safety of vaccine strategies against various diseases deserve attention.1555. Overview of mRNA loading into LNPs for SARS-CoV-2 vaccines
Unlike DNA-based vaccines, mRNA-based vaccines have the advantage of reducing the risk of gene mutation because they are not integrated into the host gene. In addition, since the encoded protein becomes translated as mRNA reaches the cytoplasm, it has the advantage of easy delivery, unlike DNA, which must reach the inside of the nucleus. Therefore, the development of mRNA vaccines for various diseases that have been difficult to treat is being actively studied. The types of mRNA used for RNA vaccines can be divided into self-amplifying mRNA (SAM) vaccines and conventional mRNA-based vaccines, which are nonreplicating mRNAs. Conventional mRNA-based vaccines encode only the gene of a target antigen and cannot replicate it. Therefore, antigen expression at this time is proportional to the number of mRNA transcripts successfully transferred during vaccination, and a large amount of administration and repeated administration are performed as needed. These vaccines include Pfizer's BNT162b2 and Moderna's mRNA-1273, which are the currently available SARS-CoV-2 mRNA vaccines. In self-amplifying mRNA (SAM) vaccines, a replicon developed from a positive-sense, single-stranded alphavirus can be directly translated into host cells, similar to mRNA. RNA called Replicon can self-replicate and amplify in cells through RNA template amplification in host cells. Accordingly, the expression of an antigenic gene may be increased even at a small dose.156,157These nucleic acids are polyvalent anionic and hydrophilic; they are difficult to be taken up by cells but easily degraded by nucleases in the blood plasma. Therefore, matching with the delivery system is important to keep an mRNA-based vaccine intact and deliver it effectively. The most optimal mRNA delivery systems are LNPs such as liposomes, CNEs, and NLCs composed of cationic lipids. Cationic lipids, which are ionized at a low pH of 4.0 and have a positive charge, form a complex with mRNA. An anionic cell membrane neutralizes the charge of the cationic LNP, thereby interfering with the electrostatic interaction between cationic LNPs and mRNA, resulting in the release of the mRNA into cells.158 In addition to mRNA vaccine delivery, the efficiency of nucleic acid delivery for transfection is higher when a cationic liposome is used than when a cationic polymer is used.159,160
The research and development of mRNA vaccines and drugs with LNP vectors is ongoing for various diseases, such as tuberculosis, influenza, and cancer. For example, Patisiran developed by ONPATTRO is a drug for the treatment of polyneuropathy caused by transthyretin-mediated amyloidosis (hATTR). It expresses the effect of transthyretin-inhibiting double-stranded siRNA encapsulated in LNPs and becomes delivered to hepatocytes.161,162 Also, all vaccine candidates are related to mRNA vaccines in the prM-E gene region, which expresses the precursor membrane (prM) protein and envelope (E) protein among the viral proteins of Zika virus.163 Recently, clinical trials are underway by encapsulating nucleoside-modified mRNA for the prM-E gene region by using LNP-containing ionizing cationic lipids.164 When injected into mice and rhesus monkeys, they elicit an immune response that protects them from viral attack.165 In addition, Moderna, Inc. is conducting phase 1 clinical trials for mRNA-1325 and mRNA-1895, which are also Zika virus vaccine candidates.
Among LNPs that encapsulate mRNA vaccines, liposomes have been widely explored probably because they are being used as vectors of SARS-CoV-2 mRNA vaccines. In March 2020, the director of the World Health Organization (WHO) declared the coronavirus pandemic.166 Since then, vaccines against SARS-CoV-2 have been rapidly developed. Among these vaccines, the LNP-encapsulated mRNA-1273 vaccine (Moderna) that encodes the prefusion stabilized full-length spike protein of SARS-CoV-2 shows 94.1% efficacy from a phase 3 randomized placebo-controlled multicenter study focusing on participants aged ≥18 years.167 The two-dose regimen of BNT162b2 (Pfizer-BioNTech), which is another mRNA vaccine and lipid nanoparticle-formulated nucleoside-modified RNA vaccine that encodes a prefusion stabilized membrane-anchored SARS-CoV-2 full-length spike protein, shows 95% protection against COVID-19 in people aged ≥16 years with a good safety profile.69
6. Conclusion
With the coronavirus disease-19 (COVID-19) pandemic, studies have focused on lipid-based nanoparticles because of their ability to deliver therapeutics, such as mRNA. In this review, we reported nucleic acid-based therapeutics and lipid nanoparticle type as a carrier of nucleic acid therapeutics. Nucleic acid therapeutics, such as DNA and RNA, control the expression of a specific protein related to disease and immune activity. Generally, nucleic acid therapeutics regulate sequence-specific recognition to inhibit mRNA translation that induces disease-related protein expression. Thus, the activated nucleic acid sequence, which interacted with therapeutic nucleic acids, represents therapeutic efficacy by inhibition of translation. Therefore, the adequate delivery of nucleic acid therapeutics has been applied to solve problems of nucleic acid-based drugs. Therefore, we introduced a diverse type of LNPs, such as liposomes, CNEs, SLNs, and NLCs. Liposomes, representative LNPs, could accommodate hydrophilic and hydrophobic drugs with an enhanced delivery capacity on tumor cells. CNEs were developed to overcome the limitation of liposomes, such as particle aggregation. SLNs are specialized in maintaining the stability of drugs conjugated with them and controlled drug release.Because of the characteristics of lipid-based nanoparticles, LNPs have been utilized to deliver SARS-CoV-2 mRNA vaccines. Many clinical trials have reported that LNP uptake into mammalian cells promotes effective mRNA delivery and leads to the expression of the SARS-CoV-2 antigen that activates innate immune responses for the secretion of the neutralization antibody against the SARS-CoV-2 antigen.
