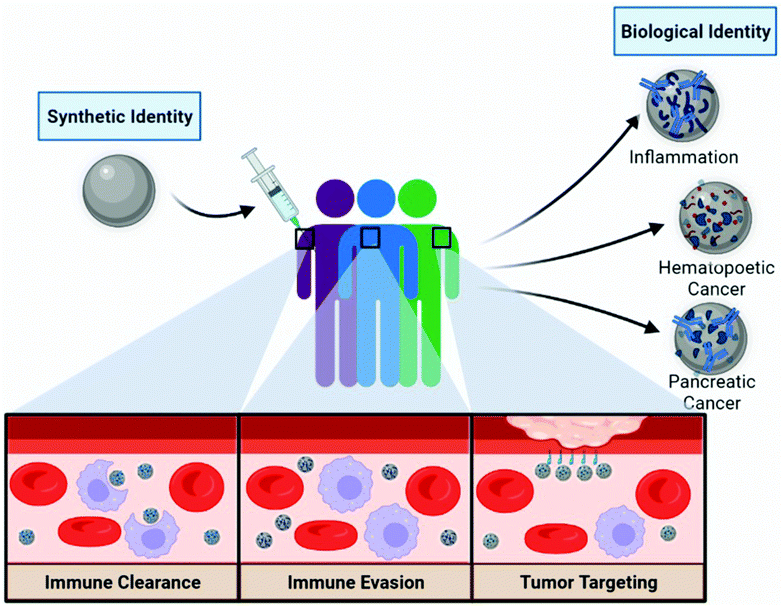
Nanoparticle personalized biomolecular corona: implications of pre-existing conditions for immunomodulation and cancer
Jacob
Shaw
 a and
Ryan M.
Pearson
a and
Ryan M.
Pearson
 *abc
*abc
aDepartment of Microbiology and Immunology, University of Maryland School of Medicine, 685 W. Baltimore Street, Baltimore, MD 21201, USA. E-mail: rpearson@rx.umaryland.edu
bDepartment of Pharmaceutical Sciences, University of Maryland School of Pharmacy, 20 N. Pine Street, Baltimore, MD 21201, USA
cMarlene and Stewart Greenebaum Comprehensive Cancer Center, University of Maryland School of Medicine, 22 S. Greene Street, Baltimore, MD 21201, USA
First published on 21st April 2022
Abstract
Nanoparticles (NPs) have demonstrated great promise as immunotherapies for applications ranging from cancer, autoimmunity, and infectious disease. Upon encountering biological fluids, NPs rapidly adsorb biomolecules, forming the “biomolecular corona” (BC), and the altered character of NPs due to their newly acquired biological identity can impact their in vivo fate. Recently, it has been shown that the NP–BC is person-specific, and even minute differences in the biomolecule composition can give rise to altered immune recognition, cellular interactions, pharmacokinetics, and biodistribution. Given the current rise in the development of NP-based therapeutics, it is of utmost importance to better understand how pre-existing conditions, that result in the formation of a personalized BC, can be leveraged to aid in the prediction of the therapeutic outcomes of NPs. In this minireview, we will discuss the formation of the BC, implications of the BC for NP-biological interactions, and its clinical importance in the context of immunomodulation and cancer therapeutics.
1. Introduction
Nanotechnology has cemented itself in the field of medicine as a robust platform for improving diagnostic and therapeutic systems. In the past decade, sixteen nanoparticle (NP) formulations have gained Food and Drug Administration (FDA) approval and over 1000 new clinical trials have begun.1,2 Among the FDA approved nanoformulations, eight are clinically used for cancer imaging or therapy. Despite the extensive literature on characterizing NP formulations, several biological barriers exist that collectively pose a challenge to their clinical translation.3,4 A meta-analysis showed a potential reason for the lack of success in the field of cancer nanomedicine was that an estimated 0.7% of injected NPs reach solid tumor tissue, whereas the vast majority accumulated in non-tumor sites including the liver and spleen.5 One aspect that has been identified as a confounding variable is the interaction between synthetic materials and biological media. When NPs are exposed to biological fluids, such as plasma, opsonins and other biomolecules rapidly adsorb to the surface.6–8 This coating, mainly consisting of proteins, lesser amounts of lipids, and other biomolecules, is referred to as the “biomolecular corona” (BC). The formation of the BC represents a transformation of the NP by altering its “synthetic identity” away from design features such as composition, charge, topology, and surface functionalization as it acquires a new “biological identity”.7 Accordingly, NP biological identity has been linked to a number of deleterious biological effects including altered immune cell activation, increased blood clearance, and altered tumor biodistribution.9 The adsorption of opsonins (such as immunoglobulins, complement factors, and lectins) to NPs has been well understood to promote phagocytic clearance, but the effects of additional endogenous biomolecule adsorption on nano-bio interactions is a promising new field of research.Recently, it has been shown that the BC fingerprint is not only unique to specific NP formulations, but also the individuals’ disease-state and plasma composition can give rise to alterations in BCs independent of the NP's synthetic identify. The term “personalized protein corona” or “personalized biomolecular corona” (PBC) has been developed to account for these disease state-dependent effects of BC formation on NPs (Fig. 1).10 Different disease-states have significant effects on the concentration and composition of biomolecules in the bloodstream. Furthermore, genetic background, lifestyle, and geographical origin play important roles in PBC changes between healthy individuals. These unique differences between individuals can directly affect the BC composition and subsequent NP biological fate. Given the recent advances in characterizing the PBC, the notion that synthetically identical nanomaterials would elicit similar effects in all patient populations should be reconsidered. Understanding the PBC's impact on NP-driven therapeutic outcomes is imperative to the development and clinical translation of personalized medicines. This minireview will highlight recent examples of the implications of the BC on NP cancer therapeutics, including commonly identified biomolecules in the corona and their receptors, BC-dependent immune cell activation, recent advances in understanding disease-specific BC innate immune and cancer cell targeting, and its clinical relevance in cancer nanomedicine.
