Open Access Article
Open Access ArticleEmerging investigator series: the role of phage lifestyle in wastewater microbial community structures and functions: insights into diverse microbial environments
Jeseth
Delgado Vela
 * and
Mitham
Al-Faliti
* and
Mitham
Al-Faliti

Department of Civil and Environmental Engineering, Howard University, 2300 Sixth St NW, 3030A, Washington, DC 20059, USA. E-mail: jeseth.delgadovela@howard.edu; Tel: +1 202 806 6630
First published on 2nd June 2023
Abstract
For over a century, environmental engineers have attempted to control the prokaryotic community biological wastewater treatment processes, but there is growing interest in both understanding and harnessing the activity of phages in wastewater bioprocesses. While phages are known to be present and abundant, their ecological role, potential benefits, and impacts on wastewater biological processes are not fully understood. Fundamental knowledge on how phages infect host cells from relatively simple pure culture studies alongside environmental studies from marine and soil systems can be used to predict the potential impact of phages in diverse and dynamic wastewater environments. This frontier review is focused on what is known about the molecular mechanisms by which phages infect bacteria and how that could apply to biological process control and operation within wastewater treatment systems. Here, we specifically focus on highlights from studies on the molecular mechanisms that drive lysis and lysogeny within phage cells and the impacts on the dissemination of antibiotic resistance genes and nutrient removal within a biological wastewater process.
Water impactThis review summarizes the rapidly emerging research on how phages survive and their potential role in biological wastewater treatment systems, with a specific focus on the impact of phages on antibiotic resistance and nutrient removal systems. Continued research in this area can lead to targeted control strategies for environmental engineering processes. |
Introduction
Microbial communities represent a complex balance among eukaryotic, prokaryotic (bacteria and archaea), and viral populations that maintain a habitat through biogeochemical interactions. Environmental engineers attempt to control these microbial communities to enable pollutant degradation. Within those microbial communities, phages (viruses that infect bacteria) are the most abundant biological entity on earth, with estimates of over 1030 phage particles on earth.1 Understanding the role of phages in the environment may allow engineers to manipulate phage–bacteria interactions for the benefit of engineered processes.In general, knowledge of phages and their role in wastewater treatment within activated sludge processes is nascent, but phage abundance as measured by flow cytometry correlates with a variety of wastewater performance factors such as concentration of effluent, COD, and ammonia.2 There have been several reviews on the impact of phages on biological wastewater systems that include methods and potential impacts of phages on microbial ecology.3,4 Examples of studies on phages that are not explicitly covered in this review are the application of phages to manage sludge bulking,5 control biofilms,6,7 or enhance disinfection;8,9 and the use of phages as surrogates for viral pathogens.10–12 There are relatively fewer studies on phages in activated sludge compared with the wealth of literature available on phages in natural environments. In this frontier review, we will first present the state of knowledge on phage lifestyles (virulent versus temperate) and phage molecular signals. We will then provide analysis on the potential impact of these aspects of phage ecology and function on biological wastewater treatment, specifically focused on phage genetic compositions related to antibiotic resistance and nutrient removal. We conclude with a vision of how understanding phage lifestyles can ultimately be used to benefit environmental engineering to provide ‘bottom-up’ control of microbial communities.13
Phage lifestyles
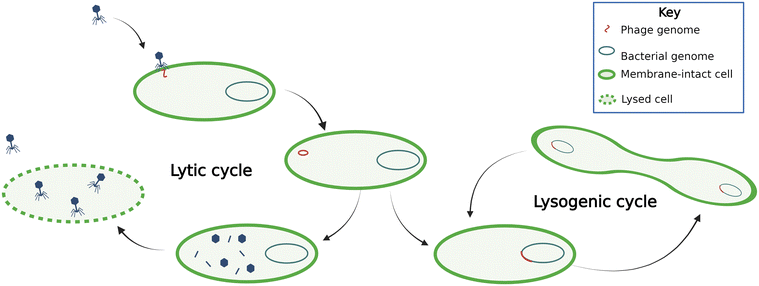
Phages can be either virulent or temperate (Fig. 1). Virulent phages attach to the host cell, inject their genetic material into the cell, hijack the host cell machinery to reproduce, and lyse the cell. This process is termed a lytic cycle. Temperate phages, or prophages, use lysogenic cycles, whereby the phages integrate their nucleic acids into the host organism's genome. During lysogeny, the proliferation of the phage genetic material depends on host reproduction. Phage-mediated movement of genetic information between bacteria is termed transduction. Phages conduct transduction through several mechanisms, which are often thought of as mistakes in packaging host DNA or excising phage genetic materials and are therefore traditionally thought to be rare.14 Overall, the phage dynamics and life cycle impact their host organism's population dynamics and subsequently ecosystem processes.Lytic phages are known to alter microbial communities significantly. In the deep ocean, phage infection of bacteria directly regulates carbon fluxes, the so-called viral shunt occurs because phage lysis supports heterotrophic community growth on cell debris,15 illustrated by proportional heterotrophic bacteria and phage abundance in deep-sea sediments.16 This phenomenon is not surprising considering phages are estimated to be about fifteen times more abundant than prokaryotes in the ocean; some studies have showed estimates up to 160 times, though the relationship between phages and prokaryotes is nonlinear and estimates depend on the location, methods, temperature, and depth.15,17,18 Some researchers have postulated that phage lytic activity supports prokaryotic growth by providing organic substrate in anaerobic digesters19 and there has been evidence of this phenomenon in sediments,20 although typical wastewaters are not carbon-limited compared with the deep ocean. To our knowledge, there haven't been studies on the role of phage lysis in altering carbon fluxes in activated sludge processes.
Lytic phages have been shown to carry genes associated with bacterial metabolism, auxiliary metabolic genes (AMGs). It is thought that phages carry these genes to alter the metabolic state of the host; AMGs provide fitness advantages, such as allowing expression of important metabolic pathway components during infection.21 The most commonly found AMGs in phages are related to DNA synthesis, illustrating that when phages hijack the host machinery, they can shift metabolism towards phage synthesis. Some have proposed that phages carry these genes to mimic nutrient-limited metabolism, allowing host metabolism to promote phage replication.22 One challenge with the evaluation of AMGs in environmental communities is the degree of confidence that the gene identified in an environmental sequencing is, in fact, on a viral fragment, and not a gene within a bacterial cell. Researchers have published guidelines for viromics, including the classification and identification of AMGs in sequencing datasets that should be applied for sequencing studies in wastewater microbial communities.23
Temperate or lysogenic phages can change the way host cells behave24–26 and are a vital part of the microbial community population ecology. Temperate phages can have evolutionary impacts on their host cells, can be sources of genetic variability, and can serve as weapons against competing organisms. Temperate phages can also be a defense mechanism for the cell because bacteria will use temperate phages that are in their genome as a tool preventing the spread of obligate lytic phages within a bacterial population. When obligate lytic phages infect a bacterial cell, bacteria induce a temperate phage (i.e., excising the phage genome from the bacterial genome and allowing phages to infect the cell). As a result, bacteria commit “altruistic suicide” because this induction process prevents the spread of the lytic phage in the population.27 Alternatively, the temperate phages also have diverse and varied mechanisms to exclude other phages from infecting the host cell, termed superinfection exclusion.28 The lifestyles and survival strategies within temperate phages are diverse and the mechanisms by which temperate phages affect wastewater microbial ecology warrant continued study.
The prevalence of both lytic and temperate phages has led to the development of two competing hypotheses in the literature on how phage activity affects microbial ecology. One hypothesis is that phages affect microbial community diversity by “killing the winner.” This mechanism is thought to help maintain microbial community diversity by increasing the availability of resources to less abundant organisms. There have been several demonstrations of this hypothesis in experimental microcosms, including in wastewater derived communities, showing that more abundant hosts are more likely to be lysed by phages (e.g., ref. 29–31). Alternatively, a lytic to lysogenic switch in lifestyle is the basis for a competing hypothesis, “piggyback the winner.” One study that included a meta-analysis of diverse ecosystems has shown that communities with a higher microbial density have lower viral abundance and increased lysogeny.32 In this instance, lysogenic phages confer a competitive advantage—superinfection exclusion—to their more abundant hosts, reducing overall microbial community diversity. These two frameworks are seemingly at odds and are the subject of much debate in the literature. Most likely, both frameworks are at play in environmental communities,33 and in combination may be able to describe the numerous ecological roles that phages have within wastewater microbial communities.
While it is known that both lytic and temperate phages are present in wastewater treatment plants, there aren't any available estimates of the percentage of prokaryotic cells that contain prophages within their genome.34 Further, while characterizing phages as either lytic or temperate is convenient, studies of isolates have shown that these exist in parallel.35,36 A study that isolated 117 phages from the ocean found that infection patterns did not necessarily reduce the cell density, even in isolates that lacked known lysogenic genes.36 These studies support “continuum of infection strategies” such that chronic viral infection is an additional lifestyle, where lysis and lysogeny are both occurring within the host population at once.37 While considering an individual phage as lytic or lysogenic is convenient and can help environmental engineers develop bottom-up control approaches—it is important to recognize that at the population level, ‘phage therapy’ is more challenging given that lysis and lysogeny are likely occurring in parallel.
Several factors dictate whether a phage is temperate or lytic
In general, the switch between a temperate and lytic lifestyle is varied and phage dependent. There are complex regulatory processes that dictate if a phage is temperate or lytic; some of these are regulatory protein systems and others are signaling molecules (see section “Phage–host signaling & phage–phage molecular signaling”). The best studied lysis–lysogeny regulatory system is that of lambda phages. In this case, regulatory proteins (CI, CII, and Cro) dictate the transition from lysogenic to lytic cycles.38,39 Regulation of lysis versus lysogeny includes inputs of the physiological state of the host cell (e.g., growth rate or DNA damage) and cell density or the multiplicity of infection (MOI).38Additional physiological inputs that regulate prophage induction includes stress response; phage induction has been documented as a response to heavy metals, potassium cyanide toxicity, and low pH in a pure culture of ammonia oxidizing bacteria.40 In contrast, in a study of contaminated chromium soils, there was higher abundance of lysogenic phages than lytic phages in more contaminated samples and lysogenic phages carried metal resistance genes.41 This indicates a selective pressure, as the prophage provided metal resistance to the bacteria. Together, these studies indicate that the relationship between bacterial stress and phage induction likely varies based on the individual type of phages and the functional capability the phages provide their host organism.
Phage host range in wastewater systems is unknown
The phage host range indicates the diversity of hosts that a phage can infect. A phage with a broad host range has the machinery and capability to infect many different bacterial species. Conversely, most studied phages have a narrow host range indicating that an individual phage can only infect one host.42 Host switching is where each individual phage can only infect one host, but the phage quasispecies can infect multiple species.42 One meta-analysis across multiple environments suggests that there is a weak negative correlation between the phage virulence and host range; phages with a broad host range are less virulent.43 Little is known about the host range for phages that dominate wastewater environments. One study indicates that phage isolates from wastewater have a broad host range, but this research was limited to a handful of isolates.44 This finding was supported by a more recent metagenomic survey of several wastewater treatment plants that suggests the phage community could infect a diverse set of hosts,45 and similarly a survey of sewage indicates a broad host range.46 While there is a significant need to isolate and identify phages from wastewater in order to build databases,45,47 culture-based methods for evaluating the host range are cumbersome and miss the abundant uncultured prokaryotic species in wastewater. Emerging bioinformatic methods including those that incorporate machine learning are very promising for evaluating host range (e.g., ref. 48 and 49) and could be applied to wastewater environments.Phage–host signaling & phage–phage molecular signaling
There are several molecular signals used by phages to signal one another and establish a lytic or lysogenic lifestyle. Specific phage communication systems that warrant further discussion are: 1) lysis inhibition, 2) synchronized lysis inhibition collapse, 3) high multiplicity lysogeny decisions (HMDLs), 4) arbitrium systems, and 5) autoinducer-associated prophage induction (AAPI), summarized in Table 1.| Lysis inhibition | Synchronized lysis inhibition collapse | HMDLs | Arbitrium systems | AAPI | |
|---|---|---|---|---|---|
| Phages | T-even phages | T-even phages | Lambda phages | Phages phi3T and SPbeta | Vibriophage VP882 |
| Type of communication | Phage–phage | Phage–phage | Phage–phage | Phage–phage | Phage–host |
| Life cycle pathway | Delayed lytic cycle | Accelerated lytic cycle | Lysogeny | Lysogeny | Lytic cycle |
| Molecule type | Re-adsorption of secondary phages | Adsorbing phages in lysis-inhibited populations | Multiple infecting phages | aimP peptide | DPO peptide |
| Ref. | 50–55 | 50 and 56 | 39, 50 and 57–62 | 63–66 | 67–69 |
Lysis inhibition delays lysis
Lysis inhibition is the oldest known form of phage-to-phage communication,50 first described almost 75 years ago in a study investigating the lysis of E. coli infected with T-even phages.51 Lysis inhibition delays the lysis of infected bacteria to ensure that there are enough bacteria for phages to infect. Basically, at low bacterial abundance, lysis is delayed. The lysis inhibition system is carried out when the newly released phages from a lysed bacterial cell get re-adsorbed into an already infected bacterial cell.50,51 The adsorption reaction of the phages delays the lysis of the infected bacterial cell. Delaying the lytic cycle results in larger burst sizes (more phages released) from the secondarily infected bacteria.51To understand how lysis inhibition occurs, it is important to first understand the mechanism by which phages lyse cells. One mechanism, termed lysis from within, uses several proteins including holin and endolysin proteins.50 The holin protein is responsible for making holes in the bacterial cell's membrane and allowing the endolysin, located in the cell's cytoplasm, to degrade the bacterial cell wall.50,52 Lysis from within occurs when the holin protein is activated.50 The activation of lysis inhibition in T-even phages occurs when secondary virions get adsorbed on the cell wall of an infected bacterial cell, triggering a signal from the R1 protein in the periplasm to the holin protein in the plasma membrane.50,53,54 The R1 protein thus functions as an anti-holin. An additional anti-holin in the cytoplasm, the RIII protein, also acts as a stabilizer for the RI and holin complex.50,55
Synchronized lysis inhibition collapse signals the beginning of lysis
The synchronized lysis inhibition collapse system is related to lysis inhibition; it is the accelerated lysis of phage-infected bacteria that were under the influence of lysis inhibition.50 In other words, it is the lysis of previously lysis-inhibited cells. This signaling system was first observed in a study investigating the lysis of lysis-inhibited T4 phage-infected cells.56 The study's authors proposed two mechanisms potentially governing synchronized lysis inhibition collapse. The first mechanism is initiated from the accumulation of adsorbed secondary virions to the lysis-inhibited bacteria, resulting in the degradation of the cell walls and a process the study's authors termed lysis from without. Lysis from without is the result of membrane disruptions that further propel lysis from within in the lysis-inhibited bacteria. The second proposed mechanism that is suggested by the study's authors is secondary traumatization, which could also cause disruptions in the plasma membrane chemiosmotic barrier that could lead to lysis from within. While the mechanisms are not known, the overall effect of synchronized lysis inhibition collapse is accelerating lysis.HMDLs lead to lysogeny when there is high MOI for lambda phages
A molecular signaling system that is observed within lambda phages is the HMDLs.50,57 HMDLs are a result of multiple phage infections to bacteria and results in lysogeny. HMDLs are different from lysis inhibition where the latter leads to lytic life cycles, and it is a result of secondary virion adsorption, not phage infections.50,57 Several experimental studies have observed this phenomenon, in which multiple lambda phage infections lead to a lysogenic life cycle.58–62 To briefly describe the mechanism for the switch between lysis and lysogeny in lambda phages, it is regulated by the CI and Cro proteins and three promoters, PR, PL, and PRM.44 Cro controls the PR and PL promoters (strong promoters) that induce the lytic cycle.44 This occurs because Cro dimers bind to operators with high affinities that eventually results in the blockage of the CI synthesis and initiates the lytic cycle in the lambda phages.44 In the lysogenic cycle, the opposite occurs and the CI dimers have a high affinity to bind with operator proteins that that will repress the promoters for the synthesis of the Cro proteins.44 Together, these communication signals ensure the initiation and maintenance of the lysogenic life cycle in lambda phages at high MOIs.44Arbitrium system leads to lysogeny
An arbitrium system is one of the more recently discovered forms of phage–phage communication systems summarized in Fig. 2.63 The arbitrium system uses a phage-specific peptide code that directs the phage life cycle to lysogeny, similar to HMDLs. The system is encoded by three phage genes which are: aimP, aimR, and aimX. The gene aimP produces the peptide signal that gets received by the AimR receptor in phages, leading to lysogeny. The AimX receptor is an inhibitor of lysogeny, and when it is active the phage displays lytic decisions. At low cell densities, the AimR receptor as a dimer activates the AimX protein leading to the lytic cycle (inhibition of lysogeny).63,64 During that time, AimP gets secreted into the medium and after multiple cycle infections it reaches a threshold concentration enabling it to bind to the AimR receptor which in turn inactivates the AimR dimer and results in the silencing of AimX leading to lysogeny.63,64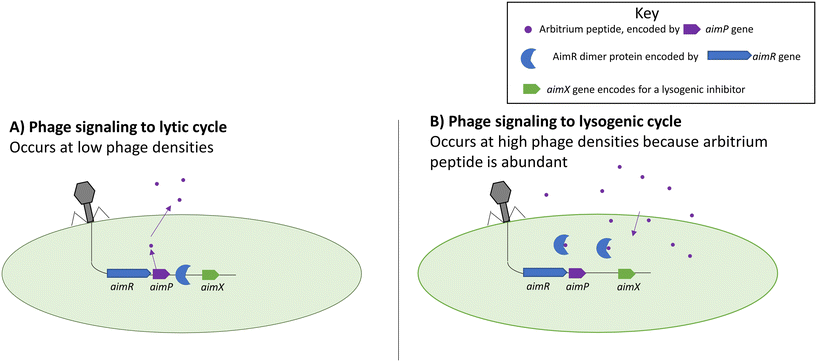 | ||
| Fig. 2 Functions of the aim complexes during lytic (left) and lysogenic (right) cycles. Arbitrium peptides prevent the AimR protein from binding to the genome and prevent lysogeny. Adapted with permission from ref. 63 Springer Nature, copyright 2017. | ||
It was shown that different phages produce different peptide signals.63,64 The peptide signals from phages Phi3T and SPbeta are SAIRGA and GMPRGA, respectively. For the Phi3T phage, the peptide signal, SAIRGA, results in conformational changes to its AimR receptor leading to its dissociation and lysogeny. However, in the SPbeta phage, the peptide signal GMPRGA stabilizes the AimR receptor in a dimeric state leading to lysis.63,64 These differences show how different phages could use the same molecular signaling system for distinct outcomes.