LNPs with varying structures and characteristics have been constructed for stable material delivery. In this review, we outline future possibilities for further applications of LNPs as flexible carriers in immuno-therapeutics and anti-cancer reagents.
Author contributions
YC and JC (Jonghoon Choi) supervised the study. MSK, SAB, CY, JH, HH, JC (Jayoung Chae), SL, and YC designed the study and performed the literature survey. JH, HH, and YC prepared the graphics. MT and JC (Jonghoon Choi) provided a discussion on the clinical perspectives and applications. MSK, SAB, CY, JH, HH, JC (Jayoung Chae), SL, and YC performed data analyses. All authors wrote the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the Chung-Ang University Research Grants in 2021. This work was supported by the National Research Foundation of Korea (NRF) grants funded by the Korean government (MSIT) (No. 2020R1A5A1018052), and the Korea Environment Industry & Technology Institute (KEITI) funded by the Korea Ministry of Environment (MOE) (No. 2022002980003).References
- J. H. Erasmus, A. P. Khandhar, J. Guderian, B. Granger, J. Archer, M. Archer, E. Gage, J. Fuerte-Stone, E. Larson and S. Lin, Mol. Ther., 2018, 26, 2507–2522 CrossRef CAS PubMed.
- M. J. Byun, J. Lim, S. N. Kim, D. H. Park, T. H. Kim, W. Park and C. G. Park, BioChip J., 2022, 16, 128–145 CrossRef CAS PubMed.
- S. Kim and J. H. Lee, BioChip J., 2022, 16, 376–396 CrossRef CAS PubMed.
- W.-J. Lee, K.-J. Kim, M. K. Hossain, H.-Y. Cho and J.-W. Choi, BioChip J., 2022, 16, 33–40 CrossRef CAS.
- K. K. Lee, S. C. Lee, H. Kim and C.-S. Lee, BioChip J., 2022, 1–8 Search PubMed.
- Y. Ha and I. Kim, BioChip J., 2022, 16, 351–365 CrossRef CAS PubMed.
- G. Eom, A. Hwang, H. Kim, J. Moon, H. Kang, J. Jung, E.-K. Lim, J. Jeong, H. G. Park and T. Kang, BioChip J., 2021, 15, 348–355 CrossRef CAS.
- M. Athar and A. J. Das, Adv. Mater. Rev., 2014, 1, 25–37 Search PubMed.
- R. Jahanban-Esfahlan, M. de la Guardia, D. Ahmadi and B. Yousefi, J. Cell. Physiol., 2018, 233, 2019–2031 CrossRef CAS PubMed.
- J. Shi, P. W. Kantoff, R. Wooster and O. C. Farokhzad, Nat. Rev. Cancer, 2017, 17, 20 CrossRef CAS PubMed.
- D. Lee, Y. Seo, M. S. Khan, J. Hwang, Y. Jo, J. Son, K. Lee, C. Park, S. Chavan and A. A. Gilad, Biotechnol. Bioprocess Eng., 2018, 23, 1–10 CrossRef CAS.
- D. Chenthamara, S. Subramaniam, S. G. Ramakrishnan, S. Krishnaswamy, M. M. Essa, F.-H. Lin and M. W. Qoronfleh, Biomater. Res., 2019, 23, 1–29 CrossRef PubMed.
- K. Cho, X. Wang, S. Nie and D. M. Shin, Clin. Cancer Res., 2008, 14, 1310–1316 CrossRef CAS PubMed.
- T. Sun, Y. S. Zhang, B. Pang, D. C. Hyun, M. Yang and Y. Xia, Angew. Chem., Int. Ed., 2014, 53, 12320–12364 CAS.
- M. Khan, J. Hwang, K. Lee, Y. Choi, J. Jang, Y. Kwon, J. Hong and J. J. N. Choi, Nanomaterials, 2019, 9, 48 CrossRef PubMed.
- G. Chauhan, M. J. Madou, S. Kalra, V. Chopra, D. Ghosh and S. O. Martinez-Chapa, ACS Nano, 2020, 14, 7760–7782 CrossRef CAS PubMed.
- E. Musielak, A. Feliczak-Guzik and I. Nowak, Materials, 2022, 15, 682 CrossRef CAS PubMed.
- B. Mikolaskova, M. Jurcik, I. Cipakova, M. Kretova, M. Chovanec and L. Cipak, Curr. Genet., 2018, 64, 971–983 CrossRef CAS PubMed.
- C. J. Wraight and P. J. White, Pharmacol. Ther., 2001, 90, 89–104 CrossRef CAS PubMed.
- K. Sridharan and N. J. Gogtay, Br. J. Clin. Pharmacol., 2016, 82, 659–672 CrossRef PubMed.
- B. M. Cavagnari, Arch. Argent. Pediatr., 2011, 109, 237–244 Search PubMed.
- V. Sardone, H. Zhou, F. Muntoni, A. Ferlini and M. S. Falzarano, Molecules, 2017, 22, 563 CrossRef PubMed.
- K. Dhuri, C. Bechtold, E. Quijano, H. Pham, A. Gupta, A. Vikram and R. Bahal, J. Clin. Med., 2020, 9, 2004 CrossRef CAS PubMed.