2. Personalized biomolecular corona
An often-overlooked factor when it comes to discussions about BC formation is patient plasma composition and the respective concentration of biomolecules. Biomolecule binding to nanomaterials is determined by a variety of factors including, NP size, hydrophobicity, charge, topology, shape, and the biological media composition it is exposed to. Historically, BC formation is understood to be driven by the ability of the NP's physicochemical properties to draw in molecules through noncovalent forces such as van der Waals, hydrophobic interactions, and hydrogen bonding.11 These interactions leading to BC formation are ubiquitous among NPs formulations, although some groups have developed coatings such as hydrophilic and zwitterionic coatings to diminish this phenomena.12,13 Conjugation of poly(ethylene glycol) (PEG) chains on the surface of NPs has long been considered an efficient strategy to limit corona formation and promote immune evasion, but recent proteomic profiling suggests that while PEGylation diminishes total protein abundance, it has only been shown to have a minor effect on altering total BC composition.14 Furthermore, anti-PEG antibodies have been shown to opsonize NPs to further promote phagocytic clearance. These findings are contrary to the so-called “stealth effect” where PEGylation was thought to impart improved NP pharmacokinetic properties leading researchers to develop PEG alternatives and anti-PEG antibody accommodations.15,16 Given this, there is a need to characterize the BC influence in both pristine and sterically-coated NP formulations. In addition to NP physicochemical properties, several parameters have a significant influence on the composition of the BC, including biomolecule binding affinities, relative concentration, and exposure time.17 According to the Vroman effect, the most abundant biomolecules in the media are readily adsorbed to the surface at the early stages of BC formation, but they are dynamically replaced by higher affinity species as time proceeds.18 Thus, disease-specific alterations in plasma biomolecule compositions and concentrations influence BC formation and subsequent nano-bio interactions.Extensive research has gone into understanding the impact of NP modifications on BC formation, but the role of blood composition as a function of interindividual and disease state variations has only recently been evaluated. Hajipour et al. documented differences in the protein corona fingerprint of polystyrene NPs incubated with plasmas from varying individuals, diseases, and disorders.10 Whole protein analysis using SDS-PAGE identified patient-specific BC compositions; these differences were elevated in the cases of hematological conditions (such as hypercholesterolemia, hemophilia, hyperfibrinogenemia). Interestingly, differences in protein fingerprints were observed between healthy individuals as well. Furthermore, BC composition may be dependent on the type, period, and severity of the disease. Table 1 lists commonly adsorbed BC molecules, their prospective receptor interactions, impact on NP therapeutics, and examples comorbidities associated with respective biomolecule upregulation in the blood. Given the BC composition has been implicated in NP biological fate, it is essential to understand the role of the BC on NP-based therapeutics.
| Protein | Biological function | Receptor targeting | Impact on nanoparticles | Associated comorbidities | Ref. |
|---|---|---|---|---|---|
| Alpha-2-macroglobulin | Humoral defense by binding foreign peptides and particles | Alpha2-macroglobulin receptor and low-density lipoprotein receptor-related protein | Immune activation, MPS clearance | Diabetes, nephrosis, infection | 55, 61 and 62 |
| Anti-thrombin | Inhibits thrombin and regulates the blood coagulation cascade | Heparin receptors and glycosaminoglycans | MPS clearance | Diabetes | 54 and 63 |
| Apolipoprotein A-I, A-II, A-IV, A-V, B-100, C-I, C-II, C-III, C-IV, D, E, L1 | Lipid transport and metabolism | Low density lipoprotein receptors (LDLR), low-density lipoprotein-associated receptors | Immune evasion, liver sequestration, blood–brain barrier penetration | Cardiovascular disease, various cancers | 52 and 60 |
| Complement, C1, C3, C4, C8 | Complement cascade for innate immune surveillance | Complement receptors | Immune activation, MPS clearance | Infection, various cancers | 56 and 67 |
| Fibrinogen | Cleaved into fibrin to polymerize and stimulate blood clotting | Glycoprotein (gp) IIb/IIIa integrin, MAC-1 | Immune activation, MPS clearance, coagulation induction | Infection, inflammation | 53 and 64 |
| Fibronectin | Extracellular matrix glycoprotein that binds integrins | Integrins | MPS clearance | Cardiovascular disease, various cancers | 50 and 63–66 |
| Hemoglobin | Oxygen transport | CD163 | Immune evasion | Cardiovascular disease, pulmonary fibrosis | 57 and 68 |
| Immunoglobulin | Humoral immunity elimination of foreign pathogens | Fc receptors and complement binding | Immune activation, MPS clearance | Infection, inflammation, various cancers | 59 and 69 |
| Prothrombin | Component of the blood coagulation cascade | Protease-activated receptor 1 | MPS clearance, coagulation induction | Cardiovascular disease, liver cirrhosis | 51, 64 and 70 |
| Serine protease | Moieties in the lectin pathway of complement | Protease-activated receptor 2 | MPS clearance | Pulmonary fibrosis | 31 and 71 |
| Serum albumin | Most abundant component of plasma necessary for homeostasis | Gp18, gp30, p60, SPARC | Immune evasion, vasculature targeting | Inflammation, malnutrition | 45 and 72 |
| Vitronectin | Cell adhesion and spreading factor | ανβ3 integrin | Immune activation, MPS clearance | Cardiovascular disease, various cancers | 42, 73 and 74 |
| Von Willebrand factor | Component of the blood coagulation cascade | GpIbα and gpIIb/IIIa integrin | Immune activation, MPS clearance, coagulation induction | Cardiovascular disease, anemia | 58, 64 and 75–77 |
3. Biomolecular corona-mediated immune activation
The adsorption of proteins on the NP surface can induce alterations in the structures of adsorbed proteins, which can affect immune responses. Apart from the nanotoxicological implications associated with opsonization and complement-mediated MPS clearance, NPs may induce additional adverse effects by inducing conformational changes, unfolding, and fibrillation (the process of misfolded proteins forming large linear aggregates) of the adsorbed proteins. The misfolding or denaturation of the protein tertiary structure may lead to exposure of new potentially immunogenic, cryptic epitopes that can cause self-protein immunogenicity and subsequent autoimmune reactions. The phenomenon of plasma proteins unfolding upon interaction with NP surfaces has been well reported. Generally, it has been observed that NPs with high hydrophobicity or surface charge density can more readily denature protein conformations. It is important to understand not just the BC fingerprint, but also the conformational statuses of the proteins themselves in order to fully understand how the BC affects NP-immune recognition.Designing NP physicochemical properties to modulate BC denaturation can allow for the fine tuning of immune phenotypes. Recently, Deng et al. showed that fibrinogen bound to poly(acrylic acid)-conjugated gold NPs undergoes denaturation, activates the integrin receptor Mac-1, and stimulates downstream NF-κB expression of inflammatory cytokines IL-8 and TNFα.19 Fibrinogen binding was observed to be dependent on NP surface charge, indicating an avenue for modulating this immune response. In another study, Park et al. characterized the protein structure and immune responses of PEGylated carbon nanotubes pre-coated with common plasma proteins.20 Interestingly, they observed an elevation in reactive oxygen species levels and proinflammatory cytokine release in IgG and α-1-acid glycoprotein (AGP) coronas, but not for fibronectin or vitronectin in human macrophage cell lines. Structural analysis of these coronas indicated denaturation in the IgG and AGP proteins, which were associated with proinflammatory phenotypes. Additionally, in vivo intravenous administration confirmed immune stimulation through the elevation of splenic neutrophils, natural killer (NK) cells, and CD8+ T cells in the nanotube protein corona treatment groups compared to soluble protein controls. It was hypothesized that modulating this immune stimulation could be favorable in the treatment of solid tumors because of the increased infiltration of NK cells and CD8+ T cells associated with the innate and adaptive immune clearance of tumor tissue, respectively. Thus, designing NPs to leverage plasma protein alterations could be used to promote immune cell infiltration into the tumor microenvironment. Although there is a deficit in the literature describing the impact of the PBC on immune regulation, this is a promising opportunity to evaluate if altered serum protein abundances can be used to modulate this phenomenon.
