Increase in germline methyltransferases governing the methylation of histone H3K9 is associated with transgenerational nanoplastic toxicity in Caenorhabditis elegans†
Le
Zhang
a,
Shuting
Wang
a,
Yue
Zhao
a,
Kun
Bi
b and
Dayong
Wang
 *ac
*ac
aKey Laboratory of Environmental Medicine Engineering of Ministry of Education, Medical School, Southeast University, Nanjing 210009, China. E-mail: dayongw@seu.edu.cn
bSchool of Biological Science and Medical Engineering, Southeast University, Nanjing 210096, China
cJiangsu Provincial Key Laboratory of Critical Care Medicine, Medical School, Southeast University, Nanjing 210009, China
First published on 25th November 2021
Abstract
In Caenorhabditis elegans, long-term exposure to 20 nm polystyrene nanoparticles (PS-NPs) caused transgenerational toxicity. However, the underlying mechanisms for induction of this transgenerational PS-NP toxicity remain largely unknown. We here determined the role of germline histone methyltransferases (HMTs) in controlling transgenerational PS-NP toxicity in nematodes. Among the germline genes encoding HMTs, PS-NP (1–100 μg L−1) exposure dysregulated met-2, ttll-12, set-5, set-6, set-12, and set-23 expressions. Among the germline genes encoding HMTs, only germline RNAi knockdown of met-2 and set-6, two genes required for methylation of H3K9, affected PS-NP toxicity at the P0 generation. Additionally, PS-NP exposure resulted in a transgenerational increase in the expressions of met-2 and set-6, and the transgenerational PS-NP toxicity was prevented in met-2(RNAi) and set-6(RNAi) nematodes. Therefore, an increase in the expression of MET-2 and SET-6, two HMTs required for methylation of H3K9, mediated the induction of transgenerational toxicity after PS-NP exposure. Moreover, in germline cells, MET-2 regulated the transgenerational PS-NP toxicity by increasing the expression and function of two downstream targets, PAT-12 and WRT-3, which provided an important basis for germline MET-2 in controlling transgenerational PS-NP toxicity in nematodes.
Environmental significanceNanoplastic particles are widely distributed in the environment, and potentially cause toxic effects on environmental animals. In Caenorhabditis elegans, the toxicity of polystyrene nanoparticles (PS-NPs) can not only be observed at the P0 generation but also be detected in offspring. Using this animal model, we further found that the transgenerational toxicity induced by PS-NPs in the range of μg L−1 was associated with the increase in both germline expressions of met-2 and set-6 encoding methyltransferases governing the methylation of histone H3K9 and methylation status of histone H3K9. Moreover, germline MET-2 controlled the transgenerational PS-NP toxicity by activating two downstream targets, PAT-12 and WRT-3, which provides the underlying mechanism for the function of germline MET-2. Our data highlight the value of the increase of germline HMTs required for methylation of H3K9 to detect the transgenerational toxicity of nanoplastics at low doses. |
Introduction
Nanoplastics with a size of 1–100 nm can be detected in different ecosystems, such as marine and aquatic ecosystems.1,2 Nanoplastic exposure contributes to global plastic pollution by causing hazards to environmental organisms.3 Polystyrene particles have the potential to be applied in adhesives, textiles, food containers, and packaging.4–6 Polystyrene nanoparticles (PS-NPs) are usually used as an example of nanoplastics to assess their possible toxic effects on organisms. The potential of PS-NPs to cross the gastrointestinal barrier so as to be accumulated in different organs has been frequently observed.7 Meanwhile, exposure to PS-NPs resulted in several aspects of toxicity to organisms, such as cellular toxicity, neurotoxicity, and immunotoxicity.8–10After exposure to different environmental pollutants, Caenorhabditis elegans exhibited a high sensitivity.11,12 In C. elegans, PS-NP exposure resulted in both neurotoxicity and reproductive toxicity.13,14 In addition, damage to intestinal development and metabolism was also observed in PS-NP exposed nematodes.15,16 Considering that C. elegans is a powerful model for the study of molecular toxicology,17 some important molecular signals, such as neurotransmission related signals, Gα signals, methylation related signals, and long non-coding RNAs, have been found to regulate PS-NP toxicity.18–21
Due to a very short life-cycle, C. elegans is also an important animal model for the study of transgenerational toxicity.11 More recently, PS-NP exposure could further lead to transgenerational toxicity to locomotion and reproduction in nematodes.22,23 This observed transgenerational PS-NP toxicity was size-dependent and under control of certain chemical modifications (such as amino modification).24,25 However, the molecular basis for the formation of transgenerational PS-NP toxicity is still largely unclear in organisms.