- L. C. Bock, L. C. Griffin, J. A. Latham, E. H. Vermaas and J. J. Toole, Nature, 1992, 355, 564–566 CrossRef CAS PubMed.
- Q. Zhu, G. Liu and M. Kai, Molecules, 2015, 20, 20979–20997 CrossRef CAS PubMed.
- T. T. T. Tran, A. Delgado and S. Jeong, BioChip J., 2021, 15, 109–122 CrossRef CAS.
- Z.-X. Liao, E.-Y. Chuang, C.-C. Lin, Y.-C. Ho, K.-J. Lin, P.-Y. Cheng, K.-J. Chen, H.-J. Wei and H.-W. Sung, J. Controlled Release, 2015, 208, 42–51 CrossRef CAS PubMed.
- C. Guerrier-Takada, K. Gardiner, T. Marsh, N. Pace and S. Altman, Cell, 1983, 35, 849–857 CrossRef CAS PubMed.
- S. DeWeerdt, Nature, 2019, 574, S2 CrossRef CAS.
- J. C. Burnett and J. J. Rossi, Chem. Biol., 2012, 19, 60–71 CrossRef CAS PubMed.
- J.-z. Xu, J.-l. Zhang and W.-g. Zhang, J. Zhejiang Univ., Sci., B, 2018, 19, 739–749 CrossRef PubMed.
- R. C. Lee, R. L. Feinbaum and V. Ambros, Cell, 1993, 75, 843–854 CrossRef CAS PubMed.
- B. Wightman, I. Ha and G. Ruvkun, Cell, 1993, 75, 855–862 CrossRef CAS PubMed.
- L.-A. MacFarlane and P. R Murphy, Curr. Genomics, 2010, 11, 537–561 CrossRef CAS PubMed.
- S. M. Johnson, H. Grosshans, J. Shingara, M. Byrom, R. Jarvis, A. Cheng, E. Labourier, K. L. Reinert, D. Brown and F. J. Slack, Cell, 2005, 120, 635–647 CrossRef CAS PubMed.
- C. D. Johnson, A. Esquela-Kerscher, G. Stefani, M. Byrom, K. Kelnar, D. Ovcharenko, M. Wilson, X. Wang, J. Shelton and J. Shingara, Cancer Res., 2007, 67, 7713–7722 CrossRef CAS PubMed.
- N. S. Asli, M. E. Pitulescu and M. Kessel, Curr. Mol. Med., 2008, 8, 698–710 CrossRef CAS PubMed.
- S. Kannappan, J. Chang, P. R. Sundharbaabu, J. H. Heo, W.-k. Sung, J. C. Ro, K. K. Kim, J. B. B. Rayappan and J. H. Lee, BioChip J., 2022, 16, 490–500 CrossRef CAS.
- M. H. Son, S. W. Park, H. Y. Sagong and Y. K. Jung, BioChip J., 2022, 1–24 Search PubMed.
- D. Kim, J. Lee, S. Park, J. Park, M.-J. Seo, W. J. Rhee and E. Kim, BioChip J., 2022, 16, 422–432 CrossRef CAS.
- J. Krützfeldt, N. Rajewsky, R. Braich, K. G. Rajeev, T. Tuschl, M. Manoharan and M. Stoffel, Nature, 2005, 438, 685–689 CrossRef PubMed.
- H. Dana, G. M. Chalbatani, H. Mahmoodzadeh, R. Karimloo, O. Rezaiean, A. Moradzadeh, N. Mehmandoost, F. Moazzen, A. Mazraeh and V. Marmari, Int. J. Biomed. Sci., 2017, 13, 48 Search PubMed.
- A. J. Hamilton and D. C. Baulcombe, Science, 1999, 286, 950–952 CrossRef CAS PubMed.
- B. Hu, L. Zhong, Y. Weng, L. Peng, Y. Huang, Y. Zhao and X.-J. Liang, Signal Transduction Targeted Ther., 2020, 5, 1–25 CrossRef PubMed.
- A. S. Lewin and W. W. Hauswirth, Trends Mol. Med., 2001, 7, 221–228 CrossRef CAS PubMed.
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818–822 CrossRef CAS PubMed.
- M. Homann and H. U. Göringer, Nucleic Acids Res., 1999, 27, 2006–2014 CrossRef CAS PubMed.
- K. Sefah, D. Shangguan, X. Xiong, M. B. O'donoghue and W. Tan, Nat. Protoc., 2010, 5, 1169 CrossRef CAS PubMed.
- K. Germer, M. Leonard and X. Zhang, Int. J. Biochem. Mol. Biol., 2013, 4, 27 CAS.
- J.-H. Lee, M. D. Canny, A. De Erkenez, D. Krilleke, Y.-S. Ng, D. T. Shima, A. Pardi and F. Jucker, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 18902–18907 CrossRef CAS PubMed.
- F. Gros, H. Hiatt, W. Gilbert, C. G. Kurland, R. Risebrough and J. D. Watson, Nature, 1961, 190, 581–585 CrossRef CAS PubMed.
- S. Brenner, F. Jacob and M. Meselson, Nature, 1961, 190, 576–581 CrossRef CAS PubMed.
- J. A. Wolff, R. W. Malone, P. Williams, W. Chong, G. Acsadi, A. Jani and P. L. Felgner, Science, 1990, 247, 1465–1468 CrossRef CAS PubMed.
- G. F. Jirikowski, P. P. Sanna, D. Maciejewski-Lenoir and F. E. Bloom, Science, 1992, 255, 996–998 CrossRef CAS PubMed.
- G. Maruggi, C. Zhang, J. Li, J. B. Ulmer and D. Yu, Mol. Ther., 2019, 27, 757–772 CrossRef CAS PubMed.
- J. Z. Xu, J. L. Zhang and W. G. Zhang, J. Zhejiang Univ., Sci., B, 2018, 19, 739–749 CrossRef PubMed.
- K. Asha, P. Kumar, M. Sanicas, C. A. Meseko, M. Khanna and B. Kumar, J. Clin. Med., 2018, 8, 1–24 CrossRef PubMed.
- S. Philippou, N. P. Mastroyiannopoulos, N. Makrides, C. W. Lederer, M. Kleanthous and L. A. Phylactou, Mol. Ther.-Nucleic Acids, 2018, 10, 199–214 CrossRef CAS PubMed.
- J. Kaur and K. Tikoo, Oncogene, 2015, 34, 5216–5228 CrossRef CAS PubMed.
- G. Maruggi, C. Zhang, J. Li, J. B. Ulmer and D. Yu, Mol. Ther., 2019, 27, 757–772 CrossRef CAS PubMed.
- E. S. Lander, L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle and W. FitzHugh, Nature, 2001, 409, 860–921 CrossRef CAS PubMed.
- J. C. Venter, M. D. Adams, E. W. Myers, P. W. Li, R. J. Mural, G. G. Sutton, H. O. Smith, M. Yandell, C. A. Evans and R. A. Holt, Science, 2001, 291, 1304–1351 CrossRef CAS PubMed.
- F. S. Collins, A. Patrinos, E. Jordan, A. Chakravarti, R. Gesteland and L. Walters, Science, 1998, 282, 682–689 CrossRef CAS PubMed.
- F. Emmert-Streib, M. Dehmer and O. Yli-Harja, Front. Genet., 2017, 8, 184 CrossRef PubMed.
- D. Hobernik and M. Bros, Int. J. Mol. Sci., 2018, 19, 3605–3632 CrossRef PubMed.
- M. A. Liu, Vaccines, 2019, 7, 37–56 CrossRef CAS PubMed.
- A. Tiptiri-Kourpeti, K. Spyridopoulou, A. Pappa and K. Chlichlia, Pharmacol. Ther., 2016, 165, 32–49 CrossRef CAS PubMed.
- M. S. Gebre, L. A. Brito, L. H. Tostanoski, D. K. Edwards, A. Carfi and D. H. Barouch, Cell, 2021, 184, 1589–1603 CrossRef CAS PubMed.
- F. P. Polack, S. J. Thomas, N. Kitchin, J. Absalon, A. Gurtman, S. Lockhart, J. L. Perez, G. Perez Marc, E. D. Moreira, C. Zerbini, R. Bailey, K. A. Swanson, S. Roychoudhury, K. Koury, P. Li, W. V. Kalina, D. Cooper, R. W. Frenck Jr, L. L. Hammitt, O. Tureci, H. Nell, A. Schaefer, S. Unal, D. B. Tresnan, S. Mather, P. R. Dormitzer, U. Sahin, K. U. Jansen, W. C. Gruber and C. C. T. Group, N. Engl. J. Med., 2020, 383, 2603–2615 CrossRef CAS PubMed.
- L. R. Baden, H. M. El Sahly, B. Essink, K. Kotloff, S. Frey, R. Novak, D. Diemert, S. A. Spector, N. Rouphael, C. B. Creech, J. McGettigan, S. Khetan, N. Segall, J. Solis, A. Brosz, C. Fierro, H. Schwartz, K. Neuzil, L. Corey, P. Gilbert, H. Janes, D. Follmann, M. Marovich, J. Mascola, L. Polakowski, J. Ledgerwood, B. S. Graham, H. Bennett, R. Pajon, C. Knightly, B. Leav, W. Deng, H. Zhou, S. Han, M. Ivarsson, J. Miller, T. Zaks and C. S. Group, N. Engl. J. Med., 2021, 384, 403–416 CrossRef CAS PubMed.
- L. M. Molle, C. H. Smyth, D. Yuen and A. P. R. Johnston, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2022, 14, e1809 CAS.
- B. N. Aldosari, I. M. Alfagih and A. S. Almurshedi, Pharmaceutics, 2021, 13, 206 CrossRef CAS PubMed.
- X. Hou, T. Zaks, R. Langer and Y. Dong, Nat. Rev. Mater., 2021, 6, 1078–1094 CrossRef CAS PubMed.
- R. Tenchov, R. Bird, A. E. Curtze and Q. Zhou, ACS Nano, 2021, 15, 16982–17015 CrossRef CAS PubMed.
- M. B. Laurens, Hum. Vaccines Immunother., 2020, 16, 480–489 CrossRef CAS PubMed.
- Y. Y. Syed, Drugs Ther. Perspect., 2022, 38, 373–381 CrossRef PubMed.
- N. Wang, M. Chen and T. Wang, J. Controled Release, 2019, 303, 130–150 CrossRef CAS PubMed.
- R. A. Feldman, R. Fuhr, I. Smolenov, A. Mick Ribeiro, L. Panther, M. Watson, J. J. Senn, M. Smith, Ö. Almarsson, H. S. Pujar, M. E. Laska, J. Thompson, T. Zaks and G. Ciaramella, Vaccine, 2019, 37, 3326–3334 CrossRef CAS PubMed.
