Open Access Article
Open Access ArticleEffectiveness of passive sampling for the detection and genetic characterization of human viruses in wastewater†
Cristina
Mejías-Molina
 ab,
Anna
Pico-Tomàs
cd,
Andrea
Beltran-Rubinat
ab,
Anna
Pico-Tomàs
cd,
Andrea
Beltran-Rubinat
 a,
Sandra
Martínez-Puchol
a,
Lluís
Corominas
c,
Marta
Rusiñol
a,
Sandra
Martínez-Puchol
a,
Lluís
Corominas
c,
Marta
Rusiñol
 *ab and
Sílvia
Bofill-Mas
ab
*ab and
Sílvia
Bofill-Mas
ab
aLaboratory of Viruses Contaminants of Water and Food, Departament de Genètica, Microbiologia i Estadística, Universitat de Barcelona, Barcelona, Catalonia, Spain. E-mail: mrusinol@ub.edu
bThe Water Research Institute (IdRA), Universitat de Barcelona, Barcelona, Catalonia, Spain
cCatalan Institute for Water Research (ICRA), Girona, Catalonia, Spain
dUniversitat de Girona, Girona, Catalonia, Spain
First published on 28th February 2023
Abstract
Wastewater-based epidemiology is a promising tool to anticipate potential viral outbreaks not only at city-scale but also at precincts-scale or even buildings. Passive samplers (PS) and, particularly, torpedo-shaped devices containing electronegative membranes, are cost-effective alternatives to traditional sampling methods to trace the circulation of pathogens amongst a community through wastewater. Here we evaluated the effectiveness of torpedo devices to detect eight different viral targets in wastewater from an urban WWTP and from a nursing home (NH), in comparison to 24 h composite active sampling (AS). The use of one or two membranes within the torpedo devices, their potential use for semi-quantification analysis and the performance of torpedoes to conduct studies of the wastewater virome was examined. Two membranes analysis provided more certain results than one single membrane. Considering PS as a semi-quantitative approach, it produced equivalent sensitivity to AS at the NH, but lower sensitivities at the WWTP. Less viral sequences were detected with torpedoes as compared to AS when analyzing the WWTP samples while no differences at the level of viral families detected were obtained at NH when applying both approaches. Our results suggest that PS is a powerful tool for viral detection and characterization at a building scale.
Water impactPassive sampling approaches are becoming promising tools in wastewater-based epidemiology. Torpedo devices fitted with electronegative membranes are useful, affordable and practical tools to monitor viral pathogens in small scale scenarios (e.g. nursing homes). They can be used to study the presence of a diversity of viruses as well as to characterize the wastewater virome. |
1. Introduction
Viruses that infect humans are shed through multiple body secretions. Wastewater surveillance is advantageous because it can be used to estimate disease burden without having to collect individual clinical specimens. This approach is simple, cost-efficient, non-invasive and allows us to obtain rapid results. Wastewater-based epidemiology (WBE) for virus surveillance has been used since the 1980s to determine the spread of poliovirus, the etiological agent for poliomyelitis.1–3 It has recently reemerged as an important tool for public health monitoring to trace the communal circulation of SARS-CoV-2 and to anticipate potential COVID-19 outbreaks caused by pre-symptomatic or asymptomatic infected individuals.4Most WBE studies focus on wastewater collected from wastewater treatment plants (WWTPs) and provide useful city-scale or suburban-scale information.5 Most studies conducted at WWTPs rely on automatic samplers that are deployed at the inlet of the WWTP. Autosamplers collect a composite sample proportional to the daily flow and/or proportional to a given time interval6 and give a representative view of the spread of a virus in the population served. Studies focusing on small sewerage systems such as nursing homes (NH), schools, hospitals or university campuses are important for early warning to target early actions in vulnerable communities. The use of autosamplers at smaller settings are limited by their cost, the need of a power supply and, in some cases, the difficulty to deploy and operate such complex devices in small drainage systems.7 Passive samplers have thus been proposed as affordable and easy-to-use alternative for virus surveillance in these smaller scale scenarios.8 Their deployment and further collection are easy and fast, they operate without electricity, so they can be used in any point of the sewer system.
The first passive sampler described was a folded gauze with a string, known as the “Moore swab” that was immersed for 48 h in a sewer for the detection of Salmonella typhi.9 Moore swabs have been used to detect several bacterial and viral pathogens such as coxsackieviruses, Vibrio cholera, E. coli O157:H7, norovirus, and poliovirus.10–18 During the COVID-19 pandemic, Moore swabs and other passive samplers were used for wastewater surveillance in both large and small catchments.19 Schang and coworkers developed a 3D-printed sampler unit that resembles a torpedo.5 Among gauzes, electronegative membranes, and cottons swabs, electronegative membranes were reported to be the best collecting material to be placed within the torpedoes.4,5 According to the available literature at the time of writing, a deployment time of 24 hours was recommended to allow viruses to concentrate into the membranes.20 Since the beginning of the COVID-19 pandemics, passive samplers have been proposed as affordable and easy-to-use alternative for virus surveillance in small scale scenarios.8
Next generation sequencing (NGS) techniques are excellent tools to monitor and identify viral pathogens circulating among the population. Sewage contains a high number of viruses and viral metagenomics (i.e., viromics) is a very promising tool that could be paired to WBE for a more complete description of viruses circulating within a population as well as for the study of viral (re)emergences and discovery.21,22
The hypothesis of this study was that passive sampling (PS) could complement and even replace data obtained from the analysis of composite samples actively collected (AS) for the presence of human viruses in wastewater. To do that, a comparison between 24 h wastewater samples from a WWTP and a NH were collected using AS and PS. The variability and semi quantification of replicate samples was evaluated. The second aim of this study was to evaluate the potential of the torpedo devices to study the sewage virome.