In C. elegans, methylation of certain forms of histone H3 (H3K36me, H3K27me, H3K9me, and H3K4me) is one important form of epigenetic regulation of gene expressions.26 This process can be controlled by some histone methyltransferases (HMTs).27 Among the identified genes encoding HMTs in nematodes,27 31 of them can be expressed in the germline (https://wormbase.org/). Our previous study has further demonstrated the function of germline met-2 encoding a HMT to control PS-NP toxicity at the P0 generation.18 The research aims here were to examine the response of germline HMTs to PS-NPs transgenerationally and to identify germline HMTs required for controlling transgenerational PS-NP toxicity in nematodes. Moreover, we focused on germline MET-2 to determine the underlying mechanism for its function in controlling transgenerational PS-NP toxicity. Our results demonstrated an important function of alteration in germline HMTs in mediating transgenerational PS-NP toxicity. Moreover, our finding also highlights the importance of germline MET-2-mediated signaling cascades in controlling transgenerational PS-NP toxicity.
Experimental
PS-NP properties
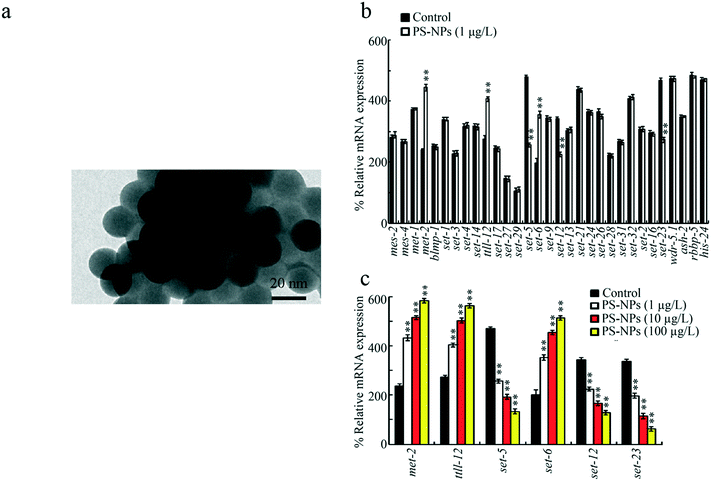
20 nm PS-NPs were from Janus New-Materials Co. (Nanjing, China). The transmission electron microscopy (TEM) image of PS-NPs is shown in Fig. 1a, which indicates their spherical morphology. The size of PS-NPs was confirmed as 20.45 ± 3.2 nm based on TEM analysis. The zeta potential of PS-NPs was −5.198 ± 0.897 mV based on a DLS assay. The other aspects of PS-NP properties (such as the Fourier transform infrared spectroscopy (FTIR) spectrum) were published previously.25C. elegans maintenance
Information on C. elegans strains is provided in Table S1.† Strain maintenance was performed as described.28To perform PS-NP exposure, an L1-larvae population was needed to be prepared. For this aim, enough gravid hermaphrodite nematodes were treated with a bleaching solution (0.45 M NaOH and 2% HOCl).29 After that, a large amount of eggs were released and transferred onto the surface of new NGM plates fed with Escherichia coli OP50, a food source for nematodes. These eggs were allowed to develop into synchronized L1-larvae.
Exposure for the assay of transgenerational PS-NP toxicity
To examine the transgenerational PS-NP toxicity, the exposure was performed basically as described in a previous study,25 that is, nematodes were exposed to suspensions of PS-NPs (1–100 μg L−1) for 6 days (from L1-larvae to adult day-3) at the parental generation (P0). We refreshed the PS-NP suspensions daily during the exposure process by transferring examined nematodes with a glass capillary into new PS-NP suspensions. In the PS-NP suspensions, OP50 was added to a concentration of ∼4 × 106 colony-forming units. Sonication was performed for the PS-NP suspensions before exposure, and the sonication conditions were 100 W and 40 kHz for 30 min. From the first filial generation (F1), the nematodes were allowed to develop on normal NGM plates with OP50.Assessment endpoints
Both inhibition in locomotion behavior and suppression in reproductive capacity could be observed in the progeny of PS-NP exposed nematodes.22,24 Locomotion behavior reflected by body bending and head thrashing and brood size were used as endpoints for the assay of transgenerational PS-NP toxicity.30Two forms of locomotion behavior were analyzed as described by Liu et al. (2021).31 Brood size was analyzed as described by Yang et al. (2021).32 After the exposure, the nematodes were firstly washed with M9 buffer three times. After a 1 min recovery on NGM plates without feeding with OP50, the locomotion behavior was examined. The frequency of body bending was counted as the change of direction for bending at the mid-body. The frequency of head thrashing was counted as the change of direction for the posterior bulb (y-axis), if we considered the direction of swimming of the nematodes as the x-axis. The brood size was counted as the total number of offspring beyond the eggs. For each exposure, 50 nematodes were examined for the assay of locomotion, and 30 nematodes were examined for the assay of brood size. Three replicates were performed for the assays of both locomotion behavior and brood size.
Quantitative real-time polymerase chain reaction (qRT-PCR)
The reagent TRIzol was used to extract the total RNA of adult nematodes. The obtained RNAs were assessed for their quality from ratio of OD260/280 using a Nanodrop One, and then used for cDNA synthesis on a Mastercycler gradient PCR system (Eppendorf, USA). In a SYBR Green qRT-PCR master mix, alteration in gene expressions was analyzed on a StepOnePlus real-time PCR system. The comparative cycle threshold method was used for quantification of examined genes after normalization with the expression of a reference gene (tba-1).33 Three replicates were performed. Table S2† shows the designed primers for qRT-PCR analysis.RNA interference (RNAi)
To inhibit the expression of gene(s), RNAi knockdown was performed by feeding L1-larvae with E. coli HT115 with the expression of dsRNA of certain gene(s).34 The progeny of nematodes on RNAi plates were used for the PS-NP exposure. Nematodes fed with HT115 expressing the empty vector L4440 was used as control for RNAi. RNAi efficiency was determined by qRT-PCR.Histone H3K9 methylation quantification
Histone H3K9 methylation was determined as described in a previous study.35 The adults (approximately 500 animals) at the P0, F1, F2, and F3 generations after exposure to PS-NPs (1 μg L−1) were used for extraction of total histone using an EpiQuick total histone kit (Farmingdale, USA). Antibodies against histone H3 with the corresponding modified lysine site were used to detect certain fractions of histone H3. The quantification was performed by reading the absorbance on a micro-plate spectrophotometer colorimetrically. Three replicates were carried out for each treatment.Data analysis
Using SPSS Statistics 19.0 Software, we performed statistical analysis. Data were presented as mean ± standard derivation (SD). The significance of differences between treatments was examined using one-way analysis of variance (ANOVA) or two-way ANOVA (for comparing multiple factors) followed by post hoc tests.Results
Effect of PS-NP exposure on the expressions of germline genes encoding HMTs
Among the 31 germline genes encoding HMTs, exposure to PS-NPs (1 μg L−1, a predicted environmental concentration)36 significantly increased the expressions of met-2, ttll-12, and set-6, and decreased the expressions of set-5, set-12, and set-23 (Fig. 1b). Moreover, in 1–100 μg L−1 PS-NP exposed nematodes, the increase in met-2, ttll-12, and set-6 expressions and the decrease in set-5, set-12, and set-23 expressions were concentration dependent (Fig. 1c).Requirement of germline MET-2 and SET-6 for the control of PS-NP toxicity at the P0 generation
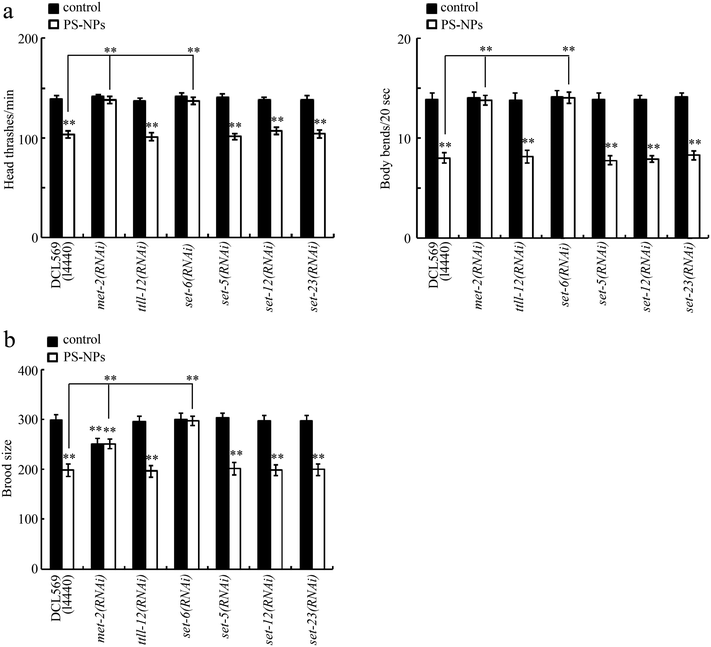