4. Leveraging innate immune cell interactions
A major hurdle that alters nanomedicine effectiveness is MPS clearance. Intravenously injected NPs are generally recognized as foreign materials and processed as such. This manifests as the adsorption of opsonins, activation of the complement system, and subsequent macrophage clearance, resulting in undesirable nanotoxicity. Interestingly, Tavares et al. evaluated the effect of depleting liver macrophages (Kupffer cells) on NP delivery efficiency to the tumor by avoiding sequestration, but this strategy was unable to increase delivery efficiency above 2%, providing evidence that other factors were major contributors.21 Depending on patient co-morbidities and cancer pathology, elevations in plasma opsonin levels can influence BC-dependent immune cell targeting.22 Many researchers have investigated the role of physicochemical modifications to reduce NP opsonization to achieve immune evasion; however, selectively targeting immune cells by leveraging the PBC is an attractive opportunity for immunomodulation. Administration of NPs in an opsonin-rich environment may allow for the specific targeting of phagocytic immune cells and delivery of immunomodulatory therapies. Ultimately, a better understanding of the PBC and its composition is necessary for researchers to achieve specific targeting or evasion of immune cells. Within this section, we will highlight examples of BC components and their impact on innate immune cell interactions and immunomodulation.(a) Classical complement
Complement proteins have long been identified as elevated markers that play a dual role in cancer immunosurveillance and enhanced tumor proliferation.23 Recently, Ren et al. demonstrated that gadolinium metallofullerenol NPs, previously shown to promote tumor clearance through inducing a proinflammatory phenotype of M1 macrophages and Th1 T cells, bound to systemically upregulated complement 1q (C1q) proteins in lung cancer patient plasma. The classical complement pathway is activated when the complement protein complex C1q binds to IgG molecules that have recognized and bound to a foreign surface leading to enhanced MPS clearance. Understandably, elevated C1q corona content was associated with increased macrophage targeting, phagocytosis, and a subsequent increase in proinflammatory cytokines TNFα and IL-1β (Fig. 2).24 Further, the enhancement in cytokine secretion was higher for C1q-coated compared to C1q alone. A follow-up investigation examined the influence of healthy patient PBC on immune cell uptake of PEGylated liposomal doxorubicin (Doxil/Caelyx), the first FDA approved liposomal nanomedicine for various solid tumors. Twenty-three healthy patient-derived PBCs were evaluated and identified to have unique uptake profiles by both macrophages and B cells.25 Given these results, leveraging elevated complement factors in relevant cancer patient plasmas, in addition to appropriate NP choice, can not only allow for macrophage targeting immunotherapies but also has implications in B cell targeting for influencing immune cell phenotypes or the delivery of vaccines.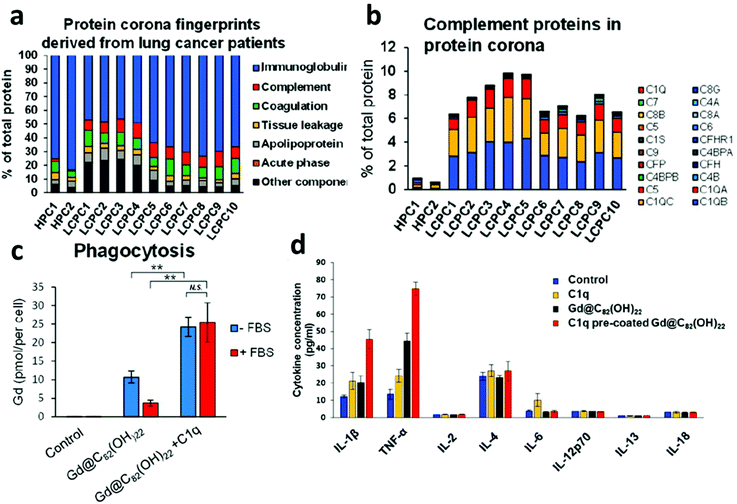 | ||
| Fig. 2 Personalized complement factor corona in lung cancer patients. (a) Classification of the personalized protein corona of healthy human patient corona (HPC), lung cancer patient corona (LCPC). (b) Relative abundance of complement factors are specific to lung cancer coronas. (c) NP uptake by human macrophages is elevated in C1q precoated samples. (d) Cytokine expression modulation with C1q precoated NPs. Adapted with permission from ref. 24. Copyright 2019 American Chemical Society. | ||
(b) Immunoglobulin
Among opsonins, immunoglobulins play an important role in foreign material clearance through phagocyte targeting of Fc receptors. In the process of characterizing the PBC of Doxil/Caelyx in murine lung and melanoma cancers, Hadjidemetriou et al. observed an elevation in BC-associated pathogen clearance proteins, immunoglobulin fragments, and mucin.26 Similarly, Colapicchioni et al. examined the protein corona fingerprint of the NP formulation Ambisome, in breast, gastric and pancreatic cancers by SDS-PAGE.27 Ambisome consists of liposome encapsulated antifungal amphotericin B, clinically administered prophylactically before chemotherapy to prevent infections. An increased abundance of 37 kDa proteins associated with immunoglobulin alpha (IgA) and immunoglobulin gamma (IgG) heavy chains were identified in pancreatic cancers patients but not others. It is thought that IgG binding to phosphatidylcholine/phosphatidylglycerol-based liposomes, like Ambisome, is primarily controlled by non-specific adsorption. It has been observed that many different cancer types induce an elevated production of IgG and IgA autoantibodies as immune-defenses against tumors.28 Elevated concentrations of IgG in cancer patients have been linked to a proportional elevation in BC-bound IgG.29 These findings are of particular interest because elevated adsorption of antibodies is correlated with complement system clearance of NPs, reduced systemic circulations, innate immune activation, and nanotoxicity.(c) Lectins
The lectin pathway of the complement system, similar to the classical pathway, is an integral component of the innate immune system recognizing and neutralizing pathogens through patterns of carbohydrates on their surfaces. Although not all NPs contain surface-exposed sugar moieties, many engineered NPs have been observed to provoke lectin pathway complement clearance.30 Digiacomo et al. observed an elevations in mannan-binding lectin serine protease 1 (MASP1) and ficolin-3 liposomal corona adsorption.31 MASP1 and ficolin-family proteins are known to be important components of the lectin complement pathway for targeted pathogen clearance.32 The phenomena of BC-driven targeting to innate immune cell populations is important in understanding nanotoxicity and may open up new therapeutic opportunities for immunotherapeutics by developing nanomaterials that preferentially adsorb opsonins for active immune cell targeting (Fig. 3).33,34 Ultimately, it is crucial to characterize the PBC influence on NPs biological outcomes in order to improve therapeutic efficiency and nanotoxicity profiles in downstream clinical applications.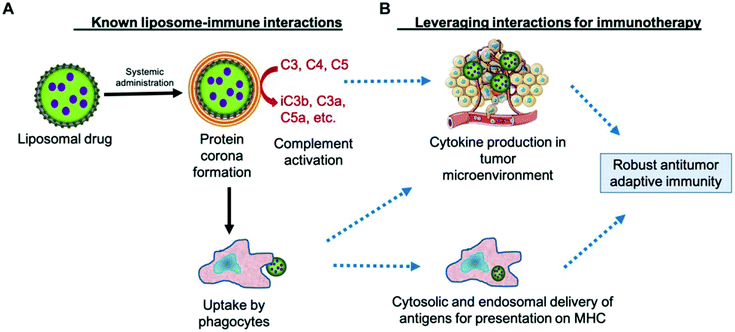 | ||
| Fig. 3 Leveraging the complement protein biomolecular corona for immune system targeting in cancer. A. Known mechanism of NP clearance by complement-induced phagocyte uptake. B. Potential utilization of complement activation to deliver tumor antigens and inducing a proinflammatory phenotype in tumor resident immune cells. Adapted with permission from ref. 33. Copyright 2019 Frontiers in Pharmacology. | ||
5. Inherent cancer cell targeting
The inherent heterogeneity in the physiological effects of cancer can cause variations in PBC composition and NP cancer cell uptake mechanism and specificity.35,36 Cancer cells often have elevated consumption of plasma proteins such as apolipoproteins (Apo), vitronectin, serum albumin, and fibrinogen. The presence of these plasma proteins in the BC can function as pseudo-active targeting ligands to specific cell types. A succeeding study from Hajipour et al.'s 2014 paper examined the effect of PBC on graphene oxide (GO) sheet uptake in MCF-7 and MDA-MB-231 breast cancer cell lines.35 GO sheets were opsonized with plasma from a broad range of patient backgrounds (healthy, pregnancy, diabetes, hypercholesterolemia, leukemia, hypofibrinogenemia, thalassemia, rheumatism) and their differential cellular responses were studied in vitro. Interestingly, GO sheet uptake and toxicity were determined to be both BC-dependent and cell line-dependent. Current evidence suggests that PBC formation can enhance cancer cell targeting through the adsorption of endogenous molecules. Here, we will discuss how changes in the disease-specific plasma components and the resulting BCs promote cancer cell targeting of NPs in the absence of other engineered targeting ligands.(a) Apolipoprotein
Previously, researchers have exploited NP targeting strategies utilizing the protein corona by binding specific plasma proteins (albumin, apolipoproteins, and fibrinogen) to the surface of NPs. In particular, apolipoprotein E (ApoE) is a glycoprotein associated with cholesterol metabolism, cellular proliferation, angiogenesis and metastasis of cancers (e.g. gastric, lung, prostate, thyroid, ovarian, endometrial cancer and glioblastoma).37 Furthermore, liposomal NP formulations are found to preferentially recruit ApoE in their BCs and apolipoprotein-enriched BCs shifted their cellular uptake mechanism from micropinocytosis to clathrin-dependent endocytosis.38 Chen et al. demonstrated the recruitment of ApoE in their liposomal BC significantly enhanced NPs transfection efficiency of hepatocellular carcinoma HepG2 cells compared to ApoE absent NPs.39 Furthermore, work from Akinc et al. demonstrated ApoE-dependent cellular uptake of GalNAc lipid nanoparticles in HeLa cells through the low-density lipoprotein receptor (LDLR).40 These results were further recapitulated in ApoE−/− knockout mice that demonstrated reduced NP efficacy. Taken together, these findings demonstrate that the systemic elevation of ApoE in cancers and BC recruitment can allow researchers to target LDLR overexpressing cancer types. That being said, precaution must be taken given the endogenous expression of LDLR in hepatocyte tissue, which can lead to off-target toxicity.(b) Vitronectin
In a thorough proteomic analysis of cationic liposomes opsonized with healthy human plasma, Palchetti et al. identified key proteins associated with NP uptake in HeLa cells. Correlations between the BC's relative protein abundance and cellular uptake allowed the identification of eight key proteins (vitronectin, ApoA1, ApoA2, ApoB, ApoC2, immunoglobulin heavy chain V-III region BRO, vitamin K-dependent protein, and integrin β3) associated with NP increased uptake.41 Apolipoproteins are known to bind apolipoprotein receptors and LDLRs (Table 1), which are often upregulated in certain cancers. Vitronectin is one of the major cell adhesion glycoproteins in plasma and was found to be abundant in the liposomal BC. Vitronectin is specifically recognized by ανβ3 integrins, which are overexpressed on many solid tumors and tumor vasculature.42 These results support previous findings that the preferential BC recruitment of vitronectin may promote tumor cell targeting through elevated ανβ3 integrin expression.43(c) Albumin
Among BC proteins, albumin is the most abundant serum proteins and is associated with dysopsonin-mediated immune evasion, prolonged systemic circulation, and tumor vascular endothelium targeting. Tumor targeting is mediated by binding to the receptor gp60 on vascular endothelium and the reliance of poor lymphatic drainage in susceptible tumor tissues.44 Tumor albumin accumulation has been demonstrated in several solid tumor models (such as sarcoma, ovarian carcinoma, and Novikoff hepatoma).45 To leverage albumin's utility, researchers have coated paclitaxel NPs with albumin and observed an enhancement in tumor targeting of B16F10 melanoma cells expressing secreted protein acidic and rich in cysteine (SPARC).46 Given this, disease-specific elevations in PBC albumin levels may lead to the specific targeting of tumor vasculature. BC formed from pancreatic patient plasma was identified to have a minor elevation in serum albumin adsorbed to cationic liposomes.47 Interestingly, Caputo et al. observed a statistically significant decrease in serum albumin adsorption in pancreatic patient compared to health control in GO sheets.48 These findings are exemplary to how different nanoformulations can recruit unique proteomic fingerprints in the same disease condition. Although NP-bound albumin has been demonstrated to allow for the increased targeting of therapies to tumor tissues, further research is required to examine if personalized upregulation of serum albumin can allow for the targeting of NPs to tumor vasculature.6. Clinical implications and future directions
Recent advances in NP-based therapeutics have resulted in a surge of research in the field. Despite the extensive research, the clinical translation of cancer nanotherapeutics has been bottlenecked by the failure to recapitulate in vivo findings. Recent work suggests that an important aspect underlying this lack of translatability is due to our insufficient understanding of the nano-bio interface. The BC around NPs has been identified as a fundamental factor in defining NPs’ biological fate. Although the nano-bio interface poses several challenges for nanomaterials beyond the BC, an increased understanding of PBC interactions may provide an opportunity to refine our drug delivery strategy. Further research in identifying patient populations that have increased affinity for particular BC fingerprints of interest would enable increased control over biomolecule binding, thereby providing potential for development of inherently immune-targeted cancer therapies with improved tumor tissue accumulation, reduced nanotoxicity, and increased NP therapeutic efficacy. Importantly, progress is being made around machine learning and meta-analysis with respect to predicting NP–BC compositions and biological implications. For example, Ban et al. developed a computational methodology to predict with high certainty the protein corona composition and cell recognition associated with NP–BC.49 More accurate prediction models in concert with characterizing NPs in environments that replicate in vivo conditions is expected to yield significant advancements in NP therapeutics. With the advances in the understanding of the PBC, the view that synthetically identical nanomaterials will elicit identical biological effects on all patient populations should be reconsidered. Thus, characterizing the PBC in heterogeneous diseases such as cancer, among others, may help in the prediction of the nano-bio interactions of NPs, speed up clinical translation, and improve therapeutic efficacy.7. Conclusion
The PBC has been demonstrated to play an important role in determining the biological fate of NPs. Systemically, the BC has been shown to impact NP toxicity, immune recognition, targeting capability, biodistribution, intracellular uptake and drug release. To date, the majority of investigations characterizing the BC formation have overlooked the contribution of the physiological environment, instead focusing on the effects of the NPs’ physicochemical properties. Differences in plasma biomolecule concentration and composition may be responsible for conflicting NP therapeutic results obtained between cancer cell lines, patient populations, varying disease-states, and unsuccessful clinical translation of promising formulations. Although there has been an increase in studies understanding the PBC, information on the corona-dependent cellular interactions is still limited. We envision that a better understanding of the relationship between the PBC and NP physicochemical properties will act to guide the design of future experiments and potentially leverage the heterogeneous corona composition in cancers for active cell targeting applications and immunotherapies.Author contributions
Conceptualization, J. S. and R. M. P.; methodology, J. S. and R. M. P.; investigation, J. S.; writing—original draft preparation, J. S.; writing—review and editing, J. S. and R. M. P.; visualization, J. S.; funding acquisition, R. M. P. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Research reported in this publication was supported by Startup funds provided by the University of Maryland, Baltimore (UMB), the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R35GM142752, the UMGCC P30 grant under award number P30CA134274 from the National Cancer Institute, NIH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.References
- A. C. Anselmo and S. Mitragotri, Nanoparticles in the clinic: An update, Bioeng. Transl. Med., 2019, 4, e10143 Search PubMed.
- A. C. Anselmo and S. Mitragotri, Nanoparticles in the clinic: An update post COVID-19 vaccines, Bioeng. Transl. Med., 2021, 6, e10246 CAS.
- R. M. Pearson, H. Hsu, J. Bugno and S. Hong, Understanding nano-bio interactions to improve nanocarriers for drug delivery, MRS Bull., 2014, 39, 227–237 CrossRef CAS.
- T. J. Anchordoquy, Y. Barenholz, D. Boraschi, M. Chorny, P. Decuzzi, M. A. Dobrovolskaia, Z. S. Farhangrazi, D. Farrell, A. Gabizon, H. Ghandehari, B. Godin, N. M. La-Beck, J. Ljubimova, S. M. Moghimi, L. Pagliaro, J.-H. Park, D. Peer, E. Ruoslahti, N. J. Serkova and D. Simberg, Mechanisms and Barriers in Cancer Nanomedicine: Addressing Challenges, Looking for Solutions, ACS Nano, 2017, 11, 12–18 CrossRef CAS PubMed.
- S. Wilhelm, A. J. Tavares, Q. Dai, S. Ohta, J. Audet, H. F. Dvorak and W. C. W. Chan, Analysis of nanoparticle delivery to tumours, Nat. Rev. Mater., 2016, 1, 1–12 Search PubMed.
- T. Cedervall, I. Lynch, S. Lindman, T. Berggård, E. Thulin, H. Nilsson, K. A. Dawson and S. Linse, Understanding the nanoparticle–protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2050–2055 CrossRef CAS PubMed.
- M. P. Monopoli, C. Åberg, A. Salvati and K. A. Dawson, Biomolecular coronas provide the biological identity of nanosized materials, Nat. Nanotechnol., 2012, 7, 779–786 CrossRef CAS PubMed.
- R. M. Pearson, V. V. Juettner and S. Hong, Biomolecular Corona on Nanoparticles: A Survey of Recent Literature and Its Implications in Targeted Drug Delivery, Front. Chem., 2014, 2, 108 Search PubMed.
- D. Chen, S. Ganesh, W. Wang and M. Amiji, Protein Corona-Enabled Systemic Delivery and Targeting of Nanoparticles, AAPS J., 2020, 22, 83 CrossRef CAS PubMed.
- M. J. Hajipour, S. Laurent, A. Aghaie, F. Rezaee and M. Mahmoudi, Personalized protein coronas: a “key” factor at the nanobiointerface, Biomater. Sci., 2014, 2, 1210–1221 RSC.
- P. del Pino, B. Pelaz, Q. Zhang, P. Maffre, G. Ulrich Nienhaus and W. J. Parak, Protein corona formation around nanoparticles – from the past to the future, Mater. Horiz., 2014, 1, 301–313 RSC.
