Metal–DNA coordination based bioinspired hybrid nanospheres for in situ amplification and sensing of microRNA†
Yijing
Jia‡
a,
Xiaotong
Shen‡
b,
Feifei
Sun
a,
Na
Na
 a and
Jin
Ouyang
a and
Jin
Ouyang
 *a
*a
aKey Laboratory of Theoretical and Computational Photochemistry, Ministry of Education, College of Chemistry, Beijing Normal University, Beijing 100875, China. E-mail: jinoyang@bnu.edu.cn
bSchool of Life Science, Beijing Institute of Technology, Beijing 100081, China
First published on 4th November 2020
Abstract
Sufficient delivery of biomolecules into cells with high loading efficiency and easy cleavability would be significant for the visualization of biomolecules in living cells. Herein, a facile approach based on nano-wire balls (NWs) for efficient loading, intracellular delivery of nucleic acids and in situ targeted miRNA bioimaging is proposed, by feeding of Zn ions for generating DNA–inorganic hybrid structures with large surface areas and good stability. Given that the versatile and robust hybridization chain reaction (HCR) amplification strategy combines DNA assembly with intracellular assay, the resulting NWs without any complicated modification are capable of enhanced signals for the targeted imaging of cancer cells. This method realized a linear detection range of 100 fM to 10 nM, with a low detection limit of 83.6 fM in vitro, and could be used to effectively differentiate the expression levels of miRNA-21 in living cells. Due to its high loading efficiency, excellent biocompatibility and low toxicity, this system can be used to construct a coordination-based delivery nanoplatform for in situ enzyme-free amplified imaging of miRNAs, expanding the application of DNA-based nanomaterials for cellular delivery and intracellular molecule analysis.
Introduction
Metal–ligand coordination chemistry has demonstrated great potential for the development of more efficient and functional nanomaterials for biosensing and bioimaging applications which potentially provide novel insights into the effects and mechanism of medical diagnosis and therapy.1–7 As the genetic material, DNA molecules have emerged as excellent substrates for the rational design and synthesis of DNA nanoparticles (NPs) in the areas of materials science, biotechnology and nanotechnology due to their good biocompatibility, high programmability, and thermodynamic stability.8–18 In addition, DNA molecules have been explored for self-assembled coordination materials with metal ions due to their multiple metal binding sites.19 For biomedical applications, biocompatible metal ions have been chosen for the construction of functional coordination polymers, including zinc, copper, and iron.20 Most recently, coordination-driven self-assembly of metal–DNA has proven to be a pretty attractive strategy for the synthesis of functional DNA nanoarchitectures with a well-defined structure and specific morphologies.21,22 Featured with favorable properties, such as versatile DNA building blocks and excellent efficiency and capacity for self-assembly, metal-coordination-directed DNA nanostructures enable applications in the selective encapsulation and delivery of imaging probes and therapeutic agents.23,24 Despite recent efforts, development of simple, novel and applicable approaches for the synthesis of functional DNA-based nanostructures with specificity toward targets for intracellular biomarker detection and imaging is a major focus in the fields of precision nanomedicine and drug delivery.With recent advancements in DNA-based nanomaterials, a plethora of intracellular environment responsive biomimetic nanoplatforms have been designed as nanocarriers for functional biomacromolecules, which are significant and fundamental for biological research and disease diagnostics. For instance, a cationic liposomal system has emerged as a promising strategy to deliver therapeutic cargos into cells for various applications, but still suffers from limitations such as sophisticated material synthesis and engineering methods.25–28 A universal approach for the synthesis of DNA-based multifunctional nanoarchitectures via coordination-driven self-assembly can be tailored for efficient DNA encapsulation and delivery with enhanced biological function. However, the assembled DNA nanostructures are not universally suitable for efficient in situ detection of biomarkers.22,24
Fluorescence in situ hybridization (FISH) with fluorescence labeled probes has proven to be a well-accepted way for intracellular delivery and visualization of gene expression by forming distinguishable luminous nanostructures at the locations of target microRNAs (miRNAs).29–32 However, due to their short length, trace amounts in cells and tissues, and the high similarity among homogeneous sequence, more sensitive approaches are required for the accurate detection of miRNAs in individual cells.33–37 Signal amplification detection strategies have attracted great interest in the accurate identification of RNA with low expression levels, which are momentous and fundamental for the early diagnosis and timely treatment of disease.38 Recently, a transcription aptasensor has been used for the on-site detection of pathogens by using a light-up RNA aptamer with efficient amplification rates.39 In addition, strand displacement reaction has been developed rapidly in recent years to detect endogenous mRNAs.40 However, most of these strategies rely on RNA engineering and the involvement of multiple proteases, which require sophisticated RNA design and enzymatic reaction optimization processes.41 At present, some non-enzymatic amplification techniques, in particular the HCR, are currently widely applied in intracellular assays due to the advantages of simple operation, efficient amplification capability and temperature independence.42–46 Therefore, it is highly desirable to design a HCR amplification strategy as a linker of programmed DNA assembly and intracellular amplification assay to expand the application field of DNA-based nanomaterials for enhanced imaging of miRNAs and intracellular gene delivery for improving diagnostic accuracy and efficiency.