Both locomotion behavior and brood size were selected as endpoints. Using the DCL569 strain, we found that the germline RNAi knockdown of ttll-12, set-5, set-12, and set-23 did not affect the toxicity of PS-NPs at the P0 generation (Fig. 2a and b). However, at the P0 generation, the germline RNAi knockdown of met-2 or set-6 suppressed the PS-NP toxicity (Fig. 2a and b). Fig. S1† shows the efficiency of the germline RNAi knockdown of met-2, ttll-12, set-6, set-5, set-12, and set-23.Transgenerational expressions of germline MET-2 and SET-6 after PS-NP exposure
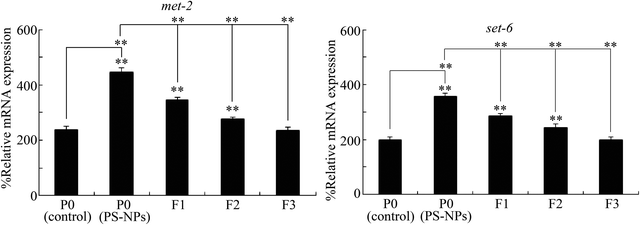
To determine whether germline MET-2 and SET-6 were involved in the control of transgenerational PS-NP toxicity, we investigated the transgenerational expressions of germline MET-2 and SET-6. After exposure to PS-NPs (1 μg L−1), an increase in the MET-2 and SET-6 expressions was further detected in the F1 and F2 generations (Fig. 3). At the F3 generation, the expressions of MET-2 and SET-6 were recovered to the control level (Fig. 3), which suggested the dynamic transgenerational expressions of MET-2 and SET-6.Effect of germline RNAi knockdown of met-2 or set-6 on transgenerational PS-NP toxicity
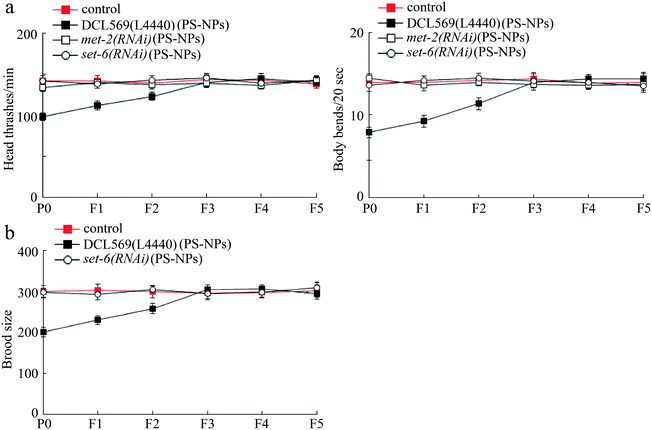
After exposure to PS-NPs (1 μg L−1), germline RNAi knockdown of met-2 or set-6 inhibited the transgenerational PS-NP toxicity that decreases locomotion behavior (Fig. 4a). Additionally, germline RNAi knockdown of set-6 also suppressed the transgenerational PS-NP toxicity that reduces brood size (Fig. 4b). Considering that the met-2(RNAi) nematodes showed a severe deficit in brood size, we did not further analyze the brood size phenotype in PS-NP exposed met-2(RNAi) nematodes. Therefore, germline MET-2 and SET-6 were required for the induction of transgenerational PS-NP toxicity.Identification of PAT-12 and WRT-3 as potential targets of germline MET-2 in regulating transgenerational PS-NP toxicity
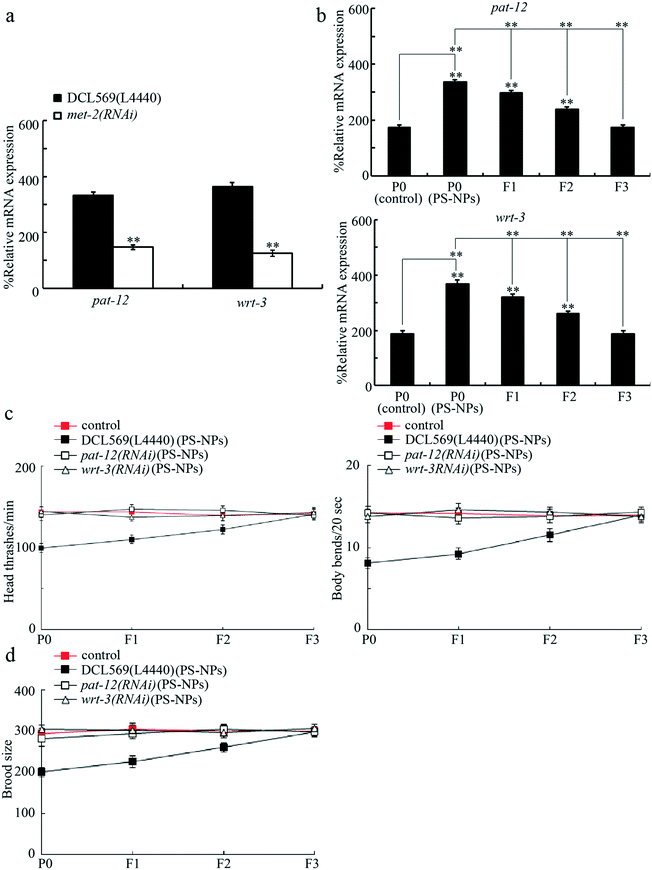
Our previous study has indicated that germline MET-2 acted upstream of PAT-12, a component of hemidesmosomes, and WRT-3, a homolog of PPIL-2, to regulate the response to 100 nm PS-NPs.18 Next, we focused on MET-2 to determine the underlying mechanism for its role in regulating transgenerational PS-NP toxicity. In PS-NP (20 nm) exposed DCL569 nematodes, the germline RNAi knockdown of met-2 decreased the pat-12 and wrt-3 expressions (Fig. 5a). Meanwhile, exposure to PS-NPs (1 μg L−1) increased the pat-12 and wrt-3 expressions, and the increase in the pat-12 and wrt-3 expressions was further observed in the F2 and F3 generations (Fig. 5b). Moreover, the germline RNAi knockdown of pat-12 or wrt-3 obviously inhibited transgenerational PS-NP toxicity that decreases locomotion behavior and reduces brood size (Fig. 5c and d). Therefore, during the control of transgenerational PS-NP toxicity, PAT-12 and WRT-3 functioned as the potential targets of MET-2 in the germline. Fig. S2† shows the efficiency of the germline RNAi knockdown of pat-12 and wrt-3.Genetic interaction between germline MET-2 and PAT-12 or WRT-3 in regulating transgenerational PS-NP toxicity
