The effect of metalation on antimicrobial piscidins imbedded in normal and oxidized lipid bilayers†
Received
17th March 2023
, Accepted 2nd June 2023
First published on 7th June 2023
Introduction
Antibiotic-resistant infections (ARIs) in high-risk patients contribute to significant excess healthcare costs.1 Natural or synthetic antimicrobial peptides (AMPs) have been explored to supplement the immune response.2 These promising alternatives are considered safe with a low risk of tissue accumulation and bacterial resistance.3 AMPs are typically cationic with fewer than fifty amino acids4 and classified based on their secondary structure (α-helical, β-sheet, loop, or extended).5–10 AMPs strongly bind to bacterial membranes rich in anionic phospholipids but have lower affinity to mammalian cells which have predominantly zwitterionic headgroups. AMP binding is proposed to cause cell leakage and death2,5 by altering membrane thickness, promoting phospholipid headgroup clustering, or translocating the membrane to target specific intracellular molecules or processes.5,6,11–16 Metal binding is believed to enhance antimicrobial potency.17–20 Many AMPs also have complex immunomodulatory functions and synergistic interactions with conventional antibiotics6 and several have been approved for clinical use.2,21
Piscidins, α-helical AMPs found in fish mast cells,22 efficiently kill bacteria by permeating and weakening the membrane in a concentration-dependent manner.23–29 Piscidins 1 and 3 (P1 and P3, Fig. 1)22,30 share 68% sequence identity conserving three histidine residues (His3, His4, and His11).23,27,31 An additional His at position 17 of P1 has been proposed to enhance its effectiveness against cancer and HIV.23,32–35 Piscidin's N-terminal Amino Terminal Cu(II)- and Ni(II)-binding (ATCUN) XXH motif36 in particular has a high affinity for binding metals through coordination to the terminal NH2, backbone nitrogens of residues 2 and His3, and His3 imidazole-Nδ. ATCUN-M2+ derivatives have been investigated for applications to antitumor activity,37 enzyme inhibition,38 water oxidation,39,40 nitrite reduction,41 and imaging agents.42 ATCUN-Cu2+-containing peptides often have nuclease, protease, and lipase activity which has been harnessed in protein design.43–45 The P1 and P3 ATCUN motifs differ at position 2 (Phe vs. Ile, Fig. 1B), but each binds Cu2+ and Ni2+.17–19,26,43,44 The enhanced antimicrobial activity of P1/P3:M2+ complexes has been attributed to a one-unit reduction in the overall peptide charge through deprotonation of the N-terminal ammonium group (the M2+ charge is balanced by the deprotonation of two backbone amides, Fig. 1C).17,46 The effect of metalation has been tested using dye leakage assays on synthetic lipid vesicles31 with one case resulting in a five-fold increase in permeabilization effectiveness.47
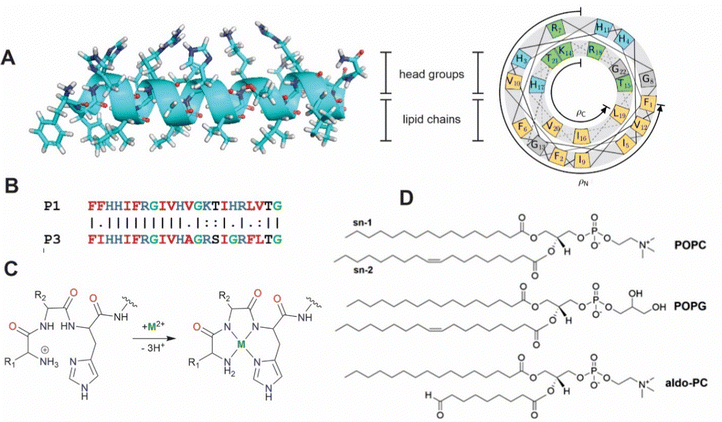 |
| | Fig. 1 (A) Secondary structure and helical wheel representation (https://helix.perrinresearch.com/wheels/) of P1. The upper polar half helix interacts with the headgroup domain with the non-polar residues of the lower half helix extending into the lipid domain. (B) Sequence alignment of P1 and P3. Basic residues = blue, nonpolar residues = red, Gly = green. (C) Metalation of the ATCUN motif. The net charge of the peptide decreases by one. (D) Phospholipids: POPC = 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, POPG = 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol, aldo-PC = 1-palmitoyl-2-(9′-oxo-nonanoyl)-sn-glycero-3-phosphocholine. | |
Previous molecular dynamics (MD) simulations of the apo peptide piscidin embedded in lipid bilayer mimics of bacterial membranes have found bilayer-dependent orientations and insertion modalities.27,31,48–52 Additionally, bilayers containing oxidized phospholipids (oxPLs) have been suggested as a novel molecular target for piscidins.53,54 The shortened acyl chains of oxPLs terminate in aldehyde or carboxylate groups and can alter the bulk properties of the membrane to increase water content and/or passive permeability.55–57 Alternatively, metalated piscidins could generate reactive oxygen species (ROS) locally to oxidize membrane lipids.38,53,55,58–61 However, this process may also protect bacteria through changes in membrane structure, biofilm formation, and motility.59–61 The behavior of piscidins in these membranes representative of conditions under oxidative stress has not yet been investigated computationally. In this study, MD simulations examine the influence of metalation and oxPLs on the structural behavior of the embedded piscidins P1 and P3 in 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 POPC/POPG and 2.6
1 POPC/POPG and 2.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.4 POPC/POPG/aldo-PC bilayers (POPC = 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, POPG = 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol; aldo-PC = 1-palmitoyl-2-(9′-oxo-nonanoyl)-sn-glycero-3-phosphocholine, Fig. 1D). The ability of P1 to form cation–π and CH–π interactions in the metalated ATCUN motif may contribute to the sequence-dependent performance of these peptides. Chain reversal observed for oxPL may contribute to loss of membrane integrity in the presence of metalated piscidins.
0.4 POPC/POPG/aldo-PC bilayers (POPC = 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, POPG = 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol; aldo-PC = 1-palmitoyl-2-(9′-oxo-nonanoyl)-sn-glycero-3-phosphocholine, Fig. 1D). The ability of P1 to form cation–π and CH–π interactions in the metalated ATCUN motif may contribute to the sequence-dependent performance of these peptides. Chain reversal observed for oxPL may contribute to loss of membrane integrity in the presence of metalated piscidins.
Computational methods
MD simulations were performed on apo and Ni2+-bound P1 and P3 in 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 POPC/POPG and 2.6
1 POPC/POPG and 2.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.4 POPC/POPG/aldo-PC lipid bilayers with a ratio of one peptide per 40 lipids in each bilayer leaflet (P/L = 1
0.4 POPC/POPG/aldo-PC lipid bilayers with a ratio of one peptide per 40 lipids in each bilayer leaflet (P/L = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40) using AMBER18.62–64 Model design follows that of Perrin et al. using CHARMM.27 Simulations of apo peptide-containing bilayers provided a benchmark for AMBER simulations with metalated peptides. The 10% aldo-PC mixture has been suggested to be physiologically relevant under conditions of oxidative stress.65,66 Simulations of peptide-free bilayers were performed to provide a reference for peptide-induced changes to the membrane. Peptide-bilayer systems were assembled using the Membrane Builder module within the CHARMM-GUI interface.67,68 Initial structures were generated as previously described for free piscidins.27 The center of mass (COM) of the peptide backbone atoms were aligned ∼20 Å above and below the center of the bilayer. Each peptide was rotated along its helical axis to face the hydrophobic residues toward the bilayer interior. Systems were solvated with 16 Å water layers above each leaflet (∼56 waters/lipid), 15–20 sodium ions, and 0–3 chloride ions to neutralize the models and add a slight salt concentration. Simulations were performed using the ff14SB69 and Lipid17 force fields.64 The aldo-PC force field was derived using the Antechamber module within AMBER18. ATCUN-Ni2+ parameters were generated using the Python-based Metal Binding Protein Builder (MCPB) module.70 Peptides were modeled with neutral His side chains based upon previously reported pKa values.31 Solvent and counterions used the TIP3P water model71 and Joung–Cheatham monovalent ion parameters.72
40) using AMBER18.62–64 Model design follows that of Perrin et al. using CHARMM.27 Simulations of apo peptide-containing bilayers provided a benchmark for AMBER simulations with metalated peptides. The 10% aldo-PC mixture has been suggested to be physiologically relevant under conditions of oxidative stress.65,66 Simulations of peptide-free bilayers were performed to provide a reference for peptide-induced changes to the membrane. Peptide-bilayer systems were assembled using the Membrane Builder module within the CHARMM-GUI interface.67,68 Initial structures were generated as previously described for free piscidins.27 The center of mass (COM) of the peptide backbone atoms were aligned ∼20 Å above and below the center of the bilayer. Each peptide was rotated along its helical axis to face the hydrophobic residues toward the bilayer interior. Systems were solvated with 16 Å water layers above each leaflet (∼56 waters/lipid), 15–20 sodium ions, and 0–3 chloride ions to neutralize the models and add a slight salt concentration. Simulations were performed using the ff14SB69 and Lipid17 force fields.64 The aldo-PC force field was derived using the Antechamber module within AMBER18. ATCUN-Ni2+ parameters were generated using the Python-based Metal Binding Protein Builder (MCPB) module.70 Peptides were modeled with neutral His side chains based upon previously reported pKa values.31 Solvent and counterions used the TIP3P water model71 and Joung–Cheatham monovalent ion parameters.72
Models were prepared for production simulations by initial minimization followed by heating from 0 to 313 K using the Langevin thermostat in four stages over 150 ps. Constant volume and weak restraints (force constants of 100 and 10 kcal mol−1 Å−2 on peptides and lipids, respectively) were applied during the first stage (0 K to 100 K). The remainder of the heating phases (from 100 K to 200 K (2), 200 K to 250 K (3), and 250 K to 313 K (4)) were conducted under constant pressure using semi-isotropic Berendsen regulation with a relaxation time of 2 ps. Production runs were performed at constant pressure (1 atm) and temperature (313 K) in a monoclinic periodic box with zero surface tension using the PMEMD routines.73–75 SHAKE constraints were applied to bonds to hydrogen atoms.76,77 Electrostatic interactions were treated with the particle mesh Ewald method (cutoff = 10 Å). Since the timescale of peptide-induced defect formation can be from microseconds to minutes,78 the MD simulations were performed for 2 μs with a 1 fs time step after 100 ns of equilibration.
Trajectory analysis was performed using Python scripts and the AmberTools CPPTRAJ routines.79 Area per lipid (AL), volume per lipid (VL), and isothermal compressibility modulus (KA) were averaged over duplicate 2.0 μs simulations (Tables 1 and 3). AL, the average area each phospholipid occupies in the x- and y-plane (eqn (1)), was computed from the dimensions of the periodic box (Lx and Ly, respectively) and the number of lipids per leaflet (mlipid).80
| | 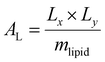 | (1) |
Computational
AL values were compared to experimental data which can vary depending upon the measurement technique.
VL was calculated (
eqn (2)) from the simulation box volume (
Vbox), the number of waters (
nw), the volume of a water molecule (
Vw for TIP3P water = 30.53 Å
3),
81 and the total number of lipids (
nlipid).
80| | 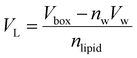 | (2) |
Peptide-induced changes to the fluidity of the bilayer were measured using
KA (
eqn (3)) where
kB is Boltzmann's constant,
T is temperature, 〈
AL〉 is the average
AL, and
σA2 is the variance in
AL.
80| | 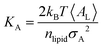 | (3) |
Average electron density profiles for all systems were decomposed into contributions from the following groups: phosphate (PO
4), methylenes (CH
2), terminal methyls (CH
3), and peptide backbone (pepBB) using the density command in CPPTRAJ. The headgroup-to-headgroup (
h(P–P)) and hydrophobic (
h(C2–C2)) bilayer thicknesses were determined as the average of the distances between the centers of mass of the phosphate phosphorus and the carbon of the C2 methylene groups from the
sn-2 chain of each leaflet, respectively. Deuterium order parameters (
SCD,
eqn (4))
82 of the lipid chain carbons reflect the chain mobility at single carbon positions as a measure of the orientation of C–D bonds relative to the
z-axis.