Herein, miRNA-21 (miR-21) was selected as the research model, as it is emerging as a promising tumor biomarker for early cancer diagnosis and clinical management. Based on the metal–DNA coordination and HCR amplification strategy, the NWs were synthesized, exploiting the delicate structure and superior biocompatibility of functionalized HCR-based DNA hybrid nanospheres, to achieve favorable stability, high loading efficiency, and low toxicity. A one-pot synthesis method for DNA molecules and Zn ions of the system provides a suitable choice by virtue of its simplicity and generality. The versatility of this approach allows us to make the hybrid nanostructures more attractive for applications in biological research and disease diagnosis, allowing for simple operation, high loading efficiency, favorable biocompatibility, good specificity, efficient amplification capability without the involvement of multiple proteases, and robust stability at physiological temperature. In addition, due to its efficient non-toxic delivery of nucleic acids into cells, the proposed biosensor was applied for in situ enzyme-free signal amplification for intracellular fluorescence imaging of miR-21 in living cells, indicating the significant potential of Zn–DNA coordination as a tool for bioimaging, the delivery of therapeutic nucleic acids, and combination therapy.
Results and discussion
Design of NW-based HCR
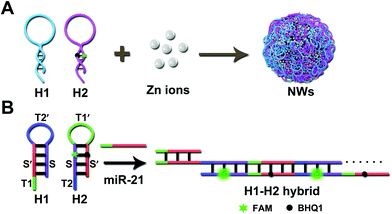
As shown in Scheme 1A, a pair of HCR programmable hairpin oligonucleotide probes (H1, H2) were designed and specifically self-assembled to form fluorescent duplex structures responsive to miR-21; the efficient loading of H1 and H2 was achieved by simple winding during the synthesis. With the aid of Zn ions, H1 and self-quenched H2 co-assembled into uniform coordination NPs without any complicated modification through a one-pot reaction. According to the typical procedure,10 first, each functional hairpin, H1 and H2, was diluted in HEPES buffer, respectively, heated to 95 °C for 5 min and allowed to cool to room temperature. Then, an aqueous solution of Zn ions was added and left undisturbed at 95 °C for 3 h; the resulting solution was centrifuged and washed with deionized water. The collected NWs were finally redispersed in deionized water for further use. Controlling the size and morphology of nanoparticles rather than large aggregates is the key to the synthesis of nanomaterials. For example, a supramolecular gel formed by the coordination of Zn2+ with adenosine monophosphate (AMP) was regulated by the base structure and Zn2+ during gel formation.5 Similar to the formation of Fe-DNA NPs,22,24 the spherical shape, size and uniformity of the coordination materials were related to the metal ion/nucleotide molar ratios during the growth and stability process of the hybrid particles. In this work, uniform spherical NWs with an average diameter of approximately 52 ± 3 nm were successfully synthesized. The multifunctional NWs with simple synthetic strategy, high loading efficiency, good specificity, and favorable biocompatibility can cause intracellular amplification for the sensitive detection of miRNAs.In this strategy, H1 was composed of a hairpin structure ST2′S′ and toehold T1 (complementary to T1′ in H2). H2 was composed of toehold T2 (complementary to T2′ in H1) and a hairpin structure (ST1′S′). The signal probe H2 was labeled with 5-carboxyfluorescein (FAM) dye quenched by a black hole quencher (BHQ1), resulting in a considerably low fluorescence background signal in the absence of miR-21 (the sequences are listed in Table S1, ESI†). Interaction of the initiator with a mixture of H1 and H2 triggered the cascade hybridization of H1 and H2, and a long double-stranded DNA (dsDNA) nano-assembly polymers with multiple repeated units were formed (Scheme 1B). The ideal structures and thermodynamic consequences of H1 and H2 have been further analyzed using the OligoAnalyzer software (IDT) (Fig. S1, ESI†).
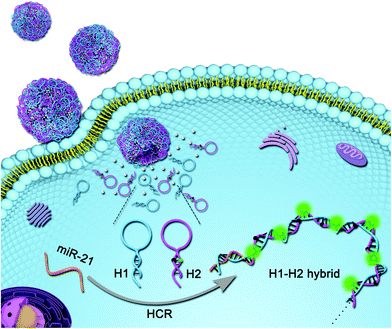
As illustrated in Scheme 2, the NW-based HCR strategy was used to amplify the visualization of miR-21 in living cells. On encountering target cancer cells, the intracellular target miR-21 could specifically hybridize with capture-H1 to expose the stem section as a toehold initiator, leading to the self-assembly of these building blocks and thus the FAM fluorophores are instantly separated from BHQ1 quenchers. Consequently, the increase in fluorescence intensity of FAM can be employed to sensitively detect miRNAs in living cells, providing a potential platform for the intracellular delivery of nucleic acids and low abundant biomarker imaging.
