Open Access Article
Open Access ArticleMixture effect assessment applying in vitro bioassays to in-tissue silicone extracts of traditional foods prepared from beluga whale blubber†
Beate I.
Escher
 *ab,
Matthew J.
Binnington‡
c,
Maria
König
a,
Ying D.
Lei
c and
Frank
Wania
*ab,
Matthew J.
Binnington‡
c,
Maria
König
a,
Ying D.
Lei
c and
Frank
Wania
 *c
*c
aDepartment of Cell Toxicology, Helmholtz Centre for Environmental Research – UFZ, Leipzig, Germany. E-mail: beate.escher@ufz.de; Tel: +49-342 235 1244
bEnvironmental Toxicology, Department of Geosciences, Eberhard Karls University Tübingen, Tübingen, Germany
cDepartment of Physical and Environmental Sciences, University of Toronto Scarborough, 1265 Military Trail, Toronto, Ontario M1C 1A4, Canada. E-mail: frank.wania@utoronto.ca; Tel: +1-416 287 7225
First published on 31st May 2023
Abstract
We complement an earlier study on the nutrient and environmental contaminant levels in Arctic beluga whale traditional foods by mixture effect assessment using in vitro bioassays. Mixtures were extracted by in-tissue sampling of raw blubber and several traditional food preparations including Muktuk and Uqsuq using silicone (polydimethylsiloxane, PDMS) as sampler. PDMS extracts persistent and degradable neutral organic chemicals of a wide range of hydrophobicity with defined lipid-PDMS partition ratios. The solvent extracts of PDMS were dosed in various reporter gene assays based on human cell lines. Cytotoxicity was consistent across all cell lines and was a good indicator of overall chemical burden. No hormone-like effects on the estrogen receptor, the progesterone receptor and the glucocorticoid receptor were observed but a few samples activated the androgen receptor, albeit with low potency. The peroxisome-proliferator activated receptor (PPARγ) was the most sensitive endpoint followed by activation of oxidative stress response and activation of the arylhydrocarbon (AhR) receptor. The detected pollutants only explained a small fraction of the experimental mixture effects, indicating additional bioactive pollutants. The effect levels of the extracted mixtures were higher than those observed in blubber extracts of dugongs living off the shore of Australia. Roasting over an open fire or food preparation near a smokehouse led to increased PAH levels that were reflected in increased oxidative stress response and activation of the AhR. So far in vitro assays have only been used to quantify persistent dioxin-like chemicals in food and feed but this pilot study demonstrates a much broader potential for food safety evaluations complementing chemical analytical monitoring.
Environmental significanceTraditional food from Arctic beluga whale comes with great nutritional and cultural value but is also a source of exposure to environmental pollutants. Individual persistent organic pollutants have been well investigated, also in relation to the food preparation method. We complement this evaluation of dietary exposure risk by assessing the mixture effects of persistent and nonpersistent organic pollutants with in vitro bioassays to capture the full picture of possible contamination by persistent and nonpersistent bioactive chemicals. The approach introduced here could potentially play an important role in a wide range of dietary risk assessments, complementing methods based on concentrations measured in extracted lipids. |
1. Introduction
The Inuvialuit of the Western Canadian Arctic have been using traditional food (TF) prepared from beluga whales, qilalukkat (Delphinapterus leucas) for many centuries. The beluga whale hunt is not only of tremendous cultural and social value, but also provides an important local source of nutrition.1The numerous benefits of such TFs need to be weighed against the health risks posed by the presence of anthropogenic contaminants in the tissues of the whales. As long-lived, fish-eating marine mammals, beluga whales have been exposed to, and have accumulated, persistent organic pollutants (POP) such as organochlorine pesticides (OCPs), polychlorinated biphenyls (PCBs), polybrominated diphenyl ethers (PDBEs) and other dioxin-like chemicals2,3 but might also be affected by more near-field pollution of less persistent chemicals. TFs derived from the lipid-rich blubber, such as Muktuk and Uqsuq are of particular concern because of the lipophilicity of many organic contaminants.
In an earlier study, we determined the chemical burden of various types of TFs prepared from outer and inner beluga blubber and compared it to that in the raw blubber.3 Most POPs, including OCPs, PCBs and PDBEs were detected and the concentrations did not change much with food preparation.3 If the food was roasted or prepared near a smokehouse, levels of polycyclic aromatic hydrocarbons (PAHs) increased substantially.3 Human health risk assessment performed in this earlier study3 indicated that several individual POPs exceeded their minimum risk levels. This assessment was clearly incomplete, as it did not account for the occurrence of POPs in mixtures and because it was limited to the chemicals that were part of the analytical target list and had been detected. In reality, (i) chemicals exert toxic effects in concert, (ii) many more chemicals, congeners and transformation products are likely to be present in the samples than were targeted by the analytical techniques and (iii) chemicals below the analytical detection limit can still contribute to mixture effects.
In vitro bioassays may provide more direct and more comprehensive information on the health risks posed by contaminants in foods. Such assays based on the aryl hydrocarbon receptor (AhR) have been applied for decades in the monitoring of food and feed for dioxin-like chemicals.4–6
Here we propose an approach to a more comprehensive assessment of the health risk posed by contaminants in food, that combines a greatly expanded battery of bioassays with in-tissue extraction with a silicone polymer (polydimethyl-siloxane, PDMS) and illustrate its application using the beluga TF samples from the study by Binnington et al.3 The battery of bioassays we rely upon (Fig. 1)7 covers several relevant steps along the cellular toxicity pathway, with representative endpoints from xenobiotic metabolism, hormone receptor binding, and adaptive stress response to apical effects such as cell viability. Thereby this test battery covers a wide range of health-relevant toxicity endpoints.
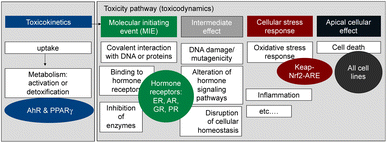 | ||
| Fig. 1 Battery of in vitro assay applied to measure the effects of the extracted mixtures. AhR = aryl hydrocarbon receptor, PPARγ = peroxisome proliferator-activated receptor gamma, ER = estrogen receptor, AR = androgen receptor, GR = glucocorticoid receptor, PR = progesterone receptor, keap-Nrf2-ARE pathway for adaptive stress response to reactive oxygen species and oxidative stress. Figure adapted from Neale et al. 2017.7 | ||
Given that PCBs, PAHs and OCPs were detected in the beluga TF samples, and they are known to activate the AhR in addition to dioxin-like chemicals, we included a highly specific AhR-CALUX assay8 (Table 1). Another biological endpoint relevant for the development of metabolic disorders is the activation of the peroxisome proliferator-activated receptor γ (PPARγ), which was quantified with the PPARγ-bla assay7 (Table 1). The AREc32 assay for oxidative stress response has been previously applied to PDMS extracts of blubber samples and found very active.9 The oxidative stress response is activated directly by electrophilic chemicals but also indirectly by chemicals that cause imbalance of the redox status of the cell or by carcinogenic chemicals.10,11 Chemicals that trigger the oxidative stress response were already previously observed in marine mammals.12
| Mode of toxic action | Endpoint | Cell line/bioassay | Reference compound (QA/QC) | Method detection limit (MDL) | Reference compound (BEQbio and iceberg modelling) |
|---|---|---|---|---|---|
| Metabolism | |||||
| Induction of the arylhydrocarbon receptor AhR | Induction of reporter gene encoding for luciferase (EC10) | AhR-CALUX (H4L1.1c4 rat)8 | 2,3,7,8-Tetrachlorodibenzodioxin (TCDD) EC10 = 0.57 ng L−1 | 0.19 ngTCDD L−1 (TCDD-EQ of 70 pgTCDD glip−1) | Benzo[a]pyrene (B[a]P) EC10 = 1.23 μg L−1 (ref. 33) |
| Peroxisome proliferator activated receptor γ | Induction of reporter gene for PPARγ (EC10) | PPARγ-bla (CellSensor PPARγ-UAS-BLA293-H)7 | Rosiglitazone EC10 = 308 ± 6 ng L−1 | 276 ngRosiglitazone L−1 (rosiglitazone-EQ of 102 ngRosiglitazone glip−1) | Rosiglitazone EC10 = 308 ± 6 ng L−1 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Specific (receptor-mediated) toxicity | |||||
| Estrogenicity (ER) | Induction of the estrogen receptor ERα with reporter gene encoding for β-lactamase (EC10) | ERα-bla (CellSensor ERα UAS BLA GRIPTITE)13 | 17β Estradiol (E2) EC10 = 6.8 ± 0.2 ng L−1 | 1.6 ngE2 L−1 (EEQ of 0.6 ngE2 glip−1) | n/a |
| Androgenicity (AR) | Induction of androgen receptor (AR) with reporter gene encoding for β-lactamase (EC10) | AR-bla (CellSensor AR UAS BLA GRIPTITE)13 | Metribolone (R1881) EC10 = 67.3 ± 2.1 ng L−1 | 20 ngR1881 L−1 (R1881-EQ of 7.4 ngR1881 glip−1) | n/a |
| Glucocorticoid receptor (GR) | Induction of GR with reporter gene encoding for β-lactamase (EC10) | GR-bla (CellSensor GR-UAS-BLA HEK293T)13 | Dexamethasone EC10 = 327 ± 13 ng L−1 | 72 ngdexamethasone L−1 (dexamethasone-EQ of 27 ngdexamethasone glip−1) | n/a |
| Progesterone receptor (PR) | Induction of PR with reporter gene encoding for β-lactamase (EC10) | PR-bla (CellSensor PR-UAS-BLA HEK293T)13 | Promegestone EC10 = 48.1 ± 2.9 ng L−1 | 6.4 ngpromegestone L−1 (promegestone-EQ of 2.4 ngpromegestone glip−1) | n/a |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Adaptive stress response | |||||
| Oxidative stress | Induction of Nrf2 protein (ECIR1.5) | AREc32 gene reporter assay (based on MCF7)43 | t-Butylhydroquinone (tBHQ) ECIR1.5 = 0.48 ± 0.1 mg L−1 | 250 μgtBHQ L−1 (tBHQ-EQ of 93 μgtBHQ glip−1) | Benzo[a]pyrene (B[a]P) EC10 = 41.6 μg L−1 (ref. 33) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Non-specific toxicity (baseline toxicity) | |||||
| Cytotoxicity | Cell viability (IC10) with imaging (only for AhR)19 and with ToxBLAzer7 for all other assays | All mammalian cell lines above | n/a | n/a | |
Furthermore, the battery includes bioassays for activation of hormone receptors13 (estrogen receptor (ER-bla), androgen receptor (AR-bla), glucocorticoid receptor (GR-bla), progestogenic receptor (PR-bla), Table 1) to evaluate if endocrine disruption effects could be elicited and also for their connection to the oxidative stress response pathway.14 In addition, cytotoxicity was assessed in all cell lines. The test battery is not comprehensive with respect to toxicological effects but covers a much wider range of chemicals than group-specific chemical analysis and can therefore be viewed as a bioanalytical test battery that quantifies risk-scaled body burden by mixtures of known and unknown chemicals. Mixture models were used to compare the effects predicted from the detected chemicals with those directly measured with bioassays.15
2. Methods
The study design with all experiments and how they relate to each other, and the research questions are given in the ESI, Fig. S1.†2.1 Beluga blubber samples
Samples were from two male beluga whales (HI-14-06 37 years old, HI-14-11 24 years old) caught in 2014 by local Inuvialuit hunters off Hendrickson Island (69° 30′N, 133° 35′W) in the Beaufort Sea.3 From the blubber of each whale, 11 beluga TF types were prepared:3 The outer blubber and skin (called Muktuk) was sampled at various stages of the preparation process. Samples were collected following initial drying on the ground (Muktuk Air Dry) or hang drying (Muktuk Hang Dry), while additional samples were isolated after subjecting dried Muktuk to boiling in a drum (Muktuk Boil Large Drum) or a pot (Muktuk Boil Pot), roasting over an open flame (Muktuk Roast) or ageing for 2d (Muktuk Age 2 Days) or 5d (Muktuk Age 5 Days) in raw Uqsuq. Uqsuq is composed of the inner blubber and the sample “Uqsuq Baseline” represents the raw sample before any food preparation. Uqsuq was then fermented for several days, and samples were taken after 2d (Uqsuq Age 2 Days) and 5d (Uqsuq Age 5 Days). At 5 days an oil had separated that was collected (Uqsuq Oil) and tested separately.The 22 TF samples were characterised in detail by Binnigton et al.3 for nutrients and chemicals. Direct sample extraction with dichloromethane using accelerated solvent extraction followed by clean-up with gel permeation chromatography3 yielded the concentrations of 26 OCPs, 11 PAHs, 9 PCBs and 7 PBDEs. For convenience the detected concentrations are reprinted from Binnington et al.3 in Table S2.† In addition to this previously published chemical characterization of direct extracts,3 a few selected compounds were also quantified in in-tissue PDMS extracts.16
2.2 PDMS-extraction
In-tissue PDMS extraction was performed according to previously published methods.17 In brief, circular 18 mm diameter thin-film discs were cut from PDMS sheets (SSP-M823, 380 μm thickness, Elasto Proxy Inc, Boisbriand, QC) using a hollow punch and pre-cleaned overnight with acetone via Soxhlet extraction. Prior to insertion in blubber samples, discs were briefly air-dried on clean lint-free tissues. A scalpel was used to cut slots into blubber tissues, into which single discs were immersed for 1 day. For blubber (raw) and Muktuk samples, disc slots were consistently cut in the outer layer of fat, within 1–2 cm of the skin surface, to minimize environmental contaminant concentration variability between blubber layers.18 Since Uqsuq sampling necessitated randomly collecting inner blubber strips from ageing buckets, this type of approach to limit variability in PDMS sampling sites was not possible. However, Uqsuq samples all originated from the same larger blubber chunk, such that POP concentration variability between body regions was avoided.To slow down tissue decay during the 24 h equilibration period, disc-containing samples were wrapped in aluminium foil and stored in a refrigerator (4 °C). Discs were then removed from the blubber and thoroughly wiped using lint-free tissues to eliminate any remaining lipid on the disc surface. Following cleaning, discs were stored in pre-cleaned glass test tubes.
For the bioassay work, 11 Beluga TF samples were extracted with 25 PDMS discs each and 11 samples were extracted with 10 PDMS discs each, amounting to approximately 2.2 to 2.4 g and 0.9 to 1 g of PDMS per sample, respectively (Table S1†). 40 Blank PDMS discs were used. This set of discs was sent to Germany for extraction and bioassay testing. Blanks were additionally prepared at UFZ. All blank disks and sample disks were extracted in ethyl acetate and run in the bioassays as described in Section 2.3 below.
For chemical analysis, PDMS extraction was performed in triplicate, such that each blubber sample contained 3 replicates of 5 discs each for a total of 15 discs; except aged Muktuk and Uqsuq samples (n = 4) from whale HI-14-11, which contained 3 replicates of 4 discs each. This set of discs was extracted for chemical analysis as described in Section 2.4 below.
The weight gain of the PDMS during the equilibration ranged from 0.89 to 2.89% (Table S1†), which necessitated the correction of the lipid–PDMS partition constant Klipid/PDMS by the lipid taken up into a PDMS disk with a mass of mPDMS. The true mean Klipid/PDMS for bioassays was derived by Jin et al.9 and amounted to 37 gPDMS glipid−1. The measured mass gain was assumed to be equal to the mass of co-extracted lipid mcoextracted lipid. The lipid corrected distribution ratio Dlipid/PDMS+coextracted lipid was calculated with eqn (1) (Table S1†).
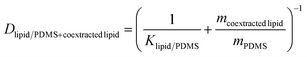 | (1) |
2.3 In vitro bioassays
The details of the used cell lines are given in Table 1. The AhR CALUX assay used the novel (third generation) more sensitive H4L1.1c4 rat cell line.8 The AhR-CALUX was performed as described by Neale et al.7 with modifications of the cytotoxicity assessment described by Escher et al.19 The AREc32 and PPARγ-bla assays were performed as described by Neale et al.7 using the ToxBLAzer as cytotoxicity indicator. The hormone receptor GeneBLAzer assays were described by König et al.20 Materials, media and cell cultures were described previously in detail.21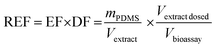 | (2) |
The PDMS were extracted with two times 15 mL ethyl acetate (EA), blown down and redissolved in EA yielding an EF of 1 kgPDMS LEA−1. An appropriate aliquot (typically approximately 50 μL) of that extract was then blown down to dryness and resolubilized with 120 μL of bioassay medium (e.g., DF = 2.4 in the dosing vial). This medium stock of the PDMS extract was diluted two-fold 11 times and 10 μL each of the resulting dilution series transferred in duplicate to a 384 well plate that contained 2500–5000 cells in 30 μL medium, resulting in an approximate highest REF in the bioassay of 0.1 kgPDMS Lbioassay−1. Exposed cells were incubated in 5% CO2 atmosphere at 37 °C for 24 h before detection.
For the detection of the activity of the β-lactamase in the GeneBlazer assays 8 μL of ToxBLAzer™ substrate (Thermo-Fisher Scientific) were added to each well of the plate and fluorescence was measured immediately for autofluorescence correction and after 2 h incubation using a Tecan Infinite® M1000 plate reader. Fluorescence was excited at 409 nm emission measured at 460 nm (blue) and 530 nm (green) to derive the blue/green ratio as a measure of β-lactamase concentration and thus indirectly reporter gene activation. Cytotoxicity was assessed by fluorescence, excitation 590 nm, emission 665 nm.
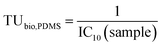 | (3) |
These TUbio, PDMS in units of Lbioassay kgPDMS−1 were converted to lipid-based TUbio (Lbioassay kglipid−1) using the lipid corrected Dlipid/PDMS+coextracted lipid calculated with eqn (1).
| TUbio = Dlipid/PDMS+coextracted lipid × TUbio, PDMS | (4) |
For activation of reporter genes, a linear, low effect-level model is preferred for complex extracts because cytotoxicity masks activation at high concentrations22 but for the AhR-CALUX also a log–logistic CRC model had to be used for reasons given below. For both types of CRC, only concentrations up to IC10 were used and the benchmark concentration derived from the CRC is the EC10 for all cell lines with the exception of AREc32, where no maximum effect could be attained and for which the ECIR1.5 describes the 50% increase in induction ratio (IR) over the control.
The bioanalytical equivalent concentrations (BEQbio) were calculated by dividing the effect concentration EC10 or ECIR1.5 of a reference chemical by the EC10 or ECIR1.5 of the sample.
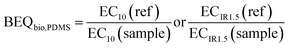 | (5) |
The conversion to lipid-based BEQbio (eqn (6)) was performed analogously to eqn (4).
 | (6) |
The reference chemicals for quality assurance/control (QA/QC), listed in Table 1, were run on each plate for quality control. Most are identical with the reference chemicals for calculating the BEQbio but for AhR-CALUX and AREc32 we used benzo[a]pyrene (B[a]P) as reference chemical for BEQbio (Table 1). The BEQ are then defined accordingly as B[a]P-EQbio, Rosiglitazone-EQbio, estradiol-EQbio (EEQbio), R1881-EQbio and dexamethasone-EQbio (Table 1).
2.4 Chemical analysis of PDMS extracts
Pooled PDMS discs destined for chemical analysis were immersed overnight in 10 mL of acetone spiked with internal standard (50 μL of 200 pg μL−1 13C12-PCBs, in isooctane). Additionally, 5 discs were extracted as a corresponding blank for each sample [4 for the aged Muktuk/Uqsuq samples from whale HI-14-11]. PDMS extracts were subjected to clean-up prior to analysis by pipetting them onto silica gel columns pre-cleaned with hexane and containing from bottom to top glass wool, Na2SO4, SiO2 and Na2SO4. Chemicals were eluted using hexane and DCM, and then concentrated first by rotary evaporation to 2 mL, then exchanged to iso-octane as a keeper and further evaporated under a gentle stream of high-purity nitrogen gas to 1 mL. Note that no such clean-up was performed on PDMS extracts destined for the bioassays to avoid any loss of nonpersistent chemicals. This does not limit the comparison between bioassays and chemical analysis with iceberg modelling (Section 2.5) but some of the missing predicted mixture effect from chemical analysis will be due to removal during clean-up.Quantification of three indicator OCPs [HCB, o,p′-DDT, p,p′-DDT, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KOW range 5.64–6.39],43 and seven indicator PCB congeners [PCB-28, −52, −101, −118, −138, −153, and 180, log
KOW range 5.64–6.39],43 and seven indicator PCB congeners [PCB-28, −52, −101, −118, −138, −153, and 180, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KOW range 5.66–7.19]23 was achieved using an Agilent 7890 gas chromatograph (GC) coupled to a 7000 triple-quad MS/MS operated in negative ionization mode using the method described in Binnington et al.3 A DB-5 column (60 m, 0.25 mm internal diameter, 0.3 μm film thickness) was used for separation. Quality control and quality assurance was described by Binnington.16
KOW range 5.66–7.19]23 was achieved using an Agilent 7890 gas chromatograph (GC) coupled to a 7000 triple-quad MS/MS operated in negative ionization mode using the method described in Binnington et al.3 A DB-5 column (60 m, 0.25 mm internal diameter, 0.3 μm film thickness) was used for separation. Quality control and quality assurance was described by Binnington.16
The concentration of chemicals in lipid (CLip) was calculated by multiplying the measured PDMS concentrations (CPDMS) with Klipid/PDMS (eqn (7)).
| CLip = CPDMS × Klipid/PDMS | (7) |
2.5 Iceberg modelling
With iceberg modelling, we refer to the comparison of measured mixture effects BEQbio with the mixture effects BEQchem predicted for the detected chemicals.15 The mixture effects of the detected chemicals BEQchem can be predicted with eqn (8), provided there are single chemical effect data EC10(i) or ECIR1.5(i) available for the detected chemicals (i) with concentration Ci, from which the relative effect potency (REPi) can be calculated in relation to the reference compound's EC10(ref) or ECIR1.5(ref) (eqn (9)).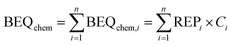 | (8) |
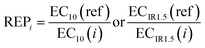 | (9) |
The summation of BEQchem,i to BEQchem (eqn (8)) follows the mixture toxicity concepts of concentration addition and independent action at <10% effect levels,24 which has been demonstrated to be well applicable to environmental mixtures25,26 and dugong blubber.9
3. Results
3.1 Cytotoxicity of the blubber extracts
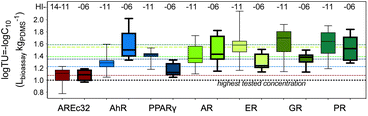
As the extracts contained complex mixtures of chemicals, the specific effects were often masked by cytotoxicity. Only non-cytotoxic concentrations of the extracts (<10% cytotoxicity) were evaluated for the specific endpoints but cytotoxicity itself is an indicator of the overall burden of chemicals acting together as mixtures.The toxic units for 10% cytotoxicity related to the PDMS-based concentration TUbio, PDMS (eqn (3)) varied more between different samples (coefficient of variation 10.6%) than the TUbio (eqn (4)) (coefficient of variation 6.3%) due to the variable amount of co-extracted lipid, that was accounted for by the sample-specific Dlipid/PDMS+coextracted lipid (Table S1†).
TUbio varied by less than a factor of ten (1.9 to 9.5) between individual samples (Fig. 2 and Table S3†). The TUbios of each sample type were not paired by food preparation between TF prepared from the two beluga whales HI-14-11 and HI-14-06 (paired T-test, p = 0.123). TUbio were very similar between the two whales with exception of the samples “Muktuk Roast” and “Muktuk Age 5 days”, which had a higher TUbio in whale HI-14-11 (Fig. S2†).
According to sensitivity distributions of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TUbio (Fig. S3†), the most sensitive cell lines were ER-bla, GR-bla and PR-bla, followed by AR-bla, which was equipotent to PPARγ-bla. AhR-CALUX followed and AREc32 was the least sensitive cell line for cytotoxicity. The GeneBLAzer reporter gene assays are all based on HEK293T and HEK293H cells and the assay is run in medium supplemented with only 2% fetal bovine serum (FBS). These assays were therefore expected to be more sensitive than the cell lines supplemented with 10% FBS (AhR-CALUX derived from H4IIe and AREc32 derived from MCF7 cells (Table 1)) because chemicals bind to serum proteins, which reduces their bioavailability to cells. This was essentially confirmed but the MCF7 cell line additionally seems to be intrinsically more robust than the other cell lines.
TUbio (Fig. S3†), the most sensitive cell lines were ER-bla, GR-bla and PR-bla, followed by AR-bla, which was equipotent to PPARγ-bla. AhR-CALUX followed and AREc32 was the least sensitive cell line for cytotoxicity. The GeneBLAzer reporter gene assays are all based on HEK293T and HEK293H cells and the assay is run in medium supplemented with only 2% fetal bovine serum (FBS). These assays were therefore expected to be more sensitive than the cell lines supplemented with 10% FBS (AhR-CALUX derived from H4IIe and AREc32 derived from MCF7 cells (Table 1)) because chemicals bind to serum proteins, which reduces their bioavailability to cells. This was essentially confirmed but the MCF7 cell line additionally seems to be intrinsically more robust than the other cell lines.
Given the consistency of cytotoxicity across cell lines and since the variability of the cytotoxicity measure is relatively high due to the nature of the endpoint, we averaged the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TUbio of all cell lines with exception of AREc32. The coefficient of variation of the mean was 1% to 10% confirming the good comparability of cytotoxicity between cell lines (Table S6†).
TUbio of all cell lines with exception of AREc32. The coefficient of variation of the mean was 1% to 10% confirming the good comparability of cytotoxicity between cell lines (Table S6†).
All samples varied in the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TUbio by a maximum of 0.5
TUbio by a maximum of 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log units (factor of 3), which indicates that food preparation had no impact on the cytotoxicity of the pollutants and bioactive endogenous compounds extractable with PDMS.
log units (factor of 3), which indicates that food preparation had no impact on the cytotoxicity of the pollutants and bioactive endogenous compounds extractable with PDMS.
3.2 Specific bioassay responses
All samples showed activation of AhR with the EC10 ranging from REF 1.1 to 4.5 glipid Lbioassay−1 (Table S4†). The % effect levels never reached 100% because cytotoxicity always kicked in before the effect reached a maximum in the CRCs (Fig. S5 and S6†). Because linear evaluation of the CRC unexpectedly did not yield a good fit, the EC10 was derived from a log–logistic fit.
The EC10 values for the activation of AhR were often very close to IC10 for cytotoxicity indicating a low specificity of the assay for the extracted mixtures. The largest specificity ratio (SR = IC10/EC10)21 of 1.9 was observed for HI-14-11 Muktuk Roast, which contained high concentrations of PAHs, that are specifically acting on the AhR, followed by HI-14-06 Muktuk Roast with a SR of 1.7. Whereas one normally would exclude any sample with a SR < 1 as “not specifically acting”, we reported EC10 for all samples since only one sample had a SR of 0.87, and four had a SR between 0.95 and 0.99. At an SR ≤ 1, the activation occurs at the same concentration as cytotoxicity, which means that the effect is not specific.
The EC10 were converted to benzo[a]pyrene equivalent concentrations (B[a]P-EQbio) because PAHs were analysed in the samples and therefore the direct comparison in iceberg modelling described below is facilitated (Table S7†).
All samples were compared with the baseline Uqsuq sample before food preparation in Fig. 3. Several preparation methods reduced the B[a]P-EQbio. B[a]P-EQbio of both specimens were significantly reduced in the Boil Drum preparation (One-way Anova, Dunnett's multiple comparison test p = 0.0176), while others showed differences between both specimens. For instance, drying (AirDry) reduced the B[a]P-EQbio, while hanging Muktuk for drying did not change the B[a]P-EQbio for H14-11 but increased it for HI-14-06.
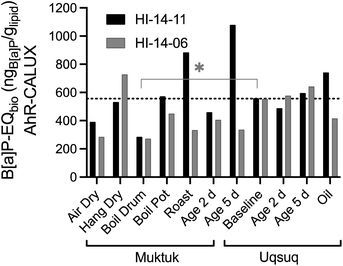 | ||
| Fig. 3 Comparison of benzo[a]pyrene equivalent concentrations B[a]P-EQbio in the AhR GeneBLAzer assay between the two animals HI-14-11 (black bars) and HI-14-06 (grey bars). Data in Table S7.† The dotted line refers to the mean of the baseline samples. | ||
Clearly, direct roasting increased the B[a]P-EQbio presumably by producing PAHs consistent with the analytical data of PAHs.3 Muktuk Age 5 Day and Uqsuq Oil of HI-14-11 were also increased over baseline which had also been observed for the total concentration of PAHs. The difference between the two specimens was explained by different setups of the food preparation with HI-14-11 aged within the smokehouse and HI-14-06 smoked far away from a smokehouse.
Although the pattern was similar to the chemical analysis of PAHs, one main difference was that the B[a]P-EQbio were already high in the samples unaffected by smoke, indicating that chemicals other than PAHs contribute to the B[a]P-EQbio. Iceberg modelling below will further investigate those contributions.
In order to facilitate comparison with data reported in the literature, we also calculated the more common TCDD-EQbio which ranged from 126 to 500 pgTCDD glipid−1. The baseline values were very similar in the two beluga whales (257 and 259 pgTCDD glipid−1 for HI-14-06 and HI-14-11, respectively). The experimental TCDD-EQbio were up to five times higher than the median TCDD-EQbio detected in Australian dugongs that ranged from 16 to 230 pgTCDD glipid−1.9 The distribution in dugongs was statistically different from the beluga baseline (Wilcoxon signed rank test, p = 0.001).
The baseline samples were again consistent for both specimens with a mean Rosiglitazone-EQ of 4.85 ngRosiglitazone glipid−1 (Fig. 4). Food preparation generally did not change or decrease the effect with the exception of the oil sample, which was higher for HI-14-11 and much lower for HI-14-06.
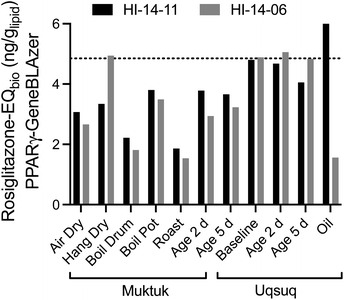 | ||
| Fig. 4 Comparison of Rosiglitazone-EQbio in the PPARγ GeneBLAzer assay between the two animals HI-14-11 (black bars) and HI-14-06 (grey bars). Data in Table S7.† The dotted line refers to the mean of the baseline samples. | ||
Since PPARγ plays a role in lipid metabolism and is also activated by long-chain alkane carboxylic acids,28 it is possible that coextracted lipids would interfere if also fatty acids were coextracted. To evaluate this possibility, the % weight gain was also plotted against the Rosiglitazone-EQ (Fig. S9†) but there was no positive association. Therefore, we deem the contribution of natural fatty acids as negligible, also because they would be fully charged and charged chemicals do not partition to PDMS.29
Five samples activated the AR-bla (Fig. S12 and S13†), namely both Muktuk samples dried on the ground (Air Dry) and also Muktuk Roast, Uqsuq Age 5 Days and Uqsuq Oil of HI-14-06. The low androgenic effects with EC10 ranging from 0.7 to 2.7 glipid Lbioassay−1 (Table S4†) were presumably not caused by chemicals in the whales, where effects were absent, but introduced by contamination during food preparation. In contrast no activation was observed in GR-bla (Fig. S14 and S15†) and PR-bla (Fig. S16 and S17†).
Environmental pollutants are known to interfere with the hormone systems of marine mammals,30 but the effects might be much more subtle than the direct binding to a hormone receptor.
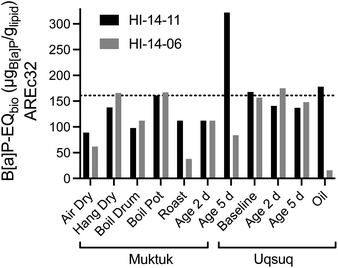 | ||
| Fig. 5 Comparison of B[a]P-EQbio in the AREc32 assay between the two animals HI-14-11 (black bars) and HI-14-06 (grey bars). Data in Table S7.† The dotted line refers to the mean of the baseline samples. | ||
Blubber, liver, kidney and brain-tissues of harbour porpoises, harbour seals, ringed seals and orcas from the North Sea and the Baltic Sea were also evaluated with AhR-CALUX, PPARγ-bla and AREc32.31,32 While effects were not converted to lipid concentrations in these studies,31,32 the direct comparison of the effect concentrations based on PDMS indicated that the mixture effect levels were very similar between the Canadian belugas and the marine mammals from European seas (Fig. S20b†).
3.3 Comparison of concentrations of analytes in lipids from direct extraction and via PDMS extraction
Ideally, chemical concentrations used for iceberg modelling should apply to the same type of in-tissue-PDMS extracts (converted into lipid-based concentrations using eqn (7)) as the extracts used in the bioassays. Most chemical analyses on the beluga blubber samples had, however, been done on direct lipid extracts.3 The analysis of a selection of chemicals (exclusively POPs) in in-tissue-PDMS extracts allows for a comparison of the final lipid-based concentrations obtained by either direct lipid extraction (Table S2†) or PDMS extraction (Table S5†). Excellent agreement between both methods (shown in Fig. S21†) justifies using the analytical data from direct extraction for the iceberg modelling. We caution that the PDMS extracts used for chemical analysis underwent an acid silica clean-up, while the bioassays were subjected to raw extract to capture persistent and non-persistent chemicals.3.4 Iceberg modelling
34 Chemicals were quantified in the TF samples but EC10 values for AhR-CALUX were available for only 14 of them (Table S6†). The hexachlorocyclohexanes, naphthalene, acenaphthylene, acenaphthene and fluorene were too volatile to be tested in a plate-based bioassays because their predicted bioassay medium-air partition constants Kmedium/air was below the threshold of 104 (Fig. S22†).19 Hexachlorobenzene was newly characterized for the present study because its concentrations were fairly high but its Kmedium/air is also close to the threshold (Fig. S22†), which was also the case for mirex, which had measured effect concentrations that are likely to be rather uncertain due to potential losses of chemical from the assay plate during the 24 h incubation at 37 °C. Of the remaining chemicals 11 were either inactive or cytotoxic before activation started or they precipitated before any activity could be determined. A comparison with the predicted baseline toxicity IC10 (Fig. S23†) showed that many of the active chemicals were close to baseline toxicity.33 Highly specific in AhR-CALUX were PCB118, chrysene and B[a]P (Fig. S23a†).These and other PAHs contributed a large fraction to B[a]P-EQchem in HI-14-11 Muktuk Roast (Fig. 6, and S24a†). In contrast, the consistently high B[a]P-EQchem in HI-14-06 stemmed rather from PCBs and PBDEs (Fig. 6, and S24b†). If Fig. 3 and 6 are superimposed, it is evident that their patterns are very similar, although the B[a]P-EQchem is very much lower than the B[a]P-EQbio.
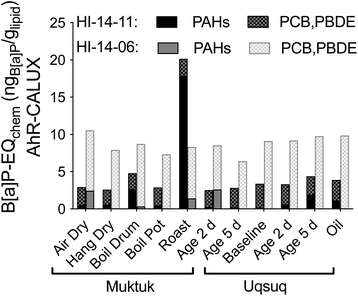 | ||
| Fig. 6 Contribution of nonpersistent PAHs and persistent PCBs and PDBEs to B[a]P-EQchem in the AhR-CALUX assay. Data in Table S7.† | ||
The B[a]P-EQchem of the detected chemicals explained >1% of the AhR-activating effect (B[a]P-EQbio) in all samples of HI-14-06, with highest percentage effect explained by HI-14-06 Muktuk Air Dry (3.7%), Muktuk Boil Large Drum (3.2%) and Muktuk Roast (2.5%), while HI-14-11 preparations had much lower explained fractions B[a]P-EQchem/B[a]P-EQbio with exception of Muktuk Boil Large Drum and Roast of HI-14-11 (Fig. S25a†). This seems a small fraction but is not implausible given that there are thousands of chemicals in complex mixtures.
In PPARγ-bla only phenanthrene, fluoranthene, PCB28 and PDBE47 were active (Table S6†) and their Rosiglitazone-EQchem explained <0.00018% of the Rosiglitazone-EQbio (Fig. S25b and Table S7†).
In AREc32, the B[a]P-EQchem of the six active chemicals, HCB, endosulfan, anthracene, fluoranthene, chrysene and B[a]P (Table S6†) explained <0.013% (Fig. S25c and Table S7†) and here the PAHs were the largest contributors to B[a]P-EQchem.
Many different PAHs have been detected in marine mammals.1 Hence it is conceivable that some of the gap between B[a]P-EQchem and B[a]P-EQbio could be closed by additional PAHs, such as alkylated and otherwise substituted PAHs. The accompanying study3 had not included polychlorinated dibenzodioxins (PCDD) and dibenzofurans (PCDF) as target analytes in the analytical methods. If PCDD and PCDFs were included in the analysis, almost 100% of the effect in AhR-CALUX were explained by PCDD/PCDFs in a previous study with extracts from dugong blubber.9 However for the activation of oxidative stress response more than 98% of the effect still remained unexplained when PCDD and PCDFs were included in iceberg modelling in dugong blubber.9
Iceberg modelling was also performed on PDMS extracts from various tissues of harbour porpoises, harbour seals, ringed seals and orcas from European seas.32 The BEQchem for the AhR-CALUX were dominated by the PCBs and BEQchem often explained more of the BEQbio than in the present study (Fig. S25a†). The picture was very similar for PPARγ-bla between the present study and ref. 32 with ≪0.1% of BEQbio explained by BEQchem. As for AhR-CALUX, the BEQchem of AREc32 were much more variable in ref. 32 but often explained a higher fraction of BEQbio than in the present study.
4. Discussion
Assessing the risk of contaminant exposure arising from the dietary intake of TFs is exceptionally challenging because it has to be weighed against the enormous social, cultural and nutritional benefits of TFs for indigenous populations. Such risk assessment relies typically on the quantification of a selection of individual contaminants in solvent extracts of TFs and the comparison of their concentrations with threshold values separating acceptable from unacceptable exposures. Limitations of this approach include that only a small fraction of the bioactive compounds present in TFs are being quantified and their ability to exert toxic effect in concert is generally not taken in account.A bioassay-based quantification of mixture effects using a PDMS-based extraction procedure as introduced here allows for a more comprehensive and unbiased chemical risk assessment of food contaminants, because it does not overlook any chemicals but accounts for the concerted action of both, persistent and nonpersistent bioactive chemicals. The iceberg modelling revealed that the contaminant concentrations in the exhaustive lipid extracts of different beluga TFs reported previously3 can only explain a tiny fraction of the observed mixture effects. The PDMS-extraction method combined with bioassays has previously been applied to tissues from other marine mammals such as dugongs,34 porpoises, whales and seals31 and also for human adipose tissue.35 However, it is applied here for the first time in the context of food in general and TFs in particular.
Bioassays have found use in dietary risk assessment previously. Specifically, the AhR-CALUX assay has been widely used to assess dioxin-like residues in food and feed for 20 years,36–38 and recently detailed recommendations have been given for the use of AhR-CALUX assays to quantify dioxins and PCBs in EU-regulated foods.4,39,40
These bioassay methods are typically only applied to exhaustive lipid extracts, from which lipids were removed through a clean-up process that is targeted to remove not only lipids but also nonpersistent organics. This is appropriate if one is only interested in dioxin-like chemicals, but bioactivity can also be introduced by nonpersistent chemicals. Our approach is unbiased with respect to the extraction of neutral organic chemicals, although the extraction efficacy of ionizable organic chemicals is admittedly limited.29
The maximum level of mixtures of PCDD/Fs and dioxin-like PCBs accepted in pork for consumption is defined as 1.25 pgWHO-PCDD/F-PCB-TEQ glipid−1 and the action levels is 0.75 pgWHO-PCDD/F-TEQ glipid−1.6 These values were derived for mixtures of 7 PCDDs, 10 PCDF and 12 planar PCBs from chemical analysis and would be equivalent to TCDD-EQchem accounting only for these 29 POPs. The TCDD-EQchem were shown to agree reasonably well with the TCDD-EQbio of POPs extracted from pork with exhaustive extraction/cleanup using different AhR-CALUX assays, among them also the cell line we used.6 Pork meat contained 0.30 to 5.29 pgWHO-PCDD/F-TEQ glipid−1, which would be 49 to 860 timed lower than the TCDD-EQbio found in the baseline beluga whale samples.
This comparison is an indication that a lot of BEQbio detected in beluga stemmed from nonpersistent organics but could partially be contributed by a higher POP level in beluga than in pork. As the food consumption thresholds are defined only for POPs, future work should also include the analysis of PCDD/Fs and should apportion the effect contribution from persistent and non-persistent mixture components.
A concern raised when using whole extract testing is that the bioassays also respond to endogenous compounds present in the extracted mixtures, thereby raising the prospect of false positives. For this reason, a certain background level of effect must be considered acceptable, although this acceptable background effect, which will be bioassay-specific, still needs to be defined. Small quantities of coextracted endogenous lipids decrease the sensitivity of the assays by lowering the bioavailability of the dosed chemicals.41 This phenomenon is well characterized and a model has been developed to account for this decrease in sensitivity.41 Applying this model, we could demonstrate that at the concentration of EC10 the fraction of coextracted lipid was too low to cause any artifacts.
Presently, no effect-based trigger (EBT) values are available differentiating between acceptable and unacceptable exposure based on bioassay-based results, that would take the place of Minimum Risk Levels (MRL) for individual chemicals and the action levels related to WHO-PCDD/F-TEQ for animal food. The first experiences with application of effect-based methods for the quantification of mixture effects in the present study will need to be expanded to eventually develop a sufficiently large database to derive EBTs.
An analogy can be drawn to in vitro bioassays in water quality assessment, where initially mainly comparative assessments were performed, e.g., to assess the treatment efficacy of a wastewater treatment plant or a drinking water plant and later the tools were also applied for surface water quality monitoring by using EBT values.15
Such comparative assessment can also be done in the present case, such as the comparison of effects seen in the extracts from Australian dugongs and Canadian Arctic belugas mentioned throughout the results section. The approach also allows for a comparison between mixture effects observed in extracts from different types of TFs and in the tissues of the two different whales. Overall, this work indicated that the observed effects are not strongly influenced by the TF preparation method, although drying near a smokehouse or roasting the Muktuk increases the AhR-activating effect. The findings thereby confirmed the chemical analysis, which noted the similar contamination levels in the different beluga TFs, but also the introduction of PAHs to roasted Muktuk and a sample aged close to a smokehouse. When comparing the two whales, we find that the BEQbio for the activation of the AhR were similar for corresponding samples from the two specimens but the effects in the older HI-14-06 was driven by classic POPs, while the effects in the younger HI-14-11 was driven by PAHs and unknown contaminants. Therefore, no single experimental approach is superior to the other, only in combination can chemical analysis and in vitro bioassays give us the full picture of environmental and food contamination.42
5. Conclusions
This pilot study clearly demonstrated the usefulness of in vitro bioassays for obtaining a full picture of contamination with environmental pollutants and for comprehensively assessing the chemical risk of TF preparations. The measurement of mixture effects is useful to complement chemical analysis and can clearly demonstrate differences between sample types and preparation methods, but at this stage the evaluation is more comparative than absolute because acceptable effect levels (EBTs) are still missing.In future work it is recommended to run the AhR-CALUX assay in duplicate, one without clean-up to evaluate the entire bioactive mixture but also one with clean-up to assess the fraction of persistent organic pollutants in the mixture. We would expect then that the PAHs are destroyed with clean-up and what remains are persistent pollutants.
Author contributions
Beate Escher: conceptualisation, methodology, formal analysis, visualization, writing – original draft. Matt Binnington: methodology and investigation, writing – review & editing. Maria König: methodology and investigation, writing – review & editing. Ying Duan Lei: methodology, Frank Wania: conceptualisation, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Lucky and James Pokiak for the provision of the beluga food samples (scientific research licence no. 2525 of the Aurora Research Institute) and the Tuktoyaktuk Hunter and Trappers Committee for the permission to perform bioassay-based investigations on those samples (licence no. 5471). We also thank Annika Jahnke, Eva Reiter and Andreas Baumer for extraction of the PDMS. The experiments were performed using the platform CITEPro (Chemicals in the Environment Profiler) funded by the Helmholtz Association with co-funding by the States of Saxony and Saxony-Anhalt. FW's research is supported through a Discovery Grant of the Natural Sciences and Engineering Research Council of Canada.References
- R. A. Lourenço, S. Taniguchi, J. da Silva, F. D. C. Gallotta and M. C. Bicego, Polycyclic aromatic hydrocarbons in marine mammals: a review and synthesis, Mar. Pollut. Bull., 2021, 171, 112699 CrossRef PubMed.
- T. J. O'Shea and S. Tanabe, in Toxicology of Marine Mammals, ed. J. G. Vos, T. J. O'Shea, M. Fournier and G. D. Bossart, CRC Press, 2002, pp. 99–134, DOI:10.1201/9780203165577.pt2.
- M. J. Binnington, Y. D. Lei, L. Pokiak, J. Pokiak, S. K. Ostertag, L. L. Loseto, H. M. Chan, L. W. Y. Yeung, H. Huang and F. Wania, Effects of preparation on nutrient and environmental contaminant levels in Arctic beluga whale (Delphinapterus leucas) traditional foods, Environ. Sci.: Processes Impacts, 2017, 19, 1000–1015 RSC.
- J. Hadrich, C. Stumpf and M. S. Denison, Bioanalytical screening for Dioxins and PCBs in EU-regulated foods: new analytical criteria adopted by the European Union in Commission Regulation (EU) 2017/644-Part 1: Introduction, Dtsch. Lebensm.-Rundsch., 2018, 114, 99–108 Search PubMed.
- G. Otarola, H. Castillo and S. Marcellini, Aryl hydrocarbon receptor-based bioassays for dioxin detection: thinking outside the box, J. Appl. Toxicol., 2018, 38, 437–449 CrossRef CAS PubMed.
- J. Haedrich, C. Stumpf and M. S. Denison, Bioanalytical screening of low levels of dioxins and dioxin-like PCBs in pig meat (pork) for checking compliance with EU maximum and action levels using highly sensitive “third generation” recombinant H4L7.5c2 rat hepatoma cells, Environ. Sci. Eur., 2021, 33, 33 CrossRef CAS PubMed.
- P. A. Neale, R. Altenburger, S. Ait-Aissa, F. Brion, W. Busch, G. de Aragão Umbuzeiro, M. S. Denison, D. Du Pasquier, K. Hilscherova, H. Hollert, D. A. Morales, J. Novac, R. Schlichting, T.-B. Seiler, H. Serra, Y. Shao, A. J. Tindall, K. E. Tollefsen, T. D. Williams and B. I. Escher, Development of a bioanalytical test battery for water quality monitoring: fingerprinting identified micropollutants and their contribution to effects in surface water, Water Res., 2017, 123, 734–750 CrossRef CAS PubMed.
- J. C. Brennan, G. He, T. Tsutsumi, J. Zhao, E. Wirth, M. H. Fulton and M. S. Denison, Development of Species-Specific Ah Receptor-Responsive Third Generation CALUX Cell Lines with Enhanced Responsiveness and Improved Detection Limits, Environ. Sci. Technol., 2015, 49, 11903–11912 CrossRef CAS PubMed.
- L. Jin, C. Gaus and B. I. Escher, Adaptive Stress Response Pathways Induced by Environmental Mixtures of Bioaccumulative Chemicals in Dugongs, Environ. Sci. Technol., 2015, 49, 6963–6973 CrossRef CAS PubMed.
- M. C. Jaramillo and D. D. Zhang, The emerging role of the Nrf2-Keap1 signaling pathway in cancer, Genes Dev., 2013, 27, 2179–2191 CrossRef CAS PubMed.
- M. Y. Song, D. Y. Lee, K. S. Chun and E. H. Kim, The Role of NRF2/KEAP1 Signaling Pathway in Cancer Metabolism, Int. J. Mol. Sci., 2021, 22(9), 4376 CrossRef CAS PubMed.
- M. Kanerva, H. Routti, Y. Tamuz, M. Nyman and M. Nikinmaa, Antioxidative defense and oxidative stress in ringed seals (Pusa hispida) from differently polluted areas, Aquat. Toxicol., 2012, 114, 67–72 CrossRef PubMed.
- R. L. Huang, M. H. Xia, M. H. Cho, S. Sakamuru, P. Shinn, K. A. Houck, D. J. Dix, R. S. Judson, K. L. Witt, R. J. Kavlock, R. R. Tice and C. P. Austin, Chemical genomics profiling of environmental chemical modulation of human nuclear receptors, Environ. Health Perspect., 2011, 119, 1142–1148 CrossRef CAS PubMed.
- L. Y. Chew, H. Zhang, J. Z. He and F. W. Yu, The Nrf2-Keap1 pathway is activated by steroid hormone signaling to govern neuronal remodeling, Cell Rep., 2021, 36(5), 109466 CrossRef CAS PubMed.
- B. Escher, P. Neale and F. Leusch, Bioanalytical Tools in Water Quality Assessment, WA Publishing, London, UK, 2nd edn, 2021, https://www.iwapublishing.com/books/9781789061970/bioanalytical-tools-water-quality-assessment-2nd-edition Search PubMed.
- M. J. Binnington, Modeling and Measuring Environmental Contaminant Exposure Among Canadian Arctic Indigenous Humans and Wildlife, Thesis, Doctor of Philosophy, University of Toronto, 2016 Search PubMed.
- A. Jahnke, P. Mayer, M. S. McLachlan, H. Wickström, D. Gilbert and M. MacLeod, Silicone passive equilibrium samplers as ‘chemometers’ in eels and sediments of a Swedish lake, Environ. Sci.: Processes Impacts, 2014, 16, 464–472 RSC.
- M. M. Krahn, D. P. Herman, G. M. Ylitalo, C. A. Sloan, D. G. Burrows, R. C. Hobbs, B. A. Mahoney, G. K. Yanagida, J. Calamokidis and S. E. Moore, Stratification of lipids, fatty acids and organochlorine contaminants in blubber of white whales and killer whales, J. Cetacean Res. Manage., 2004, 6, 175–189 CrossRef.
- B. I. Escher, L. Glauch, M. Konig, P. Mayer and R. Schlichting, Baseline Toxicity and Volatility Cutoff in Reporter Gene Assays Used for High-Throughput Screening, Chem. Res. Toxicol., 2019, 32, 1646–1655 Search PubMed.
- M. König, B. I. Escher, P. A. Neale, M. Krauss, K. Hilscherová, J. Novák, I. Teodorović, T. Schulze, S. Seidensticker, M. A. Kamal Hashmi, J. Ahlheim and W. Brack, Impact of untreated wastewater on a major European river evaluated with a combination of in vitro bioassays and chemical analysis, Environ. Pollut., 2017, 220, 1220–1230 CrossRef PubMed.
- B. I. Escher, L. Henneberger, R. Schlichting and F. C. Fischer, Cytotoxicity burst or baseline toxicity? Differentiating specific from nonspecific effects in reporter gene assays, Environ. Health Perspect., 2020, 128, 077007 CrossRef CAS PubMed.
- B. Escher, P. A. Neale and D. Villeneuve, The advantages of linear concentration-response curves for in vitro bioassays with environmental samples, Environ. Toxicol. Chem., 2018, 37, 2273–2280 CrossRef CAS PubMed.
- U. Schenker, M. MacLeod, M. Scheringer and K. Hungerbühler, Improving data quality for environmental fate models: a least-squares adjustment procedure for harmonizing physicochemical properties of organic compounds, Environ. Sci. Technol., 2005, 39, 8434–8441 CrossRef CAS PubMed.
- B. I. Escher, G. Braun and C. Zarfl, Exploring the concepts of concentration addition and independent action using a linear low-effect mixture model, Environ. Toxicol. Chem., 2020, 39, 2552–2559 CrossRef CAS PubMed.
- B. I. Escher, C. van Daele, M. Dutt, J. Y. M. Tang and R. Altenburger, Most oxidative stress response in water samples comes from unknown chemicals: the need for effect-based water quality trigger values, Environ. Sci. Technol., 2013, 47, 7002–7011 CrossRef CAS PubMed.
- P. A. Neale, G. Braun, W. Brack, E. Carmona, R. Gunold, M. Konig, M. Krauss, L. Liebmann, M. Liess, M. Link, R. B. Schafer, R. Schlichting, V. C. Schreiner, T. Schulze, P. Vormeier, O. Weisner and B. I. Escher, Assessing the Mixture Effects in In Vitro Bioassays of Chemicals Occurring in Small Agricultural Streams during Rain Events, Environ. Sci. Technol., 2020, 54, 8280–8290 CrossRef CAS PubMed.
- R. Judson, K. Houck, M. Martin, A. M. Richard, T. B. Knudsen, I. Shah, S. Little, J. Wambaugh, R. W. Setzer, P. Kothya, J. Phuong, D. Filer, D. Smith, D. Reif, D. Rotroff, N. Kleinstreuer, N. Sipes, M. H. Xia, R. L. Huang, K. Crofton and R. S. Thomas, Analysis of the Effects of Cell Stress and Cytotoxicity on In Vitro Assay Activity Across a Diverse Chemical and Assay Space, Toxicol. Sci., 2016, 152, 323–339 CrossRef CAS PubMed.
- C. D. Rummel, B. I. Escher, O. Sandblom, M. M. Plassmann, H. P. H. Arp, M. MacLeod and A. Jahnke, Effects of Leachates from UV-Weathered Microplastic in Cell-Based Bioassays, Environ. Sci. Technol., 2019, 53, 9214–9223 CrossRef CAS PubMed.
- L. Niu, L. Henneberger, J. Huchthausen, M. Krauss, A. Ogefere and B. I. Escher, pH-Dependent Partitioning of Ionizable Organic Chemicals between the Silicone Polymer Polydimethylsiloxane (PDMS) and Water, ACS Environ. Au, 2022, 2(3), 253–262 CrossRef CAS PubMed.
- M. Gregory and D. Cyr, in Toxicology of Marine Mammals, ed. J. G. Vos, T. J. O'Shea, M. Fournier and G. D. Bossart, CRC Press, 2002, pp. 67–81, DOI:10.1201/9780203165577.ch4.
- E. B. Reiter, B. I. Escher, U. Siebert and A. Jahnke, Activation of the xenobiotic metabolism and oxidative stress response by mixtures of organic pollutants extracted with in-tissue passive sampling from liver, kidney, brain and blubber of marine mammals, Environ. Int., 2022, 165, 107337 CrossRef CAS PubMed.
- E. B. Reiter, B. I. Escher, E. Rojo-Nieto, H. Nolte, U. Siebert and A. Jahnke, Characterizing the marine mammal exposome by iceberg modeling, linking chemical analysis and in vitro bioassays, Environ. Sci.: Processes Impacts, 2023 10.1039/D3EM00033H.
- J. Lee, G. Braun, L. Henneberger, M. König, R. Schlichting, S. Scholz and B. I. Escher, Critical Membrane Concentration and Mass-Balance Model to Identify Baseline Cytotoxicity of Hydrophobic and Ionizable Organic Chemicals in Mammalian Cell Lines, Chem. Res. Toxicol., 2021, 34, 2100–2109 Search PubMed.
- L. Jin, L. van Mourik, C. Gaus and B. I. Escher, Applicability of passive sampling to bioanalytical screening of bioaccumulative chemicals in marine wildlife, Environ. Sci. Technol., 2013, 47, 7982–7988 CrossRef CAS PubMed.
- A. Baumer, S. Jäsch, N. Ulrich, I. Bechmann, J. Landmann, A. Stöver and B. I. Escher, Chemical mixtures in human post-mortem tissues assessed by a combination of chemical analysis and in vitro bioassays after extraction with silicone, Environ. Int., 2021, 157, 106867 CrossRef CAS PubMed.
- M. Nording, S. Sporring, K. Wiberg, E. Björklund and P. Haglund, Monitoring dioxins in food and feedstuffs using accelerated solvent extraction with a novel integrated carbon fractionation cell in combination with a CAFLUX bioassay, Anal. Bioanal. Chem., 2005, 381, 1472–1475 CrossRef CAS PubMed.
- G. Gizzi, L. A. P. Hoogenboom, C. Von Holst, M. Rose and E. Anklam, Determination of dioxins (PCDDs/PCDFs) and PCBs in food and feed using the DR CALUX (R) bioassay: Results of an international validation study, Food Addit. Contam., 2005, 22, 472–481 CrossRef CAS PubMed.
- L. Hoogenboom, L. Goeyens, S. Carbonnelle, J. van Loco, H. Beernaert, W. Baeyens, W. Traag, T. Bovee, G. Jacobs and G. Schoeters, The CALUX bioassay: current status of its application to screening food and feed, TrAC, Trends Anal. Chem., 2006, 25, 410–420 CrossRef CAS.
- J. Haedrich, C. Stumpf and M. S. Denison, Bioanalytical screening for Dioxins and PCBs in EU-regulated foods: New analytical criteria adopted by the European Union in Commission Regulation (EU) 2017/644. Part 3: Assay criteria, validation and quality control, reporting of results, implementation, Dtsch. Lebensm.-Rundsch., 2018, 114, 215–228 Search PubMed.
- J. Haedrich, C. Stumpf and M. S. Denison, Bioanalytical screening for Dioxins and PCBs in EU-regulated foods: new analytical criteria adopted by the European Union in Commission Regulation (EU) 2017/644. Part 2: sample analysis criteria and the nature of bioanalytical results, Dtsch. Lebensm.-Rundsch., 2018, 114, 167–175 Search PubMed.
- E. B. Reiter, A. Jahnke, M. Konig, U. Siebert and B. I. Escher, Influence of Co-Dosed Lipids from Biota Extracts on the Availability of Chemicals in In Vitro Cell-Based Bioassays, Environ. Sci. Technol., 2020, 54, 4240–4247 CrossRef CAS PubMed.
- B. I. Escher, H. M. Stapleton and E. L. Schymanski, Tracking Complex Mixtures of Chemicals in our Changing Environment, Science, 2020, 397, 388–392 CrossRef PubMed.
- X. J. Wang, J. D. Hayes and C. R. Wolf, Generation of a stable antioxidant response element–driven reporter gene cell line and its use to show redox-dependent activation of Nrf2 by cancer chemotherapeutic agents, Cancer Res., 2006, 66, 10983–10994 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3em00076a |
| ‡ Present address: Mitacs, 100 College Street, Toronto, Ontario, Canada, M5G 1L5. |
| This journal is © The Royal Society of Chemistry 2023 |