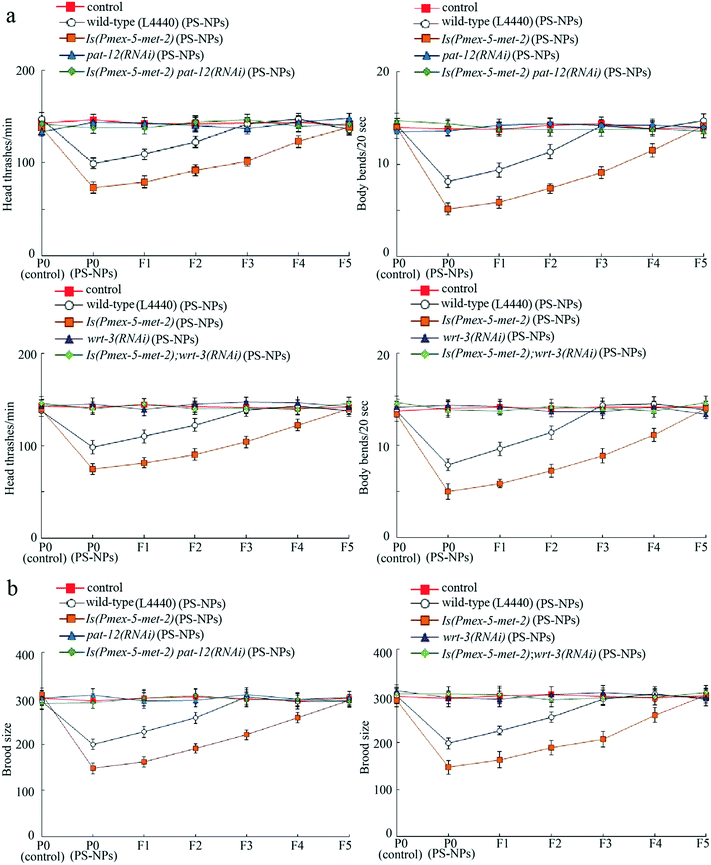
To further confirm the role of PAT-12 and WRT-3 as targets of germline MET-2 in controlling transgenerational PS-NP toxicity, we examined the genetic interaction between germline MET-2 and PAT-12 or WRT-3. In the strain Is(Pmex-5-met-2) overexpressing germline MET-2,18 susceptibility to transgenerational PS-NP toxicity that decreases locomotion behavior and reduces brood size was detected (Fig. 6a and b). Moreover, RNAi knockdown of pat-12 or wrt-3 obviously suppressed this susceptibility to transgenerational PS-NP toxicity in Is(Pmex-5-met-2) nematodes (Fig. 6a and b). Therefore, germline MET-2 regulated the transgenerational PS-NP toxicity by activating downstream PAT-12 and WRT-3. Fig. S3† shows the RNAi knockdown efficiency of pat-12 and wrt-3 in wild-type nematodes.Transgenerational alteration in the methylation status of histone H3K9 after PS-NP exposure
In C. elegans, both MET-2 and SET-6 are HMTs required for methylation of histone H3K9.37,38 We also examined the changes in the methylation status of histone H3K9 transgenerationally after exposure to PS-NPs (1 μg L−1). At the P0 generation, PS-NP exposure increased the level of global methylation in histone H3K9 (Fig. S4†). The increase in global methylation in histone H3K9 was also detected in the F1 and F2 generations, which recovered at the F3 generation of PS-NP exposed nematodes (Fig. S4†).Discussion
In this study, we tried to identify germline HMTs required for the control of transgenerational toxicity of 20 nm PS-NPs in nematodes. Our previous study has indicated the potential of exposure to 20 nm PS-NPs to cause severe transgenerational toxicity,25 which was the reason for the selection of 20 nm PS-NPs. Our data here demonstrated the important function of the germline HMTs MET-2 and SET-6 required for methylation of histone H3K9 in controlling transgenerational PS-NP toxicity.Among germline HMT genes, we first found that exposure to PS-NPs in the range of μg L−1 dysregulated the expressions of 6 HMT genes (Fig. 1b and c). Nevertheless, among these 6 candidate HMT genes, only the germline RNAi knockdown of met-2 and set-6 affected PS-NP toxicity at the P0 generation (Fig. 2). This implied that the germline HMT genes ttll-2, set-5, set-12, and set-23 may be involved in other aspects of biological processes, although their response to PS-NP exposure could be observed in nematodes. So far, the exact biological functions of TTLL-2, SET-5, SET-12, and SET-23 are still largely unclear.
The nematodes with germline RNAi knockdown of met-2 or set-6 exhibited a resistance to 20 nm PS-NP toxicity at the P0 generation (Fig. 2). Meanwhile, exposure to 20 nm PS-NPs in the range of μg L−1 increased the met-2 and set-6 expressions (Fig. 1b and c). These observations demonstrated that alterations in the germline MET-2 and SET-6 expressions mediated the toxicity induction of 20 nm PS-NPs, which is different from the observation in 100 nm PS-NP exposed nematodes. In nematodes exposed to 100 nm PS-NPs in the range of μg L−1, the germline met-2 expression was decreased,18 which reflected a protective response to PS-NPs. Exposure to 1–100 nm PS-NPs for 72 h also reduced met-2 expression.23 These findings further support the size-dependent property of PS-NP toxicity in organisms.
In nematodes, MET-2 and SET-6 have the biological function of methylation of the histone H3K9 residue.37,38 Our data further suggested the important role of the increase in MET-2 and SET-6 expressions in causing the formation of transgenerational PS-NPs toxicity. Firstly, the germline RNAi knockdown of met-2 or set-6 could also suppress the transgenerational PS-NP toxicity (Fig. 4). The transgenerational reproductive toxicity of PS-NPs was also inhibited by RNAi knockdown of met-2 in wild-type nematodes.23 Secondly, the increase in met-2 and set-6 expressions caused by PS-NP exposure could be further detected in the offspring, such as the F1 and F2 generations (Fig. 3). Moreover, we also observed the increase in the methylation status of histone H3K9 at both the P0 generation and the offspring, such as the F1 and F2 generations (Fig. S4†). These observations also suggested the sensitivity of the methylation status of histone H3K9 and related molecular signals (such as MET-2 and SET-6) to PS-NPs in the range of μg L−1.
Previous studies have also suggested the involvement of MET-2 and SET-6 in controlling stress response.35,37–40 The expression of MET-2 was also dysregulated by exposure to 1-ethyl-3-methylimidazolium bromide, CuO nanoparticles, and crude oil.35,39,40 Transgenerational deficit in reproduction caused by crude oil exposure was inhibited by met-2 mutation.35 Neuronal SET-6 inhibited behavioral deterioration during aging by accelerating the expression of mitochondrial genes and mitochondrial functions.38 Moreover, the induction of mitochondrial unfolded protein response required the activity of MET-2.37 Nevertheless, it is still unclear whether germline MET-2 and SET-6 control transgenerational PS-NP toxicity by affecting certain downstream mitochondrial signals.
Methylation of H3K9 is usually considered to be associated with transcriptional repression of gene expressions.41 However, we further found that the germline MET-2 controlled the transgenerational PS-NP toxicity by activating the downstream targets PAT-12 and WRT-3. An increase in the expression of germline pat-12 and wrt-3 was detected in both P0 and the offspring, such as the F1 and F2 generations (Fig. 5b). More importantly, the susceptibility of nematodes overexpressing germline MET-2 to transgenerational PS-NP toxicity was inhibited by RNAi knockdown of pat-12 and wrt-3 (Fig. 6). These findings provide the underlying important basis for the function of germline MET-2 in controlling transgenerational PS-NP toxicity. Previous studies have suggested the important function of germline signaling cascades of MET-2–WRT-3 and MET-2–PAT-12 in regulating PS-NP toxicity at the P0 generation. Thus, germline signaling cascades of MET-2–WRT-3 and MET-2–PAT-12 are required for control of both PS-NP toxicity at the P0 generation and transgenerational PS-NP toxicity. Further elucidation of the mechanisms for germline PAT-12 and WRT-3 functions will be helpful for understanding the molecular basis of germline HMTs required for methylation of histone H3K9 in controlling transgenerational PS-NP toxicity. In C. elegans, WRT-3 is a hedgehog (Hh) ligand. The potential transgenerational interaction between WRT-3 and its Hh receptors will provide important clues for elucidating the mechanism of WRT-3 function in controlling transgenerational PS-NP toxicity.
Conclusions
Together, we determined the association of alterations in germline HMTs with the induction of transgenerational PS-NP toxicity. In nematodes, PS-NPs in the range of μg L−1 induced an increase in both the expression of met-2 and set-6 and methylation status of histone H3K9 transgenerationally. Germline RNAi knockdown of met-2 or set-6 encoding two HMTs required for methylation of histone H3K9 effectively inhibited the induction of transgenerational PS-NP toxicity. Moreover, in the germline, HMT/MET-2 regulated transgenerational PS-NP toxicity by activating two downstream targets, PAT-12 and WRT-3. Our data highlight the association of the increase in germline HMTs required for methylation of H3K9 with the induction of transgenerational nanoplastic toxicity in organisms.Ethical statement
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Southeast University and approved by the Animal Ethics Committee of Southeast University.Conflicts of interest
There are no conflicts to declare.References
- J. Wan, Q. Chu, Y. Kok and C. Lee, Rev. Environ. Contam. Toxicol., 2018 DOI:10.1007/398_2018_14.
- C. G. Alimba and C. Faggio, Environ. Toxicol. Pharmacol., 2019, 68, 61–74 CrossRef PubMed.
- D. M. Mitrano, P. Wick and B. Nowack, Nat. Nanotechnol., 2021, 16, 491–500 CrossRef PubMed.
- M. A. Abdallah, M. Sharkey, H. Berresheim and S. Harrad, Chemosphere, 2018, 199, 612–616 CrossRef PubMed.
- H. Gelbke, M. Banton, C. Block, G. Dawkins, R. Eisert, E. Leibold, M. Pemberton, I. M. Puijk, A. Sakoda and A. Yasukawa, Food Chem. Toxicol., 2019, 124, 151–167 CrossRef PubMed.
- P. Sen, Y. Xiong, Q. Zhang, S. Park, W. You, H. Ade, M. W. Kudenov and B. T. O'Connor, ACS Appl. Mater. Interfaces, 2018, 10, 31560–31567 CrossRef PubMed.
- L. Peng, D. Fu, H. Qi, C. Q. Lan, H. Yu and C. Ge, Sci. Total Environ., 2020, 698, 134254 CrossRef PubMed.
- M. Prust, J. Meijer and R. H. S. Westerink, Part. Fibre Toxicol., 2020, 17, 24 CrossRef PubMed.
- N. Hirt and M. Body-Malapel, Part. Fibre Toxicol., 2020, 17, 57 CrossRef PubMed.
- A. Banerjee and W. L. Shelver, Sci. Total Environ., 2021, 755, 142518 CrossRef PubMed.
- D.-Y. Wang, Exposure Toxicology in Caenorhabditis elegans, Springer Nature Singapore Pte Ltd., 2020 Search PubMed.
- Y. H. Yang, Q.-L. Wu and D.-Y. Wang, Environ. Sci.: Nano, 2021, 8, 1019–1028 RSC.
- M. Qu, Y. Kong, Y.-J. Yuan and D.-Y. Wang, Environ. Sci.: Nano, 2019, 6, 2591–2601 RSC.
- M. Qu, Y.-X. Qiu, Y. Kong and D.-Y. Wang, Environ. Pollut., 2019, 254, 112978 CrossRef CAS PubMed.
- L. Lei, S. Wu, S. Lu, M. Liu, Y. Song, Z. Fu, H. Shi, K. M. Raley-Susman and D. He, Sci. Total Environ., 2018, 619–620, 1–8 CrossRef CAS PubMed.
- H. M. Kim, D. K. Lee, N. P. Long, S. W. Kwon and J. H. Park, Environ. Pollut., 2019, 246, 578–586 CrossRef CAS.
- D.-Y. Wang, Molecular Toxicology in Caenorhabditis elegans, Springer Nature Singapore Pte Ltd., 2019 Search PubMed.
- S.-T. Wang, R.-J. Zhang and D.-Y. Wang, Chemosphere, 2021, 271, 129589 CrossRef CAS.
- S.-T. Wang, H.-L. Liu, M. Qu and D.-Y. Wang, Ecotoxicol. Environ. Saf., 2021, 217, 112239 CrossRef PubMed.
- Y.-Y. Zhao, R.-R. Xu, X. Chen, J. Wang, Q. Rui and D.-Y. Wang, Ecotoxicol. Environ. Saf., 2021, 212, 111976 CrossRef PubMed.
- Y.-H. Yang, Q.-L. Wu and D.-Y. Wang, Ecotoxicol. Environ. Saf., 2021, 225, 112732 CrossRef PubMed.
- L. Zhao, M. Qu, G. Wong and D.-Y. Wang, Environ. Sci.: Nano, 2017, 4, 2356–2366 RSC.
- C. W. Yu, T. C. Luk and V. H. Liao, J. Hazard. Mater., 2021, 412, 125173 CrossRef PubMed.
- L.-M. Sun, K. Liao and D.-Y. Wang, Sci. Total Environ., 2021, 768, 144362 CrossRef PubMed.
- H.-L. Liu, L.-J. Tian, S.-T. Wang and D.-Y. Wang, Sci. Total Environ., 2021, 790, 148217 CrossRef PubMed.
- W. G. Kelly, Epigenet. Chromatin, 2014, 7, 6 CrossRef PubMed.
- D. Wenzel, F. Palladino and M. Jedrusik-Bode, Genesis, 2011, 49, 647–661 CrossRef CAS.
- S. Brenner, Genetics, 1974, 77, 71–94 CrossRef CAS PubMed.
- H.-L. Liu, L.-J. Tian, M. Qu and D.-Y. Wang, J. Hazard. Mater., 2021, 411, 125035 CrossRef CAS PubMed.
- Y.-H. Yang, W.-T. Dong, Q.-L. Wu and D.-Y. Wang, Chem. Res. Toxicol., 2021, 34, 1308–1318 Search PubMed.
- H.-L. Liu, Y.-Y. Zhao, K. Bi, Q. Rui and D.-Y. Wang, Ecotoxicol. Environ. Saf., 2021, 212, 112018 CrossRef CAS PubMed.
- Y.-H. Yang, W.-T. Dong, Q.-L. Wu and D.-Y. Wang, Nanoscale Adv., 2021, 3, 1997–2006 RSC.
- H.-L. Liu, Y.-X. Qiu and D.-Y. Wang, Chemosphere, 2021, 273, 129686 CrossRef CAS PubMed.
- L.-M. Sun, D. Li, Y.-J. Yuan and D.-Y. Wang, Sci. Rep., 2021, 11, 1997 CrossRef CAS.
- J. Yang, N. Chatterjee, Y. Kim, J. Y. Roh, J. H. Kwon, M. S. Park and J. Choi, Chemosphere, 2018, 200, 358–365 CrossRef CAS.
- R. Lenz, K. Enders and T. G. Nielsen, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E4121–E4122 CrossRef CAS.
- Y. Tian, G. Garcia, Q. Bian, K. K. Steffen, L. Joe, S. Wolff, B. J. Meyer and A. Dillin, Cell, 2016, 165, 1197–1208 CrossRef CAS.
- J. Yuan, S. Chang, S. Yin, Z. Liu, X. Cheng, X. Liu, Q. Jiang, G. Gao, S. Lin, X. Kang, S. Ye, Z. Chen, J. Yin, P. Hao, L. Jiang and S. Cai, Nature, 2020, 579, 118–122 CrossRef CAS.
- C. Wei, P. Yen, A. Chaikritsadakarn, C. Huang, C. Chang and V. H. Liao, Ecotoxicol. Environ. Saf., 2020, 203, 111001 CrossRef CAS PubMed.
- W. Yue, L. Mo and J. Zhang, Sci. Total Environ., 2021, 765, 144334 CrossRef CAS PubMed.
- S. A. Bassett and M. P. Barnett, Nutrients, 2014, 6, 4273–4301 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1en00835h |
| This journal is © The Royal Society of Chemistry 2022 |