Characterization of NWs
To confirm the successful synthesis of NWs, characterization of morphology was performed using several characterization methods. As shown in Fig. 1A and B, the morphological feature of the as-synthesized uniform spherical NWs with an average diameter of approximately 52 ± 3 nm were clearly observed using scanning electron microscopy (SEM) and transmission electron microscopy (TEM). Meanwhile, the results coincided with those of dynamic light scattering (DLS) measurements of the NWs as shown in Fig. 1C. Zeta potential measurement showed that the NWs had a negatively charged surface, −24.9 mV (Fig. S2, ESI†). Furthermore, the high-angle annular dark-field scanning TEM (HAADF-STEM) EDS mapping clearly confirmed that phosphorus and nitrogen from nucleic acids and Zn ions were evenly distributed in the NWs (Fig. 1D).24 The UV-vis absorption spectra are shown in Fig. 1E, the characteristic DNA absorbance peak possessed a slight shift to around 275 nm, indicating the successful encapsulation of DNA into the NW structures.47 As shown in Fig. 1F, Fourier transform infrared (FTIR) spectra showed that the characteristic asymmetric stretching vibrations of the backbone phosphate group in DNA shifted from 1213 cm−1 to 1194 cm−1, indicating the successful synthesis of the DNA embedded NWs through coordination with Zn ions.22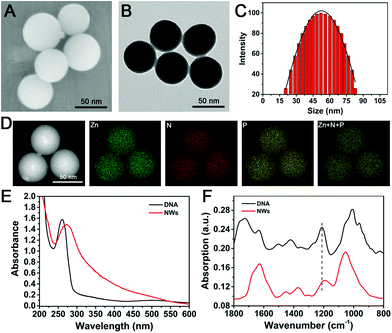 | ||
| Fig. 1 Nanoparticle characterizations. (A) SEM, (B) TEM, (C) DLS and (D) HAADF-STEM images and the corresponding elemental mapping images of NWs. (E) UV-vis absorption spectra. (F) FTIR spectra. | ||
Feasibility and reliability of NW-based HCR system
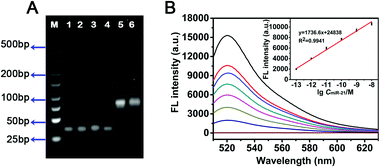
To validate the whole working principle as described, the miR-21 was converted to miR-21-DNA, where the uracil (U) sequences were replaced with thymine (T) sequences. The designed miR-21-DNA was used for the study of in vitro amplification by the NW-based HCR method to investigate the feasibility. The fluorescence spectra were acquired after treatment with different concentrations of target DNA for 2 h in the subsequent experiments. As shown in Fig. S3A (ESI†), significant fluorescence signals were obtained following the HCR reaction, and the green fluorescence intensity was significantly enhanced along with the elevated miR-21-DNA concentration, revealing the obvious concentration-dependent effect of the NW-based HCR system. As depicted in Fig. S3B (ESI†), a good linear correlation was found between the fluorescence change and the logarithm of target miR-21-DNA concentration ranging from 100 fM to 10 nM (R2 = 0.9866). The limit of detection (LOD) was estimated to be 27.8 fM based on the traditional 3σ calculation approach (S/N = 3). The performance of NW-based HCR system for the successful detection of miR-21-DNA confirmed the feasibility of amplifications in vitro.The in vitro feasibility of the miR-21 triggered HCR was further characterized using 4% agarose gel electrophoresis (AGE). As depicted in Fig. 2A, no new band was observed for only HCR reactants (lanes 1–4), which implies that the hairpin mixtures coexist stably without the formation of H1–H2 hybrid in the absence of a corresponding initiator. In contrast, a bright new band with lower electrophoretic migration stuck on the wells in lane 5 when miR-21 was introduced into the HCR mixture, which could be attributed to the amplification products H1–H2. These results confirmed that the robust NW-based HCR system could only be specifically triggered by its initiators with negligible background signals. Compared to the HCR control system, a NW-based HCR system was employed for miR-21 detection, generating H1–H2 hybrid products with a high molecular weight (lane 6). A negligible difference between lane 5 and lane 6 was observed in terms of band positions, indicating that a bright band was obtained on incubating miR-21 with two DNA probes H1 and H2 (traditional HCR) or with the nano-assembly carrying both H1 and H2 (NW-based HCR). The results of the gel electrophoresis experiments clearly proved the successful formation of our NW-based HCR platform with efficient signal amplification effect as anticipated.
