Open Access Article
Open Access ArticleFlow cytometry for on-line microbial regrowth monitoring in a membrane filtration plant: pilot-scale case study for wastewater reuse†
Thomas
Pluym
 ab,
Cristina
García-Timermans
ab,
Cristina
García-Timermans
 ab,
Sander
Vervloet
c,
Riet
Cornelissen
c,
Nico
Boon
ab and
Bart
De Gusseme
*ab
ab,
Sander
Vervloet
c,
Riet
Cornelissen
c,
Nico
Boon
ab and
Bart
De Gusseme
*ab
aCenter for Microbial Ecology and Technology (CMET), Department of Biotechnology, Ghent University, Coupure Links 653, B-9000 Ghent, Belgium. E-mail: Bart.DeGusseme@UGent.be
bCenter for Advanced Process Technology for Urban Resource Recovery (CAPTURE), Frieda Saeysstraat 1, B-9000 Ghent, Belgium
cPantarein Water, Egide Walschaertsstraat 22L, B-2800 Mechelen, Belgium
First published on 28th June 2023
Abstract
The use of advanced membrane processes, such as reverse osmosis (RO) are of great importance for process water and drinking water production. As the water quality is bound to certain imposed microbiological standards, the removal of unwanted bacteria is vital. Reverse osmosis filtration should in theory retain all, or most, bacteria. However, due to operational choices or malfunctions (e.g., leaking glue lines, oxidation of the membrane, loss of O-seals…) bacterial breakthrough can occur. Moreover, certain ultra-small species of bacteria are also able to pass the membrane. Currently, the microbiology and breakthrough are respectively monitored by looking at indicator organisms (e.g., Escherichia coli) via plating methods and conductivity monitoring. In this case-study on-line flow cytometry was evaluated as a tool to monitor membrane integrity and microbiological quality of a pilot-scale RO membrane system placed at the Citrique Belgium, Citribel site (Tienen, Belgium). Over the course of 20 days, cell density and flow cytometric fingerprinting data were gathered. With the use of these phenotypic fingerprints and conductivity data, we could showcase that the cell densities found in the permeate were due to regrowth within the piping network, rather then bacterial breakthrough of the membrane. This demonstrates the added-value of flow cytometry for microbial and membrane integrity monitoring of large-scale membrane filtration plants.
Water impactAs reverse osmosis (RO) is typically used as the final barrier in multi-barrier water reuse schemes, it is of great importance for operators to monitor the integrity and impact of operational choices. The research conducted demonstrates the added-value of flow cytometry for membrane integrity of RO membranes and microbiological regrowth monitoring. |
Introduction
The production of potable water from various water sources (wastewater, groundwater, surface water, seawater,…) using advanced membrane processes is of great importance for multiple industry sectors, such as drinking water and process water production.1 The water quality of this treated water is bound to certain regulated chemical, physical and microbiological standards by governments.2 For the microbiological quality of this water in particular, it is of great importance that during water treatment the unwanted bacteria are removed, as some of these can seriously affect human health, water quality and lead to biofouling, microbial induced corrosion and result in an increase of operational and downstream costs.3 Given the fact that these microorganisms can survive or even establish themselves within the biofilm in low-nutrient environments, it is of utmost importance to remove as many microorganisms as possible before the water is distributed or used as process water.4 The use of both filtration techniques and, in some cases, chemical disinfection and/or UV treatment are designed to remove these bacteria for the production of potable water.5Today, water treatment processes partially rely on the use of membrane processes, such as microfiltration (MF), ultrafiltration (UF), nanofiltration (NF) and reverse osmosis.6,7 Reverse osmosis in particular is important for the desalination of water, production of drinking water and the re-use of water from industrial- and wastewater. With a pore size ranging between 0.02 and 10 μm, MF should retain the majority of bacteria.8 The other three membrane processes (UF, NF and RO) are designed to retain all bacteria, as they have a pore size ranging between 10 to 100 nm. Nevertheless, a breakthrough of bacteria larger than the pore size of RO membranes has been reported.9 This may be due to some operational choices or problems with the installation, such as the loss of integrity of the RO membrane by abrasive components in the feed stream. Other potential breakthrough pathways could be due to the f malfunctioning of components that are essential for the RO installation, such as oxidation of the membrane, leaking glue lines and the loss of O-seals, connectors and other fittings.10,11 This can cause breaches in the membrane ranging between 20 to 30 nm.12 These malfunctions can then ultimately influence the quality of the reverse osmosis permeate.13 Other operational actions, such as intensive use of the membrane, with fouling as a result, and chemical damage from cleaning agents may cause alterations in pore size.14,15
On the other hand, the microbiology in particular can also influence the functionality of the RO installation, as some bacteria might penetrate the membrane. It has been seen that bacteria themselves can change shape and size depending on the environment.16 For example, based on nutrient availability, certain bacterial cells may reduce in size to survive.17 Given that the intake water for these processes often comprises unculturable bacteria (less than 1% of the diversity for aquatic environments) of whose not much is known about the size, it is possible that certain bacteria can pass the membrane.18 Recently, ultra-small, uncultivated bacteria, with a size of 0.009 ± 0.002 μm3 have been discovered in groundwater.19 Despite these uncertainties, it has been seen that the mechanisms of bacteria passing through the membrane cannot solely be attributed to differences in size.20 An approach where the bacterial species before and after the RO membrane are identified would be essential to shift towards more effective RO elements for bacterial removal.20
As of today, to evaluate the integrity of the RO membrane installation, online measurements, such as pH, total organic carbon (TOC), conductivity and temperature, are the standard methods. Although all these parameters can be monitored online and can give an indication of how well the membrane is behaving, they do not reveal any direct information about a possible bacterial breakthrough. Integrity strategies using conductivity and TOC are only able to asses bacterial removals up to log reduction values (LRV) of 2.10 Conventional techniques, such as heterotrophic plate counting and selective plating to screen for the presence of indicator organisms, such as E. coli, are both time-consuming (incubation times of 2 to 3 days) and lack a certain accuracy, as these methods are based on the cultivation of bacteria. So, given the fact that a large proportion of the bacteria in aquatic environments are not yet culturable, these are not representable for the microbiological composition of the system.18,21,22 The use of fast and high-throughput techniques, such as online flow cytometry has an enormous potential to be used on large scale industrial systems.23 With the use of flow cytometry, the bacterial cells can quickly be quantified and based on the cellular properties (size, shape, etc.) a phenotypic fingerprint can be generated. With the use of this fingerprint, it is possible to compare the microbial community in different samples.24 This flow cytometric approach can reveal, in comparison to the above mentioned conventional microbial monitoring techniques, a lot more information about the bacterial community within the system. Especially, because shifts in phenotypic community composition and increases in cell numbers can precisely be monitored. The use of flow cytometry in combination with the possibility to generate high-temporal-resolution fingerprint data still presents a lot of potential to monitor the microbial community within RO systems.25 Flow cytometry has already been applied in several studies and has shown to be a valuable tool when it comes to monitoring the microbial quality of water.26,27 In this pilot-scale study, the aim is to use flow cytometry as a tool to gather high-resolution microbiological data of the RO feed and permeate and to not only look into bacterial breakthrough, but based on the bacterial community determine if the RO membrane is leaking or bacterial regrowth is happening within the pipes.
Material and methods
Pilot installation
A pilot-scale membrane filtration installation from Pantarein was placed at the Citrique Belge site (Tienen, Belgium), in order to reuse their wastewater by means of ultrafiltration (UF) followed by reverse osmosis (RO). The final product was intended for reuse in the production process. This means that it should be compliant with drinking water standards. The wastewater used for this specific study was effluent from an aerobic wastewater treatment system (CAS system). The feed and permeate flowrate, conductivity, pH, temperature and redox values can be found on the GitHub page. After ultrafiltration (UF) treatment, the UF filtrate was used to feed a two-stage RO system with two pressure vessels in stage one and one pressure vessel in stage two. Every pressure vessel contained 4 Dow Filmtec XFRLE-400/34i membranes elements (DOW, USA). These brackish water membrane elements have an active filtration area of 37 m2 and a salt retention of minimum 99.2%, and are especially designed for the treatment of challenging water with high biological and organic fouling potential such as wastewater. The pilot-scale RO system was operated at a flow rate of 11.9 m3 h−1 for the feed and 8.8 m3 h−1 for the permeate.To monitor the microbial dynamics, the RO feed and the RO permeate were simultaneously sampled for online flow cytometric analyses over a 20 day period (from the 6th of May 2021 until the 25th of May 2021). High-resolution operational (sensor) data was also recorded for the following parameters; RO feed temperature (°C), pH, redox potential, RO feed flow (m3 h−1), RO permeate flow (m3 h−1), RO concentrate flow (m3 h−1), RO concentrate recirculation flow (m3 h−1), RO HP pressure (bar), RO differential pressure st1 (bar), RO differential pressure st2 (bar), conductivity of the RO feed (μS cm−1), conductivity of the RO permeate (μS cm−1), conductivity of the RO concentrate (μS cm−1), the RO recovery (%) and flow of the CIP pump (m3 h−1). The operational data was correlated with the corresponding sampling times of flow cytometry measurements, from the 6th of May, 2021 on, which was considered the start of the trial. When the data from the sensors does not have an exact (at seconds scale) time match with the flow cytometry measurements, matching was performed to the closest time point. All operational events during the monitoring time period were registered.
During this 20 day monitoring period, RO downtime, membrane flushes and an ultrasound treatment were registered. Three subperiods were specifically selected to determine the effect of certain operational events on the microbial community. During the first period, between the 14th and 17th of May 2021 (Day 9 till day 12), two membrane flushes were registered. A membrane flush is a cleaning procedure where clean tap water or permeate are put through the RO membrane for a short period of time. During the second subperiod, between the 22nd and 25th of May 2021 (Day 17 till day 20), a non-scheduled RO membrane downtime due to a power outage (Day 17, Day 17) and a membrane flush was monitored. The third and final period of specific interest, ranging from focuses on the influence of the ultrasound treatment on the bacterial cell densities and bacterial community.
Flow cytometry
To examine the effect of the RO membrane and environmental parameters/operational events on the dynamics of the microbial community and bacterial cell densities, the feed, permeate stream and permeate buffer tank were monitored with on-line flow cytometry (Fig. 1). Samples from these streams were automatically taken every 30 minutes, after which they were immediately stained and incubated for 20 minutes at 37 °C prior to measurement. As the samples were collected directly from the piping and low cell concentrations were expected, no pretreatment or dilution was performed. During the monitoring period, between the 6th of May and 25th of May, 999 flow cytometric samples were taken for each sampling point with a time interval of 36 minutes. To achieve continuous and automated measurements, an onCyt© (onCyt Microbiology AG, Switzerland) autosampler was coupled to an Accuri™ C6 Plus flow cytometer (BD Biosciences, Belgium). Between every measurement the onCyt autosampler performs a sample line cleaning using a chlorine and Sodium Thiosulfate solution.28 The flow cytometer was equipped with a blue (20 mW, 488 nm), a red laser (12.5 mW, 640 nm), two scatter detectors configured on the blue laser, and four fluorescence detectors with bandpass filters. Three of the bandpass filters are for the blue laser emission (FL1: 533/30 nm, FL2: 585/40 nm, and FL3: 670 LP) and one is for the red laser emission (FL4: 675/25 nm). The lower detection limit of flow cytometry in this type of application is in the order of 103 cells per mL. For each 93 samples, three MilliQ samples were measured, to ensure a correct gating strategy. As an additional control, samples were manually collected and measured on an Accuri flow cytometer in the lab. The cell concentrations, especially in the permeate buffer tank, were sometimes lower than the detection limit. Thus, as accurate and precise quantification of cell concentrations was lacking for the permeate buffer tank, these results were not considered in the statistical analysis. MilliQ (Merck, Belgium) was used as sheath fluid. Staining was performed using a 1000× dilution of SYBR® Green I concentrate (Invitrogen, Belgium) in TRIS buffer with pH 8.2, with a 10 vol% final concentration. Flow cytometric samples were run in fixed volume mode (25 μL). A Cavro® XCalibur Pump (Tecan Trading AG, Switzerland) with 12 channels connects the necessary fluidics, air and waste with the three chambers in the onCyt© robot. The pump regime is adapted to the desired sampling frequency (36 min interval) and size (800 μL total volume) using the onCyt© software.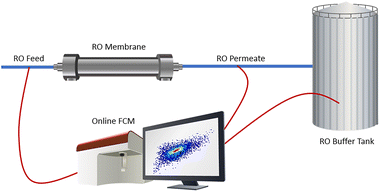 | ||
| Fig. 1 Simplistic schematic overview of the installation and different sampling points (RO feed, RO permeate & RO tank). | ||
Data analysis
The Flow Cytometry Standard (.fcs) files that were generated by the flow cytometric measurements were imported into R (v 4.1.2) using the flowCore package (v.2.8.0.). By manually drawing a gate (based on the MilliQ and manual measurements) on the FL1 and FL3 fluorescence data, background noise caused by artefacts was removed. The combination of these two parameters results in the most optimal signal and noise separation for water samples. To correctly select the bacterial population, gating was done as described in Props et al. (2016) (Gating strategy supplied in ESI,† Fig. S1).29 Further data processing was done using the Phenoflow package (v.1.1.2). To examine the capabilities of flow cytometry to safeguard membrane integrity, the bacterial cell densities were extracted by means of the Flowcore package. In a second stage, to determine the changes in the microbial communities in both streams, phenotypic community analysis by the use of flow cytometric fingerprinting was performed. In this analysis, the flow cytometry data of every sample is transformed, discretized and concatenated into a one-dimensional vector that serves as basis for further phenotypic community analysis. From these fingerprints, beta diversity analysis and a classical multidimensional scaling analysis calculations were executed using the vegan package (v.2.6–4). To evaluate the cause of certain differences in cell density and community fingerprint, the operational events, as well as metadata such as temperature, pressure, flow rate and conductivity were linked to specific measurement timepoints (ggplot2 (v.3.3.6)).A classical multidimensional scaling (cmdscale function in stats package (v.4.2.1)) analysis on the Bray–Curtis distances of the single-cell physiological data was performed to generate a non-metrical multidimensional scaling (NMDS). This method reduces the loss of information and increases interpretability by reducing the dimensionality, which was of great importance for this case-study, given the amount of parameters that were taken into consideration. This analysis was specifically chosen to evaluate the differences between the microbial community of the different samples and to easily quantify and visualize the differences between the flow cytometric measurements. The outcome of this NMDS analysis is a so-called phenotypic fingerprint. Flow cytometric fingerprinting relies on physiological data acquired through the rapid optical characterization of the cells. On the Bray–Curtis matrix that was used for the NMDS analysis, the multivariate homogeneity of group dispersions (variances) was calculated (betadispervisity). In general, the distance of each sample to the centroid of its group is calculated and tested whether the variances of these distances are significantly different between the two sample groups (ESI† S2). Furthermore, a PERMANOVA analysis was performed. This was used to evaluate if the sampling point has a significant influence on the Bray–Curtis distance matrix and thus how different the phenotypic communities are in each stream. The PERMANOVA partitions the variation in a distance matrix between groups to test for significant differences, while the betadispervisity directly tests for differences in dispersion between groups using the dissimilarity matrix.
All files and code can be found at: https://github.ugent.be/thpluym/Citrique_Belgium_05_2021.git.
Results & discussion
Bacterial cell densities during pilot testing
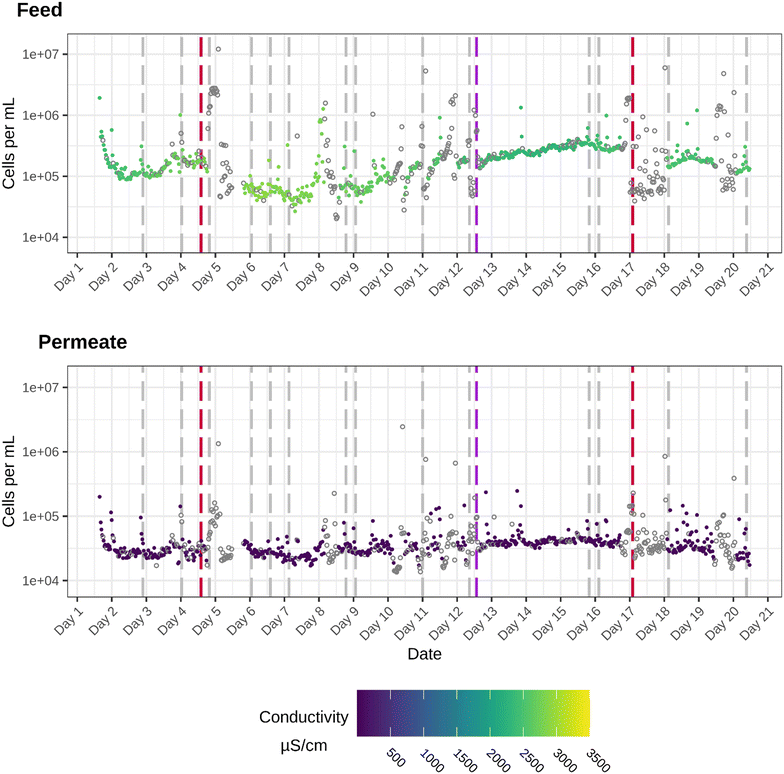
The pilot-scale RO membrane filtration unit was operated for 20 days to test its performance for wastewater reuse. The RO unit was operated at an average permeate flow rate of 8.8 m3 h−1 and online microbial monitoring was performed during a 20 day sampling campaign using flow cytometry. A general overview of the operational conditions in that 20 day period is given in Fig. 2, together with the measured cell concentrations and conductivity measurements. If the pilot is not in operation conductivity was not measured and therefor given in grey. When there is no operation (and thus no conductivity data), increasing cells are monitored because of stagnant water.The bacterial cell densities show that for both the RO feed and permeate each measurement exceeds the limit of detection (103 cells per mL), which is not expected after RO treatment. In the feed stream, the cell densities fluctuate between 2 × 104 cell per mL and 1.2 × 107 cells per mL. In the first days of operation, the concentration drops from 1.9 × 106 cells per mL to 1.3 × 105 cells per mL. After the number of bacterial cells reaches 1.3 × 105 cells per mL, conductivity increases. Despite the high bacterial cell density in the permeate, there is a significant difference between the cell densities in the feed and permeate (Wilcoxon test, P-value: 2.2 × 10−16). Moreover, a pairwise Wilcoxon comparison for each sample was performed and showed a significant difference for all timepoints. In general, great variability in cell density can be seen during the monitoring period for both the permeate as well as the feed stream. This was evaluated with the sign difference test, showing multiple periods of significant increase/decrease in cell density (ESI†). The downtime of the RO has resulted in an altered microbial water quality, as after the RO downtime of day 4, a slight increase in cell density in both sampling points was noticed. For example, a stop in the production resulted in an increase in cell density (day 4), with measurements exceeding 106 cells per mL for the permeate and 107 cells per mL for the feed. These increases may be due to the loss of flow and stagnant water in the pipes. After the peak in cell density around day 4 and 5 (during the power out), the cell densities remain relatively low until day 8, after which a steady increase can be observed.
When focusing on the removal efficiency, only a maximum log reduction value (LRV) of two can be detected between the feed and the permeate. For reverse osmosis filtration, in which most bacterial cells should be retained by the membrane, this LRV is on the lower side. Spikes in cell density can be linked to RO washes at day 3, day 4, day 7, day 8, day 9, day 11 and day 17. Cell density peaks detected in the permeate stream at these timepoints exceed 105 and even 106 cells per mL, which are high microbial loads for a RO permeate stream. However, these values are similar to what is seen in drinking water.
The use of standard microbiological tests (e.g. Plating tests) can provide a definite answer to the question if harmful bacteria are present, something which flow cytometry alone cannot. After the 20 day monitoring period a standard drinking water analysis was performed and met the standard drinking water regulations. These results showed that in the permeate zero coliforms and zero E. coli colony forming units (CFU) per 100 mL were found. This suggests that at that point the RO permeate was of excellent microbiological quality (conform drinking water standards) (report in ESI,† Fig. S3).
These cell densities do not necessarily mean that there is a risk for human health, but from an operational point of view, could cause problems with the piping, such as microbial induced corrosion and biofouling for example3,30 To ensure that no pathogenic bacteria are present, regular microbiological tests are a must. Except for the single drinking water analysis performed at the end of the experiment, no coliform or bacteriological tests were performed during the 20 day run.
The conductivity values, which are currently the state-of-the-art to monitor membrane integrity, show that the membrane is actually functioning according to the current standards, as most of the permeate values are lower than 100 μS cm−1 with peaks up to 400 μS cm−1. The conductivity values in the feed range between 1000 and 3000 μS cm−1.
Effect of operational events on the RO performance and resulting microbial water quality
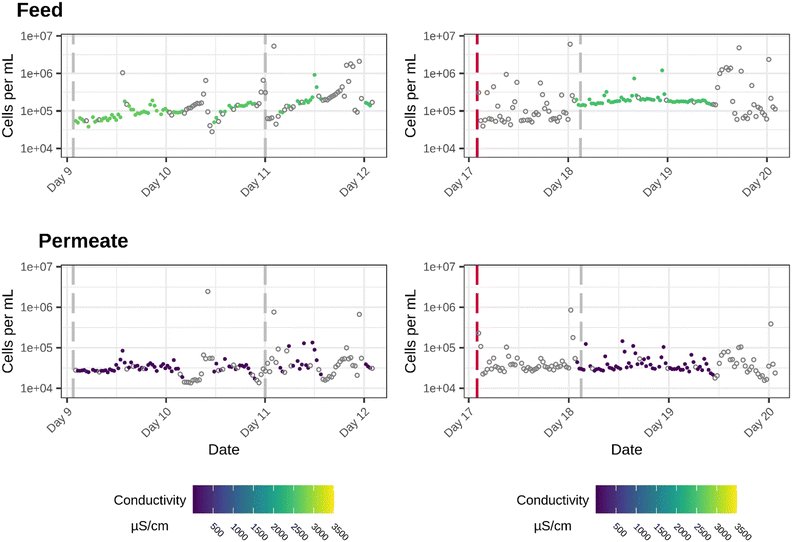
For the sake of clarity, three specific shorter periods will be highlighted, because these are the most representative to link changes in cell density or bacterial population dynamics to a specific operational event or environmental parameter:(1) a 3 day period with stable RO performance and only one flush, which will be used to examine the microbial water quality before and after the flushing event. (between day 9 and day 13 of the experiment) (Fig. 3).
(2) a 3 day period in which the effect of an RO downtime was examined on the bacterial cell density and community composition, allowing to evaluate the effect of RO downtimes and related stagnant water in the pilot with changes in microbial quality (between day 17 and day 20 of the experiment). In this period only one flush is performed (Fig. 3).
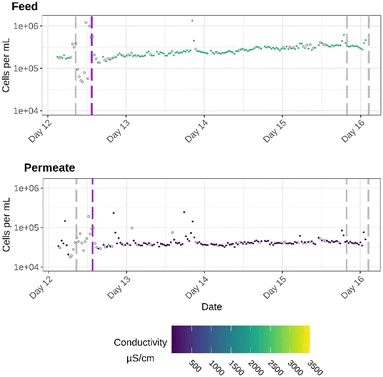
(3) a 4 day period in which the effect of an ultrasound treatment was examined. This ultrasound treatment was used to reduce biofilm growth (Fig. 4).
When focusing on the bacterial cell densities of both the feed and permeate for the period between day 9 and day 12, and between day 17 and day 20 of the experiment, relatively high cell densities for the permeate were registered. Nevertheless, they are still significantly different from the cell densities in the feed (Period 1: p-value = 0.002961; period 2: p-value = 1.115 × 10−7). After the flush cycle on day 9, the cell densities in both the feed and permeate remain stable. It can be seen that after the flush on day 11 there is a slight decrease in cell densities in the feed stream, but in the permeate stream a great variability in cell densities is induced.
During the second downtime of the membrane at day 17, cell densities increase, which can be explained by the fact that the water remained stagnant for a this period of time. The fact that the filtration plant was down water was stagnant for multiple longer periods also strengthens the hypothesis that there is bacterial regrowth in the piping network after the RO filtration. During the downtime on day 17, the cell densities and conductivity values in the feed show a lot of variability (evaluated by the Sign test, ESI†) between the different measurements. One day later, after the flush cycle on day 18, both the conductivity, as well as the cell densities stabilize more or less for one and a half day while the system is in operation. From day 19 on, due to an unforeseen shorter downtime, the cell densities in the feed start to vary again, which is reflected by a similar pattern in the permeate stream. During the entire monitoring period, the system underwent 14 flushes and two longer downtimes. During these operational events, the water is stagnant in the piping. These conditions, as they also stimulate particle accumulation, favor bacterial growth.31 This influence of stagnant water on the bacterial cell densities can clearly be seen during and after the longer RO downtimes (day 9 and day 17). An increase in flow rate, during the flushing steps, a sudden increase in flowrate after start up and during intensive use of the plant, can, on the other hand cause the detachment of bacteria from the biofilm.32 Therefore, the combination of remaining nutrients and variable hydraulic conditions could also influence the regrowth in the piping after the RO membrane filtration.
In the third period of specific attention (Fig. 4), an ultrasound treatment was applied on the RO feed to prevent and reduce biofilm formation and follow biofouling on the RO membranes. After the start of the ultrasound treatment, there is a small steady increase in cell densities of the RO feed. For the RO permeate there is no noticeable increase in cell density, but the cell densities remain relatively stable after the treatment, until the flushes on day 15 and 16. The days after the treatment, the pilot plant is operated continuously without flushes or RO downtimes, which causes the system to stabilize. This might explain why the cell densities in the feed and especially in the permeate remain stable. For the feed, it can be observed that, despite being more stable, a higher overall cell density is reached after the start of the treatment compared to the days before the treatment. These increases in cell densities can be explained by the fact that more cells are kept in the bulk phase, because an ultrasound treatment mitigates biofilm formation.33
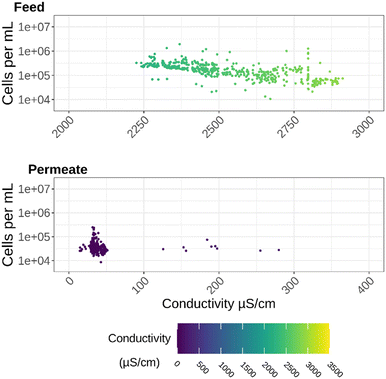
The on-line flowcytometry measurements from this case-study provide us with high resolution data regarding the bacterial cells that can be detected after the reverse osmosis filtration. For this specific case, a maximum log reduction value of two for the bacterial cell densities could be detected between the feed and the permeate, while conductivity values show expected values for RO filtration. To determine if flow cytometry could be a valuable addition to the current state-of-the-art and is thus an adequate technique to measure the bacterial cells passing through the membrane, the correlation between the cell densities, measured by the flow cytometer, and the measured conductivity data, was plotted (Fig. 5). This shows that for this specific case no correlation can be found between increases/decreases in cell densities and conductivity for the permeate. To evaluate this, a Pearson's product–moment correlation was performed. For the permeate there was no significant correlation between the conductivity values and the cell density (P-value = 0.2691). For the feed, however, there is a significant negative correlation (0.3827592) between the conductivity values and bacterial cell density (P-value = 2.2 × 10−16). The lower the conductivity value in the feed, the higher the bacterial cell densities are. This means that caution is needed when interpreting conductivity data, as this case-study shows that an increase in bacterial cell densities does not result in an increase in the conductivity that was measured.
In general and based on the current state-of-the-art, it can be concluded that the reverse osmosis membrane is doing an adequate job, as conductivity values in the permeate decrease to values ranging between zero and maximum peak values of 400 μS cm−1 (standard drinking water values), with an average conductivity of 22 μS cm−1. An average of 22 μS cm−1 is a normal value for a well-functioning RO membrane. Based on this conductivity data alone, it can be assumed that the reverse osmosis membrane is working as expected, as there is a reduction in conductivity of around 98%. It is believed that the relatively high cell densities in the permeate could be caused by regrowth in the piping system after the RO filtration. After RO filtration a 99.5% bacterial retention is expected, with low bacterial cell densities varying around 15 cells per mL in the permeate.34,35 For this case-study, an average bacterial retention of 76.98% was calculated. This could be due to the fact that measurements were not taken immediately after the RO membrane element, but further down the pipe. Previous studies have shown that even with a 99% bacterial retention, cell densities could reach 102, 103 cells per mL due to regrowth.36 This regrowth hypothesis can be put on effect by the fact that the flow cytometric measurements were taken two to three meters after the outlet of the RO membrane and the sampling point for the conductivity. It is possible that what was measured with the flow cytometer is bacterial (re)growth in the piping. This hypothesis was further examined using flow cytometric fingerprinting.
Flow cytometric fingerprinting to evaluate the cause of microbial regrowth
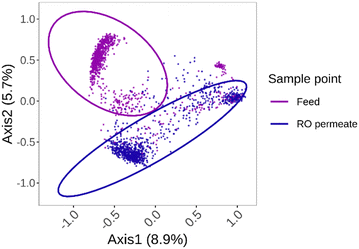
Using the physiological data of each cell, a non-metrical multidimensional scaling (NMDS) calculation was executed for all samples taken from the feed and the permeate (Fig. 6). The stress value of 0.1487618 suggests that the NMDS algorithm was able to represent the distances between the data points reasonably well in a lower-dimensional space. The NMDS plot visually shows that there is a difference in phenotypic fingerprint between the feed and permeate stream. This difference was confirmed by the significant effect of the sampling point (Feed or Permeate) on the Bray–Curtis dissimilarity. This means that there is a significant difference between the feed and permeate communities (F = 659.37, p < 0.001). The fact that for this case-study there are significant different phenotypic communities in the feed and permeate indicates that both water streams are inhabited by slightly different microbial communities (Fig. 6). Since a different phenotypic microbial community can be found in the permeate stream compared to the one in the feed stream, the hypothesis that there is regrowth after the RO filtration is strengthened. If there would be bacterial breakthrough of any kind, the phenotypic community in the feed and permeate would be similar. This showcases that with the use of fingerprinting techniques there is the possibility to evaluate the microbial community composition in a fast and on-line manner. If the system is continuously monitored using flow cytometry a slight increase or decrease in cell density or change in community composition can be monitored. Cytometers equipped with more detectors might have a higher fingerprinting sensitivity, which could result in an even better fingerprinting resolution and thus better display minor differences.37 The disadvantage of using this fingerprinting and in general flow cytometric approach results in fact that it provides no data on the type of organisms which are present. Other conventional (plating) and/or sequencing techniques are still required to gain insights into the presence of pathogenic or unwanted bacteria.It can be argued that the different communities that were seen in the feed and permeate stream can be due to the fact that there is breakthrough of specific species of bacteria, as it has been seen that small bacteria can pass the membrane.20 Given the fact that the water source in this case is actually treated wastewater, there is a possibility that certain unknown and potential unculturable (small) species of bacteria are present and were able to break through the membrane.19,38 Because of the lack of other (conventional) microbiology monitoring methods in this case (such as CCA plating, Colilert,…) it is impossible to completely exclude full bacterial breakthrough of certain species. But, the differences in fingerprint in combination with the normal conductivity values, ensure that complete bacterial breakthrough, and thus, a faulty membrane can be refuted. The placement of the membranes and the installation of the pressure vessels and the piping all happened in a non-sterile environment. It is thus very likely that the bacteria were introduced during commissioning of the system. Although the RO membrane filtration should 99.5% of the bacteria, not all nutrients are retained, as only 90% of the TOC and 80% of the assimilable organic carbon (AOC) is rejected.36 Since no system is completely sterile, these remaining nutrients are available for any present bacteria and can cause (re)growth in the RO permeate.39 The fact that certain compounds can still pass the membrane results in biologically unstable water in the permeate.40 This is reflected in the phenotypic fingerprint.
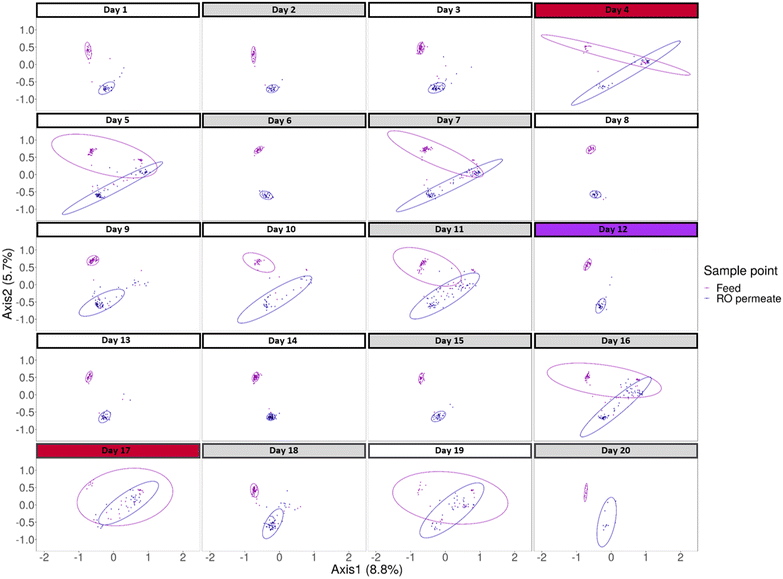
In the general NMDS plot, it is clear that different clusters are formed for each stream. In the cluster of the permeate stream, two different microbial populations can be noticed. To determine why a certain shift in the community happened, the general NMDS plot was split up for each day separately to determine temporal changes (Fig. 7). This allowed for the examination of the impact of certain operational events on the microbial community composition. To evaluate the impact of certain events a PERMANOVA and betadispervisity analysis were performed. By looking into the average distance to the center of the group, an estimation of the heterogeneity and variability within the community was made. The larger the average distance to the median, the more variable and heterogenic the community within that group is. At the start of the experiment, on day 1, two distinct populations can be detected for the feed and the permeate stream (P-value = 0.000999). During the first days of operation the average distance to the center varies between 0.1186 and 0.1211 for the feed and between 0.1286 and 0.1501 for the permeate. On day 4 of the experiment, during the first RO membrane downtime, there is a clear change of the microbial community fingerprint of both the feed and permeate, as there is an increase in variability in both the permeate (average distance to median = 0.3329) and feed community (average distance to median = 0.3927). For the second RO downtime, on the day 17, a similar increase can be detected (average distance to median feed = 0.3825, permeate = 0.2465). On day 19, there was a shorter downtime of the RO installation and this also results in a similar fingerprint and distance to the median (Feed = 0.3939, Permeate = 0.2580). On both day 4 and day 17 the filtration plant was down for the entire day. Since the system was not in operation, the changes in community composition can thus not be attributed to bacterial breakthrough. This can be confirmed by the fact that, despite the increases in variability, the community in the permeate remains significantly different from the community in the feed on the days when the RO was down (Day 4: P-value = 0.000999, F-value = 8.0211; Day 17: P-value = 0.000999, F-value = 7.0461). The stagnant water, associated with these downtimes, creates a perfect environment for bacterial growth within the system.41 Since during this downtime, the water in both the feed and permeate is stagnant, the microbial community in the system is influenced, giving specific bacteria the growth advantage. It is possible that similar bacteria in both streams are benefitting from these environmental conditions and thus a more diverse community can be found.
It can also be seen that the flushing events impact the microbiological composition of feed and the permeate, as the fingerprint changes on day 5, 7, 11 and 16 when flushing cycles were reported. These flushes result in a more variable microbial community in both streams. On day 5 for example, the average distance to the median, increases to 0.3424 for the feed and 0.3082 for the permeate. After the ultrasound treatment, which was executed on day 12, two significantly distinct communities can be detected (P-value = 0.000999, average distance to median feed = 0.1131, permeate = 0.1331). During the first days after the ultrasound treatment, this difference in fingerprint between the feed and permeate remains constant. The average distance to the median for day 13, 14 and 15 is respectively 0.1284, 0.1162 and 0.1182 for the feed and 0.1426, 0.1186 and 0.1191 for the permeate. After the ultrasound treatment, there were also no operational events registered, so the installation could continue to operate steadily for a couple of days. The phenotypic fingerprint changed again after the CIP cycle of day 15, indicated by an increase in the average distance to the median to 0.3588 for the feed and 0.2675 for the permeate. After, again less differences between the microbial community of the permeate and the feed can be seen. The fact that two distinct communities were formed after the ultrasound treatment shows that when biofilm formation is mitigated and (parts of) the biofilm are disrupted, the bacteria measured by the flow cytometer in the feed and permeate stream are different.42 This indicates that in the feed and permeate stream a different bacterial composition is present and a different biofilm is growing. The use of the phenotypic fingerprint and analysis of the variability within in each stream for each day specifically allows for a more detailed analysis of the effect of operational events on the microbiology and strengthens the hypothesis that regrowth is happening after the membrane filtration.
Conclusions
In this case-study, flow cytometry was evaluated as a technique to monitor RO membrane functionality online and its application on a larger scale pilot installation for wastewater reuse. Current standard methods such as conductivity measurements reveal information about membrane integrity but lack the possibility to monitor microbiological quality problems or regrowth within the system. Flow cytometry is a valuable and sensitive tool that can be applied to follow up membrane filtration plants, capable of collecting continuous, high-resolution data, which not only generates cell counts, but allows for phenotypic fingerprinting. The use of flow cytometry for this particular case, shows that bacterial cells are present above the expected level after RO filtration. However, the cell densities do not exceed standard drinking water values. More specifically, the flow cytometric fingerprint data showed that different microbial populations were found in the feed and permeate, and thus proves that bacterial breakthrough of the membrane was excluded, together with the expected conductivity values of the RO. Therefore, the application of flow cytometry does not only reveal information on the bacterial cell densities, but also enables the characterization of the community composition through phenotypic fingerprinting. Without the use of flow cytometry, the regrowth in the RO permeate would possibly have remained unnoticed, as based on conductivity and other indirect measurements no clear indication of bacterial (re)growth would be present. The phenotypic fingerprint enabled us to make a distinction between breakthrough of the membrane or regrowth within the system.Based on these findings, we suggest that not only the cell densities, but also the phenotypic fingerprint could be considered as a microbial parameter for a complete monitoring of the microbiological quality of RO feed and especially permeate. In this respect, flow cytometry could serve as an additional microbiological and RO membrane integrity monitoring tool to reveal information about the microbiology within the system, especially for large-scale process operation. However, in practice, the use of flow cytometry as an additional method to monitor (small-scale) RO installations might still be too expensive and complex, especially compared to standard microbiological monitoring methods. Yet, for installations with a certain scale or for reuse applications where microbial monitoring is very important, such as the food and beverage industry, we have shown that flow cytometry definitely has an added-value as a tool to monitor microbiological quality and regrowth, online and in a higher resolution compared to conventional methods.
Author contributions
T. Pluym wrote the paper with contributions from C. García Timermans, S. Vervloet, N. Boon and B. De Gusseme. Data was collected by R. Props, F.-M. Kerckhof, S. Decroo, S. Vervloet & R. Cornelissen. Data analysis was performed by T. Pluym, C. García-Timermans & F.-M. Kerckhof. The study was designed by B. De Gusseme. All the authors read and approved the final version of the manuscript.Conflicts of interest
There are no conflicts to declare. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. All the authors read and approved the final version of the manuscript.Acknowledgements
This work was part of the project RepEAT, supported by Flanders' Food and the Flanders Innovation & Entrepreneurship Agency VLAIO. Thomas Pluym and Cristina Garcia Timermans are funded by the Research Foundation – Flanders (FWO) in the FWO-SBO Biostable project [grant number S006221N]. The work was logistically supported by Citrique Belge. The authors would like to acknowledge Emmanuel Raskin of Citrique Belge, Ruben Props and Frederiek-Maarten Kerckhof of KYTOS and Piet De Langhe of Pantarein for their intellectual contribution to this project. Also, we would like to thank Sam Decroo of CMET for the technical assistance and practical information during the pilot study.Notes and references
- C. Y. Tang, Z. Yang, H. Guo, J. J. Wen, L. D. Nghiem and E. Cornelissen, Potable Water Reuse through Advanced Membrane Technology, Environ. Sci. Technol., 2018, 52(18), 10215–10223 CrossRef CAS PubMed.
- B. G. Oliver, Guidelines for drining-water quality, volume 1, Adv. Water Resour., 1984, 7(4), 200 CrossRef.
- H. C. Flemming, Biofouling in water systems – Cases, causes and countermeasures, Appl. Microbiol. Biotechnol., 2002, 59(6), 629–640 CrossRef CAS PubMed.
- E. Torvinen, S. Suomalainen, M. J. Lehtola, I. T. Miettinen, O. Zacheus and L. Paulin, et al. Mycobacteria in Water and Loose Deposits of Drinking Water Distribution Systems in Finland, Appl. Environ. Microbiol., 2004, 70(4), 1973–1981 CrossRef CAS PubMed.
- W. A. M. Hijnen, E. F. Beerendonk and G. J. Medema, Inactivation credit of UV radiation for viruses, bacteria and protozoan (oo)cysts in water: A review, Water Res., 2006, 40(1), 3–22 CrossRef CAS PubMed.
- S. Jiang, Y. Li and B. P. Ladewig, A review of reverse osmosis membrane fouling and control strategies, Sci. Total Environ., 2017, 595, 567–583 CrossRef CAS PubMed.
- M. Elimelech and W. A. Phillip, The future of seawater desalination: Energy, technology, and the environment, Science, 2011, 333(6043), 712–717 CrossRef CAS PubMed.
- U. T. D. Behnke, Brock: Membrane Filtration. A User's Guide and Reference Manual. 381 Seiten, 115 Abb., 27 Tab. Springer-Verlag, Berlin, Heidelberg, New York, Tokyo 1983, Preis : 87,-DM, Nahrung, 1983, 27(10), 1025 Search PubMed.
- S. B. S. Ghayeni, P. J. Beatson, A. J. Fane and R. P. Schneider, Bacterial passage through microfiltration membranes in wastewater applications, J. Membr. Sci., 1999, 153(1), 71–82 CrossRef.
- M. Kitis, J. C. Lozier, J. H. Kim, B. Mi and B. J. Mariñas, Microbial removal and integrity monitoring of RO and NF membranes, J. - Am. Water Works Assoc., 2003, 95(12), 105–119 CrossRef CAS.
- G. F. Crozes, S. Sethi, B. Mi, J. Curl and B. Marias, Improving membrane integrity monitoring indirect methods to reduce plant downtime and increase microbial removal credit, Desalination, 2002, 149(1–3), 493–497 CrossRef CAS.
- E. R. Ostarcevic, J. Jacangelo, S. R. Gray and M. J. Cran, Current and emerging techniques for high-pressure membrane integrity testing, Membranes, 2018, 8(3), 60 CrossRef PubMed.
- P. M. Biesheuvel, S. Porada, M. Elimelech and J. E. Dykstra, Tutorial review of reverse osmosis and electrodialysis, J. Membr. Sci., 2022, 647 DOI:10.1016/j.memsci.2021.120221.
- M. F. A. Goosen, S. S. Sablani, H. Al-Hinai, S. Al-Obeidani, R. Al-Belushi and D. Jackson, Fouling of reverse osmosis and ultrafiltration membranes: A critical review, Sep. Sci. Technol., 2004, 39(10), 2261–2297 CrossRef CAS.
- F. I. Hai, T. Riley, S. Shawkat, S. F. Magram and K. Yamamoto, Removal of pathogens by membrane bioreactors: A review of the mechanisms, influencing factors and reduction in chemical disinfectant dosing, Water, 2014, 6(12), 3603–3630 CrossRef.
- A. C. Chien, N. S. Hill and P. A. Levin, Cell size control in bacteria, Curr. Biol., 2012, 22(9), R340–R349 CrossRef CAS PubMed.
- T. Egli, How to live at very low substrate concentration, Water Res., 2010, 44(17), 4826–4837 CrossRef CAS PubMed.
- I. Douterelo, J. B. Boxall, P. Deines, R. Sekar, K. E. Fish and C. A. Biggs, Methodological approaches for studying the microbial ecology of drinking water distribution systems, Water Res., 2014, 65, 134–156 CrossRef CAS PubMed.
- B. Luef, K. R. Frischkorn, K. C. Wrighton, H. Y. N. Holman, G. Birarda and B. C. Thomas, et al. Diverse uncultivated ultra-small bacterial cells in groundwater, Nat. Commun., 2015, 6, 6372 CrossRef CAS PubMed.
- T. Fujioka and S. Boivin, Assessing bacterial infiltration through reverse osmosis membrane, Environ. Technol. Innov., 2020, 19, 100818 CrossRef.
- M. J. Allen, S. C. Edberg and D. J. Reasoner, Heterotrophic plate count bacteria – What is their significance in drinking water?, Int. J. Food Microbiol., 2004, 92(3), 265–274 CrossRef PubMed.
- R. I. Amann, W. Ludwig and K. H. Schleifer, Phylogenetic identification and in situ detection of individual microbial cells without cultivation, Microbiol. Rev., 1995, 59(1), 143–169 CrossRef CAS PubMed.
- B. Buysschaert, L. Vermijs, A. Naka, N. Boon and B. De Gusseme, Online flow cytometric monitoring of microbial water quality in a full-scale water treatment plant, npj Clean Water, 2018, 1(16) DOI:10.1038/s41545-018-0017-7.
- K. De Roy, L. Clement, O. Thas, Y. Wang and N. Boon, Flow cytometry for fast microbial community fingerprinting, Water Res., 2012, 46(3), 907–919 CrossRef CAS PubMed.
- S. Van Nevel, S. Koetzsch, C. R. Proctor, M. D. Besmer, E. I. Prest and J. S. Vrouwenvelder, et al. Flow cytometric bacterial cell counts challenge conventional heterotrophic plate counts for routine microbiological drinking water monitoring, Water Res., 2017, 113, 191–206 CrossRef CAS PubMed.
- J. Favere, B. Buysschaert, N. Boon and B. De Gusseme, Online microbial fingerprinting for quality management of drinking water: Full-scale event detection, Water Res., 2020, 170, 115353 CrossRef CAS PubMed.
- B. Buysschaert, J. Favere, L. Vermijs, V. Baetens, A. Naka and N. Boon, et al. Flow cytometric fingerprinting to assess the microbial community response to changing water quality and additives, Environ. Sci.: Water Res. Technol., 2019, 5(10), 1672–1682 RSC.
- J. Favere, F. Waegenaar, N. Boon and B. De Gusseme, Online microbial monitoring of drinking water: How do different techniques respond to contaminations in practice?, Water Res., 2021, 202, 117387 CrossRef CAS PubMed.
- R. Props, P. Monsieurs, M. Mysara, L. Clement and N. Boon, Measuring the biodiversity of microbial communities by flow cytometry, Methods Ecol. Evol., 2016, 7(11), 1376–1385 CrossRef.
- A. Reyes, M. V. Letelier, R. De la Iglesia, B. González and G. Lagos, Microbiologically induced corrosion of copper pipes in low-pH water, Int. Biodeterior. Biodegrad., 2008, 61(2), 135–141 CrossRef CAS.
- G. Liu, M. C. Lut, J. Q. J. C. Verberk and J. C. Van Dijk, A comparison of additional treatment processes to limit particle accumulation and microbial growth during drinking water distribution, Water Res., 2013, 47(8), 2719–2728 CrossRef CAS PubMed.
- M. J. Lehtola, M. Laxander, I. T. Miettinen, A. Hirvonen, T. Vartiainen and P. J. Martikainen, The effects of changing water flow velocity on the formation of biofilms and water quality in pilot distribution system consisting of copper or polyethylene pipes, Water Res., 2006, 40(11), 2151–2160 CrossRef CAS PubMed.
- L. Mathieu, A. Keraval, N. F. Declercq and J. C. Block, Assessment of a low-frequency ultrasound device on prevention of biofilm formation and carbonate deposition in drinking water systems, Ultrason. Sonochem., 2019, 52, 41–49 CrossRef CAS PubMed.
- T. Fujioka, R. Makabe, N. Mori, S. A. Snyder and M. Leddy, Assessment of online bacterial particle counts for monitoring the performance of reverse osmosis membrane process in potable reuse, Sci. Total Environ., 2019, 667, 540–544 CrossRef CAS PubMed.
- T. Fujioka, A. T. Hoang, T. Ueyama and L. D. Nghiem, Integrity of reverse osmosis membrane for removing bacteria: New insight into bacterial passage, Environ. Sci.: Water Res. Technol., 2019, 5(2), 239–245 RSC.
- S. K. Park and J. Y. Hu, Assessment of the extent of bacterial growth in reverse osmosis system for improving drinking water quality, J. Environ. Sci. Health, Part A: Toxic/Hazard. Subst. Environ. Eng., 2010, 45(8), 968–977 CrossRef CAS PubMed.
- P. Rubbens, R. Props, C. Garcia-Timermans, N. Boon and W. Waegeman, Stripping flow cytometry: How many detectors do we need for bacterial identification?, Cytometry, Part A, 2017, 91(12), 1184–1191 CrossRef PubMed.
- A. Bruno, A. Sandionigi, E. Rizzi, M. Bernasconi, S. Vicario and A. Galimberti, et al. Exploring the under-investigated “microbial dark matter” of drinking water treatment plants, Sci. Rep., 2017, 7, 44350 CrossRef CAS PubMed.
- R. Liikanen, I. Miettinen and R. Laukkanen, Selection of NF membrane to improve quality of chemically treated surface water, Water Res., 2003, 37(4), 864–872 CrossRef CAS PubMed.
- S. Okabe, T. Kokazi and Y. Watanabe, Biofilm formation potentials in drinking waters treated by different advanced treatment processes, Water Sci. Technol.: Water Supply, 2002, 2(4), 97–104 CAS.
- J. Yi, J. Lee, H. Jung, P. K. Park and S. H. Noh, Reduction of bacterial regrowth in treated water by minimizing water stagnation in the filtrate line of a gravity-driven membrane system, Environ. Eng. Res., 2019, 24(1), 17–23 CrossRef.
- N. Vyas, K. Manmi, Q. Wang, A. J. Jadhav, M. Barigou and R. L. Sammons, et al. Which Parameters Affect Biofilm Removal with Acoustic Cavitation? A Review, Ultrasound Med. Biol., 2019, 45(5), 1044–1055 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ew00921h |
| This journal is © The Royal Society of Chemistry 2023 |