The phi3T phage also exhibits a second communication system called the Rapφ–Phrφ system.65 This bacterial quorum sensing mediated system works together with the arbitrium system to weaken the host defense systems during the lytic cycle. Rapφ is a receptor that can inhibit the ComA-P and Spo0F-P regulators in Bacillus. The ComA-P regulator is responsible for the production of antimicrobials, whereas the Spo0F-P regulator is responsible for sporulation or biofilm formation. The downregulation of the host defense through Rapφ enhances the efficiency of the lytic cycle. It is also postulated that during lysogeny, this communication system is used to enhance the fitness of the host bacteria by not wasting resources in the production of biofilm or antimicrobials.
Autoinducer-associated prophage induction (AAPI)
The autoinducer-associated prophage induction (AAPI) system is a bacteria–virus communication system rather than a virus–virus communication system presented in previous sections. It was shown that vibriophage VP882 encodes a receptor that detects an autoinducer from Vibrio cholerae which then controls the life cycle decision for the phage.67V. cholerae has a well-characterized quorum sensing system that uses the autoinducer 3,5-dimethylpyrazin-2-ol (DPO) and a protein receptor VqmA,68 needed for virulence and biofilm formation.68,69 Phage VP882 has a similar receptor to that of V. cholerae called VqmAphage.68,69There are two pathways for induction in phage VP882. The first pathway is through the activation of the VqmAphage receptor by binding with bacterial autoinducer DPO. This will upregulate the quorum-triggered inactivator of the CI protein (Qtip), which will bind to the CI repressor protein and initiate the lytic life cycle through the activation of the Q antiterminator protein. The second pathway is DNA damage. This will result in the cleavage of the CI repressor in a RecA-dependent matter that leads to the upregulation of the Q antiterminator protein. VqmAphage can bind to promoters of its own and of the host, while VqmA only binds to its own promoters.48,50 In essence, phage VP882 can use the QS system used by its host and also uses the phage's own pathways to infer the decisions on lysis or lysogeny.48,50
Similar mechanisms of bacterial QS-dependent phage induction have been demonstrated with other hosts and in the environment. For instance, a study of groundwater microbial communities found increasing abundance of viruses, and correspondingly decreased abundance of bacteria, with the additional acyl homoserine lactones (AHLs), indicating that the mixture of AHLs in the study led to phage induction.70 That study identified the mechanisms of AHL-mediated phage induction in an E. coli lambda phage, both with exogenously added AHLs and in a co-culture with AHL-producing P. aeruginosa cultures. There are several additional examples of phages carrying QS system genes,71–73 but the role of these genes in guiding lysis and lysogeny decisions is unknown. As indicated with the vibriophage and phi3T communication systems mentioned above, it would be beneficial for phages to use the host QS signals to guide lysis or lysogenic cycles. An additional example is related to QS degradation; one study identified acylhydrolase genes, which encode enzymes that degrade AHLs, within a phage genome.74 Again, the fitness benefit of carrying these genes by phages is unknown. The carrying costs of these functions within a small genome indicates that phages likely derive a benefit from either ‘listening in’ or directly manipulating bacterial QS signals.
Though we still lack an understanding of phage diversity and its role in wastewater treatment systems, these varied and complex signaling systems are likely important for understanding phage lifestyle. Phages are likely using bacterial signals and phage–phage signaling systems in tandem with varied outcomes. Understanding what is regulating phage behavior is critical for applying phages to enhance wastewater treatment and other environmental engineered processes.
Role of phage lifestyle and signaling in wastewater systems
Phage within wastewater systems carry antibiotic resistance genes
The importance of phage–bacteria interactions as drivers for human and environmental health is only recently appreciated. Phages impact human health because they can carry genes that encode antimicrobial resistance (AMR) and transduction events can lead to the spread of resistance in the environment.75,76 Transduction can be generalized, specialized, and lateral.14 In generalized transduction, host DNA is mistakenly taken for viral DNA and mispackaged; any portion of the host genome can be packaged this way. Specialized transduction is rarer because it is the result of aberrant excision that joins a segment of viral and bacterial DNA; only portions of the host genome would undergo specialized transduction. Both are the result of missteps that are uncommon; therefore, there are generally low frequencies of host gene transfer due to generalized and specialized transduction. For this reason, transduction was thought to not be the dominant mechanism of horizontal gene transfer events. The recent discovery of a third mechanism of transduction, lateral transduction,77 has added more complexity to the role of phages in horizontal gene transfer events because it results in much more frequent gene transfer events, at least 1000-times more frequent than generalized and specialized transduction.78 Lateral transduction is the result of a delayed excision of viral DNA—replication occurs before excision. Studies of lateral transduction are nascent and limited to model systems, so the role of this mechanism in the environment is unknown.Though the role of lateral transduction in environmental systems is unknown, high frequency transduction is of particular concern for the case of AMR, especially if resistance genes are transferred from non-pathogenic environmental bacteria to disease causing pathogenic bacteria. It is important to note that transduction is not the only, or primary, potential form of transfer of AMR, and one analysis of all forms of mobile genetic elements identified transposable elements as dominant carriers of antibiotic resistance genes.79 The same analysis found resistance genes to be significantly enriched in phage-like elements. Given our new understanding of the mechanisms of transduction, consideration of phages for AMR spread is needed because although AMR is a natural bacterial defense mechanism, this function can have deadly consequences on human health when acquired by pathogenic organisms.
Wastewater treatment plants are thought to be an important potential transmission route for AMR in the environment.80–82 This is further exacerbated by the constant influx of antibiotics going into wastewater treatment plants at sub-lethal concentrations.83,84 In a study of phage isolates from wastewater, half of the isolated phages were found to carry beta-lactamase resistance. Twenty percent of the isolated phages were not only carrying beta-lactamase genes but were transducing phages and were able to infect a variety of E. coli indicator strains.85 This study represents only a snapshot of culturable phages that infect E. coli, but sequencing tools are expected to uncover a broader scope of the potential phages that harbor AMR. Another qPCR-based study found beta-lactamase resistance to be in some instances more prevalent (detected in more samples) in the phage DNA fraction than in the bacterial DNA fraction.86 Thus, phages may play an important role in the spread of AMR in wastewater treatment systems.
Sequencing is a powerful tool to develop an understanding of the diversity of AMR in a sample. However, particularly within the phage fraction, calculating the relative abundance of resistance genes in samples is challenging. Within bacteria, gene read counts can be normalized to a single copy bacterial gene. But, within a phage sequencing pool there is a lack of standardization in protocols to make the data quantitative or semi-quantitative. The application of internal standards holds promise in this regard,87 but for the time being many studies have focused on the richness of resistance genes found within the phage fraction. For instance, one study of phage diversity within swine wastewater confirmed a potential diverse set of resistance genes within phage DNA, including the mcr-1 gene which confers resistance to a last resort antimicrobial, colistin.88 Another study identified tetracycline resistance within the phage sequences of sewers.46 Though it is known that phages along with other mobile genetic elements can play a role in horizontal gene transfer, the relative importance of phages compared to these other mechanisms is unknown. The discovery of new and more frequent transduction mechanisms underscores the need to revisit the role of phages in AMR transfer in the environment. The adoption of quantitative sequencing methods for phages, along with benchmarking standards for accurately characterizing a gene to be in the bacterial versus viral fractions,23 could help us understand the relative importance of phages compared with other agents for the spread of AMR in the environment.
Phages are known to carry genes related to nutrient removal and related processes
Molecular evidence suggests that abundant phages in marine and soil environments can contain AMGs related to nitrogen,89,90 sulfur,90–92 and carbon15,93–95 cycling. Given the relative genome sizes of phages, it is somewhat counterintuitive that there is a fitness benefit to carrying AMGs related to nutrient metabolism and shuttling. Although cycling of these nutrients by bacteria is well-studied, the role of phages on nutrient cycling is not as well characterized. Moreover, few studies have evaluated the AMG content of phages in wastewater environments, but it has been shown that organisms related to nutrient cycling (sulfur, nitrogen, and phosphorus accumulating organisms) are all prominent hosts for phages.45Sulfur cycling organisms are consistently found to host phages within activated sludge systems. Notably in one study, all sampled wastewater treatment plants (6) had phages with hosts related to sulfate reduction, and an organism related to sulfide-based denitrification (Thiothrix), indicating that phages have a prominent impact on sulfur cycling in the treatment plants from that study.45 This was supported by another metagenomic survey,96 and a more recent study also identified carbon and sulfur-related AMGs within the phage genome in sewer metagenomes.46 The prominence of phages with sulfur cycling hosts and AMGs within wastewater is in line with a study of publicly available datasets illustrating that phages carrying sulfur cycling-related AMGs were widespread in many ecosystems.92 That study illustrated that in some cases, phage-directed sulfur oxidation was expressed at higher levels than host sulfur oxidation. This demonstrates the potential importance of phages in biogeochemical cycling of sulfur. Understanding whether this aspect extends to other nutrients (nitrogen and phosphorus) is a research area ripe for further study.
Harnessing phages to advance wastewater treatment processes requires understanding of phage genetic potential and lifestyle
The use of phage therapy for controlling environmental engineering systems has long been proposed and could provide human health or environmental benefits. For instance, there is a growing body of work in human systems to harness phages to kill antibiotic resistant pathogenic bacteria,97 but this form of bottom-up control in environmental systems is still quite challenging due to our lack of knowledge of phage diversity and host-range in environmental systems. Applying knowledge from both pure culture studies and studies of phages in other environments can help us develop hypotheses for the role of phages in activated sludge, which may lead to more targeted approaches to leverage phages for wastewater treatment. For instance, applications could include inducing lysogeny of phage carrying nitrogen cycling related AMGs to help engineers respond to pulse pollutant loads or increase rates of removal; or using phage to target lysis of filamentous or pathogenic bacteria. Understanding phages and their role in environmental engineering systems is nascent; a research frontier within this space is to develop tools to control lysogenic and lytic phage life cycles within engineered processes by harnessing the molecular signals they emit, a form of “bottom-up” control of engineered systems.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The project was supported by the National Science Foundation, Division of Chemical, Bioengineering, Environmental, and Transport Systems (CBET) (grant 1955034). The authors acknowledge the Office of Faculty Development (OFD) at Howard University.References
- S. Chibani-chennoufi, A. Bruttin, M. Dillmann and H. Brüssow, J. Bacteriol., 2004, 186, 3677–3686 CrossRef CAS PubMed.
- M. R. Brown, J. C. Baptista, M. Lunn, D. L. Swan, S. J. Smith, R. J. Davenport, B. D. Allen, W. T. Sloan and T. P. Curtis, Water Res., 2019, 152, 264–273 CrossRef CAS PubMed.
- R. Liu, Z. Li, G. Han, S. Cun, M. Yang and X. Liu, Appl. Microbiol. Biotechnol., 2021, 105, 5299–5307 CrossRef CAS PubMed.
- V. Runa, J. Wenk, S. Bengtsson, B. V. Jones and A. B. Lanham, Front. Microbiol., 2021, 12 DOI:10.3389/fmicb.2021.730071.
- S. M. Kotay, T. Datta, J. Choi and R. Goel, Water Res., 2011, 45, 694–704 CrossRef CAS PubMed.
- A. Lisac, E. Birsa and A. Podgornik, Microb. Biotechnol., 2022, 15(9), 2450–2463 CrossRef CAS PubMed.
- P. Zuo, J. Metz, P. Yu and P. J. J. Alvarez, Water Res., 2022, 224, 119070 CrossRef CAS PubMed.
- B. Reyneke, S. Khan, P. Fernández-Ibáñez and W. Khan, Environ. Sci.: Water Res. Technol., 2020, 6, 87–102 RSC.
- T. O. Worley-Morse and C. K. Gunsch, Water Res., 2015, 68, 627–636 CrossRef CAS PubMed.
- A. H. Havelaar, T. J. Nieuwstad, C. C. E. Meulemans and M. van Olphen, Water Sci. Technol., 1991, 24, 347–352 CrossRef.
- E. Stachler, C. Kelty, M. Sivaganesan, X. Li, K. Bibby and O. C. Shanks, Environ. Sci. Technol., 2017, 51, 9146–9154 CrossRef CAS PubMed.
- Y. Ye, R. M. Ellenberg, K. E. Graham and K. R. Wigginton, Environ. Sci. Technol., 2016, 50, 5077–5085 CrossRef CAS PubMed.
- C. E. Lawson, W. R. Harcombe, R. Hatzenpichler, S. R. Lindemann, F. E. Löffler, M. A. O'Malley, H. G. Martín, B. F. Pfleger, L. Raskin, O. S. Venturelli, D. G. Weissbrodt, D. R. Noguera and K. D. McMahon, Nat. Rev. Microbiol., 2019, 17, 725–741 CrossRef CAS PubMed.
- Y. N. Chiang, J. R. Penadés and J. Chen, PLoS Pathog., 2019, 15(8) DOI:10.1371/journal.ppat.1007878.
- C. A. Suttle, Nat. Rev. Microbiol., 2007, 5, 801–812 CrossRef CAS PubMed.
- R. Danovaro, A. Dell'Anno, C. Corinaldesi, M. Magagnini, R. Noble, C. Tamburini and M. Weinbauer, Nature, 2008, 454, 1084–1087 CrossRef CAS PubMed.
- E. Lara, D. Vaqué, E. L. Sà, J. A. Boras, A. Gomes, E. Borrull, C. Díez-Vives, E. Teira, M. C. Pernice, F. C. Garcia, I. Forn, Y. M. Castillo, A. Peiró, G. Salazar, X. Anxelu, G. Morán, R. Massana, T. S. Catalá, G. M. Luna, S. Agustí, M. Estrada, J. M. Gasol and C. M. Duarte, Sci. Adv., 2017, 3(9) DOI:10.1126/sciadv.1602565.
- C. H. Wigington, D. Sonderegger, C. P. D. Brussaard, A. Buchan, J. F. Finke, J. A. Fuhrman, J. T. Lennon, M. Middelboe, C. A. Suttle, C. Stock, W. H. Wilson, K. E. Wommack, S. W. Wilhelm and J. S. Weitz, Nat. Microbiol., 2016, 1 DOI:10.1038/nmicrobiol.2015.24.
- J. Zhang, Q. Gao, Q. Zhang, T. Wang, H. Yue, L. Wu, J. Shi, Z. Qin, J. Zhou, J. Zuo and Y. Yang, Microbiome, 2017, 5, 57 CrossRef PubMed.
- D. Pan, R. Watson, D. Wang, Z. H. Tan, D. D. Snow and K. A. Weber, ISME J., 2014, 8, 1691–1703 CrossRef CAS PubMed.
- M. Breitbart, C. Bonnain, K. Malki and N. A. Sawaya, Nat. Microbiol., 2018, 3, 754–766 CrossRef CAS PubMed.
- B. L. Hurwitz and J. M. U'Ren, Curr. Opin. Microbiol., 2016, 31, 161–168 CrossRef CAS PubMed.
- A. A. Pratama, B. Bolduc, A. A. Zayed, Z. P. Zhong, J. Guo, D. R. Vik, M. C. Gazitúa, J. M. Wainaina, S. Roux and M. B. Sullivan, PeerJ, 2021, 9, e11447 CrossRef PubMed.
- Y. Xu, R. Zhang, N. Wang, L. Cai, Y. Tong, Q. Sun, F. Chen and N. Jiao, Environ. Microbiol., 2018, 20(8), 2974–2989 CrossRef CAS PubMed.
- E. Harrison and M. A. Brockhurst, BioEssays, 2017, 39(12) DOI:10.1002/bies.201700112.
- C. Howard-Varona, M. M. Lindback, G. E. Bastien, N. Solonenko, A. A. Zayed, H. B. Jang, B. Andreopoulos, H. M. Brewer, T. G. del Rio, J. N. Adkins, S. Paul, M. B. Sullivan and M. B. Duhaime, ISME J., 2020, 14(4), 881–895 CrossRef PubMed.
- D. Refardt, T. Bergmiller and R. Kümmerli, Proc. R. Soc. B, 2013, 280 DOI:10.1098/rspb.2012.3035.
- J. Bondy-Denomy, J. Qian, E. R. Westra, A. Buckling, D. S. Guttman, A. R. Davidson and K. L. Maxwell, ISME J., 2016, 10, 2854–2866 CrossRef PubMed.
- B. Koskella and M. A. Brockhurst, FEMS Microbiol. Rev., 2014, 38, 916–931 CrossRef CAS PubMed.
- M. A. Brockhurst, A. Fenton, B. Roulston and P. B. Rainey, BMC Ecol., 2006, 6, 1–7 CrossRef PubMed.
- O. H. Shapiro, A. Kushmaro and A. Brenner, ISME J., 2010, 4, 327–336 CrossRef PubMed.
- B. Knowles, C. B. Silveira, B. A. Bailey, K. Barott, V. A. Cantu, A. G. Cobian-Guëmes, F. H. Coutinho, E. A. Dinsdale, B. Felts, K. A. Furby, E. E. George, K. T. Green, G. B. Gregoracci, A. F. Haas, J. M. Haggerty, E. R. Hester, N. Hisakawa, L. W. Kelly, Y. W. Lim, M. Little, A. Luque, T. McDole-Somera, K. McNair, L. S. de Oliveira, S. D. Quistad, N. L. Robinett, E. Sala, P. Salamon, S. E. Sanchez, S. Sandin, G. G. Z. Silva, J. Smith, C. Sullivan, C. Thompson, M. J. A. Vermeij, M. Youle, C. Young, B. Zgliczynski, R. Brainard, R. A. Edwards, J. Nulton, F. Thompson and F. Rohwer, Nature, 2016, 531, 466–470 CrossRef CAS PubMed.
- X. Chen, M. G. Weinbauer, N. Jiao and R. Zhang, Sci. Bull., 2021, 66, 871–874 CrossRef CAS PubMed.
- O. H. Shapiro and A. Kushmaro, Curr. Opin. Biotechnol., 2011, 22, 449–455 CrossRef CAS PubMed.
- A. N. Shkoporov, E. V. Khokhlova, C. B. Fitzgerald, S. R. Stockdale, L. A. Draper, R. P. Ross and C. Hill, Nat. Commun., 2018, 9, 1–8 CrossRef CAS PubMed.
- H. H. Buchholz, M. L. Michelsen, L. M. Bolaños, E. Browne, M. J. Allen and B. Temperton, ISME J., 2021, 1585–1598 CrossRef CAS PubMed.
- J. S. Weitz, G. Li, H. Gulbudak, M. H. Cortez and R. J. Whitaker, Virus Evol., 2019, 5, 1–9 Search PubMed.
- A. B. Oppenheim, O. Kobiler, J. Stavans, D. L. Court and S. Adhya, Annu. Rev. Genet., 2005, 39, 409–429 CrossRef CAS PubMed.
- S. Lee, D. E. A. Lewis and S. Adhya, J. Mol. Biol., 2018, 430, 58–68 CrossRef CAS PubMed.
- J. Choi, S. M. Kotay and R. Goel, Water Res., 2010, 44, 4550–4558 CrossRef CAS PubMed.
- D. Huang, P. Yu, M. Ye, C. Schwarz, X. Jiang and P. J. J. Alvarez, Microbiome, 2021, 9, 1–15 CrossRef PubMed.
- P. A. de Jonge, F. L. Nobrega, S. J. J. Brouns and B. E. Dutilh, Trends Microbiol., 2019, 27, 51–63 CrossRef CAS PubMed.
- F. Molina, A. Simancas, M. Ramírez, R. Tabla, I. Roa and J. E. Rebollo, Front. Microbiol., 2021, 12, 1–14 Search PubMed.
- M. A. Khan, H. Satoh, H. Katayama, F. Kurisu and T. Mino, Water Res., 2002, 36, 3364–3370 CrossRef CAS PubMed.
- Y. Chen, Y. Wang, D. Paez-Espino, M. F. Polz and T. Zhang, Nat. Commun., 2021, 12, 1–11 CrossRef PubMed.
- K. Gulino, J. Rahman, M. Badri, J. Morton, R. Bonneau and E. Ghedin, mSystems, 2020, 5(3) DOI:10.1128/msystems.00876-19.
- B. Hegarty, Z. Dai, L. Raskin, A. Pinto, K. Wigginton and M. Duhaime, Water Res., 2022, 218, 118484 CrossRef CAS PubMed.
- S. Roux, A. P. Camargo, F. H. Coutinho, S. M. Dabdoub, B. E. Dutilh, S. Nayfach and A. Tritt, PLoS Biol., 2023, 21(4), e3002083 CrossRef CAS PubMed.
- S. Roux, S. J. Hallam, T. Woyke and M. B. Sullivan, eLife, 2015, 4, 1–20 CrossRef PubMed.
- S. T. Abedon, Viruses, 2019, 11, 951 CrossRef CAS PubMed.
- A. H. Doermann, J. Bacteriol., 1948, 55, 257–276 CrossRef CAS PubMed.
- I.-N. Wang, D. L. Smith and R. Young, Annu. Rev. Microbiol., 2000, 54, 799–825 CrossRef CAS PubMed.
- P. Paddison, S. T. Abedon, H. K. Dressman, K. Gailbreath, J. Tracy, E. Mosser, J. Neitzel, B. Guttman and E. Kutter, Genetics, 1998, 148, 1539–1550 CrossRef CAS PubMed.
- T. A. T. Tran, D. K. Struck and R. Young, J. Bacteriol., 2007, 189, 7618–7625 CrossRef CAS PubMed.
- E. Ramanculov and R. Young, Mol. Microbiol., 2001, 41, 575–583 CrossRef CAS PubMed.
- S. T. Abedon, J. Bacteriol., 1992, 174, 8073–8080 CrossRef CAS PubMed.
- D. Wadhwa, BioRxiv, 2020, preprint, DOI:10.1101/146308.
- J. S. K. Boyd, J. Pathol. Bacteriol., 1951, 63, 445–457 CrossRef CAS PubMed.
- M. Lieb, J. Bacteriol., 1953, 65, 642–651 CrossRef CAS PubMed.
- P. Kourilsky, Mol. Gen. Genet., 1973, 122, 183–195 CrossRef CAS PubMed.
- D. B. Hoffman Jr and I. Rubenstein, J. Mol. Biol., 1968, 35, 375–399 CrossRef CAS PubMed.
- R. I. Joh and J. S. Weitz, PLoS Comput. Biol., 2011, 7, e1002006 CrossRef CAS PubMed.
- Z. Erez, I. Steinberger-Levy, M. Shamir, S. Doron, A. Stokar-Avihail, Y. Peleg, S. Melamed, A. Leavitt, A. Savidor and S. Albeck, Nature, 2017, 541, 488–493 CrossRef CAS PubMed.
- C. Dou, J. Xiong, Y. Gu, K. Yin, J. Wang, Y. Hu, D. Zhou, X. Fu, S. Qi and X. Zhu, Nat. Microbiol., 2018, 3, 1285–1294 CrossRef CAS PubMed.
- C. Bernard, Y. Li, P. Lopez and E. Bapteste, ISME J., 2021, 15, 545–549 CrossRef CAS PubMed.
- N. Aframian, S. O. Bendori, S. Kabel, P. Guler, A. Stokar-Avihail, E. Manor, K. Msaeed, V. Lipsman, I. Grinberg, A. Mahagna and A. Eldar, Nat. Microbiol., 2022, 7, 145–153 CrossRef CAS PubMed.
- J. E. Silpe and B. L. Bassler, Cell, 2019, 176, 268–280 CrossRef CAS PubMed.
- K. Papenfort and B. L. Bassler, Nat. Rev. Microbiol., 2016, 14, 576–588 CrossRef CAS PubMed.
- K. L. Maxwell, Cell, 2019, 176, 7–8 CrossRef CAS PubMed.
- D. Ghosh, K. Roy, K. E. Williamson, S. Srinivasiah, K. E. Wommack and M. Radosevich, Appl. Environ. Microbiol., 2009, 75, 7142–7152 CrossRef CAS PubMed.
- K. R. Hargreaves, A. M. Kropinski and M. R. J. Clokie, PLoS One, 2014, 9(1) DOI:10.1371/journal.pone.0085131.
- S. F. Lan, C. H. Huang, C. H. Chang, W. C. Liao, I. H. Lin, W. N. Jian, Y. G. Wu, S. Y. Chen and H. C. Wong, Appl. Environ. Microbiol., 2009, 75, 2659–2667 CrossRef CAS PubMed.
- M. Boyer, J. Haurat, S. Samain, B. Segurens, F. Gavory, V. González, P. Mavingui, R. Rohr, R. Bally and F. Wisniewski-Dyé, Appl. Environ. Microbiol., 2008, 74, 861–874 CrossRef CAS PubMed.
- C. Leblanc, A. Caumont-Sarcos, A. M. Comeau and H. M. Krisch, Environ. Microbiol. Rep., 2009, 1, 499–509 CrossRef CAS PubMed.
- S. R. Modi, H. H. Lee, C. S. Spina and J. J. Collins, Nature, 2013, 499, 219–222 CrossRef CAS PubMed.
- D. Debroas and C. Siguret, ISME J., 2019, 13, 2856–2867 CrossRef CAS PubMed.
- J. Chen, N. Quiles-Puchalt, N. Chiang, R. Bacigalupe, A. Fillol-Salom, M. Su, J. Chee, J. R. Fitzgerald and J. R. Penadés, Genome hypermobility by lateral transduction, 2018, vol. 362 Search PubMed.
- S. Humphrey, A. Fillol-Salom, N. Quiles-Puchalt, R. Ibarra-Chávez, A. F. Haag, J. Chen and J. R. Penadés, Nat. Commun., 2021, 12 DOI:10.1038/s41467-021-26004-5.
- S. Khedkar, G. Smyshlyaev, I. Letunic, O. M. Maistrenko, L. P. Coelho, A. Orakov, S. K. Forslund, F. Hildebrand, M. Luetge, T. S. B. Schmidt, O. Barabas and P. Bork, Nucleic Acids Res., 2022, 50, 3155–3168 CrossRef CAS PubMed.
- T. U. Berendonk, C. M. Manaia, C. Merlin, D. Fatta-Kassinos, E. Cytryn, F. Walsh, H. Bürgmann, H. Sørum, M. Norström, M. N. Pons, N. Kreuzinger, P. Huovinen, S. Stefani, T. Schwartz, V. Kisand, F. Baquero and J. L. Martinez, Nat. Rev. Microbiol., 2015, 13 Search PubMed.
- N. Makowska, R. Koczura and J. Mokracka, Chemosphere, 2016, 144, 1665–1673 CrossRef CAS PubMed.
- N. Fahrenfeld, Y. Ma, M. O'Brien and A. Pruden, Front. Microbiol., 2013, 4, 1–10 Search PubMed.
- J. Guo, J. Li, H. Chen, P. L. Bond and Z. Yuan, Water Res., 2017, 123, 468–478 CrossRef CAS PubMed.
- F. Ju, B. Li, L. Ma, Y. Wang, D. Huang and T. Zhang, Water Res., 2016, 91, 1–10 CrossRef CAS PubMed.
- G. U. Gunathilaka, V. Tahlan, A. I. Mafiz, M. Polur and Y. Zhang, Int. J. Antimicrob. Agents, 2017, 50, 678–683 CrossRef CAS PubMed.
- A. Zhang, D. R. Call, T. E. Besser, J. Liu, L. Jones, H. Wang and M. A. Davis, Water Res., 2019, 161, 335–340 CrossRef CAS PubMed.
- K. Langenfeld, B. Hegarty, S. Vidaurri, E. Crossette, M. B. Duhaime and K. R. Wigginton, bioRxiv, 2022, preprint, DOI:10.1101/2022.07.08.499345.
- M. Wang, W. Xiong, P. Liu, X. Xie, J. Zeng, F. Walsh and J. L. Balcazar, Front. Microbiol., 2018, 9, 1–10 CrossRef PubMed.
- N. A. Ahlgren, C. A. Fuchsman, G. Rocap and J. A. Fuhrman, ISME J., 2019, 13, 618–631 CrossRef CAS PubMed.
- S. Roux, J. R. Brum, B. E. Dutilh, S. Sunagawa, M. B. Duhaime, A. Loy, B. T. Poulos, N. Solonenko, E. Lara, J. Poulain, S. Pesant, S. Kandels-Lewis, C. Dimier, M. Picheral, S. Searson, C. Cruaud, A. Alberti, C. M. Duarte, J. M. Gasol, D. Vaqué, P. Bork, S. G. Acinas, P. Wincker and M. B. Sullivan, Nature, 2016, 537, 689–693 CrossRef CAS PubMed.
- K. Anantharaman, M. B. Duhaime, J. A. Breier, K. Wendt, B. M. Toner and G. J. Dick, Science, 2014, 344, 757–760 CrossRef CAS PubMed.
- K. Kieft, Z. Zhou, R. E. Anderson, A. Buchan, B. J. Campbell, S. J. Hallam, M. Hess, M. B. Sullivan, D. A. Walsh, S. Roux and K. Anantharaman, Nat. Commun., 2021, 12, 1–16 CrossRef PubMed.
- G. Trubl, H. B. Jang, S. Roux, J. B. Emerson, N. Solonenko, D. R. Vik, L. Solden, J. Ellenbogen, A. T. Runyon, B. Bolduc, B. J. Woodcroft, S. R. Saleska, G. W. Tyson, K. C. Wrighton, M. B. Sullivan and V. I. Rich, mSystems, 2018, 3(5), 1–21 CrossRef PubMed.
- Y. Kuzyakov and K. Mason-Jones, Soil Biol. Biochem., 2018, 127, 305–317 CrossRef CAS.
- B. L. Hurwitz, S. J. Hallam and M. B. Sullivan, Genome Biol., 2013, 14 DOI:10.1186/gb-2013-14-11-r123.
- H. Tamaki, R. Zhang, F. E. Angly, S. Nakamura, P. Y. Hong, T. Yasunaga, Y. Kamagata and W. T. Liu, Environ. Microbiol., 2012, 14, 441–452 CrossRef CAS PubMed.
- A. C. Anna, I. Dabrawksa, K. P. Gregorczyk and Z. Wyzewski, Curr. Microbiol., 2017, 74, 277–283.
| This journal is © The Royal Society of Chemistry 2023 |