In vitro analysis of NW-based HCR
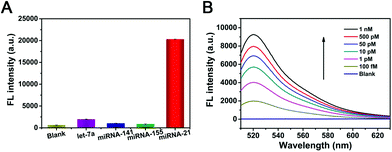
To verify the feasibility of the as-prepared NW-based system for in vitro sensing of target miR-21, the fluorescence spectra of the NWs with different concentrations of target miRNAs were compared. Fig. 2B depicted the fluorescence spectral responses of the developed miR-21-targeting strategy upon analyzing various concentrations of miR-21. The fluorescence intensity obviously increased with increasing concentrations of miR-21 from 100 fM to 10 nM, which is also perfectly consistent with the result of the miR-21-DNA assay. As shown in the inset of Fig. 2B, there exists a good linear correlation between fluorescence intensity and the logarithm of miR-21 concentration with a fine correlation coefficient R2 = 0.9941. Based on the NWs amplifier, the detection limit of miR-21 was estimated to be 83.6 fM based on the 3σ/slope rule. Thus, the NW-based HCR scheme could be considered as a universal amplification tool for sensing in a variety of applications.The universal sensing performance of the NW-based HCR system relies on its high amplifying capacity, high selectivity and desirable specificity. The selectivity of NW-based HCR-amplified miR-21 assay was investigated by comparing the fluorescence responses from a series of control miRNAs, including let-7a, miR-141, and miR-155 miRNAs. As presented in Fig. 3A, a significant fluorescence readout was acquired from miR-21, while a negligible fluorescence response was obtained for these interfering miRNAs, indicating a desirable selectivity for the detection of miR-21 assay. For specificity experiments, the NW-based system was then investigated using the control hybridization experiments for three even one-base mismatched target miRNAs (miR-21-a/miR-21-b/miR-21-c). As shown in Fig. S4 (ESI†), the single-base mismatched strands presented a slight fluorescence intensity that was approximately equal to that of the blank control, while only the miR-21 target exhibited a strong fluorescence signal, demonstrating a satisfactory specificity of the constructed miR-21 amplified sensing system.
To further investigate the capability of the proposed NW-based strategy for the detection of miR-21 in a complicated biological environment, a series of different amounts of miR-21 were spiked in 10% diluted real human serum and then detected using the synthesized NWs (Fig. 3B). As shown in Table S2 (ESI†), the recovery efficiency ranged from 95.4% to 106.8%, and the relative standard deviation (RSD) ranged from 3.8% to 6.4%, which showed a good accordance with the previous fluorescence measurement as expected. The results illustrated that the established NW-based miR-21-detecting strategy showed little interference in complex biological systems.
Cell imaging of NWs
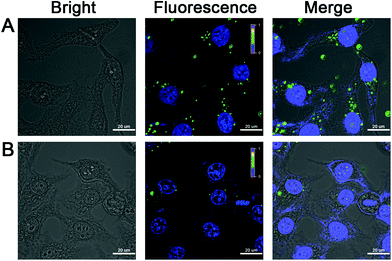
After establishing the discerning ability of the target miRNA using our NW-based method through spectroscopy, we attempted to apply the NW-based strategy to detect target miRNA in living cells. HepG2 cells with high miR-21 expression were employed for the in situ evaluation of the performance of the intracellular miR-21-targeting NW-based imaging system. The toxicity and stability of NWs were major concerns in applying the NW-based strategy for the detection of miR-21 in live cells. Initially, a standard CCK-8 assay was conducted, and the results showed that the NWs exhibited low in vitro cytotoxicity toward HepG2 cells, and the cell viability was maintained above 87% after incubation for 24 h (Fig. S5, ESI†). Next, to determine the NW concentration, we incubated HepG2 cells with different concentrations of NWs ranging from 50 to 100 μg mL−1, after which images of the cells were obtained using confocal laser scanning microscopy (CLSM). On using a NW-based strategy for cell imaging, strong green fluorescence signal was observed for the miR-21 assay (Fig. S6, ESI†), confirming the successful implementation of the fluorescent NW-based sensing system with excellent imaging capability in living cells. Furthermore, HepG2 cells were incubated with NWs for different time periods ranging from 1 h to 4 h to optimize the treatment time between NWs and cells (Fig. S7, ESI†). A dramatically improved intracellular amplification efficiency of the fluorescent NW-based imaging was indirectly testified by optimizing the NW concentration and the incubation time in this system, which is in good agreement with the results of the previous fluorescence experiments. The confocal imaging response and efficient cytosolic delivery of the nanoassembly NW-based strategy provided robust evidence of the possibility for ultrasensitive imaging of miR-21 in living cells.The specificity and reliability of the FAM signal by the NW-based system were then investigated for the in situ miR-21 imaging in HepG2 cells with varying target concentrations. We designed miR-21 inhibitor oligonucleotide (anti-miR-21) to reduce the intracellular target miR-21 concentration, proving the resulting fluorescence readout was thoroughly produced by the intracellular miR-21 rather than other miRNAs. As shown in Fig. 4A, after incubation with NWs for 2 h, HepG2 cells displayed a strong green fluorescence, demonstrating that HepG2 cells possessed a significantly higher expression level of miR-21. As a control, a slight fluorescence signal was obtained in the miR-21 inhibitor treated HepG2 cells compared to in untreated cells (Fig. 4B), implying that the green fluorescence signal was captured by intracellular target miR-21, which correlated well with the miR-21 expression level. Furthermore, RT-PCR was employed to identify the expression level of miR-21 in HepG2 cells without and with miR-21 inhibitor. As shown in Fig. S8 (ESI†), compared with the data of untreated HepG2 cells, HepG2 cells with miR-21 inhibitor showed that the maximum slope of the FL curve and the time corresponding to the inflection point were delayed. Therefore, the concentration of miR-21 in HepG2 cells treated with the miR-21 inhibitor decreased, which is consistent with the intracellular analysis by the NW-based imaging system. These results collectively verified the specificity of the proposed nanomachine for the specific miRNA target, indicating that the NW-based strategy was capable of discriminating miR-21 of varied expression levels in living cancer cells.
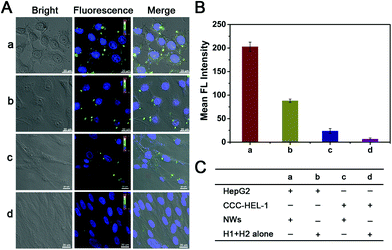
The versatility and universality of our NW-based imaging system were further verified by monitoring miR-21 in tumor and normal cells (Fig. 5); it is known that a lower miR-21 expression occurs in normal cells compared with cancer cells.48–50 We selected HepG2 cells as the positive cells and CCC-HEL-1 normal cells as the negative cells in our experiment. Statistical histogram analysis of the mean fluorescence intensity (MFI) of the miR-21 amplified imaging system is shown in Fig. 5B, and the reactants in each above cell samples is displayed in Fig. 5C. As expected, bright green fluorescence imaging signals for miR-21 are observed in HepG2 cells under confocal microscopy after 2 h of incubation with NWs (sample a in Fig. 5A and B), whereas weak fluorescence signals were obtained in CCC-HEL-1 normal cells (sample c in Fig. 5A and B), which are well corresponding with the miR-21 expression level. The results revealed that the proposed method could accurately distinguish cancer cells from normal cells based on inherent gene expression. Furthermore, the ability of the NW-based imaging system for intracellular signal amplification was then investigated. All cell types were sequentially treated with H1 and H2 alone for comparison. A stronger green fluorescence intensity was observed in the HepG2 cells incubating with NWs compared to that of those incubating with H1 + H2 alone (sample b of Fig. 5A and B), indicating that the signal amplification of the NW-based method could work efficiently in the intracellular environment. As shown in Fig. 5A-c and A-d, a similar phenomenon was also observed in CCC-HEL-1 normal cells, further verifying that NW-based HCR reaction can significantly enhance the fluorescence intensity and the generation of fluorescence can be attributed to the expression level of miR-21. Meanwhile, the miR-21 expression level of HepG2 cells and CCC-HEL-1 cells was determined using RT-PCR (Fig. S8, ESI†). The traditional RT-PCR analysis of the above cell lines indicated that HepG2 cells show a relatively high expression level of miR-21, while CCC-HEL-1 cells show a low expression level of miR-21, which is in good accordance with the intracellular imaging result. These experimental results demonstrated that the developed strategy could discriminate different cells based on the distinct expression level of intracellular target miRNA, confirming that this strategy has the potential for quantifying the miRNA expression level in various living cells.
Conclusions
In summary, we have successfully developed a simple and facile strategy for the construction of a biomimetic nanoplatform with low background interference for the sufficient delivery of nucleic acid into living cells and in situ intracellular targeted miRNA bioimaging. In virtue of their versatile DNA building blocks and excellent capacity to self-assemble, the metal–DNA nanostructures possess uniform sizes, high loading efficiency, high programmability, and robust stability. The NW-based strategy was further demonstrated to exhibit intracellular enzyme-free signal amplification in living cells at physiological temperature, allowing for simple operation and efficient amplification capability without the involvement of multiple proteases. Importantly, the proposed self-assembly system with efficient encapsulation capability provides a widely applicable approach for in situ visualization of intracellular low-abundance nucleic acids and the assessment of gene expression, which may have great potential as a nanocarrier for delivery strategies and combined therapy.Experimental section
Chemicals and materials
Zinc chloride, anhydrous, 99.95% (metals basis) was purchased from Alfa (Beijing, China). All of the synthetic oligonucleotide sequences used in this study were synthesized by Sangon Biotechnology Co., Ltd (Shanghai, China) and purified using HPLC. The sequence information is listed in Table S1 (ESI†). Diethyl pyrocarbonate (DEPC)-treated water, 1 × TAE electrophoresis buffer (40 mM Tris–acetate, 1 mM EDTA, pH 8.0–8.4) and agarose were obtained from Shanghai Sangon Biotechnology Co., Ltd (Shanghai, China). Hoechst 33342 and 10 mM phosphate buffered saline (PBS, pH 7.2–7.4) were acquired from Solarbio (Beijing, China), Cell Counting Kit-8 (CCK-8 reagent) and 10 mM HEPES buffer (100 mM NaCl, 20 mM MgCl2, pH 7.2) were purchased from Beijing BioDee Biotechnology Co., Ltd (Beijing, China). Fetal bovine serum (FBS), Dulbecco's Modified Eagle Medium (DMEM), 0.25% trypsin-ethylenediamine tetraacetic acid (EDTA) solution (containing 0.02% EDTA) and penicillin–streptomycin were obtained from Gibco (USA). Methanol was obtained from Beijing Chemical Co. (Beijing, China). The HepG2 cells and CCC-HEL-1 cells were obtained from Peking Union Medical College Hospital. All reagents were purchased and used without further purification, deionized water obtained from a Milli-Q water purification system (18.2 MΩ cm−1, Milli-Q, Millipore) was utilized in all the experiments.Instrumentation
Fluorescence spectra were recorded using a RF-6000 fluorescence spectrophotometer (Shimadzu, Japan). UV-vis spectra were collected using a UV-2450 spectrophotometer (Shimadzu, Japan). Dynamic light scattering (DLS) analysis was performed using a Nano-ZS Zetasizer ZEN3600 instrument (Malvern Instruments Ltd, UK). The infrared spectra were recorded using an IRAffinity-1 Fourier transform infrared spectrometer (Shimadzu, Japan). Transmission electron microscopy (TEM) images were measured using an FEI Talos F200S electron microscope (FEI, America). The morphology of the products was determined using an SEM (FESEM, S-8010, Hitachi). Fluorescence imaging was performed using a confocal laser scanning microscope (Nikon ALR, Japan). The vitro cytotoxicity assay was recorded on BioTek Epoch (BioTek Instruments, USA) at room temperature using 96-microwell plates.Synthesis of self-assembly of NWs
Each functional hairpin, H1 and H2, was diluted to 10 μM in HEPES buffer (10 mM, 100 mM NaCl, 20 mM MgCl2, pH 7.2), respectively, then heated to 95 °C for 5 min and then allowed to slowly cool down to room temperature within 2 h before use. Then, 30 μL of 4 mM ZnCl2 in Millipore water was added to 570 μL of an aqueous solution of 2.5 μM H1, and 2.5 μM H2 in 1.5 mL centrifuge tubes. After vigorous vortexing for 20 s, the mixture was kept at 95 °C. After being left undisturbed for 3 h, the resulting solution was centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min and washed twice with deionized water. The collected NWs were finally redispersed in deionized water for further use. The Fourier transform infrared (FTIR) spectroscopy and the ultraviolet absorbance spectroscopy were carried out to characterize the NW hybrid materials synthesized.
000 rpm for 15 min and washed twice with deionized water. The collected NWs were finally redispersed in deionized water for further use. The Fourier transform infrared (FTIR) spectroscopy and the ultraviolet absorbance spectroscopy were carried out to characterize the NW hybrid materials synthesized.
Gel electrophoresis verification
The resulting amplification products were recorded by performing 4% agarose gel electrophoresis. The agarose gel electrophoresis samples including HCR amplicons NWs (400 nM) and target miR-21 (T1, 100 nM) were incubated in HEPES buffer at 37 °C for 2 h. Then 10 μL of resulting amplification products was mixed with 2 μL of loading buffer and loaded into wells in an agarose gel (4%). The agarose gel electrophoresis was performed at a constant potential of 120 V in 1 × TAE buffer (40 mM Tris–acetate, 1 mM EDTA, pH 8.0–8.4) at room temperature for 50 min. After electrophoresis, the gel was photographed using a gel imaging system (Vilber Lourmat, France).In vitro fluorescence assay
The miR-21-triggered HCR strategy was verified using a fluorescence assay in vitro. The NW-based HCR reaction was carried out in HEPES buffer (10 mM, 100 mM NaCl, 20 mM MgCl2, pH 7.2). The target miR-21 was added in different concentrations into the 200 nM NW mixture solution to initiate the autonomous nonenzymatic DNA-assembled HCR reaction at 37 °C for 2 h in the HEPES buffer. The fluorescence spectrum of each sample was obtained at a fixed emission wavelength of 520 nm with excitation at 490 nm at room temperature. In the case of selectivity assay, other miRNAs were used instead of target miR-21.Cell culture
The HepG2 cells and CCC-HEL-1 cells were used in the experiments. All cell lines were cultured in the culture dishes with DMEM culture medium including 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin, and grown at 37 °C in a fully-humidified incubator containing 5% CO2.Real-time RT-PCR analysis of miRNA in living cells
The total RNA was extracted in HepG2 cells, CCC-HEL-1 cells, and HepG2 cells treated with miR-21 inhibitor oligonucleotide using a Total RNA Extractor Kit (Trizol) according to the manufacturer's instructions. cDNA was prepared by using miRNA First Strand cDNA Synthesis Kit (Sangon Biotech) according to the indicated protocol. Real-time PCR analysis was performed using the MicroRNAs qPCR Kit (Sangon Biotech). Real-time PCR amplification was performed using the ABI Quant Studio 6Flex system under the following conditions: an initial temperature of 95 °C for 30 s followed by 40 cycles at 95 °C for 5 s and 60 °C for 30 s.In vitro cytotoxicity assay