- S. M. Hoy, Drugs, 2018, 78, 1625–1631 CrossRef CAS PubMed.
- K. Bahl, J. J. Senn, O. Yuzhakov, A. Bulychev, L. A. Brito, K. J. Hassett, M. E. Laska, M. Smith, O. Almarsson, J. Thompson, A. M. Ribeiro, M. Watson, T. Zaks and G. Ciaramella, Mol. Ther., 2017, 25, 1316–1327 CrossRef CAS PubMed.
- M. Brazzoli, D. Magini, A. Bonci, S. Buccato, C. Giovani, R. Kratzer, V. Zurli, S. Mangiavacchi, D. Casini, L. M. Brito, E. De Gregorio, P. W. Mason, J. B. Ulmer, A. J. Geall and S. Bertholet, J. Virol., 2016, 90, 332–344 CrossRef CAS PubMed.
- S. Calabro, E. Tritto, A. Pezzotti, M. Taccone, A. Muzzi, S. Bertholet, E. De Gregorio, D. T. O'Hagan, B. Baudner and A. Seubert, Vaccine, 2013, 31, 3363–3369 CrossRef CAS PubMed.
- A. L. Robson, P. C. Dastoor, J. Flynn, W. Palmer, A. Martin, D. W. Smith, A. Woldu and S. Hua, Front. Pharmacol., 2018, 9, 80 CrossRef PubMed.
- M. S. Lee, J. W. Lee, S. J. Kim, O. V. Pham-Nguyen, J. Park, J. H. Park, Y. M. Jung, J. B. Lee and H. S. Yoo, Macromol. Biosci., 2021, 21, e2000413 CrossRef PubMed.
- C. Weber, M. Voigt, J. Simon, A. K. Danner, H. Frey, V. Mailander, M. Helm, S. Morsbach and K. Landfester, Biomacromolecules, 2019, 20, 2989–2999 CrossRef CAS PubMed.
- V. P. Torchilin, Nat. Rev. Drug Discovery, 2005, 4, 145 CrossRef CAS PubMed.
- A. Akbarzadeh, R. Rezaei-Sadabady, S. Davaran, S. W. Joo, N. Zarghami, Y. Hanifehpour, M. Samiei, M. Kouhi and K. Nejati-Koshki, Nanoscale Res. Lett., 2013, 8, 1–9 CrossRef PubMed.
- N. Monteiro, A. Martins, R. L. Reis and N. M. Neves, J. R. Soc., Interface, 2014, 11, 20140459 CrossRef PubMed.
- W. Mehnert and K. Mäder, Adv. Drug Delivery Rev., 2012, 64, 83–101 CrossRef.
- A. Wagner and K. Vorauer-Uhl, J. Drug Delivery, 2011, 2011 Search PubMed.
- M. S. Khan, J. Hwang, Y. Seo, K. Shin, K. Lee, C. Park, Y. Choi, J. W. Hong and J. Choi, Artif. Cells, Nanomed., Biotechnol., 2018, 1–10, DOI:10.1080/21691401.2018.1492420.
- M. Li, C. Du, N. Guo, Y. Teng, X. Meng, H. Sun, S. Li, P. Yu and H. Galons, Eur. J. Med. Chem., 2019, 164, 640–653 CrossRef CAS PubMed.
- P. Liu, G. Chen and J. Zhang, Molecules, 2022, 27, 1372–1394 CrossRef CAS PubMed.
- D. Guimaraes, A. Cavaco-Paulo and E. Nogueira, Int. J. Pharm., 2021, 601, 120571 CrossRef CAS PubMed.
- F. Rommasi and N. Esfandiari, Nanoscale Res. Lett., 2021, 16, 95 CrossRef CAS PubMed.
- U. Bulbake, S. Doppalapudi, N. Kommineni and W. Khan, Pharmaceutics, 2017, 9, 12 CrossRef PubMed.
- J. Lao, J. Madani, T. Puértolas, M. Álvarez, A. Hernández, R. Pazo-Cid, Á. Artal and A. Antón Torres, J. Drug Delivery, 2013, 2013 Search PubMed.
- M. Li, Y. Li, S. Li, L. Jia, H. Wang, M. Li, J. Deng, A. Zhu, L. Ma, W. Li, P. Yu and T. Zhu, Eur. J. Med. Chem., 2022, 227, 113910 CrossRef CAS PubMed.
- W. Ho, M. Gao, F. Li, Z. Li, X. Q. Zhang and X. Xu, Adv. Healthcare Mater., 2021, 10, e2001812 CrossRef PubMed.
- F. Martinon, S. Krishnan, G. Lenzen, R. Magné, E. Gomard, J. G. Guillet, J. P. Lévy and P. Meulien, Eur. J. Immunol., 1993, 23, 1719–1722 CrossRef CAS PubMed.
- A. Sesarman, L. Tefas, B. Sylvester, E. Licarete, V. Rauca, L. Luput, L. Patras, S. Porav, M. Banciu and A. Porfire, Drug Delivery Transl. Res., 2019, 9, 260–272 CrossRef CAS PubMed.
- J. Li, C. Guo, F. Feng, A. Fan, Y. Dai, N. Li, D. Zhao, X. Chen and Y. Lu, Sci. Rep., 2016, 6, 38787 CrossRef CAS PubMed.
- L. M. Kranz, M. Diken, H. Haas, S. Kreiter, C. Loquai, K. C. Reuter, M. Meng, D. Fritz, F. Vascotto and H. Hefesha, Nature, 2016, 534, 396–401 CrossRef PubMed.
- D. Lu, R. Benjamin, M. Kim, R. Conry and D. Curiel, Cancer Gene Ther., 1994, 1, 245–252 CAS.
- W.-Z. Zhou, D. Hoon, S. Huang, S. Fujii, K. Hashimoto, R. Morishita and Y. Kaneda, Hum. Gene Ther., 1999, 10, 2719–2724 CrossRef CAS PubMed.
- A. Allison and G. Gregoriadis, Nature, 1974, 252, 252 CrossRef CAS PubMed.
- P. A. Bovier, Expert Rev. Vaccines, 2008, 7, 1141–1150 CrossRef CAS PubMed.
- K. Asadi and A. Gholami, Int. J. Biol. Macromol., 2021, 182, 648–658 CrossRef CAS PubMed.
- W. W. Andrew, O Eleanor, Z. Hua, W. S. Jai, M. M. Lisa, F. C. Charles and W. F. Katherine, Am. J. Nucl. Med. Mol. Imaging, 2013, 3, 134–160 Search PubMed.
- E. Samaridou, J. Heyes and P. Lutwyche, Adv. Drug Delivery Rev., 2020, 154–155, 37–63 CrossRef CAS PubMed.
- T. Michel, D. Luft, M. K. Abraham, S. Reinhardt, M. L. Salinas Medina, J. Kurz, M. Schaller, M. Avci-Adali, C. Schlensak, K. Peter, H. P. Wendel, X. Wang and S. Krajewski, Mol. Ther.-Nucleic Acids, 2017, 8, 459–468 CrossRef CAS PubMed.
- L. A. Brito, M. Chan, C. A. Shaw, A. Hekele, T. Carsillo, M. Schaefer, J. Archer, A. Seubert, G. R. Otten, C. W. Beard, A. K. Dey, A. Lilja, N. M. Valiante, P. W. Mason, C. W. Mandl, S. W. Barnett, P. R. Dormitzer, J. B. Ulmer, M. Singh, D. T. O'Hagan and A. J. Geall, Mol. Ther., 2014, 22, 2118–2129 CrossRef CAS PubMed.
- Q. Cheng, T. Wei, L. Farbiak, L. T. Johnson, S. A. Dilliard and D. J. Siegwart, Nat. Nanotechnol., 2020, 15, 313–320 CrossRef CAS PubMed.
- J. H. Felgner, R. Kumar, C. Sridhar, C. J. Wheeler, Y. J. Tsai, R. Border, P. Ramsey, M. Martin and P. L. Felgner, J. Biol. Chem., 1994, 269, 2550–2561 CrossRef CAS PubMed.
- Y. Mai, J. Guo, Y. Zhao, S. Ma, Y. Hou and J. Yang, Cell. Immunol., 2020, 354, 104143 CrossRef CAS PubMed.
- R. Tada, A. Hidaka, H. Kiyono, J. Kunisawa and Y. Aramaki, BMC Res. Notes, 2018, 11, 472 CrossRef PubMed.
- E. J. Sayour, H. R. Mendez-Gomez and D. A. Mitchell, Int. J. Mol. Sci., 2018, 19, 2890–2902 CrossRef PubMed.
- N. A. Nagy, A. M. de Haas, T. B. H. Geijtenbeek, R. van Ree, S. W. Tas, Y. van Kooyk and E. C. de Jong, Front. Immunol., 2021, 12, 674048 CrossRef CAS PubMed.
- Y. Granot and D. Peer, Semin. Immunol., 2017, 34, 68–77 CrossRef CAS PubMed.
- X. Han, H. Zhang, K. Butowska, K. L. Swingle, M. G. Alameh, D. Weissman and M. J. Mitchell, Nat. Commun., 2021, 12, 7233 CrossRef CAS PubMed.
- M. A. Oberli, A. M. Reichmuth, J. R. Dorkin, M. J. Mitchell, O. S. Fenton, A. Jaklenec, D. G. Anderson, R. Langer and D. Blankschtein, Nano Lett., 2017, 17, 1326–1335 CrossRef CAS PubMed.
- L. Tan and X. Sun, Nano Res., 2018, 11, 5338–5354 CrossRef CAS.
- J. A. Kulkarni, P. R. Cullis and R. van der Meel, Nucleic Acid Ther., 2018, 28, 146–157 CrossRef CAS PubMed.
- H. F. Teixeira, F. Bruxel, M. Fraga, R. S. Schuh, G. K. Zorzi, U. Matte and E. Fattal, Int. J. Pharm., 2017, 534, 356–367 CrossRef CAS PubMed.
- M. Fraga, F. Bruxel, D. Diel, T. G. de Carvalho, C. A. Perez, R. Magalhães-Paniago, Â. Malachias, M. C. Oliveira, U. Matte and H. F. Teixeira, J. Controlled Release, 2015, 209, 37–46 CrossRef CAS PubMed.
- S. Parvez, G. Yadagiri, M. R. Gedda, A. Singh, O. P. Singh, A. Verma, S. Sundar and S. L. Mudavath, Sci. Rep., 2020, 10, 12243 CrossRef CAS PubMed.
- R. Paliwal, S. R. Paliwal, R. Kenwat, B. D. Kurmi and M. K. Sahu, Expert Opin. Ther. Pat., 2020, 30, 179–194 CrossRef CAS PubMed.