θ is the angle between the C–H vector and bilayer normal (
z-axis) from the simulations, such that an
SCD value of 1 infers a parallel position and 0 suggests total arbitrary motion.
| | 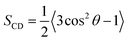 | (4) |
Peptide tilt (
τ) and azimuthal rotational (
ρ) angles (
Fig. 1A) were determined at 10 ns intervals relative to a reference structure consisting of α-helix
φ/
ψ angles of −61°/−45° and Euler angles of zero. Peptide insertion depths (
z) were calculated as the distance between the COM of the peptide backbone heavy atoms and the plane of the phosphorus atoms of the lipid headgroups.
τ,
ρ and
z were calculated separately for the N-terminal (residues 5–10) and C-terminal (residues 14–20) ends due to fraying of terminal residues, metalation of the N-terminus, and previously reported kinking at a flexible helix turn that includes G13.
27
Table 1 Average structural properties of the POPC/POPG bilayer model (VL = volume per lipid, AL = area per lipid, KA = isothermal area expansion modulus, h(P–P) = headgroup-to-headgroup thickness, h(C2–C2) = hydrophobic thickness)
| POPC/POPG |
V
L (Å3) |
A
L (Å2) |
K
A (mN m−1) |
h(P–P) (Å) |
h(C2–C2) (Å) |
|
The experimental value for pure POPC at 298 K.83
The experimental value for pure POPC at 303 K.84
The experimental value for pure POPC at 313 K.85
|
| Exp |
1256
|
64.3b, 65.3
, 68.3b |
180–330a |
37.0
|
|
|
|
1265
|
67.1 ± 0.4 |
|
36.0c |
|
|
|
1206 ± 0 |
|
|
37.5 ± 0.2 |
27.4 ± 0.2 |
| MD |
|
|
254 ± 114 |
39.0 ± 1.0
|
28.9 ± 0.7
|
| +P1 |
1290 ± 0 |
72.1 ± 0.5 |
208 ± 87 |
37.3 ± 0.4 |
27.4 ± 0.4 |
|
|
|
|
332 ± 42
|
37.4 ± 1.2
|
28.0 ± 1.2
|
| +P3 |
1287 ± 1 |
71.8 ± 0.6 |
218 ± 120 |
37.2 ± 0.3 |
27.3 ± 0.4 |
|
|
|
|
248 ± 22
|
37.0 ± 1.1
|
27.5 ± 1.1
|
| +P1:Ni |
1290 ± 0 |
71.9 ± 0.4 |
202 ± 98 |
37.4 ± 0.3 |
27.5 ± 0.3 |
| +P3:Ni |
1286 ± 0 |
71.4 ± 0.4 |
250 ± 98 |
37.5 ± 0.3 |
27.6 ± 0.3 |
Results and discussion
MD simulations of apo piscidins in POPC/POPG bilayers
MD simulations of peptide-free, P1-, and P3-bound 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
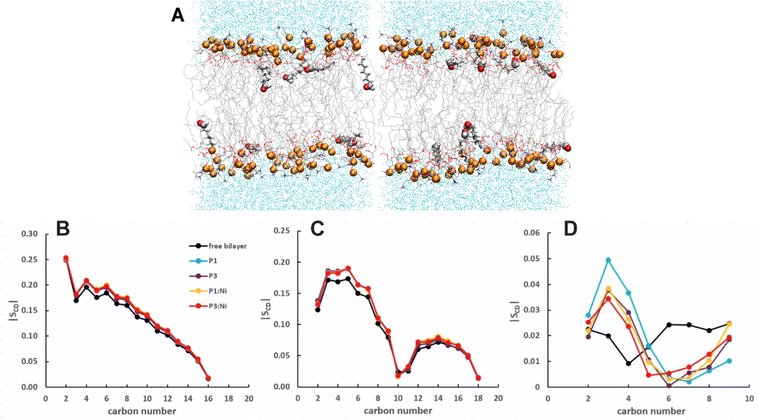
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 POPC/POPG bilayers performed with AMBER are generally consistent with previous models in CHARMM.48 Embedded P1 and P3 maintain their α-helical structure throughout simulations with the average orientation of their N-termini canted slightly toward the headgroups (τN ≈ 95°) and the C-terminal sequences nearly parallel to the surface (τC ≈ 89°/88°, Fig. 2 and S1 (ESI†), Table 2).27 The peptide backbones for P1 and P3 float on average ∼7–8 Å below the P atoms, just under the C2 atoms of the lipid sn-2 acyl chain (Fig. 3 and Table 2). P1 inserts slightly deeper in the bilayer versus P3 but both experience similar differences in the zN and zC insertion depth values (Δz ∼ 0.1 Å). P3 is slightly more flexible than P1 based on the backbone heavy-atom RMSD of the first 20 residues (0.98 Å vs. 0.80 Å, respectively).
1 POPC/POPG bilayers performed with AMBER are generally consistent with previous models in CHARMM.48 Embedded P1 and P3 maintain their α-helical structure throughout simulations with the average orientation of their N-termini canted slightly toward the headgroups (τN ≈ 95°) and the C-terminal sequences nearly parallel to the surface (τC ≈ 89°/88°, Fig. 2 and S1 (ESI†), Table 2).27 The peptide backbones for P1 and P3 float on average ∼7–8 Å below the P atoms, just under the C2 atoms of the lipid sn-2 acyl chain (Fig. 3 and Table 2). P1 inserts slightly deeper in the bilayer versus P3 but both experience similar differences in the zN and zC insertion depth values (Δz ∼ 0.1 Å). P3 is slightly more flexible than P1 based on the backbone heavy-atom RMSD of the first 20 residues (0.98 Å vs. 0.80 Å, respectively).
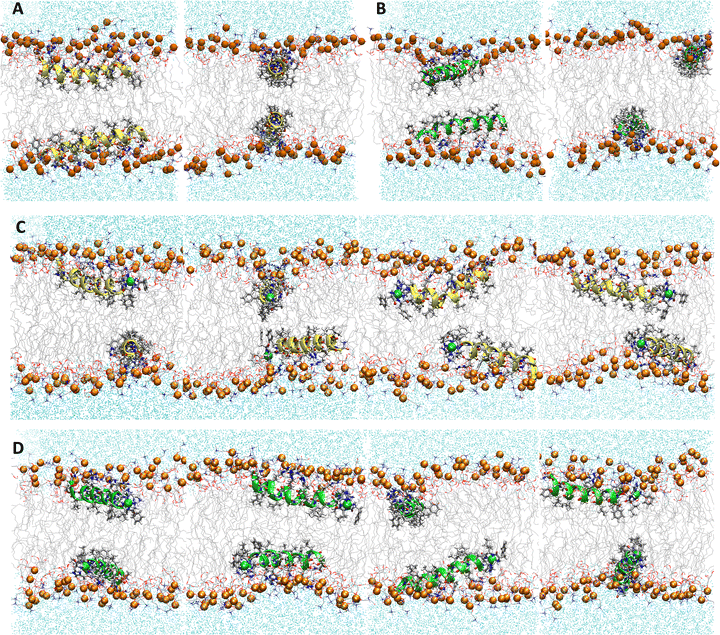 |
| | Fig. 2 Snapshots from MD simulations of (A) P1, (B) P3, (C) P1:Ni, and (D) P3:Ni in the POPC/POPG bilayer model. Peptides are generally localized at the headgroup/lipid sublayer interface and do not interact with one another. Peptides are represented as yellow (P1 isoforms) or green (P3 isoforms) ribbons and licorice. Phosphorus atoms are depicted as orange spheres while the rest of phospholipids atoms except hydrogen are depicted as lines colored as follows: carbon – gray, oxygen – red, and nitrogen – blue. Water molecules are represented as cyan points. | |
Table 2 Average tilt angles-τ, azimuthal rotation angles ρ (Fig. 1A), and depths of insertion z for the backbone atoms of N-(residues 5–10), C-(residues 14–20) helical segments, and the full peptide in POPC/POPG bilayer models
|
|
τ
N, ° |
τ
C, ° |
Δτ = (τN − τC), ° |
| P1 |
95 ± 1 |
88 ± 1 |
7.5 ± 1 |
| P3 |
95 ± 1 |
89 ± 1 |
6.1 ± 0 |
| P1:Ni |
90 ± 1 |
83 ± 1 |
7.0 ± 1 |
| P3:Ni |
86 ± 2 |
82 ± 2 |
4.0 ± 1 |
|
|
ρ
N, ° |
ρ
C, ° |
Δρ = (ρN – ρC), ° |
| P1 |
261 ± 2 |
241 ± 1 |
19.5 ± 1 |
| P3 |
248 ± 1 |
233 ± 2 |
15.3 ± 0 |
| P1:Ni |
251 ± 1 |
235 ± 1 |
16.0 ± 1 |
| P3:Ni |
237 ± 1 |
224 ± 1 |
12.7 ± 1 |
|
|
z
N, Å |
z
C, Å |
Δz = (zN − zC), Å |
z
pep, Å |
| P1 |
−8.2 ± 0.2 |
−8.3 ± 0.3 |
0.1 ± 0.2 |
−7.8 ± 0.3 |
| P3 |
−7.4 ± 0.1 |
−7.5 ± 0.3 |
0.1 ± 0.2 |
−7.0 ± 0.2 |
| P1:Ni |
−9.1 ± 0.2 |
−8.0 ± 0.3 |
−1.2 ± 0.1 |
−8.2 ± 0.2 |
| P3:Ni |
−9.1 ± 0.4 |
−7.1 ± 0.2 |
−2.0 ± 0.5 |
−7.9 ± 0.2 |
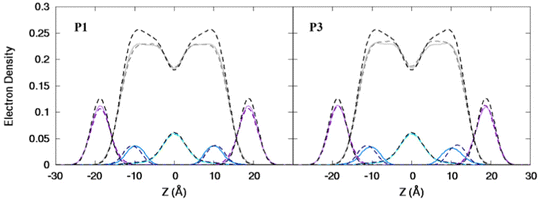 |
| | Fig. 3 Electron density profiles for P1 and P3 in the 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 POPC/POPG bilayer. Profiles are shown for the phosphates (purple), peptide backbones (blue), CH2 groups (gray), and terminal CH3 (cyan). Electron density profiles for apo or metalated piscidin-bound bilayer are in dashed or solid lines, respectively, whereas the bilayer-only systems are in dashed black lines. 1 POPC/POPG bilayer. Profiles are shown for the phosphates (purple), peptide backbones (blue), CH2 groups (gray), and terminal CH3 (cyan). Electron density profiles for apo or metalated piscidin-bound bilayer are in dashed or solid lines, respectively, whereas the bilayer-only systems are in dashed black lines. | |
Attractive interactions with the PO4 groups may contribute to the asymmetric widening of their density peaks found in both the current and previous simulations (Fig. 3).48AL, VL, and KA values indicate that the free bilayer is equilibrated in the liquid phase and consistent with experimental measurements of a single component POPC bilayer (Table 1).83–85 Interactions of the peptides with the phospholipid headgroups and upper chain regions induce increases in AL and VL as well as greater bilayer fluidity as measured by KA, which contrasts with previous results that found that KA decreased in the presence of P3 but increased upon addition of P1.48 Looser packing of aliphatic lipid chains (lower KA) was also observed in the MD simulations of fentanyl in a 1,2-dioleoyl-sn-glycero-phosphocholine (DOPC) bilayer.81 P1 and P3 display similar ρC angles with a smaller ρN for the latter (248° vs. 261°, Table 2), indicating that the P3 N-terminal sequence distributes its hydrophobic and hydrophilic residues more unevenly in the bilayer. Similar results have been reported for P1 and P3 in DMPC/DMPG and POPE/POPG bilayers (Δτ ∼ 6–8° and Δρ ∼ 15–20°).27 The differential rotation of the two piscidin segments maximizes hydrophobic interactions with the nonpolar half helix settled into the lipid sublayer. Residues of the polar half helix are solvent-exposed through a gap in the headgroup sublayer with the sidechains of Lys and Arg extended to salt bridge with anionic phosphates. Water molecules penetrate into the headgroup sublayer but are rarely found below the peptide within the nonpolar region of the bilayer. Examples of “funneling” as observed in previous studies31 were short-lived and not accompanied by movement of water through the bilayer, although frames with single water molecules in the nonpolar domain could be occasionally observed.
The apo piscidins induce a slight thinning of the bilayer (< 0.3 Å) as calculated by the h(P–P) distances with little effect on the h(C2–C2) thickness (Table 1). In contrast, the h(C2–C2) values for P1 and P3 decrease by 0.9 Å and 1.4 Å, respectively, in CHARMM simulations48 where the standard errors (±1.1–1.2 Å) suggest that piscidins cause large variations in the hydrophobic thickness of the membrane. However, the free bilayer in our AMBER simulations is on average ∼1.5 Å thinner than those modeled with the CHARMM force fields (i.e., h(P–P) ∼37.5 Å and ∼39.0 Å, respectively, Table 1).48 This difference in results could be attributed to short simulation times in CHARMM (100 ns) and/or a force field dependence on the results of membrane-bound AMP simulations.86 In either case, the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 model with the placement of peptides on both sides of the leaflet may be too small and symmetric to observe bulk distortions that could be attributed to membrane disruption. Therefore, in the following discussion, we instead focus on the local effects of metalation on the peptide and bilayer.
40 model with the placement of peptides on both sides of the leaflet may be too small and symmetric to observe bulk distortions that could be attributed to membrane disruption. Therefore, in the following discussion, we instead focus on the local effects of metalation on the peptide and bilayer.
MD simulations of metalated piscidins in POPC/POPG bilayers
Nickelated P1 and P3 embed into the 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 POPC/POPG bilayer similarly to the apo peptides maintaining their highly helical structure (RMSD = 1.07 Å and 1.13 Å, respectively) with similar h(P–P) and h(C2–C2) thickness to the apo peptide simulations (Table 1). The metalated ATCUN motif is less dynamic (RMSD = 0.19 Å and 0.22 Å, respectively, Table 2) than the apo piscidin N-termini due to the rigid Ni2+ coordination sphere. A decrease in KA for P1:Ni suggests an increase in membrane fluidity, where P3:Ni shows little change compared to the bilayer alone. Charge neutralization of the ATCUN-Ni2+ motif contributes to deeper embedding of the N-termini relative to the apo peptide by ∼0.9 Å for P1:Ni and ∼1.7 Å for P3:Ni (|zN − zC|, 1–2 Å (metalated) vs. 0.1 Å (apo), Table 2 and Fig. S1, ESI†). The mean peptide distance from the bilayer center increases as P1:Ni < P3:Ni < P1 < P3 with P1:Ni displaying a density maximum at ∼9.9 Å. Similar bilayer behavior is also observed upon cholesterol incorporation into the membrane.87,88 The N-terminus is more parallel to the bilayer surface for P1:Ni and slightly canted toward the bilayer center (τN = 86°) in P3:Ni with the C-termini tilted toward the bilayer core (Table 2). Nickelated P1/P3 adopt ρN smaller values by 10° and 11°, respectively, relative to the apo peptides, with Δρ (ρN − ρC) lower by only ∼3.5° upon metal-binding for the following trend in Δρ: P1 > P1:Ni ≈ P3 > P3:Ni (Table 2). These distribution angles allow better positioning of Phe1 and Phe2/Ile2 for interaction with the lipid hydrophobic core and of His3 with the water-bilayer interface. However, attractions between the hydrophilic residues of the polar half helix and the solvent and headgroups prevent a lower depth of insertion. This enhanced N-terminal insertion may contribute to the higher toxicity of metalated piscidins toward planktonic bacteria.26 The SCD order parameter of the piscidin-embedded bilayers (Fig. 4) indicate increased order in the acyl sn-1 and sn-2 chains nearest the headgroups. This restricted motility induced by the peptides decreases along the chains but does not discriminate between isoforms (P1 or P3) or metalation state (apo or Ni2+-bound).
1 POPC/POPG bilayer similarly to the apo peptides maintaining their highly helical structure (RMSD = 1.07 Å and 1.13 Å, respectively) with similar h(P–P) and h(C2–C2) thickness to the apo peptide simulations (Table 1). The metalated ATCUN motif is less dynamic (RMSD = 0.19 Å and 0.22 Å, respectively, Table 2) than the apo piscidin N-termini due to the rigid Ni2+ coordination sphere. A decrease in KA for P1:Ni suggests an increase in membrane fluidity, where P3:Ni shows little change compared to the bilayer alone. Charge neutralization of the ATCUN-Ni2+ motif contributes to deeper embedding of the N-termini relative to the apo peptide by ∼0.9 Å for P1:Ni and ∼1.7 Å for P3:Ni (|zN − zC|, 1–2 Å (metalated) vs. 0.1 Å (apo), Table 2 and Fig. S1, ESI†). The mean peptide distance from the bilayer center increases as P1:Ni < P3:Ni < P1 < P3 with P1:Ni displaying a density maximum at ∼9.9 Å. Similar bilayer behavior is also observed upon cholesterol incorporation into the membrane.87,88 The N-terminus is more parallel to the bilayer surface for P1:Ni and slightly canted toward the bilayer center (τN = 86°) in P3:Ni with the C-termini tilted toward the bilayer core (Table 2). Nickelated P1/P3 adopt ρN smaller values by 10° and 11°, respectively, relative to the apo peptides, with Δρ (ρN − ρC) lower by only ∼3.5° upon metal-binding for the following trend in Δρ: P1 > P1:Ni ≈ P3 > P3:Ni (Table 2). These distribution angles allow better positioning of Phe1 and Phe2/Ile2 for interaction with the lipid hydrophobic core and of His3 with the water-bilayer interface. However, attractions between the hydrophilic residues of the polar half helix and the solvent and headgroups prevent a lower depth of insertion. This enhanced N-terminal insertion may contribute to the higher toxicity of metalated piscidins toward planktonic bacteria.26 The SCD order parameter of the piscidin-embedded bilayers (Fig. 4) indicate increased order in the acyl sn-1 and sn-2 chains nearest the headgroups. This restricted motility induced by the peptides decreases along the chains but does not discriminate between isoforms (P1 or P3) or metalation state (apo or Ni2+-bound).
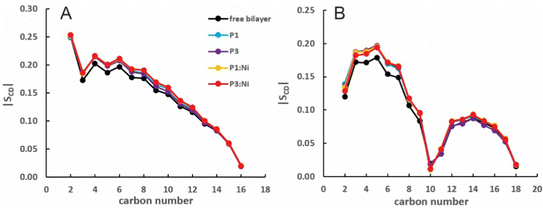 |
| | Fig. 4 Average |SCD| for the (A) sn-1 and (B) sn-2 lipid acyl chains for the free 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 POPC/POPG bilayer and in the presence of P1 (cyan), P3 (purple), P1 1 POPC/POPG bilayer and in the presence of P1 (cyan), P3 (purple), P1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ni (orange), and P3 Ni (orange), and P3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ni (red). Ni (red). | |
MD simulations of free POPC/POPG/aldo-PC bilayers
The 2.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.4 POPC/POPG/aldo-PC bilayers are thinner by ∼1 Å (both h(P–P) and h(C2–C2)) for a smaller VL (1183 Å3) (compare Tables 1 and 3) relative to POPC/POPG due to the shortened oxPL chains. AL is similar to the 3
0.4 POPC/POPG/aldo-PC bilayers are thinner by ∼1 Å (both h(P–P) and h(C2–C2)) for a smaller VL (1183 Å3) (compare Tables 1 and 3) relative to POPC/POPG due to the shortened oxPL chains. AL is similar to the 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 POPC/POPG bilayer with a lower KA (243 ± 88 vs. 254 ± 114), consistent with greater fluidity due to the 4 short aldo-PC chains per leaflet. A more fluid membrane (lower KA and SCD) containing kinked lipid tails (smaller SCD) is consistent with the potential for oxPLs to disrupt membrane bulk proprieties for enhanced permeabilization.55–57 The oxPL chains can occasionally be found oriented parallel to the headgroup region to allow the aldehyde tails to interact with the more polar headgroups (Fig. 5A). While these chain reversals may contribute to membrane instability, they did not disrupt bilayers in simulations of up to 25% oxPL.54
1 POPC/POPG bilayer with a lower KA (243 ± 88 vs. 254 ± 114), consistent with greater fluidity due to the 4 short aldo-PC chains per leaflet. A more fluid membrane (lower KA and SCD) containing kinked lipid tails (smaller SCD) is consistent with the potential for oxPLs to disrupt membrane bulk proprieties for enhanced permeabilization.55–57 The oxPL chains can occasionally be found oriented parallel to the headgroup region to allow the aldehyde tails to interact with the more polar headgroups (Fig. 5A). While these chain reversals may contribute to membrane instability, they did not disrupt bilayers in simulations of up to 25% oxPL.54
Table 3 Average structural properties of the POPC/POPG/aldo-PC bilayer model (VL = volume per lipid, AL = area per lipid, KA = isothermal area expansion modulus, h(P–P) = headgroup-to-headgroup thickness, h(C2–C2) = hydrophobic thickness)
|
|
V
L (Å3) |
A
L (Å2) |
K
A (mN m−1) |
h(P–P) (Å) |
h(C2–C2) (Å) |
|
The experimental value for pure POPC at 303 K.84
The experimental value for pure POPC at 313 K.85
|
| Exp |
1256
|
|
|
37.0
|
|
|
1265
|
64.3a, 65.3
68.3
|
180–330a |
36.0b |
|
| MD |
1183 ± 0 |
67.7 ± 0.6 |
243 ± 88 |
36.4 ± 0.2 |
26.3 ± 0.3 |
| +P1 |
1267 ± 0 |
72.5 ± 0.5 |
185 ± 81 |
36.3 ± 0.3 |
26.3 ± 0.5 |
| +P3 |
1264 ± 0 |
72.1 ± 0.5 |
198 ± 95 |
36.3 ± 0.4 |
26.4 ± 0.5 |
| +P1:Ni |
1267 ± 0 |
72.2 ± 0.4 |
222 ± 93 |
36.5 ± 0.3 |
26.6 ± 0.3 |
| + P3:Ni |
1264 ± 0 |
71.9 ± 0.5 |
212 ± 86 |
36.5 ± 0.3 |
26.5 ± 0.4 |
 |
| | Fig. 5 Snapshots from MD simulations of POPC/POPG/aldo-PC illustrating that oxPL chains can organize parallel to the headgroup region. Aldo sn-2 acyl chains are represented as licorice and the carbonyl group as spheres. Average |SCD| for the (B) sn-1, (C) sn-2 lipid, and (D) oxPL acyl chains for the free POPC/POPG/aldo-PC bilayer and in the presence of P1 (cyan), P3 (purple), P1:Ni (orange), and P3:Ni (red). | |
MD simulations of apo- and metalated piscidins in POPC/POPG/aldo-PC bilayers
The α-helices of the apo and metalated piscidins settle below the phosphate plane of the aldo-PC-containing bilayer (RMSD = 0.75/0.78 Å (P1/P1:Ni) and 0.83/0.89 Å (P3/P3:Ni)) with low RMSD values for the ATCUN residues (0.20 Å (P1:Ni) and 0.22 Å (P3:Ni)) (Fig. 6). The apo piscidins orient their N-termini toward the headgroups (τ = 95°/93) with ρ values of 259° (P1) and 249° (P3) (Table 4). The increased AL for the apo piscidin-embedded bilayer, with a slightly more pronounced expansion for the P1 isoform (Table 3), could be associated with a more dynamic diffusion of the peptide molecules through the bilayer. VL is slightly smaller relative to the POPC/POPG bilayers due to the shortened oxPL chain (Tables 1 and 3). The embedded peptides disrupt phospholipid packing for a ∼9–24% drop in fluidity as measured by KA (Table 3) with apo peptides lower than the metalated piscidins and P1:Ni having the lowest decrease in KA relative to the POPC/POPG bilayer. While the average number of hydrogen bonds between the PC and PG headgroups is generally lower in the presence of piscidin, P1:Ni (and P3:Ni in POPC/POPG) induced only a slight decrease in hydrogen bonding (Fig. S3, ESI†).
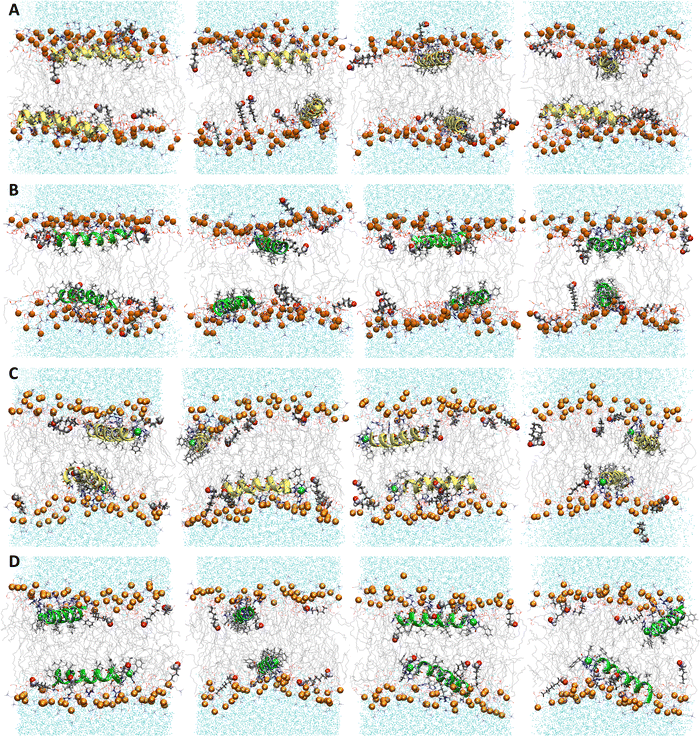 |
| | Fig. 6 Snapshots from MD simulations of (A) P1, (B) P3, (C) P1:Ni and (D) P3:Ni in POPC/POPG/aldo-PC. Peptides are generally localized at the headgroup/lipid sublayer interface and do not interact with one another. Peptides are represented as yellow (P1 isoforms) or green (P3 isoforms) ribbons and licorice. Ni2+ ions and phosphorous atoms are depicted as green and orange spheres, respectively, while the rest of the phospholipid atoms except for hydrogen are depicted as lines colored as follows: carbon – gray, oxygen – red, and nitrogen – blue. Aldo sn-2 acyl chains are represented as licorice, and water molecules as cyan points. | |
Table 4 Average tilt angles τ, azimuthal rotation angles ρ (Fig. 1A), and depths of insertion z for the backbone atoms of N-(residues 5–10), C-(residues 14–20) helical segments, and the full peptide in POPC/POPG/aldo-PC bilayer models
|
|
τ
N, ° |
τ
C, ° |
Δτ = (τN – τC), ° |
| P1 |
95 ± 1 |
88 ± 1 |
7.6 ± 1 |
| P3 |
93 ± 0 |
89 ± 1 |
4.5 ± 1 |
| P1:Ni |
90 ± 1 |
83 ± 1 |
6.6 ± 1 |
| P3:Ni |
85 ± 1 |
83 ± 1 |
2.1 ± 1 |
|
|
ρ
N, ° |
ρ
C, ° |
Δρ = (ρN − ρC), ° |
| P1 |
259 ± 2 |
240 ± 2 |
19.0 ± 1 |
| P3 |
249 ± 3 |
228 ± 4 |
21.0 ± 2 |
| P1:Ni |
251 ± 1 |
234 ± 2 |
16.3 ± 1 |
| P3:Ni |
232 ± 4 |
216 ± 2 |
16.0 ± 6 |
|
|
z
N, Å |
z
C, Å |
Δz = (zN − zC), Å |
z
pep, Å |
| P1 |
−8.1 ± 0.2 |
−8.2 ± 0.4 |
0.2 ± 0.2 |
−7.7 ± 0.2 |
| P3 |
−7.6 ± 0.1 |
−7.6 ± 0.1 |
0.0 ± 0.0 |
−7.2 ± 0.1 |
| P1:Ni |
−9.2 ± 0.3 |
−8.0 ± 0.3 |
−1.2 ± 0.1 |
−8.4 ± 0.3 |
| P3:Ni |
−8.7 ± 0.2 |
−6.9 ± 0.2 |
−1.8 ± 0.2 |
−7.6 ± 0.1 |
The N-terminus of P1:Ni generally orients parallel to the headgroups with P3:Ni slightly canted toward the bilayer core (τN = 85°) for (Table 4 and Fig. S2, ESI†). The C-termini tilt toward the bilayer center for the Ni2+-bound piscidins or parallel to the surface for the apo piscidins. The P1 rotation angles ρ are affected by metal binding as P1:Ni has similar values in both lipid mixtures. The P3:Ni ρ values are more responsive to metalation and oxPL as ρN and ρC decrease by 5° and 8°, respectively, and sample a broader range (Tables 2 and 4, and Fig. S2, ESI†) for a more uneven distribution of the C-terminal hydrophobic and hydrophilic residues in the bilayer. P3 is the only isoform where Δρ is appreciably different in the presence of aldo-PC (+5.7° and +3.3° for the apo and Ni2+-bound piscidin, respectively) with the following Δρ trend for the studied peptides: P3 > P1 > P1:Ni ≈ P3:Ni.
The peptides are located at similar depths to the POPC/POPG bilayer (Table 4) and sample a relatively narrow zN range. P1 inserts embeds ∼0.5 Å deeper into the oxPL membrane than P3, similar to the behavior in POPC/POPG bilayer reported in this study and previously by Perrin et al.48 (Table 4). Metalation increases the insertion of the N-termini by ∼1.1 Å such that zN of P1:Ni settles deeper in the bilayer than P3:Ni (zN ∼ 9.2 Å vs. 8.7 Å). Meanwhile, the peptides’ C-termini, particularly of P3:Ni, insert less with smaller |Δz| than their apo counterparts. The incorporation of oxPL into the bilayers does not significantly affect the electron density profiles relative to the POPC/POPG systems (Fig. 7) except for a larger decrease in the PO4 density peak in the POPC/POPG/aldo-PC bilayers. The maximum density of P1:Ni places it closest to the bilayer center (∼9.3 Å vs. ∼9.9 Å in POPC/POPG). The h(P–P) and h(C2–C2) bilayer thicknesses are similar to those observed for POPC/POPG membranes.48 Where the oxPL-containing bilayer is slightly thinner with embedded apo piscidins based on the h(P–P) distance, metalation leads to a slight thickening in C2–C2 because deeper insertion reverses the contraction of the bilayer due to the lower volume of oxPL (Table 3). The trends in the |SCD| of the sn-1 and sn-2 chains (Fig. 5) are similar to the POPC/POPG bilayer results and not significantly influenced by the presence of the aldo-functionalized sn-2 tails. These shortened chains occasionally snorkel toward the water layer to interact with the positively charged trimethylamino groups of the headgroup domain when piscidins are embedded (i.e., Fig. 5B–D) as indicated by the change in pattern of |SCD| relative to the free bilayer.
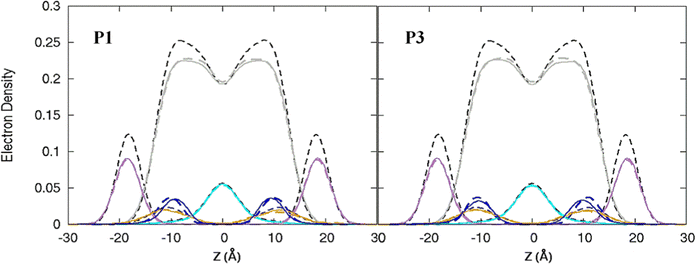 |
| | Fig. 7 Electron density profiles for P1 and P3 in POPC/POPG/aldo-PC. Profiles are shown for the phosphates (purple), peptides (blue), CH2 acyl chains (gray), and terminal CH3 (cyan). Electron density profiles containing membrane and apo or metalated piscidin are in dashed or solid lines, respectively, whereas the systems with only the membrane are in dashed black lines. | |
The PG headgroup region responds more strongly to P1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ni in the presence of aldo-PC and at the 1
Ni in the presence of aldo-PC and at the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 peptide-to-lipid ratio in 31P NMR studies.47 In simulations, the interactions between piscidin and PC/PG headgroups assessed through hydrogen bond analysis do not necessarily reflect a peptide preference for a specific headgroup. Nonetheless, the average number of PG-peptide hydrogen bonding interactions increases slightly for P1:Ni, but not for P3:Ni, in the presence of aldo-PC, in agreement with the experimental data (Fig. S4, ESI†).47Apo P1 increases its hydrogen bonding with the PC headgroups, and reduces those with PG, whereas P3 follows an opposite trend. Altogether, P1:Ni interacts more readily with the anionic PG headgroups, which correlates well with the enhanced membrane activity of the peptide on the POPC/POPG/aldo-PC lipid mixture.47
20 peptide-to-lipid ratio in 31P NMR studies.47 In simulations, the interactions between piscidin and PC/PG headgroups assessed through hydrogen bond analysis do not necessarily reflect a peptide preference for a specific headgroup. Nonetheless, the average number of PG-peptide hydrogen bonding interactions increases slightly for P1:Ni, but not for P3:Ni, in the presence of aldo-PC, in agreement with the experimental data (Fig. S4, ESI†).47Apo P1 increases its hydrogen bonding with the PC headgroups, and reduces those with PG, whereas P3 follows an opposite trend. Altogether, P1:Ni interacts more readily with the anionic PG headgroups, which correlates well with the enhanced membrane activity of the peptide on the POPC/POPG/aldo-PC lipid mixture.47
Conformation dynamics of the ATCUN:Ni2+ motif
The ATCUN motif binds metals through the N-terminal amine, the deprotonated backbone amides of residues 2 and His3, and the His3 imidazole group. Metal binding allows the piscidins to embed deeper in the bilayer and decreases the conformational dynamics of the N-terminus. The Ni2+ ion adopts square-planar coordination which leaves two potential open coordination sites. In simulations, a fifth site is occupied by the His4 imidazole facing the headgroup region in 94% of frames leaving the sixth site embedded in and generally exposed to the nonpolar sublayer. The aromatic sidechains of the P1 and P3 ATCUN motif have been proposed to stabilize the Ni2+ ion through cation–π interactions at the sixth coordination site to aid insertion of the metal-bound N-termini into the bilayer.89 The COM of the P1 Phe2 phenyl ring is positioned ∼6.5 Å from the metal for 96% of frames while Phe1 makes closer contacts of ∼5.2 Å for 59% (Fig. 8). Although these distances are within the range expected for cation–π interactions,90,91 their actual orientations are generally not consistent with formation of a strong attractive interaction with the metal. A fully extended conformation with long COM-Ni2+ distances for both residues (∼6.3 for 37% of frames) occurs when the Phe1 sidechain protrudes into the lipid bilayer and Phe2 folds back to interact with Phe6. However, Phe1 and Phe2 are found to intermittently make a much closer contact of ∼4.0 Å and ∼4.2 Å, respectively, for ∼3% and ∼2.5% of frames in POPC/POPG, respectively, consistent with partial shielding of the metal by aromatic sidechains.89 The populations of these close contacts increase to ∼4% (Phe1) and ∼8% (Phe2) in POPC/POPG/aldo-PC due to the greater fluidity of the oxPL-containing bilayer. Short Phe2-Ni2+ distances can be divided into a potential cation–π-type interactions (a in Fig. 8E) and close contacts due to a CH–π interaction with Phe1 (b in Fig. 8E). In DFT calculations (see Fig. S5, ESI†), the Phe2–Phe1 CH–π interaction (b) is 12.9 kcal mol−1 more stable than the conformation with only a close Phe2–Ni2+ contact (a). The conformation with a Phe1–Ni2+ cation–π interaction is 9.2 kcal mol−1 more stable than the weaker close Phe2–Ni2+ contact [note: previous DFT calculations89 that identified a cation–π interaction for only Phe2 erroneously used the wrong chirality for Phe1 which prevented the formation of the Phe1⋯Ni2+ interaction observed here]. In simulations of P3, Phe1 is prevented from forming close contacts with the nickel center due to steric interactions with Ile2. The formation of cation–π interactions with Ni2+ could contribute to the lack of change in the insertion depth of the N-terminal end (zN ∼ 9.1 Å) when Phe2 (in P1:Ni) is replaced by Ile2 (in P3:Ni). Similar cation–π interactions, if present in cuprated piscidins, could also contribute to the lower nuclease activity26 of the P1:Cu2+ relative to P3:Cu2+ if the interactions protect the metal from redox events.
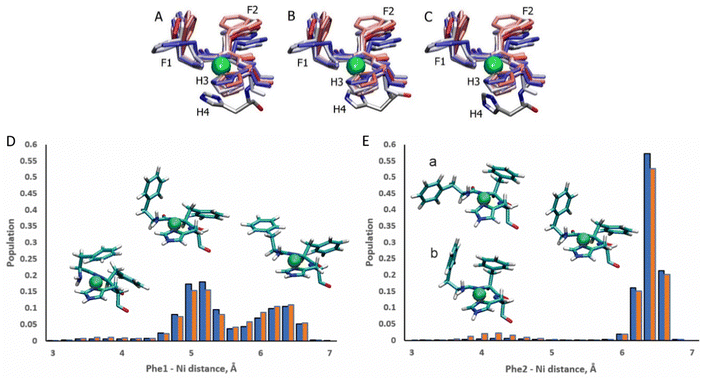 |
| | Fig. 8 Snapshots of the P1:Ni ATCUN residues 1–3 in POPC/POPG taken from the MD simulations at intervals of 200 ns (0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 209). Color-coded with red at the beginning of the trajectory and blue at the end. H4 residue is included at t = 1100 ns (A), 1700 ns (B), and 2100 ns (C). Population distributions with representative structures of the ATCUN motif for the distances between the Ni2+ ion and (D) the COM of the Phe1 phenyl ring and (E) the COM of the Phe2 phenyl ring of P1 in the POPC/POPG (blue) and POPC/POPG/aldo-PC (red) bilayer models. Structures labeled (a) and (b) are representative of snapshots with short Phe2–Ni2+ distances. Structure (a) is consistent with a Phe2–Ni2 209). Color-coded with red at the beginning of the trajectory and blue at the end. H4 residue is included at t = 1100 ns (A), 1700 ns (B), and 2100 ns (C). Population distributions with representative structures of the ATCUN motif for the distances between the Ni2+ ion and (D) the COM of the Phe1 phenyl ring and (E) the COM of the Phe2 phenyl ring of P1 in the POPC/POPG (blue) and POPC/POPG/aldo-PC (red) bilayer models. Structures labeled (a) and (b) are representative of snapshots with short Phe2–Ni2+ distances. Structure (a) is consistent with a Phe2–Ni2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) cation–π interaction. In structure (b) Phe2 has a short contact with Ni2+, but a cation–π interaction with Phe1. cation–π interaction. In structure (b) Phe2 has a short contact with Ni2+, but a cation–π interaction with Phe1. | |
Conclusions
The novel antimicrobial and antiviral therapeutic properties of piscidins are enhanced by metal binding to its ATCUN motif. All-atom molecular dynamics simulations examined the effect of metalation and oxidized lipid incorporation on the structural properties of the embedded peptides. Piscidins are found at distinct depths dependent on the isoform (P1 vs. P3) and state (free or Ni2+-bound). Apo P1 tends to insert deeper than P3 with no preference for a particular terminal end, while the N-termini of Ni2+-bound piscidins become more tilted into the bilayers due to charge neutralization of the ATCUN motif, confirming earlier predictions. Metal coordination also constrains the conformation dynamics of the N-terminus to position the nonpolar ATCUN sidechains in the lipid sublayer and His3 toward the headgroup sublayer. Cation–π interactions between Phe1 and the bound Ni2+ and CH–π interactions between Phe1 and Phe2 are found intermittently in the P1:Ni simulation and may contribute to protection against redox events at the metal center. These interactions in metalated P3 are blocked by the bulky sidechain of Ile2, suggesting a potential mechanism for the observed differential antimicrobial properties of metalated P1 and P3. This sequence dependence could be harnessed for engineering of new antimicrobial piscidins. Piscidins induce expansion and increased fluidity of the bilayer that could precede complete membrane disruption and bacteria death, but larger leaflet models are required to observe these events. Chain reversal of the oxPL chains into the aqueous phase reduces the number of hydrogen bonds between the headgroup constituents and may contribute to disruption of the bulk properties of the aldo-PC-containing membrane.
Conflicts of interest
There are no conflicts to declare.
Acknowledgements
This project was funded by NIH grant 1R15GM126527-01A1 (PI, Myriam Cotten, College of William and Mary). The Python script to calculate the τ and ρ angles was written by Alexander Greenwood. Simulations were performed on high performance clusters maintained by ODU Information Technology Services.
References
- R. E. Nelson, K. M. Hatfield, H. Wolford, M. H. Samore, R. D. Scott, S. C. Reddy, B. Olubajo, P. Paul, J. A. Jernigan and J. Baggs, National Estimates of Healthcare Costs Associated With Multidrug-Resistant Bacterial Infections Among Hospitalized Patients in the United States, Clin. Infect. Dis., 2021, 72(Suppl 1), S17–S26, DOI:10.1093/cid/ciaa1581.
- G. S. Dijksteel, M. M. W. Ulrich, E. Middelkoop and B. K. H. L. Boekema, Review: Lessons Learned From Clinical Trials Using Antimicrobial Peptides (AMPs), Front. Microbiol., 2021, 12, 616979, DOI:10.3389/fmicb.2021.616979.
- O. V. Makhlynets and G. A. Caputo, Characteristics and Therapeutic Applications of Antimicrobial Peptides, Biophys. Rev., 2021, 2(1), 011301, DOI:10.1063/5.0035731.
- A. Di Somma, A. Moretta, C. Canè, A. Cirillo and A. Duilio, Antimicrobial and Antibiofilm Peptides, Biomolecules, 2020, 10(4), 652, DOI:10.3390/biom10040652.
- M. Mahlapuu, C. Björn and J. Ekblom, Antimicrobial Peptides as Therapeutic Agents: Opportunities and Challenges, Crit. Rev. Biotechnol., 2020, 40(7), 978–992, DOI:10.1080/07388551.2020.1796576.
- D. Parai, P. Dey and S. K. Mukherjee, Antimicrobial Peptides: An Approach to Combat Resilient Infections, Curr. Drug. Discovery Technol., 2020, 17(4), 542–552, DOI:10.2174/1570163816666190620114338.
- J. Mwangi, X. Hao, R. Lai and Z.-Y. Zhang, Antimicrobial Peptides: New Hope in the War against Multidrug Resistance, Zool. Res., 2019, 40(6), 488–505, DOI:10.24272/j.issn.2095-8137.2019.062.
- P. Kumar, J. N. Kizhakkedathu and S. K. Straus, Antimicrobial Peptides: Diversity, Mechanism of Action and Strategies to Improve the Activity and Biocompatibility In Vivo, Biomolecules, 2018, 8(1), 4, DOI:10.3390/biom8010004.
- I. Zelezetsky and A. Tossi, Alpha-Helical Antimicrobial Peptides—Using a Sequence Template to Guide Structure–Activity Relationship Studies, Biochem. Biophys. Acta Biomembr., 2006, 1758(9), 1436–1449, DOI:10.1016/j.bbamem.2006.03.021.
- T. Rončević, D. Vukičević, N. Ilić, L. Krce, G. Gajski, M. Tonkić, I. Goić-Barišić, L. Zoranić, Y. Sonavane, M. Benincasa, D. Juretić, A. Maravić and A. Tossi, Antibacterial Activity Affected by the Conformational Flexibility in Glycine–Lysine Based α-Helical Antimicrobial Peptides, J. Med. Chem., 2018, 61(7), 2924–2936, DOI:10.1021/acs.jmedchem.7b01831.
- M. R. Yeaman and N. Y. Yount, Mechanisms of Antimicrobial Peptide Action and Resistance, Pharmacol. Rev., 2003, 55(1), 27–55, DOI:10.1124/pr.55.1.2.
- L. T. Nguyen, E. F. Haney and H. J. Vogel, The Expanding Scope of Antimicrobial Peptide Structures and Their Modes of Action, Trends Biotechnol., 2011, 29(9), 464–472, DOI:10.1016/j.tibtech.2011.05.001.
-
R. M. Epand, Anionic Lipid Clustering Model, in Antimicrobial Peptides: Basics for Clinical Application, ed. K. Matsuzaki, Advances in Experimental Medicine and Biology, Springer, Singapore, 2019, pp. 65–71 DOI:10.1007/978-981-13-3588-4_5.
- K. Lohner, Membrane-Active Antimicrobial Peptides as Template Structures for Novel Antibiotic Agents, Curr. Top. Med. Chem., 2017, 17(5), 508–519 CrossRef CAS PubMed.
- R. E. W. Hancock and H.-G. Sahl, Antimicrobial and Host-Defense Peptides as New Anti-Infective Therapeutic Strategies, Nat. Biotechnol., 2006, 24(12), 1551–1557, DOI:10.1038/nbt1267.
-
K. Matsuzaki, Membrane Permeabilization Mechanisms, in Antimicrobial Peptides: Basics for Clinical Application, ed. K. Matsuzaki, Advances in Experimental Medicine and Biology, Springer, Singapore, 2019, pp. 9–16 DOI:10.1007/978-981-13-3588-4_2.
- C. Harford and B. Sarkar, Amino Terminal Cu(II)- and Ni(II)-Binding (ATCUN) Motif of Proteins and Peptides: Metal Binding, DNA Cleavage, and Other Properties, Acc. Chem. Res., 1997, 30(3), 123–130, DOI:10.1021/ar9501535.
- J. C. Joyner and J. A. Cowan, Target-Directed Catalytic Metallodrugs, Braz. J. Med. Biol. Res., 2013, 46, 465–485, DOI:10.1590/1414-431X20133086.
- S. Melino, C. Santone, P. Di Nardo and B. Sarkar, Histatins: Salivary Peptides with Copper(II)- and Zinc(II)-Binding Motifs, FEBS J., 2014, 281(3), 657–672, DOI:10.1111/febs.12612.
- M. Jeżowska-Bojczuk and K. Stokowa-Sołtys, Peptides Having Antimicrobial Activity and Their Complexes with Transition Metal Ions, Eur. J. Med. Chem., 2018, 143, 997–1009, DOI:10.1016/j.ejmech.2017.11.086.
- J. L. Lau and M. K. Dunn, Therapeutic Peptides: Historical Perspectives, Current Development Trends, and Future Directions, Bioorg. Med. Chem., 2018, 26(10), 2700–2707, DOI:10.1016/j.bmc.2017.06.052.
- U. Silphaduang and E. J. Noga, Peptide Antibiotics in Mast Cells of Fish, Nature, 2001, 414(6861), 268–269, DOI:10.1038/35104690.
- R. M. Hayden, G. K. Goldberg, B. M. Ferguson, M. W. Schoeneck, M. D. J. Libardo, S. E. Mayeux, A. Shrestha, K. A. Bogardus, J. Hammer, S. Pryshchep, H. K. Lehman, M. L. McCormick, J. Blazyk, A. M. Angeles-Boza, R. Fu and M. L. Cotten, Complementary Effects of Host Defense Peptides Piscidin 1 and Piscidin 3 on DNA and Lipid Membranes: Biophysical Insights into Contrasting Biological Activities, J. Phys. Chem. B, 2015, 119(49), 15235–15246, DOI:10.1021/acs.jpcb.5b09685.
- Y. Shai, Mode of Action of Membrane Active Antimicrobial Peptides, Pept. Sci., 2002, 66(4), 236–248, DOI:10.1002/bip.10260.
- S. Guha, J. Ghimire, E. Wu and W. C. Wimley, Mechanistic Landscape of Membrane-Permeabilizing Peptides, Chem. Rev., 2019, 119(9), 6040–6085, DOI:10.1021/acs.chemrev.8b00520.
- M. D. J. Libardo, A. A. Bahar, B. Ma, R. Fu, L. E. McCormick, J. Zhao, S. A. McCallum, R. Nussinov, D. Ren, A. M. Angeles-Boza and M. L. Cotten, Nuclease Activity Gives an Edge to Host-Defense Peptide Piscidin 3 over Piscidin 1, Rendering It More Effective against Persisters and Biofilms, FEBS J., 2017, 284(21), 3662–3683, DOI:10.1111/febs.14263.
- B. S. Perrin, Jr., Y. Tian, R. Fu, C. V. Grant, E. Y. Chekmenev, W. E. Wieczorek, A. E. Dao, R. M. Hayden, C. M. Burzynski, R. M. Venable, M. Sharma, S. J. Opella, R. W. Pastor and M. L. Cotten, High-Resolution Structures and Orientations of Antimicrobial Peptides Piscidin 1 and Piscidin 3 in Fluid Bilayers Reveal Tilting, Kinking, and Bilayer Immersion, J. Am. Chem. Soc., 2014, 136(9), 3491–3504, DOI:10.1021/ja411119m.
- E. Y. Chekmenev, B. S. Vollmar and M. Cotten, Can Antimicrobial Peptides Scavenge around a Cell in Less than a Second?, Biochem. Biophys. Acta Biomembr., 2010, 1798(2), 228–234, DOI:10.1016/j.bbamem.2009.08.018.
- E. Y. Chekmenev, B. S. Vollmar, K. T. Forseth, M. N. Manion, S. M. Jones, T. J. Wagner, R. M. Endicott, B. P. Kyriss, L. M. Homem, M. Pate, J. He, J. Raines, P. L. Gor’kov, W. W. Brey, D. J. Mitchell, A. J. Auman, M. J. Ellard-Ivey, J. Blazyk and M. Cotten, Investigating Molecular Recognition and Biological Function at Interfaces Using Piscidins, Antimicrobial Peptides from Fish, Biochim. Biophys. Acta Biomembr., 2006, 1758(9), 1359–1372, DOI:10.1016/j.bbamem.2006.03.034.
- X. Lauth, H. Shike, J. C. Burns, M. E. Westerman, V. E. Ostland, J. M. Carlberg, J. C. V. Olst, V. Nizet, S. W. Taylor, C. Shimizu and P. Bulet, Discovery and Characterization of Two Isoforms of Moronecidin, a Novel Antimicrobial Peptide from Hybrid Striped Bass, J. Biol. Chem., 2002, 277(7), 5030–5039, DOI:10.1074/jbc.M109173200.
- M. Mihailescu, M. Sorci, J. Seckute, V. I. Silin, J. Hammer, B. S. Perrin, Jr., J. I. Hernandez, N. Smajic, A. Shrestha, K. A. Bogardus, A. I. Greenwood, R. Fu, J. Blazyk, R. W. Pastor, L. K. Nicholson, G. Belfort and M. L. Cotten, Structure and Function in Antimicrobial Piscidins: Histidine Position, Directionality of Membrane Insertion, and PH-Dependent Permeabilization, J. Am. Chem. Soc., 2019, 141(25), 9837–9853, DOI:10.1021/jacs.9b00440.
- W. S. Sung, J. Lee and D. G. Lee, Fungicidal Effect of Piscidin on Candida Albicans: Pore Formation in Lipid Vesicles and Activity in Fungal Membranes, Biol. Pharm. Bull., 2008, 31(10), 1906–1910, DOI:10.1248/bpb.31.1906.
- J. Menousek, B. Mishra, M. L. Hanke, C. E. Heim, T. Kielian and G. Wang, Database Screening and in Vivo Efficacy of Antimicrobial Peptides against Methicillin-Resistant Staphylococcus Aureus USA300, Int. J. Antimicrob. Agents, 2012, 39(5), 402–406, DOI:10.1016/j.ijantimicag.2012.02.003.
- G. Wang, K. M. Watson, A. Peterkofsky and R. W. Buckheit, Identification of Novel Human Immunodeficiency Virus Type 1-Inhibitory Peptides Based on the Antimicrobial Peptide Database, Antimicrob. Agents Chemother., 2010, 54(3), 1343–1346, DOI:10.1128/AAC.01448-09.
- H.-J. Lin, T.-C. Huang, S. Muthusamy, J.-F. Lee, Y.-F. Duann and C.-H. Lin, Piscidin-1, an Antimicrobial Peptide from Fish (Hybrid Striped Bass Morone Saxatilis x M. Chrysops), Induces Apoptotic and Necrotic Activity in HT1080 Cells, Zool. Res., 2012, 29(5), 327–332, DOI:10.2108/zsj.29.327.
- R. Sankararamakrishnan, S. Verma and S. Kumar, ATCUN-like Metal-Binding Motifs in Proteins: Identification and Characterization by Crystal Structure and Sequence Analysis, Proteins, 2005, 58(1), 211–221, DOI:10.1002/prot.20265.
- E. Kimoto, H. Tanaka, J. Gyotoku, F. Morishige and L. Pauling, Enhancement of Antitumor Activity of Ascorbate against Ehrlich Ascites Tumor Cells by the Copper:Glycylglycylhistidine Complex, Cancer Res., 1983, 43(2), 824–828 CAS.
- N. H. Gokhale, S. Bradford and J. A. Cowan, Catalytic Inactivation of Human Carbonic Anhydrase I by a Metallopeptide−Sulfonamide Conjugate Is Mediated by Oxidation of Active Site Residues, J. Am. Chem. Soc., 2008, 130(8), 2388–2389, DOI:10.1021/ja0778038.
- D. Deng, L. Liu, Y. Bu, X. Liu, X. Wang and B. Zhang, Electrochemical Sensing Devices Using ATCUN-Cu(II) Complexes as Electrocatalysts for Water Oxidation, Sens. Actuators, B, 2018, 269, 189–194, DOI:10.1016/j.snb.2018.04.177.
- B. Kandemir, L. Kubie, Y. Guo, B. Sheldon and K. L. Bren, Hydrogen Evolution from Water under Aerobic Conditions Catalyzed by a Cobalt ATCUN Metallopeptide, Inorg. Chem., 2016, 55(4), 1355–1357, DOI:10.1021/acs.inorgchem.5b02157.
- Y. Guo, J. R. Stroka, B. Kandemir, C. E. Dickerson and K. L. Bren, Cobalt Metallopeptide Electrocatalyst for the Selective Reduction of Nitrite to Ammonium, J. Am. Chem. Soc., 2018, 140(49), 16888–16892, DOI:10.1021/jacs.8b09612.
- B. K. Maiti, N. Govil, T. Kundu and J. J. G. Moura, Designed Metal-ATCUN Derivatives: Redox- and Non-Redox-Based Applications Relevant for Chemistry, Biology, and Medicine, iScience, 2020, 23(12) DOI:10.1016/j.isci.2020.101792.
- J. C. Joyner, J. Reichfield and J. A. Cowan, Factors Influencing the DNA Nuclease Activity of Iron, Cobalt, Nickel, and Copper Chelates, J. Am. Chem. Soc., 2011, 133(39), 15613–15626, DOI:10.1021/ja2052599.
- K. P. Neupane, A. R. Aldous and J. A. Kritzer, Metal-Binding and Redox Properties of Substituted Linear and Cyclic ATCUN Motifs, J. Inorg. Biochem., 2014, 139, 65–76, DOI:10.1016/j.jinorgbio.2014.06.004.
- C. Wende and N. Kulak, Fluorophore ATCUN Complexes: Combining Agent and Probe for Oxidative DNA Cleavage, Chem. Commun., 2015, 51(62), 12395–12398, 10.1039/C5CC04508H.
- R. K. Rai, A. De Angelis, A. Greenwood, S. J. Opella and M. L. Cotten, Metal-Ion Binding to Host Defense Peptide Piscidin 3 Observed in Phospholipid Bilayers by Magic Angle Spinning Solid-State NMR, Chem. Phys. Chem., 2019, 20(2), 295–301, DOI:10.1002/cphc.201800855.
- S. D. Paredes, S. Kim, M. T. Rooney, A. I. Greenwood, K. Hristova and M. L. Cotten, Enhancing the Membrane Activity of Piscidin 1 through Peptide Metallation and the Presence of Oxidized Lipid Species: Implications for the Unification of Host Defense Mechanisms at Lipid Membranes, Biochim. Biophys. Acta Biomembr., 2020, 1862(7), 183236, DOI:10.1016/j.bbamem.2020.183236.
- B. S. Perrin, A. J. Sodt, M. L. Cotten and R. W. Pastor, The Curvature Induction of Surface-Bound Antimicrobial Peptides Piscidin 1 and Piscidin 3 Varies with Lipid Chain Length, J. Membr. Biol., 2015, 248(3), 455–467, DOI:10.1007/s00232-014-9733-1.
- B. S. Perrin, R. Fu, M. L. Cotten and R. W. Pastor, Simulations of Membrane-Disrupting Peptides II: AMP Piscidin 1 Favors Surface Defects over Pores, Biophys. J., 2016, 111(6), 1258–1266, DOI:10.1016/j.bpj.2016.08.015.
- F. Mehrnejad and M. Zarei, Molecular Dynamics Simulation Study of the Interaction of Piscidin 1 with DPPC Bilayers: Structure-Activity Relationship, J. Biomol. Struct. Dyn., 2010, 27(4), 551–559, DOI:10.1080/07391102.2010.10507338.
- T. Yuan, X. Zhang, Z. Hu, F. Wang and M. Lei, Molecular Dynamics Studies of the Antimicrobial Peptides Piscidin 1 and Its Mutants with a DOPC Lipid Bilayer, Biopolymers, 2012, 97(12), 998–1009, DOI:10.1002/bip.22116.
- A. Rahmanpour, M. M. Ghahremanpour, F. Mehrnejad and M. E. Moghaddam, Interaction of Piscidin-1 with Zwitterionic versus Anionic Membranes: A Comparative Molecular Dynamics Study, J. Biomol. Struct. Dyn., 2013, 31(12), 1393–1403, DOI:10.1080/07391102.2012.737295.
- J.-P. Mattila, K. Sabatini and P. K. J. Kinnunen, Oxidized Phospholipids as Potential Molecular Targets for Antimicrobial Peptides, Biochem. Biophys. Acta Biomembr., 2008, 1778(10), 2041–2050, DOI:10.1016/j.bbamem.2008.03.020.
- H. Khandelia and O. G. Mouritsen, Lipid Gymnastics: Evidence of Complete Acyl Chain Reversal in Oxidized Phospholipids from Molecular Simulations, Biophys. J., 2009, 96(7), 2734–2743, DOI:10.1016/j.bpj.2009.01.007.
- A. Ayala, M. F. Muñoz and S. Argüelles, Lipid Peroxidation: Production, Metabolism, and Signaling Mechanisms of Malondialdehyde and 4-Hydroxy-2-Nonenal, Ox. Med. Cell. Longev., 2014, 2014, e360438, DOI:10.1155/2014/360438.
- M. Wallgren, L. Beranova, Q. D. Pham, K. Linh, M. Lidman, J. Procek, K. Cyprych, P. K. J. Kinnunen, M. Hof and G. Gröbner, Impact of Oxidized Phospholipids on the Structural and Dynamic Organization of Phospholipid Membranes: A Combined DSC and Solid State NMR Study, Faraday Discuss., 2013, 161, 499–513, 10.1039/C2FD20089A.
- H. L. Smith, M. C. Howland, A. W. Szmodis, Q. Li, L. L. Daemen, A. N. Parikh and J. Majewski, Early Stages of Oxidative Stress-Induced Membrane Permeabilization: A Neutron Reflectometry Study, J. Am. Chem. Soc., 2009, 131(10), 3631–3638, DOI:10.1021/ja807680m.
- S. V. Avery, Molecular Targets of Oxidative Stress, Biochem. J., 2011, 434(2), 201–210, DOI:10.1042/BJ20101695.
- A. R. Moravec, A. W. Siv, C. R. Hobby, E. N. Lindsay, L. V. Norbash, D. J. Shults, S. J. K. Symes and D. K. Giles, Exogenous Polyunsaturated Fatty Acids Impact Membrane Remodeling and Affect
Virulence Phenotypes among Pathogenic Vibrio Species, Appl. Environ. Microbiol., 2017, 83(22), e01415–e01417, DOI:10.1128/AEM.01415-17.
- L. Y. Baker, C. R. Hobby, A. W. Siv, W. C. Bible, M. S. Glennon, D. M. Anderson, S. J. Symes and D. K. Giles, Pseudomonas Aeruginosa Responds to Exogenous Polyunsaturated Fatty Acids (PUFAs) by Modifying Phospholipid Composition, Membrane Permeability, and Phenotypes Associated with Virulence, BMC Microbiol., 2018, 18(1), 117, DOI:10.1186/s12866-018-1259-8.
- L.-H. Wang, X.-A. Zeng, M.-S. Wang, C. S. Brennan and D. Gong, Modification of Membrane Properties and Fatty Acids Biosynthesis-Related Genes in Escherichia Coli and Staphylococcus Aureus: Implications for the Antibacterial Mechanism of Naringenin, Biochem. Biophys. Acta Biomembr., 2018, 1860(2), 481–490, DOI:10.1016/j.bbamem.2017.11.007.
- D. A. Pearlman, D. A. Case, J. W. Caldwell, W. S. Ross, T. E. Cheatham, S. DeBolt, D. Ferguson, G. Seibel and P. Kollman, AMBER, a Package of Computer Programs for Applying Molecular Mechanics, Normal Mode Analysis, Molecular Dynamics and Free Energy Calculations to Simulate the Structural and Energetic Properties of Molecules, Comput. Phys. Commun., 1995, 91(1), 1–41, DOI:10.1016/0010-4655(95)00041-D.
- D. A. Case, T. E. Cheatham III, T. Darden, H. Gohlke, R. Luo, K. M. Merz Jr., A. Onufriev, C. Simmerling, B. Wang and R. J. Woods, The Amber Biomolecular Simulation Programs, J. Comput. Chem., 2005, 26(16), 1668–1688, DOI:10.1002/jcc.20290.
-
AMBER 18, 2018 Search PubMed.
- P. K. J. Kinnunen, K. Kaarniranta and A. K. Mahalka, Protein-Oxidized Phospholipid Interactions in Cellular Signaling for Cell Death: From Biophysics to Clinical Correlations, Biochem. Biophys. Acta Biomembr., 2012, 1818(10), 2446–2455, DOI:10.1016/j.bbamem.2012.04.008.
- M. Wallgren, M. Lidman, Q. D. Pham, K. Cyprych and G. Gröbner, The Oxidized Phospholipid PazePC Modulates Interactions between Bax and Mitochondrial Membranes, Biochem. Biophys. Acta Biomembr., 2012, 1818(11), 2718–2724, DOI:10.1016/j.bbamem.2012.06.005.
- E. L. Wu, X. Cheng, S. Jo, H. Rui, K. C. Song, E. M. Dávila-Contreras, Y. Qi, J. Lee, V. Monje-Galvan, R. M. Venable, J. B. Klauda and W. Im, CHARMM-GUI Membrane Builder toward Realistic Biological Membrane Simulations, J. Comput. Chem., 2014, 35(27), 1997–2004, DOI:10.1002/jcc.23702.
- S. Jo, T. Kim, V. G. Iyer and W. Im, CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM, J. Comput. Chem., 2008, 29(11), 1859–1865, DOI:10.1002/jcc.20945.
- J. A. Maier, C. Martinez, K. Kasavajhala, L. Wickstrom, K. E. Hauser and C. Simmerling, ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB, J. Chem. Theory Comput., 2015, 11(8), 3696–3713, DOI:10.1021/acs.jctc.5b00255.
- P. Li and K. M. Merz, Jr., MCPB.Py: A Python Based Metal Center Parameter Builder, J. Chem. Inf. Model., 2016, 56(4), 599–604, DOI:10.1021/acs.jcim.5b00674.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, Comparison of Simple Potential Functions for Simulating Liquid Water, J. Chem. Phys., 1983, 79(2), 926–935, DOI:10.1063/1.445869.
- I. S. Joung and T. E. I. Cheatham, Determination of Alkali and Halide Monovalent Ion Parameters for Use in Explicitly Solvated Biomolecular Simulations, J. Phys. Chem. B, 2008, 112(30), 9020–9041, DOI:10.1021/jp8001614.
- A. W. Götz, M. J. Williamson, D. Xu, D. Poole, S. Le Grand and R. C. Walker, Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 1. Generalized Born, J. Chem. Theory Comput., 2012, 8(5), 1542–1555, DOI:10.1021/ct200909j.
- R. Salomon-Ferrer, A. W. Götz, D. Poole, S. Le Grand and R. C. Walker, Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald, J. Chem. Theory Comput., 2013, 9(9), 3878–3888, DOI:10.1021/ct400314y.
- S. Le Grand, A. W. Götz and R. C. Walker, SPFP: Speed without Compromise—A Mixed Precision Model for GPU Accelerated Molecular Dynamics Simulations, Comput. Phys. Commun., 2013, 184(2), 374–380, DOI:10.1016/j.cpc.2012.09.022.
- J.-P. Ryckaert, G. Ciccotti and H. J. C. Berendsen, Numerical Integration of the Cartesian Equations of Motion of a System with Constraints: Molecular Dynamics of n-Alkanes, J. Comput. Phys., 1977, 23(3), 327–341, DOI:10.1016/0021-9991(77)90098-5.
- W. F. van Gunsteren and H. J. C. Berendsen, Algorithms for Macromolecular Dynamics and Constraint Dynamics, Mol. Phys., 1977, 34(5), 1311–1327, DOI:10.1080/00268977700102571.
- G. E. Fantner, R. J. Barbero, D. S. Gray and A. M. Belcher, Kinetics of Antimicrobial Peptide Activity Measured on Individual Bacterial Cells Using High-Speed Atomic Force Microscopy, Nat. Nanotechnol., 2010, 5(4), 280–285, DOI:10.1038/nnano.2010.29.
- D. R. Roe and T. E. Cheatham, PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data, J. Chem. Theory Comput., 2013, 9(7), 3084–3095, DOI:10.1021/ct400341p.
- C. J. Dickson, L. Rosso, R. M. Betz, R. C. Walker and I. R. Gould, GAFFlipid: A General Amber Force Field for the Accurate Molecular Dynamics Simulation of Phospholipid, Soft Matter, 2012, 8(37), 9617–9627, 10.1039/C2SM26007G.
- L. Rosso and I. R. Gould, Structure and Dynamics of Phospholipid Bilayers Using Recently Developed General All-Atom Force Fields, J. Comput. Chem., 2008, 29(1), 24–37, DOI:10.1002/jcc.20675.
- L. S. Vermeer, B. L. de Groot, V. Réat, A. Milon and J. Czaplicki, Acyl Chain Order Parameter Profiles in Phospholipid Bilayers: Computation from Molecular Dynamics Simulations and Comparison with 2H NMR Experiments, Eur. Biophys. J., 2007, 36(8), 919–931, DOI:10.1007/s00249-007-0192-9.
- H. Binder and K. Gawrisch, Effect of Unsaturated Lipid Chains on Dimensions, Molecular Order and Hydration of Membranes, J. Phys. Chem. B, 2001, 105(49), 12378–12390, DOI:10.1021/jp010118h.
- N. Kučerka, S. Tristram-Nagle and J. F. Nagle, Structure of Fully Hydrated Fluid Phase Lipid Bilayers with Monounsaturated Chains, J. Membr. Biol., 2006, 208(3), 193–202, DOI:10.1007/s00232-005-7006-8.
- N. Kučerka, M.-P. Nieh and J. Katsaras, Fluid Phase Lipid Areas and Bilayer Thicknesses of Commonly Used Phosphatidylcholines as a Function of Temperature, Biochim. Biophys. Acta Biomembr., 2011, 1808(11), 2761–2771, DOI:10.1016/j.bbamem.2011.07.022.
- Å. A. Skjevik, B. D. Madej, C. J. Dickson, C. Lin, K. Teigen, R. C. Walker and I. R. Gould, Simulation of Lipid Bilayer Self-Assembly Using All-Atom Lipid Force Fields, Phys. Chem. Chem. Phys., 2016, 18(15), 10573–10584, 10.1039/C5CP07379K.
- F. de Meyer and B. Smit, Effect of Cholesterol on the Structure of a Phospholipid Bilayer, Proc. Natl. Acad. Sci. U. S. A., 2009, 106(10), 3654–3658, DOI:10.1073/pnas.0809959106.
- T. Róg, M. Pasenkiewicz-Gierula, I. Vattulainen and M. Karttunen, Ordering Effects of Cholesterol and Its Analogues, Biochim. Biophys. Acta Biomembr., 2009, 1788(1), 97–121, DOI:10.1016/j.bbamem.2008.08.022.
- R. Fu, M. T. Rooney, R. Zhang and M. L. Cotten, Coordination of Redox Ions within a Membrane-Binding Peptide: A Tale of Aromatic Rings, J. Phys. Chem. Lett., 2021, 12(18), 4392–4399, DOI:10.1021/acs.jpclett.1c00636.
- J. P. Gallivan and D. A. Dougherty, Cation–π Interactions in Structural Biology, Proc. Natl. Acad. Sci. U. S. A., 1999, 96(17), 9459–9464, DOI:10.1073/pnas.96.17.9459.
- H. Yorita, K. Otomo, H. Hiramatsu, A. Toyama, T. Miura and H. Takeuchi, Evidence for the Cation–π Interaction between Cu2+ and Tryptophan, J. Am. Chem. Soc., 2008, 130(46), 15266–15267, DOI:10.1021/ja807010f.
Footnote |
| † Electronic supplementary information (ESI) available: Distribution of tilt angles (τ), azimuthal rotation angles (ρ), depths of insertion, and hydrogen bonding analysis. Details of density functional theory calculations. See DOI: https://doi.org/10.1039/d3cb00035d |
|
| This journal is © The Royal Society of Chemistry 2023 |
 Open Access Article
Open Access Article and
Craig A.
Bayse
and
Craig A.
Bayse
 *
*
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 POPC/POPG and 2.6
1 POPC/POPG and 2.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.4 POPC/POPG/aldo-PC bilayers (POPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine: POPG, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol; aldo-PC, 1-palmitoyl-2-(9′-oxo-nonanoyl)-sn-glycero-3-phosphocholine) bilayer models. Nickel(II) binding decreases the conformation dynamics of the ATCUN motif and lowers the charge of the N-terminus to allow it to embed deeper in the bilayer without significantly changing the overall depth due to interactions of the charged half-helix of the peptide with the headgroups. Phe1⋯Ni2+ cation–π and Phe2–Phe1 CH–π interactions contribute to a small fraction of structures within the nickelated P1 simulations and may partially protect a bound metal from metal-centered chemical activity. The substitution of Phe2 for Ile2 in P3 sterically blocks conformations with cation–π interactions offering less protection to the metal. This difference between metalated P1 and P3 may indicate a mechanism by which peptide sequence can influence antimicrobial properties. Any loss of bilayer integrity due to chain reversal of the oxidized phospholipid chains of aldo-PC may be enhanced in the presence of metalated piscidins.
0.4 POPC/POPG/aldo-PC bilayers (POPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine: POPG, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol; aldo-PC, 1-palmitoyl-2-(9′-oxo-nonanoyl)-sn-glycero-3-phosphocholine) bilayer models. Nickel(II) binding decreases the conformation dynamics of the ATCUN motif and lowers the charge of the N-terminus to allow it to embed deeper in the bilayer without significantly changing the overall depth due to interactions of the charged half-helix of the peptide with the headgroups. Phe1⋯Ni2+ cation–π and Phe2–Phe1 CH–π interactions contribute to a small fraction of structures within the nickelated P1 simulations and may partially protect a bound metal from metal-centered chemical activity. The substitution of Phe2 for Ile2 in P3 sterically blocks conformations with cation–π interactions offering less protection to the metal. This difference between metalated P1 and P3 may indicate a mechanism by which peptide sequence can influence antimicrobial properties. Any loss of bilayer integrity due to chain reversal of the oxidized phospholipid chains of aldo-PC may be enhanced in the presence of metalated piscidins.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 POPC/POPG and 2.6
1 POPC/POPG and 2.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.4 POPC/POPG/aldo-PC bilayers (POPC = 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, POPG = 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol; aldo-PC = 1-palmitoyl-2-(9′-oxo-nonanoyl)-sn-glycero-3-phosphocholine, Fig. 1D). The ability of P1 to form cation–π and CH–π interactions in the metalated ATCUN motif may contribute to the sequence-dependent performance of these peptides. Chain reversal observed for oxPL may contribute to loss of membrane integrity in the presence of metalated piscidins.
0.4 POPC/POPG/aldo-PC bilayers (POPC = 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, POPG = 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol; aldo-PC = 1-palmitoyl-2-(9′-oxo-nonanoyl)-sn-glycero-3-phosphocholine, Fig. 1D). The ability of P1 to form cation–π and CH–π interactions in the metalated ATCUN motif may contribute to the sequence-dependent performance of these peptides. Chain reversal observed for oxPL may contribute to loss of membrane integrity in the presence of metalated piscidins.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 POPC/POPG and 2.6
1 POPC/POPG and 2.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.4 POPC/POPG/aldo-PC lipid bilayers with a ratio of one peptide per 40 lipids in each bilayer leaflet (P/L = 1
0.4 POPC/POPG/aldo-PC lipid bilayers with a ratio of one peptide per 40 lipids in each bilayer leaflet (P/L = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40) using AMBER18.62–64 Model design follows that of Perrin et al. using CHARMM.27 Simulations of apo peptide-containing bilayers provided a benchmark for AMBER simulations with metalated peptides. The 10% aldo-PC mixture has been suggested to be physiologically relevant under conditions of oxidative stress.65,66 Simulations of peptide-free bilayers were performed to provide a reference for peptide-induced changes to the membrane. Peptide-bilayer systems were assembled using the Membrane Builder module within the CHARMM-GUI interface.67,68 Initial structures were generated as previously described for free piscidins.27 The center of mass (COM) of the peptide backbone atoms were aligned ∼20 Å above and below the center of the bilayer. Each peptide was rotated along its helical axis to face the hydrophobic residues toward the bilayer interior. Systems were solvated with 16 Å water layers above each leaflet (∼56 waters/lipid), 15–20 sodium ions, and 0–3 chloride ions to neutralize the models and add a slight salt concentration. Simulations were performed using the ff14SB69 and Lipid17 force fields.64 The aldo-PC force field was derived using the Antechamber module within AMBER18. ATCUN-Ni2+ parameters were generated using the Python-based Metal Binding Protein Builder (MCPB) module.70 Peptides were modeled with neutral His side chains based upon previously reported pKa values.31 Solvent and counterions used the TIP3P water model71 and Joung–Cheatham monovalent ion parameters.72
40) using AMBER18.62–64 Model design follows that of Perrin et al. using CHARMM.27 Simulations of apo peptide-containing bilayers provided a benchmark for AMBER simulations with metalated peptides. The 10% aldo-PC mixture has been suggested to be physiologically relevant under conditions of oxidative stress.65,66 Simulations of peptide-free bilayers were performed to provide a reference for peptide-induced changes to the membrane. Peptide-bilayer systems were assembled using the Membrane Builder module within the CHARMM-GUI interface.67,68 Initial structures were generated as previously described for free piscidins.27 The center of mass (COM) of the peptide backbone atoms were aligned ∼20 Å above and below the center of the bilayer. Each peptide was rotated along its helical axis to face the hydrophobic residues toward the bilayer interior. Systems were solvated with 16 Å water layers above each leaflet (∼56 waters/lipid), 15–20 sodium ions, and 0–3 chloride ions to neutralize the models and add a slight salt concentration. Simulations were performed using the ff14SB69 and Lipid17 force fields.64 The aldo-PC force field was derived using the Antechamber module within AMBER18. ATCUN-Ni2+ parameters were generated using the Python-based Metal Binding Protein Builder (MCPB) module.70 Peptides were modeled with neutral His side chains based upon previously reported pKa values.31 Solvent and counterions used the TIP3P water model71 and Joung–Cheatham monovalent ion parameters.72
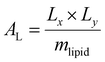
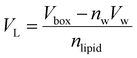
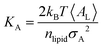
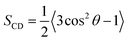
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 POPC/POPG bilayers performed with AMBER are generally consistent with previous models in CHARMM.48 Embedded P1 and P3 maintain their α-helical structure throughout simulations with the average orientation of their N-termini canted slightly toward the headgroups (τN ≈ 95°) and the C-terminal sequences nearly parallel to the surface (τC ≈ 89°/88°, Fig. 2 and S1 (ESI†), Table 2).27 The peptide backbones for P1 and P3 float on average ∼7–8 Å below the P atoms, just under the C2 atoms of the lipid sn-2 acyl chain (Fig. 3 and Table 2). P1 inserts slightly deeper in the bilayer versus P3 but both experience similar differences in the zN and zC insertion depth values (Δz ∼ 0.1 Å). P3 is slightly more flexible than P1 based on the backbone heavy-atom RMSD of the first 20 residues (0.98 Å vs. 0.80 Å, respectively).
1 POPC/POPG bilayers performed with AMBER are generally consistent with previous models in CHARMM.48 Embedded P1 and P3 maintain their α-helical structure throughout simulations with the average orientation of their N-termini canted slightly toward the headgroups (τN ≈ 95°) and the C-terminal sequences nearly parallel to the surface (τC ≈ 89°/88°, Fig. 2 and S1 (ESI†), Table 2).27 The peptide backbones for P1 and P3 float on average ∼7–8 Å below the P atoms, just under the C2 atoms of the lipid sn-2 acyl chain (Fig. 3 and Table 2). P1 inserts slightly deeper in the bilayer versus P3 but both experience similar differences in the zN and zC insertion depth values (Δz ∼ 0.1 Å). P3 is slightly more flexible than P1 based on the backbone heavy-atom RMSD of the first 20 residues (0.98 Å vs. 0.80 Å, respectively).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 model with the placement of peptides on both sides of the leaflet may be too small and symmetric to observe bulk distortions that could be attributed to membrane disruption. Therefore, in the following discussion, we instead focus on the local effects of metalation on the peptide and bilayer.
40 model with the placement of peptides on both sides of the leaflet may be too small and symmetric to observe bulk distortions that could be attributed to membrane disruption. Therefore, in the following discussion, we instead focus on the local effects of metalation on the peptide and bilayer.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 POPC/POPG bilayer similarly to the apo peptides maintaining their highly helical structure (RMSD = 1.07 Å and 1.13 Å, respectively) with similar h(P–P) and h(C2–C2) thickness to the apo peptide simulations (Table 1). The metalated ATCUN motif is less dynamic (RMSD = 0.19 Å and 0.22 Å, respectively, Table 2) than the apo piscidin N-termini due to the rigid Ni2+ coordination sphere. A decrease in KA for P1:Ni suggests an increase in membrane fluidity, where P3:Ni shows little change compared to the bilayer alone. Charge neutralization of the ATCUN-Ni2+ motif contributes to deeper embedding of the N-termini relative to the apo peptide by ∼0.9 Å for P1:Ni and ∼1.7 Å for P3:Ni (|zN − zC|, 1–2 Å (metalated) vs. 0.1 Å (apo), Table 2 and Fig. S1, ESI†). The mean peptide distance from the bilayer center increases as P1:Ni < P3:Ni < P1 < P3 with P1:Ni displaying a density maximum at ∼9.9 Å. Similar bilayer behavior is also observed upon cholesterol incorporation into the membrane.87,88 The N-terminus is more parallel to the bilayer surface for P1:Ni and slightly canted toward the bilayer center (τN = 86°) in P3:Ni with the C-termini tilted toward the bilayer core (Table 2). Nickelated P1/P3 adopt ρN smaller values by 10° and 11°, respectively, relative to the apo peptides, with Δρ (ρN − ρC) lower by only ∼3.5° upon metal-binding for the following trend in Δρ: P1 > P1:Ni ≈ P3 > P3:Ni (Table 2). These distribution angles allow better positioning of Phe1 and Phe2/Ile2 for interaction with the lipid hydrophobic core and of His3 with the water-bilayer interface. However, attractions between the hydrophilic residues of the polar half helix and the solvent and headgroups prevent a lower depth of insertion. This enhanced N-terminal insertion may contribute to the higher toxicity of metalated piscidins toward planktonic bacteria.26 The SCD order parameter of the piscidin-embedded bilayers (Fig. 4) indicate increased order in the acyl sn-1 and sn-2 chains nearest the headgroups. This restricted motility induced by the peptides decreases along the chains but does not discriminate between isoforms (P1 or P3) or metalation state (apo or Ni2+-bound).
1 POPC/POPG bilayer similarly to the apo peptides maintaining their highly helical structure (RMSD = 1.07 Å and 1.13 Å, respectively) with similar h(P–P) and h(C2–C2) thickness to the apo peptide simulations (Table 1). The metalated ATCUN motif is less dynamic (RMSD = 0.19 Å and 0.22 Å, respectively, Table 2) than the apo piscidin N-termini due to the rigid Ni2+ coordination sphere. A decrease in KA for P1:Ni suggests an increase in membrane fluidity, where P3:Ni shows little change compared to the bilayer alone. Charge neutralization of the ATCUN-Ni2+ motif contributes to deeper embedding of the N-termini relative to the apo peptide by ∼0.9 Å for P1:Ni and ∼1.7 Å for P3:Ni (|zN − zC|, 1–2 Å (metalated) vs. 0.1 Å (apo), Table 2 and Fig. S1, ESI†). The mean peptide distance from the bilayer center increases as P1:Ni < P3:Ni < P1 < P3 with P1:Ni displaying a density maximum at ∼9.9 Å. Similar bilayer behavior is also observed upon cholesterol incorporation into the membrane.87,88 The N-terminus is more parallel to the bilayer surface for P1:Ni and slightly canted toward the bilayer center (τN = 86°) in P3:Ni with the C-termini tilted toward the bilayer core (Table 2). Nickelated P1/P3 adopt ρN smaller values by 10° and 11°, respectively, relative to the apo peptides, with Δρ (ρN − ρC) lower by only ∼3.5° upon metal-binding for the following trend in Δρ: P1 > P1:Ni ≈ P3 > P3:Ni (Table 2). These distribution angles allow better positioning of Phe1 and Phe2/Ile2 for interaction with the lipid hydrophobic core and of His3 with the water-bilayer interface. However, attractions between the hydrophilic residues of the polar half helix and the solvent and headgroups prevent a lower depth of insertion. This enhanced N-terminal insertion may contribute to the higher toxicity of metalated piscidins toward planktonic bacteria.26 The SCD order parameter of the piscidin-embedded bilayers (Fig. 4) indicate increased order in the acyl sn-1 and sn-2 chains nearest the headgroups. This restricted motility induced by the peptides decreases along the chains but does not discriminate between isoforms (P1 or P3) or metalation state (apo or Ni2+-bound).

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 POPC/POPG bilayer and in the presence of P1 (cyan), P3 (purple), P1
1 POPC/POPG bilayer and in the presence of P1 (cyan), P3 (purple), P1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ni (orange), and P3
Ni (orange), and P3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ni (red).
Ni (red).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.4 POPC/POPG/aldo-PC bilayers are thinner by ∼1 Å (both h(P–P) and h(C2–C2)) for a smaller VL (1183 Å3) (compare Tables 1 and 3) relative to POPC/POPG due to the shortened oxPL chains. AL is similar to the 3
0.4 POPC/POPG/aldo-PC bilayers are thinner by ∼1 Å (both h(P–P) and h(C2–C2)) for a smaller VL (1183 Å3) (compare Tables 1 and 3) relative to POPC/POPG due to the shortened oxPL chains. AL is similar to the 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 POPC/POPG bilayer with a lower KA (243 ± 88 vs. 254 ± 114), consistent with greater fluidity due to the 4 short aldo-PC chains per leaflet. A more fluid membrane (lower KA and SCD) containing kinked lipid tails (smaller SCD) is consistent with the potential for oxPLs to disrupt membrane bulk proprieties for enhanced permeabilization.55–57 The oxPL chains can occasionally be found oriented parallel to the headgroup region to allow the aldehyde tails to interact with the more polar headgroups (Fig. 5A). While these chain reversals may contribute to membrane instability, they did not disrupt bilayers in simulations of up to 25% oxPL.54
1 POPC/POPG bilayer with a lower KA (243 ± 88 vs. 254 ± 114), consistent with greater fluidity due to the 4 short aldo-PC chains per leaflet. A more fluid membrane (lower KA and SCD) containing kinked lipid tails (smaller SCD) is consistent with the potential for oxPLs to disrupt membrane bulk proprieties for enhanced permeabilization.55–57 The oxPL chains can occasionally be found oriented parallel to the headgroup region to allow the aldehyde tails to interact with the more polar headgroups (Fig. 5A). While these chain reversals may contribute to membrane instability, they did not disrupt bilayers in simulations of up to 25% oxPL.54
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ni in the presence of aldo-PC and at the 1
Ni in the presence of aldo-PC and at the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 peptide-to-lipid ratio in 31P NMR studies.47 In simulations, the interactions between piscidin and PC/PG headgroups assessed through hydrogen bond analysis do not necessarily reflect a peptide preference for a specific headgroup. Nonetheless, the average number of PG-peptide hydrogen bonding interactions increases slightly for P1:Ni, but not for P3:Ni, in the presence of aldo-PC, in agreement with the experimental data (Fig. S4, ESI†).47Apo P1 increases its hydrogen bonding with the PC headgroups, and reduces those with PG, whereas P3 follows an opposite trend. Altogether, P1:Ni interacts more readily with the anionic PG headgroups, which correlates well with the enhanced membrane activity of the peptide on the POPC/POPG/aldo-PC lipid mixture.47
20 peptide-to-lipid ratio in 31P NMR studies.47 In simulations, the interactions between piscidin and PC/PG headgroups assessed through hydrogen bond analysis do not necessarily reflect a peptide preference for a specific headgroup. Nonetheless, the average number of PG-peptide hydrogen bonding interactions increases slightly for P1:Ni, but not for P3:Ni, in the presence of aldo-PC, in agreement with the experimental data (Fig. S4, ESI†).47Apo P1 increases its hydrogen bonding with the PC headgroups, and reduces those with PG, whereas P3 follows an opposite trend. Altogether, P1:Ni interacts more readily with the anionic PG headgroups, which correlates well with the enhanced membrane activity of the peptide on the POPC/POPG/aldo-PC lipid mixture.47







![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) cation–π interaction. In structure (b) Phe2 has a short contact with Ni2+, but a cation–π interaction with Phe1.
cation–π interaction. In structure (b) Phe2 has a short contact with Ni2+, but a cation–π interaction with Phe1.