A standard CCK-8 assay was used to assess the cell toxicity using HepG2 cells. For the CCK-8 assay, HepG2 cells were seeded on a 96-well plate at a density of 1 × 104 cells per well and cultured in DMEM culture medium supplemented with 10% FBS and 1% penicillin–streptomycin at 37 °C under 5% CO2 for 12 h. Then, the DMEM medium was carefully discarded, and the HepG2 cells were washed three times with PBS. The medium was replaced with fresh cell culture medium containing different concentrations of NWs. After incubating with NWs for another 24 h, the HepG2 cells were washed and incubated with 100 μL fresh culture medium containing 10% CCK-8. After incubating for another 2 h, the absorbance of each well was measured at 450 nm using a microplate reader.Cellular uptake assay and imaging analysis
HepG2 cells were seeded in 1 mL DMEM medium in confocal microscopy Petri dishes and kept at 37 °C in a 5% CO2 atmosphere. After incubation for 24 h, the medium was carefully removed, and the miR-21-targeting NWs (400 nM) were added to fresh DMEM medium (500 μL). After incubation for predetermined time periods (1 h, 2 h and 4 h), the cells were washed three times with PBS to remove the uninternalized NPs. Then, the cells were incubated with Hoechst 33342 for 20 min at 37 °C for cell nuclei counterstain followed subsequently by three washes with PBS. Finally, the cells were mixed with 1 mL chilled methanol at −20 °C for 10 min and rinsed three times with PBS. The fluorescence imaging analysis was then performed using a confocal laser scanning microscope.Abbreviations
| NWs | Nano-wire balls |
| miRNAs | microRNAs |
| HCR | Hybridization chain reaction |
| NPs | Nanoparticles |
| FISH | Fluorescence in situ hybridization |
| miR-21 | miRNA-21 |
| BHQ1 | Black hole quencher |
| FAM | 5-Carboxyfluorescein |
| dsDNA | Double-stranded DNA |
| LOD | Limit of detection |
| RSD | Relative standard deviation |
| CCK-8 | Cell Counting Kit-8 |
| FBS | Fetal bovine serum |
| DMEM | Dulbecco's modified Eagle medium |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
J. Ouyang gratefully acknowledges the support from the National Natural Science Foundation of China (21974010 and 21675014) and the Fundamental Research Funds for the Central Universities. N. Na gratefully acknowledges the support from the National Natural Science Foundation of China (21874012 and 21675015).Notes and references
- H. Furukawa, K. E. Cordova, M. O’Keeffe and O. M. Yaghi, Science, 2013, 341, 1230444 CrossRef.
- A. Kuschel, M. Luka, M. Wessig, M. Drescher, M. Fonin, G. Kiliani and S. Polarz, Adv. Funct. Mater., 2010, 20, 1133–1143 CrossRef CAS.
- G. Du, L. Lou, S. Guan, Y. Peng, H. Qiao, P. Liu and D. Wu, J. Mater. Chem. B, 2019, 7, 4177–4183 RSC.
- H. C. Zhou, J. R. Long and O. M. Yaghi, Chem. Rev., 2012, 112, 673–674 CrossRef CAS.
- H. Liang, Z. Zhang, Q. Yuan and J. Liu, Chem. Commun., 2015, 51, 15196–15199 RSC.
- P. Mi, H. Cabral and K. Kataoka, Adv. Mater., 2020, 32, 1902604 CrossRef CAS.
- Y. Ma, X. Li, A. Li, P. Yang, C. Zhang and B. Tang, Angew. Chem., Int. Ed., 2017, 56, 13752–13756 CrossRef CAS.
- J. B. Lee, J. Hong, D. K. Bonner, Z. Poon and P. T. Hammond, Nat. Mater., 2012, 11, 316–322 CrossRef CAS.
- H. J. Vaughan, J. J. Green and S. Y. Tzeng, Adv. Mater., 2020, 32, 1901081 CrossRef CAS.
- B. Liu, J. Zhang and L. Li, Chem. – Eur. J., 2019, 25, 13452–13457 CrossRef CAS.
- S. M. Douglas, I. Bachelet and G. M. Church, Science, 2012, 335, 831–834 CrossRef CAS.
- M. Wang, D. Kong, D. Su, Y. Liu and X. Su, Nanoscale, 2019, 11, 13903–13908 RSC.
- C. Lu, Z. Huang, B. Liu, Y. Liu, Y. Ying and J. Liu, Angew. Chem., Int. Ed., 2017, 129, 6304–6308 CrossRef.
- C. Huang, J. Zheng, D. Ma, N. Liu, C. Zhu, J. Li and R. Yang, J. Mater. Chem. B, 2018, 6, 6424–6430 RSC.
- M. R. Jones, N. C. Seeman and C. A. Mirkin, Science, 2015, 347, 1260901 CrossRef.
- Y. Yang, G. Mao, X. Ji and Z. He, J. Mater. Chem. B, 2020, 8, 9–17 RSC.
- M. Wang, Y. Chen, W. Cai, H. Feng, T. Du, W. Liu, H. Jiang, A. Pasquarelli, Y. Weizmann and X. Wang, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 308–316 CrossRef CAS.
- Z. Hu, G. Sun, W. Jiang, F. Xu, Y. Zhang, M. Xia, X. Pan, Z. Xing, S. Zhang and X. Zhang, Anal. Chem., 2019, 91, 5980–5986 CrossRef CAS.
- J. An, S. J. Geib and N. L. Rosi, J. Am. Chem. Soc., 2009, 131, 8376–8377 CrossRef CAS.
- A. Lopez and J. Liu, ChemNanoMat, 2017, 3, 670–684 CrossRef CAS.
- Y. Li, Z. Liu, G. Yu, W. Jiang and C. Mao, J. Am. Chem. Soc., 2015, 137, 4320–4323 CrossRef CAS.
- M. Li, C. Wang, Z. Di, H. Li, J. Zhang, W. Xue, M. Zhao, K. Zhang, Y. Zhao and L. Li, Angew. Chem., Int. Ed., 2019, 58, 1350–1354 CrossRef CAS.
- W. Ning, Z. Di, Y. Yu, P. Zeng, C. Di, D. Chen, X. Kong, G. Nie, Y. Zhao and L. Li, Small, 2018, 14, 1703812 CrossRef.
- B. Liu, F. Hu, J. Zhang, C. Wang and L. Li, Angew. Chem., Int. Ed., 2019, 58, 8804–8808 CrossRef CAS.
- X. Gong, J. Li, T. Tan, Z. Wang, H. Wang, Y. Wang, X. Xu, Z. Zhang and Y. Li, Adv. Funct. Mater., 2019, 29, 1903441 CrossRef CAS.
- C. Chu, E. Ren, Y. Zhang, J. Yu, H. Lin, X. Pang, Y. Zhang, H. Liu, Z. Qin, Y. Cheng, X. Wang, W. Li, X. Kong, X. Chen and G. Liu, Angew. Chem., Int. Ed., 2019, 58, 269–272 CrossRef CAS.
- D. Putnam, Nat. Mater., 2006, 5, 439–451 CrossRef CAS.
- Y. Wang, S. Gao, W. H. Ye, H. S. Yoon and Y. Y. Yang, Nat. Mater., 2006, 5, 791–796 CrossRef CAS.
- M. J. Levesque, P. Ginart, Y. Wei and A. Raj, Nat. Methods, 2013, 10, 865–867 CrossRef CAS.
- X. Shen, Y. Wang, Y. Zhang, J. Ouyang and N. Na, Adv. Funct. Mater., 2018, 1803286 CrossRef.
- J. Chen, W. Yin, Y. Ma, H. Yang, Y. Zhang, M. Xu, X. Zheng, Z. Dai and X. Zou, Chem. Commun., 2018, 54, 13981–13984 RSC.
- M. Xiang, N. Li, J. Liu, R. Yu and J. Jiang, Nanoscale, 2020, 12, 8727–8731 RSC.
- L. Yang, Q. Wu, Y. Chen, X. Liu, F. Wang and X. Zhou, ACS Sens., 2019, 4, 110–117 CrossRef CAS.
- Y. Shen, T. Wu, Q. Tian, Y. Mao, J. Hu, X. Luo, Y. Ye, H. Y. Chen and J. J. Xu, Anal. Chem., 2019, 91, 7879–7886 CrossRef CAS.
- F. Yang, Y. Cheng, Y. Cao, H. Dong, H. Lu, K. Zhang, X. Meng, C. Liu and X. Zhang, Chem. Sci., 2019, 10, 1709–1715 RSC.
- J. Wang, D. Wang, J. Ma, Y. Wang and D. Kong, Chem. Sci., 2019, 10, 9758–9767 RSC.
- Y. Wang, N. Wu, F. Guo, R. Gao, T. Yang and J. Wang, J. Mater. Chem. B, 2019, 7, 7566–7573 RSC.
- X. Ren, K. Zhang, R. Deng and J. Li, Chem, 2019, 5, 2571–2592 CAS.
- L. Sheng, Y. Lu, S. Deng, X. Liao, K. Zhang, T. Ding, H. Gao, D. Liu, R. Deng and J. Li, Chem. Commun., 2019, 55, 10096–10099 RSC.
- Y. Li, X. Teng, K. Zhang, R. Deng and J. Li, Anal. Chem., 2019, 91, 3989–3996 CrossRef CAS.
- K. Zhang, R. Deng, H. Gao, X. Teng and J. Li, Chem. Soc. Rev., 2020, 49, 1932–1954 RSC.
- M. Liu, Q. Zhang, D. Chang, J. Gu, J. D. Brennan and Y. Li, Angew. Chem., Int. Ed., 2017, 129, 6238–6242 CrossRef.
- H. Wang, H. Wang, Q. Wu, M. Liang, X. Liu and F. Wang, Chem. Sci., 2019, 10, 9597–9604 RSC.
- H. M. T. Choi, V. A. Beck and N. A. Pierce, ACS Nano, 2014, 8, 4284–4294 CrossRef CAS.
- Z. Qing, J. Xu, J. Hu, J. Zheng, L. He, Z. Zou, S. Yang, W. Tan and R. Yang, Angew. Chem., Int. Ed., 2019, 58, 11574–11585 CrossRef CAS.
- Z. Cheglakov, T. M. Cronin, C. He and Y. Weizmann, J. Am. Chem. Soc., 2015, 137, 6116–6119 CrossRef CAS.
- Y. Lyu, Y. Guo, R. Cai, R. Peng, C. Hong, X. Chen, W. Hou, X. Li, J. Tan, Y. Zou, X. Zhang, Q. Liu and W. Tan, Chem, 2019, 5, 913–928 CAS.
- L. Xian, H. Ge, F. Xu, N. Xu, J. Fan, K. Shao and X. Peng, Chem. Sci., 2019, 10, 7111–7118 RSC.
- K. Zhang, S. Song, L. Yang, Q. Min, X. Wu and J. J. Zhu, Chem. Commun., 2018, 54, 13131–13134 RSC.
- H. Wang, H. Wang, Q. Wu, M. Liang, X. Liu and F. Wang, Chem. Sci., 2019, 10, 9597–9604 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0tb02315a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |