Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Magnetic particle imaging: tracer development and the biomedical applications of a radiation-free, sensitive, and quantitative imaging modality
Stanley
Harvell-Smith
 ab,
Le Duc
Tung
ab and
Nguyen Thi Kim
Thanh
ab,
Le Duc
Tung
ab and
Nguyen Thi Kim
Thanh
 *ab
*ab
aBiophysics Group, Department of Physics and Astronomy, University College London, Gower Street, London WC1E 6BT, UK. E-mail: ntk.thanh@ucl.ac.uk
bUCL Healthcare Biomagnetic and Nanomaterials Laboratories, University College London, 21 Albemarle Street, London W1S 4BS, UK
First published on 30th December 2021
Abstract
Magnetic particle imaging (MPI) is an emerging tracer-based modality that enables real-time three-dimensional imaging of the non-linear magnetisation produced by superparamagnetic iron oxide nanoparticles (SPIONs), in the presence of an external oscillating magnetic field. As a technique, it produces highly sensitive radiation-free tomographic images with absolute quantitation. Coupled with a high contrast, as well as zero signal attenuation at-depth, there are essentially no limitations to where that can be imaged within the body. These characteristics enable various biomedical applications of clinical interest. In the opening sections of this review, the principles of image generation are introduced, along with a detailed comparison of the fundamental properties of this technique with other common imaging modalities. The main feature is a presentation on the up-to-date literature for the development of SPIONs tailored for improved imaging performance, and developments in the current and promising biomedical applications of this emerging technique, with a specific focus on theranostics, cell tracking and perfusion imaging. Finally, we will discuss recent progress in the clinical translation of MPI. As signal detection in MPI is almost entirely dependent on the properties of the SPION employed, this work emphasises the importance of tailoring the synthetic process to produce SPIONs demonstrating specific properties and how this impacts imaging in particular applications and MPI's overall performance.
1. Introduction
MPI is a recently developed tracer-based modality which has emerged as a promising diagnostic and therapeutic tool with wide ranging potential applications. It can generate 2D projection images or true 3D tomographic images that are easily interpretable, stemming from their ‘positive contrast’, a trademark of other tracer-based techniques like positron emission tomography (PET), single-photon emission computed tomography (SPECT), and optical imaging techniques. The tracer employed by MPI are SPIONs. MPI utilises a gradient field with strong gradients and weak field strengths, and the unique, intrinsic, non-linear magnetic response of these SPIONs to the gradient field is directly detected to generate an image.MPI is the first new imaging modality in 30 years. It was first introduced by Gleich and Weizenecker in 2005 at the Philips Research Laboratory (Germany).31 Then from 2007, Conolly and Goodwill at the University of California, Berkeley, developed a series of prototype alternative MPI scanners based on the same basic MPI principles but different reconstruction approaches and scanning techniques.32,33 Philips later licensed the production of their MPI systems to Bruker BioSpin AG (Switzerland), who subsequently released the world's first pre-clinical scanner to the market in 2013.34 Around 2014, Magnetic Insight Inc. (USA) was founded, later becoming the second company to release a commercial pre-clinical MPI scanner in 2016. Other academic labs and companies across the world have also contributed to the development of this technology.35–40 Until now, MPI has not been implemented clinically, but multiple groups are working on human clinical systems.41–43
Application of MPI has many benefits. SPIONs are a sensitive, safe, and biocompatible tracing material with potentially long physical half-lives, where the MPI signal can remain constant over long time-periods, enabling longer-term imaging of labelled cells.18 Both the SPION tracers and the scanners themselves have an excellent safety profile, remaining ionising radiation-free. Additionally, MPI is highly applicable in vivo as it provides unambiguous depth-independent detection of SPIONs, with the magnetic field capable of passing transparently through any anatomical tissue or bone without any signal attenuation.44 Also, without the presence of endogenous background signal, MPI has an essentially infinite contrast, and it can produce highly sensitive and specific images,45 with detection limits as low as ∼200 labelled cells, containing a total of 5.4 ng of iron.26 Furthermore, MPI is truly linearly quantitative, meaning there is a strong linear relationship between the signal intensity produced and the iron content, and this close relationship holds true for even very small quantities of iron, and at any depth.46,47 The coefficient of determination indicates the relationship is almost perfectly linear (R2 = 0.99),44 thus permitting estimation of SPION concentration in target tissues, based on just the signal intensity. Moreover, the temporal resolution of MPI is very high (<1 s),48 allowing real-time in vivo imaging and the potential for immediate assistance during medical interventions.49,50
These well-established characteristics make MPI greatly suitable for many applications of clinical relevance, including those previously inaccessible using other imaging modalities. For successful imaging, tailored SPIONs with specific properties are required, as the signal detection in MPI is almost entirely dependent on the properties of the SPION tracer employed. As a consequence, there has been extensive research on the syntheses of novel monodispersed SPIONs, controlling the size, shape, and crystallinity of the core, whether there is any surface modification, and the aggregation state.51–54
Because of the fundamental importance of optimising the characteristics of the SPION for performance in MPI and its applications, the bulk of this review will comprise a comprehensive study of the current research directions in the production of MPI-tailored SPIONs, as well as an in-depth discussion on the up-to-date literature for developments in the current and promising biomedical applications of this rapidly advancing technique. The key areas of focus are divided into applications in theranostics, cell tracking, and perfusion imaging. Prior to this discussion, the simplified principles of image generation in MPI are explained, along with a basic description of the physics and hardware operating within the system. Also included is a detailed comparison between the fundamental properties of MPI and other common imaging modalities with their relative advantages and disadvantages, and a detailing on the recent progress in MPI's clinical translation. Though research in MPI is still in the early stages, we hope this discussion on the major advancements and research directions in this rapidly advancing field over the past 5 years will encourage further exploration into the applications of MPI, as well as in the development of its SPION tracers.
2. MPI background and theory
2.1. Hardware and basic imaging principles
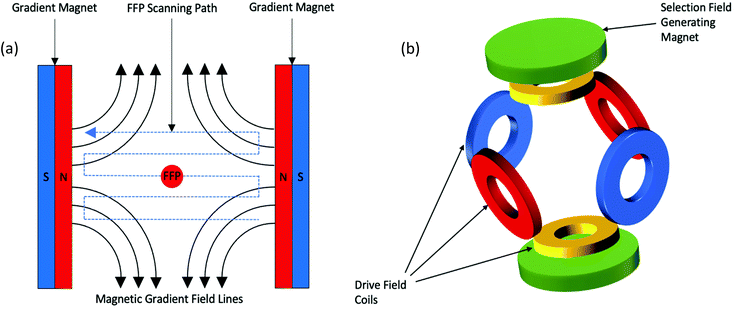
In this section, a simplified description of MPI's hardware and the basic principles for image acquisition and reconstruction is provided. Detailed descriptions of these methods can be found elsewhere.31,46,55,56 To utilise the magnetic properties of SPIONs for imaging, a standard MPI system consists of three major components: a selection field, a drive field, and a receiving coil. In the first MPI scanner developed in 2005,31 the selection field was generated through two permanent magnets positioned such that their field lines are pointing directly towards each other. This alignment produces a strong magnetic field gradient with a sensitive point located in the centre, known as the field free region (FFR), or in this case, the field free point (FFP) (Fig. 1a). The FFP is an area that is void of any magnetic field. This original scanner was able to generate magnetic field gradients of ∼3.4 T m−1, however, the hardware setup in current preclinical scanners, permits gradients of up to 7 T m−1 to be reached.57–59 The second component, the drive field, is an alternating magnetic field (AMF). In the original scanner, it is generated by three opposing pairs of drive field coils, one for each respective direction in space (Fig. 1b), each with an amplitude of 10 mT.31 The AMF causes changes in the oscillations of the SPION tracers, which results in variation in their magnetisation that is subsequently detected by the receiving coil. However, the strong magnetic field gradients produced by the selection field saturate all SPIONs except those within the FFP, thus inhibiting the effect of the AMF. Therefore, just SPIONs within the FFP respond to the AMF. Here, it is important to consider the unique magnetic properties of SPIONs in the presence of an external magnetic field.31 First, the two most commonly encountered SPION systems for MPI, single- and multi-core SPIONs, must be described, where their structural differences significantly alter the magnetic properties, resulting in nanoparticles with differing magnetic behaviour in the external AMFs applied in MPI. “Single-core” have just one magnetic core per particle, whereas “multi-core” contain several closely aggregated magnetic cores per cluster, where the cores are most often linked through dipole–dipole interactions.60,61 Single-cores have a permanent magnetic moment, whereas, multi-cores acquire a magnetic moment in the presence of strong applied fields. Within an MPI system, when SPIONs are exposed to the strong magnetic selection fields in proximity to the magnets, both types of SPIONs are fully magnetised to a state of saturation, and therefore, do not generate an MPI signal. However, when exposed to small or no magnetic field, as in the FFP, the nanoparticles are randomly oriented and constantly oscillate. The FFP is shifted over the entire field-of-view (FOV) via rapid variation of the drive field. Whenever it crosses a SPION, the magnetic dipole of the nanoparticle flips orientation instantaneously to become aligned with the field lines, and in accordance with Faraday's law of induction, induces a voltage that is detected by the sensitive receiver coils. Single-core SPIONs simply orient their non-zero permanent magnetic moment in the direction of the applied field, whereas under the same conditions, multi-core SPIONs first become magnetised and then orient their induced magnetic moment in the direction of the applied field.This concept can be utilised to generate a 2D projection image of SPION distribution, allowing for spatial encoding of the particle signal, indicating the presence and location of SPIONs within the FOV.62,63 This spatial reconstruction process requires complex algorithms, the most well-established of which, are system function reconstruction (SFR) based processes,31,50,56,64–69 and x-space processes.32,44,46,55,57,59,70 These two processes rely on the same basic hardware and imaging principles, assigning the signal to a location that corresponds to the respective location of the FFP within the region of interest. The voltages induced in the coil are also linearly proportional to the concentration of SPIONs at the instantaneous location of the FFP, enabling their quantification. This linearity of signal intensity with the density of SPIONs has been confirmed in theory.44 Furthermore, through rotation of the system around the sample, it is possible to capture 2D projection images at multiple angles, where these data can then be converted into a 3D image.
When MPI was first established, SFR-based processes were primarily employed for image reconstruction.31,56,64,65,67–69 These processes require a pre-characterisation of the SPIONs, whose signal response is formulated into a system matrix containing Fourier harmonics of the temporal signal for all possible locations of a point source.32 The system matrix is typically measured physically using a SPION sample,50 but may also be estimated through application of a model.67 Reconstruction of the image is achieved using matrix inversion and regularisation techniques. However, this inversion is often complex since the matrix is large, comprising millions of elements. Additionally, the system matrix is greatly specific to the SPION sample in solution, and thus the accuracy of reconstruction will be less if the SPION behaves differently in tissue, or if the model is inaccurate. Nowadays, x-space processes have received more research attention and are more frequently implemented.22,71–73 In these processes, a fast reconstruction algorithm computes the MPI image without any requirement for pre-characterisation, modelling, and matrix inversions,32 resulting in robust, real-time imaging. An x-space MPI image is reconstructed from a raw MPI signal through a simple two-step process of velocity compensation of the received signal, followed by gridding of said signal to the instantaneous position of the FFP.70 Velocity must be compensated in the received signal as the induced signal is proportional to the instantaneous velocity of the FFP.
Recent developments have shown that MPI sensitivity can be significantly improved through application of a more expansive spatial encoding scheme. Instead of scanning the FOV with an FFP, a field free line (FFL) can be utilised.37,74,75 An FFP integrates signal from just a small area, whereas an FFL allows for spatial encoding along a line, integrating signal from an area ∼10 times greater. This yields a correspondent theoretical sensitivity increase of a factor of 10, and an increased signal-to-noise ratio (SNR).76 Additionally, a lower level of power consumption is required.77 The first experimental setup of an FFL was presented by Knopp et al., where the line is generated by two orthogonal Maxwell coil pairs.78 The magnetic fields generated by the opposing coils flow in exact opposite directions, thus generating an FFP between each coil pair as before. However, superposition of the generated magnetic fields leads to production of an FFL centered along the bore axis of the scanner. Top et al. recently engineered the first open-sided FFL prototype MPI scanner.79 Along with the ability to electronically scan the FOV to generate tomographic images, this open-sided design permits interaction with the subject for potential real-time interventional procedures. The results from initial 2D imaging experiments have shown that high quality images with comparatively low resolutions of 2.5 mm can be produced using very low gradient levels (0.6 T m−1).
2.2. The Langevin model
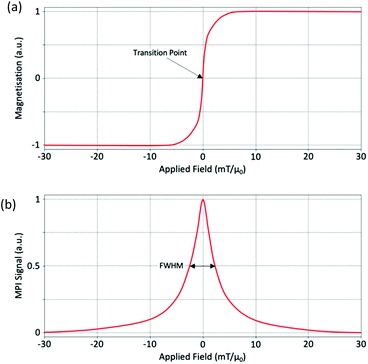
In a simplified model, this non-linear magnetisation response of SPIONs in the presence of an AMF follows the classic Langevin magnetisation curve (Fig. 2a), so long as anisotropy, hysteresis effects, and any particle interactions are disregarded.80 To explain this in terms of an ideal system of single-core SPIONs, or multi-core SPIONs, when the applied field is strong in a particular direction to one side of the particles, the magnetisation starts in a saturated state with the SPIONs aligned in the direction of the field. As the applied field is shifted across the particle, the magnetisation desaturates, eventually to a state where the applied field is ∼0 mT, and the SPIONs become randomly oriented. SPIONs have zero remanence and zero coercivity, so as the applied field is further shifted across the particles, there is a linear transition in the curve, shown in Fig. 2a, and the orientation of the nanoparticles flips rapidly with respect to the externally applied AMF. Following this transition, and with a still shifting applied field, the SPIONs re-saturate to the same intensity as before once the field strength has passed a certain threshold, but with reverse polarisation and alignment. The exact opposite process occurs when shifting the applied field in the opposite direction.In x-space reconstruction, the imaging effect can be described by a point spread function (PSF).55 A PSF is generated from the differential of this Langevin behavior and provides important information about the signal produced (Fig. 2b). The PSF of a particular SPION is a measure of the change in magnetisation as a function of the applied drive field. There are two important parameters to consider when looking at a PSF: the signal intensity, which reflects the sensitivity of the nanoparticle, and the full-width at half-maximum (FWHM), which is related to the effective spatial dimensions of the signal, and thus the spatial resolution of the nanoparticle. The FWHM is often referred to as the ‘nanoparticle resolution’.81 By dividing the value for the FWHM of the PSF by the imaging gradient strength, it is possible to estimate the overall MPI spatial image resolution.32,55 It is also possible to compare the FWHM, and thus resolutions, for two different SPIONs. To do so it is necessary to first normalise the signal obtained by the SPION concentration in the sample.
2.3. SPION relaxation
Upon application of the externally applied drive field, the dynamics of SPION magnetisation changes with the relaxation time constant, τ−1 = τBrownian−1 + τNéelian−1,82 which is influenced by the Néel time constant (τNéelian), and the Brownian time constant (τBrownian). Therefore, it can be stated that the magnetic moments of SPIONs relax to align with the external field through joint Néel and Brownian processes.83–85 The Néel time constant describes the internal flip in magnetisation of the particles from one orientation to another without physical rotation of the particle. This occurs on a timescale of nanoseconds. Whereas the Brownian time constant describes the physical rotation of the particle in space without a change in the internal magnetisation of the particle. This rotation happens on a scale of microseconds. Both time constants are affected by different parameters.83,84 The Néel relaxation process is primarily affected by temperature fluctuation, the particle's composition and size, effective magnetic anisotropy, and any interdomain interactions within the particles. Conversely, the Brownian relaxation process, whilst also effected by temperature fluctuation and particle size, is influenced by the hydrodynamic volume of the SPIONs, the local microenvironment and the liquid medium viscosity of the immediate surrounding. Though both relaxation mechanisms coexist and often take place simultaneously, in general, SPIONs with smaller core sizes exhibit Néel relaxation to guide the dynamic magnetic responses of SPIONs and produce their signal in MPI, whereas larger core size SPIONs are instead Brownian relaxation dominant.83,85 For further information on these processes, the interested reader should refer to a review written by Krishnan.863. Comparison of MPI to other in vivo imaging modalities
Each in vivo imaging technique has its own advantages and disadvantages depending on the application scenario. Table 1 illustrates a basic comparison of the qualities of MPI with other widely applied clinical and pre-clinical modalities. Some of the primary comparative advantages of MPI that can be inferred are its true quantification, high sensitivity and temporal resolution, and radiation-free labelling.| Modality | Contrast agents/tracer | Type of labelling | Sensitivity | Spatial resolution | Temporal resolution | Quantitation | Patient risk | Cost |
|---|---|---|---|---|---|---|---|---|
| a Ultrasound. | ||||||||
| MPI | SPIONs | Hot spot | 0.1 μM | <1 mm | <1 Second | Yes | Heating and peripheral nerve stimulation | Medium |
| 1H MRI | Gd, Mn, SPIONs | Contrast | mM | 25–100 μm | Seconds to hours | No | Heating and peripheral nerve stimulation | High |
| PET | Radionuclides (e.g., 18F, 68Ga) | Hot spot | pM | 2–4 mm | Minutes | Yes | Ionising radiation | High |
| SPECT | radionuclides (e.g., 111In, 99mTc) | Hot spot | pM | 3–10 mm | Minutes | Yes | Ionising radiation | Medium |
| CT | iodine | Contrast | mM | 50–200 μm | Seconds | Yes | Ionising radiation | Medium |
| USa | microbubbles | Contrast | mM | 1 mm | <1 Second | No | Heating and cavitation | Low |
3.1. Nuclear medicine
One of the more popular non-invasive imaging modalities that draws easy comparisons to MPI in terms of its properties is nuclear medicine.87–89 SPECT and PET scans are the two most frequently applied nuclear medicine imaging modalities, with significant clinical potential in that they are highly quantitative, and show great tissue penetration capability.90,91 Both MPI and nuclear medicine are highly sensitive and operate through a ‘hot spot’ detection mechanism of their tracers within a sample. The primary difference between MPI and these techniques, however, is the tracing modality. MPI makes use of SPION tracers, whereas nuclear medicine detects radioactive tracer agents or isotopes. Whilst both MPI and nuclear medicine are highly sensitive with no background signal nor signal attenuation from the tissues, the radionuclides used in PET and SPECT have shorter half-lives on the order of minutes to hours (e.g., PET tracer: t1/2(18fluorodeoxyglucose (FDG)) = 2 h; SPECT tracer: t1/2(99mTc) = 6 h),92 in comparison to that of SPIONs which has enabled researchers to track the location of SPION-labelled cells for longer time periods (see section 4.2).26 Additionally, SPIONs do not produce harmful ionising radiation as with radionuclides.87,89,92 The shelf life of SPIONs are also orders of magnitude longer, obviating the need for preparation of the tracers immediately before patient use.93 It's also worth noting that the production costs of SPIONs are significantly lower than those of radionuclides.933.2. Magnetic resonance imaging (MRI)
Another prevalent modality easily comparable to MPI is MRI. It is an anatomical technique that operates through measurement of tissue-dependent proton-spin relaxation times, showing great soft tissue contrast and high values of spatial resolution (25–100 μm).77,91,94,95 One of the major differences between MRI and MPI is in the physics required for signal generation.46,91,96 MRI utilises a strong field strength, weak gradients, and images across a high uniform magnetic field, whereas MPI utilises a weak field strength, strong gradients and images in the previously described FFR, as generated by the gradient field. Visualising the change in magnetisation via Faraday's law with a receiver coil in MPI is not dissimilar to the process for image generation in MRI. However, unlike MRI, the magnetisation change in MPI is of electronic, rather than nuclear magnetisation.97 This contributes to a higher sensitivity in MPI, as the electronic magnetisation of iron detected in MPI is 22 × 106 times stronger than that of the nuclear magnetisation of water detected in MRI.92,98 On another note, where the SPIONs employed by MPI act as a tracing modality detected via ‘hot spots’, the SPIONs that may be employed by MRI act as contrast agents, where the contrast generated is from proton density and the relaxation effects of protons in the vicinity of the particles.99MRI contrast agents are generally differentiated as either “positive” T1-weighted contrast agents or “negative” T2 (or T2*)-weighted contrast agents, where each class manifests proton-spin relaxation times in different ways.100,101T1-weighted contrast agents shorten longitudinal spin–lattice relaxation times, generating an overall bright image. Conversely, T2-weighted agents shorten transverse spin–spin relaxation times, generating an overall dark image. SPIONs have many favourable chemical and physical properties that benefit application in MRI, including great magnetic characteristics, targeting capability, limited toxicity, and a unique biodistribution and pharmacokinetic profile. The development of these nanoparticles as MRI contrast agents results in better, safer alternatives to the conventional, toxic, gadolinium-based paramagnetic agents.102–106 A number of different parameters including core shape, hydrodynamic diameter, aggregation, and coating choice influence the longitudinal and transverse relaxation times of SPIONs, but the most important determination as to whether SPIONs can be implemented as T1- or T2/T2*-weighted contrast agents is their core size.107 Generally, larger SPIONs (≥5 nm) function as T2/T2* contrast agents, whereas smaller SPIONs (≤4 nm) function as T1 contrast agents. The properties of SPIONs ideal for MPI is discussed further in section 4.1. Generally, SPIONs that are used as T2/T2* contrast agents can also function as MPI tracers, however they may not be ideal for MPI performance.
SPIONs in MRI are most implemented as T2 (or T2*) contrast agents, rather than T1 contrast agents. Despite them being the most effective T2 MRI contrast agents to date, with excellent T2 field-dependent relaxivities surpassing 100 m M−1 s−1,108 they have several drawbacks. As a negative contrast agent in MRI, SPIONs create ‘black holes’, obscuring the underlying anatomical tissue structures.12,99,109 Additionally, other endogenous sources of contrast may be mistaken for the exogenous SPIONs, such as haemorrhagic tissue or air-tissue interfaces (i.e., in lung, skin surface, and bowel studies). As a result of this, and since they are not detected directly but instead indirectly, it is not possible to reliably quantify the concentration of SPIONs in targeted tissues.12,99 This is in stark contrast to positively-contrasted MPI, which allows efficient quantitation.24,46,47 There have been several studies demonstrating the potential for SPIONs in positively-contrasted T1 MRI, yet research in this field remains uncommon.110–119 In a recent paper, Thanh et al. synthesised monodisperse SPIONs, with sizes ≤5 nm, through application of a millifluidic multistage flow reactor.120 These flow-synthesised SPIONs generated great values for enhancement of the T1 contrast, with longitudinal relaxivities (r1) greater than 10 mM−1 s−1, and transversal relaxivities (r2) reduced to just 20.5 mM−1 s−1. On a final note, scanning and imaging in MPI is much more rapid and straightforward than in MRI, with no specialised training required to acquire or interpret the images.
3.3. Multi-modal imaging with MPI
A key disadvantage of modalities like MPI and nuclear medicine are that the images produced do not provide any anatomical information. They are only able to generate morphological information from contrasted structures. Therefore, to provide context about where the SPIONs have accumulated in the animal, MPI requires an anatomic reference. MRI and CT are examples of anatomical imaging modalities. They may provide complementary images of the structural information of a sample, with which an MPI signal can be referred against.Dual modality MPI systems are currently being investigated, looking to combine the advantages of two techniques within one system, similar to how PET/MRI or PET/CT systems complement each other.121–123 MPI is most frequently co-registered with MRI as it is possible to employ the same type of SPION for both techniques, and because both modalities utilise magnetic fields to generate their images. Typically, the imaging is performed in their respective devices and the information is combined via post-processing procedures to produce a 3D tomographic image of the sample.124,125 The first in vivo studies for co-registered MRI/MPI images were conducted by Kaul et al., in which measurements with preclinical 7 T MRI were performed before and after MPI scan.124
Recently there has been lots of work on the construction of hybrid MPI/MRI scanners to help ease the co-registration process.122,123 High spatial registration accuracy can be achieved as MRI and MPI modalities share the same FOV. However, both modalities require very different magnetic field topologies, and the field strength required for MRI is so great that the SPION tracers become fully saturated. Consequently, simultaneous imaging is difficult to realise, and acquisition of data sequentially has been employed instead. The first instrument setup to combine both modalities within a single system was introduced by Franke et al. in 2013, wherein, all magnetic components are arranged concentrically and with an identical magnetic centre.126 The subject can therefore be sequentially imaged with both modalities without the need for repositioning or transportation of the subject. From this initial magnet design, the first fully integrated pre-clinical MPI/MRI system for static 3D imaging was presented.122,127 Successful initial phantom measurements were taken, demonstrating the feasibility of this system. The first in vivo results from this new methodology were gathered later by the same group, presenting a rapid, quantitative, and non-invasive in situ cardiovascular assessment of a beating rat's heart.128 In this work, 3D anatomical information obtained through MRI is combined with 3D information on the in situ location of SPIONs from MPI, following a bolus tail vein injection. These results demonstrate the potential of this system for future clinical applications.
The benefits of imaging simultaneously rather than sequentially have been appreciated with other hybrid modalities such as PET/CT.129–131 As established above; this mode of imaging is difficult to realise with MPI/MRI hybrids. However, Vogel et al. have developed a hybrid MPI/CT scanner that can indeed provide simultaneous data acquisitions.132 The MPI component is based upon a unique concept for generating a static FFL, working through the integration of Halbach rings into a rotating gantry.72 This offers an open design, providing a ‘window’ for direct feedthrough of X-rays and thus, CT imaging. As a result, the quantitative SPION distribution as well as the anatomical information of the surrounding tissue material can be visualised simultaneously and rapidly, providing a potential basis for improving diagnostic accuracy in pre-clinical imaging.132
4. Development of SPIONs as MPI tracers
4.1. Introduction to SPIONs
Signal detection and imaging quality in MPI is almost entirely dependent on the specific SPION tracer used.53,133 This stems from direct detection of the non-linear magnetisation of SPIONs by the system for signal generation, without any background interference from tissues or signal noise. Hence, for improved MPI performance, tailored tracers with specific properties are required. Generally, SPIONs used in MPI comprise a spherical crystalline core, typically of maghemite (Fe2O3) or magnetite (Fe3O4) crystals, which are colloidally stabilised using biocompatible magnetically neutral polymeric coatings like polyethylene glycol (PEG) or carboxydextran. These particles tend to have a hydrodynamic diameter between 50 and 100 nm.134 At this size, iron oxide nanoparticles show superparamagnetic behaviour, with zero remanent magnetisation and coercivity following removal of the field, in the case of multi-core SPIONs or a system of single-core SPIONs.135,136 This intrinsic property of SPIONs, together with the non-linear Langevin behavior detailed above,80 allows for signal differentiation and detection of SPIONs in MPI.The application of SPIONs as a tracing material is highly advantageous, not just in MPI, but in other SPION-compatible imaging modalities also. This was demonstrated in section 3.2, through a discussion of how MRI performance can be enhanced through implementation of SPION contrast agents. Generally, SPIONs are widely available, easy to handle, and relatively inexpensive compared to other commonly used tracers.62 Furthermore, SPIONs are non-radioactive, and their signal does not decay over time. This enables effective longitudinal tracking studies of cell-based therapeutics.26 Outside of MPI, SPIONs have been implemented in a wide variety of applications. They have shown promising clinical indication in iron supplementation therapy for anaemic candidates,137,138 cell separation,139,140 drug delivery,141 hyperthermia,142 mapping of lymph node metastases,143,144 diagnosis of liver cancers,145,146 and as a T1 agent for angiographic MRI,147 to name a few. There are several SPIONs that have either received approval from the USA Food & Drug Administration (FDA) for clinical applications or are in/close to a clinical trial.148–153 A selection of these formulations, as well as some well-established pre-clinical SPIONs that could potentially serve as tracers in MPI, are presented in Table 2.
| Name | Company | Coating | Hydrodynamic diameter (nm) | Core diameter (nm) | Applications | Market status |
|---|---|---|---|---|---|---|
| a Locations where commercially available. b Poly(maleic anhydride alt-1-octadecene). | ||||||
| Ferucarbotran:82 Resovist®/VivoTrax™ (USA, Japan, EU)a; Cliavist® (France) | Bayer AG (Resovist®/Cliavist®); Magnetic Insight (VivoTrax™) | Carboxydextran | 62 | Multi-core, ∼4 each core | MRI contrast agent & MPI | Clinically approved (Resovist® in EU, Japan) |
| Ferumoxytol:155 Feraheme® (USA); Rienso® (EU) | AMAG Pharmaceuticals (Feraheme®); Takeda Pharma (Rienso®) | Carboxymethyl-dextran | 30 | 3–4 | Iron supplementation therapy & MRI angiography | Clinically approved (USA) |
| Feruglose:156 Clariscan™ | GE Healthcare | PEGylated starch | 20 | 5–7 | MRI blood pool agent | Clinical trial |
| FeraSpin® XXL157 | Miltenyi Biotec | Carboxydextran | 65 | Multi-core, 5–7 each core | MRI blood pool agent | Pre-clinical |
| LS-0083 | LodeSpin Laboratories | PMAOb–PEG | 80 | 25 | MPI blood pool agent | Pre-clinical |
| Perimag®158 | MicroMod | Dextran | 130 | Multi-core, ∼5.5 each core | MRI contrast agent | Pre-clinical |
| PrecisionMRX®159 | Imagion Biosystems | mPEG | 41 | 24–25 | MRI contrast agent & MPI | Pre-clinical |
| Synomag®-D160 | MicroMod | Dextran | 56 | Multi-core, 5–15 each core | Hyperthermia & MPI | Pre-clinical |
A particularly important feature of SPIONs, which is true of all nanomedicine, is that there is great modularity and flexibility possible when altering the structure and properties of the nanoparticle. This tunability is particularly valuable in MPI where the advantages of a particular SPION in a particular application is dependent on so many factors, including the size and shape of the iron oxide core, the type of surface coating, and whether there is any additional surface modification.18,153,154 As a result, researchers can produce meticulously engineered tracers in MPI with different physical and biological properties specific for different applications (Fig. 3).
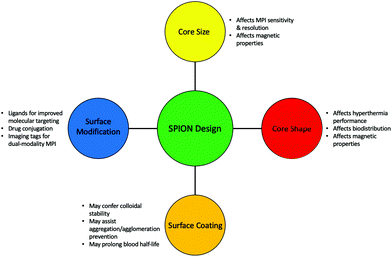 | ||
| Fig. 3 Important structural modifications that could be considered when synthesising a SPION tailored for different MPI applications. | ||
At the beginning of MPI exploration, SPION tracers that were designed as T2* or T1-weighted MRI contrast agents were evaluated for their MPI performance.134 However, since MRI and MPI have totally different physics, it was determined that these existing tracers were not ideal for reliable application in MPI and that new SPIONs would have to be tailored to MPI's unique set-up.51,63,161
Multi-core SPIONs are far more common than their single-core counterparts.5,157,162,163 Many of the frequently implemented commercial SPIONs have multi-core structures, as demonstrated in Table 2. It is, however, very complex to control the structural uniformity of clustered cores, as there are many structural variables that need to be considered, including, the number of cores per cluster, the cluster diameter, shape, density, and the inter-core distances and spatial distribution.60 Any alteration in these parameters will lead to considerable changes in the packing arrangements, and this can significantly affect the dipole–dipole or exchange-interactions between the tightly associated cores, which strongly influence the magnetic behaviour of the SPIONs.164–166 Because of this, in general, multi-core SPIONs have less uniform magnetic properties than standard single-core formulations, and produce poorer signals in MPI. Despite this, the presence of multiple cores can also be advantageous, if there is proper control of the structure during the synthesis.
Many recent studies on the synthesis of single-core SPIONs for MPI have been focussed on the effect of core size on MPI performance and sensitivity. One of the principal reasons for the poor performance of MRI-tailored SPIONs in MPI is the small size of their magnetic iron oxide cores (generally <10 nm), which results in lower magnetic moments for the nanoparticles.53,167
Both core structures hold equally important roles in MPI applications. Individually coated single-core SPIONs have demonstrated significantly increased blood half-lives in vivo,168 favouring their use in perfusion imaging.58,169,170 Additionally, the structural uniformity is beneficial for the targeted delivery of SPIONs to a therapeutic site, since core size is a major determinant of a SPIONs pharmacokinetic behaviour, and therefore its biodistribution.171 On the other hand, multi-core SPIONs have also been implicated as greatly beneficial for many biomedical applications. The presence of the magnetic coupling interactions between the clustered cores is particularly advantageous in magnetic hyperthermia applications.164 The improved magnetic moment renders advantages in standard magnetic fluid hyperthermia (MFH) techniques,172–176 as well as MPI-coupled MFH.177–180 The benefits of MPI–MFH is discussed in depth in section 5.2.2.
The shape of the core, or cores in the case of a multi-core formulation, is another factor that demonstrates strong influence over SPION performance in MPI. To date, most SPION cores developed for MPI, and its applications, have a spherical shape. However, it is known that non-spherical MNPs can offer significant advantages in many different biomedical applications, as a consequence of altered physical properties, and the potential for improved magnetic characteristics, including Ms, magnetic anisotropy,181–183 and heating properties.184,185 Additionally, as alternative MNP shapes present larger available surface areas for cell interactions, as compared with equivalently sized spherical nanoparticles, they often demonstrate greater cellular uptake.186 As well as this, they have potential for enhanced blood circulation half-lives.187
In theory, the potential for improved magnetic properties, through the synthesis of non-spherical SPIONs, should be an effective way enhance MPI sensitivity and spatial resolution. As a result, such work has attracted the attention of various research groups. In particular, there has been sustained recent interest in the implementation of cubic SPIONs in MPI.18,188 These nanocubes have a lower proportion of disordered spins at their surface and smaller surface anisotropies in comparison to equivalently sized spherical SPIONs.189 This results in higher values for Ms and magnetic susceptibility, and consequently improved MPI performance, at certain sizes.190 Along with this enhanced magnetic performance, the tendency of cubic SPIONs to spontaneously form chain-like arrangements has led to improvements in the performance of SPIONs in MPI–MFH.13,190,191 A recent study from Avugadda et al. showcased the advantages of SPIONs, comprising a controlled number of cubic cores, on MPI, and potential MPI–MFH performance.5 Among the variety of magnetic assemblies synthesised in this study (Fig. 4a), multi-core dimer and trimer structures exhibited the greatest MPI properties (Fig. 4b and c). The other structures investigated were larger multi-core clusters of nanocubes, and individual single-core nanocubes, all synthesised using the same polymer coating. The enhanced performance is attributed to the beneficial uniaxial magnetic dipolar coupling present in the chain-like smaller multi-core assemblies.192
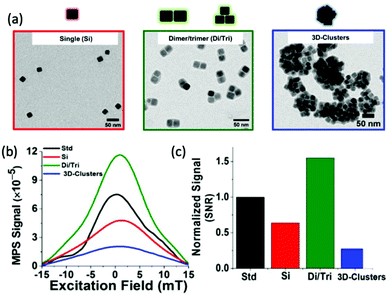 | ||
| Fig. 4 Magnetic assemblies comprising a controlled number of cubic cores, designed for optimal MPI performance. (a) Transmission electron microscopy (TEM) images of the different SPION assemblies made from iron oxide nanocubes encapsulated in a poly(styrene-co-maleic anhydride) coating. The assemblies synthesised were single nanocubes (Si), short-chain dimers/trimers (Di/Tri), and various 3D-cluster configurations. (b) PSF demonstrating the MPI signal obtained for each synthesised sample, in comparison to that of the reference, VivoTrax (Std). (c) Histograms of the corresponding SNRs for each synthesised sample and the commercial reference. Reproduced from ref. 5 with permission from the Multidisciplinary Digital Publishing Institute. This is an open access article distributed under the terms of the CC BY License (https://creativecommons.org/licenses/by/4.0/). Copyright 2020, the authors. Article can be found at: https://www.mdpi.com/2079-4991/11/1/62/htm. No changes were made to the original figure. | ||
Another key consideration in the design of SPIONs for MPI applications is the choice of biocompatible surface coating. As a general trend, a coating must confer colloidal stability to the particle, assisting aggregation prevention under various complex physiological environments, whilst also mediating any interactions with biological entities. This further stability can also potentially prolong half-life in the bloodstream, help prevent agglomeration during storage or application, and counteract possible oxidation.107,193 It is also important to ensure the coatings promote cellular uptake yet preserve the optimal magnetic response of the SPIONs when dispersed in the acidic endosomal environment. There are many common choices of coating, each with their own advantages and functions depending on their molecular structure. Polymers are among the most popular coatings for SPIONs.194,195 Early MPI research typically relied on multi-core ferucarbotran (Resovist, Bayer AG), a SPION originally clinically approved as an MRI contrast agent in the liver. Ferucarbotran nanoparticles are coated by a carboxydextran polymer.50 More recently, PEG and dextran polymer coatings have been employed most frequently. This is because they are not rapidly recognised by macrophages in the liver and spleen when administered intravenously and consequently have enhanced circulation time.196,197 They have also been generally recognised as safe by the FDA.107 The biodistribution of PEG- and carboxydextran-coated SPIONs was studied by Keselman et al.198 While the carboxydextran-modified SPIONs are cleared rapidly to the liver, the PEG-coated particles are sustained for a relatively long blood half-life of 4.2 h before eventual excretion through the reticuloendothelial system (RES), illustrating the benefit of PEG-coated SPIONs for in vivo studies.
It is also worth noting that coatings can be altered to tailor their biochemical properties towards a specific physiological application. Examples of such alteration are that the coating may be grafted differently (i.e., it can be chemisorbed, physisorbed, covalently bonded etc.), or that different graft densities and molecular weights of coating may be applied. Guzy et al. demonstrated the importance of polymer coating choice, and coating molecular weight, on SPIONs undergoing biodegradation.81 SPIONs can undergo a variety of physicochemical changes as they degrade, which generally results in detrimental effects on MPI signal properties and ‘nanoparticle resolution’. It was found that larger polymers with a greater molecular weight will degrade more slowly in harsher endosomal conditions, such as at a tumor site, and thus their MPI signal will remain for longer.
The surface coating can also act as a structural support for additional surface modification, with the potential to conjugate a huge variety of possible functional molecules like drugs and ligands to improve molecular targeting, and proteins, antibodies, or aptamers for highly specific chemical interactions with complex biological systems. It can also provide a platform for imaging tags, specifically molecules that allow dual modality imaging with MPI, for example fluorophores for fluorescent microscopy.4,199 Following successful surface-modification, the resulting SPIONs may be employed for MPI.
4.2. Synthetic methods
The modularity in SPION design in Section 4.1 is incredibly useful. To ensure production of SPIONs with desired properties, it is fundamentally important to choose the most appropriate method of synthesis. The choice has a strong influence on key physical characteristics of SPIONs such as crystal structure, core size, and size distribution, and consequently, on the magnetic properties of the nanoparticles. A comparison between five of the most employed synthetic methods are briefly summarised in Table 3, demonstrating their specific advantages and downsides. Besides those outlined in the table, SPIONs can be effectively prepared by various other techniques, including sonochemical and electrochemical deposition, microwave irradiation, laser pyrolysis, and reduction methods.194,200–202| Synthetic method | Synthesis complexity | Reaction temperature (°C) | Reaction length | Solvent | Size distribution | Shape control | Yield |
|---|---|---|---|---|---|---|---|
| Co-precipitation | Very simple, ambient conditions | 20–90 | Minutes | Water | Relatively narrow | Poor | High, scalable |
| Thermal decomposition | Very complicated, inert atmosphere | 100–320 | Hours-days | Organic compound | Very narrow | Very good | High, scalable |
| Hydrothermal | Simple, high pressure | 150–220 | Hours-days | Water–ethanol | Relatively narrow | Very Good | Medium, scalable |
| Microemulsion | Complicated, ambient conditions | 20–50 | Hours | Organic compound | Relatively broad | Good | Low, not scalable |
| Sol–gel | Complicated, ambient conditions | 25–200 | Hours | Water–ethanol | Relatively broad | Good | Medium, scalable |
Co-precipitation and thermal decomposition and are preferred for the synthesis of SPIONs for MPI.203 Co-precipitation works by the simultaneous precipitation of Fe3+ and Fe2+ aqueous salts following addition of a basic solution.204 It is a cost-effective and simple process, capable of high-yielding, scalable syntheses. Additionally, the synthesis does not require an organic solvent and the precursors used are generally environmentally friendly.205 Despite these beneficial qualities, the nanoparticles formed are often relatively polydisperse with a low degree of crystallinity.206,207 This can result in an appreciably weakened MPI signal. Differently, SPIONs with a very narrow size distribution and excellent crystallinity can be prepared through thermal decomposition.208,209 In this method, SPIONs are synthesised through the decomposition of organoiron precursors in organic solvents with high-boiling points, in the presence of stabilising surfactants. These surfactants bind to the growing nanocrystals, controlling their nucleation and growth. However, despite formation of high-quality samples, this technique requires expensive, and generally toxic reagents and solvents, and is therefore not environmentally friendly.107 Furthermore, as a hydrophobic coating is formed on the surface of the SPIONs during synthesis, an additional surface modification step is required to obtain the biocompatible, water dispersible SPIONs to be used in biomedical applications. An in-depth description of all syntheses techniques is not within the scope of this review, thus, for further description of the other techniques, interested readers should refer to reviews by Thanh et al.210,211
4.3. Recent developments in SPION research
Similar to when SPION contrast agents were first realised for MRI in the 1980s,212 research on the development and synthesis of monodispersed novel MPI-tailored SPIONs has recently become an important area of research.5,63,188,213–216 These new particles can be functionalised and optimised for improved performance in specific applications, like for increased circulation time or for more efficient cell targeting. Whilst MPI-optimised tracers will have to undergo a lengthy evaluation before clinical approval, the exploration and development of better performing SPIONs should spur further work towards clinical applications.The development of tracers with long circulation times is crucial for many applications. Typically, when nanoparticles are administered into bloodstream circulation, there is just a narrow time window to image the particles before they accumulate in the liver and spleen for excretion, where the concentration of the particles decreases such that MPI can no longer get a meaningful signal.102 Studies on the circulation time of carboxydextran-coated multi-core ferucarbotran (Resovist), which is generally considered the ‘gold-standard’ of MPI tracers, show that following administration to rabbits, the MPI signal decreased to ∼12% of the initial intensity after just 15 min, and within 30 min, the signal had disappeared entirely.217 Another commercially available tracer with potential use in MPI is dextran-coated Synomag-D (MicroMod), where the MPI performance shows better circulation times (t1/2, ∼1 h) as compared to Resovist.20 Khandhar et al. developed a new single-core nanoparticle known as LS-008 (LodeSpin Lab) through a post-synthesis oxidation method.218 This MPI-tailored tracer of core diameter, 25 nm, produced outstanding resolutions (1.6 mm at a 7 T m−1 μ0−1 field gradient) and was designed for long blood circulation times (t1/2, ∼105 min in mice), with an exceptionally stable PMAO–PEG coating. However, for many of the potential applications of MPI, much longer half-lives are required.3,58,92,169,170,219 Additionally, the availability of longer-circulating tracers will reduce the quantity of tracer required and/or the number of tracer administrations for treatment.
New MPI tracers with long blood circulation half-lives of 7 h were obtained by Liu et al., termed RL-1, as shown in Fig. 5a.20 These single-core SPIONs were developed using a semi-batch thermal decomposition process with molecular oxygen addition. This was followed with an optimised PEG–silane ligand exchange process, producing SPIONs with high values for spatial resolution (∼2 mm at 5.7 T m−1) and sensitivities greater than multi-core Synomag-D, and ∼3 times greater than multi-core ferucarbotran (Resovist) (Fig. 5b). Another long-circulating tracer was developed by Song et al.,28 composed of a Janus iron oxide@semiconducting polymer nanostructure (Fig. 5c) synthesised through a nanoprecipitation method reported prior by the same group, shown in Fig. 5d, where the semiconducting polymers employed demonstrate great biocompatiblity.12 These particles provide a nanoplatform for ultrasensitive multi-modal imaging (MPI/MRI/photoacoustic/fluorescence, termed MMPF nanoparticles) of tumor xenografts in living mice, possessing exceptionally long-term blood circulation times (t1/2, ∼49 h) and consequently very high tumor uptake.28 This half-life permits the tracer to be tracked and quantified in the mice longitudinally for up to 85 d (Fig. 5e).
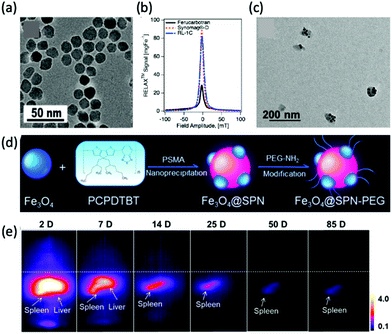 | ||
| Fig. 5 MPI-tailored SPIONs, designed for long blood circulation half-lives. (a) TEM image of RL-1 SPIONs. (b) PSF demonstrating the MPI signal obtained for the RL-1 SPIONs, in comparison to that of the references, Synomag-D and ferucarbotran SPIONs. Reproduced from ref. 20 with permission from Ivyspring International Publisher. This is an open access article distributed under the terms of the CC BY License (https://creativecommons.org/licenses/by/4.0/). Copyright 2021, the authors. Article can be found at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8040827/. No changes were made to the original figure. (c) TEM image of MMPF SPIONs. (d) Schematic demonstrating the synthetic preparation route of MMPF nanoparticles, where PCPDTBT is poly[2,6-(4,4-bis(2-ethylhexyl)-4H-cyclopenta[2,1-b;3,4-b′]dithiophene)-alt-4,7(2,1,3-benzothiadiazole)], and PSMA is poly[styrene-co-maleic anhydride]. (e) Longitudinal MPI images of mice injected with MMPF SPIONs, demonstrating effective imaging for up to 85 d. Reproduced with permission from ref. 28. Copyright 2019 American Chemical Society. | ||
An alternative approach to increasing circulation times includes entrapping the SPIONs into human red blood cells (RBCs).221 Through nuclear magnetic resonance measurements, such loaded RBCs were demonstrated to circulate for over 12 d in mouse models before an obvious reduction in concentration could be detected.222 For RBCs loaded with Resovist, transmission electron microscopy images show that the particles have a spatially uniform distribution within the cells, without any discernible indication of particle aggregation.223 Utilising Resovist-loaded RBCs, Rahmer et al. presented the first evidence that SPION-loaded cells could be imaged in vivo with MPI, showing clear imaging of the blood pool in mice several hours following injection.224 This observation was supported with magnetic particle spectroscopy (MPS) measurements, performed to determine the concentration of iron in samples of blood extracted from the mice at different time points following injection. Antonelli et al. also performed a study encapsulating different commercially available SPIONs, Synomag-D and Perimag (MicroMod) into RBCs through hypotonic dialysis.225 COOH-functionalised Perimag loaded RBCs proved to be viable cells, while the magnetic signal of the equivalently functionalised Synomag-D loaded cells dropped sharply. Therefore, just the Perimag-loaded RBCs have potential for MPI diagnostic applications, showing potential for longer blood retention times than the equivalent free nanoparticles. Successful application of MPI to the imaging of pathological diseases depends on the quantity of nanoparticles that accumulate at a diseased site relative to other sites. Hence, another important area for MPI-tailored tracer development is in the synthesis of nanoparticles with coatings functionalised towards the active targeting of specific pathophysiologies.226
One of the more prevalent functional targeting applications is the targeting of nanoparticles towards cancerous cells/tumors. Cancer is one of the global leading causes of death, accounting for almost 10 million deaths worldwide in 2020 alone.227 Interest in its effective targeting is therefore necessary. Arami et al. conjugated the glioma-targeting glycoprotein, lactoferrin, to the PMAO–PEG surface coatings of their optimised single-core MPI tracers with diameters of 25–27 nm.4 Very high-resolution 3D tomographic multi-modal images (MPI/CT/X-ray) demonstrate an enhanced uptake of the functionalised SPIONs in brain cancer xenografts in mice (Fig. 6a and b). This is due to the fact that lactoferrin molecules can pass through the blood brain barrier (BBB) with ease, through a receptor-mediated transcytosis mechanism, for active targeting.199 Another work in this field, by Tomitaka et al., investigated the functionalisation of gold-coated multi-core SPIONs of ∼30 nm with transferrin (Fig. 6c).30 Like lactoferrin, transferrin is a brain glioma targeting ligand, which has exhibited great specificity due to the high expression of transferrin receptors on the surface of brain glioma and capillary endothelial cells. They demonstrated a high functionalisation efficiency of 58% for the SPION with the targeting ligand, using the procedure in Fig. 6d, and these functionalised particles showed great biocompatibility also.
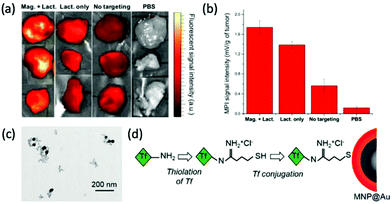 | ||
| Fig. 6 MPI-tailored SPIONs, designed for efficient targeting towards cancerous cells/tumors. (a) Near infra-red fluorescent images and (b) MPI signal intensities of tumor xenografts excised from mice, under different conditions. ‘Mag. + Lact.’ corresponds to the injection of Cy5.5-lactoferrin conjugated SPIONs, using magnetic targeting, ‘Lact. only’ to the injection of conjugated SPIONs, without using magnetic targeting, ‘No targeting’ to the injection of non-conjugated Cy5.5-labelled SPIONs, and ‘PBS’, to the use of phosphate buffered solution, as a control. Reproduced with permission of the Royal Society of Chemistry, ref. 4; permission conveyed through Copyright Clearance Center, Inc. (c) TEM image of transferrin-conjugated SPIONs. (d) Schematic demonstrating the process for transferrin conjugation onto MNP@Au, where Tf is transferrin. Reproduced with permission of the Royal Society of Chemistry, from ref. 30; permission conveyed through Copyright Clearance Center, Inc. | ||
Another targeting application, which is receiving increased interest, is the targeting of active myeloperoxidase (MPO), a potential inflammatory marker of vulnerable atherosclerotic plaque. Tong et al. developed novel single-core multi-modal SPIONs, as termed 5HFeC nanoparticles conjugated with 5-hydroxytryptamine and a cyanine 7 N-hydroxysuccinimide ester, that can specifically target MPO and identify these high-risk plaques in vivo.228 These 21 nm nanoprobes image the active MPO in an atherosclerosis mouse model with high sensitivity, thus enabling quantitative evaluation of the severity of inflammation and monitoring of the MPO activity.
4.4. Effect of SPIONs on spatial resolution and sensitivity in MPI
Traditionally, the most significant technical weakness of MPI is the relatively poor spatial resolution (typically ∼1 mm).134 To compete with preclinical CT and MRI, improving these resolutions to the submillimetre range would be an enabling advance. MPI spatial resolution can be defined as the ability to clearly distinguish two signals with the same intensity in space.97 From a physical and radiologic viewpoint, this depends on two major factors. First is the magnetic gradient strength of the instrument, more specifically, the stronger the gradient, the narrower the FFR, allowing assignment of the generated SPION signal to a narrower space, and thus a greater spatial resolution.55,56 The second factor comes from the physical and magnetic properties of the SPION utilised. This is based on effects to the FWHM of the PSF, where nanoparticles with narrower PSFs produce higher achievable resolutions. Along with a higher quality image, utilising a SPION with better spatial resolutions could significantly reduce instrument cost. A 10-fold improvement in resolution from MPI-tailored SPIONs could potentially reduce the cost of a clinical instrument by up to 100-fold, as spatial resolution can be exchanged for lower MPI gradients which would be required to be high in a clinical system.229Sensitivity is another important parameter in determining overall MPI performance. A high sensitivity permits the detection of very small amounts of tracer,26 and this is enabling for many biomedical applications, and is particularly important in cell tracking. It is characterised by the height of the Langevin curve at the transition point, where a taller curve indicates a higher sensitivity.230 Consequently, a greater signal intensity in a PSF, also signifies a better sensitivity.231 As with spatial resolution, progressing to the theoretical limits of MPI for sensitivity involves work on both the hardware and tracers.44,232 In terms of instrumentation, advancements in sensitivity have most frequently been from developments in coil design.37,74,78,79,233,234 Regarding tracer development for enhanced sensitivity, the signal intensity is governed by the physical and inherent magnetic properties of the tracer, as with spatial resolution, but is mostly dependent on the Ms.16,18,55 Although the detection sensitivity of MPI is strong currently, through significant advancement it has the potential to become comparable with that of exceptionally sensitive nuclear imaging techniques.233
Given what has been discussed, the development of SPIONs with optimal sensitivity and spatial resolution performance has become a crucial part of MPI research. Sustained work in this area will enable high-performing, cost-effective, and safe MPI for humans. Tailoring of the structural parameters of SPIONs, such as their shape and crystallinity, can increase resolution and signal strength through enhancing of Ms.18,51,52,183 One of the most important factors for improved performance is the size of the magnetic core. For single-core SPIONs, resolution and sensitivity increase cubically with core size, stemming primarily from higher Ms values.235,236 Unfortunately, Tay et al. demonstrated that the improvement in performance is limited with respect to size, as a result of increased Brownian relaxation blurring and a gradual shift from the superparamagnetic to ferromagnetic regime as sizes increase.159 This has been described as the Langevin wall, and this phenomena reduces Ms, and as a result, hampers performance. An empirical ideal core size with respect to both resolution and sensitivity performance has been estimated at 24–28 nm for single-core SPIONs.159,237 Gevaert et al. examined the MPI performance of several commercially available SPION tracers, confirming the resolution and sensitivity dependence of SPIONs to core size.238
Recently, a new strategy for tracer formation has been established based on strongly interacting particle–particle magnetic dipole interactions which subsequently form nanoparticle chains in fluid, on application of an external magnetic field (Fig. 7a).239 This chaining was found to amplify the applied field 10-fold, resulting in very sharp PSFs. Through this method, Tay et al. demonstrated the potential of a 40-fold boost in sensitivity, and unprecedented 10-fold improvements in spatial resolution (Fig. 7b).14,229 These properties theoretically allow the tracking of single cells in vivo. There have been several other studies testing the ‘chaining hypothesis’, specifically, examining the parameters that effect chain formation times. Colson et al. outlined that for optimised chain formation time, the conditions required are low media viscosities, as viscous solvents can block chain formation, and high nanoparticle concentrations, as the smaller the inter-particle separations, the easier it is for dipoles to interact.240
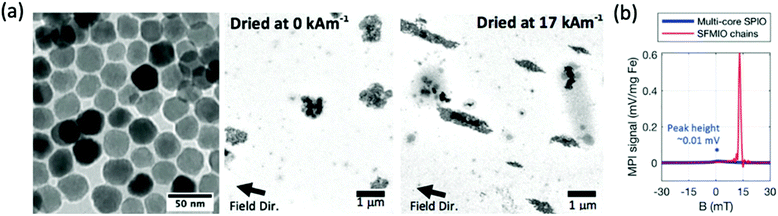 | ||
| Fig. 7 Microscale linear chain structures, designed for optimal MPI spatial resolution and sensitivity. (a) TEM images showing clear formation of microscale chain structures following application of an external magnetic field. (b) PSF demonstrating order-of-magnitude taller and sharper signal peaks for the chains over standard multi-core ferucarbotran, where SFMIO is a superferromagnetic iron oxide chain. Reproduced from ref. 14 with permission from Wiley-VCH. | ||
The formation of these ‘chained’ tracers has also been investigated and modelled computationally.241 Zhao et al. employed simulations to evaluate the effect of the magnetic dipole–dipole interactions on the MPI performance and dynamic magnetisation of individual particles within the chain.242 The results illustrate similar MPI signal intensity and resolution enhancements to those demonstrated by Tay et al., for interacting chains of ≥2 particles.14 They also suggest a large parameter space for design that can be used to tailor these chains towards optimised MPI performance, including, the number of particles in the chain and their separation distances, the composition of the SPIONs, and the viscosity of the solution.
Outside of novel ‘chaining’ approaches, there are other more established methods to improve spatial resolution and sensitivity through tracer synthesis, one of which is through the synthesis of SPIONs with improved monodispersity.47,243 Monodisperse SPIONs with uniform magnetic domains have smaller FWHMs and higher values of Ms in the PSF, resulting in nanoparticles with higher spatial resolutions and signal intensities. Dadfar et al. describe a straightforward co-precipitation synthesis and following sequential centrifugation protocol to obtain monodisperse single-core particles of different sizes from a polydisperse SPION starting formulation.244 Resulting from the narrow size distribution (polydispersity index (PDI) below 0.1), these optimised dispersions showed substantially improved Ms values, and thus performance in MPI, MRI and MFH, up to 7 times greater in comparison to the polydisperse starting formulation, as well as to commercially recognised SPIONs, such as Resovist. However, this is a two-step process and is not feasible for scale-up, nor is environmentally friendly. In a different work, Unni et al. synthesised monodisperse SPIONs through a modified thermal decomposition process involving the controlled addition of molecular oxygen.16 The particles synthesised in oxygen presence demonstrate greater magnetic properties (Fig. 8a), and exhibit a better MPI performance (Fig. 8b) than those in the absence of oxygen, with greater Ms values (74 Am2 kg−1vs. 17 Am2 kg−1, respectively), and consequently improved values for FWHM (12.0 mT vs. 15.2 mT, respectively) and signal intensity (39.0 mL mg(Fe)−1vs. 13.1 mL mg(Fe)−1, respectively). This improvement is attributed to appreciably more uniform magnetic and physical domain sizes, and fewer structural defects.
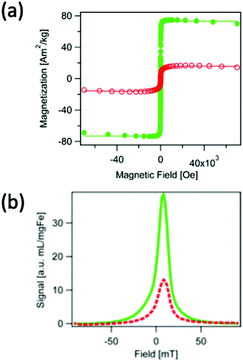 | ||
| Fig. 8 SPIONs synthesised through a modified thermal decomposition reaction, in the presence and absence of molecular oxygen, designed for optimal MPI spatial resolution and sensitivity. (a) Hysteresis curves displaying the field-dependent magnetism of SPIONs synthesised in the presence of molecular oxygen (closed, green points), and absence (open, red circles). (b) PSF demonstrating the MPI signal for the SPIONs synthesised in the presence (full, green line), and absence (dashed, red line) of molecular oxygen. Reproduced with permission from ref. 16. Copyright 2017 American Chemical Society. | ||
The choice of coating should also not be neglected when optimising SPIONs for higher image resolutions and sensitivities. In one study, Horvat et al. reported the positive impact of preparing SPIONs with cross-linked over non-crosslinked polymeric coatings on performance in MPI.245 The cross-linked single-core SPIONs of average core size 18 nm, termed PF127DAPG, displayed a superior SNR, and consequently higher spatial resolution than their uncross-linked equivalents, PF127. Resulting from this, an iron quantity of 5.3 μg was required in PF127 SPIONs to produce the same spatial resolution values as 1.3 μg of iron in PF127DAPG SPIONs.
4.5. Development of alternatives to SPIONs as MPI tracers
Whilst almost all tracers designed for MPI have been based on iron oxide species, there has, in recent years, been evidence to suggest the potential of alternative MNPs to SPIONs for MPI, however research in this area remains underdeveloped. Theoretically, any MNP that is both highly biocompatible, and can display superparamagnetic behaviour with favourable Ms values, could potentially be employed as an effective MPI contrast agent.Almost pure iron nanoparticles have generated exceptionally high values for Ms, up to 176 emu g−1, at sizes of 13 nm.246 However, as with many metal MNPs, they suffer from poor chemical stability and this has limited their application in imaging.247 Such nanoparticles are rapidly oxidised to iron oxide following exposure to air, and therefore require stabilisation for application in MPI. Gloag et al. prepared superparamagnetic single-core nanoparticles with zero valent iron cores, coated with an iron oxide shell and a strongly binding brush co-polymer (Fig. 9a and b).1 The iron oxide coating prevents the rapid oxidation of the metallic core, and the additional polymeric layer provides high colloidal stability, where the nanoparticles could remain water-dispersed for over 8 wk. These nanoparticles show an excellent Ms of 166 emu g−1, at a size of 14 nm (Fig. 9c). At this size, the coated nanoparticles achieve an MPI signal intensity that is ∼80% that of the much larger multi-core VivoTrax (Fig. 9d), whilst also having a very similar spatial resolution. In comparison, SPIONs of a similar size could not generate recognisable signals in MPI, due to their weak values for Ms. Strong MPI properties for tracers at this size is significant, opening the door to many potential MPI applications within cells and the brain, that were not possible with larger nanoparticles.
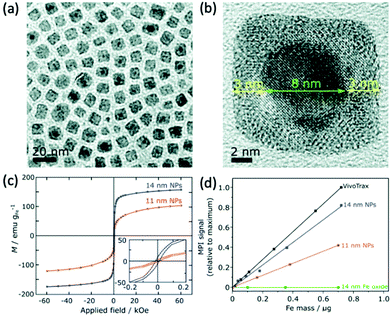 | ||
| Fig. 9 Stable Fe(0) nanoparticles, designed for optimal MPI performance. (a) Low and (b) high resolution TEM (HRTEM) images of the cubic 14 nm nanoparticles. (c) Hysteresis curves displaying the field-dependent magnetism of 14 and 11 nm Fe(0) nanoparticles. (d) Plot of the MPI signal of the synthesised samples in comparison to that of the reference, VivoTrax, as a function of core mass. Reproduced from ref. 1 with permission from the Royal Society of Chemistry. | ||
Metal alloy MNPs have also become of interest for MPI and its applications. FeCo alloyed nanoparticles in particular, demonstrate almost unmatched Ms values up to 215 emu g−1.248 On top of this, they exhibit superparamagnetic behaviour at sizes less than 20 nm, indicating their potential for MPI.249 However, as with iron nanoparticles, they must be stabilised to prevent oxidation on exposure to air. In a recent study, Song et al. synthesised 10 nm FeCo nanoparticles (Fig. 10a) coated with a layer of graphitic carbon and PEG. These exhibit exceptional MPI signal intensities, 6.08 times higher than VivoTrax for equivalent molar core concentrations (Fig. 10b).10 The graphitic carbon coating prevents rapid oxidation of the unstable FeCo core. Many more alloyed nanoparticles exhibit promising properties for MPI application, including FePt (Ms = 100 emu g−1) and Fe5C2 (Ms = 125 emu g−1) particles, but none, as of now, have been investigated for MPI.181,250–252
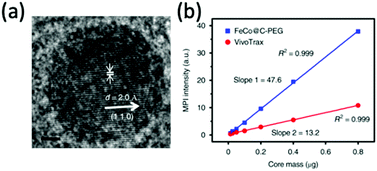 | ||
| Fig. 10 Stable FeCo@C–PEG nanoparticles, designed for MPI and magnetic hyperthermia performance. (a) HRTEM image of the FeCo@C–PEG nanoparticles. (b) Plot of the MPI signal of the FeCo@C–PEG nanoparticles in comparison to that of the reference, VivoTrax, as a function of core mass. Reproduced with permission of Nature Research, from ref. 10; permission conveyed through Copyright Clearance Center, Inc. | ||
Ferrites of the MxFe3−xO4 general formula, display a spinel structure where M could theoretically be any divalent transition metal ion. In the standard Fe3O4 SPION structure, M is Fe2+. Doping of this structure with appropriate metal ions can improve the MPI performance of the particles by altering their magnetic properties.54 Upon doping, the Fe2+ ion is substituted to a desired extent with the new divalent cation dopant (e.g., Zn2+, Co2+, or Mn2+). However, for potential use in biomedical applications, biocompatibility and toxicity of the ferrites is a concern.253
In one recent study, Silvestri et al. synthesised cubic ferrite nanoparticles with a tunable quantity of doped Co and Zn, which were then analysed for their MPI capability.13 Firstly, they synthesised high quality Co-ferrite through a non-hydrolytic synthetic procedure (Fig. 11a). By altering the metal precursors used in this process, the cobalt ions could either be partially substituted with zinc ions, producing mixed Zn–Co-ferrite (Fig. 11b), or totally substituted, producing Zn-ferrite (Fig. 11c). All the cubes were synthesised within a similar size range below 15 nm. Among the different synthesised ferrites, the most impressive MPI properties were achieved with the superparamagnetic Zn-ferrite nanocubes, demonstrating the narrowest FWHM and greatest SNR (Fig. 11d and e). Notably, compared to VivoTrax, the Zn-ferrite nanocubes showed 3-fold higher values for SNR, whereas the Co-ferrite nanocubes exhibited almost zero signal in MPI. These results, combined with the fact that the Zn-ferrite composition is generally deemed more biocompatible than Co-ferrite, with much higher recommended daily intake quantities for Zn than Co,254 indicate the potential of just the Zn-ferrites as effective MPI tracers. These impressive properties of Zn-ferrites in MPI have been echoed in other studies in the literature.190,213,215
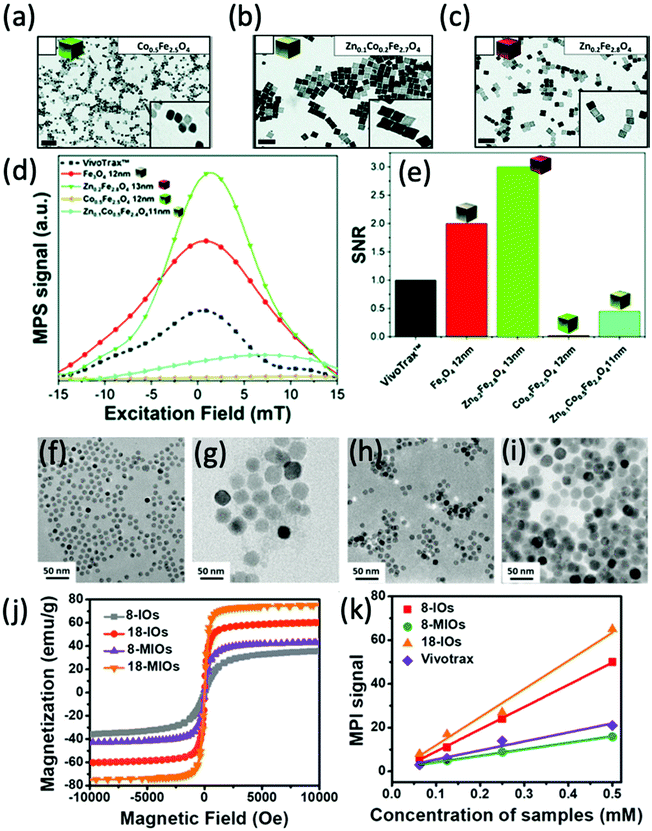 | ||
| Fig. 11 Ferrite nanoparticles, designed for optimal MPI performance. TEM images of different ferrite nanocubes synthesised. The nanoparticles synthesised were (a) Co0.5Fe2.5O4 nanocubes, (b) Zn0.1Co0.2Fe2.7O4 nanocubes, and (c) Zn0.2Fe2.8O4 nanocubes. (d) PSF demonstrating the MPI signal obtained for each synthesised sample, in comparison to that of references, VivoTrax, and Fe3O4 SPIONs. (e) Histograms of the corresponding SNRs for each synthesised sample, the commercial reference, and Fe3O4 SPIONs. Reproduced with permission of the Royal Society of Chemistry, from ref. 13; permission conveyed through Copyright Clearance Center, Inc. TEM images of different SPIONs and Mn-ferrites (MIOs) synthesised. The nanoparticles synthesised were (f) 8 nm SPIONs, (g) 18 nm SPIONs, (h) 8 nm MIOs, and (i) 18 nm MIOs. (j) Hysteresis curves displaying the field-dependent magnetism of each synthesised nanoparticle. (k) Plot of the MPI signal of the SPIONs synthesised in comparison to that of reference, VivoTrax, as a function of sample concentration. Reproduced with permission from ref. 24. Copyright 2019 American Chemical Society. | ||
Other ferrite nanoparticles that have been investigated for their MPI properties include Ni- and Mn-ferrites. Irfan et al. synthesised small Ni-ferrite nanoparticles (9–12 nm), functionalised with either a citric acid (CA) or polyacrylic acid (PAA) coating.255 The MPI performance of the superparamagnetic NiFe2O4@CA and NiFe2O4@PAA nanoparticles was evaluated, both showcasing an improved FWHM over commercially available VivoTrax and Perimag SPIONs. In a different study, Du et al. designed monodisperse single-core Mn-ferrite nanoparticles, with the intention of producing a highly sensitive tracer with good multi-modal MRI/MPI performance and impressive MFH properties.24 Along with both 8 nm, and 18 nm superparamagnetic Mn-ferrite nanoparticles, they also synthesised single-core SPIONs of equivalent sizes (Fig. 11f–i). Although the Mn-ferrite nanoparticles generally exhibit a greater Ms (Fig. 11j) and reduced magnetocrystalline anisotropies in comparison to SPIONs of equivalent core size, they demonstrate a poor MPI signal (Fig. 11k).
5. Primary biomedical applications of MPI
5.1. Cell tracking
One of the earliest applications in MPI was the systematic tracking of cells, as with MRI.256–258 MPI benefits in vivo cell tracking in several ways. The most advantageous characteristics are the superior cellular sensitivity and contrast, and that it allows direct quantification of the SPION content and number of labelled cells at any depth.8 These factors contribute to an in vivo cell detection limit down to ∼200 labelled cells after implantation,26 far surpassing the standard detection limits in MRI.259 There has however been evidence to the capability of MRI to track single cells in vivo,260–262 but as discussed in section 4.4, with advancements in the sensitivity of MPI scanners and tracers, the theoretical detection limit may also be just a single cell.26 Nonetheless, the current properties of MPI permit efficient monitoring of the fate of cell-based therapies and diagnostics, as well as their biodistribution, clearance and retention. There are two primary cell-labelling techniques for cell tracking with MPI.263 The first is in situ labelling, where SPIONs are administered intravenously, and phagocytic cells take up the nanoparticles in situ to deliver them to their target site (Fig. 12a). The second is ex vivo labelling, where the relevant cells are collected from the animal, with subsequent incubation/labelling with the nanoparticles in vitro, and injection back into the animal in vivo (Fig. 12b). Currently, many cells and cell-based therapeutics are tracked for biomedical applications, including cancer cells, stem cells for disease treatment, or immune cells for immunotherapeutic cancer treatment.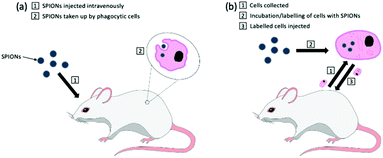 | ||
| Fig. 12 The two primary cell-labelling techniques for cell tracking with MPI: (a) in situ labelling, (b) ex vivo labelling. | ||
One of the most important advantages of SPIONs for this application is their biocompatibility. They have a favourable profile of toxicity, and in vitro, generally showing no, or very little short- or long-term effects on cell viability, differentiation or proliferation, in a range of cellular cultures.264–267 This indicates the great potential for in vivo applications, especially cell tracking, and the clinical translation of MPI.235 One study revealed that rodents can tolerate iron oxide concentrations of up to 3 mmol Fe kg−1.268 A further phase I clinical study on the commonly used SPION tracer, ferucarbotran (Resovist), demonstrated safe use of the tracer at doses of 5–40 μmol Fe kg−1.151 As a further point, SPIONs are completely biodegradable and will remain in the bloodstream until they are metabolised and finally excreted, just like endogenous iron, through the =RES.102 Within this system they are primarily cleared by the liver (80%), spleen (5–8%), and the bone marrow (1–2%).269
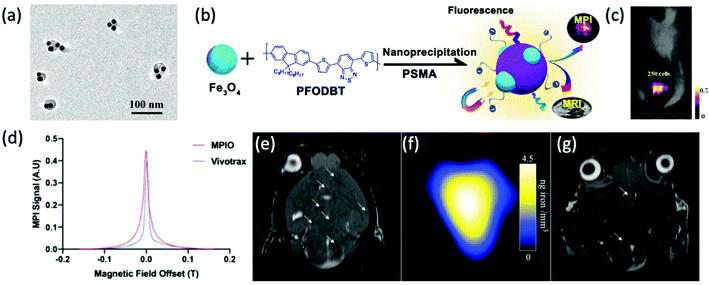 | ||
| Fig. 13 The application of SPIONs in cancerous cell tracking with MPI. (a) TEM image of Fe3O4@PFODBT–COOH Janus nanoparticles, where PFODBT is poly[2,7-(9,9-dioctylfluorene)-alt-4,7-bis (thiophen-2-yl)benzo-2,1,3-thiadiazole]. (b) Schematic demonstrating the synthetic preparation route of Fe3O4@PFODBT–COOH Janus nanoparticles. (c) Overlay of a 2D projection MPI and white light image of a mouse implanted with 250 nanoparticle-labelled cells, following background subtraction. Reproduced with permission from ref. 12. Copyright 2018 American Chemical Society. (d) PSF demonstrating the MPI signal obtained for MPIO, in comparison to that of the reference, VivoTrax. The (e) MRI, and (f) MPI of a mouse brain injected with 5 × 104 4T1BR5 cells labelled with VivoTrax. The (g) MRI, of a mouse brain injected with 5 × 104 4T1BR5 cells labelled with MPIO. There was no MPI signal detected in the mouse brains injected with 5 × 104 4T1BR5 cells labelled with VivoTrax. Reproduced with permission from ref. 29. Copyright 2020 the authors. | ||
Recently, there has been momentum towards studying patient-derived xenograft (PDX) models, supplanting traditional cell lines in cancer research as they better represent the tumor heterogeneity observed in the original tumor.270 Studied results are, therefore, more clinically relevant. The ability to produce quality images and longitudinally track and study the fate of PDXs in vivo would be very valuable. Knier et al. demonstrated the first method for the efficient iron-labelling of PDX cells, which was also the first successful iron-labelling of breast cancer cells derived from patient brain metastases.271 Utilising bioluminescence imaging (BLI) to evaluate cell viability, and MPI for detection and quantification of SPION content, they demonstrated sensitive longitudinal tracking of pre-labelled F2–7 PDX cells with MPIOs, where the signals indicate cell viability and tumor proliferation.
In another work developing imaging tools to target and label primary and metastatic lesions, Parkins et al. looked to exploit the tumor self-homing phenomenon.272 This describes where circulating tumor cells (CTCs) that break away from the primary tumor into the blood stream, preferentially colonise established tumors rather than non-malignant tissues, accelerating metastatic disease. They demonstrate for the first time, the ability of MPI to sensitively detect systemically administered SPION-labelled CTCs and visualise self-homing. This was done in a murine model bearing pre-established human breast cancer lesions, and the labelling demonstrated no detrimental effects on cell viability. The results provide invaluable information about off-target tumor accumulation, as well as the efficiency of CTC infiltration, proliferation, and survival inside tumors, and the potential mechanisms driving self-homing within the body.
 | ||
| Fig. 14 Schematic representation of the TME. Reproduced from ref. 6, with permission from Elsevier. | ||
The most abundant immune cell type in the TME are tumor-associated macrophages (TAMs). The presence of TAMs has been correlated with many protumoral effects, therefore enhancing and contributing to cancer progression.276 Due to their role, they can act as a biomarker to quantifiably monitor both cancer detection and prognosis through imaging. In addition to aiding therapeutic development, the ability to label and image TAMs effectively through SPION phagocytosis will assist in the understanding of their recruitment and infiltration in tumors and could be utilised to predict how aggressive a tumor may be.277
Previous studies have utilised cellular MRI to indirectly image the spatial distribution of TAMs, through in situ SPION uptake.278,279 More recently, Makela et al. demonstrated the first use of MPI in the tracking and detection of SPION-labelled TAMs in vivo, which are intravenously injected in a murine model of a 4T1 breast cancer tumor.277 The images provide quantitative information on the in vivo iron labelling of TAMs for macrophage migration and their association with tumors, showcasing the advantages over equivalent MRI scans. Gerosa et al. also demonstrated MPI imaging of in situ labelled TAMs, where the MPI tumor signal could be detected at doses of just 3 mg kg−1 of Synomag-D.280 Mansfield et al. built on these works by investigating the applicability of MPI to non-invasively track TAMs in response to a CD47 mAb immunotherapy treatment.281 CD47 is a cell surface glycoprotein often overexpressed by cancer cells that prevents the immune system from recognising the cancer.282 Administration of the antibody during treatment blocks CD47, therefore modulating the phagocytosis of macrophages in cancer, and potentially triggering elimination of the cancerous cells. MPI images were acquired 1, 3, and 7 d after tracer injection.281 The increase in MPI signal at the tumor site over this time, in comparison to an untreated control tumor, implies a higher accumulation of TAMs, therefore indicating increased phagocytosis and treatment success.
There is accelerated interest in tracking adoptive T cell cancer immunotherapies with MPI. Adoptive cellular therapy (ACT) is an effective strategy to boost the immune response against many cancers, but faces challenges treating solid tumors and invasive central nervous system malignancies like malignant gliomas.283 An important step in the success of ACT is achieving efficient trafficking and persistence of T cells in solid tumors. Rivera-Rodriguez et al. demonstrate the first application of MPI to track ferucarbotran-labelled T cells in vivo, where the labelling did not affect T cell viability, phenotype, or cytotoxic function.283 The labelled T cells were intraventricularly administered in a mouse model of invasive brain cancer, and their biodistribution and localisation over time was monitored successfully, despite a low uptake of nanoparticles by T cells observed (∼1 pg Fe per cell). The non-invasive quantitative tracking of adoptively transferred T cell biodistributions will assist in the development of new and effective ACT strategies.
In a recent example, Mangarova et al. implemented MPI in the ex vivo monitoring of vascular inflammation in abdominal aortic aneurysms (AAAs).286 AAAs are defined by a weakening and dilatation in the abdominal aorta, usually in the infrarenal portion of the artery, and are currently one of the leading causes of death in developed countries. In this study, the inflammatory response in the aneurysmal wall of an Angiotensin II-infused ApoE −/− mouse model was assessed with in vivo MRI, ex vivo MPI and ex vivo MPS.286 The ex vivo MPI was performed 24 h post-intravenous administration of ferucarbotran (Resovist), following 3–4 wk of Angiotensin II perfusion. The results reveal abundant iron concentration within AAAs following their uptake by macrophages, demonstrating the feasibility of sensitive ex vivo MPI for the detection of vascular inflammation in AAA. These measurements correlated strongly with the results of ex vivo MPS, confirming the inflammatory activity and resulting SPION accumulation in the aneurysmal wall.
The non-invasive tracking of white blood cell (WBC) distributions can also be utilised in the monitoring and diagnosis of inflammation. Traditionally, tracking has been performed using WBCs directly labelled with radionuclides for scintigraphy or SPECT.17 However, this is not ideal for general screening due to the extensive costs, tracer radioactivity, and high dosage of tracer required. In a very recent work, Chandrasekharan et al. explored for the first time, MPI, for the sensitive and radiation-free tracking of WBCs to sites of inflammation and infection.17 For this, they used the commercially available antibody-conjugated multi-core anti-Ly6G SPION (Miltenyi Biotec GmBH, Fig. 15a) for in situ labelling and tracking, which is specific to the Ly6G antigen expressed on neutrophils. Prior to in vivo experiments, the MPI capabilities of this SPION were determined, with anti-Ly6G demonstrating improved values for MPI spatial resolution (1.26 mm vs. 1.49 mm, respectively) and sensitivity (∼1.8 times greater) over multi-core VivoTrax (Fig. 15b and c). The in vivo studies were performed in a murine model of lipopolysaccharide-induced myositis, and the nanoparticles were intravenously injected following inflammation induction. MPI was used to image the biodistribution and demonstrated highly sensitive targeting and detection of inflammation at the sites of myositis, with strong contrast-to-noise ratios between 8 and 13 (Fig. 15d).
 | ||
| Fig. 15 The application of anti-Ly6G SPIONs in the imaging of localised inflammation with MPI. (a) TEM image of the anti-Ly6G SPIONs. PSFs demonstrating the MPI signal in saline, and blood, obtained for (b) anti-Ly6G SPIONs, in comparison to that of the reference, (c) VivoTrax. (d) Inflammation MPI images in three different mouse subjects undergoing myositis, with administered anti-Ly6G–SPIONs. Reproduced from ref. 17 with permission from Ivyspring International Publisher. This is an open access article distributed under the terms of the CC BY License (https://creativecommons.org/licenses/by/4.0/). Copyright 2021, the authors. Article can be found at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7893534/. No changes were made to the original figure. | ||
Mesenchymal stem cells (MSCs), which are found in many types of tissue and are multipotent, have shown particularly promising therapeutic results, where MSC-based therapies have shown success in treating many diseases, like stroke, myocardial infarction, and cancer.296 Importantly, previous MRI studies displayed no decrease in cell viability, proliferation, or differentiation of MSCs after SPION-labelling.297 However, targeted MSC delivery remains a challenge. Commonly, intravenous deliveries of MSCs become entrapped in lung microvasculature instead of the target tissue.298 Zheng et al. sought to better understand this observation as they demonstrated, for the first time, the dynamic tracking of MSC administrations in vivo in a rat model with MPI, employing Resovist to label MSCs.296 The labelled cells were intravenously administered, and the transplantation, dynamic biodistribution, and clearance, were monitored over a 12 d period. A 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) viability assay was carried out, and there was no considerable difference in the cell viabilities of unlabelled and labelled hMSC populations. The MPI images, co-registered with CT, confirm that labelled MSC injections become immediately entrapped in lung tissue and are mostly cleared to the liver within one day. Most significantly though, these results demonstrate that MPI can longitudinally track intravenously administered MSCs safely and quantitatively.
In a different work, Nejadnik et al. utilised dual MPI/MRI to track the status of an implanted MSC scaffold over 14 d.11 The MSCs were labelled with very small FDA-approved ferumoxytol (Feraheme, AMAG Pharmaceuticals) and ferucarbotran (VivoTrax) SPIONs. These labelled MSCs were successfully quantified with MPI at day 1 and 14 (Fig. 16a), with results indicating that MPI was sensitive to changes in cell number of labelled cells at the transplant site over time, with a significant decrease in SPION content observed at the later timepoints (Fig. 16b). Similarly, Sehl et al. implemented a trimodal imaging approach, utilising iron-based 1H MRI, and quantitative MPI and 19F MRI, to monitor the fate of transplanted ferumoxytol-labelled MSCs and the ensuing inflammation, which is inferred through the tracking of infiltrating macrophages at the transplanted sites in vivo.19 This is the first time these three modalities have been combined to monitor cell populations in vivo. Significantly, the viability of these cells was unchanged following MSC labelling, where a 97% viability was determined pre- and post-labelling. These labelled cells were implanted within the hind limb muscle of C57BI/6 mice. A perfluorocarbon agent was also administered intravenously for uptake by phagocytic macrophages in situ. The labelled MSCs were detected using 1H MRI and MPI, and perfluorocarbon-labelled macrophages were detected using 19F MRI. The MPI signal decreased over a 12 d period, which is consistent with the death and clearance of the MSCs, whereas 19F signal persisted (Fig. 16c), suggesting the continuous infiltration of perfluorocarbon-labelled macrophages, and thus inflammation.
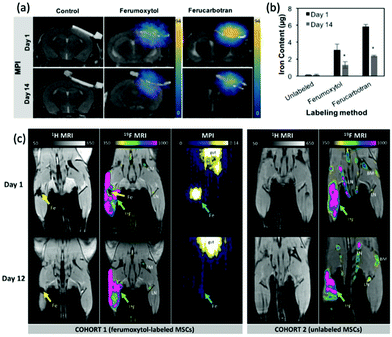 | ||
| Fig. 16 The in vivo tracking of MSCs with MPI. (a) In vivo MPI images at days 1 and 14 following the implantation of ferumoxytol- or ferucarbotran-labelled therapeutic MSCs, and unlabelled MSCs as a control, in calvarial defects of experimental mouse subjects. (b) Iron content in the calvarial defects at days 1 and 14 following implantation of the differently labelled MSCs, as calculated from the MPI signals. Reproduced with permission of Springer Nature, from ref. 11; permission conveyed through Copyright Clearance Center, Inc. (c) In vivo1H/19F MRI and MPI images at days 1 and 12 following the implantation of ferumoxytol-labelled therapeutic MSCs, and unlabelled MSCs as a control, in experimental mouse subjects. Reproduced from ref. 19 with permission from the Multidisciplinary Digital Publishing Institute. This is an open access article distributed under the terms of the CC BY-NC-ND License (https://creativecommons.org/licenses/by-nc-nd/4.0/). Copyright 2019, the authors. Article can be found at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6935990/. No changes were made to the original figure. | ||
In addition to using established SPIONs in the MPI tracking of labelled MSCs,299 several groups have synthesised their own optimised tracers. Lemaster et al. synthesised hybrid poly(lactide-co-glycolide acid) (PLGA)-based iron oxide nanobubbles (Fig. 17a), labelled with a fluorophore, that show potential as a trimodal imaging tracer (MPI/photoacoustic/US) for MSC labelling.8 The PLGA coating facilitates the US signal, the iron oxide core enables the MPI signal, and the fluorophore (DiR, 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindotricarbocyanine iodide) increases the photoacoustic signal (Fig. 17b). They confirmed that there were no adverse effects on cell treatment with nanobubbles in vitro, and hence the nanobubble-labelled cells were injected intramyocardially into live mice for real-time imaging. This multi-modal tracer was shown to track MSCs effectively. In another study, Wang et al. designed a series of specialised cubic tracers with differing edge lengths.18 The tracer with the most beneficial properties for MPI has an edge length of 22 nm, termed CIONs-22 (Fig. 17c). In comparison to the other synthesised tracers of differing sizes, CIONs-22 exhibit good Ms values (Fig. 17d) and high resolution and sensitivity (∼2500 cells) for MPI, (Fig. 17e). With their efficient cellular uptake and labelling, these particles enable the long-term and real-time tracking of bone MSCs in vivo for MPI, exhibiting accurate tracking of their migration and distribution pattern once transplanted to hindlimb ischemia mice.
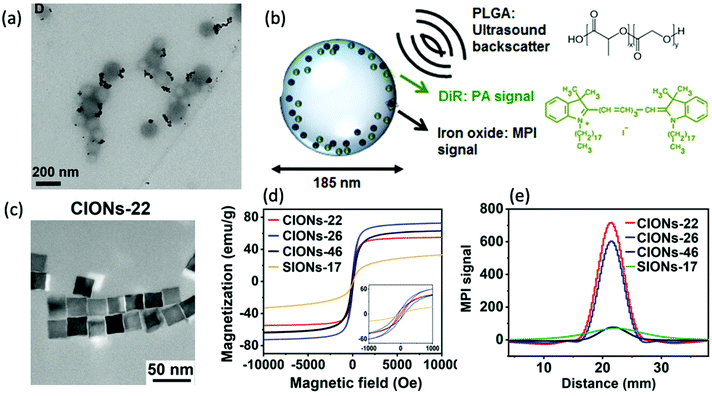 | ||
| Fig. 17 MPI-tailored nanostructures, designed for the tracking of MSCs. (a) TEM image of PLGA-based iron oxide nanobubbles. (b) Schematic of the nanobubbles demonstrating which structural elements generate the different trackable signals. Reproduced with permission from ref. 8. Copyright 2018 American Chemical Society. (c) TEM image of 22 nm cubic CIONs-22 nanoparticles. (d) Hysteresis curves displaying the field-dependent magnetism of each synthesised nanoparticle, where CIONs refers to cubic iron oxide nanoparticles, and SIONs refers to spherical iron oxide nanoparticles. The number following, refers to the edge length. (e) PSF demonstrating the MPI signal obtained for each synthesised sample. Reproduced with permission from ref. 18. Copyright 2018 American Chemical Society. | ||
MPI has also demonstrated promise in the monitoring of neural progenitor cell (NPC) and neural stem cell grafts. Progenitor cells are descendants of stem cells that further differentiate to create specialised cell types. NPCs have been implicated in the treatment of several serious neurodegenerative disorders, such as brain ischemia, epilepsy, and Parkinson's and Alzheimer's disease.48,107 The high sensitivity in MPI could facilitate the quantification of dynamic events, such as graft movement, in such brain disease models. In an important in vivo study, Zheng et al. implanted SPION-labelled NPCs into the forebrain of rats with forebrain ischemia and monitored the in vivo fate of the implanted cells longitudinally with MPI.26 First, the NPCs were differentiated from human embryonic stem cells and labelled with ferucarbotran (Resovist). Following injection, it was demonstrated that the labelled cell grafts could be sensitively and quantitatively detected, and that the particles measured nonsignificant signal decay over a period of 87 d (Fig. 18a and b). The authors also demonstrated a detection sensitivity of 200 cells in their custom-built machine. This study therefore exemplifies the feasibility and advantages of long-term sensitive tracking of NPC and stem cell transplants with MPI.
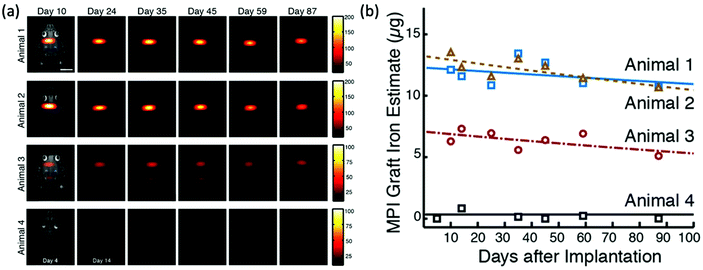 | ||
| Fig. 18 The in vivo tracking of NPCs with MPI. (a) In vivo MPI images at days 1 and 14 following the implantation of 5 × 105 ferucarbotran-labelled NPCs in the forebrain cortex (animals 1–2), near lateral ventricle (animal 3), and with ferucarbotran only, in the forebrain cortex as control (animal 4). (b) Total iron estimation from MPI for the in vivo cell grafts, as a function of time. Reproduced from ref. 26 with permission from Springer Nature. This is an open access article distributed under the terms of the CC BY License (https://creativecommons.org/licenses/by/4.0/). Copyright 2015, the authors. Article can be found at: https://www.nature.com/articles/srep14055. No changes were made to the original figure. | ||
Transplantation of stem cell-derived islet organoids is a promising approach for the treatment of type 1 diabetes.300,301 However, there has traditionally been no appropriate imaging technique for the accurate monitoring of graft outcomes after transplantation. Wang et al. demonstrated the use of MPI in monitoring transplanted islets in animal models for the first time.302 Pancreatic islets were isolated and labelled with VivoTrax SPIONs before transplantation either under the kidney capsule or in the liver of NOD/scid mice. MPI successfully images and quantifies the islets in these models, post-mortem, at days 1 and 14 following transplantation. At the World Molecular Imaging Congress in 2020, Sun described the feasibility of in vivo tracking of transplanted stem cell-derived islet organoids using MPI.303 Human induced pluripotent stem cells were differentiated to islet organoids, which were subsequently labelled with VivoTrax following 21 d of differentiation. The labelled organoids were then similarly transplanted into the left kidney capsule of NOD/scid mice, which were examined by MPI/CT up to 28 d post transplantation. There is a strong signal initially detected in the kidney, but the intensity decreases over the study, which is to be expected with organoid clearance. Additionally, they performed an artificial intelligence analysis of the MPI images, to assist with total iron value prediction of the transplanted cells on different days, using an accurate machine learning algorithm established by the same team of researchers.304 MPI, assisted by this machine learning algorithm analysis, was shown to accurately monitor islet organoids labelled with SPIONs post transplantation and provide quantitative information on their presence in vivo. These results demonstrate the potential for future imaging of cell transplantation and therapy for type 1 diabetes.
5.2. Theranostics
Theranostics, from a clinical and translational viewpoint, refers to an intimate combination of therapeutic and diagnostic interventions. MPI may open unprecedented opportunities for exploring new nanoparticle-based theranostic applications, stemming from the imaging and therapeutic capabilities of SPIONs and the intrinsic properties of MPI itself. In these approaches, diagnostic interventions are most used in combination with targeted MFH or magnetic drug delivery.Often, the SPION tracer and therapeutic molecules are co-loaded into the same nanoparticle. In this context, the SPION shell is commonly modified with pharmacologically active compounds. This can either be done via covalent bonds, or with physical intermolecular interactions, like π–π stacking.107 SPIONs have been conjugated to many different therapeutics. Most frequently, anti-cancer agents have been employed, from standard chemotherapeutic molecules like doxorubicin (DOX),307 to more complex therapeutic compounds like siRNA.308
In recent years, there have been multiple attempts to assemble ‘smart’ multifunctional nanoparticles for theranostic applications, owing to their improved therapeutic and imaging capabilities.309 A common approach here is in the development of nanoparticles with stimuli-responsive polymeric coatings. These coatings typically undergo reversible phase transitions in response to environmental stimuli alterations, most frequently temperature, pH, and enzymatic action. Zhu et al. designed the first ‘smart’ activatable probe for MPI drug delivery applications.15 In this work, they prepared a nanocluster composed of a clustered SPION core with a PLGA coating. By loading the clinically relevant chemotherapeutic drug, DOX, into the PLGA matrix, this nanocluster (labelled SPNCD) can serve as both a dual drug delivery system and an MPI tracer (Fig. 20a). The PLGA shell degrades under mildly acidic biological environments (pH = 6.5), such as at a tumor site, permitting simultaneous DOX release and disassembly of the clustered SPIONs in the core (Fig. 20b). As the SPIONs are released, the MPI signal intensity increases as they recover their Brownian relaxation (Fig. 20c). This allows accurate quantitation and monitoring of drug release in vitro and in vivo (Fig. 20d and e).
 | ||
| Fig. 20 MPI-tailored nanostructures, designed for drug delivery. (a) TEM image of SPNCD nanoclusters. (b) Schematic of the nanoclusters, demonstrating how degradation of the PLGA shell permits simultaneous DOX release and disassembly of the SPIONs clustered in the core. (c) In vitro MPI signal change as a function of time for SPNCD-labelled MDA-MB-231 cells, following degradation of the PLGA shell. (d) In vitro relative DOX release percentages as a function of time from SPNCD-labelled MDA-MB-231 cells, following degradation of the PLGA shell. (e) Correlation between DOX release percentage and MPI signal in cells. The curve exhibits perfectly linear behaviour. Reproduced with permission from ref. 15. Copyright 2019 American Chemical Society. (f) TEM image of DOX-loaded SPION-labelled nanocarriers. (g) Percent release of the nanocarriers as a function of time at 37 °C and varying pH values. Values of 4, 6, and 7.4 were selected for pH, representing the lysosomal, intratumoral, and vascular environments, respectively. Reproduced from ref. 21, with permission from Elsevier. (h) Schematic demonstrating the synthetic preparation, drug binding, and NIR-triggered drug release procedure of Tenofovir disoproxil fumarate (TDF)-loaded gold-coated SPION (MNP@Au) nanostars, where NIR is near-infrared radiation. (i) TEM image of the MNP@Au nanostars. (j) Drug release percentage from the nanostars following illumination with NIR for 5–30 min. Reproduced from ref. 25 with permission from Springer Nature. This is an open access article distributed under the terms of the CC BY License (https://creativecommons.org/licenses/by/4.0/). Copyright 2015, the authors. Article can be found at: https://www.nature.com/articles/s41598-020-66706-2. No changes were made to the original figure. | ||
Fuller et al. also composed magnetic nanocarriers combining high DOX loading and MPI capability (Fig. 20f).21 Unlike SPNCD, these nanocarriers contain the drug within the hydrophobic core, along with the SPIONs, and are coated with a PEG-block-poly(lactic acid) block copolymer, for water solubility and colloidal stability. The release rate of SPIONs and DOX from the nanocarriers was also dependent on environmental pH (Fig. 20g). A different ‘smart’ agent based on the near-infrared-responsive plasmonic properties of tuned nanostars was developed by Tomitaka et al. (Fig. 20h).25 These engineered nanoparticles are composed of a SPION core and a star-shaped plasmonic shell, made up of high-aspect-ratio gold branches (Fig. 20i). Model drug molecules (TDF) are bound to the gold shell and can be triggered to release upon near-infrared illumination because of the photothermal effect in the shell (Fig. 20j). Quantitative MPI can be used to monitor the drug release.
Around 30% of breast cancer tumors will metastasise, most commonly to the liver, lungs, brain and bone.310 Brain metastases are of a particular concern as they are typically fatal. In the treatment of these brain metastases, targeted therapeutics may not be able to, or will have limited penetration, when attempting to cross the BBB, resulting in reduced treatment efficacy.311 Extracellular vesicles (EVs) are small particles released by cells and are implicated in numerous biological processes, including metastasis. These particles may cross the BBB and can therefore be exploited to deliver therapeutic agents to brain metastases.311,312 However, before further progress in this field, it is important to understand the activity, localisation, and accumulation of EVs during metastatic progression. Toomajian et al. demonstrated the tracking of SPION-labelled EVs within brain metastases using MPI.313 For in vivo experiments, mice received an intracardiac injection of 4T1-BR Fluc/GFP cells, a breast cancer cell line that forms brain metastases and expresses firefly luciferase, which allows for BLI imaging of the metastases. 4T1 SPION-labelled EVs were subsequently injected at either early or late stages of metastatic progression and imaged using MPI and BLI to determine the timing of EV accumulation at the metastatic site. This multi-modal experiment demonstrated that EVs can successfully cross the BBB and accumulate in metastatic sites, implicating their therapeutic potential. With the successful tracking of EVs, this experiment also shows the potential for MPI monitoring of treatment.
Exosomes are a type of membrane-bound cell-derived EV, with diameters ranging from 30 to 200 nm.314 They had previously been investigated as nanoparticle carriers for therapeutic delivery, but never imaged with MPI.311,312 Jung et al. demonstrated in vivo MPI imaging of exosomes for the first time, where the exosomes were isolated from MDA-MB-231 human breast cancer cells and labelled with SPIONs and Olaparib.314 Olaparib is an inhibitor of the poly (ADP-ribose) polymerase (PARP) enzyme involved in DNA repair, and is an established cancer therapy. This platform successfully monitors and targets therapeutic delivery towards hypoxic tumors, demonstrating theranostic potential. Imaging and treating these hypoxic regions is a crucial clinical challenge for effective cancer therapy as they are an important factor in therapeutic resistance.
The first proposed use of magnetic hyperthermia with iron oxide particles was as early as 1957,318 yet it remains a prevalent research field today with significant work on the development of novel hyperthermia-tailored nanocarriers.142,319 Interest as a therapeutic treatment can be attributed to multiple key advantages, including: no fundamental depth limitation, synergy with many other therapies (e.g., chemotherapies), and internalisation of the ‘fluidic’ heat source.320 The most frequently used MFH application paradigms are whole-body MFH (Fig. 21a), where homogeneous AMF-generating alternating current (AC) coils scan the whole-body for sites of accumulation, and local MFH (Fig. 21b), where surface AC coils target localised areas of accumulation.320 Despite the advantages, both approaches face several challenges towards further development and eventual clinical translation. Local MFH struggles with deep-seated treatment and therefore can only be used for treatments at the surface of the body. The most pressing issue for the clinical translation of the more commonly applied whole-body MFH is in the localisation of SPIONs at the target site. Systematically administered SPIONs, even if targeted, will end up in other critical organs like the liver and spleen, as well as the target site. This is challenging since accumulation of SPIONs in off-target organs will increase their temperature during AMF application, leading to serious side effects.315 Experiments in mice with systemically administered SPIONs and whole-body AMF show elevated liver enzymes, liver and spleen necrosis, and even death.321
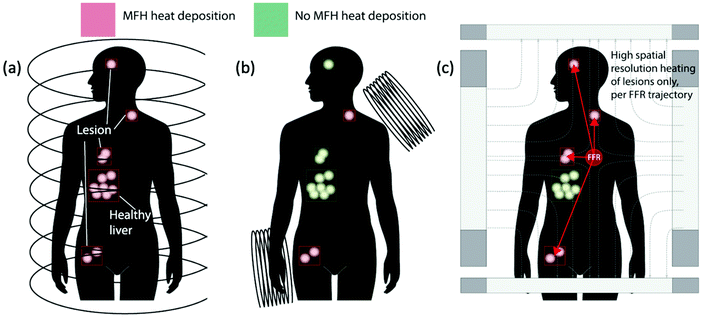 | ||
| Fig. 21 (a) Whole-body MFH targets all tracer, including healthy sites of accumulation. (b) Local MFH targets tracers near the surface only. (c) MPI–MFH can selectively target tracer anywhere, including those deep in the body, while avoiding tracer accumulations in healthy sites. Reproduced with permission of IOP Publishing, from ref. 23; permission conveyed through Copyright Clearance Center, Inc. | ||
To address these important technical challenges, several research groups have demonstrated theoretically and experimentally the benefits of the combined MFH and MPI approach, MPI–MFH (Fig. 21c).177,178,322 The same principles for image generation in MPI, as described in section 2.1, can be modified to spatially select and localise thermal heat deposition to a desired region in biological tissue. MPI uses low sinusoidal excitation frequencies in the drive field, in the order of ∼20 kHz, but by exciting SPIONs to higher frequencies of >300 kHz, heat can be generated.320 The particles under the influence of the selection field are locked, and the particles in the FFR are free to rotate, as before, thereby restricting the SPION heating to the FFR alone. Combination with MPI also allows robust treatment planning. It provides precise quantitative imaging of the SPION distribution within the sample before heating, which is essential for quantification of particle accumulation at the target site and thus accurate SAR prediction and appropriate treatment.315 As a result of this, and because the MPI-based gradients specifically limit the location of the hyperthermia to only a small, adjustable FFR in the sample, MPI–MFH may overcome the previous limitations with off-target heating experienced with whole-body MPI. The established physics of MPI also benefit MFH by providing high resolution targeting anywhere in the body and without depth limitation, addressing the primary problem encountered with local MFH.320 Recently, newer designs for the FFR are being investigated where its size can be varied through alteration of the gradient field strength, thereby allowing correlation of treatment and dosage to the size of the lesion.323
As the physics germane to and exploited by MPI and MFH is similar, the same SPIONs can be used effectively for both. The techniques may also be integrated together in a single device for simultaneous MPI–MFH, that can seamlessly switch between imaging and heating modes through modulation of the AC excitation field magnitude, whilst the subject remains in the scanner. This provides opportunities for real-time diagnostic image monitoring and feedback. Hensley et al. developed the first combined MPI–MFH system, which establishes high heating resolution in phantoms.23 In this work, they applied a 2.35 T m−1 magnetic gradient and a 353 kHz, 13 mT excitation frequency, which demonstrates on-demand selective heating of target components in phantoms, separated by just 3 mm (7 mm centre-to-centre distance). They were also able to repeatedly target these components for heating at will, achieving SAR deposition rates of up to 150 W g−1 and heating rates of 0.4 °C s−1. This system was adopted by Tay et al. for in vivo studies.179 Through this approach, they could localise thermal dose deposition in dual-tumor murine models with a spatio-thermal resolution of 7 mm, and with negligible toxic effects to nearby clearance organs such as the liver. Treatment efficacy and specificity were confirmed with BLI, with a 6-fold decrease in activity shown in the treated tumor and no damage to off-target tissue. More precise localisation of ∼2.3 mm can be achieved by improving the gradient from 2.35 to 7 T m−1, based on previous imaging and theory results.324
From these promising initial studies, commercial MPI–MFH instruments have been recently developed, one of which is the HYPER system (Magnetic Insight Inc., 0.5–2.6 T m−1 gradient field, scan at 340 kHz, 15 mT).325 Gaudet et al. showcased the strong in vitro and in vivo results of this system, demonstrating precise and localised heating of the target.325
Over the last few years, there has been considerable work designing MPI- and hyperthermia-tailored SPIONs with high resolution and sensitivity, as well as efficient heating performance.13,326 Generally, for better heating characteristics, SPIONs should have colloidal stability, phase purity, a narrow size distribution, and optimal magnetic properties.327 In a recent study, Bleul et al. demonstrated the use of a micromixer synthesis platform (Fig. 22a) for the size-controlled synthesis of monodisperse single-core SPIONs, with a core diameter of ∼30 nm.9 These particles show promising theranostic capabilities with high signal amplitudes in MPI, and SAR values of up to 1 kW g(Fe)−1, which exceeds the comparable SAR value of Resovist by more than a factor of three (Fig. 22b). Du et al. also designed monodisperse single-core SPIONs with improved hyperthermia therapy.24 The hyperthermia performance can also be attributed to SPION surface functionalisation with CREKA, a peptide that actively targets fibrin–fibronectin complexes overexpressed in tumor interstitium, therefore improving delivery uniformity and enabling more effective cancer ablation.
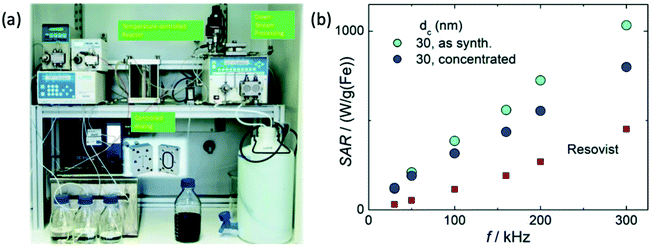 | ||
| Fig. 22 MPI-tailored single-core SPIONs, designed for MPI–MFH. (a) Photograph of the micromixer system used for the synthesis of 30 nm monodisperse single-core SPIONs. (b) SAR values of the synthesised SPIONs as a function of frequency (f), in comparison to that of the reference, Resovist. Reproduced with permission of the Royal Society of Chemistry, from ref. 9; permission conveyed through Copyright Clearance Center, Inc. | ||
Another factor that may affect the heating performance is the choice of coating material. Jordan et al. first noticed a SAR difference between dextran-coated and aminosilan-coated SPIONs, with aminosilan particles having a 1.2-fold greater SAR in identical particles.328 Liu et al. then performed a systemic analysis of the SAR produced by SPIONs coated in different PEG molecular weights, observing a SAR increase with thinner coatings.329 This is attributed to the dominance of Brownian relaxation based heat losses. However, it is noted that reducing the thickness may also affect the colloidal stability of the SPIONs, which detrimentally affects SAR. Differently, shape anisotropy in SPIONs has also been shown to improve hyperthermia performance, as is described in section 4.1. Khurshid et al. reported a 1.4-fold improvement in the particle SAR for cubic SPIONs over spherical SPIONs.330 Bauer et al. also reported the high performance of cubic SPIONs, this time selectively doped with anisotropic zinc, increasing the overall anisotropy.190 They observed a 2-fold enhancement of MPI signal and a 5-fold improvement in SAR, in comparison with equivalent undoped single-core spherical SPIONs.
5.3. Perfusion imaging
Perfusion imaging is a technique whereby the passage of a tracer is monitored continuously through the capillary tissue bed, capturing any temporal changes in the tracer. It has seen extensive application in the diagnosis and detection of various pathophysiology's related to blood perfusion and vascular changes. Stemming from the short scanning time and high temporal resolution of MPI, it has become possible to monitor perfusion and blood-flow in real-time.3 The speed at which images are captured would substantially benefit diseases for which a rapid assessment of the vasculature and perfusion are mandatory for the treatment. Another beneficial property of MPI for perfusion imaging is the high contrast-to-noise, permitting higher accuracy imaging that compares favourably to the traditionally applied techniques of contrast-enhanced CT or MRI, which produce unreliable perfusion images.334Neuropathological diseases such as ischemic stroke, and traumatic brain injury (TBI) are severe conditions requiring immediate medical attention and extensive monitoring following treatment. Specifically stroke, with 17 million cases worldwide every year, is one of the leading annual causes of death and disability.170 Rapid high quality cerebral perfusion imaging is fundamental in successful stroke diagnosis and management, as it allows for early assessment of penumbra volume and location, as well as in predicting which patients may benefit from cerebral revascularisation therapies.335 The low temporal resolution of perfusion MRI and CT can cause variations in the penumbra volume estimate, which may affect optimal stroke treatment.336,337 In a pivotal study, Ludewig et al. demonstrated the potential of MPI in the real-time perfusion imaging and diagnosis of acute stroke in a rodent model.170 Cerebral ischemia was induced in the internal carotid artery of C57BL/6 mice, and within seconds and a single injection of long-circulating LS-008 SPIONs, cerebral perfusions could be assessed by MPI. The signal lowered in the ischemic hemisphere, allowing precise detection of an ischemic stroke within a few cubic millimetres. The SPIONs persist in the bloodstream for hours following injection, without any leakage into the brain interstitium.
Long-circulating tracers are crucial in most perfusion imaging applications, enabling blood flow and blood volume studies. Orendorff et al. exhibited their importance in the imaging of TBI, where a closed-head rat model was monitored longitudinally to study cerebral bleeding caused by an impact, as well as changes in the blood pool as the wound heals.169 This work utilised single-core LS-13 nanoparticles (LodeSpin Labs) that show a circulation half-life of approximately 4–6 h. Such times allowed infiltration into the blood pools in the interstitial space, and thus successful monitoring of TBI with MPI over long time scales, without significant signal loss.
In addition to TBI imaging, MPI has shown great promise for various other blood pool imaging applications, including the detection of gastrointestinal (GI) bleeding. GI bleeding is a serious clinical issue associated with the haemorrhaging of organs in the digestive system, yet its diagnosis remains very challenging. Traditionally, diagnosis is achieved using scintigraphy techniques in which a red blood cell (RBC) is tagged using a radioisotope like 99mTc.338 However, the bleeding site must be located quickly for rapid diagnosis and intervention, and the long hot chemistry preparation times for 99mTc RBCs are not ideal.339 An accurate and timely diagnosis method is therefore essential to reduce morbidity and mortality, and for the determination of transfusion requirements. Yu et al. demonstrated that MPI could be utilised for the safe and quantitative detection of GI bleeding, without any hot chemistry preparation time.58 This work was applied to a murine model that is genetically predisposed to polyp development in the GI lumen (ApcMin/+), with subsequent heparin injection to induce acute GI bleeding. Following the injection of long-circulating MPI-tailored SPIONs (single-core LS-017, LodeSpin Laboratories) through the tail vein, dynamic MPI projection images could be analysed to show tracer accumulation in the lower GI tract. These images illustrate that acute GI bleeding could be detected with high sensitivity using MPI in vivo, with bleed rates as slow as 1 to 5 μL min−1.
Accurate imaging of lung perfusion is required for the diagnosis of conditions like pulmonary embolism (PE), as many patients experience mild or non-specific symptoms. PE carries a 30% mortality rate when left untreated, however following time-sensitive detection and treatment, this mortality rate may reduce to ∼8%.27 Zhou et al. reported the first MPI-based lung perfusion study in vivo in a healthy rat model.27 In this work, they utilised novel multi-core SPIONs that are conjugated to macroaggregated albumin (MAA), aptly termed MAA–SPIONs (Fig. 23a). These are administered intravenously where the particles effectively pass through the lung capillaries after 15 min as the MAA–SPION size is larger than that of the capillary diameter, becoming entrapped in the lung capillary bed (Fig. 23b). This enables the accurate and reliable diagnosis of any vascular related pulmonary defects. In comparison, non-conjugated SPIONs alone cannot target the lung and are instead immediately cleared to the liver and spleen (Fig. 23c). 3D MPI lung perfusion images are comparable in quality to that of traditionally used scintigraphy and SPECT with MMA-conjugated 99mTc, whilst also demonstrating advantages over many modalities due to no air-tissue interface artifacts nor ionising radiation. In a different study, Tay et al. showed that inhaled multi-core SPIONs can be tracked and quantified with MPI in mice, with accuracies comparable to radiolabelled aerosols.158 When taken with the results from Zhou et al.,27 the authors demonstrated a proof-of-concept for MPI-based perfusion–ventilation mapping.340 This mapping has been applied clinically using other modalities, for the diagnosis of PE and in preoperative evaluation of the lungs.
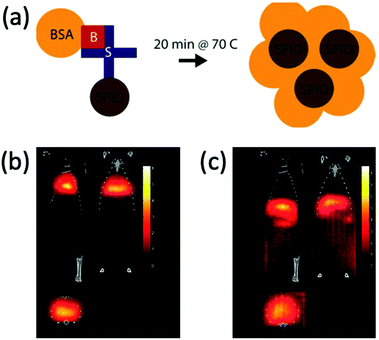 | ||
| Fig. 23 MPI-tailored MAA–SPIONs, designed for lung perfusion imaging. (a) Schematic demonstrating the synthetic preparation of MAA–SPIONs, where biotinylated albumin is conjugated to streptavidin-functionalized SPIONs, and then heated with stirring to form macroaggregates, and B is biotin, S is streptavidin, and BSA is bovine serum albumin. In vivo 3D MPI scans of rats 10 min following intravenous injection, with coronal, sagittal and axial maximum intensity projections (MIPs) shown, where the distribution of (b) MAA–SPIONs are compared with that of the reference, (c) unconjugated SPIONs. Reproduced with permission of IOP Publishing, from ref. 27; permission conveyed through Copyright Clearance Center, Inc. | ||
For tumour visualisation, SPIONs must first become localised within the tumor. Currently this process has relied on the enhanced permeability and retention (EPR) effect, otherwise known as ‘passive targeting’.343,344 This is a phenomenon observed in cancer where the improper alignment of endothelial cells, poor drainage of the lymphatic system, and leaky vasculature of a growing tumor allow larger sized therapeutic agents, like SPIONs or antibodies, access to the interstitial portion of the tumor, to some extent (Fig. 24). This leads to substantial non-targeted accumulation of such agents, at concentrations several folds higher than in the plasma.343
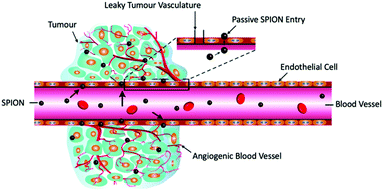 | ||
| Fig. 24 Schematic representation of the EPR effect. Reproduced with permission of the Royal Society of Chemistry, from ref. 7; permission conveyed through Copyright Clearance Center, Inc. | ||
Yu et al. demonstrated the EPR phenomenon with MPI for the detection of triple negative breast cancer xenograft tumors in an immune deficient murine model.92 Long-circulating LS-008 tracers were administered intravenously through the tail vein, and MPI exhibited quantitative visualisation of the tracer dynamics in the tumor over time, showing initial EPR wash-in into the tumors, and delayed wash-out 48 h later. Notably, the tumor can clearly be detected with MPI, even with large cardiac and liver signals, highlighted with a tumor-to-background ratio of up to 50.
There have also been various examples of non-targeted SPIONs that have been specifically developed to exploit the EPR effect for MPI, demonstrating significant increases in accumulation and MPI signal in tumors based on passive targeting alone.10,28 Additionally, to improve the specificity and uptake of SPIONs for cancer detection and imaging, and as passive targeting is not possible in all tumors, there have also been many cases of researchers binding tumor biomarker targeting moieties to SPIONs for MPI.4,24,30 This active targeting should further improve the capability of MPI for early cancer diagnosis.
Kaul et al. demonstrated the improvement in angiographic imaging quality of single-core LS-008, a tracer tailored towards angiography and blood pool imaging in MPI, over MRI-tailored multi-core Resovist.3 For in vivo experiments, the tracers were injected into equivalent healthy mice. Whilst both tracers could visualise the propagation of the bolus through the inferior vena cava (Fig. 25a and b), LS-008 clearly displayed fewer temporally fluctuating artifacts, and the signal modulation in the caval vein, resulting from periodic cardiac and respiratory motion, could be obviously depicted. Additionally, the aorta was clearly distinguishable from the caval vein (Fig. 25c), and several further vessel structures and processes could be observed and monitored that were missing with Resovist. These results demonstrate that regarding the quality of delineation and number of visualised vessels, tailored LS-008 outperforms Resovist in angiographic and perfusion MPI.
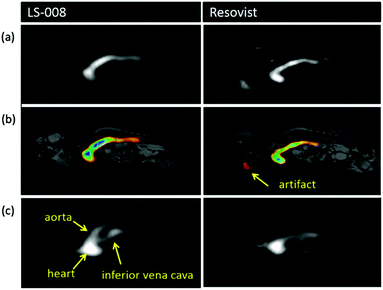 | ||
| Fig. 25 A comparison between angiographic images produced in mice with MPI, using MPI- and angiography-tailored SPIONs, LS-008 (left column), and a reference, Resovist (right column). Propagation of the bolus through the inferior vena cava to the heart for (a) just the MPI signal, and (b) the MPI signal co-registered with MRI. (c) MPI image clearly displaying the aorta leaving the heart when implementing LS-008, but not when implementing Resovist. Reproduced with permission of IOP Publishing, from ref. 3; permission conveyed through Copyright Clearance Center, Inc. | ||
In a similar study, Mohtashamdolatshahi et al. compared the in vivo imaging quality of angiographic MPI for newly developed multi-core particles (MCP 3),206 and Resovist, in the inferior vena cava and aorta.346 The tracers were administered intravenously into the tail veins of rat models at doses of 0.1, 0.05 and 0.025 mmol Fe kg−1. This was followed by serial MPI acquisition using a commercial MPI system. The MCP 3 MPI images demonstrated a significantly higher image quality than those with Resovist. Following administration of MCP 3 at dosages of 0.1, and clinically acceptable 0.05 mmol Fe kg−1, morphological features such as vessel lumen diameters of the inferior vena cava and abdominal aorta could be assessed, which was not possible with Resovist images at any dosage. Additionally, there are fewer severe background noise artifacts than with Resovist images. It can therefore be said that MCP 3 increased the visibility of vessel lumens in vivo in MPI, towards the possible detection of vascular abnormalities.
Differently, Kaul et al. demonstrated the potential of MPI for quantifiable measurements of in vivo blood-flow velocities.347 This study was performed in the inferior vena cava of healthy mice, and the images obtained with MPI were compared with those from MRI. Analysis revealed good agreement between the in vivo velocities, with MRI at 4.0 ± 1.5 cm s−1 and MPI at 4.8 ± 1.1 cm s−1. They also performed an in vitro study, where a phantom setup mimicking the flow within the inferior vena cava concluded that velocities of up to 21 cm s−1 can be captured by MPI as they occur.
Recently, Molwitz et al. obtained the first angiographic MPI images from organs of human-size.348 This work was performed using a multi-modal ex vivo porcine kidney perfusion system compatible with MPI and well-established magnetic resonance angiography (MRA). This developed system practically evaluates MPI's potential for human-sized angiography, which is especially relevant as current angiography-suited MPI scanners still suffer from subject size and spatial resolution restrictions. All the visible vessels in MRA and MPI were counted and compared, where 33% of all vessels imaged in MRA were visible in MPI. This difference is likely a result of the restricted spatial resolution in MPI. Despite these current limitations, this work does demonstrate MPIs capability for detecting vessels within a single human-sized organ and will be useful for improving MPI's angiographic potential.
Chiu-lam et al. reported the first successful example of the cryopreservation and nanowarming process in a whole organ, using an mCPA solution.350 3D MPI was chosen as an appropriate non-invasive and quantitative technology to evaluate the distribution of the perfused mCPA within the organ. MPI demonstrated that a stable mCPA solution containing a VS55 cryopreservation agent, and especially formulated non-toxic SPIONs that have exceptional stability within VS55, can uniformly perfuse whole rat hearts. Once this uniformity was determined, the hearts were successfully vitrified, cryopreserved in liquid nitrogen for up to 1 wk, and rapidly nanowarmed in a uniform fashion to room temperature using an AMF. These formulated SPIONs demonstrated ultrafast nanowarming rates (>320 °C min−1), that can be controlled with AMF amplitude. To complete the process, the SPIONs were subsequently perfused out, at a 95% success rate as determined by MPI. In this work, there was no evidence of macroscopic damage or stress to the hearts after the processes of vitrification, cryopreservation, and nanowarming. These results support the potential of nanowarming, utilising MPI to visualise mCPA distribution, as a strategy for biobanking tissues for transplantation, potentially significantly enhancing the availability of viable donor organs.
6. Multi-colour MPI
Recently, it has been demonstrated that MPI can take advantage of SPIONs with differential magnetic relaxation or harmonic response behaviors to generate multiple contrasts, enabling ‘multi-colour’ imaging and the ability to measure distinct signals corresponding to a specific particle subtype.352,353 This enablement of MPI to simultaneously measure and separate the signal generated by different SPION types would be useful for many applications in the fields of intervention, therapy, and medical imaging.Multi-colour imaging was first experimentally realised using x-space and SFR-based approaches. Hensley et al. demonstrated that x-space reconstruction is capable of differentiating between the relaxation behaviors of different SPIONs, with multiple measurements at different drive field amplitudes.354 Rahmer et al. demonstrated that different SPIONs could be differentiated based on the differences in their harmonic response through the SFR approach, where an extensive calibration procedure was performed separately for each SPION type.38 Through the x-space reconstruction process, the work was first extended to in vivo imaging, where the images successfully differentiate between MAA–SPION tracers in the lungs and Chemicell tracers in the liver of a murine model.355 However, the x-space and SFR approaches rely on extensive calibrations or measurements to capture the differences in the relaxation behaviors or harmonic responses of the nanoparticles. To negate this, Muslu et al. proposed a calibration-free method for x-space MPI, where no prior information about the SPIONs is required.356 This technique can generate a multi-colour relaxation time constant map of different SPION types from a single back and forth scan of the FOV, at a single drive field amplitude.
There has been significant progress in using multi-colour MPI for catheter tracking during vascular interventions.357 In such applications, one SPION type is injected into the blood stream for vessel visualisation, whilst a different type of SPION is used to coat a catheter. Multi-colour MPI can discriminate between the particles impregnated in the catheter and those flowing in the vessels. In a recent work, following the staining of a blood vessel phantom and catheter with different SPIONs, Rahmer et al. demonstrated how 3D real-time multi-colour MPI feedback with an online reconstruction can be used to steer the catheter into a desired vessel phantom, following implementation of an external magnetic field.358
With multi-colour MPI, it is not only possible to distinguish between particles with different characteristics, but also identical particles in different environments.359 Multi-colour MPI can therefore be used to map information relating to the local microenvironment. There are many potential environmental factors where changes could produce different MPI signals. Essentially any factor that causes change in the particle spectrum could form a basis for discrimination. For instance, the temperature mapping capability of multi-colour MPI is well documented, with identical SPIONs in differently heated environments able to be differentiated.360,361 Recently, Paysen et al. were even able to discriminate between free and cell-bound SPIONs by their relaxation behavior, forming multi-colour images (Fig. 26a and b).2 They quantified the dynamic changes that occur when free SPIONs come into contact with and are internalised by cells in vitro over time, without any damage to the cells.
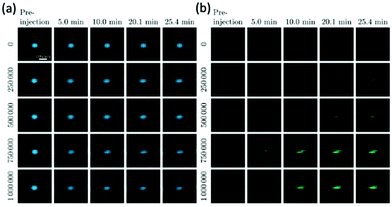 | ||
| Fig. 26 Multi-colour MPI images that distinguish between free and cell-bound SPIONs. Reconstructed MPI images displaying the distribution of (a) free and (b) cell-bound SPIONs in contact with THP-1 cells, as a function of time. Number of cells injected are displayed along the left-hand side of each figure. Following injection, there is an increasing intensity of cell-bound SPIONs in (b) over time as they become internalised. Reproduced from ref. 2 with permission from Springer Nature. This is an open access article distributed under the terms of the CC BY License (https://creativecommons.org/licenses/by/4.0/). Copyright 2020, the authors. Article can be found at: https://www.nature.com/articles/s41598-020-58853-3. No changes were made to the original figure. | ||
There has been sustained work in viscosity mapping with multi-colour MPI.362 Certain diseases, like cancer and atherosclerosis, are known to significantly increase the levels of cellular viscosity. These diseases can be potentially probed with MPI, through tissue viscosity measurements at the locations of SPION accumulation.22 Möddel et al. reported a method that allowed them to experimentally determine the viscosity of a small sample using a novel multi-colour reconstruction approach.363 This approach was adapted from a previously reported method for temperature mapping with MPI.361 A series of samples with differing viscosities were prepared with glycerol/distilled water mixtures of varying proportions and a fixed concentration of Resovist. Using these samples, they were able to determine the viscosity of the particle environment within a range of 1.0–51.8 mPa s, with a methodological error of 6%. More recently, Utkur et al. demonstrated the results of relaxation-based multi-colour MPI for viscosity mapping.22 An imaging phantom that contained SPION samples at five different viscosity levels was prepared (Fig. 27a), again using water/glycerol mixtures of varying proportions. The resulting samples covered the biologically relevant viscosity levels, with values between 0.89 mPa s and 5.04 mPa s. Using the relaxation time constant estimation technique outlined by the same group,356 they showed that relaxation-based experiments can distinguish multi-core SPIONs within the biologically relevant viscosity range (Fig. 27b–d).
 | ||
| Fig. 27 Multi-colour MPI images for viscosity mapping. (a) An imaging phantom of 5 different samples of SPIONs in increasing viscosity levels from 0.89 mPa s to 5.04 mPa s. (b) The MPI images for each sample, with (c) their corresponding relaxation (τ) maps. (d) The color overlay between the τ maps and MPI images for each sample. Reproduced with permission of IOP Publishing, from ref. 22; permission conveyed through Copyright Clearance Center, Inc. | ||
7. Clinical translation
Commercial pre-clinical MPI scanners are only available through Magnetic Insight Inc. and Bruker GmbH. Pre-clinical system specifications are slowly improving as researchers have been highly active in designing improved software for image acquisition and reconstruction.35,37,39,40,364–372 For example, von Gladiss et al. recently demonstrated the advantages of a hybrid system matrix for efficient calibration and image reconstruction.373 To scale-up MPI hardware to human-size, there are several notable safety considerations and ergonomic challenges that must be overcome. Most notably, issues regarding potential allergic reactions to SPION injections,374–376 and the higher cost and power consumption requirements for the larger field amplitudes needed for sufficient image resolution at a clinical scale.80 Initially, tissue heating and peripheral nerve stimulation resulting from time-varying magnetic fields was also thought be an issue. However, studies have demonstrated that the physical properties of the magnetic fields required for scanning human-sized volumes in MPI can be tuned as to not cross the limit at which these effects are generated, without having a negative effect on performance.377–379Despite these obstacles, there has been sustained work on the translation and hardware scale-up.42,43,79,234,380,381 In the design of a functional MPI brain imager, Mason et al. developed simulation studies that demonstrated promising capabilities for human-scale systems.382 In an alternative approach Graeser et al. successfully presented a human-sized MPI hardware set-up, tailored for brain applications, that has low technical requirements for fast and flexible operation in a clinical environment.383 For a different application, Mason et al. also developed a highly sensitive small-bore 2D FFL MPI projection imager, that can rapidly image the distribution of tumors in excised breast tissue, from intravenously injected SPIONs, during breast-conserving surgery treatments for breast cancer.384 This work shows the potential for MPI as a clinical solution to issues with positive margins often seen in such surgeries.
Another vital area for focus in advancement towards clinical MPI is in the development and commercialisation of SPIONs tailored towards MPI physics. Currently, no such SPIONs have been approved for clinical treatment, yet it has already been demonstrated that enhanced MPI performance can be realised through specific SPION design, with improvements in for example, MPI spatial resolution and sensitivity.14,16,218,242,244,245 Notably, further optimisation of the SPIONs for spatial resolution can be traded-off for lower MPI gradients, lowering the overall cost of MPI implementation and easing the scale-up of hardware to human size.229 With different cores, coatings, and the potential conjugation of targeting and therapeutic moieties, it is anticipated that SPIONs will be designed for many different applications and diseases. Particularly, there has been increased interest in the design of smart multifunctional nanoparticle assemblies for drug delivery applications.15,21,25,180,332,333 This work could be extended to designing activatable nanomaterials where the breakdown and release of SPIONs is triggered by enzyme activation. Such particles could be used to measure enzymatic levels in an in vivo area through enzymatic sensing.
8. Conclusions and perspectives
MPI is an emerging radiation-free imaging modality that utilises sensitive, safe, and biocompatible SPIONs as its tracing material. It demonstrates totally unambiguous depth-independent detection of these tracers, negligible background signal, and superb image sensitivity, which enables an essentially infinite contrast and the production of highly specific images with very low detection limits. MPI is also truly linearly quantitative, demonstrating an almost perfectly linear relationship between MPI signal and iron content, allowing estimation of SPION concentration in target tissues, based on just the signal intensity. This quantitation can even be demonstrated in regions that are challenging for other modalities, like the lungs or bone marrow. These great advantages enable a variety of applications of clinical relevance, as well as unprecedented new applications that were inaccessible using other modalities. Recently, impressive progress has been made in pre-clinical animal studies, in areas such as MFH, drug delivery, inflammation imaging, cell tracking, and perfusion imaging.For advancing the biomedical applications of MPI, the design of optimal SPIONs and rigorously controlled syntheses have also become important topics of research. Great progress has been made, with many new SPIONs, and other MNPs, demonstrating impressive improvements in general MPI performance,5,10,14 and performance in specific MPI applications.15,190,218 Critical parameters to control include the size and shape of the core, the specific functionalisation, and of upmost importance, assessing whether a single- or multi-core is appropriate for an intended application.
Despite the sustained research, there is plenty of space for the further expansion of MPI. The development of multi-colour MPI and its reconstruction approaches would drive the modality forward, extracting information relating to the local nanoenvironment that could be used in therapy and diagnostics. Viscosity mapping for example, should be applied in the detection of blood coagulation or diseases that are related to viscosity changes. In addition, MPI has been only recently applied to the nanowarming of cryopreserved organs, where it is implemented to quantitatively assess SPION loading in an organ before vitrification and after nanowarming.350 To further this, nanowarming could potentially be coupled with the capability of MPI to control the location of SPION heating.179 This would facilitate great control over the resulting temperature distribution during the rewarming process. There has also been encouraging progress in the design and development of hybrid scanning systems where MPI is merged with complementary imaging techniques to improve imaging quality, such as in MPI/MRI and MPI/CT.122,123,126,127,132
With multiple promising approaches for clinical translation, improved SPION design, and sustained research into the great number of biomedical applications of clinical promise, it can be anticipated that MPI will be a crucial complimentary clinical diagnostic and therapeutic tool in the near future.
Author contributions
Stanley Harvell-Smith: Investigation, visualization, writing – original draft, writing – review & editing. Le Duc Tung: Writing – review & editing. Nguyen T. K. Thanh: Conceptualization, supervision, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Nguyen T. K. Thanh thanks EPSRC (EP/M015157/1); AOARD (FA2386-17-1-4042 award). Stanley Harvell-Smith thanks EPSRC for his PhD studentship.References
- L. Gloag, M. Mehdipour, M. Ulanova, K. Mariandry, M. A. Nichol, D. J. Hernandez-Castillo, J. Gaudet, R. R. Qiao, J. Zhang, M. Nelson, B. Thierry, M. A. Alvarez-Lemus, T. T. Tan, J. J. Gooding, N. Braidy, P. S. Sachdev and R. D. Tilley, Chem. Commun., 2020, 56, 3504–3507 RSC.
- H. Paysen, N. Loewa, A. Stach, J. Wells, O. Kosch, S. Twamley, M. R. Makowski, T. Schaeffter, A. Ludwig and F. Wiekhorst, Sci. Rep., 2020, 10, 1922 CrossRef CAS PubMed.
- M. G. Kaul, T. Mummert, C. Jung, J. Salamon, A. P. Khandhar, R. M. Ferguson, S. J. Kemp, H. Ittrich, K. M. Krishnan, G. Adam and T. Knopp, Phys. Med. Biol., 2017, 62, 3454–3469 CrossRef CAS PubMed.
- H. Arami, E. Teeman, A. Troksa, H. Bradshaw, K. Saatchi, A. Tomitaka, S. S. Gambhir, U. O. Hafeli, D. Liggitt and K. M. Krishnan, Nanoscale, 2017, 9, 18723–18730 RSC.
- S. K. Avugadda, S. Wickramasinghe, D. Niculaes, M. Ju, A. Lak, N. Silvestri, S. Nitti, I. Roy, A. C. S. Samia and T. Pellegrino, Nanomaterials, 2020, 11, 62 CrossRef PubMed.
- P. Prajapati and D. W. Lambert, J. Bone Oncol., 2016, 5, 128–131 CrossRef PubMed.
- Y. Dai, C. Xu, X. Sun and X. Chen, Chem. Soc. Rev., 2017, 46, 3830–3852 RSC.
- J. E. Lemaster, F. Chen, T. Kim, A. Hariri and J. V. Jokerst, ACS Appl. Nano Mater., 2018, 1, 1321–1331 CrossRef CAS PubMed.
- R. Bleul, A. Baki, C. Freese, H. Paysen, O. Kosch and F. Wiekhorst, Nanoscale Adv., 2020, 2, 4510–4521 RSC.
- G. Song, M. Kenney, Y. S. Chen, X. Zheng, Y. Deng, Z. Chen, S. X. Wang, S. S. Gambhir, H. Dai and J. Rao, Nat. Biomed. Eng., 2020, 4, 325–334 CrossRef CAS PubMed.
- H. Nejadnik, P. Pandit, O. Lenkov, A. P. Lahiji, K. Yerneni and H. E. Daldrup-Link, Mol. Imaging Biol., 2019, 21, 465–472 CrossRef CAS PubMed.
- G. Song, M. Chen, Y. Zhang, L. Cui, H. Qu, X. Zheng, M. Wintermark, Z. Liu and J. Rao, Nano Lett., 2018, 18, 182–189 CrossRef CAS PubMed.
- N. Silvestri, H. Gavil, P. Guardia, R. Brescia, S. Fernandes, A. C. S. Samia, F. J. Teran and T. Pellegrino, Nanoscale, 2021, 13, 13665–13680 RSC.
- Z. W. Tay, S. Savliwala, D. W. Hensley, K. L. B. Fung, C. Colson, B. D. Fellows, X. Zhou, Q. Huynh, Y. Lu, B. Zheng, P. Chandrasekharan, S. M. Rivera-Jimenez, C. M. Rinaldi-Ramos and S. M. Conolly, Small Methods, 2021, 2100796 CrossRef PubMed.
- X. Zhu, J. Li, P. Peng, N. H. Nassab and B. R. Smith, Nano Lett., 2019, 19, 6725–6733 CrossRef CAS PubMed.
- M. Unni, A. M. Uhl, S. Savliwala, B. H. Savitzky, R. Dhavalikar, N. Garraud, D. P. Arnold, L. F. Kourkoutis, J. S. Andrew and C. Rinaldi, ACS Nano, 2017, 11, 2284–2303 CrossRef CAS PubMed.
- P. Chandrasekharan, K. L. B. Fung, X. Y. Zhou, W. Cui, C. Colson, D. Mai, K. Jeffris, Q. Huynh, C. Saayujya, L. Kabuli, B. Fellows, Y. Lu, E. Yu, Z. W. Tay, B. Zheng, L. Fong and S. M. Conolly, Nanotheranostics, 2021, 5, 240–255 CrossRef PubMed.
- Q. Y. Wang, X. B. Ma, H. W. Liao, Z. Y. Liang, F. Y. Li, J. Tian and D. S. Ling, ACS Nano, 2020, 14, 2053–2062 CrossRef CAS PubMed.
- O. C. Sehl, A. V. Makela, A. M. Hamilton and P. J. Foster, Tomography, 2019, 5, 367–376 CrossRef PubMed.
- S. Liu, A. Chiu-Lam, A. Rivera-Rodriguez, R. DeGroff, S. Savliwala, N. Sarna and C. M. Rinaldi-Ramos, Nanotheranostics, 2021, 5, 348–361 CrossRef PubMed.
- E. G. Fuller, G. M. Scheutz, A. Jimenez, P. Lewis, S. Savliwala, S. Liu, B. S. Sumerlin and C. Rinaldi, Int. J. Pharm., 2019, 572, 118796 CrossRef CAS PubMed.
- M. Utkur, Y. Muslu and E. U. Saritas, Phys. Med. Biol., 2017, 62, 3422–3439 CrossRef CAS PubMed.
- D. Hensley, Z. W. Tay, R. Dhavalikar, B. Zheng, P. Goodwill, C. Rinaldi and S. Conolly, Phys. Med. Biol., 2017, 62, 3483–3500 CrossRef PubMed.
- Y. Du, X. Liu, Q. Liang, X. J. Liang and J. Tian, Nano Lett., 2019, 19, 3618–3626 CrossRef CAS PubMed.
- A. Tomitaka, H. Arami, A. Ahmadivand, N. Pala, A. J. McGoron, Y. Takemura, M. Febo and M. Nair, Sci. Rep., 2020, 10, 10115 CrossRef CAS PubMed.
- B. Zheng, T. Vazin, P. W. Goodwill, A. Conway, A. Verma, E. U. Saritas, D. Schaffer and S. M. Conolly, Sci. Rep., 2015, 5, 14055 CrossRef PubMed.
- X. Y. Zhou, K. E. Jeffris, E. Y. Yu, B. Zheng, P. W. Goodwill, P. Nahid and S. M. Conolly, Phys. Med. Biol., 2017, 62, 3510–3522 CrossRef CAS PubMed.
- G. Song, X. Zheng, Y. Wang, X. Xia, S. Chu and J. Rao, ACS Nano, 2019, 13, 7750–7758 CrossRef CAS PubMed.
- K. P. Melo, A. V. Makela, A. M. Hamilton and P. J. Foster, bioRxiv, 2020 DOI:10.1101/2020.07.12.197780 , preprint.
- A. Tomitaka, S. Ota, K. Nishimoto, H. Arami, Y. Takemura and M. Nair, Nanoscale, 2019, 11, 6489–6496 RSC.
- B. Gleich and R. Weizenecker, Nature, 2005, 435, 1214–1217 CrossRef CAS PubMed.
- P. W. Goodwill and S. M. Conolly, IEEE Trans. Magn., 2011, 30, 1581–1590 Search PubMed.
- P. W. Goodwill, PhD Thesis, UC Berkeley, 2010.
- Bruker BioSpin GmbH, https://ir.bruker.com/press-releases/press-release-details/2013/Bruker-Announces-the-Worlds-First-Preclinical-Magnetic-Particle-Imaging-MPI-System/default.aspx, (accessed March 2021).
- H. Bagheri and M. E. Hayden, J. Magn. Magn. Mater., 2020, 498, 166021 CrossRef CAS.
- K. Murase, T. Konishi, Y. Takeuchi, H. Takata and S. Saito, Radiol. Phys. Technol., 2013, 6, 399–414 CrossRef PubMed.
- K. Murase, S. Hiratsuka, R. X. Song and Y. Takeuchi, Jpn. J. Appl. Phys., 2014, 53, 067001 CrossRef CAS.
- J. Rahmer, A. Halkola, B. Gleich, I. Schmale and J. Borgert, Phys. Med. Biol., 2015, 60, 1775–1791 CrossRef CAS PubMed.
- V. Schulz, M. Straub, M. Mahlke, S. Hubertus, T. Lammers and F. Kiessling, IEEE Trans. Magn., 2015, 51, 6501804 Search PubMed.
- M. Straub and V. Schulz, IEEE Trans. Magn., 2018, 37, 1192–1203 Search PubMed.
- J. Borgert, J. D. Schmidt, I. Schmale, C. Bontus, B. Gleich, B. David, J. Weizenecker, J. Jockram, C. Lauruschkat, O. Mende, M. Heinrich, A. Halkola, J. Bergmann, O. Woywode and J. Rahmer, Biomed. Tech., 2013, 58, 551–556 Search PubMed.
- J. Rahmer, C. Stehning and B. Gleich, PLoS One, 2018, 13, e0193546 CrossRef PubMed.
- C. B. Top, S. Ilbey and H. E. Güven, Med. Phys., 2017, 44, 6225–6238 CrossRef PubMed.
- E. U. Saritas, P. W. Goodwill, L. R. Croft, J. J. Konkle, K. Lu, B. Zheng and S. M. Conolly, J. Magn. Reson., 2013, 229, 116–126 CrossRef CAS PubMed.
- B. Zheng, E. Yu, R. Orendorff, K. Lu, J. J. Konkle, Z. W. Tay, D. Hensley, X. Y. Zhou, P. Chandrasekharan, E. U. Saritas, P. W. Goodwill, J. D. Hazle and S. M. Conolly, Mol. Imaging Biol., 2017, 19, 385–390 CrossRef PubMed.
- P. W. Goodwill, J. J. Konkle, B. Zheng, E. U. Saritas and S. M. Conolly, IEEE Trans. Med. Imaging, 2012, 31, 1076–1085 Search PubMed.
- A. P. Khandhar, R. M. Ferguson, H. Arami and K. M. Krishnan, Biomaterials, 2013, 34, 3837–3845 CrossRef CAS PubMed.
- A. Meola, J. Rao, N. Chaudhary, G. Song, X. Zheng and S. D. Chang, World Neurosurg., 2019, 125, 261–270 CrossRef PubMed.
- J. Haegele, J. Rahmer, B. Gleich, J. Borgert, H. Wojtczyk, N. Panagiotopoulos, T. M. Buzug, J. Barkhausen and F. M. Vogt, Radiology, 2012, 265, 933–938 CrossRef PubMed.
- J. Weizenecker, B. Gleich, J. Rahmer, H. Dahnke and J. Borgert, Phys. Med. Biol., 2009, 54, L1–L10 CrossRef CAS PubMed.
- R. M. Ferguson, A. P. Khandhar and K. M. Krishnan, J. Appl. Phys., 2012, 111, 7B318-317B3185 CrossRef PubMed.
- R. M. Ferguson, K. R. Minard, A. P. Khandhar and K. M. Krishnan, Med. Phys., 2011, 38, 1619–1626 CrossRef PubMed.
- R. M. Ferguson, K. R. Minard and K. M. Krishnan, J. Magn. Magn. Mater., 2009, 321, 1548–1551 CrossRef CAS PubMed.
- C. Lu, L. B. Han, J. N. Wang, J. C. Wan, G. S. Song and J. H. Rao, Chem. Soc. Rev., 2021, 50, 8102–8146 RSC.
- P. W. Goodwill and S. M. Conolly, IEEE Trans. Med. Imaging, 2010, 29, 1851–1859 Search PubMed.
- J. Rahmer, J. Weizenecker, B. Gleich and J. Borgert, BMC Med. Imaging, 2009, 9, 4 CrossRef PubMed.
- T. M. Buzug and J. Borgert, Magnetic Particle Imaging, Springer Proceedings in Physics, New York, 2012 Search PubMed.
- E. Y. Yu, P. Chandrasekharan, R. Berzon, Z. W. Tay, X. Y. Zhou, A. P. Khandhar, R. M. Ferguson, S. J. Kemp, B. Zheng, P. W. Goodwill, M. F. Wendland, K. M. Krishnan, S. Behr, J. Carter and S. M. Conolly, ACS Nano, 2017, 11, 12067–12076 CrossRef CAS PubMed.
- P. W. Goodwill, L. R. Croft, J. J. Konkle, K. Lu, E. U. Saritas, B. Zheng and S. M. Conolly, A 7 T/M 3D X-space MPI mouse and rat scanner, IEEE, New York, 2013 Search PubMed.
- L. Gutierrez, R. Costo, C. Gruttner, F. Westphal, N. Gehrke, D. Heinke, A. Fornara, Q. A. Pankhurst, C. Johansson, S. Veintemillas-Verdaguer and M. P. Morales, Dalton Trans., 2015, 44, 2943–2952 RSC.
- J. Wells, N. Lowa, H. Paysen, U. Steinhoff and F. Wiekhorst, J. Magn. Magn. Mater., 2019, 475, 421–428 CrossRef CAS.
- L. C. Wu, Y. Zhang, G. Steinberg, H. Qu, S. Huang, M. Cheng, T. Bliss, F. Du, J. Rao, G. Song, L. Pisani, T. Doyle, S. M. Conolly, K. M. Krishnan, G. Grant and M. Wintermark, Am. J. Neuroradiol., 2019, 40, 206–212 CrossRef CAS PubMed.
- R. M. Ferguson, A. P. Khandhar, S. J. Kemp, H. Arami, E. U. Saritas, L. R. Croft, J. Konkle, P. W. Goodwill, A. Halkola, J. Rahmer, J. Borgert, S. M. Conolly and K. M. Krishnan, IEEE Trans. Med. Imaging, 2015, 34, 1077–1084 Search PubMed.
- B. Gleich, J. Weizenecker and J. Borgert, Phys. Med. Biol., 2008, 53, N81–N84 CrossRef CAS PubMed.
- J. Weizenecker, J. Borgert and B. Gleich, Phys. Med. Biol., 2007, 52, 6363–6374 CrossRef CAS PubMed.
- T. F. Sattel, T. Knopp, S. Biederer, B. Gleich, J. Weizenecker, J. Borgert and T. M. Buzug, J. Phys. D: Appl. Phys., 2009, 42, 022001 CrossRef.
- T. Knopp, S. Biederer, T. F. Sattel, J. Rahmer, J. Weizenecker, B. Gleich, J. Borgert and T. M. Buzug, Med. Phys., 2010, 37, 485–491 CrossRef PubMed.
- T. Knopp, T. F. Sattel, S. Biederer, J. Rahmer, J. Weizenecker, B. Gleich, J. Borgert and T. M. Buzug, IEEE Trans. Med. Imaging, 2010, 29, 12–18 Search PubMed.
- T. Knopp, S. Biederer, T. Sattel, J. Weizenecker, B. Gleich, J. Borgert and T. M. Buzug, Phys. Med. Biol., 2009, 54, 385–397 CrossRef CAS PubMed.
- P. W. Goodwill, K. Lu, B. Zheng and S. M. Conolly, Rev. Sci. Instrum., 2012, 83, 033708 CrossRef PubMed.
- S. Kurt, Y. Muslu and E. U. Saritas, IEEE Trans. Med. Imaging, 2020, 39, 3441–3450 Search PubMed.
- M. Weber, J. Beuke, A. Gladiss, K. Gräfe, P. Vogel, V. C. Behr and T. M. Buzug, Int. J. Magn. Part. Imaging, 2018, 4, 1811004 Search PubMed.
- E. Yagiza, A. R. Cagila and E. U. Saritasa, Int. J. Magn. Part. Imaging, 2020, 6, 2006001 Search PubMed.
- M. Erbe, T. Knopp, T. F. Sattel, S. Biederer and T. M. Buzug, Med. Phys., 2011, 38, 5200–5207 CrossRef PubMed.
- K. Nomura, K. Yamauchi, T. Matsuda, S. Tonooka, K. Yoshida, Y. Susumu, Y. Okada and S. Sato, presented in part of World Molecular Imaging Congress (WMIC), Virtual, October 2020.
- J. Weizenecker, B. Gleich and J. Borgert, J. Phys. D: Appl. Phys., 2008, 41, 105009 CrossRef.
- M. H. Pablico-Lansigan, S. F. Situ and A. C. Samia, Nanoscale, 2013, 5, 4040–4055 RSC.
- T. Knopp, M. Erbe, T. F. Sattel, S. Biederer and T. M. Buzug, Appl. Phys. Lett., 2010, 97, 092505 CrossRef.
- C. B. Top and A. Gungor, IEEE Trans. Med. Imaging, 2020, 39, 4164–4173 Search PubMed.
- A. C. Bakenecker, M. Ahlborg, C. Debbeler, C. Kaethner, T. M. Buzug and K. Ludtke-Buzug, Innovative Surg. Sci., 2018, 3, 179–192 Search PubMed.
- J. Guzy, S. Chakravarty, F. J. Buchanan, H. Chen, J. M. Gaudet, J. M. L. Hix, C. L. Mallett and E. M. Shapiro, ACS Appl. Nano Mater., 2020, 3, 3991–3999 CrossRef CAS PubMed.
- P. Chandrasekharan, Z. W. Tay, X. Y. Zhou, E. Yu, R. Orendorff, D. Hensley, Q. Huynh, K. L. B. Fung, C. C. VanHook, P. Goodwill, B. Zheng and S. Conolly, Br. J. Radiol., 2018, 91, 20180326 CrossRef PubMed.
- R. E. Rosensweig, J. Magn. Magn. Mater., 2002, 252, 370–374 CrossRef CAS.
- R. J. Deissler, Y. Wu and M. A. Martens, Med. Phys., 2014, 41, 012301 CrossRef PubMed.
- P. W. Goodwill, A. Tamrazian, L. R. Croft, C. D. Lu, E. M. Johnson, R. Pidaparthi, R. M. Ferguson, A. P. Khandhar, K. M. Krishnan and S. M. Conolly, Appl. Phys. Lett., 2011, 98, 262502 CrossRef.
- K. M. Krishnan, IEEE Trans. Magn., 2010, 46, 2523–2558 CAS.
- G. Hong, A. L. Antaris and H. Dai, Nat. Biomed. Eng., 2017, 1, 0010 CrossRef CAS.
- H. Hong, Y. Yang, Y. Zhang and W. Cai, Curr. Top. Med. Chem., 2010, 10, 1237–1248 CrossRef CAS PubMed.
- E. T. Ahrens and J. W. Bulte, Nat. Rev. Immunol., 2013, 13, 755–763 CrossRef CAS PubMed.
- W. Cai, G. Niu and X. Chen, Curr. Pharm. Des., 2008, 14, 2943–2973 CrossRef CAS PubMed.
- T. F. Massoud and S. S. Gambhir, Genes Dev., 2003, 17, 545–580 CrossRef CAS PubMed.
- E. Y. Yu, M. Bishop, B. Zheng, R. M. Ferguson, A. P. Khandhar, S. J. Kemp, K. M. Krishnan, P. W. Goodwill and S. M. Conolly, Nano Lett., 2017, 17, 1648–1654 CrossRef CAS PubMed.
- N. Panagiotopoulos, R. L. Duschka, M. Ahlborg, G. Bringout, C. Debbeler, M. Graeser, C. Kaethner, K. Lüdtke-Buzug, H. Medimagh, J. Stelzner, T. M. Buzug, J. Barkhausen, F. M. Vogt and J. Haegele, Int. J. Nanomed., 2015, 10, 3097–3114 CrossRef CAS PubMed.
- R. Hachani, M. Lowdell, M. Birchall and N. T. Thanh, Nanoscale, 2013, 5, 11362–11373 RSC.
- Q. A. Pankhurst, N. K. T. Thanh, S. K. Jones and J. Dobson, J. Phys. D: Appl. Phys., 2009, 42, 224001 CrossRef.
- A. R. Kherlopian, T. Song, Q. Duan, M. A. Neimark, M. J. Po, J. K. Gohagan and A. F. Laine, BMC Syst. Biol., 2008, 2, 74 CrossRef PubMed.
- N. Talebloo, M. Gudi, N. Robertson and P. Wang, J. Magn. Reson. Imaging, 2020, 51, 1659–1668 CrossRef PubMed.
- J. F. Schenck, Med. Phys., 1996, 23, 815–850 CrossRef CAS PubMed.
- J. W. M. Bulte, P. Walczak, B. Gleich, J. Weizenecker, D. E. Markov, H. C. J. Aerts, H. Boeve, J. Borgert and M. Kuhne, Proc. SPIE-Int. Soc. Opt. Eng., 2011, 7965, 79650z CrossRef PubMed.
- E. B. Ehlerding, P. Grodzinski, W. B. Cai and C. H. Liu, ACS Nano, 2018, 12, 2106–2121 CrossRef CAS PubMed.
- G. Kandasamy and D. Maity, Mater. Sci. Eng., C, 2021, 127, 112199 CrossRef CAS PubMed.
- Y. X. Wang, Quant. Imaging Med. Surg., 2011, 1, 35–40 Search PubMed.
- A. B. Nayak, A. Luhar, M. Hanudel, B. Gales, T. R. Hall, J. P. Finn, I. B. Salusky and J. Zaritsky, Pediatr. Nephrol., 2015, 30, 515–521 CrossRef PubMed.
- R. A. Revia and M. Q. Zhang, Mater. Today, 2016, 19, 157–168 CrossRef CAS PubMed.
- E. A. Neuwelt, B. E. Hamilton, C. G. Varallyay, W. R. Rooney, R. D. Edelman, P. M. Jacobs and S. G. Watnick, Kidney Int., 2009, 75, 465–474 CrossRef CAS PubMed.
- S. Caspani, R. Magalhaes, J. P. Araujo and C. T. Sousa, Materials, 2020, 13, 2586 CrossRef CAS PubMed.
- S. M. Dadfar, K. Roemhild, N. I. Drude, S. von Stillfried, R. Knuchel, F. Kiessling and T. Lammers, Adv. Drug Delivery Rev., 2019, 138, 302–325 CrossRef CAS PubMed.
- J. W. Bulte, J. Vymazal, R. A. Brooks, C. Pierpaoli and J. A. Frank, J. Magn. Reson. Imaging, 1993, 3, 641–648 CrossRef CAS PubMed.
- J. W. Bulte, AJR, Am. J. Roentgenol., 2009, 193, 314–325 CrossRef PubMed.
- Y. Wang, C. Xu, Y. Chang, L. Zhao, K. Zhang, Y. Zhao, F. Gao and X. Gao, ACS Appl. Mater. Interfaces, 2017, 9, 28959–28966 CrossRef CAS PubMed.
- W. C. Xiao, P. Legros, P. Chevallier, J. Lagueux, J. K. Oh and M. A. Fortin, ACS Appl. Nano Mater., 2018, 1, 894–907 CrossRef CAS.
- H. Groult, N. Poupard, F. Herranz, E. Conforto, N. Bridiau, F. Sannier, S. Bordenave, J. M. Piot, J. Ruiz-Cabello, I. Fruitier-Arnaudin and T. Maugard, Biomacromolecules, 2017, 18, 3156–3167 CrossRef CAS PubMed.
- S. Magnitsky, J. Zhang, D. Idiyatullin, G. Mohan, M. Garwood, N. E. Lane and S. Majumdar, Magn. Reson. Med., 2017, 78, 1900–1910 CrossRef CAS PubMed.
- J. Zhang, H. L. Ring, K. R. Hurley, Q. Shao, C. S. Carlson, D. Idiyatullin, N. Manuchehrabadi, P. J. Hoopes, C. L. Haynes, J. C. Bischof and M. Garwood, Magn. Reson. Med., 2017, 78, 702–712 CrossRef CAS PubMed.
- J. Pellico, J. Ruiz-Cabello, I. Fernandez-Barahona, L. Gutierrez, A. V. Lechuga-Vieco, J. A. Enriquez, M. P. Morales and F. Herranz, Langmuir, 2017, 33, 10239–10247 CrossRef CAS PubMed.
- L. Sandiford, A. Phinikaridou, A. Protti, L. K. Meszaros, X. Cui, Y. Yan, G. Frodsham, P. A. Williamson, N. Gaddum, R. M. Botnar, P. J. Blower, M. A. Green and R. T. de Rosales, ACS Nano, 2013, 7, 500–512 CrossRef CAS PubMed.
- F. Hu, Q. Jia, Y. Li and M. Gao, Nanotechnology, 2011, 22, 245604 CrossRef PubMed.
- M. Jeon, M. V. Halbert, Z. R. Stephen and M. Q. Zhang, Adv. Mater., 2021, 33, 1906539 CrossRef CAS PubMed.
- Z. J. Zhou, Z. H. Zhao, H. Zhang, Z. Y. Wang, X. Y. Chen, R. F. Wang, Z. Chen and J. H. Gao, ACS Nano, 2014, 8, 7976–7985 CrossRef CAS PubMed.
- M. O. Besenhard, L. Panariello, C. Kiefer, A. P. LaGrow, L. Storozhuk, F. Perton, S. Begin, D. Damien Mertz, N. T. K. Thanh and A. Gavriilidis, Nanoscale, 2021, 13, 8795–8805 RSC.
- M. S. Judenhofer, H. F. Wehrl, D. F. Newport, C. Catana, S. B. Siegel, M. Becker, A. Thielscher, M. Kneilling, M. P. Lichy, M. Eichner, K. Klingel, G. Reischl, S. Widmaier, M. Rocken, R. E. Nutt, H. J. Machulla, K. Uludag, S. R. Cherry, C. D. Claussen and B. J. Pichler, Nat. Med., 2008, 14, 459–465 CrossRef CAS PubMed.
- J. Franke, U. Heinen, H. Lehr, A. Weber, F. Jaspard, W. Ruhm, M. Heidenreich and V. Schulz, IEEE Trans. Med. Imaging, 2016, 35, 1993–2004 Search PubMed.
- P. Vogel, S. Lother, M. A. Ruckert, W. H. Kullmann, P. M. Jakob, F. Fidler and V. C. Behr, IEEE Trans. Med. Imaging, 2014, 33, 1954–1959 Search PubMed.
- M. G. Kaul, O. Weber, U. Heinen, A. Reitmeier, T. Mummert, C. Jung, N. Raabe, T. Knopp, H. Ittrich and G. Adam, RöFo, 2015, 187, 347–352 CAS.
- J. Salamon, M. Hofmann, C. Jung, M. G. Kaul, F. Werner, K. Them, R. Reimer, P. Nielsen, A. Vom Scheidt, G. Adam, T. Knopp and H. Ittrich, PLoS One, 2016, 11, e0156899 CrossRef PubMed.
- J. Franke, U. Heinen, L. Matthies, V. Niemann, F. Jaspard, M. Heidenreich and T. M. Buzug, First hybrid MPI-MRI imaging system as integrated design for mice and rats: Description of the instrumentation setup, IEEE, New York, 2013 Search PubMed.
- J. Franke, U. Heinen, H. Lehr, A. Weber, F. Jaspard, W. Ruhm, M. Heidenreich and V. Schulz, First 3D dual modality phantom measurements of a hybrid MPI-MRI system using a resistive 12 channel MPI-MRI magnet design, IEEE, New York, 2015 Search PubMed.
- J. Franke, N. Baxan, H. Lehr, U. Heinen, S. Reinartz, J. Schnorr, M. Heidenreich, F. Kiessling and V. Schulz, IEEE Trans. Med. Imaging, 2020, 39, 4335–4345 Search PubMed.
- T. C. Kwee, R. M. Kwee and R. A. Nievelstein, Blood, 2008, 111, 504–516 CrossRef CAS PubMed.
- D. Lardinois, W. Weder, T. F. Hany, E. M. Kamel, S. Korom, B. Seifert, G. K. von Schulthess and H. C. Steinert, N. Engl. J. Med., 2003, 348, 2500–2507 CrossRef PubMed.
- S. Monti, C. Cavaliere, M. Covello, E. Nicolai, M. Salvatore and M. Aiello, J. Healthcare Eng., 2017, 2017, 2634389 Search PubMed.
- P. Vogel, J. Markert, M. A. Ruckert, S. Herz, B. Kessler, K. Dremel, D. Althoff, M. Weber, T. M. Buzug, T. A. Bley, W. H. Kullmann, R. Hanke, S. Zabler and V. C. Behr, Sci. Rep., 2019, 9, 12627 CrossRef PubMed.
- P. Goodwill, K. M. Krishnan and S. M. Conolly, in Magnetic Nanoparticles: From Fabrication to Clinical Applications, ed. N. T. K. Thanh, Taylor & Francis, Oxfordshire, 1st edn, 2012, vol. 20, pp. 523–541 Search PubMed.
- X. Y. Zhou, Z. W. Tay, P. Chandrasekharan, E. Y. Yu, D. W. Hensley, R. Orendorff, K. E. Jeffris, D. Mai, B. Zheng, P. W. Goodwill and S. M. Conolly, Curr. Opin. Chem. Biol., 2018, 45, 131–138 CrossRef CAS PubMed.
- A. K. Gupta, R. R. Naregalkar, V. D. Vaidya and M. Gupta, Nanomedicine, 2007, 2, 23–39 CrossRef CAS PubMed.
- H. Ittrich, K. Peldschus, N. Raabe, M. Kaul and G. Adam, RöFo, 2013, 185, 1149–1166 CAS.
- M. H. Schwenk, Pharmacotherapy, 2010, 30, 70–79 CrossRef CAS PubMed.
- S. S. Vasanawala, K. L. Nguyen, M. D. Hope, M. D. Bridges, T. A. Hope, S. B. Reeder and M. R. Bashir, Magn. Reson. Med., 2016, 75, 2107–2111 CrossRef PubMed.
- J. He, M. Huang, D. Wang, Z. Zhang and G. Li, J. Pharm. Biomed. Anal., 2014, 101, 84–101 CrossRef CAS PubMed.
- D. W. Inglis, R. Riehn, R. H. Austin and J. C. Sturm, Appl. Phys. Lett., 2004, 5093 CrossRef CAS.
- K. Hola, Z. Markova, G. Zoppellaro, J. Tucek and R. Zboril, Biotechnol. Adv., 2015, 33, 1162–1176 CrossRef CAS PubMed.
- L. Wang, A. Hervault, P. Southern, O. Sandre, F. Couillaud and N. T. K. Thanh, J. Mater. Chem. B, 2020, 8, 10527–10539 RSC.
- L. Wu, Y. Cao, C. Liao, J. Huang and F. Gao, Eur. J. Radiol., 2011, 80, 582–589 CrossRef PubMed.
- J. J. Pouw, M. R. Grootendorst, R. Bezooijen, C. A. Klazen, W. I. De Bruin, J. M. Klaase, M. A. Hall-Craggs, M. Douek and B. Ten Haken, Br. J. Radiol., 2015, 88, 20150634 CrossRef PubMed.
- S. Namkung, C. J. Zech, T. Helmberger, M. F. Reiser and S. O. Schoenberg, J. Magn. Reson. Imaging, 2007, 25, 755–765 CrossRef PubMed.
- S. Tokunaga, M. Koda, T. Matono, T. Sugihara, T. Nagahara, M. Ueki, Y. Murawaki, S. Kakite and E. Yamashita, Br. J. Radiol., 2012, 85, 745–752 CrossRef CAS PubMed.
- N. Ruangwattanapaisarn, A. Hsiao and S. S. Vasanawala, Pediatr. Radiol., 2015, 45, 831–839 CrossRef PubMed.
- B. Bonnemain, J. Drug Targeting, 1998, 6, 167–174 CrossRef CAS PubMed.
- R. Jin, B. Lin, D. Li and H. Ai, Curr. Opin. Pharmacol., 2014, 18, 18–27 CrossRef CAS PubMed.
- M. Lu, M. H. Cohen, D. Rieves and R. Pazdur, Am. J. Hematol., 2010, 85, 315–319 CAS.
- P. Reimer and T. Balzer, Eur. Radiol., 2003, 13, 1266–1276 CrossRef PubMed.
- C. Sun, J. S. Lee and M. Zhang, Adv. Drug Delivery Rev., 2008, 60, 1252–1265 CrossRef CAS PubMed.
- Y. X. Wang, S. M. Hussain and G. P. Krestin, Eur. Radiol., 2001, 11, 2319–2331 CrossRef CAS PubMed.
- J. Weizenecker, B. Gleich, J. Rahmer and J. Borgert, Phys. Med. Biol., 2012, 57, 7317–7327 CrossRef PubMed.
- J. P. Bullivant, S. Zhao, B. J. Willenberg, B. Kozissnik, C. D. Batich and J. Dobson, Int. J. Mol. Sci., 2013, 14, 17501–17510 CrossRef PubMed.
- J. Lodhia, G. Mandarano, N. J. Ferris, P. Eu and S. F. Cowell, Biomed. Imaging Intervention J., 2010, 6, e12 CAS.
- F. Ludwig, T. Wawrzik, T. Yoshida, N. Gehrke, A. Briel, D. Eberbeck and M. Schilling, IEEE Trans. Magn., 2012, 48, 3780–3783 Search PubMed.
- Z. W. Tay, P. Chandrasekharan, X. Y. Zhou, E. Yu, B. Zheng and S. Conolly, Theranostics, 2018, 8, 3676–3687 CrossRef CAS PubMed.
- Z. W. Tay, D. W. Hensley, E. C. Vreeland, B. Zheng and S. M. Conolly, Biomed. Phys. Eng. Express, 2017, 3, 035003 CrossRef PubMed.
- P. Bender, J. Fock, C. Frandsen, M. F. Hansen, C. Balceris, F. Ludwig, O. Posth, E. Wetterskog, L. K. Bogart, P. Southern, W. Szczerba, L. Zeng, K. Witte, C. Grüttner, F. Westphal, D. Honecker, D. Gonzalez-Alonso, L. F. Barquín and C. Johansson, J. Phys. Chem. C, 2018, 122, 3068–3077 CrossRef CAS.
- K. Ludtke-Buzug, J. Haegele, S. Biederer, T. F. Sattel, M. Erbe, R. L. Duschka, J. Barkhausen and F. M. Vogt, Biomed. Tech., 2013, 58, 527–533 Search PubMed.
- H. Kratz, M. Taupitz, A. Ariza de Schellenberger, O. Kosch, D. Eberbeck, S. Wagner, L. Trahms, B. Hamm and J. Schnorr, PLoS One, 2018, 13, e0190214 CrossRef PubMed.
- D. Eberbeck, C. L. Dennis, N. F. Huls, K. L. Krycka, C. Grüttner and F. Westphal, IEEE Trans. Magn., 2013, 49, 269–274 Search PubMed.
- C. Jonasson, V. Schaller, L. J. Zeng, E. Olsson, C. Frandsen, A. Castro, L. Nilsson, L. K. Bogart, P. Southern, Q. A. Pankhurst, M. P. Morales and C. Johansson, J. Magn. Magn. Mater., 2019, 477, 198–202 CrossRef CAS.
- M. Wetegrove, K. Witte, W. Bodnar, D. E. Pfahl, A. Springer, N. Schell, F. Westphal and E. Burkel, CrystEngComm, 2019, 21, 1956–1966 RSC.
- K. Riahi, M. M. van de Loosdrecht, L. Alic and B. ten Haken, J. Magn. Magn. Mater., 2020, 514, 167238 CrossRef CAS.
- R. Dhavalikar, D. Hensley, L. Maldonado-Camargo, L. R. Croft, S. Ceron, P. W. Goodwill, S. M. Conolly and C. Rinaldi, J. Phys. D: Appl. Phys., 2016, 49, 305002 CrossRef PubMed.
- A. P. Khandhar, R. M. Ferguson, H. Arami, S. J. Kemp and K. M. Krishnan, IEEE Trans. Magn., 2015, 51, 5300304 Search PubMed.
- R. Orendorff, A. J. Peck, B. Zheng, S. N. Shirazi, R. Matthew Ferguson, A. P. Khandhar, S. J. Kemp, P. Goodwill, K. M. Krishnan, G. A. Brooks, D. Kaufer and S. Conolly, Phys. Med. Biol., 2017, 62, 3501–3509 CrossRef CAS PubMed.
- P. Ludewig, N. Gdaniec, J. Sedlacik, N. D. Forkert, P. Szwargulski, M. Graeser, G. Adam, M. G. Kaul, K. M. Krishnan, R. M. Ferguson, A. P. Khandhar, P. Walczak, J. Fiehler, G. Thomalla, C. Gerloff, T. Knopp and T. Magnus, ACS Nano, 2017, 11, 10480–10488 CrossRef CAS PubMed.
- I. Hilger and W. A. Kaiser, Nanomedicine, 2012, 7, 1443–1459 CrossRef CAS PubMed.
- L. Storozhuk, M. O. Besenhard, S. Mourdikoudis, A. P. LaGrow, M. R. Lees, L. Tung, A. Gavriilidis and N. T. K. Thanh, ACS Appl. Mater. Interfaces, 2021, 13, 45870–45880 CrossRef CAS PubMed.
- S. Dutz, M. Kettering, I. Hilger, R. Muller and M. Zeisberger, Nanotechnology, 2011, 22, 265102 CrossRef PubMed.
- S. Dutz, J. H. Clement, D. Eberbeck, T. Gelbrich, R. Hergt, R. Muller, J. Wotschadlo and M. Zeisberger, J. Magn. Magn. Mater., 2009, 321, 1501–1504 CrossRef CAS.
- G. Hemery, C. Genevois, F. Couillaud, S. Lacomme, E. Gontier, E. Ibarboure, S. Lecommandoux, E. Garanger and O. Sandre, Mol. Syst. Des. Eng., 2017, 2, 629–639 RSC.
- C. Blanco-Andujar, D. Ortega, P. Southern, Q. A. Pankhurst and N. T. K. Thanh, Nanoscale, 2015, 7, 1768–1775 RSC.
- T. Kuboyabu, M. Yamawaki, M. Aoki, A. Ohki and K. Murase, Int. J. Nanomed. Nanosurg., 2016, 2, 1–7 Search PubMed.
- E. Myrovali, N. Maniotis, T. Samaras and M. Angelakeris, Nanoscale Adv., 2020, 2, 408–416 RSC.
- Z. W. Tay, P. Chandrasekharan, A. Chiu-Lam, D. W. Hensley, R. Dhavalikar, X. Y. Zhou, E. Y. Yu, P. W. Goodwill, B. Zheng, C. Rinaldi and S. M. Conolly, ACS Nano, 2018, 12, 3699–3713 CrossRef CAS PubMed.
- N. C. V. Rost, K. Sen, S. Savliwala, I. Singh, S. Liu, M. Unni, L. Raniero and C. Rinaldi, J. Magn. Magn. Mater., 2020, 504, 166675 CrossRef CAS.
- L. H. Wu, A. Mendoza-Garcia, Q. Li and S. H. Sun, Chem. Rev., 2016, 116, 10473–10512 CrossRef CAS PubMed.
- D. L. LesliePelecky and R. D. Rieke, Chem. Mater., 1996, 8, 1770–1783 CrossRef CAS.
- R. M. Ferguson, A. P. Khandhar, H. Arami, L. Hua, O. Hovorka and K. M. Krishnan, Biomed. Eng.-Biomed. Tech., 2013, 58, 493–507 CAS.
- W. Xie, Z. Guo, F. Gao, Q. Gao, D. Wang, B. S. Liaw, Q. Cai, X. Sun, X. Wang and L. Zhao, Theranostics, 2018, 8, 3284–3307 CrossRef CAS PubMed.
- P. Guardia, R. Di Corato, L. Lartigue, C. Wilhelm, A. Espinosa, M. Garcia-Hernandez, F. Gazeau, L. Manna and T. Pellegrino, ACS Nano, 2012, 6, 3080–3091 CrossRef CAS PubMed.
- S. Behzadi, V. Serpooshan, W. Tao, M. A. Hamaly, M. Y. Alkawareek, E. C. Dreaden, D. Brown, A. M. Alkilany, O. C. Farokhzad and M. Mahmoudi, Chem. Soc. Rev., 2017, 46, 4218–4244 RSC.
- H. L. Ye, Z. Q. Shen, L. Yu, M. Wei and Y. Li, Proc. R. Soc. A, 2018, 474, 20170845 CrossRef PubMed.
- S. Ziemian, N. Lowa, O. Kosch, D. Bajj, F. Wiekhorst and G. Schutz, Nanomaterials, 2018, 8, 180 CrossRef PubMed.
- S. H. Noh, W. Na, J. T. Jang, J. H. Lee, E. J. Lee, S. H. Moon, Y. Lim, J. S. Shin and J. Cheon, Nano Lett., 2012, 12, 3716–3721 CrossRef CAS PubMed.
- L. M. Bauer, S. F. Situ, M. A. Griswold and A. C. Samia, Nanoscale, 2016, 8, 12162–12169 RSC.
- A. Lak, M. Cassani, B. T. Mai, N. Winckelmans, D. Cabrera, E. Sadrollahi, S. Marras, H. Remmer, S. Fiorito, L. Cremades-Jimeno, F. J. Litterst, F. Ludwig, L. Manna, F. J. Teran, S. Bals and T. Pellegrino, Nano Lett., 2018, 18, 6856–6866 CrossRef CAS PubMed.
- D. Niculaes, A. Lak, G. C. Anyfantis, S. Marras, O. Laslett, S. K. Avugadda, M. Cassani, D. Serantes, O. Hovorka, R. Chantrell and T. Pellegrino, ACS Nano, 2017, 11, 12121–12133 CrossRef CAS PubMed.
- T. Knopp, N. Gdaniec and M. Moddel, Phys. Med. Biol., 2017, 62, R124–R178 CrossRef CAS PubMed.
- S. F. Hasany, I. Ahmed, J. Rajan and A. Rehman, Nanosci. Nanotechnol., 2012, 2, 148–158 CrossRef.
- J. K. Oh and J. M. Park, Prog. Polym. Sci., 2011, 36, 168–189 CrossRef CAS.
- M. L. Mojica Pisciotti, E. Lima Jr., M. Vasquez Mansilla, V. E. Tognoli, H. E. Troiani, A. A. Pasa, T. B. Creczynski-Pasa, A. H. Silva, P. Gurman, L. Colombo, G. F. Goya, A. Lamagna and R. D. Zysler, J. Biomed. Mater. Res., Part B, 2014, 102, 860–868 CrossRef CAS PubMed.
- M. Yu, S. Huang, K. J. Yu and A. M. Clyne, Int. J. Mol. Sci., 2012, 13, 5554–5570 CrossRef CAS PubMed.
- P. Keselman, E. Y. Yu, X. Y. Zhou, P. W. Goodwill, P. Chandrasekharan, R. M. Ferguson, A. P. Khandhar, S. J. Kemp, K. M. Krishnan, B. Zheng and S. M. Conolly, Phys. Med. Biol., 2017, 62, 3440–3453 CrossRef CAS PubMed.
- A. Tomitaka, H. Arami, S. Gandhi and K. M. Krishnan, Nanoscale, 2015, 7, 16890–16898 RSC.
- M. P. Morales, O. Bomati-Miguel, R. P. de Alejo, J. Ruiz-Cabello, S. Veintemillas-Verdaguer and K. O'Grady, J. Magn. Magn. Mater., 2003, 266, 102–109 CrossRef CAS.
- S. H. Chaki, T. J. Malek, M. D. Chaudhary, J. P. Tailor and M. P. Deshpande, Adv. Nat. Sci.: Nanosci. Nanotechnol., 2015, 6, 035009 Search PubMed.
- A. Ali, H. Zafar, M. Zia, I. Ul Haq, A. R. Phull, J. S. Ali and A. Hussain, Nanotechnol., Sci. Appl., 2016, 9, 49–67 CrossRef CAS PubMed.
- H. Kratz, D. Eberbeck, S. Wagner, M. Taupitz and J. Schnorr, Biomed. Eng.-Biomed. Tech., 2013, 58, 509–515 CAS.
- M. C. Mascolo, Y. B. Pei and T. A. Ring, Materials, 2013, 6, 5549–5567 CrossRef CAS PubMed.
- M. O. Besenhard, A. P. LaGrow, A. Hodzic, M. Kriechbaum, L. Panariello, G. Bais, K. Loizou, S. Damilos, M. M. Cruz, N. T. K. Thanh and A. Gavriilidis, Chem. Eng. J., 2020, 399, 125740 CrossRef CAS.
- H. Kratz, A. Mohtashamdolatshahi, D. Eberbeck, O. Kosch, R. Hauptmann, F. Wiekhorst, M. Taupitz, B. Hamm and J. Schnorr, Nanomaterials, 2019, 9, 1466 CrossRef CAS PubMed.
- W. Wu, Z. H. Wu, T. Yu, C. Z. Jiang and W. S. Kim, Sci. Technol. Adv. Mater., 2015, 16, 023501 CrossRef PubMed.
- C. Kim, J. Lee, D. Schmucker and J. D. Fortner, npj Clean Water, 2020, 3, 8 CrossRef CAS.
- R. Hufschmid, H. Arami, R. M. Ferguson, M. Gonzales, E. Teeman, L. N. Brush, N. D. Browning and K. M. Krishnan, Nanoscale, 2015, 7, 11142–11154 RSC.
- N. T. K. Thanh, I. Robinson and T. L. D. Dekker, Encycl. Nanosci. Nanotechnol., 2007, 1, 1–10 Search PubMed.
- C. Blanco-Andujar, L. D. Tung and N. T. K. Thanh, Annu. Rep. Prog. Chem., Sect. A: Inorg. Chem., 2010, 106, 553–568 RSC.
- M. H. M. Dias and P. C. Lauterbur, Magn. Reson. Med., 1986, 3, 328–330 CrossRef CAS PubMed.
- S. Hildebrand, N. Löwa, H. Paysen, R. M. Fratila, L. Reverte-Salisa, T. Trakoolwilaiwan, Z. Niu, G. Kasparis, S. F. Preuss, O. Kosch, J. M. de la Fuente, N. T. K. Thanh, F. Wiekhorst and A. Pfeifer, ACS Nano, 2021, 15, 434–446 CrossRef CAS PubMed.
- V. Herynek, M. Babic, O. Kaman, H. Charvatova, M. Vesela, O. Buchholz, M. Vosmanska, D. Kubaniova, J. Kohout, U. G. Hofmann and L. Sefc, J. Nanopart. Res., 2021, 23, 52 CrossRef CAS.
- Z. Jiang, X. Han, Y. Du, Y. Li, Y. Li, J. Li, J. Tian and A. Wu, Nano Lett., 2021, 21, 2730–2737 CrossRef CAS PubMed.
- A. Baki, A. Remmo, N. Lowa, F. Wiekhorst and R. Bleul, Int. J. Mol. Sci., 2021, 22, 6235 CrossRef CAS PubMed.
- J. Haegele, R. L. Duschka, M. Graeser, C. Schaecke, N. Panagiotopoulos, K. Ludtke-Buzug, T. M. Buzug, J. Barkhausen and F. M. Vogt, Int. J. Nanomed., 2014, 9, 4203–4209 CrossRef PubMed.
- A. P. Khandhar, P. Keselman, S. J. Kemp, R. M. Ferguson, P. W. Goodwill, S. M. Conolly and K. M. Krishnan, Nanoscale, 2017, 9, 1299–1306 RSC.
- H. Kratz, A. Mohtashamdolatshahi, D. Eberbeck, O. Kosch, F. Wiekhorst, M. Taupitz, B. Hamm, N. Stolzenburg and J. Schnorr, Nanomaterials, 2021, 11, 1532 CrossRef CAS PubMed.
- A. H. Lu, E. L. Salabas and F. Schuth, Angew. Chem., Int. Ed., 2007, 46, 1222–1244 CrossRef CAS PubMed.
- Y. Takeuchi, H. Suzuki, H. Sasahara, J. Ueda, I. Yabata, K. Itagaki, S. Saito and K. Murase, Adv. Biomed. Eng., 2014, 4, 37–43 CrossRef.
- A. Antonelli, C. Sfara, L. Mosca, E. Manuali and M. Magnani, J. Nanosci. Nanotechnol., 2008, 8, 2270–2278 CrossRef CAS PubMed.
- D. E. Markov, H. Boeve, B. Gleich, J. Borgert, A. Antonelli, C. Sfara and M. Magnani, Phys. Med. Biol., 2010, 55, 6461 CrossRef CAS PubMed.
- J. Rahmer, A. Antonelli, C. Sfara, B. Tiemann, B. Gleich, M. Magnani, J. Weizenecker and J. Borgert, Phys. Med. Biol., 2013, 58, 3965–3977 CrossRef CAS PubMed.
- A. Antonelli, P. Szwargulski, E. Scarpa, F. Thieben, G. Cordula, G. Ambrosi, L. Guidi, P. Ludewig, T. Knopp and M. Magnani, Nanomedicine, 2020, 15, 739–753 CrossRef CAS PubMed.
- A. Tomitaka, H. Arami, Z. Huang, A. Raymond, E. Rodriguez, Y. Cai, M. Febo, Y. Takemura and M. Nair, Nanoscale, 2017, 10, 184–194 RSC.
- World Health Organization, https://gco.iarc.fr/today/fact-sheets-cancers, (accessed May 2021).
- W. Tong, H. Hui, W. Shang, Y. Zhang, F. Tian, Q. Ma, X. Yang, J. Tian and Y. Chen, Theranostics, 2021, 11, 506–521 CrossRef CAS PubMed.
- Z. W. Tay , presented in part of World Molecular Imaging Congress (WMIC), Montreal, September 2019.
- Z. W. Tay, D. W. Hensley, P. Chandrasekharan, B. Zheng and S. M. Conolly, IEEE Trans. Med. Imaging, 2020, 39, 1724–1734 Search PubMed.
- K. P. Melo, A. V. Makela, N. N. Knier, A. M. Hamilton and P. J. Foster, Magnetic Resonance in Medicine, 2021 Search PubMed.
- T. Knopp, S. Biederer, T. F. Sattel, M. Erbe and T. M. Buzug, IEEE Trans. Med. Imaging, 2011, 30, 1284–1292 Search PubMed.
- M. Graeser, T. Knopp, P. Szwargulski, T. Friedrich, A. von Gladiss, M. Kaul, K. M. Krishnan, H. Ittrich, G. Adam and T. M. Buzug, Sci. Rep., 2017, 7, 6872 CrossRef PubMed.
- H. Paysen, O. Kosch, J. Wells, N. Loewa and F. Wiekhorst, Phys. Med. Biol., 2020, 65, 235031 CrossRef CAS PubMed.
- P. W. Goodwill, E. U. Saritas, L. R. Croft, T. N. Kim, K. M. Krishnan, D. V. Schaffer and S. M. Conolly, Adv. Mater., 2012, 24, 3870–3877 CrossRef CAS PubMed.
- R. Dhavalikar and C. Rinaldi, J. Appl. Phys., 2014, 115, 074308 CrossRef.
- C. Shasha, E. Teeman and K. M. Krishnan, Biomed. Phys. Eng. Express, 2019, 5, 055010 CrossRef.
- J. J. Gevaert, N. D. Calvert, O. C. Sehl, K. P. Melo, A. J. Shuhendler and P. J. Foster, presented in part of World Molecular Imaging Congress (WMIC), Virtual, October 2020.
- Z. W. Tay, PhD Thesis, UC Berkeley, 2018.
- C. Colson, Z. W. Tay, K. L. B. Fung, D. W. Hensley, S. Savliwala, B. D. Fellows, X. Y. Zhou, Y. Lu, P. Chandrasekharan, C. Rinaldi and S. M. Conolly, presented in part of World Molecular Imaging Congress (WMIC), Virtual, October 2020.
- C. Saayujya , presented in part of World Molecular Imaging Congress (WMIC), Virtual, October 2020.
- Z. Zhao and C. Rinaldi, Phys. Med. Biol., 2020, 65, 185013 CrossRef CAS PubMed.
- S. Arsalani, N. Lowa, O. Kosch, P. Radon, O. Baffa and F. Wiekhorst, Phys. Med. Biol., 2021, 66, 015002 CrossRef CAS PubMed.
- S. M. Dadfar, D. Camozzi, M. Darguzyte, K. Roemhild, P. Varvarà, J. Metselaar, S. Banala, M. Straub, N. Güvener, U. Engelmann, I. Slabu, M. Buhl, J. Leusen, P. Kögerler, B. Hermanns-Sachweh, V. Schulz, F. Kiessling and T. Lammers, J. Nanobiotechnol., 2020, 18, 22 CrossRef CAS PubMed.
- S. Horvat, P. Vogel, T. Kampf, A. Brandl, A. Alshamsan, H. A. Alhadlaq, M. Ahamed, K. Albrecht, V. C. Behr, A. Beilhack and J. Groll, ChemNanoMat, 2020, 6, 755–758 CrossRef CAS.
- A. J. McGrath, S. Cheong, A. M. Henning, J. J. Gooding and R. D. Tilley, Chem. Commun., 2017, 53, 11548–11551 RSC.
- T. J. Yoon, H. Lee, H. L. Shao and R. Weissleder, Angew. Chem., Int. Ed., 2011, 50, 4663–4666 CrossRef CAS PubMed.
- W. S. Seo, J. H. Lee, X. M. Sun, Y. Suzuki, D. Mann, Z. Liu, M. Terashima, P. C. Yang, M. V. McConnell, D. G. Nishimura and H. J. Dai, Nat. Mater., 2006, 5, 971–976 CrossRef CAS PubMed.
- J. M. Liu, K. Wu and J. P. Wang, AIP Adv., 2016, 6, 056126 CrossRef.
- Y. L. Hou, H. Kondoh, T. Kogure and T. Ohta, Chem. Mater., 2004, 16, 5149–5152 CrossRef CAS.
- J. Yu, C. Yang, J. D. S. Li, Y. C. Ding, L. Zhang, M. Z. Yousaf, J. Lin, R. Pang, L. B. Wei, L. L. Xu, F. G. Sheng, C. H. Li, G. J. Li, L. Y. Zhao and Y. L. Hou, Adv. Mater., 2014, 26, 4114–4120 CrossRef CAS PubMed.
- J. H. Gao, G. L. Liang, J. S. Cheung, Y. Pan, Y. Kuang, F. Zhao, B. Zhang, X. X. Zhang, E. X. Wu and B. Xu, J. Am. Chem. Soc., 2008, 130, 11828–11833 CrossRef CAS PubMed.
- A. Rivera-Rodriguez and C. M. Rinaldi-Ramos, Annu. Rev. Chem. Biomol. Eng., 2021, 12, 163–185 CrossRef CAS PubMed.
- S. B. Goldhaber, Regul. Toxicol. Pharmacol., 2003, 38, 232–242 CrossRef CAS PubMed.
- M. Irfan, N. Dogan, A. Bingolbali and F. Aliew, J. Magn. Magn. Mater., 2021, 537, 168150 CrossRef CAS.
- J. W. Bulte, M. W. De Jonge, R. L. Kamman, K. G. Go, F. Zuiderveen, B. Blaauw, J. A. Oosterbaan, T. H. The and L. de Leij, Magn. Reson. Med., 1992, 23, 215–223 CrossRef CAS PubMed.
- J. W. Bulte, L. D. Ma, R. L. Magin, R. L. Kamman, C. E. Hulstaert, K. G. Go, T. H. The and L. de Leij, Magn. Reson. Med., 1993, 29, 32–37 CrossRef CAS PubMed.
- T. C. Yeh, W. Zhang, S. T. Ildstad and C. Ho, Magn. Reson. Med., 1993, 30, 617–625 CrossRef CAS PubMed.
- P. K. Nguyen, J. Riegler and J. C. Wu, Cell Stem Cell, 2014, 14, 431–444 CrossRef CAS PubMed.
- E. M. Shapiro, K. Sharer, S. Skrtic and A. P. Koretsky, Magn. Reson. Med., 2006, 55, 242–249 CrossRef PubMed.
- C. Heyn, J. A. Ronald, S. S. Ramadan, J. A. Snir, A. M. Barry, L. T. MacKenzie, D. J. Mikulis, D. Palmieri, J. L. Bronder, P. S. Steeg, T. Yoneda, I. C. MacDonald, A. F. Chambers, B. K. Rutt and P. J. Foster, Magn. Reson. Med., 2006, 56, 1001–1010 CrossRef PubMed.
- E. M. Shapiro, S. Skrtic, K. Sharer, J. M. Hill, C. E. Dunbar and A. P. Koretsky, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 10901–10906 CrossRef CAS PubMed.
- A. V. Makela , presented in part of World Molecular Imaging Congress (WMIC), Montreal, September 2019.
- B. Laffon, N. Fernandez-Bertolez, C. Costa, F. Brandao, J. P. Teixeira, E. Pasaro and V. Valdiglesias, Adv. Exp. Med. Biol., 2018, 1048, 199–213 CrossRef CAS PubMed.
- M. Geppert, M. C. Hohnholt, S. Nurnberger and R. Dringen, Acta Biomater., 2012, 8, 3832–3839 CrossRef CAS PubMed.
- M. Geppert, M. C. Hohnholt, K. Thiel, S. Nurnberger, I. Grunwald, K. Rezwan and R. Dringen, Nanotechnology, 2011, 22, 145101 CrossRef PubMed.
- C. Costa, F. Brandao, M. J. Bessa, S. Costa, V. Valdiglesias, G. Kilic, N. Fernandez-Bertolez, P. Quaresma, E. Pereira, E. Pasaro, B. Laffon and J. P. Teixeira, J. Appl. Toxicol., 2016, 36, 361–372 CrossRef CAS PubMed.
- R. Weissleder, D. D. Stark, B. L. Engelstad, B. R. Bacon, C. C. Compton, D. L. White, P. Jacobs and J. Lewis, AJR, Am. J. Roentgenol., 1989, 152, 167–173 CrossRef CAS PubMed.
- S. Laurent, J. L. Bridot, L. V. Elst and R. N. Muller, Future Med. Chem., 2010, 2, 427–449 CrossRef CAS PubMed.
- M. Hidalgo, F. Amant, A. V. Biankin, E. Budinska, A. T. Byrne, C. Caldas, R. B. Clarke, S. de Jong, J. Jonkers, G. M. Maelandsmo, S. Roman-Roman, J. Seoane, L. Trusolino and A. Villanueva, Cancer Discovery, 2014, 4, 998–1013 CrossRef CAS PubMed.
- N. N. Knier, V. P. Dubois, Y. Chen, J. A. Ronald and P. J. Foster, J. Biol. Methods, 2021, 8, e154 Search PubMed.
- K. M. Parkins, K. P. Melo, Y. Chen, J. A. Ronald and P. J. Foster, Nanoscale, 2021, 13, 6016–6023 RSC.
- T. L. Whiteside, Oncogene, 2008, 27, 5904–5912 CrossRef CAS PubMed.
- C. Fink, M. Smith, O. C. Sehl, J. M. Gaudet, T. C. Meagher, N. A. Sheikh, J. D. Dikeakos, M. J. Rieder, P. J. Foster and G. A. Dekaban, Diagn. Interv. Imaging, 2020, 101, 577–588 CrossRef CAS PubMed.
- R. Rohani, S. N. Chickera, C. Willert, Y. Chen, G. A. Dekaban and P. J. Foster, Mol. Imaging Biol., 2011, 13, 679–694 CrossRef PubMed.
- E. Obeid, R. Nanda, Y. X. Fu and O. I. Olopade, Int. J. Oncol., 2013, 43, 5–12 CrossRef CAS PubMed.
- A. V. Makela, J. M. Gaudet, M. A. Schott, O. C. Sehl, C. H. Contag and P. J. Foster, Mol. Imaging Biol., 2020, 22, 958–968 CrossRef CAS PubMed.
- A. V. Makela, J. M. Gaudet and P. J. Foster, Sci. Rep., 2017, 7, 42109 CrossRef CAS PubMed.
- A. V. Makela and P. J. Foster, Magn. Reson. Med., 2018, 80, 1138–1147 CrossRef CAS PubMed.
- M. Gerosa, G. Ren, Y. Zhang, P. W. Goodwill, J. Mansfield, P. Marzola and M. Wintermark, Int. J. Magn. Part. Imaging, 2020, 6, 2009029 Search PubMed.
- J. Mansfield , presented in part of World Molecular Imaging Congress (WMIC), Virtual, October 2020.
- I. Baccelli, A. Schneeweiss, S. Riethdorf, A. Stenzinger, A. Schillert, V. Vogel, C. Klein, M. Saini, T. Bäuerle, M. Wallwiener, T. Holland-Letz, T. Höfner, M. Sprick, M. Scharpff, F. Marmé, H. P. Sinn, K. Pantel, W. Weichert and A. Trumpp, Nat. Biotechnol., 2013, 31, 539–544 CrossRef CAS PubMed.
- A. Rivera-Rodriguez, L. B. Hoang-Minh, A. Chiu-Lam, N. Sarna, L. Marrero-Morales, D. A. Mitchell and C. M. Rinaldi-Ramos, Nanotheranostics, 2021, 5, 431–444 CrossRef PubMed.
- S. Lefevre, D. Ruimy, F. Jehl, A. Neuville, P. Robert, C. Sordet, M. Ehlinger, J. L. Dietemann and G. Bierry, Radiology, 2011, 258, 722–728 CrossRef PubMed.
- S. G. Ruehm, C. Corot, P. Vogt, S. Kolb and J. F. Debatin, Circulation, 2001, 103, 415–422 CrossRef CAS PubMed.
- D. B. Mangarova, J. Brangsch, A. Mohtashamdolatshahi, O. Kosch, H. Paysen, F. Wiekhorst, R. Klopfleisch, R. Buchholz, U. Karst, M. Taupitz, J. Schnorr, B. Hamm and M. R. Makowski, Sci. Rep., 2020, 10, 12410 CrossRef CAS PubMed.
- W. Zakrzewski, M. Dobrzynski, M. Szymonowicz and Z. Rybak, Stem Cell Res. Ther., 2019, 10, 68 CrossRef CAS PubMed.
- M. F. Kircher, S. S. Gambhir and J. Grimm, Nat. Rev. Clin. Oncol., 2011, 8, 677–688 CrossRef CAS PubMed.
- M. Zhang, X. Liu, J. Huang, L. Wang, H. Shen, Y. Luo, Z. Li, H. Zhang, Z. Deng and Z. Zhang, Nanomedicine, 2018, 14, 2475–2483 CrossRef CAS PubMed.
- M. Edmundson, N. T. Thanh and B. Song, Theranostics, 2013, 3, 573–582 CrossRef PubMed.
- F. Chen, M. Ma, J. Wang, F. Wang, S. X. Chern, E. R. Zhao, A. Jhunjhunwala, S. Darmadi, H. Chen and J. V. Jokerst, Nanoscale, 2017, 9, 402–411 RSC.
- M. Hofmann, K. C. Wollert, G. P. Meyer, A. Menke, L. Arseniev, B. Hertenstein, A. Ganser, W. H. Knapp and H. Drexler, Circulation, 2005, 111, 2198–2202 CrossRef PubMed.
- F. Fidler, M. Steinke, A. Kraupner, C. Grüttner, K. H. Hiller, A. Briel, F. Westphal, H. Walles and P. M. Jakob, IEEE Trans. Magn., 2015, 51, 5100704 Search PubMed.
- J. W. M. Bulte, P. Walczak, M. Janowski, K. M. Krishnan, H. Arami, A. Halkola, B. Gleich and J. Rahmer, Tomography, 2015, 1, 91–97 CrossRef CAS PubMed.
- K. Lüdtke-Buzug, D. H. Rapoport and D. Schneider, AIP Conf. Proc., 2010, 1311, 244–248 CrossRef.
- B. Zheng, M. P. von See, E. Yu, B. Gunel, K. Lu, T. Vazin, D. V. Schaffer, P. W. Goodwill and S. M. Conolly, Theranostics, 2016, 6, 291–301 CrossRef CAS PubMed.
- H. S. Kim, S. Y. Oh, H. J. Joo, K. R. Son, I. C. Song and W. K. Moon, NMR Biomed., 2010, 23, 514–522 CrossRef CAS PubMed.
- S. Schrepfer, T. Deuse, H. Reichenspurner, M. P. Fischbein, R. C. Robbins and M. P. Pelletiera, Transplant. Proc., 2007, 39, 573–576 CrossRef CAS PubMed.
- O. C. Sehl and P. J. Foster, Sci. Rep., 2021, 11, 22198 CrossRef CAS PubMed.
- A. A. Dayem, S. B. Lee, K. Kim, K. M. Lim, T. I. Jeon and S. G. Cho, BMB Rep., 2019, 52, 295–303 CrossRef CAS PubMed.
- Y. Wang, C. Blanco-Andujar, Z. L. Zhi, P. W. So, N. T. Thanh and J. C. Pickup, Chem. Commun., 2013, 49, 7255–7257 RSC.
- P. Wang, P. W. Goodwill, P. Pandit, J. Gaudet, A. Ross, J. Wang, E. Yu, D. W. Hensley, T. C. Doyle, C. H. Contag, S. Conolly and A. Moore, Quant. Imaging Med. Surg., 2018, 8, 114–122 CrossRef PubMed.
- A. Sun , presented in part of World Molecular Imaging Congress (WMIC), Virtual, October 2020.
- H. Hayat, A. Sun, H. Hayat, S. Liu, N. Talebloo, C. Pinger, J. O. Bishop, M. Gudi, B. F. Dwan, X. Ma, Y. Zhao, A. Moore and P. Wang, Mol. Imaging Biol., 2021, 23, 18–29 CrossRef PubMed.
- T. Kuboyabu, A. Ohki, N. Banura and K. Murase, Open J. Med. Imaging, 2016, 6, 33–41 CrossRef.
- Y. Zhang , presented in part of World Molecular Imaging Congress (WMIC), Virtual, October 2020.
- P. C. Liang, Y. C. Chen, C. F. Chiang, L. R. Mo, S. Y. Wei, W. Y. Hsieh and W. L. Lin, Int. J. Nanomed., 2016, 11, 2021–2037 CAS.
- K. Li, H. Nejadnik and H. E. Daldrup-Link, Drug Discovery Today, 2017, 22, 1421–1429 CrossRef CAS PubMed.
- B. K. Lee, Y. H. Yun and K. Park, Chem. Eng. Sci., 2015, 125, 158–164 CrossRef CAS PubMed.
- C. Aversa, V. Rossi, E. Geuna, R. Martinello, A. Milani, S. Redana, G. Valabrega, M. Aglietta and F. Montemurro, Breast, 2014, 23, 623–628 CrossRef CAS PubMed.
- G. Jia, Y. Han, Y. An, Y. Ding, C. He, X. Wang and Q. Tang, Biomaterials, 2018, 178, 302–316 CrossRef CAS PubMed.
- T. Yang, P. Martin, B. Fogarty, A. Brown, K. Schurman, R. Phipps, V. P. Yin, P. Lockman and S. Bai, Pharm. Res., 2015, 32, 2003–2014 CrossRef CAS PubMed.
- V. Toomajian, E. E. Ural, C. H. Contag and A. V. Makela, presented in part of World Molecular Imaging Congress (WMIC), Virtual, October 2020.
- K. O. Jung, H. Jo, J. H. Yu, S. S. Gambhir and G. Pratx, Biomaterials, 2018, 177, 139–148 CrossRef CAS PubMed.
- P. Chandrasekharan, Z. W. Tay, D. Hensley, X. Y. Zhou, B. K. L. Fung, C. Colson, Y. Lu, B. D. Fellows, Q. Huynh, C. Saayujya, E. Yu, R. Orendorff, B. Zheng, P. W. Goodwill, C. Rinaldi and S. M. Conolly, Theranostics, 2020, 10, 2965–2981 CrossRef CAS PubMed.
- A. Hervault and N. T. Thanh, Nanoscale, 2014, 6, 11553–11573 RSC.
- S. Laurent, S. Dutz, U. O. Hafeli and M. Mahmoudi, Adv. Colloid Interface Sci., 2011, 166, 8–23 CrossRef CAS PubMed.
- R. K. Gilchrist, R. Medal, W. D. Shorey, R. C. Hanselman, J. C. Parrott and C. B. Taylor, Ann. Surg., 1957, 146, 596–606 CrossRef CAS PubMed.
- A. A. Sousa-Junior, S. A. Mendanha, M. S. Carriao, G. Capistrano, A. G. Prospero, G. A. Soares, E. R. Cintra, S. F. O. Santos, N. Zufelato, A. Alonso, E. M. Lima, J. R. A. Miranda, E. P. Silveira-Lacerda, C. G. Cardoso and A. F. Bakuzis, Mol. Pharm., 2020, 17, 837–851 CrossRef CAS PubMed.
- Y. Lu, A. Rivera-Rodriguez, Z. W. Tay, D. Hensley, K. L. B. Fung, C. Colson, C. Saayujya, Q. Huynh, L. Kabuli, B. Fellows, P. Chandrasekharan, C. Rinaldi and S. Conolly, Int. J. Hyperthermia, 2020, 37, 141–154 CrossRef CAS PubMed.
- C. Kut, Y. Zhang, M. Hedayati, H. Zhou, C. Cornejo, D. Bordelon, J. Mihalic, M. Wabler, E. Burghardt, C. Gruettner, A. Geyh, C. Brayton, T. L. Deweese and R. Ivkov, Nanomedicine, 2012, 7, 1697–1711 CrossRef CAS PubMed.
- R. Dhavalikar and C. Rinaldi, J. Magn. Magn. Mater., 2016, 419, 267–273 CrossRef CAS PubMed.
- A. R. Sebastian, S. H. Ryu, H. M. Ko and S. H. Kim, IEEE Access, 2019, 7, 96094–96104 Search PubMed.
- E. Y. Yu, P. W. Goodwill and S. M. Conolly, Preliminary characterization of a laminated iron-core 6.3 T/m FFL magnet, IEEE, New York, 2015 Search PubMed.
- J. M. Gaudet , presented in part of World Molecular Imaging Congress (WMIC), Virtual, October 2020.
- M. S. A. Darwish, H. Kim, M. P. Bui, T. A. Le, H. Lee, C. Ryu, J. Y. Lee and J. Yoon, Nanomaterials, 2021, 11, 1096 CrossRef CAS PubMed.
- A. P. Khandhar, R. M. Ferguson, J. A. Simon and K. M. Krishnan, J. Biomed. Mater. Res., Part A, 2012, 100, 728–737 CrossRef PubMed.
- A. Jordan, R. Scholz, P. Wust, H. Schirra, T. Schiestel, H. Schmidt and R. Felix, J. Magn. Magn. Mater., 1999, 194, 185–196 CrossRef CAS.
- X. L. Liu, H. M. Fan, J. B. Yi, Y. Yang, E. S. G. Choo, J. M. Xue, D. D. Fana and J. Ding, J. Mater. Chem., 2012, 22, 8235–8244 RSC.
- H. Khurshid, J. Alonso, Z. Nemati, M. H. Phan, P. Mukherjee, M. L. Fdez-Gubieda, J. M. Barandiarán and H. Srikanth, J. Appl. Phys., 2015, 117, 17A337 CrossRef.
- J. F. Liu, N. Neel, P. Dang, M. Lamb, J. McKenna, L. Rodgers, B. Litt, Z. Cheng, A. Tsourkas and D. Issadore, Small, 2018, 14, e1802563 CrossRef PubMed.
- S. Maruyama, K. Shimada, K. Enmeiji and K. Murase, Int. J. Nanomed. Nanosurg., 2016, 2, 1–11 Search PubMed.
- E. G. Fuller, H. Sun, R. D. Dhavalikar, M. Unni, G. M. Scheutz, B. S. Sumerlin and C. Rinaldi, ACS Appl. Polym. Mater., 2019, 1, 211–220 CrossRef CAS.
- M. Wintermark, M. Sesay, E. Barbier, K. Borbely, W. P. Dillon, J. D. Eastwood, T. C. Glenn, C. B. Grandin, S. Pedraza, J. F. Soustiel, T. Nariai, G. Zaharchuk, J. M. Caille, V. Dousset and H. Yonas, Stroke, 2005, 36, e83–e99 CrossRef.
- R. E. Latchaw, H. Yonas, G. J. Hunter, W. T. Yuh, T. Ueda, A. G. Sorensen, J. L. Sunshine, J. Biller, L. Wechsler, R. Higashida, G. Hademenos and A. Council on Cardiovascular Radiology of the American Heart, Stroke, 2003, 34, 1084–1104 CrossRef PubMed.
- X. Huang, D. Kalladka, B. K. Cheripelli, F. C. Moreton and K. W. Muir, J. Neuroimaging, 2017, 27, 602–606 CrossRef PubMed.
- N. D. Forkert, P. Kaesemann, A. Treszl, S. Siemonsen, B. Cheng, H. Handels, J. Fiehler and G. Thomalla, AJNR Am. J. Neuroradiol., 2013, 34, 1697–1703 CrossRef CAS PubMed.
- H. Q. Dam, D. C. Brandon, V. V. Grantham, A. J. Hilson, D. M. Howarth, A. H. Maurer, M. G. Stabin, M. Tulchinsky, H. A. Ziessman and L. S. Zuckier, J. Nucl. Med. Technol., 2014, 42, 308–317 CrossRef PubMed.
- S. C. Srivastava and L. R. Chervu, Semin. Nucl. Med., 1984, 14, 68–82 CrossRef CAS.
- F. Wegner, T. M. Buzug and J. Barkhausen, Theranostics, 2018, 8, 3691–3692 CrossRef CAS PubMed.
- K. Miles, Cancer Imaging, 2011, 11, S86–S92 CrossRef PubMed.
- J. V. Frangioni, J. Clin. Oncol., 2008, 26, 4012–4021 CrossRef PubMed.
- K. Greish, J. Drug Targeting, 2007, 15, 457–464 CrossRef CAS PubMed.
- N. Schleich, C. Po, D. Jacobs, B. Ucakar, B. Gallez, F. Danhier and V. Preat, J. Controlled Release, 2014, 194, 82–91 CrossRef CAS PubMed.
- P. Vogel, M. A. Ruckert, P. Klauer, W. H. Kullmann, P. M. Jakob and V. C. Behr, Phys. Med. Biol., 2016, 61, 6620–6634 CrossRef CAS PubMed.
- A. Mohtashamdolatshahi, H. Kratz, O. Kosch, R. Hauptmann, N. Stolzenburg, F. Wiekhorst, I. Sack, B. Hamm, M. Taupitz and J. Schnorr, Sci. Rep., 2020, 10, 17247 CrossRef CAS PubMed.
- M. G. Kaul, J. Salamon, T. Knopp, H. Ittrich, G. Adam, H. Weller and C. Jung, Phys. Med. Biol., 2018, 63, 064001 CrossRef PubMed.
- I. Molwitz, H. Ittrich, T. Knopp, T. Mummert, J. Salamon, C. Jung, G. Adam and M. G. Kaul, Physiol. Meas., 2019, 40, 105002 CrossRef CAS PubMed.
- J. K. Lewis, J. C. Bischof, I. Braslavsky, K. G. Brockbank, G. M. Fahy, B. J. Fuller, Y. Rabin, A. Tocchio, E. J. Woods, B. G. Wowk, J. P. Acker and S. Giwa, Cryobiology, 2016, 72, 169–182 CrossRef PubMed.
- A. Chiu-Lam, E. Staples, C. J. Pepine and C. Rinaldi, Sci. Adv., 2021, 7, eabe3005 CrossRef PubMed.
- M. L. Etheridge, Y. Xu, L. Rott, J. Choi, B. Glasmacher and J. C. Bischof, Technology, 2014, 2, 229–242 CrossRef.
- J. Haegele, S. Vaalma, N. Panagiotopoulos, J. Barkhausen, F. M. Vogt, J. Borgert and J. Rahmer, Phys. Med. Biol., 2016, 61, N415 CrossRef CAS PubMed.
- S. H. Han, E. Cho, D. K. Lee, G. Cho, Y. R. Kim and H. Cho, Phys. Med. Biol., 2014, 59, 6521 CrossRef CAS PubMed.
- D. Hensley, P. W. Goodwill, L. Croft and S. Conolly, Preliminary experimental X-space color MPI, IEEE, New York, 2015 Search PubMed.
- D. Hensley, PhD Thesis, UC Berkeley, 2017.
- Y. Muslu, M. Utkur, O. M. Demirel and E. U. Saritas, IEEE Trans. Med. Imaging, 2018, 37, 1920–1931 Search PubMed.
- J. Haegele, N. Panagiotopoulos, S. Cremers, J. Rahmer, J. Franke, R. L. Duschka, S. Vaalma, M. Heidenreich, J. Borgert, P. Borm, J. Barkhausen and F. M. Vogt, IEEE Trans. Med. Imaging, 2016, 35, 2312–2318 Search PubMed.
- J. Rahmer, D. Wirtz, C. Bontus, J. Borgert and B. Gleich, IEEE Trans. Med. Imaging, 2017, 36, 1449–1456 Search PubMed.
- K. Murase, R. Song and S. Hiratsuka, Appl. Phys. Lett., 2014, 104, 252409 CrossRef.
- J. B. Weaver, A. M. Rauwerdink and E. W. Hansen, Med. Phys., 2009, 36, 1822–1829 CrossRef CAS PubMed.
- C. Stehning, B. Gleich and J. Rahmer, Int. J. Magn. Part. Imaging, 2016, 2, 1612001 Search PubMed.
- S. Draack, F. Ludwig, M. Schilling and T. Viereck, J. Magn. Magn. Mater., 2021, 522, 167478 CrossRef CAS.
- M. Möddel, C. Meins, J. Dieckhoff and T. Knopp, New J. Phys., 2018, 20, 083001 CrossRef.
- N. Gdaniec, P. Szwargulski and T. Knopp, Med. Phys. Lett., 2017, 44, 6456–6460 CAS.
- T. Knopp and M. Hofmann, Phys. Med. Biol., 2016, 61, N257–N267 CrossRef CAS PubMed.
- K. Lu, P. Goodwill, B. Zheng and S. Conolly, IEEE Trans. Med. Imaging, 2018, 37, 1989–1998 Search PubMed.
- M. Storath, C. Brandt, M. Hofmann, T. Knopp, J. Salamon, A. Weber and A. Weinmann, IEEE Trans. Med. Imaging, 2017, 36, 74–85 Search PubMed.
- A. Tateo, A. Iurino, G. Settanni, A. Andrisani, P. F. Stifanelli, P. Larizza, F. Mazzia, R. M. Mininni, S. Tangaro and R. Bellotti, Phys. Med. Biol., 2016, 61, 4061–4077 CrossRef CAS PubMed.
- A. Weber, F. Werner, J. Weizenecker, T. M. Buzug and T. Knopp, Phys. Med. Biol., 2016, 61, 475–487 CrossRef CAS PubMed.
- X. J. Chen, X. Han and X. Y. Tang, J. Med. Imaging Health Inf., 2021, 11, 703–711 CrossRef.
- K. Murase, Jpn. J. Appl. Phys., 2021, 60, 088001 CrossRef CAS.
- F. Lieb and T. Knopp, Med. Phys., 2021, 48, 3893–3903 CrossRef PubMed.
- A. von Gladiss, M. Graeser, A. Behrends, X. Chen and T. M. Buzug, Sci. Rep., 2020, 10, 18432 CrossRef CAS PubMed.
- H. Bernd, E. De Kerviler, S. Gaillard and B. Bonnemain, Invest. Radiol., 2009, 44, 336–342 CrossRef CAS PubMed.
- D. T. Kehagias, A. D. Gouliamos, V. Smyrniotis and L. J. Vlahos, J. Magn. Reson. Imaging, 2001, 14, 595–601 CrossRef CAS PubMed.
- A. Singh, T. Patel, J. Hertel, M. Bernardo, A. Kausz and L. Brenner, Am. J. Kidney Dis., 2008, 52, 907–915 CrossRef CAS PubMed.
- E. U. Saritas, P. W. Goodwill, G. Z. Zhang and S. M. Conolly, IEEE Trans. Med. Imaging, 2013, 32, 1600–1610 Search PubMed.
- I. Schmale, B. Gleich, J. Rahmer, C. Bontus, J. Schmidt and J. Borgert, IEEE Trans. Magn., 2015, 51, 6502604 Search PubMed.
- O. Dossel and J. Bohnert, Biomed. Tech., 2013, 58, 611–621 Search PubMed.
- M. Graeser, P. Ludewig, P. Szwargulski, F. Foerger, T. Liebing, N. D. Forkert, F. Thieben, T. Magnus and T. Knopp, Phys. Med. Biol., 2020, 65, 235007 CrossRef CAS PubMed.
- J. Pagan, C. McDonough, T. Vo and A. Tonyushkin, IEEE Trans. Magn., 2021, 57, 5300105 Search PubMed.
- E. E. Mason, C. Z. Cooley, S. F. Cauley, M. A. Griswold, S. M. Conolly and L. L. Wald, Int. J. Magn. Part. Imaging, 2017, 3, 1703008 Search PubMed.
- M. Graeser, F. Thieben, P. Szwargulski, F. Werner, N. Gdaniec, M. Boberg, F. Griese, M. Moddel, P. Ludewig, D. van de Ven, O. M. Weber, O. Woywode, B. Gleich and T. Knopp, Nat. Commun., 2019, 10, 1936 CrossRef CAS PubMed.
- E. E. Mason, E. Mattingly, K. Herb, M. Sliwiak, S. Franconi, C. Z. Cooley, P. J. Slanetz and L. L. Wald, Sci. Rep., 2021, 11, 13456 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |




![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 citations. She has been Visiting Professor at various Universities in France, Japan, Singapore. She has been invited to speak at over 270 institutes and scientific meetings. She has been chairing and organising over 45 high profile international conferences. She is Editor-in-chief of the Royal Society of Chemistry book Series, Nanoscience and Nanotechnology. She edited 7 theme issues including: (2021) Nanoscale Web themed issue on “Advanced Functional Nanomaterials for Biomedical Applications”. The Royal Society (2016), Interface Focus, “Multifunctional nanostructures for diagnosis and therapy of diseases”; The Royal Society Chemistry, RSC (2014), Faraday Discussions, “Physical Chemistry of Functionalised Biomedical Nanoparticles”; RSC (2013) Nanoscale, Special issue “Functional Nanoparticles for Biomedical Applications” and Philosophical Transactions of the Royal Society A (2010), “Nanoparticles”; MDPI (2021) Biomedicines “Advanced Functional Nanomaterials for Biomedical applications”. She is the sole editor of two seminal books on Magnetic Nanoparticles from Fabrication to Clinical Applications in 2012 (ISBN 9781439869321) and Clinical Applications of Magnetic nanoparticles in 2018 (ISBN 9781138051553). She is co-organising a Magnetic Carrier Meeting in Jun 2022 in London.
000 citations. She has been Visiting Professor at various Universities in France, Japan, Singapore. She has been invited to speak at over 270 institutes and scientific meetings. She has been chairing and organising over 45 high profile international conferences. She is Editor-in-chief of the Royal Society of Chemistry book Series, Nanoscience and Nanotechnology. She edited 7 theme issues including: (2021) Nanoscale Web themed issue on “Advanced Functional Nanomaterials for Biomedical Applications”. The Royal Society (2016), Interface Focus, “Multifunctional nanostructures for diagnosis and therapy of diseases”; The Royal Society Chemistry, RSC (2014), Faraday Discussions, “Physical Chemistry of Functionalised Biomedical Nanoparticles”; RSC (2013) Nanoscale, Special issue “Functional Nanoparticles for Biomedical Applications” and Philosophical Transactions of the Royal Society A (2010), “Nanoparticles”; MDPI (2021) Biomedicines “Advanced Functional Nanomaterials for Biomedical applications”. She is the sole editor of two seminal books on Magnetic Nanoparticles from Fabrication to Clinical Applications in 2012 (ISBN 9781439869321) and Clinical Applications of Magnetic nanoparticles in 2018 (ISBN 9781138051553). She is co-organising a Magnetic Carrier Meeting in Jun 2022 in London.