2. Materials and methods
2.1. Study site, passive and active wastewater sampling
An urban WWTP serving 1.5 million inhabitants and the sewer from a NH, hosting 300 residents and 200 workers were selected to validate the use of passive samplers for the recovery of a diversity of viruses (see next section). From January 10th till March 22nd, 2022, a total of 30 samples from the inlet of the WWTP were collected using the two sampling methodologies. Fifteen AS using an ISCO 6712FR autosampler (Teledyne, Lincoln, NE, USA), that collects 24 h influent time-proportional samples (100 ml per hour over a 24 h period), and 15 PS devices, also known as torpedo sampling units (kindly donated by David McCarthy), containing 3 electronegative membranes. Torpedoes were deployed and retrieved 24 h later. Over the same time period, 30 samples from the residence building were also collected using both methods (a HACH-Bühler 2000 autosampler (HACH-LANGE GmbH, Germany) was used for AS (at the rate of 100 ml per hour over a 24 h period)). Passive and active samples (in a final volume of 250 ml of the total 2.4 litres of wastewater collected by the autosamplers) were handled using protective equipment (lab coats, glasses, gloves) in sterile containers (sealable plastic bags or 250 ml bottles respectively) and transported into a cooler to the laboratory for processing in less than 2 h.2.2. Concentration of viruses, extraction of nucleic acids and analysis using quantitative PCR (qPCR)
Concentration of viral particles from composite wastewater samples (hereafter active sampling, AS), was performed by first removing debris by centrifuging 100 mL of the total 250 ml of sample at 4750 × g for 30 min. The resulting supernatant (80 mL) was then ultrafiltered using the automatic Concentration Pipette (CP-Select™) with 150 KDa tips (Innovaprep) into a final volume of 300 μL as previously described.23 All water samples were spiked with the bacteriophage MS2 as process control, at a final concentration of 1 × 105 GC ml−1, before any processing was carried out. Viral nucleic acids (NA) were extracted with the QIAamp Viral RNA mini kit using the QIAcube automatic system (Qiagen). The sample concentrates (140 μL) were used for the extraction in an elution volume of 70 μL. A negative control of the viral nucleic acid extraction, using PBS was added per batch of samples. PS units were dismantled on the day of retrieval. Two electronegative membranes fitted into the PS were collected and placed in a Petri dish with 0.5 ml of RNA preservative (RNA shield, Zymo research) and maintained at 4 °C until extraction on the next day. The third membrane was stored at −80 °C as a counter sample. Viral RNA extract was carried out using the RNeasy Power Microbiome Kit (Qiagen) into a final volume of 50 μL following manufacturer instructions and adding a step of bead-beating for 30 s at 4 m s−1 using FastPrep-24™ (MP Bio, USA).Specific qPCR and RT-qPCR assays were used to quantify MS2 bacteriophage,24 human adenovirus (HAdV)25,26 JC polyomavirus (JCPyV),27 human enterovirus (EV),28 rotavirus (RoV),29 norovirus genogroup I (NoV GI),30–32 and norovirus genogroup II (NoV GII)33,34 as previously described. For SARS-CoV-2, the N1 and N2 assays35 targeting the gene encoding for the viral nucleocapsid protein were selected. The TaqMan® Environmental Master Mix 2.0 (Thermofisher Scientific) and the RNA Ultrasense™ One-Step RT-qPCR System (Invitrogen) were used for DNA and RNA viruses, respectively. In exception of the synthetic SARS-CoV-2 control (control 51 from Twist Biosciences), all the qPCR standards were prepared (as described by Rusiñol et al., 2020 (ref. 36)) using synthetic gBlocks Gene fragments (IDT), quantified with a Qubit fluorometer (Thermofisher Scientific), and diluted serially from 100 to 107 copies per reaction. Quantification was performed in a QuantStudio™ Real-Time PCR System from ThermoFisher Scientific. Undiluted and 10-fold dilutions of the NA extracts were analysed, and MS2 was also assessed in wastewater samples collected by AS, to test for inhibition. Non-template controls were included in each qPCR plate. All qPCR preparations were performed in a clean laboratory and template addition was done inside a PCR cabinet. Standard qPCR curves were accepted under the following parameters: mean slope, between −3.1 and −3.5; r2 = 0.999; and mean efficiency between 85 and 110%. The limit of detection (LoD) of the qPCR method was calculated by running six replicate tenfold dilutions of target SARS-CoV-2, JCPyV, HAdV suspensions around the detection end point (2.5, 5, 25 and 50 GC per well). The concentration that produced at least 95% positive replicates was assumed to be the LoD of the assay. The limit of quantification (LoQ) was estimated using the procedure described previously.37
2.3. Target enrichment sequencing (TES)
3. Results and discussion
3.1. Determination of the number of membranes needed for a representative passive sampling
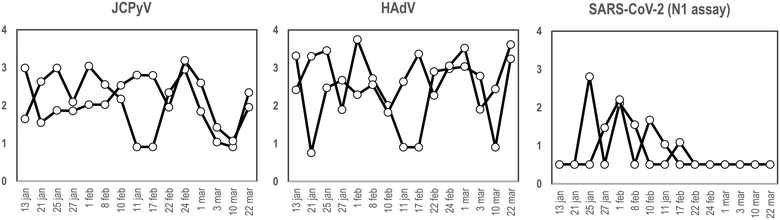
To our knowledge and since the first use of the torpedo-shaped passive samplers for SARS-CoV-2 surveillance in wastewater, only Habtewold and coworkers used more than one electronegative membrane from each passive sampler to test for the presence of viral pathogens and indicators.4 To our knowledge our study is the first one testing the rationality of using two membranes from the same sampling device to give reliable results.5,7,47,48 In the current work, two electronegative membranes were tested per torpedo device to evaluate the presence of 3 different viral targets using qPCR: two viral indicators of human faecal contamination (HAdV and JCPyV) and SARS-CoV-2 (N1). A total of 15 PS, each containing 2 membranes, were collected in parallel to two 24 h-AS replicates. Fig. 1 shows the concentration (in genome copies per litre) of the target viral indicators and SARS-CoV-2 (N1 assay) obtained from AS replicates while Fig. 2 shows the values obtained when analysing the torpedo membranes.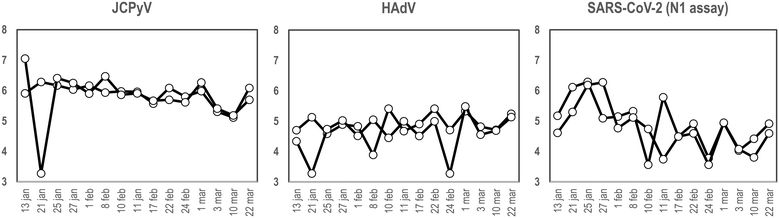 | ||
| Fig. 1 Quantification (in log10 GC L−1), of JCPyV, HAdV and SARS-CoV-2 (N1 assay), from two different wastewater replicates collected at different dates (X-axis) using AS in a WWTP. | ||
 | ||
| Fig. 2 Quantification (in log10 GC/reaction), of JCPyV, HAdV and SARS-CoV-2 (N1 assay), from two different membranes from a PS at different dates (X-axis) at the inlet of a WWTP. | ||
As shown in Fig. 1, whereas no noticeable differences were observed comparing viral GC values obtained from different AS replicates, (Pearson's correlation coefficient test 0.76, 0.61 and 0.96 for JCPyV, HAdV and SARS-CoV-2 (N1 assay) respectively) a lower correlation was obtained between the PS membranes (Pearson's correlation coefficient test below 0.33). Note that for PS membranes results are expressed in copies per reaction since the exact volume of wastewater passing through the torpedo is unknown and the calculation of viral concentrations is, thus, unattainable. Exceptional differences were detected on January 21st in JCPyV and HAdV quantifications, which may arise from errors in the analytical procedure (e.g. ultrafiltration tips, pipetting errors).
Membrane replicates showed greater variability (Fig. 2) probably due to irregular distribution of the wastewater flowing inside the torpedo. In that case a positive detection was considered when qPCR result was above the limit of detection (LoD) (10, 8 and 3 genome copies (GC) for JCPyV, HAdV and SARS-CoV-2 respectively). Considering positive and negative results, membrane replicates matched 12 out of 15 days for JCPyV and HAdV and 11 out of 15 days for SARS-CoV-2. For the further analysis in this study, it was decided to include in the analysis at least 2 membranes to obtain a representative result from each sample. A result was considered positive when at least one of the membranes resulted positive in virus detection.
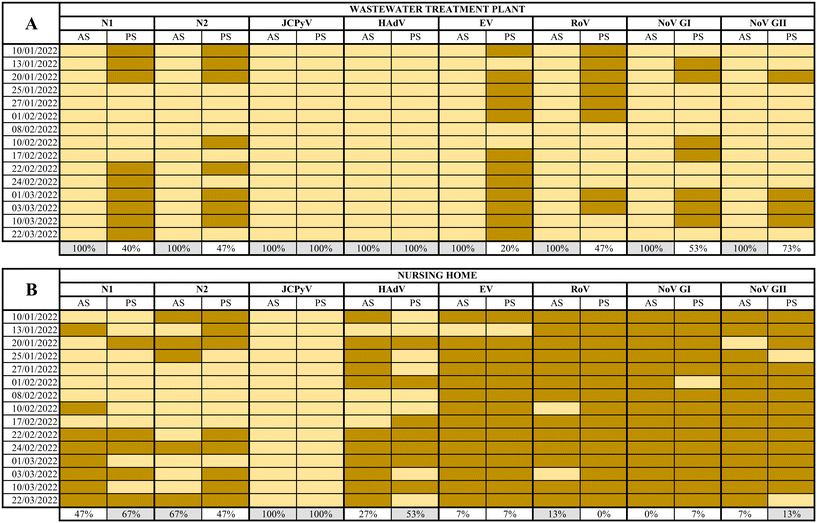
3.2. Detection of human viruses at a WWTP and a NH by applying two different sampling/detection strategies
SARS-CoV-2 (N1 and N2 assays), JCPyV, HAdV, EV, RoV, NoV GI and GII, were analysed in a total of 60 samples collected using AS and PS at both WWTP and the NH from January 10th to March 22nd, 2022.At the WWTP, all composite samples collected tested positive for all viruses analysed while the percentages of detection using PS ranged from 20% to 100% as summarized in Fig. 3. The full dataset of quantification/semi quantification values is shown in the ESI† Tables S1–S3. Inhibition was observed by quantifying JCPyV, as internal process control in 5 out of 120 total samples (Table S4†). The calculated MS2 recovery in the WW samples resulted in 40% of recovery of the seeded material (data not shown).
