Open Access Article
Open Access ArticleLipid nanoparticle-based mRNA candidates elicit potent T cell responses†
Ye
Zeng
a,
Oscar
Escalona-Rayo
a,
Renzo
Knol
a,
Alexander
Kros
 *a and
Bram
Slütter
*a and
Bram
Slütter
 *b
*b
aDepartment of Supramolecular & Biomaterials Chemistry, Leiden Institute of Chemistry, Leiden University, The Netherlands. E-mail: a.kros@chem.leidenuniv.nl
bLeiden Academic Centre for Drug Research, Biotherapeutics, Leiden University, The Netherlands. E-mail: b.a.slutter@lacdr.leidenuniv.nl
First published on 20th December 2022
Abstract
The induction of a potent T cell response is essential for successful tumor immunotherapy and protection against many infectious diseases. In the past few years, mRNA vaccines have emerged as potent immune activators and inducers of a robust T cell immune response. The recent approval of the Moderna and the Pfizer/BioNTech vaccines based on lipid nanoparticles (LNP) encapsulating antigen-encoding mRNA has revolutionized the field of vaccines. The advantages of LNPs are their ease of design and formulation resulting in potent, effective, and safe vaccines. However, there is still plenty of room for improvement with respect to LNP efficacy, for instance, by optimizing the lipid composition and tuning LNP for specific purposes. mRNA delivery is known to be strongly dependent on the lipid composition of LNPs and the efficiency is mainly determined by the ionizable lipids. Besides that, cholesterol and helper lipids also play important roles in mRNA transfection potency. Here, a panel of LNP formulations was studied by keeping the ionizable lipids constant, replacing cholesterol with β-sitosterol, and changing the fusogenic helper lipid DOPE content. We studied the ability of this LNP library to induce antigen presentation and T cell proliferation to identify superior LNP candidates eliciting potent T cell immune responses. We hypothesize that using β-sitosterol and increasing DOPE content would boost the mRNA transfection on immune cells and result in enhanced immune responses. Transfection of immortal immune cell lines and bone marrow dendritic cells (BMDCs) with LNPs was studied. Delivery of mRNA coding for the model antigen ovalbumin (OVA-mRNA) to BMDCs with a number of LNP formulations, resulted in a high level of activation, as evidenced by the upregulation of the co-stimulatory receptors (CD40 and CD86) and IL-12 in BMDCs. The enhancement of BMDC activation and T cell proliferation induced by the introduction of β-sitosterol and fusogenic DOPE lipids were cell dependent. Four LNP formulations (C12-200-cho-10%DOPE, C12-200-sito-10%DOPE, cKK-E12-cho-10%DOPE and cKK-E12-sito-30%DOPE) were identified that induced robust T cell proliferation and enhanced IFN-γ, TNF-α, IL-2 expression. These results demonstrate that T cell proliferation is strongly dependent on LNP composition and promising LNP-mRNA vaccine formulations were identified.
Introduction
Messenger RNA (mRNA) is an intermediate genetic carrier that is used by organisms as a translational template; therefore, it can serve as a tool for protein expression by introducing exogenous mRNA into target cells.1 The recent coronavirus pandemic dramatically accelerated the development of mRNA-based vaccines and also put a spotlight on other potential applications such as cancer immunotherapy, infectious disease vaccination, protein replacement, gene editing, and tissue engineering.2–5 This surging interest and development of mRNA as a vaccine is driven by the following advantages: (I) there is no potential infection or insertional mutagenesis risk as mRNA is a non-infectious, non-integrating genetic carrier;6 (II) mRNA is degraded by physiological metabolic pathways and the in vivo half-life can be tuned by the introduction of various chemical modifications and the delivery method;1,7,8 (III) in vitro transcription (IVT) enables rapid, inexpensive, and scalable industrial manufacturing of mRNA.2 Combined these advantages contribute to the great promise of mRNA-based therapies for both infectious diseases and cancer.Since mRNA is susceptible to degradation by nucleases in vivo, it needs to be protected from the environment upon administration. Furthermore, mRNA is unable to transfect cells and a drug delivery system is therefore required to overcome these problems. Thus, an ideal mRNA delivery system must protect against endonucleases, avoid rapid renal clearance, and promote cell entry of the tissue of interest.9,10 Lipid nanoparticles (LNPs) are the most advanced non-viral nucleic acid vector and the first RNA interference (RNAi) therapy, Onpattro, was approved in 2018 to treat hereditary amyloidogenic transthyretin amyloidosis (hATTRv).11 LNPs are typically composed of 4 types of lipids: ionizable lipids, helper lipids, cholesterol, and PEGylated lipids. Each of these components is required to obtain stable LNPs with control over mRNA encapsulation efficiency, particle size, charge, and stability. Ionizable lipids are required to condense and protect the genetic cargo via electrostatic interactions and their chemical structure plays a crucial role in the resulting transfection efficiency. Helper lipids, cholesterol, and PEGylated lipids are required to control the LNP size as well as colloidal stability, and to minimize protein absorption.12,13 The two LNP-based vaccines against SARS-CoV-2 approved in 2020 are a milestone in mRNA-based therapeutics and accelerated the development of LNPs as a facile drug delivery tool for any nucleic acid-based therapy.14,15 LNPs have also been studied in cancer immunotherapy3,16,17 and vaccination against infectious diseases, such as Zika virus,18,19 powassan virus,20 HIV-1,21,22 and influenza virus.23–25 In these studies, the immune response was investigated mainly as a function of mRNA dose, but there is still room for improvement by optimizing the lipid composition of LNPs to achieve the desired cytotoxic T-cell production to mediate successful immunotherapy against many viral diseases and tumors.
For instance, replacing cholesterol with its analog β-sitosterol was reported to enhance the mRNA transfection efficiency.26,27 Onpattro uses the lipid MC3 as the ionizable lipid, which was the first approved ionizable lipid for LNPs and showed effective gene silencing by delivering siRNA to hepatocytes.11,28 The two ionizable lipids, C12-200 and cKK-E12, were chosen for this study because they demonstrated to be more efficient in delivering siRNA as compared to MC3 and were also able to deliver mRNA, leading to potent immune responses in tumor immunotherapy.3,29–31 Helper lipids like 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) can increase the fusogenicity of LNPs, aiding endosomal escape and cytosolic translation.32 This is due to the fact that DOPE prefers to adopt an inverted hexagonal phase, which is assumed to be fusogenic, thereby promoting endosomal escape resulting in enhanced transfection.33,34
To evaluate the effect of lipid composition on mRNA delivery, translation into proteins, antigen-presenting ability, and ultimately T-cell activation, we designed a library of LNPs by varying the lipid composition. In this study, the amount of the ionizable lipids (C12-200 and cKK-E12) and PEGylated lipid was kept constant, the replacement of cholesterol for β-sitosterol and different ratio of fusogenic lipid DOPE were studied to optimize the mRNA delivery efficiency. All studied LNPs induced in general high transfection of immortal cell lines regardless of the exact composition when encapsulating mRNA encoding a green fluorescent protein (EGFP-mRNA). Bone marrow dendritic cells (BMDCs) with a potent antigen-presenting capacity for stimulating naive, memory, and effector T cells were employed to evaluate the immune responses. mRNA which codes for the model immunology protein chicken ovalbumin (OVA-mRNA) was encapsulated in LNPs to activate BMDCs and stimulate T cell proliferation. BMDCs were highly activated and T cells were strongly proliferated after the internalization of OVA-mRNA-LNPs in a concentration-dependent manner (Scheme 1). Based on robust T cell proliferation and cytokine expression measurements, we obtained 4 efficient LNP candidates for future in vivo studies towards the development of superior LNP-mRNA formulation. This study provides evidence that the lipid composition optimization of LNPs is beneficial for maximizing T cell immune responses.
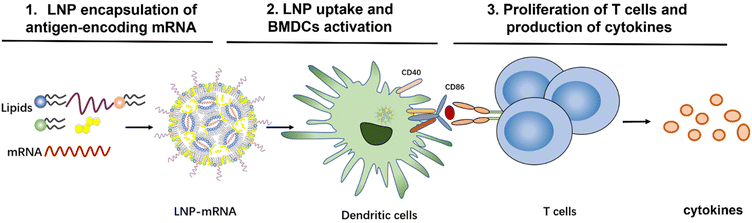 | ||
| Scheme 1 Workflow to investigate the role of mRNA-LNP composition on T-cell activation and immune response. | ||
Results
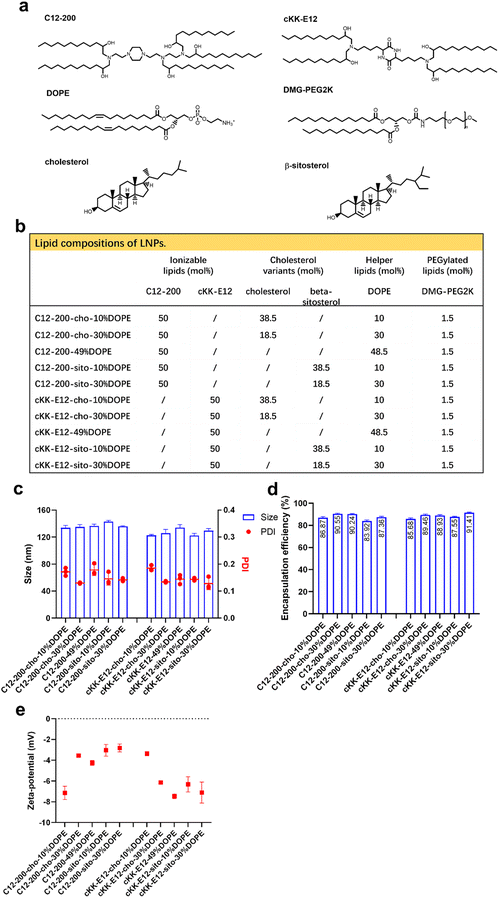
Design and characterization of LNPs
We previously showed that the silencing effect of siRNA using LNPs could be improved via tuning the internal LNP structure of the hydrophobic core from lamellar to inverse hexagonal by replacing DSPC with increasing amounts (10, 30 and 49%) of the fusogenic lipid DOPE in the LNP formulation. Inspired by this, we wondered whether mRNA transfection could also be enhanced by the addition of DOPE. In the current study, two highly efficient ionizable lipids were used: C12-200 and cKK-E12, which have shown superior transfection efficiency in siRNA delivery as the effective dose (ED50, C12-200 ∼ 0.01 mg kg−1, cKK-E12 ∼ 0.002 mg kg−1) was significantly lower than for MC3 (ED50 ∼ 0.03 mg kg−1). They also induced strong cytotoxic CD8+ T cell responses against B16F10 melanoma tumors after immunization with LNP containing OVA-mRNA, resulting in tumor shrinkage and extended overall survival of the treated mice.3,29–31 In a recent study, the cholesterol analog β-sitosterol was able to trigger enhanced mRNA transfection efficiency of LNPs in cancer cells compared to cholesterol due to the enhanced fragility, originating from an altered surface composition and shape,26 and it was therefore included in our LNP library.In this study, 10 LNP formulations were prepared, for which the ionizable lipids C12-200/cKK-E12 and the PEGylated lipid DMG-PEG2000 were kept constant at 50% and 1.5%, respectively. To study the effect of DOPE, its content varied from 10 to 49 mol% by replacing cholesterol (or its variant) (Fig. 1a and b). All resulting LNPs had a comparable diameter (∼120 nm) with a polydispersity index (PDI) < 0.20 as determined by dynamic light scattering (DLS), as well as a near-neutral surface charge (Fig. 1c and d). mRNA encapsulation efficiencies were also comparable for all LNPs and typically >80% (Fig. 1e). Thus, replacing cholesterol with β-sitosterol or increasing DOPE did not change the physicochemical characteristics of the LNPs. The LNPs were stable for at least 1 month when stored at 4 °C (ESI Fig. 1a–d†).
Cell transfection efficiency
To study mRNA delivery and translation, EGFP-mRNA encoding for green fluorescent protein (GFP) was encapsulated in the LNPs and the transfection efficiency was compared as a function of lipid composition on different cells.Previous work showed that β-sitosterol achieved higher mRNA transfection in HeLa cells.26,27 To allow a comparison between our data and previous data we included HeLa and Calu3 for cell experiments to compare the transfection efficiency of LNPs using different ionizable lipids. Both C12-200 and cKK-E12 induced enhanced mRNA transfection efficiency on HeLa and Calu-3 as compared to MC3 (ESI Fig. 2a and b†). This is not unexpected as MC3 was designed for siRNA delivery while the mRNA delivery efficiency was less efficient.11,28 Maximum transfection of HeLa cells was obtained with an EGFP-mRNA concentration of 0.5 μg mL−1 for all LNPs. In Calu-3 cells, the GFP expression of most LNPs increased with increasing EGFP-mRNA concentrations. Next, the effect of replacing cholesterol with β-sitosterol was studied. The introduction of the latter sterol significantly enhanced GFP expression for LNPs with MC3 as the ionizable lipid in both cell lines. When cKK-E12 was included in the LNPs, GFP expression was only enhanced in HeLa cells, while for C12-200-based LNP formulations no enhancement was observed at all. Finally, we studied whether increasing amounts of DOPE would enhance mRNA delivery and concomitant GFP expression. However, no general trend could be deduced from changing the DOPE ratio in LNP. Thus, we only observed a modest increase in transfection efficiency by replacing cholesterol with β-sitosterol.
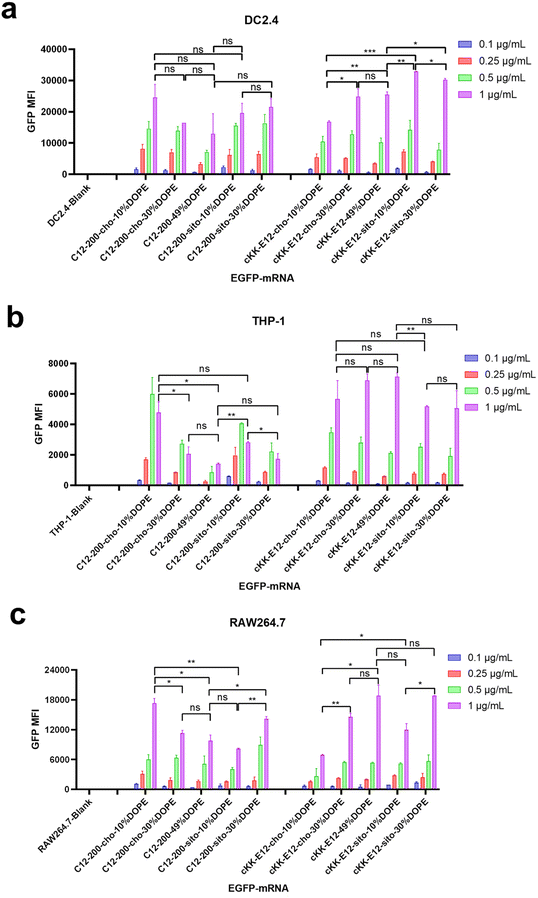
Next, antigen-presenting cells (DC2.4) and macrophage cells (THP-1 and RAW264.7) were studied to evaluate the LNP-mRNA transfection performance on immortal cells mediating immune responses. Since LNPs with C12-200 and cKK-E12 as the ionizable lipids exhibited significantly higher transfection than MC3 containing LNPs, we continued this study with C12-200 and cKK-E12 only. In general, LNPs containing cKK-E12 induced a higher GFP expression than LNPs with C12-200 in three cell lines used (Fig. 2a–c). On the other hand, introduction of β-sitosterol increased the transfection efficiency of the cKK-E12 LNPs in DC2.4 and RAW264.7, but not for C12-200 LNPs. When cholesterol was replaced by increasing amounts of DOPE, no transfection enhancement was observed for C12-200 LNPs. In contrast, enhanced transfection efficiency of cKK-E12 LNPs was observed in all tested cell lines. However, transfection enhancement was observed in DC2.4, and RAW264.7 cells when cKK-E12 LNPs contained β-sitosterol, but not in THP-1 cells (Fig. 2a–c). Confocal microscopy imaging was used to visualize GFP expression and concomitant strong fluorescence intensity, and almost every cell produced strong and uniform GFP expression. In contrast, transfection with the commercial mRNA transfection reagent lipofectamine message MAX (lipofectamine) resulted in only a few fluorescent cells in the DC2.4 cell line (Fig. S3†). In summary, all LNPs with either C12-200 or cKK-E12 as the ionizable lipid induced efficient transfection on immortal immune cell lines; however, the transfection efficiency enhancement by both the β-sitosterol replacement and the fusogenic helper lipid DOPE ratio increase was dependent on the cell line used.
BMDC transfection with EGFP-mRNA
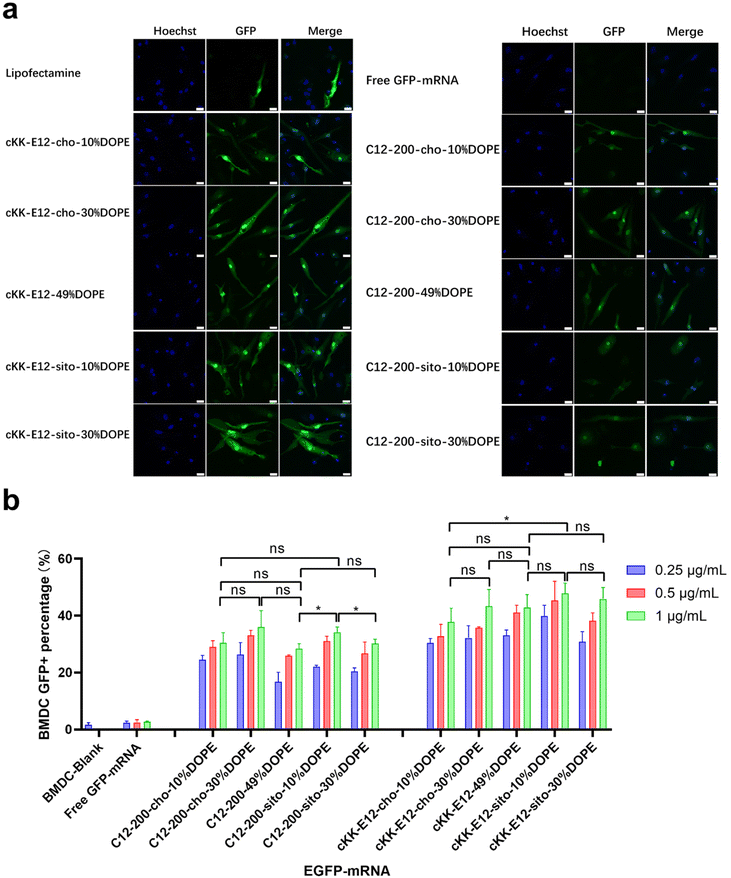
As APC cell lines provided mixed results, we next investigated primary APCs. As the next step towards both efficient intracellular antigen expression and subsequent immune cell activation to generate a robust immune response, the transfection efficiency of EGFP-mRNA loaded LNPs in BMDCs was investigated. Confocal microscopy imaging showed that all LNP formulations induced effective intracellular mRNA delivery to BMDCs, and performed better than the commercial transfection reagent Lipofectamine (Fig. 3a and ESI Fig. 4†). β-Sitosterol boosted the transfection efficiency of the LNPs with cKK-E12 as the ionizable lipids but not for C12-200 (Fig. 3b). Finally, replacing cholesterol with DOPE did not enhance the GFP expression of either C12-200 and cKK-E12 LNPs (Fig. 3b).Activation of BMDCs
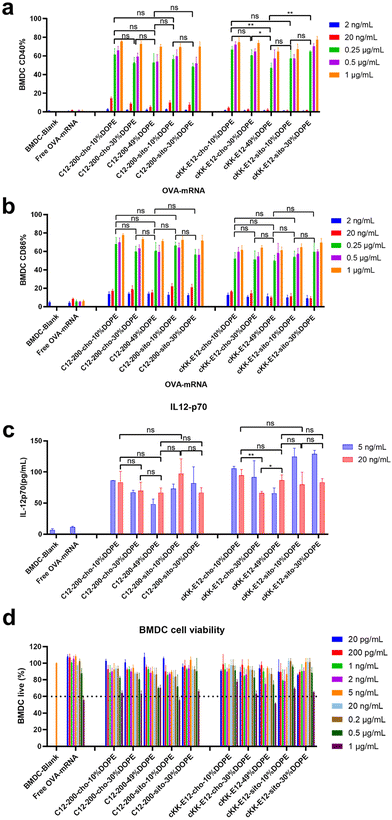
Effective mRNA vaccination demands both efficient intracellular expression and subsequent APC activation to generate a robust immune response.16 The model antigen chicken ovalbumin protein has been widely applied to evaluate the immune response, thus mRNA encoding ovalbumin (OVA-mRNA) formulated in the LNPs was employed to activate BMDCs. The activation of BMDCs results in elevated expression of surface costimulatory molecules such as CD40, and CD86. To test APC activation induced by LNPs, we treated naïve BMDCs for 24 h with LNPs containing OVA-mRNA. All LNPs managed to produce increased expression of CD40 and CD86 compared to nontreated BMDCs or incubated with free OVA-mRNA, which indicates successful BMDC activation (Fig. 4a and b). Both positive percentage and fluorescence intensity comparisons of CD40 and CD86 revealed no significant differences between C12-200 LNP formulations. Furthermore, neither the introduction of DOPE nor β-sitosterol at the expense of cholesterol boosted BMDC activation (Fig. 4a, b and ESI Fig. 5a, b†). For LNPs with cKK-E12, we observed that cKK-E12-cho-10%DOPE and cKK-E12-sito-30%DOPE induced a stronger upregulation of CD40 and CD86 expression (ESI Fig. 5a and b†).The activation of dendritic cells often promotes inflammatory cytokine gene expression. We, therefore, examined cytokine IL-12 (p70) expression in the supernatant of BMDCs. Interleukin-12 (IL-12) is a heterodimeric pro-inflammatory cytokine that regulates T helper 1 (Th1) and CD8+ T-cell responses, and is mainly produced by dendritic cells and phagocytes in response to pathogens during infection.35 Compared to blank BMDCs and free OVA-mRNA, all LNPs mediated superior IL-12 (p70) production (Fig. 4c); however, no significant differences among the tested LNPs were observed. The cell viability of BMDCs was determined to ensure that the LNPs were non-toxic at the concentrations used. All LNP formulations showed no detectable cytotoxicity (cell viability >60%), even at a high concentration (1 μg mL−1), revealing that these LNPs are indeed non-toxic (Fig. 4d).
CD8+ T-cell expansion by LNPs and cytokine production
The goal of tumor and viral vaccination is to expand antigen-specific CD8+ T cells through priming by APCs, creating a large pool of cytotoxic effector T cells that migrate through the body to clear tumors or infections.36–38 Therefore, we evaluated OVA-specific T cell expansion induced by different LNPs. BMDCs were incubated with LNPs for 4 h, followed by the addition of CD8+ (OT-I) T-cells, and the mixed cells were co-cultured for another 72 h.All LNPs induced potent OT-I proliferation in a mRNA concentration-dependent manner, which was significantly stronger than the blank and free OVA-mRNA groups (Fig. 5). T cell proliferation was negligible in the lowest concentration (200 pg mL−1), while it plateaued at ∼90% with an OVA-mRNA concentration of 20 ng mL−1. Next, we investigated the proliferation differences of LNPs in the middle mRNA concentration range. We observed that the C12-200-cho-10%DOPE, C12-200-sito-10%DOPE, cKK-E12-cho-10%DOPE and cKK-E12-sito-30%DOPE LNP formulations induced a potent OT-I T cell stimulation with 1 ng mL−1 of OVA-mRNA. This indicates these four LNP formulations are able to elicit a robust T cell proliferation even at low OVA-mRNA concentrations, which could serve as efficient LNP candidates eliciting potent T cell responses.
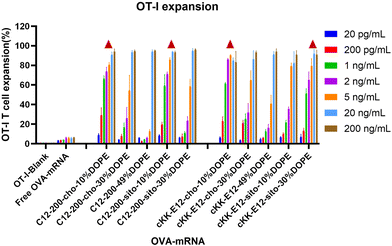 | ||
| Fig. 5 OT-I T cell expansion of LNPs with different OVA-mRNA concentrations. Triangle represents the four leading LNP candidates that induced superior OT-I responses when compared at 1 ng mL−1. | ||
Cytokine production of T cell supernatant
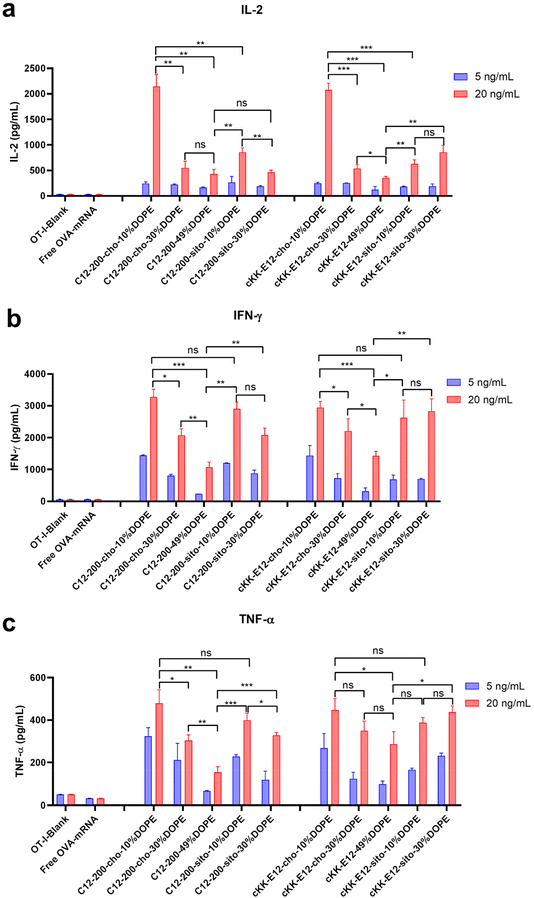
When T cells divide and differentiate into effector T cells, cytokines like IFN-γ, TNF-α, and cytotoxic proteins such as granzymes and perforin are simultaneously induced in response to acute infection.39 Proinflammatory cytokines, such as interleukin-2 (IL-2), are pivotal for the proliferation of T cells and the generation of effector and memory cells.40,41 Activated CD8+ T cells possess superior effector functions when cultured in a high concentration of IL-2 compared to cells cultured in low concentrations of this cytokine.42 Therefore, we quantified the expression of IL-2, IFN-γ, and TNF-α in OT-I cells in the culture supernatant 72 h after LNP stimulation using an ELISA assay (Fig. 6a–c). The cytokines were significantly upregulated after treating cells with LNPs as compared to non-treated or free OVA-mRNA treated cells. Differences in cytokine production among the evaluated LNPs were determined at two mRNA concentrations (5 and 20 ng mL−1). Consistent with OT-I cell proliferation data, replacing cholesterol with β-sitosterol did not boost the T cell response and cytokine production for LNPs with C12-200. The increased molar ratio of fusogenic helper lipid DOPE in C12-200 LNPs neither enhanced the T cell response nor cytokine IL-2 production. The increased molar ratio of DOPE did not enhance the cytokine IL-2 production for cKK-E12 LNPs, and it only showed some enhancement in cytokine IL-2 production at 30% DOPE but decreased at 49% DOPE for cKK-E12 LNPs using β-sitosterol. On the other hand, for Th1 cytokine (IFN-γ) and proinflammatory cytokines (TNF-α) production, we also observed that C12-200-cho-10%DOPE, C12-200-sito-10%DOPE, cKK-E12-cho-10%DOPE and cKK-E12-sito-30%DOPE LNP formulations triggered higher IFN-γ and TNF-α levels in the OT-I T cell supernatant. Taken together, these results showed that four LNP formulations (i.e. C12-200-cho-10%DOPE, C12-200-sito-10%DOPE, cKK-E12-cho-10%DOPE and cKK-E12-sito-30%DOPE) induced a potent T cell proliferation and concomitant inflammatory cytokine production.Conclusion
We investigated a small library of LNP formulations to deliver mRNA into BMDCs and evaluate T cell activation towards the development of LNP-mRNA vaccine candidates. LNPs have been validated as effective and well-tolerated on mRNA delivery and recognized as an exceptional mRNA vaccine vector. Proper LNP vaccine candidate screening is essential to identify superior LNP formulations that could boost robust T cell proliferation. It has been reported that replacing cholesterol with β-sitosterol or fusogenic DOPE could enhance transfection efficiency. We discovered that the introduction of β-sitosterol only exerted enhanced transfection in LNPs when MC3 was used as the ionizable lipid, while exhibiting varied transfection efficiency effects on different cell lines when C12-200 and cKK-E12 were used. Replacing cholesterol with DOPE resulted in mixed mRNA transfection efficiencies in different cells. This may be due to the enhanced transfection efficiency requiring different helper lipids ratios when using different ionizable lipids of LNPs, and also the mechanism of mRNA release into cytoplasm seems to be cell type-dependent. We demonstrated that the LNP-mRNA vaccine candidates can generate significant activation of BMDCs, robust T cell proliferation, and enhanced cytokine production ex vivo. We identified four LNP formulations (C12-200-cho-10%DOPE, C12-200-sito-10%DOPE, cKK-E12-cho-10%DOPE, and cKK-E12-sito-30%DOPE) which exhibit efficient T cell expansion and cytokines production and these will be tested in a future in vivo study towards the development of a cancer vaccine.Author contributions
Y. Z., O. E. R. and R. K. devised the experiments. Y. Z., and O. E. R. performed the experiments and analyzed the data. B. S. and A. K. supervised the project. Y. Z., B. S. and A. K. wrote the manuscript. All the authors contributed to the discussion of the project.Conflicts of interest
All the authors declare financial interests.Acknowledgements
We acknowledge the financial support of the Chinese Scholarship Council for a CSC grant to Y. Z., O. E. R. acknowledges the support provided by the CONACyT-Mexico [CVU/grant number: 619711/766609].References
- A. Thess, S. Grund, B. L. Mui, M. J. Hope, P. Baumhof, M. Fotin-Mleczek and T. Schlake, Sequence-engineered mRNA Without Chemical Nucleoside Modifications Enables an Effective Protein Therapy in Large Animals, Mol. Ther., 2015, 23(9), 1456–1464 CrossRef CAS.
- U. Sahin, K. Karikó and Ö Türeci, mRNA-based therapeutics—developing a new class of drugs, Nat. Rev. Drug Discovery, 2014, 13(10), 759–780 CrossRef CAS PubMed.
- M. A. Oberli, A. M. Reichmuth, J. R. Dorkin, M. J. Mitchell, O. S. Fenton, A. Jaklenec, D. G. Anderson, R. Langer and D. Blankschtein, Lipid Nanoparticle Assisted mRNA Delivery for Potent Cancer Immunotherapy, Nano Lett., 2017, 17(3), 1326–1335 CrossRef CAS.
- Q. Cheng, T. Wei, L. Farbiak, L. T. Johnson, S. A. Dilliard and D. J. Siegwart, Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing, Nat. Nanotechnol., 2020, 15(4), 313–320 CrossRef CAS PubMed.
- M. M. Billingsley, N. Singh, P. Ravikumar, R. Zhang, C. H. June and M. J. Mitchell, Ionizable Lipid Nanoparticle-Mediated mRNA Delivery for Human CAR T Cell Engineering, Nano Lett., 2020, 20(3), 1578–1589 CrossRef CAS.
- N. Pardi, M. J. Hogan, F. W. Porter and D. Weissman, mRNA vaccines—a new era in vaccinology, Nat. Rev. Drug Discovery, 2018, 17(4), 261–279 CrossRef CAS.
- S. Guan and J. Rosenecker, Nanotechnologies in delivery of mRNA therapeutics using nonviral vector-based delivery systems, Gene Ther., 2017, 24(3), 133–143 CrossRef CAS.
- K. Karikó, H. Muramatsu, F. A. Welsh, J. Ludwig, H. Kato, S. Akira and D. Weissman, Incorporation of Pseudouridine Into mRNA Yields Superior Nonimmunogenic Vector With Increased Translational Capacity and Biological Stability, Mol. Ther., 2008, 16(11), 1833–1840 CrossRef.
- S. Sabnis, E. S. Kumarasinghe, T. Salerno, C. Mihai, T. Ketova, J. J. Senn, A. Lynn, A. Bulychev, I. McFadyen, J. Chan, Ö Almarsson, M. G. Stanton and K. E. Benenato, A Novel Amino Lipid Series for mRNA Delivery: Improved Endosomal Escape and Sustained Pharmacology and Safety in Non-human Primates, Mol. Ther., 2018, 26(6), 1509–1519 CrossRef CAS PubMed.
- H. Yin, R. L. Kanasty, A. A. Eltoukhy, A. J. Vegas, J. R. Dorkin and D. G. Anderson, Non-viral vectors for gene-based therapy, Nat. Rev. Genet., 2014, 15(8), 541–555 CrossRef CAS.
- A. Akinc, M. A. Maier, M. Manoharan, K. Fitzgerald, M. Jayaraman, S. Barros, S. Ansell, X. Du, M. J. Hope, T. D. Madden, B. L. Mui, S. C. Semple, Y. K. Tam, M. Ciufolini, D. Witzigmann, J. A. Kulkarni, R. van der Meel and P. R. Cullis, The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs, Nat. Nanotechnol., 2019, 14(12), 1084–1087 CrossRef CAS PubMed.
- P. S. Kowalski, A. Rudra, L. Miao and D. G. Anderson, Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery, Mol. Ther., 2019, 27(4), 710–728 CrossRef CAS PubMed.
- S. C. Semple, A. Akinc, J. Chen, A. P. Sandhu, B. L. Mui, C. K. Cho, D. W. Y. Sah, D. Stebbing, E. J. Crosley, E. Yaworski, I. M. Hafez, J. R. Dorkin, J. Qin, K. Lam, K. G. Rajeev, K. F. Wong, L. B. Jeffs, L. Nechev, M. L. Eisenhardt, M. Jayaraman, M. Kazem, M. A. Maier, M. Srinivasulu, M. J. Weinstein, Q. Chen, R. Alvarez, S. A. Barros, S. De, S. K. Klimuk, T. Borland, V. Kosovrasti, W. L. Cantley, Y. K. Tam, M. Manoharan, M. A. Ciufolini, M. A. Tracy, A. de Fougerolles, I. MacLachlan, P. R. Cullis, T. D. Madden and M. J. Hope, Rational design of cationic lipids for siRNA delivery, Nat. Biotechnol., 2010, 28(2), 172–176 CrossRef CAS PubMed.
- C. Wang, Y. Zhang and Y. Dong, Lipid Nanoparticle–mRNA Formulations for Therapeutic Applications, Acc. Chem. Res., 2021, 54(23), 4283–4293 CrossRef CAS.
- R. Verbeke, I. Lentacker, S. C. De Smedt and H. Dewitte, The dawn of mRNA vaccines: The COVID-19 case, J. Controlled Release, 2021, 333, 511–520 CrossRef CAS PubMed.
- L. Miao, L. Li, Y. Huang, D. Delcassian, J. Chahal, J. Han, Y. Shi, K. Sadtler, W. Gao, J. Lin, J. C. Doloff, R. Langer and D. G. Anderson, Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation, Nat. Biotechnol., 2019, 37(10), 1174–1185 CrossRef CAS PubMed.
- M. Klichinsky, M. Ruella, O. Shestova, X. M. Lu, A. Best, M. Zeeman, M. Schmierer, K. Gabrusiewicz, N. R. Anderson, N. E. Petty, K. D. Cummins, F. Shen, X. Shan, K. Veliz, K. Blouch, Y. Yashiro-Ohtani, S. S. Kenderian, M. Y. Kim, R. S. O'Connor, S. R. Wallace, M. S. Kozlowski, D. M. Marchione, M. Shestov, B. A. Garcia, C. H. June and S. Gill, Human chimeric antigen receptor macrophages for cancer immunotherapy, Nat. Biotechnol., 2020, 38(8), 947–953 CrossRef CAS PubMed.
- J. M. Richner, S. Himansu, K. A. Dowd, S. L. Butler, V. Salazar, J. M. Fox, J. G. Julander, W. W. Tang, S. Shresta, T. C. Pierson, G. Ciaramella and M. S. Diamond, Modified mRNA Vaccines Protect against Zika Virus Infection, Cell, 2017, 168(6), 1114–1125 CrossRef CAS.
- N. Pardi, M. J. Hogan, R. S. Pelc, H. Muramatsu, H. Andersen, C. R. DeMaso, K. A. Dowd, L. L. Sutherland, R. M. Scearce, R. Parks, W. Wagner, A. Granados, J. Greenhouse, M. Walker, E. Willis, J.-S. Yu, C. E. McGee, G. D. Sempowski, B. L. Mui, Y. K. Tam, Y.-J. Huang, D. Vanlandingham, V. M. Holmes, H. Balachandran, S. Sahu, M. Lifton, S. Higgs, S. E. Hensley, T. D. Madden, M. J. Hope, K. Karikó, S. Santra, B. S. Graham, M. G. Lewis, T. C. Pierson, B. F. Haynes and D. Weissman, Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination, Nature, 2017, 543(7644), 248–251 CrossRef CAS.
- L. A. VanBlargan, S. Himansu, B. M. Foreman, G. D. Ebel, T. C. Pierson and M. S. Diamond, An mRNA Vaccine Protects Mice against Multiple Tick-Transmitted Flavivirus Infections, Cell Rep., 2018, 25(12), 3382–3392 CrossRef CAS.
- N. Pardi, A. J. Secreto, X. Shan, F. Debonera, J. Glover, Y. Yi, H. Muramatsu, H. Ni, B. L. Mui, Y. K. Tam, F. Shaheen, R. G. Collman, K. Karikó, G. A. Danet-Desnoyers, T. D. Madden, M. J. Hope and D. Weissman, Administration of nucleoside-modified mRNA encoding broadly neutralizing antibody protects humanized mice from HIV-1 challenge, Nat. Commun., 2017, 8(1), 14630 CrossRef.
- N. Pardi, C. C. LaBranche, G. Ferrari, D. W. Cain, I. Tombácz, R. J. Parks, H. Muramatsu, B. L. Mui, Y. K. Tam, K. Karikó, P. Polacino, C. J. Barbosa, T. D. Madden, M. J. Hope, B. F. Haynes, D. C. Montefiori, S.-L. Hu and D. Weissman, Characterization of HIV-1 Nucleoside-Modified mRNA Vaccines in Rabbits and Rhesus Macaques, Mol. Ther.–Nucleic Acids, 2019, 15, 36–47 CrossRef CAS.
- N. Pardi, K. Parkhouse, E. Kirkpatrick, M. McMahon, S. J. Zost, B. L. Mui, Y. K. Tam, K. Karikó, C. J. Barbosa, T. D. Madden, M. J. Hope, F. Krammer, S. E. Hensley and D. Weissman, Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies, Nat. Commun., 2018, 9(1), 3361 CrossRef.
- A. W. Freyn, J. Ramos da Silva, V. C. Rosado, C. M. Bliss, M. Pine, B. L. Mui, Y. K. Tam, T. D. Madden, L. C. de Souza Ferreira, D. Weissman, F. Krammer, L. Coughlan, P. Palese, N. Pardi and R. Nachbagauer, A Multi-Targeting, Nucleoside-Modified mRNA Influenza Virus Vaccine Provides Broad Protection in Mice, Mol. Ther., 2020, 28(7), 1569–1584 CrossRef CAS.
- K. Bahl, J. J. Senn, O. Yuzhakov, A. Bulychev, L. A. Brito, K. J. Hassett, M. E. Laska, M. Smith, Ö Almarsson, J. Thompson, A. Ribeiro, M. Watson, T. Zaks and G. Ciaramella, Preclinical and Clinical Demonstration of Immunogenicity by mRNA Vaccines against H10N8 and H7N9 Influenza Viruses, Mol. Ther., 2017, 25(6), 1316–1327 CrossRef CAS.
- S. Patel, N. Ashwanikumar, E. Robinson, Y. Xia, C. Mihai, J. P. Griffith, S. Hou, A. A. Esposito, T. Ketova, K. Welsher, J. L. Joyal, Ö Almarsson and G. Sahay, Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA, Nat. Commun., 2020, 11(1), 983 CrossRef CAS.
- Y. Eygeris, S. Patel, A. Jozic and G. Sahay, Deconvoluting Lipid Nanoparticle Structure for Messenger RNA Delivery, Nano Lett., 2020, 20(6), 4543–4549 CrossRef CAS PubMed.
- J. A. Kulkarni, D. Witzigmann, J. Leung, R. van der Meel, J. Zaifman, M. M. Darjuan, H. M. Grisch-Chan, B. Thöny, Y. Y. C. Tam and P. R. Cullis, Fusion-dependent formation of lipid nanoparticles containing macromolecular payloads, Nanoscale, 2019, 11(18), 9023–9031 RSC.
- K. T. Love, K. P. Mahon, C. G. Levins, K. A. Whitehead, W. Querbes, J. R. Dorkin, J. Qin, W. Cantley, L. L. Qin, T. Racie, M. Frank-Kamenetsky, K. N. Yip, R. Alvarez, D. W. Y. Sah, A. de Fougerolles, K. Fitzgerald, V. Koteliansky, A. Akinc, R. Langer and D. G. Anderson, Lipid-like materials for low-dose, in vivo gene silencing, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(5), 1864–1869 CrossRef CAS PubMed.
- Y. Dong, K. T. Love, J. R. Dorkin, S. Sirirungruang, Y. Zhang, D. Chen, R. L. Bogorad, H. Yin, Y. Chen, A. J. Vegas, C. A. Alabi, G. Sahay, K. T. Olejnik, W. Wang, A. Schroeder, A. K. R. Lytton-Jean, D. J. Siegwart, A. Akinc, C. Barnes, S. A. Barros, M. Carioto, K. Fitzgerald, J. Hettinger, V. Kumar, T. I. Novobrantseva, J. Qin, W. Querbes, V. Koteliansky, R. Langer and D. G. Anderson, Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates, Proc. Natl. Acad. Sci. U. S. A., 2014, 111(11), 3955–3960 CrossRef CAS.
- K. J. Kauffman, J. R. Dorkin, J. H. Yang, M. W. Heartlein, F. DeRosa, F. F. Mir, O. S. Fenton and D. G. Anderson, Optimization of Lipid Nanoparticle Formulations for mRNA Delivery in Vivo with Fractional Factorial and Definitive Screening Designs, Nano Lett., 2015, 15(11), 7300–7306 CrossRef CAS PubMed.
- R. Pattipeiluhu, G. Arias-Alpizar, G. Basha, K. Y. T. Chan, J. Bussmann, T. H. Sharp, M. A. Moradi, N. Sommerdijk, E. N. Harris, P. R. Cullis, A. Kros, D. Witzigmann and F. Campbell, Anionic Lipid Nanoparticles Preferentially Deliver mRNA to the Hepatic Reticuloendothelial System, Adv. Mater., 2022, 34(16), e2201095 CrossRef.
- I. M. Hafez and P. R. Cullis, Roles of lipid polymorphism in intracellular delivery, Adv. Drug Delivery Rev., 2001, 47(2), 139–148 CrossRef CAS.
- I. Koltover, T. Salditt, J. O. Rädler and C. R. Safinya, An Inverted Hexagonal Phase of Cationic Liposome-DNA Complexes Related to DNA Release and Delivery, Science, 1998, 281(5373), 78–81 CrossRef CAS.
- G. Trinchieri, Interleukin-12 and the regulation of innate resistance and adaptive immunity, Nat. Rev. Immunol., 2003, 3(2), 133–146 CrossRef CAS PubMed.
- D. O'Sullivan and E. L. Pearce, Expanding the role of metabolism in T cells, Science, 2015, 348(6238), 976–977 CrossRef.
- V. Appay, D. C. Douek and D. A. Price, CD8+ T cell efficacy in vaccination and disease, Nat. Med., 2008, 14(6), 623–628 CrossRef CAS.
- N. Zhang and M. J. Bevan, CD8+ T Cells: Foot Soldiers of the Immune System, Immunity, 2011, 35(2), 161–168 CrossRef CAS.
- B. J. Laidlaw, J. E. Craft and S. M. Kaech, The multifaceted role of CD4(+) T cells in CD8(+) T cell memory, Nat. Rev. Immunol., 2016, 16(2), 102–111 CrossRef CAS.
- O. Boyman, M. Kovar, M. P. Rubinstein, C. D. Surh and J. Sprent, Selective Stimulation of T Cell Subsets with Antibody-Cytokine Immune Complexes, Science, 2006, 311(5769), 1924–1927 CrossRef CAS.
- A. K. Abbas, E. Trotta, D. R. Simeonov, A. Marson and J. A. Bluestone, Revisiting IL-2: Biology and therapeutic prospects, Sci. Immunol., 2018, 3(25), eaat1482 CrossRef.
- M. E. Pipkin, J. A. Sacks, F. Cruz-Guilloty, M. G. Lichtenheld, M. J. Bevan and A. Rao, Interleukin-2 and Inflammation Induce Distinct Transcriptional Programs that Promote the Differentiation of Effector Cytolytic T Cells, Immunity, 2010, 32(1), 79–90 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2bm01581a |
| This journal is © The Royal Society of Chemistry 2023 |