- J. Campos, P. Severino, A. Santini, A. Silva, R. Shegokar, S. Souto and E. Souto, Nanopharmaceuticals, 2020, 1–15 Search PubMed.
- J. R.-C. Itziar Gómez-Aguado, M. Vicente-Pascual, A. Rodríguez-Gascón, A. del Pozo-Rodríguez and M. Á. S. Aspiazu, Molecules, 2020, 25, 5995–6017 CrossRef PubMed.
- S.-E. J. Sung Hee Choi, M.-K. Lee, S.-J. Lim, J.-S. Park, B.-G. Kim, W. S. Ahn and C.-K. Kim, Eur. J. Pharm. Biopharm., 2008, 68, 545–554 CrossRef PubMed.
- K. L. Swingle, A. G. Hamilton and M. J. Mitchell, Trends Mol. Med., 2021, 6, 616–617 CrossRef PubMed.
- K. Susumu, B. C. Mei and H. Mattoussi, Nat. Protoc., 2009, 4, 424–436 CrossRef CAS PubMed.
- B. Borresen, J. R. Henriksen, G. Clergeaud, J. S. Jorgensen, F. Melander, D. R. Elema, J. Szebeni, S. A. Engelholm, A. T. Kristensen, A. Kjaer, T. L. Andresen and A. E. Hansen, ACS Nano, 2018, 12, 11386–11398 CrossRef PubMed.
- M. Ghaferi, M. J. Asadollahzadeh, A. Akbarzadeh, H. Ebrahimi Shahmabadi and S. E. Alavi, Int. J. Mol. Sci., 2020, 21 Search PubMed.
- R. S. Riley, M. V. Kashyap, M. M. Billingsley, B. White, M.-G. Alameh, S. K. Bose, P. W. Zoltick, H. Li, R. Zhang and A. Y. Cheng, Sci. Adv., 2021, 7, eaba1028 CrossRef CAS PubMed.
- S. Patel, N. Ashwanikumar, E. Robinson, Y. Xia, C. Mihai, J. P. Griffith, S. Hou, A. A. Esposito, T. Ketova and K. Welsher, Nat. Commun., 2020, 11, 1–13 CrossRef PubMed.
- H. Farzaneh, M. E. Nik, M. Mashreghi, Z. Saberi, M. R. Jaafari and M. Teymouri, Int. J. Pharm., 2018, 551, 300–308 CrossRef CAS PubMed.
- M. Sun, U. J. Dang, Y. Yuan, A. M. Psaras, O. Osipitan, T. A. Brooks, F. Lu and A. J. Di Pasqua, AAPS PharmSciTech, 2022, 23, 135 CrossRef CAS PubMed.
- J. A. Kulkarni, D. Witzigmann, J. Leung, Y. Y. C. Tam and P. R. Cullis, Nanoscale, 2019, 11, 21733–21739 RSC.
- X. Cheng and R. J. Lee, Adv. Drug Delivery Rev., 2016, 99, 129–137 CrossRef CAS PubMed.
- H. Chen, X. Ren, S. Xu, D. Zhang and T. Han, Int. J. Nanomed., 2022, 17, 2893–2905 CrossRef PubMed.
- S. Daneshmand, S. Golmohammadzadeh, M. R. Jaafari, J. Movaffagh, M. Rezaee, A. Sahebkar and B. Malaekeh-Nikouei, J. Cell. Biochem., 2018, 119, 4251–4264 CrossRef CAS PubMed.
- S. S. Hallan, P. Marchetti, D. Bortolotti, M. Sguizzato, E. Esposito, P. Mariani, C. Trapella, R. Rizzo and R. Cortesi, Molecules, 2020, 25, 5655–5670 CrossRef CAS PubMed.
- S. Salatin and A. Yari Khosroushahi, J. Cell. Mol. Med., 2017, 21, 1668–1686 CrossRef CAS PubMed.
- Z. Mao, X. Zhou and C. Gao, Biomater. Sci., 2013, 1, 896–911 RSC.
- Y. Ju, H. Guo, M. Edman and S. F. Hamm-Alvarez, Adv. Drug Delivery Rev., 2020, 157, 118–141 CrossRef CAS PubMed.
- M. J. Mitchell, M. M. Billingsley, R. M. Haley, M. E. Wechsler, N. A. Peppas and R. Langer, Nat. Rev. Drug Discovery, 2021, 20, 101–124 CrossRef CAS PubMed.
- S. Behzadi, V. Serpooshan, W. Tao, M. A. Hamaly, M. Y. Alkawareek, E. C. Dreaden, D. Brown, A. M. Alkilany, O. C. Farokhzad and M. Mahmoudi, Chem. Soc. Rev., 2017, 46, 4218–4244 RSC.
- P. Foroozandeh and A. A. Aziz, Nanoscale Res. Lett., 2018, 13, 339 CrossRef PubMed.
- J. C. Leach, A. Wang, K. Ye and S. Jin, Int. J. Mol. Sci., 2016, 17, 380 CrossRef PubMed.
- M. R. Usman Ali Ashfaq, E. Yasmeen and M. Z. Yousaf, Crit. Rev. Ther. Drug Carrier Syst., 2017, 34, 317–353 CrossRef PubMed.
- G. Erel-Akbaba, L. A. Carvalho, T. Tian, M. Zinter, H. Akbaba, P. J. Obeid, E. A. Chiocca, R. Weissleder, A. G. Kantarci and B. A. Tannous, ACS Nano, 2019, 13, 4028–4040 CrossRef CAS PubMed.
- A. Le Moignic, V. Malard, T. Benvegnu, L. Lemiegre, M. Berchel, P. A. Jaffres, C. Baillou, M. Delost, R. Macedo, J. Rochefort, G. Lescaille, C. Pichon, F. M. Lemoine, P. Midoux and V. Mateo, J. Controled Release, 2018, 278, 110–121 CrossRef CAS PubMed.
- S. H. Kang, S. J. Hong, Y. K. Lee and S. Cho, Polymers, 2018, 10, 948–965 CrossRef PubMed.
- A. Bolhassani, Mol. Biotechnol., 2022, 1–30 Search PubMed.
- K. Bloom, F. van den Berg and P. Arbuthnot, Gene Ther., 2021, 28, 117–129 CrossRef CAS PubMed.
- A. R. Rappaport, S.-J. Hong, C. D. Scallan, L. Gitlin, A. Akoopie, G. R. Boucher, M. Egorova, J. A. Espinosa, M. Fidanza, M. A. Kachura, A. Shen, G. Sivko, A. Van Abbema, R. L. Veres and K. Jooss, Nat. Commun., 2022, 13, 3289 CrossRef CAS PubMed.
- I. Gomez-Aguado, J. Rodriguez-Castejon, M. Vicente-Pascual, A. Rodriguez-Gascon, M. A. Solinis and A. Del Pozo-Rodriguez, Nanomaterials, 2020, 10, 364–405 CrossRef CAS PubMed.
- J. Wang, X. Ye, H. Ni, J. Zhang, S. Ju and W. Ding, DNA Cell Biol., 2019, 38, 1048–1055 CrossRef CAS PubMed.
- H. Song, S. L. Hart and Z. Du, Int. J. Pharm., 2021, 592, 120033 CrossRef CAS PubMed.
- D. Adams, A. Gonzalez-Duarte, W. D. O'Riordan, C. C. Yang, M. Ueda, A. V. Kristen, I. Tournev, H. H. Schmidt, T. Coelho, J. L. Berk, K. P. Lin, G. Vita, S. Attarian, V. Plante-Bordeneuve, M. M. Mezei, J. M. Campistol, J. Buades, T. H. Brannagan 3rd, B. J. Kim, J. Oh, Y. Parman, Y. Sekijima, P. N. Hawkins, S. D. Solomon, M. Polydefkis, P. J. Dyck, P. J. Gandhi, S. Goyal, J. Chen, A. L. Strahs, S. V. Nochur, M. T. Sweetser, P. P. Garg, A. K. Vaishnaw, J. A. Gollob and O. B. Suhr, N. Engl. J. Med., 2018, 379, 11–21 CrossRef CAS PubMed.
- A. Akinc, M. A. Maier, M. Manoharan, K. Fitzgerald, M. Jayaraman, S. Barros, S. Ansell, X. Du, M. J. Hope, T. D. Madden, B. L. Mui, S. C. Semple, Y. K. Tam, M. Ciufolini, D. Witzigmann, J. A. Kulkarni, R. van der Meel and P. R. Cullis, Nat. Nanotechnol., 2019, 14, 1084–1087 CrossRef CAS PubMed.
- A. Pattnaik, B. R. Sahoo and A. K. Pattnaik, Vaccines, 2020, 8, 266–284 CrossRef CAS PubMed.
- N. Pardi, M. J. Hogan, R. S. Pelc, H. Muramatsu, H. Andersen, C. R. DeMaso, K. A. Dowd, L. L. Sutherland, R. M. Scearce, R. Parks, W. Wagner, A. Granados, J. Greenhouse, M. Walker, E. Willis, J. S. Yu, C. E. McGee, G. D. Sempowski, B. L. Mui, Y. K. Tam, Y. J. Huang, D. Vanlandingham, V. M. Holmes, H. Balachandran, S. Sahu, M. Lifton, S. Higgs, S. E. Hensley, T. D. Madden, M. J. Hope, K. Kariko, S. Santra, B. S. Graham, M. G. Lewis, T. C. Pierson, B. F. Haynes and D. Weissman, Nature, 2017, 543, 248–251 CrossRef CAS PubMed.
- A. Cagigi and K. Loré, Vaccines, 2021, 9, 61–74 CrossRef CAS PubMed.
- J. Hiscott, M. Alexandridi, M. Muscolini, E. Tassone, E. Palermo, M. Soultsioti and A. Zevini, Cytokine Growth Factor Rev., 2020, 53, 1–9 CrossRef CAS PubMed.
- L. A. Jackson, E. J. Anderson, N. G. Rouphael, P. C. Roberts, M. Makhene, R. N. Coler, M. P. McCullough, J. D. Chappell, M. R. Denison, L. J. Stevens, A. J. Pruijssers, A. McDermott, B. Flach, N. A. Doria-Rose, K. S. Corbett, K. M. Morabito, S. O'Dell, S. D. Schmidt, P. A. Swanson 2nd, M. Padilla, J. R. Mascola, K. M. Neuzil, H. Bennett, W. Sun, E. Peters, M. Makowski, J. Albert, K. Cross, W. Buchanan, R. Pikaart-Tautges, J. E. Ledgerwood, B. S. Graham and J. H. Beigel, N. Engl. J. Med., 2020, 383, 1920–1931 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |