Open Access Article
Open Access ArticleVesicle-based artificial cells: materials, construction methods and applications
Yao
Lu†
*,
Giulia
Allegri
and
Jurriaan
Huskens
 *
*
Molecular NanoFabrication Group, Department of Molecules & Materials, MESA+ Institute, Faculty of Science and Technology, University of Twente, P.O. Box 217, 7500 AE, Enschede, The Netherlands. E-mail: y.lu-1@utwente.nl; j.huskens@utwente.nl; y.lu@bit.edu.cn
First published on 26th November 2021
Abstract
The construction of artificial cells with specific cell-mimicking functions helps to explore complex biological processes and cell functions in natural cell systems and provides an insight into the origins of life. Bottom-up methods are widely used for engineering artificial cells based on vesicles by the in vitro assembly of biomimetic materials. In this review, the design of artificial cells with a specific function is discussed, by considering the selection of synthetic materials and construction technologies. First, a range of biomimetic materials for artificial cells is reviewed, including lipid, polymeric and hybrid lipid/copolymer materials. Biomaterials extracted from natural cells are also covered in this part. Then, the formation of microscale, giant unilamellar vesicles (GUVs) is reviewed based on different technologies, including gentle hydration, electro-formation, phase transfer and microfluidic methods. Subsequently, applications of artificial cells based on single vesicles or vesicle networks are addressed for mimicking cell behaviors and signaling processes. Microreactors for synthetic biology and cell–cell communication are highlighted here as well. Finally, current challenges and future trends for the development and applications of artificial cells are described.
1. Introduction
A cell is the basic constitutional unit of all forms of life and encompasses a series of complex and accurate working principles.1,2 In order to explore the origin of life and understand how this sophisticated cellular factory conducts the essential biological interactions to maintain our vital signs, simplified cell models are fabricated to deconvolute the complicated mechanisms in a natural cell system.3In bottom-up approaches of artificial cells, the constitution of a synthetic cell model is based on vesicles, assembled from molecular building blocks and capable of mimicking cellular behavior.4,5 The building blocks can be varied using different materials such as phospholipids,6,7 amphiphilic polymers,8,9 their hybrid composites,10,11 and cell membrane-derived native materials.12,13 Thus, a deep understanding of the individual building blocks, including their component materials, self-assembling structures and dynamic behavior, is required in order to obtain a well-defined model of artificial cells.
The concept of artificial cells was first put forward around 60 years ago.14 It indicates a closed-volume container with the encapsulation of bioactive materials such as enzymes and gene sequences that can aid to present biomimetic behavior.15 There are two important issues in the formation of artificial cells: the self-assembly of a membrane container (vesicles) and the encapsulation of functional materials. From this point of view, two basic concepts need to be elucidated before the engineering of an artificial cell: the protocell model and a minimal cell. A protocell is a hypothetical system with cellular compartment structures, which has primary properties of a cell but is not a living system, and it is geared toward the understanding of the origin of life.16 Vesicles that are assembled from amphiphiles can be regarded as the model of a protocell. Vesicles contain a closed volume of artificial cells to separate the inside from the extracellular environment using a similar membrane structure as cells.17,18 Besides serving as a physical boundary, the membrane structures also mediate the exchange of physical and chemical information between the self-assembled container and its environment in order to maintain intra- and intercellular functions.19 These properties facilitate the protocell model to become a minimal cell by meeting the criteria of cell-like behavior. A typical minimal cell is a self-reproducing system that can generate and replicate genetic and functional molecules, which help facilitate a simple metabolic reaction with energy conversion.20 In this way, the assembled system mimics the living form of cells by presenting essential bioactive features. Thus, the compartment structure of a vesicle can be regarded as a minimal cell after the encapsulation of different bioinspired materials that provide life-like functions.
Recent efforts have focused on realizing complex cellular behaviors based on such a “living” artificial container from two viewpoints: structure and function. In order to build an artificial cell containing different functions, compartmentalization is an essential step to facilitate different reactions and functions separated from others in space and time, which mimics the function of organelles in natural cells. The key challenge to realize a cell-like artificial structure is the construction of a living system with full cellular functions. Currently, it is feasible to assemble artificial cells to mimic some basic cellular behaviors, such as the expression of proteins or genes. However, these discrete functions need to be further integrated to reach a full-scene, higher-level function, in which the intercellular communication plays a key role in providing nutrients and energy for the isolated artificial cells. In this way, work on artificial cells may provide new tools to address questions and challenges in biology and medicine,21 for example, regarding the roles of signal transduction and cellular communication in healthy processes as well as the development of diseases. A well-defined artificial cell is also aimed at deconstructing the complex function of cellular components from the interplay of multiple factors, trying to provide insights into the cell system from the viewpoint of synthetic biology.2 This is, however, still challenging and may offer possibilities for future fundamental research in the fields of biotechnology and bioanalysis.
In this review, we focus primarily on the construction of artificial cells based on giant unilamellar vesicles (GUVs: diameter >1 μm),22 which have a size that is comparable to natural cells. Small unilamellar vesicles (SUVs: diameter <100 nm)23 and large unilamellar vesicles (LUVs: diameter of 100 nm–1 μm)24 are excluded here. The review first introduces the materials that can be used to self-assemble into GUVs, including liposomes, polymersomes, hybrid vesicles and vesicles made of cell membrane-derived native materials (e.g., proteoliposomes with extracted membrane proteins). The strengths and limitations of each material are illustrated to help make a selection when choosing a kind of giant vesicle as the model for an artificial cell. Thereafter, the typical fabrication methods of giant vesicles are described. To be used as an artificial cell model, giant vesicles are targeted with the bilayer structure that is similar to natural cells. Thus, the fabrication methods (the gentle hydration, electroformation, phase transfer and microfluidic methods) are described with an emphasis on the formation of GUVs. Giant vesicles with other structures, such as multilamellar vesicles (MLVs)25 and multivesicular vesicles (MVVs),26 can be obtained simultaneously with GUVs. Here, MLVs are not discussed in detail, while MVVs are regarded as a vesicle model in which SUVs/LUVs are encapsulated as organelles into a GUV to form a compartmental structure (also termed vesosomes), which will be discussed in the methods of phase transfer and microfluidics. The commonly used extrusion method for SUVs and LUVs is excluded here. In the last sections, the applications of vesicle-based artificial cells are described, separately in the form of a single giant vesicle and as a network of multiple giant vesicles. The simplified artificial cell model based on a single vesicle is used to mimic biological processes, such as cell membrane fusion, cell growth and protein/gene expression. The use of GUVs as microreactors to investigate metabolic cascade interactions is also included. The cellular communication between adjacent giant vesicles is demonstrated, presenting the potential of vesicle networks in synthetic biology.
Various reviews on artificial cells have appeared.2,27–33 Rather than being comprehensive, we aim here to highlight the strategies of building artificial cells, with a discussion on the choices for materials and methods. This way we aim to demonstrate the current applications of artificial cells based on both individual giant vesicles and multiple vesicles, and to provide an insight into the directions of this field.
2. Biomimetic materials for the self-assembly of giant vesicles
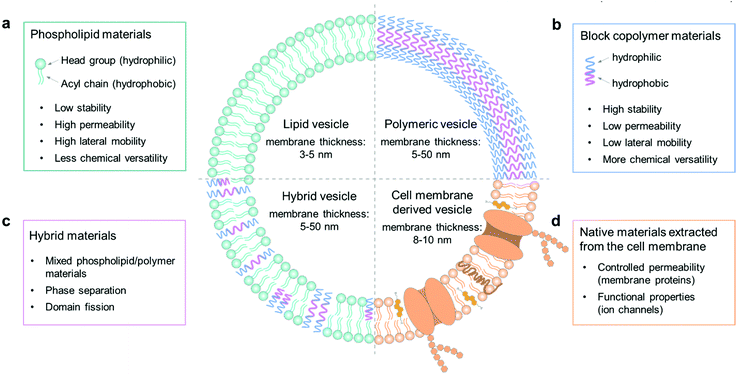
The cell membrane plays a key role in maintaining intracellular homeostasis by creating a physical boundary that separates the inside from the extracellular environment. Thus, the first essential step to build a cell model is to prepare a closed-volume container with a bilayer structure analogous to the native cell membrane. Giant unilamellar vesicles (GUVs) are commonly used as an artificial cell model because they can mimic the cellular membrane structure well. The molecular components to assemble GUVs can be varied from phospholipids, amphiphilic polymers, and hybrids thereof, to cell membrane-derived native materials (Fig. 1). The properties of each component, such as molecular weight, chemical structure, surface charge and amphiphilicity, will affect the membrane structure and the properties of the GUVs, such as their stability, permeability, and lateral fluidity, and they can therefore be used to achieve different functions and applications.2.1. Phospholipids
Phospholipid materials have good biocompatibility owing to their similarity in composition to the natural cellular system. Since the first formation of a vesicular container with lipids in the 1960s,34 phospholipids have been widely used as a biomimetic material to assemble giant vesicles, also to create artificial cells and to study the properties of cell membranes.35 The self-assembled vesicular structure with a pure and spherical lipid bilayer is termed a liposome.36 As Fig. 1a shows, a phospholipid molecule has a hydrophilic head group and two hydrophobic hydrocarbon chains,37 making it suitable to self-assemble into a giant bilayer lipid vesicle. The thickness of a lipid bilayer structure is around 3–5 nm.16 The typically used phospholipids can have different properties; some have a net charge, while others are zwitterionic at neutral pH, the gel-to-liquid phase transition temperature (Tm) can vary from −20 to +60 °C, and the chain length can range from 14 to 18 carbon atoms with or without a C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C double bond.
C double bond.
2.2. Polymers
In recent years, there has been a strong drive to use amphiphilic block copolymers as an alternative to lipids to assemble into polymer vesicles.46 As shown in Fig. 1b, the character of amphiphilic copolymers is similar to that of phospholipids. They both have a hydrophilic end (i.e., a phosphate headgroup in lipids and one hydrophilic moiety in copolymers) covalently attached to a hydrophobic end (one/two fatty acid chains in lipids and one hydrophobic moiety in copolymers).8 With this lipid-like amphiphilic structure, block copolymers can also be applied to build giant vesicles, which are called giant polymersomes.36Block copolymers have the same basic architecture as lipids but consist of distinct polymer chains covalently linked in a series of two or more segments, the molecular weights (MWs) of which can be orders of magnitude larger than those of lipids. The structural features of vesicles, as well as properties including stability, fluidity, and intermembrane dynamics, are greatly influenced by the MW.9 In general, the lipid molecules are smaller than 1 kDa, whereas polymers have a mean MW from 2 to 20 kDa, so that the thickness of polymersome membranes ranges from 5 to 50 nm (Fig. 1b).16 This parameter thus ensures some properties of polymersomes to be different from liposomes, such as permeability, viscosity and elasticity. For instance, the thicker membrane of polymersomes leads to a lower leakage with low permeability. Moreover, by changing the chemical properties of either the hydrophilic or hydrophobic portion of a block copolymer, it is possible to adjust the characteristics of polymersomes. Some commonly used copolymers can be found in a recent review of Dimova et al.16
Polymersomes also have the potential to be used for biomimetic studies, in which proteins, like enzymes or transmembrane channels, can be introduced into their polymeric membranes.47–49 It was also found that polymer GUVs could be assembled from the family of bilayer-forming polymer architectures. They have since not only provided a platform to study the physical properties of polymer bilayers, but also developed a new class of low permeability, mechanically tough, cell-sized vesicles that cannot be made from the commonly used lipid materials.50
2.3. Hybrid lipid/copolymer materials
Hybrid vesicles composed of mixed lipid/polymer materials can be regarded as an advanced vesicular structure in comparison with the forerunners of liposomes or polymersomes, since they potentially combine the favorable characteristics of both components in a single membrane structure.10 Therefore, these structures can present the biocompatibility and bio-functionality of liposomes, as well as the low permeability and functional variability conferred by polymersomes. In applications, hybrid vesicles are an advantageous tool in the study of membrane fusion and domain separation.51 In these systems, the physical and molecular factors that govern the phase separation process can be tuned by changing the hybrid lipid/copolymer ratio in membrane composition, which helps to provide an insight into the membrane events in biological processes.In the formation of stable hybrid vesicles from lipid/copolymer mixtures, a discrepancy of the chemical composition and the size of hydrophobic segments between lipids and polymers needs to be avoided by controlling the parameters of the components. For example, when forming liposomes only from lipid mixtures, the interactions between the lipid tails, composed of saturated or unsaturated fatty acid chains, does not strongly affect the assembly process.52 However, in the case of lipid/copolymer mixtures, the nature of the monomeric unit may lead to a stronger immiscibility between the hydrophobic copolymer blocks and the lipid tails. For instance, it is found in previous studies that the solubility of hydrocarbon moieties in the acyl chains of phospholipids and in the hydrophobic part of block polymers will affect the formation of vesicles; if the two are close, it is much easier to obtain good mixing and thus intact vesicles.53,54 In addition to the possible chemical incompatibility between phospholipid tails and copolymer block chains, one also needs to consider the respective dimensions of the molecules and those of the corresponding bilayers. Generally, the thickness of a lipid bilayer membrane of liposomes is around 3–5 nm, which is smaller than that of polymersomes (5–50 nm).16 For polymersomes, this parameter is directly controlled by the degree of polymerization. In general, a strong geometric difference between the lipid and polymer molecules constituting the membranes can induce a large entropic driving force towards de-mixing of the materials in the formation of vesicles.47
2.4. Extracted native biomaterials
In natural systems, the cell membrane not only serves as a boundary, but also mediates the exchange of physical and chemical information between the cell and its environment in order to maintain intra- and intercellular functions. Liposomes can mimic the structure of cells well with their similar structure of the lipid bilayer, but a pure lipid membrane cannot realize some functional properties of a real cell membrane in the absence of membrane proteins. Thus, proteoliposomes are fabricated by reconstituting membrane proteins into the bilayer structure of liposomes.55 However, the direct functionalization of vesicles by inserting membrane proteins can sometimes lead to instability due to changes in the rearrangement of lipid molecules in the bilayer structure.13 Recently, the assembly of vesicles by using materials derived from native cell membranes has become a versatile approach to endow vesicles with cell-like interface properties. The cell membrane is a thin semipermeable structure consisting of lipids, proteins, and carbohydrates. Different cells have different compositions, which are highly associated with their cellular functions.56 The native membrane preparations extracted from real cells can be used as a source material, which already contains all the natural proteins and small molecules.57–59An example of naturally derived materials are red blood cells, which can be harvested by removing cells and antigens from tissues with detergents.12 In this case, the membrane materials of red blood cells are extracted by cellular hypotonic lysing and then dissolved in chloroform for the construction of vesicles. These cell membrane-based vesicles have been used as drug carriers for biomedical applications,12 which can be regarded as a potential candidate for the assembly of giant vesicles as artificial cells in future applications.
3. Construction of giant vesicles
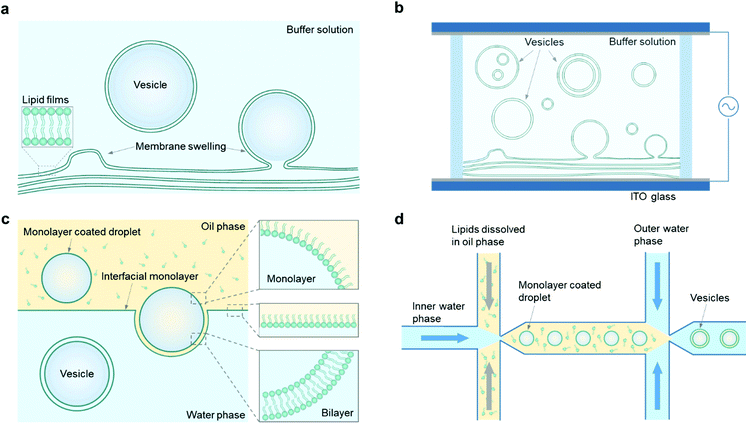
In this section, we will focus on the construction of giant vesicles with a diameter in the range of 1–100 μm, which is comparable to the size of most biological cells. The construction of giant vesicles is a self-assembly process of amphiphiles in their fluid state. Since most polymersomes can be fabricated in the same way as liposomes, depending on the solubility of block copolymers,60 the methods to make giant vesicles in this section apply to both giant liposomes and giant polymersomes, as well as hybrid lipid/copolymer vesicles.In general, there are two main ways to assemble giant vesicles.42,61 In the first, one starts with lamellae-forming amphiphiles (e.g., phospholipids and/or amphiphilic copolymers) in their dry state, often spread with the help of an organic solvent on a solid surface. The amphiphiles are then hydrated to form giant vesicles in a defined and controlled way by adding an aqueous solution. This method is called the “hydration” method (Fig. 2a). The hydration of amphiphiles can be affected in a positive way by applying an external electric field when the amphiphiles are deposited on an electrically conductive surface, which indicates the “electro-formation” method (Fig. 2b).
The other main way to engineer a giant vesicle is the emulsion-based method. This method exploits the fact that a giant vesicle is a micrometre-sized aqueous droplet in a bulk aqueous solution, which is separated by a layer of amphiphiles. To obtain such a closed-volume bilayer structure, water-in-oil droplets are first prepared, followed by a transformation of the droplets into vesicles, which is the so-called “phase transfer” method (Fig. 2c). On the basis of this droplet template from the double-emulsion state, the “microfluidic” method (Fig. 2d) is illustrated as well. To make a comparison, the advantages and disadvantages of each method are illustrated in Table 1.
| Method | Advantages | Disadvantages |
|---|---|---|
| Gentle hydration method | Simple operation | Low production rate |
| Low encapsulation rate | ||
| Inhomogeneous vesicle size | ||
| Electro-formation method | Simple procedure | Limitation to buffers with low ion concentrations |
| Easy for vesicle immobilization | Limitation on building materials (e.g., charged lipids) | |
| Phase transfer method | Good vesicle stability | Difficult operation |
| Regular vesicle shape | Inhomogeneous vesicle size | |
| Asymmetric leaflet of vesicle composition | ||
| Controllable protein orientation | ||
| Microfluidic method | Regular vesicle shape | Limited vesicle size (dependent on the size of microfluidic channel) |
| Uniform vesicle size | ||
| Asymmetric leaflet of vesicle composition | ||
| Controllable protein orientation |
3.1 Gentle hydration method
Based on the principle of membrane swelling, a commonly used procedure to obtain giant vesicles is called the gentle hydration method, which was first applied in the formation of liposomes in the 1960s.34 In general, phospholipids or amphiphilic copolymers are first dissolved in an organic solvent (typically chloroform) and evaporated under a gentle flow of argon or nitrogen gas until the solvents are dried, during which a phospholipid or copolymer film forms on a glass substrate. Then, the film is hydrated with an aqueous buffer solution (typically containing sucrose) and giant vesicles form from the surface (Fig. 2a). With further study, the yield of giant vesicle production is increased by adding a thin layer of hydrogel (e.g., agarose) on a glass surface, which improves the surface hydrophilicity and enhances the swelling of the amphiphilic films.24,62 These hydration methods are simple, productive and easy to operate, and have been widely used in studies of giant liposomes or polymersomes. However, it is difficult to control the vesicle membrane structures by the hydration process, and both unilamellar and multilamellar vesicles are obtained. In addition, the loading efficiency is limited with this method since introducing cargos can hinder the spontaneous swelling of membranes. There are also certain requirements of buffer solutions for the hydration procedure, for example, giant vesicles have a lower yield when there is a high salt concentration in the hydration solution. Yet, in some applications, ions are necessary, and the hydration method is not suitable in these cases.24 Also it is difficult to use the hydration method to obtain vesicles with hybrid lipid/copolymer materials.3.2 Electro-formation method
With the same working principle as the hydration method, the electro-formation method was developed on the basis of hydration to form giant vesicles.63,64 In this method, amphiphilic films are deposited on indium tin oxide (ITO)-coated glass, which can improve the membrane swelling efficiency by applying an alternating current (AC) electric field to the hydrated films (Fig. 2b). The applied frequency and stimulation time of the electric field necessary for vesicle production vary depending on the different components of lipids or polymers, and the typical diameter of vesicles formed with this method is approximately 5–200 μm.42 Compared with the hydration method, the size distribution of giant vesicles obtained by electro-formation is more uniform, but vesicles with different membrane structures, such as GUVs, multilamellar vesicles (MLVs) and multivesicular vesicles (MVVs), will be obtained at the same time. There are also some other restrictions of this method, for instance, it is difficult to form giant vesicles from charged phospholipids, since their surface charge affects the applied electric field on the electrodes. The working principle also sets a limitation of vesicle formation under physiological conditions in the presence of abundant ions.To solve these problems, researchers modified the electro-formation process by tuning the frequency and amplitude of the applied electric field. For example, Li et al. managed to obtain giant liposomes with a negatively charged phospholipid (at 2.5 V and 1 kHz).65 Montes et al. formed giant liposomes composed of DOPC/DPPC/cholesterol (1/1/20 mol%) under physiological conditions (25 mM HEPES, 150 mM NaCl) by increasing the frequency of the applied AC field from 10 Hz to 500 Hz.66 In addition to a single electrical stimulus, it is also possible to set a sequence of AC fields with different frequencies, amplitudes and stimulation periods in order to optimize the size and yield of liposomes based on their components and synthesis conditions. For example, in our previous work, GUVs made of mixed DOPC/TR-DEPE/Biotin-cap-PE (99.8/0.1/0.1 mol%) were successfully obtained in sucrose buffer (50 mM, pH 7.4) by applying a sequence of different AC fields.67
3.3 Phase transfer method
Giant vesicles can also be produced by transferring a lipid or polymer monolayer-coated water-in-oil droplet to an aqueous phase through an interfacial monolayer (Fig. 2c). When the droplet is transferred through the oil–water interface, the interfacial monolayer envelopes the droplet, which renders the formation of a vesicle with a bilayer. Normally, the solution inside the droplet has a relatively high density compared with the surrounding oil and aqueous phases, which allows it to transfer through the interface by the force of gravity. In order to improve the phase transfer efficiency, techniques such as centrifugation68 and micromanipulation69 are also used to drive the droplets across the water–oil interface. Different from the hydration and electro-formation methods, giant vesicles engineered from a phase transfer process can adopt an asymmetric membrane structure, in which the inner leaflet is controlled by the lipid/oil solution applied during droplet incubation, while the outer leaflet is dependent on the monolayer materials distributed at the water–oil interface.70The size of the giant vesicles can be controlled well in the incubation step to prepare the droplets, and it can also ensure complete encapsulation of cargos by pre-loading of the droplets. This property is quite useful in applications that require the loading of charged and large cargos (such as macromolecules, nanoparticles, and nanotubes), which turns to be difficult to realize using the hydration methods. An issue of this method is that not all the engineered giant vesicles can retain a single-compartment structure; two or more droplets can transfer simultaneously through the interfacial monolayer and lead to a giant vesicle with multiple compartments inside its interior. On the other hand, these multi-compartment giant vesicles can be further applied in the study of compartmentalization of artificial cells. For example, Elani et al. first made two/three small droplets with a nanoliter volume and separately loaded them with different reagents.69 Then they manually pipetted the group of droplets at a localized water–oil interface at the same time, aiming to assemble a giant vesicle with multiple compartments.
3.4 Microfluidic method
To ensure high-throughput production and precise control over giant vesicles, microfluidic methods have been developed on the basis of the double emulsion strategy resembling the phase transfer method. In a microfluidic method, the first step is to form a series of droplets from water/oil/water (w/o/w) double emulsions, and giant vesicles are assembled one by one in the microfluidic channel through the subsequent extraction of the middle oil phase (Fig. 2d).71 With advances made in microfabrication techniques, microfluidic devices with various designs have been created and applied in the formation of giant vesicles for different applications.72,73 For example, Huck and co-workers reported a microfluidic method for the robust and straightforward assembly of droplets containing monodisperse coacervates.74 By taking advantage of microfluidic flexibility and controllability, giant vesicles with a hierarchical structure were obtained, which can be used as a model to mimic protocells.Besides the phase-transfer working principle, different microfluidic methods, including micro-jetting and emulsification, have also been developed to form both liposomes and polymersomes.75 For instance, a micro-jetting flow device for the fabrication of cell-sized asymmetric liposomes was reported by Takeuchi and co-workers.76 It demonstrates that the asymmetric liposomes exhibit the behavior of lipid flip-flop in the membrane, which is analogous to apoptotic cells. The emulsification-controlled assembly was used to fabricate giant vesicles with multiple compartments by Lee et al.77 In their work, onion-like vesicles were obtained by changing the interfacial instability of the oil–water emulsion, which provides a new perspective for the design of biomedical materials. From an application point-of-view, the microfluidic method can potentially provide a versatile and miniaturized platform to produce giant vesicles by integrating them with procedures of sample preparation, analysis, and detection in a whole system-on-a-chip, which is important in further biological applications.
4. Mimicking cell functions in vesicle systems
In this section, giant vesicles are used as an artificial cell model to mimic basic biological processes of natural cells. The applications are classified into two main parts. In the first, the mimicking of molecular events at the cell membrane is discussed, for example, the membrane fusion process. The second part is related to biological functions of artificial cells that are facilitated by the encapsulation of bioactive materials inside vesicles, e.g., protein and gene expression, cell growth, (bio)chemical cascade reactions, and intercellular communication in multiple-vesicle systems.4.1 Mimicking membrane fusion
Membrane fusion is a fundamental phenomenon associated with many biological processes, such as endocytosis and exocytosis at the cell membrane,78,79 intracellular exchange of lipids and proteins at the organelle membrane,80 and infections by viruses.81,82 For example, extracellular vesicles called exosomes play a key role in these inter- and intra-cellular trafficking events, during which vesicle fusion is promoted by a membrane fusion process.83 Particularly, in neural systems, membrane fusion is the basis of the synaptic vesicle fusion process, which is regulated by Ca2+ and important for the effective release of neurotransmitters.84,85 To figure out the mechanism of membrane fusion in these natural biological systems, extensive research has been carried out by employing a simplified model of giant lipid vesicles. It is essential that this artificial container maintains good membrane fluidity and mobility with a mixture of functional lipid materials. In previous studies, we found that a lipid membrane composed of different phospholipids has good mobility and permeability, which allows membrane events such as receptor recruitment and transient pore formation to be studied.86,87 In this case, membrane changes induced by the lateral phase separation or lipid domain transition from inserted proteins can be mimicked well.88The typical membrane fusion process between transport synaptic vesicles and neuronal plasma membranes is controlled by the specific interactions of soluble N-ethylmaleimide-sensitive fusion attachment protein receptor (SNARE) complexes,89,90 during which a four-helix coiled-coil bundle is formed between the membrane-bound SNARE proteins and brings the synaptic vesicle and the plasma membrane into close proximity (Fig. 3a).84 Based on this principle, Kros and co-workers designed a model system using vesicles functionalized with two lipidated oligopeptide hybrids (LPE and LPK), which can realize functions similar to SNARE proteins.91 The formation of the LPE/LPK complex is the driving force to bring two different liposomes close together, which triggered the fusion of lipid membranes. Furthermore, a pair of DNA–lipid conjugates was used instead of SNARE proteins, in which the complementary DNA oligonucleotides were functionalized at the head groups of lipids, the sequence-specific hybridization of which induced a close contact of the vesicles to the lipid membrane, thereby facilitating the membrane fusion between vesicles and model membranes (Fig. 3b).92 Following this principle, Boxer and his co-workers proposed another system, in which the pair of complementary DNAs was anchored in the membranes of two suspended vesicles, thereby enabling vesicle fusion by complete mixing of both the lipid membranes and their internal contents (Fig. 3c).92 In addition, the fusion of individual lipid vesicles with a supported lipid bilayer was also realized by the formation of DNA duplexes between complementary lipid-anchored oligonucleotide sequences on the opposing membranes (Fig. 3d),93 during which the change of state from membrane contact to membrane fusion was found to go through docking, hemi-fusion and full-fusion. The study of membrane fusion based on these vesicle models may provide insights into important issues in basic biological fusion interactions, thus helping to understand membrane events in endocytosis and exocytosis.
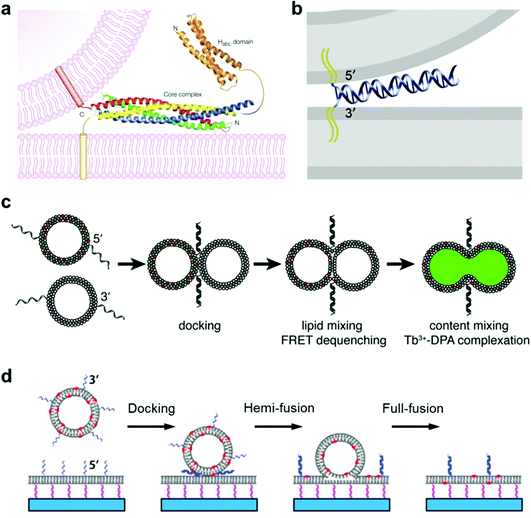 | ||
| Fig. 3 Mimicking membrane fusion based on a vesicle model. (a) Model of a synaptic vesicle that is connected with the plasma membrane by the SNARE complex, which is composed of synaptobrevin (red), syntaxin 1 (yellow, including the amino-terminal Habc domain), synaptosomal-associated protein of a 25 kDa (SNAP25) amino terminus (blue) and a SNAP25 carboxyl terminus (green).84 Copyright © 2002 by Nature Reviews Neuroscience. Reprinted with permission from Nature Reviews Neuroscience. (b) Model of a vesicle docking to a lipid membrane that is induced by a pair of complementary membrane-anchored DNAs. The docking vesicle displays the 5′-coupled conjugates, while the lipid membrane displays the 3′-coupled conjugates, in which the anti-parallel hybridization of the two complementary DNAs allows the vesicle and the lipid membrane to stay in close contact.92 Copyright © 2009 by the Proceedings of the National Academy of Sciences. Reprinted with permission from the Proceedings of the National Academy of Sciences. (c) Fusion of two suspended lipid vesicles that is controlled by a pair of complementary DNAs (3′-coupled conjugates and 5′-coupled conjugates). The two suspended vesicles demonstrate a fusion process including docking, lipid membrane merging and mixing of the contents.92 (d) Individual vesicle fusion mediated by anchored DNAs on a supported lipid bilayer. The fusion process consists of docking, hemi-fusion and full-fusion.93 Copyright © 2013 by Biophysical Journal. Reprinted with permission from Biophysical Journal. | ||
4.2 Mimicking protein and gene expression
A key machinery for artificial cells to realize a specific function is the expression of proteins from genes. In order to develop a vesicle container capable of conducting the whole transcription and translation process, complex biochemical reaction networks need to be encapsulated inside giant vesicles, the membrane composition of which facilitates a selective materials exchange between the vesicle interior and the external environment. Lipid materials are commonly used in the assembly of such vesicles with good membrane permeability.Libchaber and co-workers managed to observe the expression of enhanced green fluorescent proteins (eGFPs) by encapsulating an Escherichia coli cell-free expression system inside individual giant lipid vesicles (Fig. 4a).94 They proved that a cell-free extract can be successfully loaded inside giant lipid vesicles in spite of its complex composition. In order to reach prolonged protein expression with this lipid vesicle container, the α-hemolysin proteins were assembled into the membrane of the vesicles to increase their permeability for the feed solution.
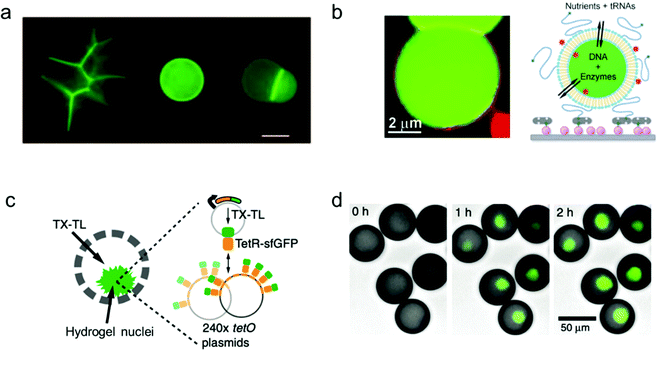 | ||
| Fig. 4 Mimicking protein and gene expression based on a vesicle model. (a) Expression of α-hemolysin-eGFPs with giant vesicle models in different states: aggregated vesicles, individual single vesicles and a dimer–vesicle system (from left to right). The scale bar indicates 20 μm.94 Copyright © 2004 by the Proceedings of the National Academy of Sciences. Reprinted with permission from the Proceedings of the National Academy of Sciences. (b) Schematic of the encapsulation of enzymes and genes inside giant vesicles for the expression of GFP protein.95 Copyright © 2012 by Angewandte Chemie International Edition. Reprinted with permission from Angewandte Chemie International Edition. (c) Schematic and (d) fluorescence images of the expression of GFP protein (green, merged with brightfield images) in hydrogel nuclei inside an artificial container made of polymers. Transcription and translation (TX–TL) reagents were added at 0 h, and the expression was detected at 1 h and 2 h.96 Copyright © 2018 by Nature Communications. Reprinted with permission from Nature Communications. | ||
Similarly, Danelon's group synthesized a vesicle-based microreactor by enclosing a minimal gene expression system of both the Escherichia coli ribosome and the bacteriophage T7 RNA polymerase (Fig. 4b).95 Instead of incorporating pore proteins into the membrane of the vesicle containers, they used short dimyristoylated phospholipids with 14-carbon atom acyl chains to build giant vesicles to enable semi-permeability for the nutrient exchange from the feed solution. In their system, purified enzymes were loaded into the giant vesicles together with DNA (translation factor) and RNA (transcription factor) templates, and protein expression could be triggered by adding nutrients of nucleotides and amino acids through the semi-permeable membrane of the vesicles.
Furthermore, cell-free gene expression was realized in a cell-like container with an artificial nucleus, which was assembled with polymers in a microfluidic system.96 Different from the previous systems, the DNA templates were pre-loaded inside the hydrogel nuclei instead of the bulk volume of the container (Fig. 4c). Thus, after sufficient supply of transcription and translation (TX–TL) reagents from the external feeding environment, the corresponding protein expression was induced locally in the artificial nuclei (Fig. 4d). Cell-free expression was successfully realized in the compartmentalized cell-like system, which promotes further study of multi-vesicular systems as nano/micro-reactors.
4.3 Mimicking cell growth
One of the important properties of an artificial cell when modelling a natural cell is the ability to realize replication. This process normally includes two parts: self-reproduction of the cellular boundary that is based on membrane replication with lipid/copolymer materials, and self-replication of the nucleus that is mediated by genome reproduction.97 Numerous synthetic biology studies have been conducted aiming to construct an artificial cell, capable of self-replication, by encapsulating enzymes and reagents into the synthetic compartments of a giant vesicle.In contrast to the spontaneous, equilibrium-driven formation process of vesicles, which is mostly dependent on the osmotic pressure difference across the membrane and the fatty acid concentration in the extra-vesicular medium, the reproduction of a vesicle-based artificial cell is promoted by realizing an out-of-equilibrium, self-supporting or basic metabolism process, which is based on protein synthesis inside vesicles.98 The first attempt to create an artificial cell system that can endogenously produce lipid membranes was set up in Luisi's group.98 In their work, four different enzymes were loaded inside a giant vesicle to consecutively transform the precursors acyl-coenzyme A (CoA) and glycerol-3-phosphate (G3P) into diacyl-glycero-phosphatidylcholine, which can be used as the new lipid components to build lipid membranes.99 Furthermore, giant lipid vesicles were synthesized by loading the enzyme palmitoyl-CoA and G3P; then another enzyme, sn-glycerol-3-phosphate-acyltransferase, was injected into the giant vesicles to trigger the production of the lipid precursor 1-palmitoyl-sn-glycerol-3-phosphate. As shown in Fig. 5a, this process facilitated the newly produced lipids to distribute into the lipid membrane, thereby inducing the formation of small vesicles inside the giant one.
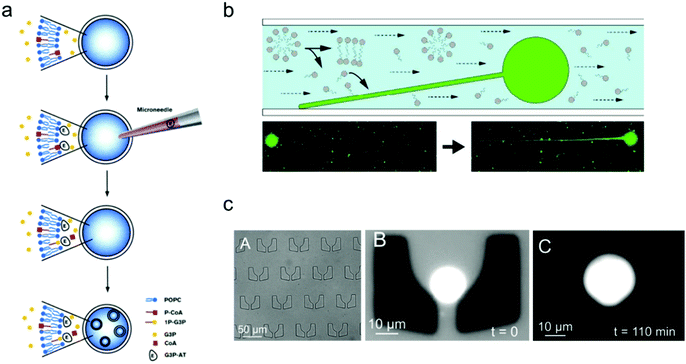 | ||
| Fig. 5 Mimicking artificial cell growth based on vesicle models. (a) Schematic representation of the production of small vesicles inside a giant vesicle by loading palmitoyl-CoA (P-CoA), sn-glycerol-3-phosphate-acyltransferase (G3P-AT) and glycerol-3-phosphate (1P-G3P).99 Copyright © 1996 by Chemistry & Biology. Reprinted with permission from Chemistry & Biology. (b) Filamentous growth of immobilized fatty acid vesicles.100 Copyright © 2014 by Langmuir. Reprinted with permission from Langmuir. (c) Trapping structures in a microfluidic channel (left), a vesicle trapped at t = 0 (middle), and the vesicle growth as the buffer solution is flushed in at t = 110 min.101 Copyright © 2017 by The European Physical Journal Plus. Reprinted with permission from The European Physical Journal Plus. | ||
The growth of artificial cells can also be realized with immobilized vesicles. Szostak and co-workers demonstrated a system in which fatty acid vesicles were immobilized on the surface of a microfluidic channel.100 As shown in Fig. 5b, the spherical vesicle gradually grew into a filamentous vesicle with a constant replenishment of fatty acid molecules induced by the flow in the microfluidic chamber. Moreover, the vesicle growth can be achieved as well by trapping vesicles in a patterned microfluidic channel. As illustrated in Fig. 5c, an individual lipid vesicle was first trapped on a microfluidic chip, and a buffer solution containing fatty acids was then pumped into the channel and continuously flown along the trapped vesicle, which induced the growth of the vesicle.101
4.4 Mimicking cascade reactions and intracellular communication
Individual giant vesicles are suited to downscale chemical and biochemical reactions in closed volumes with a size ranging from 1 μm to 10 μm in diameter. Zare and co-workers were among the first to use giant vesicles as micro-reactors. They built a giant vesicle that contained a single reagent or a complete reaction system with the hydration method, and a chemical reaction process was triggered by adding another reagent into the vesicle. For example, Danelon and co-workers realized the replication of DNA by DNA-encoded proteins that are encapsulated in a giant vesicle.102 As shown in Fig. 6a, a DNA replication cycle was realized by encapsulating the linear DNA template for Φ 29 virus proteins, during which the giant vesicle can be regarded as a micro-reactor to enable the efficient complex genome synthesis in a confined environment.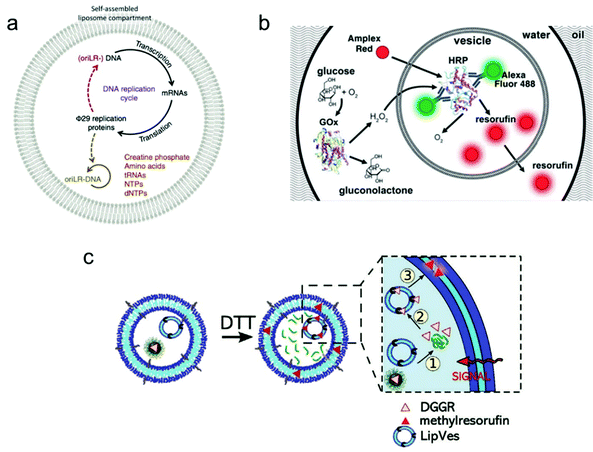 | ||
| Fig. 6 Application of giant vesicles as a micro-reactor. (a) The Φ 29 virus-inspired DNA expression is established inside the giant vesicle by maintaining a series of interactions in this confined micro-reactor.102 Copyright © 2018 by Nature Communications. Reprinted with permission from Nature Communications. (b) Cascade reactions in a micro-reactor based on a giant vesicle with multiple compartments.26 Copyright © 2017 by the Lab on a Chip. Reprinted with permission from the Lab on a Chip. (c) Schematic representation of enzymatic reactions using substrate (DGGR)-loaded nanoparticles and enzyme (lipase)-adsorbed polymersomes, which are encapsulated into a polymer GUV as subcompartments. The substrate was released from nanoparticles in the presence of DTT and transformed into a fluorescent product (methyl resorufin).103 Copyright © 2021 by Advanced Functional Materials. Reprinted with permission from Advanced Functional Materials. | ||
Another application is the use of multi-compartmental giant vesicles to realize inter-compartmental communication within the vesicle. The enclosed compartments inside giant vesicles can enable the separate storage of reagents, thereby facilitating the selective and consecutive release of chemicals to initiate coupled and complex biochemical reactions. Efficient communication between the compartments is necessary to provide a sequence of different steps and orders. Dittrich and co-workers used a microfluidic method to assemble a multivesicular system that performs biochemical reactions with an enzyme cascade assay (Fig. 6b).26 The system can potentially be applied in bio-catalysis to achieve spatially and temporally segregated enzyme cascades.
More complex cascade reactions were realized in a giant vesicle-based system by loading small vesicles or nanoparticles as sub-compartments inside a giant vesicle.103 As shown in Fig. 6c, a polymer GUV with multiple compartments was assembled from a mixture of poly(2-methyl-2-oxazoline)-b-poly(dimethyl-siloxane)-b-poly(2-methyl-2-oxazoline) (PMOXA5-b-PDMS58-b-PMOXA5) and PDMS65-b-heparin copolymers. Inside this giant polymer vesicle, there are two kinds of loaded compartments: (i) nanoparticles functionalized with the lipase substrate 1,2-di-O-lauryl-rac-glycero-3-(glutaric acid 6-methylresorufin ester) (DGGR) and (ii) small polymersomes pre-encapsulated with the enzyme lipase. Without stimulation, no fluorescence was observed inside the giant vesicle due to the spatial segregation of the substrate and enzyme in the two separate artificial organelles. However, once a signalling molecule was added, a series of intracellular communication events were initiated. As the zoom-in picture in Fig. 6c shows, first, the signalling molecule dithiothreitol (DTT) was added to the GUV, causing the release of the enzymatic substrate (DGGR) from the nanoparticles (i.e., the reduction sensitive subcompartment). This led to the interaction with the lipase inside the polymersome, during which the fluorescent product methyl resorufin was generated. By assembling a giant vesicle with an internal subcompartment architecture, the triggered and localized lipase reaction was successfully achieved by an intracellular communication process.
4.5 Mimicking artificial cell communication in a multiple-vesicle system
Constructing higher-order vesicle assemblies has discipline-spanning potential from responsive soft matter materials to artificial cell networks in synthetic biology. By assembling a spatial network of multiple vesicles, it is possible to mimic the intercellular communication occurring between adjacent vesicles, which may provide an insight into the mechanisms underlying this biological phenomenon.Recently, the Ces group has made a lot of progress on the assembly of biomimetic vesicle networks using optical tweezers.104 In their work, they demonstrated a vesicle interface membrane (VIM) system, in which a double membrane was formed at the interfaces between two adjacent giant vesicles. Thus, vesicle communication can be realized by inserting the transmembrane protein α-hemolysin (α-HL) in the conjunct part between the two vesicles (Fig. 7). To confirm successful operation of this system, a fluorescence leakage assay was conducted between two vesicles containing α-HL, which realized the communication between vesicles by the materials transfer from one to another. These vesicle networks are reconfigurable, which enables transformations of various geometries, and their morphology can be tuned using both physical and chemical variables.
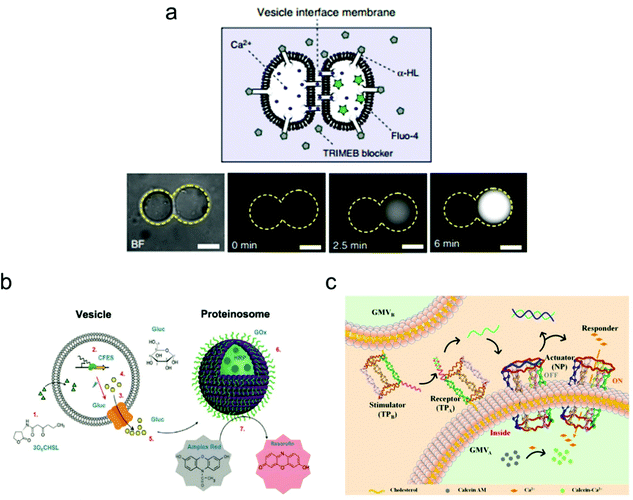 | ||
| Fig. 7 Mimicking artificial cell communication based on a vesicle network. (a) Schematic (top) of a two-vesicle system that is connected by the α-hemolysin (α-HL) protein pores. Ca2+ can selectively translocate through the pores that are not covered by channel blockers. Fluorescence images (bottom) of the two-vesicle system. When Ca2+ moves from one vesicle to another (which is pre-loaded with a Ca2+ indicator, Fluo-4), increased fluorescence was observed indicating vesicle communication. Scale bars indicate 10 μm.104 Copyright © 2018 by Nature Communications. Reprinted with permission from Nature Communications. (b) Schematic illustration of the intercellular communication between liposomes and proteinosomes.105 Copyright © 2018 by American Chemical Society. Reprinted with permission from American Chemical Society. (c) Schematic illustration of the cellular communication between GMVA and GMVB. One vesicle (GMVA) acts as a receptor and an actuator, as modified by DNA triangular prism A (TPA, receptor) and the DNA nanopore (NP, actuator), respectively. The other vesicle (GMVB) acts as a stimulator, as modified by DNA triangular prism B (TPB, stimulator).106 Copyright © 2021 by American Chemical Society. Reprinted with permission from American Chemical Society. | ||
The mimicking of cellular communication has led to the signalling and materials transmission between two separate vesicles. Different from the case in Fig. 7a, non-contact intercellular communication was achieved here. For instance, Tang and co-workers assembled a communication system based on two types of vesicles: liposomes and proteinosomes (Fig. 7b).105 In this system, the liposome was encapsulated with DNA templates and used as a transmitter, while the proteinosome was designed to be a receiver by loading it with biocatalysts that are utilized for enzyme cascade reactions. The protein expression of α-hemolysin within the liposome triggered the release of glucose molecules, which then induced the enzyme cascade reactions within the proteinosome between glucose oxidase and horseradish peroxidase. The end product of this process is the red-fluorescent resorufin.
Cell–cell communication has been successfully mimicked by designing an artificial signal transduction system based on the model of giant membrane vesicles (GMVs).106 As shown in Fig. 7c, a membrane protein-like stimulator (i.e., the DNA triangular prism B, TPB) from GMVB will stimulate a receptor (i.e., DNA triangular prism A, TPA) on another GMVA to release a piece of DNA messenger, which can activate the synthetic transmembrane channels (i.e., the DNA nanopore, NP) on GMVA, thus enabling the influx of ions. Here, the DNA nanopore used as a channel protein contains a DNA strand that blocks the entrance for molecular transport, but it will reopen once a piece of complementary DNA is released from TPA and the strand displacement reaction occurs in the nanopore. This triggered signal transduction process helps to realize the communication between two artificial cells, which can be applied potentially to the analysis of a natural biological signalling process.
5. Summary
As a simplified model fabricated by bottom-up approaches, vesicle-based artificial cells are important in the investigation of biological phenomena in natural cellular systems. An ultimate goal is to provide an insight into the origins of life. In this review, we have focused on how to prepare giant vesicles for modelling artificial cells from the view of materials, which includes phospholipids, amphiphilic copolymers, hybrid lipid/copolymer materials and materials extracted from native cell membranes. The properties of these constituting materials have been illustrated and compared, aiming to present the strengths and limitations of each material in the self-assembly of giant vesicles for different applications. For example, in studies of membrane fusion, it is feasible to use either phospholipids or amphiphilic copolymers to assemble vesicles instead of using hybrid lipid/copolymer materials, since the latter may suffer from phase separation due to incompatibility between the lipid and the block copolymer. On the other hand, when a vesicle model with both characteristics is required, i.e., biocompatibility of lipids and multi-functionality of copolymers,30 then the hybrid lipid/copolymer materials are the preferred choice for the assembly of artificial cells.Methodologies for fabricating giant vesicles have been reviewed as well. In detail, the gentle hydration and electro-formation methods based on membrane swelling, and the phase transfer and microfluidic methods based on double emulsions were discussed. The choice for a construction method is dependent on the requirements of the desired vesicle structures in the targeted application. The gentle hydration and electro-formation methods are typically used to assemble vesicles of symmetric bilayer structures from lipids and/or copolymers. However, in some specific applications, a giant vesicle with an asymmetric bilayer structure is required to realize different functions in the inner and outer leaflets of the vesicle. In that case, the phase transfer and microfluidic methods are needed to build the asymmetric vesicles. Furthermore, the microfluidic method is also commonly applied to form vesicles with subcompartments by loading small vesicles inside a giant one, which can be used as a micro-reactor for biochemical applications.
Finally, applications of vesicle-based artificial cells were illustrated. The single giant vesicle model can be used to mimic cellular behavior, such as membrane fusion, protein/gene expression, cell growth, and intracellular metabolic reactions. Furthermore, the mimicking of cell–cell communication by introduction of interactions between multiple vesicles was discussed. Some recent and illustrative examples have been highlighted in this section.
6. Future directions
In recent years, various materials have been used to fabricate vesicle-based artificial cells with different structures. However, limitations still exist for the construction of multifunctional artificial cells with complex machineries like in natural cells. In this regard, there are mainly two ways to address this issue: materials and structure.The further development of alternative materials is necessary to assemble artificial cells that can mimic the cellular behavior of natural cells with increasing levels of complexity and function. Even though it is now possible to demonstrate a vesicle-based artificial cell that can grow by adding genetic materials and enzymes, it is still a major challenge to realize a true cellular reproduction process including cell growth, division and differentiation, while effective materials exchange and energy conversion are also crucial. Likewise, we may be close to the assembly of a protocell, but still have a distance from achieving the so-called minimal cell with autonomous adaptivity and functionality. Therefore, establishing multicomponent vesicles through targeted integration of multi-material properties is necessary. In this regard, more work is needed on the assembly of vesicles by the incorporation of additional proteins into the membrane structure to ensure membrane permeability, thus facilitating the selective materials exchange with the environment and/or adjacent vesicles. Moreover, the assembly of vesicles with stimuli-responsive materials is becoming increasingly popular in the study of artificial cells as biosensors for cell-free sensing. The introduction of stimuli-responsive materials enables an artificial cell to respond to a specific external stimulus, which can potentially be applied to build a controllable and tuneable micro-reaction system.
Another challenge lies in the mimicking of cellular communication. Implementation of these important biological processes in a vesicle system needs accurate control and good adaptability of vesicles in order to realize triggered responses during remote signal transduction. In this regard, artificial cells with complex structures or artificial cell networks need to be fabricated. For intracellular communication, the focus has been on the construction of giant vesicles with multiple compartments. The subcompartments inside the vesicle are regarded as artificial organelles, in which different reactions can happen spatially independently, and the corresponding products can be released/loaded among these subcompartments to realize intracellular communication. Future work could take advantage of this spatially segmented structure to conduct a more complex metabolic reaction pathway by distributing each reaction step in separate subcompartments. For intercellular communication, recent work has been done on the assembly of two or more vesicles together to form a communication system, in which the signalling transmission can be achieved by the release of chemical stimuli or loading of materials. In order to mimic a more complex communication between cells, the specific assembly of artificial tissue would be a key issue for further study.
To construct an artificial cell with full cellular function is still challenging at this stage. Future work needs to be done to address the deconvolution of the complex cellular machinery and to establish cell-like systems in which complex biological processes are connected in an integrated manner. To achieve such multi-disciplinary goals, other fields like materials science and nanofabrication need to be combined with synthetic biology in a synergistic way to realize the precise manipulation of vesicle-based artificial cells.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Y. L. and J. H. acknowledge the financial support by the Netherlands Organization for Scientific Research (NWO-NSFC grant no. 729001032 to J. H.). G. A. acknowledges the financial support by the University of Parma for the exchange visit at the University of Twente.References
- K. Kaneko, Life: An Introduction to Complex Systems Biology, Springer, 2006, pp. 1–36 Search PubMed.
- B. C. Buddingh and J. C. van Hest, Acc. Chem. Res., 2017, 50, 769–777 CrossRef CAS.
- J. W. Szostak, D. P. Bartel and P. L. Luisi, Nature, 2001, 409, 387–390 CrossRef CAS.
- P. L. Luisi and P. Stano, The Minimal Cell: the Biophysics of Cell Compartment and the Origin of Cell Functionality, Springer Science & Business Media, 2010, pp. 123–153 Search PubMed.
- P. L. Luisi, The Emergence of Life: from Chemical Origins to Synthetic Biology, Cambridge University Press, 2016, pp. 243–267 Search PubMed.
- P. Walde, Orig. Life Evol. Biosph., 2006, 36, 109–150 CrossRef CAS.
- X. Wang, H. Du, Z. Wang, W. Mu and X. Han, Adv. Mater., 2021, 33, 2002635 CrossRef CAS.
- D. E. Discher and A. Eisenberg, Science, 2002, 297, 967–973 CrossRef CAS.
- T. M. S. Chang, Nat. Rev. Drug Discovery, 2005, 4, 221–235 CrossRef CAS.
- J. F. Le Meins, C. Schatz, S. Lecommandoux and O. Sandre, Mater. Today, 2013, 16, 397–402 CrossRef CAS.
- S. Khan, M. Li, S. P. Muench, L. J. Jeuken and P. A. Beales, Chem. Commun., 2016, 52, 11020–11023 RSC.
- X. Zhang, P. Angsantikul, M. Ying, J. Zhuang, Q. Zhang, X. Wei, Y. Jiang, Y. Zhang, D. Dehaini and M. Chen, Angew. Chem., Int. Ed., 2017, 56, 14075–14079 CrossRef CAS.
- W. L. Liu, M. Z. Zou, S. Y. Qin, Y. J. Cheng, Y. H. Ma, Y. X. Sun and X. Z. Zhang, Adv. Funct. Mater., 2020, 30, 2003559 CrossRef CAS.
- Y. Ding, F. Wu and C. Tan, Life, 2014, 4, 1092–1116 CrossRef.
- D. A. Hammer and N. P. Kamat, FEBS Lett., 2012, 586, 2882–2890 CrossRef CAS.
- R. Dimova, P. Stano, C. M. Marques and P. Walde, in The Giant Vesicle Book, ed. R. Dimova and C. M. Marques, CRC Press, 2019, pp. 1–18 Search PubMed.
- T. F. Zhu and J. W. Szostak, J. Am. Chem. Soc., 2009, 131, 5705–5713 CrossRef CAS.
- P. A. Monnard and P. Walde, Life, 2015, 5, 1239–1263 CrossRef CAS.
- N. J. Yang and M. J. Hinner, Site-specific Protein Labeling, Springer, 2015, pp. 29–53 Search PubMed.
- S. Jeong, H. T. Nguyen, C. H. Kim, M. N. Ly and K. Shin, Adv. Funct. Mater., 2020, 30, 1907182 CrossRef CAS.
- I. Ivanov, S. L. Castellanos, S. Balasbas III, L. Otrin, N. Marušič, T. Vidaković-Koch and K. Sundmacher, Annu. Rev. Chem. Biomol. Engin., 2021, 12, 287–308 CrossRef CAS.
- C. V. Kulkarni, Nanoscale, 2012, 4, 5779–5791 RSC.
- D. van Swaay and A. DeMello, Lab Chip, 2013, 13, 752–767 RSC.
- H. Stein, S. Spindler, N. Bonakdar, C. Wang and V. Sandoghdar, Front. Physiol., 2017, 8, 63 Search PubMed.
- L. Mayer, M. Hope and P. Cullis, Biochim. Biophys. Acta, Biomembr., 1986, 858, 161–168 CrossRef CAS.
- N. Nuti, P. Verboket and P. S. Dittrich, Lab Chip, 2017, 17, 3112–3119 RSC.
- R. Lentini, N. Y. Martín and S. S. Mansy, Curr. Opin. Chem. Biol., 2016, 34, 53–61 CrossRef CAS.
- S. Matosevic, BioEssays, 2012, 34, 992–1001 CrossRef CAS.
- H. Pick, A. C. Alves and H. Vogel, Chem. Rev., 2018, 118, 8598–8654 CrossRef CAS.
- E. Rideau, R. Dimova, P. Schwille, F. R. Wurm and K. Landfester, Chem. Soc. Rev., 2018, 47, 8572–8610 RSC.
- X. Wang, H. Du, Z. Wang, W. Mu and X. Han, Adv. Mater., 2021, 33, 2002635 CrossRef CAS.
- C. Xu, S. Hu and X. Chen, Mater. Today, 2016, 19, 516–532 CrossRef CAS.
- Y. Zhu, X. Guo, J. Liu, F. Li and D. Yang, Small Methods, 2020, 4, 2000406 CrossRef.
- A. D. Bangham and R. Horne, J. Mol. Biol., 1964, 8, 660–668 CrossRef CAS.
- J. P. Reeves and R. M. Dowben, J. Cell. Physiol., 1969, 73, 49–60 CrossRef CAS.
- M. Antonietti and S. Förster, Adv. Mater., 2003, 15, 1323–1333 CrossRef CAS.
- K. Roberts, B. Alberts, A. Johnson, P. Walter and T. Hunt, Molecular Biology of the Cell, New York: Garland Science, 2002, pp. 617–650 Search PubMed.
- W. Shinoda, R. DeVane and M. L. Klein, J. Phys. Chem. B, 2010, 114, 6836–6849 CrossRef CAS.
- S. Pöyry and I. Vattulainen, Biochim. Biophys. Acta, Biomembr., 2016, 1858, 2322–2333 CrossRef.
- M. Claessens, B. Van Oort, F. Leermakers, F. Hoekstra and M. C. Stuart, Biophys. J., 2004, 87, 3882–3893 CrossRef CAS.
- H. Träuble and H. Eibl, Proc. Natl. Acad. Sci. U. S. A., 1974, 71, 214–219 CrossRef.
- K. Kamiya and S. Takeuchi, J. Mater. Chem. B, 2017, 5, 5911–5923 RSC.
- F. Szoka Jr and D. Papahadjopoulos, Annu. Rev. Biophys., 1980, 9, 467–508 CAS.
- E. J. Shimshick and H. M. McConnell, Biochemistry, 1973, 12, 2351–2360 CrossRef CAS.
- P. L. Yeagle, The Membranes of Cells, Academic Press, 2016, pp. 27–56 Search PubMed.
- J. J. Green and J. H. Elisseeff, Nature, 2016, 540, 386–394 CrossRef CAS.
- P. Tanner, P. Baumann, R. Enea, O. Onaca, C. Palivan and W. Meier, Acc. Chem. Res., 2011, 44, 1039–1049 CrossRef CAS.
- C. Martino, S. H. Kim, L. Horsfall, A. Abbaspourrad, S. J. Rosser, J. Cooper and D. A. Weitz, Angew. Chem., Int. Ed., 2012, 124, 6522–6526 CrossRef.
- R. Tamate, T. Ueki and R. Yoshida, Adv. Mater., 2015, 27, 837–842 CrossRef CAS.
- C. G. Palivan, R. Goers, A. Najer, X. Zhang, A. Car and W. Meier, Chem. Soc. Rev., 2016, 45, 377–411 RSC.
- S. Khan, J. McCabe, K. Hill and P. A. Beales, J. Colloid Interface Sci., 2020, 562, 418–428 CrossRef CAS.
- M. K. Riaz, M. A. Riaz, X. Zhang, C. Lin, K. H. Wong, X. Chen, G. Zhang, A. Lu and Z. Yang, Int. J. Mol. Sci., 2018, 19, 195 CrossRef.
- T. M. Kuo and H. Gardner, Lipid Biotechnolog, CRC Press, 2002, pp. 768–796 Search PubMed.
- M. Kurata and Y. Tsunashima, in Polymer Handbook, ed. J. Brandrup, E. H. Immergut, E. A. Grulke, A. Abe and D. R. Bloch, Wiley, New York, 1999, pp. VII/1–84 Search PubMed.
- J. L. Rigaud and D. Lévy, Meth. Enzymol., 2003, 372, 65–86 CrossRef CAS.
- B. György, M. E. Hung, X. O. Breakefield and J. N. Leonard, Annu. Rev. Pharmacol. Toxicol., 2015, 55, 439–464 CrossRef.
- F. Junge, B. Schneider, S. Reckel, D. Schwarz, V. Dötsch and F. Bernhard, Cell. Mol. Life Sci., 2008, 65, 1729–1755 CrossRef CAS.
- J. Andréll and C. G. Tate, Mol. Membr. Biol., 2013, 30, 52–63 CrossRef.
- S. Zorman, M. Botte, Q. Jiang, I. Collinson and C. Schaffitzel, Curr. Opin. Struct. Biol., 2015, 32, 123–130 CrossRef CAS.
- T. Trantidou, M. Friddin, Y. Elani, N. J. Brooks, R. V. Law, J. M. Seddon and O. Ces, ACS Nano, 2017, 11, 6549–6565 CrossRef CAS.
- P. Walde, K. Cosentino, H. Engel and P. Stano, ChemBioChem, 2010, 11, 848–865 CrossRef CAS.
- Z. Peng, S. Kanno, K. Shimba, Y. Miyamoto and T. Yagi, Conf. Proc. IEEE. Eng. Med. Biol. Soc., 2020, 2198–2201 Search PubMed.
- M. I. Angelova and D. S. Dimitrov, Faraday Discuss., 1986, 81, 303–311 RSC.
- D. Dimitrov and M. Angelova, J. Electroanal. Chem. Interf. Electrochem., 1988, 253, 323–336 CrossRef.
- Q. Li, X. Wang, S. Ma, Y. Zhang and X. Han, Colloids Surf., B, 2016, 147, 368–375 CrossRef CAS.
- L. R. Montes, A. Alonso, F. M. Goni and L. A. Bagatolli, Biophys. J., 2007, 93, 3548–3554 CrossRef CAS.
- Y. Lu, W. C. De Vries, N. J. Overeem, X. Duan, H. Zhang, H. Zhang, W. Pang, B. J. Ravoo and J. Huskens, Angew. Chem., Int. Ed., 2019, 58, 159–163 CrossRef CAS.
- R. Xu, R. J. Simpson and D. W. Greening, Exosomes and Microvesicles, Springer, 2017, pp. 91–116 Search PubMed.
- Y. Elani, A. Gee, R. V. Law and O. Ces, Chem. Sci., 2013, 4, 3332–3338 RSC.
- S. Pautot, B. J. Frisken and D. Weitz, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 10718–10721 CrossRef CAS.
- C. Martino and A. J. deMello, Interface Focus, 2016, 6, 20160011 CrossRef.
- N. N. Deng, M. Yelleswarapu, L. Zheng and W. T. Huck, J. Am. Chem. Soc., 2017, 139, 587–590 CrossRef CAS.
- Y. Ai, R. Xie, J. Xiong and Q. Liang, Small, 2020, 16, 1903940 CrossRef CAS.
- N. N. Deng, M. Yelleswarapu and W. T. Huck, J. Am. Chem. Soc., 2016, 138, 7584–7591 CrossRef CAS.
- A. M. Gañán-Calvo, R. González-Prieto, P. Riesco-Chueca, M. A. Herrada and M. Flores-Mosquera, Nat. Phys., 2007, 3, 737–742 Search PubMed.
- K. Kamiya, R. Kawano, T. Osaki, K. Akiyoshi and S. Takeuchi, Nat. Chem., 2016, 8, 881 CrossRef CAS.
- M. K. Park, S. Jun, I. Kim, S. M. Jin, J. G. Kim, T. J. Shin and E. Lee, Adv. Funct. Mater., 2015, 25, 4570–4579 CrossRef CAS.
- N. Oh and J.-H. Park, Int. J. Nanomed., 2014, 9, 51 Search PubMed.
- S. C. Silverstein, R. M. Steinman and Z. A. Cohn, Annu. Rev. Biochem., 1977, 46, 669–722 CrossRef CAS.
- Y. Li-Beisson, J. Neunzig, Y. Lee and K. Philippar, Curr. Opin. Plant Biol., 2017, 40, 138–146 CrossRef CAS.
- V. Buzon, G. Natrajan, D. Schibli, F. Campelo, M. M. Kozlov and W. Weissenhorn, Pathogens, 2010, 6, e1000880 Search PubMed.
- S. C. Harrison, Nat. Struct. Mol. Biol, 2008, 15, 690 CrossRef CAS.
- G. Cevc and H. Richardsen, Adv. Drug Delivery Rev., 1999, 38, 207–232 CrossRef CAS.
- J. Rizo and T. C. Südhof, Nat. Rev. Neurosci., 2002, 3, 641–653 CrossRef CAS.
- M. Kyoung, Y. Zhang, J. Diao, S. Chu and A. T. Brunger, Nat. Protoc., 2013, 8, 1–16 CrossRef CAS.
- D. Di Iorio, Y. Lu, J. Meulman and J. Huskens, Chem. Sci., 2020, 11, 3307–3315 RSC.
- Y. Lu, J. Huskens, W. Pang and X. Duan, Mater. Chem. Front., 2019, 3, 782–790 RSC.
- R. Jahn and R. H. Scheller, Nat. Rev. Mol. Cell Biol., 2006, 7, 631–643 CrossRef CAS.
- Y. H. M. Chan, B. van Lengerich and S. G. Boxer, Biointerphases, 2008, 3, FA17–FA21 CrossRef CAS.
- H. R. Pelham, Trends Cell Biol., 2001, 11, 99–101 CrossRef CAS.
- H. Robson Marsden, N. A. Elbers, P. H. Bomans, N. A. Sommerdijk and A. Kros, Angew. Chem., Int. Ed., 2009, 121, 2366–2369 CrossRef.
- Y. H. M. Chan, B. van Lengerich and S. G. Boxer, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 979–984 CrossRef CAS.
- B. van Lengerich, R. J. Rawle, P. M. Bendix and S. G. Boxer, Biophys. J., 2013, 105, 409–419 CrossRef CAS.
- V. Noireaux and A. Libchaber, Proc. Natl. Acad. Sci. U. S. A, 2004, 101, 17669–17674 CrossRef CAS.
- Z. Nourian, W. Roelofsen and C. Danelon, Angew. Chem., Int. Ed., 2012, 124, 3168–3172 CrossRef.
- H. Niederholtmeyer, C. Chaggan and N. K. Devaraj, Nat. Comm., 2018, 9, 1–8 CrossRef CAS.
- G. Murtas, Syst. Synth. Biol., 2010, 4, 85–93 CrossRef.
- P. K. Schmidli, P. Schurtenberger and P. L. Luisi, J. Am. Chem. Soc., 1991, 113, 8127–8130 CrossRef CAS.
- R. Wick and P. L. Luisi, Chem. Biol., 1996, 3, 277–285 CrossRef CAS.
- C. Hentrich and J. W. Szostak, Langmuir, 2014, 30, 14916–14925 CrossRef CAS.
- J. Dervaux, V. Noireaux and A. Libchaber, Eur. Phys. J. Plus, 2017, 132, 1–10 CrossRef CAS.
- P. Van Nies, I. Westerlaken, D. Blanken, M. Salas, M. Mencía and C. Danelon, Nat. Comm., 2018, 9, 1–12 CrossRef CAS.
- A. Belluati, S. Thamboo, A. Najer, V. Maffeis, C. von Planta, I. Craciun, C. G. Palivan and W. Meier, Adv. Funct. Mater., 2020, 30, 2002949 CrossRef CAS.
- G. Bolognesi, M. S. Friddin, A. Salehi-Reyhani, N. E. Barlow, N. J. Brooks, O. Ces and Y. Elani, Nat. Comm., 2018, 9, 1–11 CrossRef CAS.
- T. D. Tang, D. Cecchi, G. Fracasso, D. Accardi, A. Coutable-Pennarun, S. S. Mansy, A. W. Perriman, J. R. Anderson and S. Mann, ACS Synth. Biol., 2018, 7, 339–346 CrossRef CAS.
- Q. Yang, Z. Guo, H. Liu, R. Peng, L. Xu, C. Bi, Y. He, Q. Liu and W. Tan, J. Am. Chem. Soc., 2020, 143, 232–240 CrossRef.
Footnote |
| † Present address: School of Information and Electronics, Beijing Institute of Technology, 100081, Beijing, China. |
| This journal is © The Royal Society of Chemistry 2022 |