While human faecal viral indicators, at the WWTP, showed 100% positivity for both AS and PS, the other viral pathogens analysed resulted in 100% positivity by AS between 20% and 73% when using PS. Regarding SARS-CoV-2, between 40–47% positives were reported in PS samples, in contrast to 100% positive samples from AS. This comparison is based on the different performances between PS and AS over a 24 h period while some studies have reported higher PS sensitivities compared PS samples with grab samples.5,7,19,47 High levels of suspended solids in wastewater may affect the adsorption of viruses to the electronegative membranes. Hayes and coworkers49 demonstrated that adsorption of the virus to the sampling material is highly dependent of the characteristics of the water. Retention of solids can improve RNA recovery but and excessive retention can inhibit de RNA extraction process. It should be noticed that nucleic acids analysed in the current study were extracted using kits designed to provide highly pure RNA isolation by using inhibitor removal. PS performance might also have been altered by temperature, pH, or dissolved solids concentration.50 It is also expected that different viruses show different presence patterns despite being adsorbed and concentrated with the same method, since they present different physical properties. Li and coworkers51 suggested that the different structure between non-enveloped viruses (EV, HAdV, RoV, NoV) and enveloped ones (SARS-CoV-2) could vary their behavior in the affinity for membranes or in the recovery process. All in all, our experimental design did not allow us to differentiate whether the virus adsorption or sample processing were the critical steps in the process. According to recent studies, a low viral recovery could also have been caused by a short deployment time.50,52 Li and coworkers recently demonstrated that the maximum viral accumulation using electronegative membranes was obtained after 48 h of deployment.50 Wilson and coworkers7 also concluded that sensitivities of PS could overcome grab sampling when more than 24 h of wastewater flow were considered.7 Since the concentration of viruses from AS and PS was performed using different methods for viral recovery, the direct comparison of results is risky if not impossible.
On the other hand, at the NH, the percentage of virus detection using AS ranged from 0% for RoV to 100% for JCPyV and from 0 to 100% for the same viruses when using PS (Fig. 3, the complete data set is available in ESI† Table S3). In general, nearly equivalent results were observed when applying both methods: the more frequently detected virus was JCPyV followed by SARS-CoV-2 (N1 and N2) and HAdV while EV, NoVs and RoV were rarely detected. Negative detection for SARS-CoV-2 by PS corresponded to samples collected during a period of low prevalence of infections in the assisted community53 suggesting lower sensitivity (in terms of the capacity to produce a positive detection of viral nucleic acid) than traditional detection using AS.
It is remarkable to notice that JCPyV and HAdV showed 100% positivity in both AS and PS in the WWTP and JCPyV also showed 100% positivity in the NH samples. These viruses are used as process controls and fecal indicators because of their abundance in wastewater and their correlation with fecal pollution.54 It is clear that all NH samples analyzed presented fecal contamination and that JCPyV is more appropriate as fecal indicator at small-scale sewage systems as previously proposed.55 This is a relevant output of our work since it may be used as a tool to rule out false negative samples. Unlike the WWTP, in this specific site, the presence and spread of a symptomatic viral infection within a small community is more infrequent. Consequently, the likelihood of detection of viral pathogens in wastewater is very low despite the sampling protocol. This is sustained by the high number of negative samples obtained by both methods for RoV, EV, NoV GI, and NoV GII. The obtained results confirm that PS is a very good wastewater sampling method for viral studies in small-scale scenarios where AS is not feasible.
3.3. Evaluation of the PS as semiquantitative approach
The PS method here evaluated is based on the free flow of viral particles through the membranes and since we could not assume that all the flow contacts the PS membrane, results cannot be expressed per unit volume (e.g., CG mL−1). Although most studies using this methodology report qualitative results, some authors have shown a significant correlation between the measured SARS-CoV-2 RNA concentrations in samples collected using AS and PS methods.5,20 These results suggest that PS can express semi-quantitative results for virus detection in wastewater, meaning that a higher quantity of virus in wastewater correlates with a higher accumulation of viruses in membranes and, therefore, a higher GC quantification. To perform a semiquantitative analysis, and by using all the GC values obtained in this study for the different viral species determined, we considered half of the LoD when a result was below the LoD was considered. The semiquantitative detection of JCPyV in both sampling sites presented the highest Pearson's correlation observed, being R2 = 0.42 and R2 = 0.38 at the WWTP and the NH respectively. The results indicate low correlation for SARS-CoV-2 (N1 assay) in both sites (0.13 at the WWTP and 0.18 at the NH). In other words, fluctuations in the genome copy detection at the qPCR would indicate increases/decreases in the genome copies per ml of wastewater.For SARS-CoV-2, correlations improve when considering results above the LoD, being 0.78 at the WWTP and 0.8 at the NH. Other authors reported correlations between 0.27 and 0.76, in PS collected from WWTP inlets or manholes.5,20 All in all, when viral concentrations are above the LOD, PS results correlates positively with qualitative results obtained after AS.
3.4. Virome characterization from samples obtained using PS
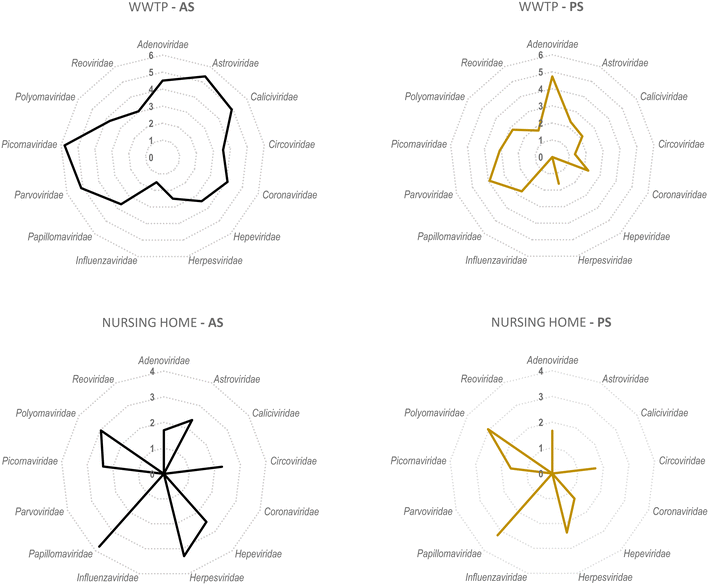
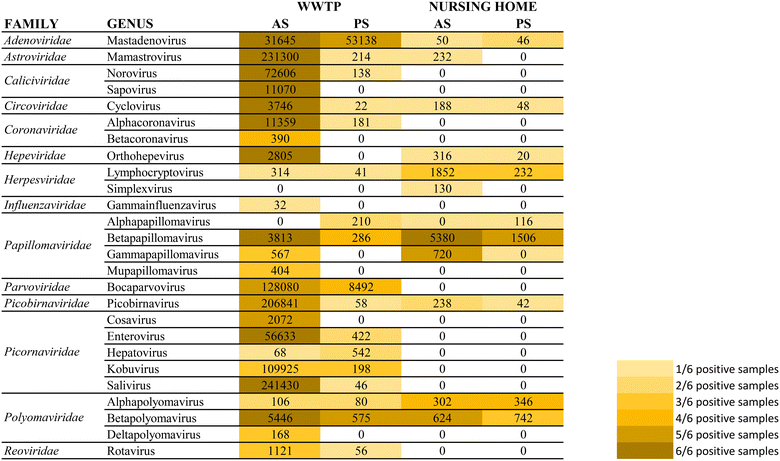
A secondary aim of this study was to evaluate the performance of torpedoes to study the sewage virome. With this aim, and to increase the sensitivity of the mass sequencing on-wards viruses that infect vertebrates, a target enrichment sequencing was conducted using Vir-CapSeq-VERT panel.At the WWTP, viral families observed were those expected and reported in other studies where the same viral enrichment method was applied.21 Results showed a higher diversity of viral families and genus present at wastewater samples when collected by AS (Fig. 4 and 5). The number of reads and the number of samples presenting viruses of specific viral groups were higher in samples collected using AS rather than PS. However equivalent viral groups were detected.
The most importantly excreted human pathogens are members of nine families: Adenoviridae, Astroviridae, Caliciviridae, Hepeviridae, Parvoviridae, Papillomaviridae, Picornaviridae, Polyomaviridae, and Reoviridae.56 All these families have been also described as members of the sewage virome21,57 and more recently also Coronaviridae and Influenzaviridae have been included in this list.58 Aligned with the bibliography,21 the Picornaviridae family, including important pathogens like enterovirus, poliovirus or hepatitis A virus, were the most abundant in raw wastewater. Reoviridae and Hepeviridae were found to be the least abundant when using AS. Although lower number of reads were obtained using PS at the WWTP (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log fold), most viral families representing the sewage virome were also found using this passive approach. Viruses belonging to the Reoviridae family were not represented when using this sampling methodology probably because of a lower sensitivity of the methodology.
log fold), most viral families representing the sewage virome were also found using this passive approach. Viruses belonging to the Reoviridae family were not represented when using this sampling methodology probably because of a lower sensitivity of the methodology.
Pathogenic viruses belonging to Adenoviridae, Astroviridae, Hepeviridae, Picornaviridae and Polyomaviridae were present at the NH. Both methodologies provided similar abundances except for Astroviridae for which the AS provided higher number of viral sequences. Interestingly, a high diversity of Herpesviridae and Papillomaviridae reads were detected in the building WW. Among those viruses herpesviruses-2 and -3 as well as human papillomaviruses β and γ, highly prevalent in older people compared to young adults,59 were found at the NH facility. The absence of Coronaviridae or Picornaviridae at NH over the sampling period could be explained because the virus was not circulating over the period of sampling in the tested community.
Passive sampling seems to be less sensitive in terms of detection and number of viral families detected by massive sequencing. Despite providing a lower number of sequences, equal diversity of viral families was obtained using both sampling methodologies. Shannon index (H′) when using AS indicates higher diversity of viruses, at the WWTP (H′ = 4.8), than at the NH (H′ = 4.1). Nevertheless, when applying NGS at building level, the diversity of viruses obtained using PS was higher than when using AS.
To our knowledge, this is the first study applying passive sampling to the successful genetic characterization of a wastewater virome, which could be of relevance in terms of viral emergence surveillance and key information for the corresponding targeted actions. The purpose of this study was to provide information that support the use of torpedo passive sampler as an useful affordable and practical tool to provide information on viral presence in the frame of wastewater-based epidemiology at building scale level. To obtain more detailed information about viral species some improvements in the protocol of the sequencing methodology should be conducted and will be the aim of further studies.
4. Conclusions
Passive sampling presents a low-cost, safe and power-free solution for smaller sewer catchments. Torpedo shaped devices fitted with electronegative membranes can be applied as an efficient method that is sensitive to a few infection cases residing in the same building. In the event of new epidemic outbreaks, the use of passive samplers can be used for targeted actions or to track back the excreted virome of specific facilities where reside or travel vulnerable population (e.g. schools, universities, or aged care facilities).Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research has been done as part of the VIRWASTE project financed by the Catalan Agency for Management of University and Research Grants (AGAUR) under the “Pandèmies 2020” call and through donations obtained during “La Marató de TV3” dedicated to COVID-19 within the project EPISARS (544/C/2021). ICRA authors acknowledge the funding provided by the Generalitat de Catalunya through the Consolidated Research Group grants ICRA-ENV 2017 SGR 1124 and ICRA-TiA 2017 SGR 1318. The authors are thankful to the sampled building administrators and their maintenance team as well as the staff from the WWTPs for their help and technical support during the sampling campaigns. Also, to David McCarthy (Monash University) and collaborators for supplying the passive samplers used in this study and to Carles Borrego for critical review of this manuscript. Sílvia Bofill-Mas is a Serra-Hunter fellow at the University of Barcelona.References
- T. Nakamura, M. Hamasaki, H. Yoshitomi, T. Ishibashi, C. Yoshiyama and E. Maeda, et al., Environmental surveillance of poliovirus in sewage water around the introduction period for inactivated polio vaccine in Japan, Appl. Environ. Microbiol., 2015, 81(5), 1859–1864 CrossRef PubMed.
- L. Roberts, Israel's silent polio epidemic breaks all the rules, Science, 2013, 342(6159), 679–680 CrossRef CAS PubMed.
- T. Hovi, L. M. Shulman, H. Van Der Avoort, J. Deshpande, M. Roivainen and E. M. De Gourville, Role of environmental poliovirus surveillance in global polio eradication and beyond, Epidemiol. Infect., 2012, 140(1), 1–13 CrossRef CAS PubMed.
- J. Habtewold, D. McCarthy, E. McBean, I. Law, L. Goodridge and M. Habash, et al., Passive sampling, a practical method for wastewater-based surveillance of SARS-CoV-2, Environ. Res., 2022, 204, 112058 CrossRef CAS PubMed.
- C. Schang, N. D. Crosbie, M. Nolan, R. Poon, M. Wang and A. Jex, et al., Passive Sampling of SARS-CoV-2 for Wastewater Surveillance, Environ. Sci. Technol., 2021, 55(15), 10432–10441 CrossRef CAS PubMed.
- G. Medema, L. Heijnen, G. Elsinga, R. Italiaander and A. Brouwer, Presence of SARS-Coronavirus-2 RNA in Sewage and Correlation with Reported COVID-19 Prevalence in the Early Stage of the Epidemic in the Netherlands, Environ. Sci. Technol. Lett., 2020, 7(7), 511–516 CrossRef CAS.
- M. Wilson, Y. Qiu, J. Yu, B. E. Lee, D. T. McCarthy and X. Pang, Comparison of Auto Sampling and Passive Sampling Methods for SARS-CoV-2 Detection in Wastewater, Pathogens, 2022, 11(3), 359 CrossRef CAS PubMed.
- P. A. Acer, Quantifying the relationship between SARS-CoV-2 wastewater concentrations and building-level COVID-19 prevalence at an isolation residence using a passive sampling approach, 2022, pp. 1–34.
- B. Moore, The Detection of Paratyphoid Carriers in Towns by means of Sewage Examination, Mon. Bull. Minist. Health Public Health Lab. Serv., 1948, 7, 241–248 Search PubMed.
- S. M. Kelly, M. E. Cark and M. B. Coleman, Demonstration of infectious agents in sewage, Am. J. Public Health, 1955, 45(11), 1438–1446 CrossRef CAS PubMed.
- M. Isaäcson, K. Clarke, G. Ellacombe, W. Smit, H. Koornhof and L. Smith, et al., The recent cholera outbreak in the South African gold mining industry. A preliminary report – PubMed, Afr. Med. J., 1974, 48, 2557–2560 Search PubMed , Available from: https://pubmed.ncbi.nlm.nih.gov/4453936/.
- T. J. Barrett, P. A. Blake, G. K. Morris, N. D. Puhr, H. B. Bradford and J. G. Wells, Use of Moore swabs for isolating Vibrio cholerae from sewage, J. Clin. Microbiol., 1980, 11(4), 385–388 CrossRef CAS PubMed.
- S. A. Sattar and J. C. N. Westwood, Isolation of apparently wild strains of poliovirus type 1 from sewage in the Ottawa area, Can. Med. Assoc. J., 1977, 116(1), 25–27 CAS.
- G. Pazzaglia, M. Lesmana, P. Tjaniadi, D. Subekti and B. Kay, Use of vaginal tampons in sewer surveys for non-O1 Vibrio cholerae, Appl. Environ. Microbiol., 1993, 59(8), 2740–2742 CrossRef CAS PubMed.
- A. Sbodio, S. Maeda, G. Lopez-Velasco and T. V. Suslow, Modified Moore swab optimization and validation in capturing E. Coli O157: H7 and Salmonella enterica in large volume field samples of irrigation water, Food Res. Int., 2013, 51(2), 654–662 CrossRef CAS.
- K. M. S. De Melo Cassemiro, F. M. Burlandy, M. R. F. Barbosa, Q. Chen, J. Jorba and E. M. Hachich, et al., Molecular and phenotypic characterization of a highly evolved type 2 vaccine-derived poliovirus isolated from seawater in Brazil, 2014, PLoS One, 2016, 11(3), 1–18 Search PubMed.
- P. Tian, D. Yang, L. Shan, Q. Li, D. Liu and D. Wang, Estimation of Human Norovirus Infectivity from Environmental Water Samples by In Situ Capture RT-qPCR Method, Food Environ. Virol., 2018, 10(1), 29–38 CrossRef CAS PubMed.
- G. Matrajt, B. Naughton, A. S. Bandyopadhyay and J. S. Meschke, A Review of the Most Commonly Used Methods for Sample Collection in Environmental Surveillance of Poliovirus, Clin. Infect. Dis., 2018, 67(Suppl 1), S90–S97 CrossRef CAS PubMed.
- P. Liu, M. Ibaraki, J. VanTassell, K. Geith, M. Cavallo and R. Kann, et al., A sensitive, simple, and low-cost method for COVID-19 wastewater surveillance at an institutional level, Sci. Total Environ., 2022, 807, 151047, DOI:10.1016/j.scitotenv.2021.151047.
- A. Bivins, D. Kaya, W. Ahmed, J. Brown, C. Butler and J. Greaves, et al., Passive sampling to scale wastewater surveillance of infectious disease: Lessons learned from COVID-19, Sci. Total Environ., 2022, 835, 155347, DOI:10.1016/j.scitotenv.2022.155347.
- S. Martínez-Puchol, M. Rusiñol, X. Fernández-Cassi, N. Timoneda, M. Itarte, C. Andrés, A. Antón, J. F. Abril, R. Girones and S. Bofill-Mas, Characterisation of the sewage virome: comparison of NGS tools and occurrence of significant pathogens, Sci. Total Environ., 2020, 713, 136604 CrossRef PubMed.
- X. Fernandez-Cassi, N. Timoneda, S. Martínez-Puchol, M. Rusiñol, J. Rodriguez-Manzano and N. Figuerola, et al., Metagenomics for the study of viruses in urban sewage as a tool for public health surveillance, Sci. Total Environ., 2018, 618, 870–880 CrossRef CAS PubMed.
- E. Forés, S. Bofill-Mas, M. Itarte, S. Martínez-Puchol, A. Hundesa, M. Calvo, C. M. Borrego, L. L. Corominas, R. Girones and M. Rusiñol, Evaluation of two rapid ultrafiltration-based methods for SARS-CoV-2 concentration from wastewater, Sci. Total Environ., 2021, 768, 144786 CrossRef PubMed.
- B. M. Pecson, Quantitative PCR for determining the infectivity of bacteriophage MS2 upon inactivation by heat, UV-B radiation, and singlet oxygen: Advantages and limitations of an enzymatic treatment to reduce false-positive results, Appl. Environ. Microbiol., 2009, 75(17), 5544–5554 CrossRef CAS PubMed.
- B. E. Hernroth, A. C. Conden-Hansson, A. S. Rehnstam-Holm, R. Girones and A. K. Allard, Environmental factors influencing human viral pathogens and their potential indicator organisms in the blue mussel, Mytilus edulis: The first Scandinavian report, Appl. Environ. Microbiol., 2002, 68(9), 4523–4533 CrossRef CAS PubMed.
- S. Bofill-Mas, N. Albinana-Gimenez, P. Clemente-Casares, A. Hundesa, J. Rodriguez-Manzano and A. Allard, et al., Quantification and stability of human adenoviruses and polyomavirus JCPyV in wastewater matrices, Appl. Environ. Microbiol., 2006, 72(12), 7894–7896 CrossRef CAS PubMed.
- A. Pal, L. Sirota, T. Maudru, K. Peden and A. M. Lewis, Real-time, quantitative PCR assays for the detection of virus-specific DNA in samples with mixed populations of polyomaviruses, J. Virol. Methods, 2006, 135(1), 32–42 CrossRef CAS PubMed.
- M. Rusiñol, A. Carratalà, A. Hundesa, A. Bach, A. Kern and A. Vantarakis, et al., Decription of a novel viral tool to identify and quantify ovine faecal pollution in the environment, Sci. Total Environ., 2013, 458–460, 355–360 CrossRef PubMed.
- S. Q. Zeng, A. Halkosalo, M. Salminen, E. D. Szakal, L. Puustinen and T. Vesikari, One-step quantitative RT-PCR for the detection of rotavirus in acute gastroenteritis, J. Virol. Methods, 2008, 153(2), 238–240 CrossRef CAS PubMed.
- S. Svraka, E. Duizer, H. Vennema, E. De Bruin, B. Van Der Veer and B. Dorresteijn, et al., Etiological role of viruses in outbreaks of acute gastroenteritis in The Netherlands from 1994 through 2005, J. Clin. Microbiol., 2007, 45(5), 1389–1394 CrossRef PubMed.
- M. Hoehne and E. Schreier, Detection of norovirus genogroup I and II by multiplex real-time RT-PCR using a 3′-minor groove binder-DNA probe, BMC Infect. Dis., 2006, 6, 1–6 CrossRef PubMed.
- A. K. Da Silva, J. C. Le Saux, S. Parnaudeau, M. Pommepuy, M. Elimelech and F. S. Le Guyader, Evaluation of removal of noroviruses during wastewater treatment, using real-time reverse transcription-PCR: Different behaviors of genogroups I and II, Appl. Environ. Microbiol., 2007, 73(24), 7891–7897 CrossRef CAS PubMed.
- T. Kageyama, S. Kojima, M. Shinohara, K. Uchida, S. Fukushi and F. B. Hoshino, et al., Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR, J. Clin. Microbiol., 2003, 41(4), 1548–1557 CrossRef CAS PubMed.
- F. Loisy, R. L. Atmar, P. Guillon, P. Le Cann, M. Pommepuy and F. S. Le Guyader, Real-time RT-PCR for norovirus screening in shellfish, J. Virol. Methods, 2005, 123(1), 1–7 CrossRef CAS PubMed.
- CDC. 2019-nCoV CDC EUA qPCR Assay, 2020, 7, pp. 1–38.
- M. Rusiñol, A. Hundesa, Y. Cárdenas-Youngs, A. Fernández-Bravo, A. Pérez-Cataluña and L. Moreno-Mesonero, et al., Microbiological contamination of conventional and reclaimed irrigation water: Evaluation and management measures, Sci. Total Environ., 2020, 710, 136298 CrossRef PubMed.
- A. Forootan, R. Sjöback, J. Björkman, B. Sjögreen, L. Linz and M. Kubista, Methods to determine limit of detection and limit of quantification in quantitative real-time PCR (qPCR), Biomol. Detect. Quantif., 2017, 12, 1 CrossRef CAS PubMed.
- X. Fernandez-Cassi, M. Rusiñol and S. Martínez-Puchol, Viral concentration and amplification from human serum samples prior to application of next-generation sequencing analysis, Methods Mol. Biol., 2018, 1838, 173–188 CrossRef CAS PubMed.
- IDseq Portal. [cited 2022 Sep 20]. Available from: https://czid.org/.
- A. Dobin, C. A. Davis, F. Schlesinger, J. Drenkow, C. Zaleski and S. Jha, et al., STAR: Ultrafast universal RNA-seq aligner, Bioinformatics, 2013, 29(1), 15–21 CrossRef CAS PubMed.
- A. M. Bolger, M. Lohse and B. Usadel, Trimmomatic: A flexible trimmer for Illumina sequence data, Bioinformatics, 2014, 30(15), 2114–2120 CrossRef CAS PubMed.
- J. G. Ruby, P. Bellare and J. L. DeRisi, PRICE: Software for the targeted assembly of components of (Meta) genomic sequence data, G3: Genes, Genomes, Genet., 2013, 3(5), 865–880 CrossRef PubMed.
- W. Li and A. Godzik, Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences, Bioinformatics, 2006, 22(13), 1658–1659 CrossRef CAS PubMed.
- Home – Nucleotide – NCBI. [cited 2022 Sep 20]. Available from: https://www.ncbi.nlm.nih.gov/nucleotide/.
- T. D. Wu and S. Nacu, Fast and SNP-tolerant detection of complex variants and splicing in short reads, Bioinformatics, 2010, 26(7), 873–881 CrossRef CAS PubMed.
- Y. Ye, J. H. Choi and H. Tang, RAPSearch: A fast protein similarity search tool for short reads, BMC Bioinf., 2011, 12, 159 CrossRef CAS PubMed.
- E. K. Hayes, C. L. Sweeney, L. E. Anderson, B. Li, G. B. Erjavec and M. T. Gouthro, et al., A novel passive sampling approach for SARS-CoV-2 in wastewater in a Canadian province with low prevalence of COVID-19, Environ. Sci.: Water Res. Technol., 2021, 7(9), 1576–1586 RSC.
- D. L. Jones, J. M. S. Grimsley, J. L. Kevill, R. Williams, C. Pellett and K. Lambert-Slosarska, et al., Critical Evaluation of Different Passive Sampler Materials and Approaches for the Recovery of SARS-CoV-2, Faecal-Indicator Viruses and Bacteria from Wastewater, Water, 2022, 14(21), 3568 CrossRef CAS.
- E. K. Hayes, C. L. Sweeney, M. Fuller, G. B. Erjavec, A. K. Stoddart and G. A. Gagnon, Operational Constraints of Detecting SARS-CoV-2 on Passive Samplers using Electronegative Filters: A Kinetic and Equilibrium Analysis, ACS EST Water, 2021, 1c00441 Search PubMed.
- J. Li, R. Verhagen, W. Ahmed, S. Metcalfe, P. K. Thai and S. L. Kaserzon, et al., In Situ Calibration of Passive Samplers for Viruses in Wastewater, ACS EST Water, 2021, 1c00406 Search PubMed.
- J. Li, W. Ahmed, S. Metcalfe, W. J. M. Smith, B. Tscharke and P. Lynch, et al., Monitoring of SARS-CoV-2 in sewersheds with low COVID-19 cases using a passive sampling technique, Water Res., 2022, 218, 118481, DOI:10.1016/j.watres.2022.118481.
- J. L. Kevill, K. Lambert-Slosarska, C. Pellett, N. Woodhall, I. Richardson-O’Neill and I. Pântea, et al., Assessment of two types of passive sampler for the efficient recovery of SARS-CoV-2 and other viruses from wastewater, Sci. Total Environ., 2022, 838, 156580 CrossRef CAS PubMed.
- A. Pico-Tomàs, C. Mejías-Molina, I. Zammit, M. Rusiñol, S. Bofill-Mas and C. M. Borrego, et al., Surveillance of SARS-CoV-2 in sewage from buildings housing residents with different vulnerability levels. Submited.
- K. Farkas, D. I. Walker, E. M. Adriaenssens, J. E. Mcdonald, L. S. Hillary and S. K. Malham, et al., Since January 2020 Elsevier has created a COVID-19 resource centre with free information in English and Mandarin on the novel coronavirus COVID-19. The COVID-19 resource centre is hosted on Elsevier Connect , the company's public news and information, 2020, (January).
- R. E. Mayer, S. Bofill-Mas, L. Egle, G. H. Reischer, M. Schade and X. Fernandez-Cassi, et al., Occurrence of human-associated Bacteroidetes genetic source tracking markers in raw and treated wastewater of municipal and domestic origin and comparison to standard and alternative indicators of faecal pollution, Water Res., 2016, 90, 265–276 CrossRef CAS PubMed.
- M. Rusiñol and R. Girones, Summary of Excreted and Waterborne Viruses, in Water and Sanitation for the 21st Century: Health and Microbiological Aspects of Excreta and Wastewater Management (Global Water Pathogen Project), ed. J. B. Rose and B. Jiménez-Cisneros, 2017. (J. S. Meschke, and R. Girones (eds), Part 3: Specific Excreted Pathogens: Environmental and Epidemiology Aspects - Section 1: Viruses) Search PubMed.
- E. M. Adriaenssens, K. Farkas, J. E. McDonald, D. L. Jones, H. E. Allison and A. J. McCarthy, Tracing the fate of wastewater viruses reveals catchment-scale virome diversity and connectivity, Water Res., 2021, 203, 117568 CrossRef CAS PubMed.
- S. Martínez-Puchol, M. Itarte, M. Rusiñol, E. Forés, C. Mejías-Molina and C. Andrés, et al., Exploring the diversity of coronavirus in sewage during COVID-19 pandemic: Don’t miss the forest for the trees, Sci. Total Environ., 2021, 800, 149562 CrossRef PubMed.
- A. J. Kombe Kombe, B. Li, A. Zahid, H. M. Mengist, G. A. Bounda and Y. Zhou, et al., Epidemiology and Burden of Human Papillomavirus and Related Diseases, Molecular Pathogenesis, and Vaccine Evaluation, Front. Public Health, 2021, 8, 1–19 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ew00867j |
| This journal is © The Royal Society of Chemistry 2023 |