- K. P. García, K. Zarschler, L. Barbaro, J. A. Barreto, W. O'Malley, L. Spiccia, H. Stephan and B. Graham, Zwitterionic-Coated “Stealth” Nanoparticles for Biomedical Applications: Recent Advances in Countering Biomolecular Corona Formation and Uptake by the Mononuclear Phagocyte System, Small, 2014, 10, 2516–2529 CrossRef PubMed.
- Y. Zou, S. Ito, F. Yoshino, Y. Suzuki, L. Zhao and N. Komatsu, Polyglycerol Grafting Shields Nanoparticles from Protein Corona Formation to Avoid Macrophage Uptake, ACS Nano, 2020, 14, 7216–7226 CrossRef CAS PubMed.
- D. Pozzi, V. Colapicchioni, G. Caracciolo, S. Piovesana, A. L. Capriotti, S. Palchetti, S. D. Grossi, A. Riccioli, H. Amenitsch and A. Laganà, Effect of polyethyleneglycol (PEG) chain length on the bio–nano-interactions between PEGylated lipid nanoparticles and biological fluids: from nanostructure to uptake in cancer cells, Nanoscale, 2014, 6, 2782–2792 RSC.
- N. J. Butcher, G. M. Mortimer and R. F. Minchin, Unravelling the stealth effect, Nat. Nanotechnol., 2016, 11, 310–311 CrossRef CAS PubMed.
- M. D. McSweeney, L. Shen, A. C. DeWalle, J. B. Joiner, E. C. Ciociola, D. Raghuwanshi, M. S. Macauley and S. K. Lai, Pre-treatment with high molecular weight free PEG effectively suppresses anti-PEG antibody induction by PEG-liposomes in mice, J. Controlled Release, 2021, 329, 774–781 CrossRef CAS PubMed.
- I. Lynch and K. A. Dawson, Protein-nanoparticle interactions, Nano Today, 2008, 3, 40–47 CrossRef CAS.
- L. Vroman, A. Adams, G. Fischer and P. Munoz, Interaction of high molecular weight kininogen, factor XII, and fibrinogen in plasma at interfaces, Blood, 1980, 55, 156–159 CrossRef CAS PubMed.
- Z. J. Deng, M. Liang, M. Monteiro, I. Toth and R. F. Minchin, Nanoparticle-induced unfolding of fibrinogen promotes Mac-1 receptor activation and inflammation, Nat. Nanotechnol., 2011, 6, 39–44 CrossRef CAS PubMed.
- J.-Y. Park, S. J. Park, J. Y. Park, S.-H. Kim, S. Kwon, Y. Jung and D. Khang, Unfolded Protein Corona Surrounding Nanotubes Influence the Innate and Adaptive Immune System, Adv. Sci., 2021, 8, 2004979 CrossRef CAS PubMed.
- A. J. Tavares, W. Poon, Y.-N. Zhang, Q. Dai, R. Besla, D. Ding, B. Ouyang, A. Li, J. Chen, G. Zheng, C. Robbins and W. C. W. Chan, Effect of removing Kupffer cells on nanoparticle tumor delivery, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E10871–E10880 CAS.
- L. Papafilippou, A. Claxton, P. Dark, K. Kostarelos and M. Hadjidemetriou, Protein corona fingerprinting to differentiate sepsis from non-infectious systemic inflammation, Nanoscale, 2020, 12, 10240–10253 RSC.
- C. Stover, Dual role of complement in tumour growth and metastasis (Review), Int. J. Mol. Med., 2010, 25, 307–313 CAS.
- J. Ren, R. Cai, J. Wang, M. Daniyal, D. Baimanov, Y. Liu, D. Yin, Y. Liu, Q. Miao, Y. Zhao and C. Chen, Precision Nanomedicine Development Based on Specific Opsonization of Human Cancer Patient-Personalized Protein Coronas, Nano Lett., 2019, 19, 4692–4701 CrossRef CAS PubMed.
- Y. Ju, H. G. Kelly, L. F. Dagley, A. Reynaldi, T. E. Schlub, S. K. Spall, C. A. Bell, J. Cui, A. J. Mitchell, Z. Lin, A. K. Wheatley, K. J. Thurecht, M. P. Davenport, A. I. Webb, F. Caruso and S. J. Kent, Person-Specific Biomolecular Coronas Modulate Nanoparticle Interactions with Immune Cells in Human Blood, ACS Nano, 2020, 14, 15723–15737 CrossRef PubMed.
- M. Hadjidemetriou, Z. Al-ahmady, M. Buggio, J. Swift and K. Kostarelos, A novel scavenging tool for cancer biomarker discovery based on the blood-circulating nanoparticle protein corona, Biomaterials, 2019, 188, 118–129 CrossRef CAS PubMed.
- V. Colapicchioni, M. Tilio, L. Digiacomo, V. Gambini, S. Palchetti, C. Marchini, D. Pozzi, S. Occhipinti, A. Amici and G. Caracciolo, Personalized liposome–protein corona in the blood of breast, gastric and pancreatic cancer patients, Int. J. Biochem. Cell Biol., 2016, 75, 180–187 CrossRef CAS PubMed.
- H. de Jonge, L. Iamele, M. Maggi, G. Pessino and C. Scotti, Anti-Cancer Auto-Antibodies: Roles, Applications and Open Issues, Cancers, 2021, 13, 813 CrossRef CAS PubMed.
- T. Zheng, N. Pierre-Pierre, X. Yan, Q. Huo, A. J. O. Almodovar, F. Valerio, I. Rivera-Ramirez, E. Griffith, D. D. Decker, S. Chen and N. Zhu, Gold Nanoparticle-Enabled Blood Test for Early Stage Cancer Detection and Risk Assessment, ACS Appl. Mater. Interfaces, 2015, 7, 6819–6827 CrossRef CAS PubMed.
- S. M. Moghimi, P. P. Wibroe, L. Wu and Z. S. Farhangrazi, Insidious pathogen-mimicking properties of nanoparticles in triggering the lectin pathway of the complement system, Eur. J. Nanomed., 2015, 7, 263–268 CAS.
- L. Digiacomo, F. Giulimondi, D. Pozzi, A. Coppola, V. La Vaccara, D. Caputo and G. Caracciolo, A Proteomic Study on the Personalized Protein Corona of Liposomes. Relevance for Early Diagnosis of Pancreatic DUCTAL Adenocarcinoma and Biomarker Detection, J. Nanotheranostics, 2021, 2, 82–93 CrossRef.
- S. Thiel, Complement activating soluble pattern recognition molecules with collagen-like regions, mannan-binding lectin, ficolins and associated proteins, Mol. Immunol., 2007, 44, 3875–3888 CrossRef CAS PubMed.
- N. M. La-Beck, X. Liu and L. M. Wood, Harnessing Liposome Interactions With the Immune System for the Next Breakthrough in Cancer Drug Delivery, Front. Pharmacol., 2019, 10, 220 CrossRef CAS PubMed.
- Z. Luo, L. Lu, W. Xu, N. Meng, S. Wu, J. Zhou, Q. Xu, C. Xie, Y. Liu and W. Lu, In vivo self-assembled drug nanocrystals for metastatic breast cancer all-stage targeted therapy, J. Controlled Release, 2022, S0168-3659(22)00192-4 Search PubMed.
- M. J. Hajipour, J. Raheb, O. Akhavan, S. Arjmand, O. Mashinchian, M. Rahman, M. Abdolahad, V. Serpooshan, S. Laurent and M. Mahmoudi, Personalized disease-specific protein corona influences the therapeutic impact of graphene oxide, Nanoscale, 2015, 7, 8978–8994 RSC.
- L. A. Liotta, M. Ferrari and E. Petricoin, Clinical proteomics: Written in blood, Nature, 2003, 425, 905–905 CrossRef CAS PubMed.
- Z. Zhao, S. Zou, X. Guan, M. Wang, Z. Jiang, Z. Liu, C. Li, H. Lin, X. Liu, R. Yang, Y. Gao and X. Wang, Apolipoprotein E Overexpression Is Associated With Tumor Progression and Poor Survival in Colorectal Cancer, Front. Genet., 2018, 9, 650 CrossRef CAS PubMed.
- L. Digiacomo, F. Cardarelli, D. Pozzi, S. Palchetti, M. A. Digman, E. Gratton, A. L. Capriotti, M. Mahmoudi and G. Caracciolo, An apolipoprotein-enriched biomolecular corona switches the cellular uptake mechanism and trafficking pathway of lipid nanoparticles, Nanoscale, 2017, 9, 17254–17262 RSC.
- D. Chen, S. Ganesh, W. Wang and M. Amiji, The role of surface chemistry in serum protein corona-mediated cellular delivery and gene silencing with lipid nanoparticles, Nanoscale, 2019, 11, 8760–8775 RSC.
- A. Akinc, W. Querbes, S. De, J. Qin, M. Frank-Kamenetsky, K. N. Jayaprakash, M. Jayaraman, K. G. Rajeev, W. L. Cantley, J. R. Dorkin, J. S. Butler, L. Qin, T. Racie, A. Sprague, E. Fava, A. Zeigerer, M. J. Hope, M. Zerial, D. W. Sah, K. Fitzgerald, M. A. Tracy, M. Manoharan, V. Koteliansky, A. de Fougerolles and M. A. Maier, Targeted Delivery of RNAi Therapeutics With Endogenous and Exogenous Ligand-Based Mechanisms, Mol. Ther., 2010, 18, 1357–1364 CrossRef CAS PubMed.
- S. Palchetti, L. Digiacomo, D. Pozzi, G. Peruzzi, E. Micarelli, M. Mahmoudi and G. Caracciolo, Nanoparticles-cell association predicted by protein corona fingerprints, Nanoscale, 2016, 8, 12755–12763 RSC.
- S. Cabodi, M. del Pilar Camacho-Leal, P. Di Stefano and P. Defilippi, Integrin signalling adaptors: not only figurants in the cancer story, Nat. Rev. Cancer, 2010, 10, 858–870 CrossRef CAS PubMed.
- G. Caracciolo, F. Cardarelli, D. Pozzi, F. Salomone, G. Maccari, G. Bardi, A. L. Capriotti, C. Cavaliere, M. Papi and A. Laganà, Selective Targeting Capability Acquired with a Protein Corona Adsorbed on the Surface of 1,2-Dioleoyl-3-trimethylammonium Propane/DNA Nanoparticles, ACS Appl. Mater. Interfaces, 2013, 5, 13171–13179 CrossRef CAS PubMed.
- E. N. Hoogenboezem and C. L. Duvall, Harnessing albumin as a carrier for cancer therapies, Adv. Drug Delivery Rev., 2018, 130, 73–89 CrossRef CAS PubMed.
- A. M. Merlot, D. S. Kalinowski and D. R. Richardson, Unraveling the mysteries of serum albumin—more than just a serum protein, Front. Physiol., 2014, 5, 299 Search PubMed.
- J. Park, B. Sun and Y. Yeo, Albumin-coated nanocrystals for carrier-free delivery of paclitaxel, J. Controlled Release, 2017, 263, 90–101 CrossRef CAS PubMed.
- R. Di Santo, L. Digiacomo, E. Quagliarini, A. L. Capriotti, A. Laganà, R. Zenezini Chiozzi, D. Caputo, C. Cascone, R. Coppola, D. Pozzi and G. Caracciolo, Personalized Graphene Oxide-Protein Corona in the Human Plasma of Pancreatic Cancer Patients, Front. Bioeng. Biotechnol., 2020, 8, 491 CrossRef PubMed.
- D. Caputo, L. Digiacomo, C. Cascone, D. Pozzi, S. Palchetti, R. Di Santo, E. Quagliarini, R. Coppola, M. Mahmoudi and G. Caracciolo, Synergistic Analysis of Protein Corona and Haemoglobin Levels Detects Pancreatic Cancer, Cancers, 2021, 13, 93 CrossRef CAS PubMed.
- Z. Ban, P. Yuan, F. Yu, T. Peng, Q. Zhou and X. Hu, Machine learning predicts the functional composition of the protein corona and the cellular recognition of nanoparticles, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 10492–10499 CrossRef CAS PubMed.
- S. Johansson, Fibronectin-integrin interactions, Front. Biosci., 1997, 2, d126–d146 CrossRef CAS PubMed.
- S. R. Coughlin, How the protease thrombin talks to cells, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 11023–11027 CrossRef CAS PubMed.
- R. W. Mahley and T. L. Innerarity, Lipoprotein receptors and cholesterol homeostasis, Biochim. Biophys. Acta, Rev. Biomembr., 1983, 737, 197–222 CrossRef CAS.
- P. Singh, C. Carraher and J. E. Schwarzbauer, Assembly of Fibronectin Extracellular Matrix, Annu. Rev. Cell Dev. Biol., 2010, 26, 397–419 CrossRef CAS PubMed.
- C. Schlömmer, A. Brandtner and M. Bachler, Antithrombin and Its Role in Host Defense and Inflammation, Int. J. Mol. Sci., 2021, 22, 4283 CrossRef PubMed.
- M. Z. Kounnas, R. E. Morris, M. R. Thompson, D. J. FitzGerald, D. K. Strickland and C. B. Saelinger, The alpha 2-macroglobulin receptor/low density lipoprotein receptor-related protein binds and internalizes Pseudomonas exotoxin A., J. Biol. Chem., 1992, 267, 12420–12423 CrossRef CAS.
- J. V. Sarma and P. A. Ward, The complement system, Cell Tissue Res., 2011, 343, 227–235 CrossRef CAS PubMed.
- M. J. Nielsen and S. K. Moestrup, Receptor targeting of hemoglobin mediated by the haptoglobins: roles beyond heme scavenging, Blood, 2009, 114, 764–771 CrossRef CAS PubMed.
- F. Peyvandi, I. Garagiola and L. Baronciani, Role of von Willebrand factor in the haemostasis, Blood Transfus., 2011, 9, s3–s8 Search PubMed.
- H. W. Schroeder and L. Cavacini, Structure and Function of Immunoglobulins, J. Allergy Clin. Immunol., 2010, 125, S41–S52 CrossRef PubMed.
- S. Borgquist, T. Butt, P. Almgren, D. Shiffman, T. Stocks, M. Orho-Melander, J. Manjer and O. Melander, Apolipoproteins, lipids and risk of cancer, Int. J. Cancer, 2016, 138, 2648–2656 CrossRef CAS PubMed.
- J. Housley, Alpha2-macroglobulin levels in disease in man, J. Clin. Pathol., 1968, 21, 27–31 CrossRef CAS PubMed.
- S. Yoshino, K. Fujimoto, T. Takada, S. Kawamura, J. Ogawa, Y. Kamata, Y. Kodera and M. Shichiri, Molecular form and concentration of serum α2-macroglobulin in diabetes, Sci. Rep., 2019, 9, 12927 CrossRef PubMed.
- H. Asakawa, K. Tokunaga and F. Kawakami, Elevation of fibrinogen and thrombin-antithrombin III complex levels of type 2 diabetes mellitus patients with retinopathy and nephropathy, J. Diabetes Complications, 2000, 14, 121–126 CrossRef CAS PubMed.
- M. Sun and A. S. Gupta, Vascular Nanomedicine: Current Status, Opportunities, and Challenges, Semin. Thromb. Hemostasis, 2020, 46, 524–544 CrossRef PubMed.
- A. B. Engin, D. Nikitovic, M. Neagu, P. Henrich-Noack, A. O. Docea, M. I. Shtilman, K. Golokhvast and A. M. Tsatsakis, Mechanistic understanding of nanoparticles’ interactions with extracellular matrix: the cell and immune system, Part. Fibre Toxicol., 2017, 14, 22 CrossRef PubMed.
- N. E. Stathakis, A. Fountas and E. Tsianos, Plasma fibronectin in normal subjects and in various disease states., J. Clin. Pathol., 1981, 34, 504–508 CrossRef CAS PubMed.
- R. Pio, L. Corrales and J. D. Lambris, The Role of Complement in Tumor Growth, Adv. Exp. Med. Biol., 2014, 772, 229–262 CrossRef CAS PubMed.
- G. Lee, S. Choi, K. Kim, J. Yun, J. S. Son, S. Jeong, S. M. Kim and S. M. Park, Association of Hemoglobin Concentration and Its Change With Cardiovascular and All–Cause Mortality, J. Am. Heart Assoc., 2018, 7, e007723 CrossRef PubMed.
- I. Peppas, G. George, S. Sollie, D. H. Josephs, N. Hammar, G. Walldius, S. N. Karagiannis and M. Van Hemelrijck, Association of Serum Immunoglobulin Levels with Solid Cancer: A Systematic Review and Meta-analysis, Cancer Epidemiol., Biomarkers Prev., 2020, 29, 527–538 CrossRef CAS PubMed.
- G. Mindrum and H. I. Glueck, Plasma prothrombin in liver disease: its clinical and prognostic significance, Ann. Intern. Med., 1959, 50, 1370–1384 CrossRef CAS PubMed.
- M. S. Twigg, S. Brockbank, P. Lowry, S. P. FitzGerald, C. Taggart and S. Weldon, The Role of Serine Proteases and Antiproteases in the Cystic Fibrosis Lung, Mediators Inflammation, 2015, 2015, 293053 Search PubMed.
- A. Eckart, T. Struja, A. Kutz, A. Baumgartner, T. Baumgartner, S. Zurfluh, O. Neeser, A. Huber, Z. Stanga, B. Mueller and P. Schuetz, Relationship of Nutritional Status, Inflammation, and Serum Albumin Levels During Acute Illness: A Prospective Study, Am. J. Med., 2020, 133, 713–722 CrossRef CAS PubMed.
- M.-H. Chen, C. Lu, J. Sun, X.-D. Chen, J.-X. Dai, J.-Y. Cai and X.-L. Chen, Diagnostic and prognostic value of serum vitronectin levels in human glioma, J. Neurol. Sci., 2016, 371, 54–59 CrossRef CAS PubMed.
- H. Ekmekci, H. Sonmez, O. B. Ekmekci, Z. Ozturk, N. Domanic and E. Kokoglu, Plasma Vitronectin Levels in Patients with Coronary Atherosclerosis are Increased and Correlate with Extent of Disease, J. Thromb. Thrombolysis, 2002, 14, 221–225 CrossRef CAS PubMed.
- T. N. Bongers, M. P. M de Maat, M. L. van Goor, V. Bhagwanbali, H. H. D. M. van Vliet, E. B. Gómez Garcia, D. W. J. Dippel and F. W. Leebeek, High von Willebrand Factor Levels Increase the Risk of First Ischemic Stroke, Stroke, 2006, 37, 2672–2677 CrossRef CAS PubMed.
- J. M. O'Sullivan, R. J. S. Preston, T. Robson and J. S. O'Donnell, Emerging Roles for von Willebrand Factor in Cancer Cell Biology, Semin. Thromb. Hemostasis, 2018, 44, 159–166 CrossRef PubMed.
- A. T. Bauer, E. A. Strozyk, C. Gorzelanny, C. Westerhausen, A. Desch, M. F. Schneider and S. W. Schneider, Cytotoxicity of silica nanoparticles through exocytosis of von Willebrand factor and necrotic cell death in primary human endothelial cells, Biomaterials, 2011, 32, 8385–8393 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |