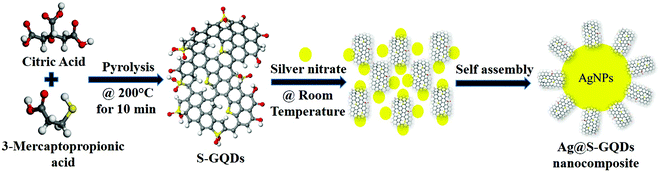
Synthesis, characterization and investigation of synergistic antibacterial activity and cell viability of silver–sulfur doped graphene quantum dot (Ag@S-GQDs) nanocomposites†
Sachin
Kadian
 a,
Gaurav
Manik
a,
Gaurav
Manik
 *a,
Neeladrisingha
Das
b,
Poonam
Nehra
c,
Rishi Pal
Chauhan
c and
Partha
Roy
b
*a,
Neeladrisingha
Das
b,
Poonam
Nehra
c,
Rishi Pal
Chauhan
c and
Partha
Roy
b
aDepartment of Polymer and Process Engineering, Indian Institute of Technology Roorkee, Uttarakhand, India. E-mail: sachinkadian68@gmail.com; skadian@pe.iitr.ac.in; manikfpt@iitr.ac.in; gauravmanik3m@gmail.com
bDepartment of Biotechnology, Indian Institute of Technology Roorkee, Uttarakhand, India
cDepartment of Physics, National Institute of Technology Kurukshetra, Haryana, India
First published on 22nd February 2020
Abstract
The excessive use of traditional antibiotic and antibacterial agents has globally increased the growth of antibiotic-resistant bacteria that poses serious health risks. Therefore, the development of new generation antibacterial or antimicrobial agents for effective inhibition of bacterial growth is highly desired. In this study, we report a facile one-step synthesis approach for the preparation of a nanocomposite composed of silver nanoparticles (AgNPs) decorated with sulfur-doped graphene quantum dots (S-GQDs). The nanocomposite was comprehensively characterized using transmission electron microscopy (TEM), X-ray diffraction (XRD), UV-vis absorption spectra, Fourier transform infrared (FTIR) spectroscopy and X-ray photoelectron spectroscopy (XPS). The characterization results demonstrated that the AgNPs were closely and uniformly surrounded by the S-GQDs, and consequently, this ensured the dispersion and stability of the so formed nanocomposite (Ag@S-GQDs). Further, the antibacterial activity of the Ag@S-GQDs nanocomposite was investigated and compared with bare S-GQDs and AgNPs against Gram-positive S. aureus (MTCC 737) and Gram-negative P. aeruginosa (MTCC 424) bacteria using macrodilution and agar well diffusion methods. Minimum inhibitory concentration (MIC) values of 70 and 35 μg mL−1 of the Ag@S-GQDs nanocomposite were found to be sufficient to hinder the growth of P. aeruginosa and S. aureus. A fractional inhibition concentration (FIC) index below 0.5 confirmed the existence of a synergistic effect between AgNPs and S-GQDs in the Ag@S-GQDs nanocomposite. In addition, the cytotoxicity of the Ag@S-GQDs nanocomposite, AgNPs and S-GQDs was also investigated using HEK 293 cell lines. Interestingly, the Ag@S-GQDs nanocomposite exhibited superior cell viability as compared to AgNPs and S-GQDs. These improved antibacterial and biocompatibility data demonstrate that the Ag@S-GQDs nanocomposite can serve as a promising antibacterial agent for industry to fabricate next-generation self-sterile textiles, antibacterial coatings and useful health care products supporting cell viability.
1. Introduction
A number of microbes are present in our surrounding environment, such as soil, air and water. Once a microbe or a bacterium adheres to a surface, it leads to the formation of a complex bacterial community enriched with antibiotic-resistant bacteria known as biofilm.1 According to a recent report from the U.S. Centers for Disease Control and Prevention, around 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 individuals suffer every year with microbial or bacterial infections that do not respond to medication due to the presence of bacteria resistant to antibiotics.2 The presence of biofilms may cause several serious diseases and hygienic problems through food pathogens involved in the food industry due to corrosion of metallic surfaces, mechanical blockage in fluid systems and disturbance in other industrial processes. Moreover, it may also cause various medical tool-associated infections.3,4 Therefore, the demand of the treatment and prevention of such diseases has drawn extensive attention towards the development of new generation antibacterial or antimicrobial agents for the effective inhibition of bacterial growth. Several antibacterial substances including tetracycline, benzyl penicillin and streptomycin have been developed to prevent bacterial infections. However, the most serious concern is that a few bacteria are easily resistant to these antibiotics and some have in due course of time developed resistance to antibiotics that have been frequently used to treat and prevent them. For instance, infections caused by Staphylococcus aureus (S. aureus) and Neisseria gonorrhoeae (N. gonorrhoeae) were generally controlled by penicillin and are now almost completely resistant to it. Therefore, it is essential to develop a next generation antimicrobial agent that can overcome the drawbacks of regular medicines and actively work against both kinds of bacterial strains. As an alternative to traditional antibacterial agents, some emerging nanomaterials including silver nanoparticles,5–8 metal oxide nanoparticles,9 nanocarbon,10 graphene and its derivatives11–14 have been introduced as novel antibacterial agents. More specifically, silver and silver-based nanocomposites have attracted significant attention since they are strong antibacterial agents and have widely served in many bactericidal applications against numerous bacteria, viruses and fungi.15,16 Although the exact mechanism behind the antibacterial activity of silver nanoparticles is not clear,17 some plausible facts, such as the inhibition of respiratory enzymes,18,19 the disruption of outer membrane15,20,21 and the denaturation of 30s ribosomal subunits,18 have been introduced to explain the same. It has been clearly reported that the bactericidal effect of silver nanoparticles is associated with their shape, size and surface charge distribution.5,22 Unfortunately, silver nanoparticles have a tendency of aggregation to minimize the surface energy during synthesis, which results in reduced antibacterial properties. To resolve this issue, it is highly desired to find a proper surface passivation or support material for silver nanoparticles to maintain their antibacterial activity with higher stability.
000 individuals suffer every year with microbial or bacterial infections that do not respond to medication due to the presence of bacteria resistant to antibiotics.2 The presence of biofilms may cause several serious diseases and hygienic problems through food pathogens involved in the food industry due to corrosion of metallic surfaces, mechanical blockage in fluid systems and disturbance in other industrial processes. Moreover, it may also cause various medical tool-associated infections.3,4 Therefore, the demand of the treatment and prevention of such diseases has drawn extensive attention towards the development of new generation antibacterial or antimicrobial agents for the effective inhibition of bacterial growth. Several antibacterial substances including tetracycline, benzyl penicillin and streptomycin have been developed to prevent bacterial infections. However, the most serious concern is that a few bacteria are easily resistant to these antibiotics and some have in due course of time developed resistance to antibiotics that have been frequently used to treat and prevent them. For instance, infections caused by Staphylococcus aureus (S. aureus) and Neisseria gonorrhoeae (N. gonorrhoeae) were generally controlled by penicillin and are now almost completely resistant to it. Therefore, it is essential to develop a next generation antimicrobial agent that can overcome the drawbacks of regular medicines and actively work against both kinds of bacterial strains. As an alternative to traditional antibacterial agents, some emerging nanomaterials including silver nanoparticles,5–8 metal oxide nanoparticles,9 nanocarbon,10 graphene and its derivatives11–14 have been introduced as novel antibacterial agents. More specifically, silver and silver-based nanocomposites have attracted significant attention since they are strong antibacterial agents and have widely served in many bactericidal applications against numerous bacteria, viruses and fungi.15,16 Although the exact mechanism behind the antibacterial activity of silver nanoparticles is not clear,17 some plausible facts, such as the inhibition of respiratory enzymes,18,19 the disruption of outer membrane15,20,21 and the denaturation of 30s ribosomal subunits,18 have been introduced to explain the same. It has been clearly reported that the bactericidal effect of silver nanoparticles is associated with their shape, size and surface charge distribution.5,22 Unfortunately, silver nanoparticles have a tendency of aggregation to minimize the surface energy during synthesis, which results in reduced antibacterial properties. To resolve this issue, it is highly desired to find a proper surface passivation or support material for silver nanoparticles to maintain their antibacterial activity with higher stability.
More recently, carbon-based nanostructures including carbon nanotubes (CNTs),23,24 graphene nanosheets,16 graphene oxide (GO),15,25 reduced graphene oxide (rGO),26 carbon dots (CDs)27 and graphene quantum dots (GQDs)21 have emerged as useful nanomaterials to synthesize silver nanocomposites with enhanced antibacterial effects. However, CNTs, GO and rGO have been described as carcinogenic materials with toxic effects against mammalian cells.25,28 Subsequently, GQDs have not shown observable toxicity and have been used in cellular imaging as well as in drug delivery applications.29–32 Recently, Habiba et al. have studied the antibacterial activity and cytotoxicity of PEGylated Ag–GQD nanocomposites.21 It is remarkable that different reducing agents, such as NaBH4, tert-butylamine, sodium citrate and citric acid, have been used to synthesize AgNPs first, and then AgNPs are grown onto the surface of the graphene-based nanostructure in the presence of thiol compounds to synthesize the Ag–GQD nanocomposites.21 In comparison to bare GQDs, the heteroatom doped GQDs can provide more active sites on their surface and better chemical interaction with AgNPs. For instance, Tian et al. have reported an environment-friendly method for the preparation of AgNPs grown on the surface of N-doped graphene.33 Liu et al. introduced the reductive activity of nitrogen-doped carbon nanodots to synthesize Ag nanoparticles for the electrochemical detection of hydrogen peroxide.34 This new strategy was identified to be effective for the easy synthesis of Ag/GQD nanocomposites with enhanced properties. Therefore, one step synthesis of Ag@S-GQDs can be easily achieved by doping GQDs with sulfur atoms because of the strong interaction between S and Ag atoms. Moreover, to the best of our knowledge, so far, there are no reports on the use of S-GQDs as reducing agents to synthesize Ag@S-GQDs nanocomposites for antibacterial applications and cytotoxicity assay.
In this work, we have synthesized a Ag@S-GQDs nanocomposite using a one-step synthesis process and their antibacterial as well as cytotoxicity properties were systematically investigated. As shown in Scheme 1, S-GQDs were synthesized by the simple pyrolysis of citric acid and 3-mercaptopropionic acid and then AgNPs were grown in situ on the surface of S-GQDs to prepare the Ag@S-GQDs nanocomposite at room temperature without an additional reducing agent. The as-synthesized nanocomposite was comprehensively characterized and its antibacterial properties investigated against Gram-positive bacteria, S. aureus, and Gram-negative bacteria, Pseudomonas aeruginosa (P. aeruginosa). The obtained antibacterial properties of the Ag@S-GQDs nanocomposite were compared with those of bare AgNPs and S-GQDs to explore a synergistic effect among AgNPs and S-GQDs. In addition, the cytotoxicity of the Ag@S-GQDs nanocomposite was also examined in HEK 293 cells and compared with S-GQDs and AgNPs.
2. Experimental section
2.1 Material and reagents
3-Mercaptopropionic acid (MPA) and citric acid (CA) were obtained from Sigma-Aldrich. Sodium borohydride (NaBH4) and silver nitrate (AgNO3) were procured from Merck India. Sodium dodecyl sulphate (SDS) was purchased from SRL Chemical India. The test bacteria, P. aeruginosa (MTCC 424) and S. aureus (MTCC 737), were obtained from the Institute of Microbial Technology (IMTECH), Chandigarh, India. The nutrient broth (NB) medium was procured from HiMedia (Mumbai, India). The cell lines (HEK 293) used in this study were purchased from the National Centre for Cell Science (NCCS), Pune, India. Dulbecco's Modified Eagle Medium High Glucose (DMEM-HG), cell culture media, dimethylsulphoxide (DMSO, cell culture grade) and MTT (3-(4,5-dimethyl-2-thiazolyl) 2,5diphenyl-2H-tetrazoliumbromide) were acquired from HiMedia (Mumbai, India). Penicillin–streptomycin antibiotics and Fetal Bovine Serum (FBS) were procured from GIBCO (Life Technologies, Massachusetts, USA). All reagents were of analytical grade and used as received. Fresh Millipore water was used to prepare all the solutions.2.2 Synthesis of sulfur-doped graphene quantum dots (S-GQDs)
S-GQDs were produced using our previously reported bottom-up approach by the simple pyrolysis of MPA and CA with minor modifications.35 Briefly, 300 μL of MPA and 2.1 g of CA were pyrolyzed at 200 °C for 10 minutes. The obtained orange colored liquid was mixed into 50 mL of freshly prepared NaBH4 solution (10 mg mL−1) under vigorous stirring overnight. Next, the pH of the mixture was maintained at 7.0 followed by filtration through a syringe filter having a pore size of 0.22 μm to eliminate the large particles and residuals. The obtained product was preserved at 4 °C for further characterization and assessment.2.3 Synthesis of silver nanoparticles (AgNPs)
Silver nanoparticles were synthesized using the simple chemical reduction of silver nitrate. For this 80 μL of 0.1 M AgNO3 was mixed with 50 mL of deionized water. Freshly prepared NaBH4 solution (2 mg mL−1) was immediately added into the obtained mixture followed by addition of the capping agent sodium dodecyl sulphate (SDS) to control the aggregation of the synthesized nanoparticles. The resulting solution was allowed to stir till the formation of a pale-yellow color solution, signifying the formation of AgNPs. The as-synthesized AgNPs were stored at 4 °C for further use and characterization.2.4 Preparation of Ag@S-GQDs nanocomposite
For the preparation of Ag@S-GQDs, a one-step in situ synthesis approach has been employed. In this method, the as-synthesized S-GQDs were used as stabilizers and reducing agents to reduce AgNO3 and form the nanocomposite. For this, 66 μL of 0.1 M AgNO3 was added into 10 mL of a S-GQD solution and left under overnight stirring to complete the reaction. Formation of a pale-yellow color solution indicated the formation of the Ag@S-GQDs nanocomposite. After neutralization at a pH of 7.0, the resultant mixture, i.e. the Ag@S-GQDs nanocomposite, was filtered through a syringe filter (pore size 0.45 μm) to eliminate the large residuals or particles. For further elimination of unreacted silver ions and to obtain the pure Ag@S-GQDs nanocomposite, the resultant solution was subjected to dialysis against water. The final solution was kept in a refrigerator for further experiments and characterization.2.5 Characterization and instrumentation
X-ray diffraction (XRD) patterns were obtained on a Bruker D8-Advance X-ray diffractometer at an accelerating potential of 40 kV with a Cu Kα radiation source. Surface morphology and particle size of S-GQDs, AgNPs and the Ag@S-GQDs nanocomposite were evaluated via transmission electron microscopy (TEM). The optical properties assessed through UV-visible spectra of S-GQDs, AgNPs and the Ag@S-GQDs nanocomposite were monitored within the wavelength range from 200 to 800 nm using a Cary 5000 UV-Vis-NIR spectrometer. The Fourier transform infrared (FTIR) spectra were determined in the range of 4000 to 500 cm−1 using a PerkinElmer C91158 spectrometer, UK. In addition, elemental composition analysis was accomplished via X-ray photoelectron spectroscopy (XPS, Physical Electronics, PHI 5000 Versa Probe III).2.6 Antibacterial test
The antibacterial activities of the as-prepared S-GQDs, AgNPs and Ag@S-GQDs nanocomposite were examined using agar well diffusion and macrodilution methods against two bacterial strains, namely P. aeruginosa (MTCC 424) and S. aureus (MTCC 737). Initially, P. aeruginosa (Gram-negative) and S. aureus (Gram-positive) were inoculated in a logarithmic phase from an overnight culture in their desired growth medium at 37 °C. The bacterial concentration was calculated through optical density (OD) measurements at 600 nm.2.7 Cell culture
The HEK 293 cell lines were cultured in DMEM-HG supplemented with 10% FBS and 1% antibiotic (100 U mL−1 penicillin and 100 μg mL−1 streptomycin) at 37 °C in a humidified incubator with 5% CO2 supply.3. Results and discussion
3.1 Characterization of Ag@S-GQDs nanocomposite
To validate the successful synthesis of S-GQDs, AgNPs and the Ag@S-GQDs nanocomposite, all samples were comprehensively characterized using different techniques. The size distribution pattern and surface morphology of freshly prepared S-GQDs, AgNPs and Ag@S-GQDs nanocomposite were first characterized using TEM, as shown in Fig. 1. The TEM image demonstrated in Fig. 1a indicates that the as-synthesized S-GQDs were uniformly dispersed and spherical in shape. The size distribution histogram (inset, Fig. 1a) reflects a narrow size distribution of S-GQDs ranging between ∼2.5 nm and 5.5 nm with an estimated average diameter of about 4 nm. The inset image of Fig. 1a depicts a fine fringe pattern with a lattice spacing of 0.35 nm, corresponding to the (002) plane of graphene.33,40Fig. 1b and the inset display the TEM image and the selected-area electron diffraction (SAED) pattern of the as-prepared AgNPs. It can be observed that the particles were spherical in shape with varying size ranging from ∼18 nm to ∼25 nm (shown in the histogram). The average diameter of AgNPs was estimated to be 22 ± 1.76 nm. The SAED pattern of AgNPs comprises four characteristic diffraction rings corresponding to the (111), (200), (220) and (300) planes, clearly indicating the crystalline structure of the as-prepared AgNPs. Further, Fig. 1c and d show the TEM images of the as-prepared Ag@S-GQDs nanocomposite. It can be seen that the particles shown with yellow color arrows correspond to S-GQDs and decorate the surface of AgNPs, and ensure the stability and dispersity of the Ag@S-GQDs nanocomposite. The visible attachment of some S-GQDs on the surface of the AgNPs suggests the formation of the Ag@S-GQDs nanocomposite, as shown in Scheme 1. Interestingly, the size distribution histogram of the Ag@S-GQDs nanocomposite demonstrates a significant change in the average particle size (from 22 ± 1.76 nm to 28 ± 1.16 nm) as compared to the bare AgNPs, preliminarily confirming the successful synthesis of the Ag@S-GQDs nanocomposite.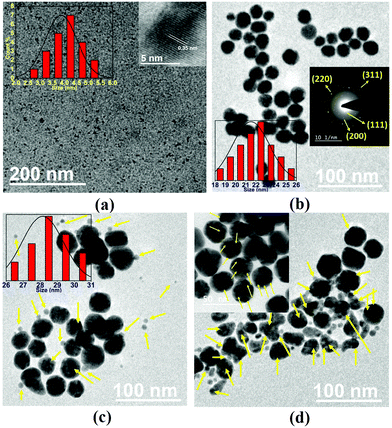 | ||
| Fig. 1 TEM image of the as-synthesized (a) S-GQDs, (b) AgNPs, and (c) the Ag@S-GQDs nanocomposite at 100 nm scale and (d) the Ag@S-GQDs nanocomposite at 100 nm with the inset image at 50 nm scale. | ||
To further confirm the formation of S-GQDs, AgNPs and the Ag@S-GQDs nanocomposite and verify the TEM results, XRD patterns were recorded to analyze the crystalline nature of their structures, as demonstrated in Fig. 2a. The XRD pattern of S-GQDs (Fig. 2a) shows a broad peak centered at 2θ = 24.4°, assigned to the (002) plane of graphene and is consistent with the corresponding TEM results discussed above. For AgNPs, the XRD peaks at 2θ = 39.8, 46.6, 68.1 and 77.6 correspond to the characteristic crystallographic planes of (111), (200), (220) and (311) of silver nanoparticles, respectively, and are in good agreement with JCPDS card no. 04-0783.33 Moreover, all these characteristic peaks observed in the XRD patterns are also in agreement with the specific diffraction rings obtained in the SAED patterns of the AgNPs, thereby confirming the synthesis of AgNPs. Further, the XRD pattern of the Ag@S-GQDs nanocomposite comprises all the signature peaks of the AgNPs as well as a broad peak corresponding to the S-GQDs, confirming the obvious presence of both AgNPs and S-GQDs in the structure of the Ag@S-GQDs nanocomposite. All these results of the XRD patterns again confirm the successful formation of S-GQDs, AgNPs and the Ag@S-GQDs nanocomposite and are consistent with the TEM results.
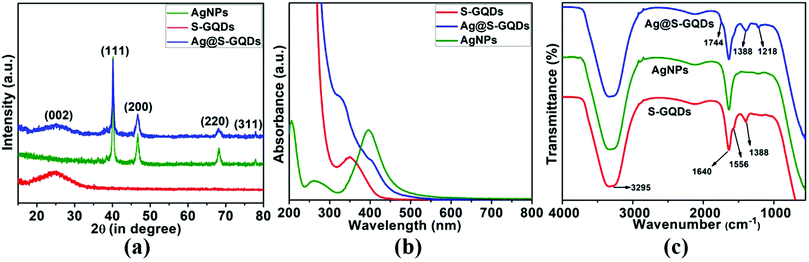 | ||
| Fig. 2 Illustration of (a) XRD patterns, (b) UV-vis absorption spectra and (c) FTIR spectra of S-GQDs (red lines), AgNPs (green lines) and the Ag@S-GQDs nanocomposite (blue lines). | ||
The UV-vis absorption spectra of S-GQDs, AgNPs and the Ag@S-GQDs nanocomposite were also recorded to investigate their optical properties, as shown in Fig. 2b. The UV-Vis absorbance spectrum of S-GQDs demonstrates a characteristic absorption band at ∼360 nm ascribed to the electronic transition (n–π*) of the existing C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups, signifying that the present oxygen-containing functional groups offer anchors for further chemical reaction and excellent aqueous dispersity.35 Next, the UV-vis absorbance spectrum of AgNPs was found to display a typical absorbance band at ∼396 nm attributed to the localized surface plasmon of spherical silver nanoparticles as reported previously, which ascertains the formation of AgNPs.41 Further, the synthesis of the Ag@S-GQDs nanocomposite was confirmed by multiple shifted shoulders at ∼405 nm (due to the characteristic band of AgNPs) and ∼332 nm (due to electronic transitions (n–π*) of C
O groups, signifying that the present oxygen-containing functional groups offer anchors for further chemical reaction and excellent aqueous dispersity.35 Next, the UV-vis absorbance spectrum of AgNPs was found to display a typical absorbance band at ∼396 nm attributed to the localized surface plasmon of spherical silver nanoparticles as reported previously, which ascertains the formation of AgNPs.41 Further, the synthesis of the Ag@S-GQDs nanocomposite was confirmed by multiple shifted shoulders at ∼405 nm (due to the characteristic band of AgNPs) and ∼332 nm (due to electronic transitions (n–π*) of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups present in S-GQDs). This noticeable shift in the absorption peak of Ag@S-GQDs from 396 to 405 nm clearly indicates the increase in particle size of the nanocomposite, which is consistent with the TEM image results where the size of Ag@S-GQDs increased from ∼22 nm to 28 nm. In addition, the shift in the absorption band from 360 to 332 nm might be ascribed to the active participation of the oxygen containing group present in the SGQD structure in the formation of AgNPs, representing the occurrence of relatively less n–π* transition. All these results support the successful synthesis of highly dispersible S-GQDs, AgNPs and the Ag@S-GQDs nanocomposite and are in agreement with the dispersibility and change in size observed in the TEM characterization results.
O groups present in S-GQDs). This noticeable shift in the absorption peak of Ag@S-GQDs from 396 to 405 nm clearly indicates the increase in particle size of the nanocomposite, which is consistent with the TEM image results where the size of Ag@S-GQDs increased from ∼22 nm to 28 nm. In addition, the shift in the absorption band from 360 to 332 nm might be ascribed to the active participation of the oxygen containing group present in the SGQD structure in the formation of AgNPs, representing the occurrence of relatively less n–π* transition. All these results support the successful synthesis of highly dispersible S-GQDs, AgNPs and the Ag@S-GQDs nanocomposite and are in agreement with the dispersibility and change in size observed in the TEM characterization results.
Further, FTIR spectra were also recorded to examine the successful formation of S-GQDs, AgNPs and the Ag@S-GQDs nanocomposite in the presence of different surface functional groups. All compounds displayed a broad and intense band centered at 3295 cm−1, ascribed to the stretching vibration of the –OH group, as demonstrated in Fig. 2c. This intense band also represents the remarkable hydrophilicity of all compounds due to the occurrence of –OH groups on their surface.35,42,43 A strong intensity band at 1640 cm−1 in all compounds indicates the presence of carboxylic moieties and a new shoulder peak at 1744 cm−1 in the Ag@S-GQDs spectra is attributed to the interaction of AgNPs with oxygen-containing groups of S-GQDs.44 The peaks at 1556 cm−1 and 1388 cm−1 are ascribed to the bending of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and stretching vibrations of S
C and stretching vibrations of S![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, respectively.35 It is well described in the literature that a spectral feature ranging between 1400 and 1050 cm−1 is observed because of sulfate, sulfone and sulfide stretching, ascribed to the existing S atom in S-GQDs.45 Another peak observed in the FTIR spectra of the Ag@S-GQDs nanocomposite at 1218 cm−1 may be assigned to the S related bond (i.e., the presence of Ag–S bonds), which also supports the formation of the nanocomposite.45 The occurrence of well-defined characteristic peaks related to different surface functional groups (hydroxyl, carboxylic, sulfide, sulfones and sulfate) in the FTIR spectra confirms the completion of synthesis and ascertains the hydrophilic nature of S-GQDs, AgNPs and the Ag@S-GQDs nanocomposite.
O, respectively.35 It is well described in the literature that a spectral feature ranging between 1400 and 1050 cm−1 is observed because of sulfate, sulfone and sulfide stretching, ascribed to the existing S atom in S-GQDs.45 Another peak observed in the FTIR spectra of the Ag@S-GQDs nanocomposite at 1218 cm−1 may be assigned to the S related bond (i.e., the presence of Ag–S bonds), which also supports the formation of the nanocomposite.45 The occurrence of well-defined characteristic peaks related to different surface functional groups (hydroxyl, carboxylic, sulfide, sulfones and sulfate) in the FTIR spectra confirms the completion of synthesis and ascertains the hydrophilic nature of S-GQDs, AgNPs and the Ag@S-GQDs nanocomposite.
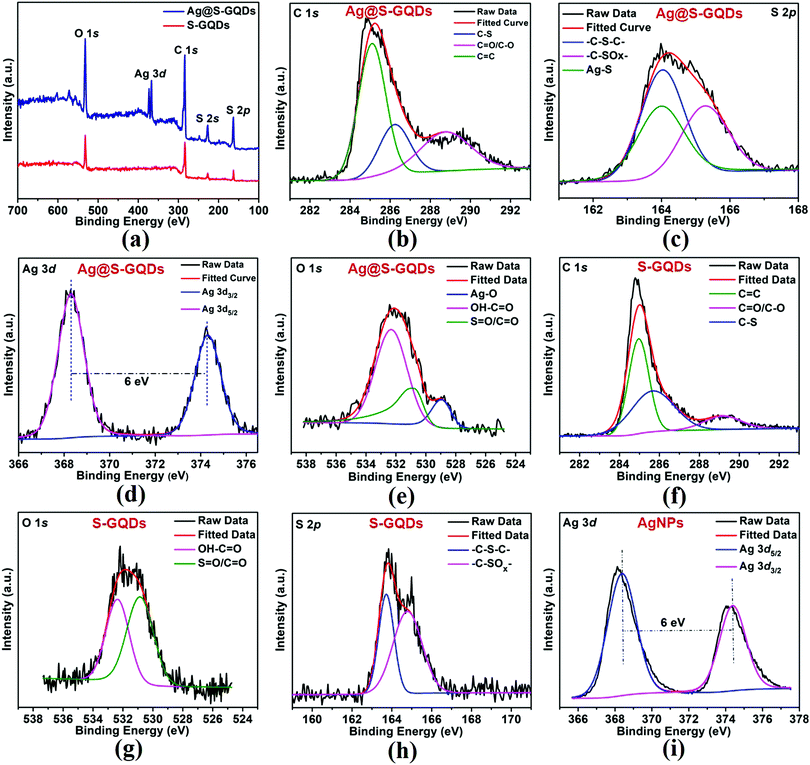
To additionally verify the results of FTIR spectra, XPS spectroscopy was used to characterize the elemental composition of S-GQDs and the Ag@S-GQDs nanocomposite. The full scan spectrum of S-GQDs (Fig. 3a) comprises four prominent peaks of C 1s, O 1s, S 2s and S 2p positioned at binding energies of 284, 531, 228 and 164 eV, respectively. After the formation of the Ag@S-GQDs nanocomposite, two new peaks at a binding energy of ∼368 and 374 eV were observed corresponding to Ag 3d5/2 and Ag 3d3/2, respectively. The chemical bonding of C, S and Ag was characterized by deconvoluting the C 1s, S 2p and Ag 3d spectra. The deconvoluted high-resolution C 1s spectrum of the Ag@S-GQDs nanocomposite (Fig. 3b) indicates three types of C atoms located at 285.07 eV (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 286.3 eV (C–S) and 288.8 eV (C
C), 286.3 eV (C–S) and 288.8 eV (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O/C–O) binding energy.35,42,46 The presence of C
O/C–O) binding energy.35,42,46 The presence of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O/C–O and C
O/C–O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C peaks justifies that the nanocomposites are enriched with carboxyl and carbonyl groups on their surface. In addition, the existence of the C–S peak reveals the successful incorporation of S atoms in the structure of S-GQDs. The deconvoluted S 2p spectra of the Ag@S-GQDs nanocomposite (Fig. 3c) show three types of S atoms with peaks located at 165.2 eV (–C–SOx–), 164.09 eV (–C–S–C–), and 163.9 eV (Ag–S) binding energy.35,47–49 On the other side, the deconvoluted S 2p spectra of S-GQDs (Fig. 3h) show only two peaks located at 164.84 eV (–C–SOx–) and 163.76 eV (–C–S–C–) binding energy. The occurrence of (–C–SOx–) and (–C–S–C–) peaks clearly indicates that S atoms are present in the framework of S-GQDs while another peak at 163.9 eV (Ag–S) demonstrates the participation of the S atom in the Ag–S bond formation that results in the successful synthesis of the nanocomposite.
C peaks justifies that the nanocomposites are enriched with carboxyl and carbonyl groups on their surface. In addition, the existence of the C–S peak reveals the successful incorporation of S atoms in the structure of S-GQDs. The deconvoluted S 2p spectra of the Ag@S-GQDs nanocomposite (Fig. 3c) show three types of S atoms with peaks located at 165.2 eV (–C–SOx–), 164.09 eV (–C–S–C–), and 163.9 eV (Ag–S) binding energy.35,47–49 On the other side, the deconvoluted S 2p spectra of S-GQDs (Fig. 3h) show only two peaks located at 164.84 eV (–C–SOx–) and 163.76 eV (–C–S–C–) binding energy. The occurrence of (–C–SOx–) and (–C–S–C–) peaks clearly indicates that S atoms are present in the framework of S-GQDs while another peak at 163.9 eV (Ag–S) demonstrates the participation of the S atom in the Ag–S bond formation that results in the successful synthesis of the nanocomposite.
Furthermore, the deconvoluted high-resolution Ag 3d spectra of the Ag@S-GQDs nanocomposite (Fig. 3d) demonstrate typical doublet peaks at 368.30 and 374.29 eV attributed to Ag 3d5/2 and Ag 3d3/2, depicting the successful reduction of Ag+ into AgNPs by S-GQDs. The binding energy splitting of the 3d doublet (shown in Fig. 3d) was found to be ∼6 eV, signifying the metallic nature of existing AgNPs in the nanocomposite.50 In contrast, the deconvoluted high-resolution Ag 3d spectra of AgNPs (Fig. 3i) show a doublet peak at a binding energy 368.38 eV and 374.38 eV attributed to Ag 3d5/2 and Ag 3d3/2, portraying the successful reduction of Ag+ into AgNPs. It has been reported in the literature that the core levels shift towards higher binding energies as the size of AgNPs decreases.51,52 This small shift of 0.08 eV between the binding energy peaks in the Ag 3d spectra of AgNPs and Ag@S-GQDs also supports the change in the diameter of AgNPs synthesized by the standard method and AgNPs synthesized by S-GQDs, as shown in the TEM image and the size distribution pattern (Fig. 1). Next, the deconvoluted O 1s spectra of the Ag@S-GQDs nanocomposite (Fig. 3e) indicate three bands at 532.34 eV (OH–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 530.84 eV (S
O), 530.84 eV (S![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O/C
O/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) and 528.94 eV (Ag–O) binding energy, whereas the deconvoluted O 1s spectra of S-GQDs (Fig. 3g) show only two bands centered at 532.35 eV (OH–C
O) and 528.94 eV (Ag–O) binding energy, whereas the deconvoluted O 1s spectra of S-GQDs (Fig. 3g) show only two bands centered at 532.35 eV (OH–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) and 530.85 eV (S
O) and 530.85 eV (S![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O/C
O/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) binding energy.53–56 It can be observed that the band at 530.84 eV (S
O) binding energy.53–56 It can be observed that the band at 530.84 eV (S![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O/C
O/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) in the O 1s spectra of Ag@S-GQDs diminished as compared to 530.85 eV (S
O) in the O 1s spectra of Ag@S-GQDs diminished as compared to 530.85 eV (S![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O/C
O/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) for S-GQDs, suggesting the active participation of oxygen containing groups present in the structure of S-GQDs for the construction of AgNPs. This active participation of the oxygen containing group present in the structure of S-GQDs was also noticed through a blue shift (360 to 332 nm) in the UV-vis absorption band of S-GQDs. In addition, the new band at 528.94 eV (Ag–O) in the O 1s spectra of Ag@S-GQDs indicates the interaction of AgNPs with oxygen-containing groups of S-GQDs.53,54 Interestingly, all these results are in accordance with the FTIR spectra of S-GQDs and the Ag@S-GQDs nanocomposite. The aforementioned characterization results enable us to conclude that the AgNPs were bounded by S atoms of S-GQDs through a Ag–S linkage, which resulted in the formation of the Ag@S-GQDs nanostructure. Since carbon is a well-known reducing agent for Ag+ ions, the reduced silver species were self-assembled into AgNPs to construct the Ag@S-GQDs nanocomposite. This chemistry of AgNPs and S-GQDs has also been displayed in a schematic diagram (Scheme 1). Consequently, it can be concluded that the Ag@S-GQDs nanocomposite has been synthesized successfully and can be used further for antibacterial applications.
O) for S-GQDs, suggesting the active participation of oxygen containing groups present in the structure of S-GQDs for the construction of AgNPs. This active participation of the oxygen containing group present in the structure of S-GQDs was also noticed through a blue shift (360 to 332 nm) in the UV-vis absorption band of S-GQDs. In addition, the new band at 528.94 eV (Ag–O) in the O 1s spectra of Ag@S-GQDs indicates the interaction of AgNPs with oxygen-containing groups of S-GQDs.53,54 Interestingly, all these results are in accordance with the FTIR spectra of S-GQDs and the Ag@S-GQDs nanocomposite. The aforementioned characterization results enable us to conclude that the AgNPs were bounded by S atoms of S-GQDs through a Ag–S linkage, which resulted in the formation of the Ag@S-GQDs nanostructure. Since carbon is a well-known reducing agent for Ag+ ions, the reduced silver species were self-assembled into AgNPs to construct the Ag@S-GQDs nanocomposite. This chemistry of AgNPs and S-GQDs has also been displayed in a schematic diagram (Scheme 1). Consequently, it can be concluded that the Ag@S-GQDs nanocomposite has been synthesized successfully and can be used further for antibacterial applications.
3.2 Antibacterial activity assay
The potential of the as-synthesized Ag@S-GQDs nanocomposite as a next-generation antibacterial agent was examined at several concentrations against S. aureus and P. aeruginosa using the macrodilution method (Fig. S1, ESI†). The investigation also included different concentrations of S-GQDs and AgNPs to compare and determine the antibacterial activity exhibited by individual components of the nanocomposite. Initially, the macrodilution method was employed to evaluate the antibacterial behavior and to calculate the MIC values followed by the calculation of FIC index values. The obtained MIC values of S-GQDs, AgNPs and Ag@S-GQDs needed to inhibit S. aureus and P. aeruginosa are summarized in Table 1. It can be seen from Table 1 that the MIC values of 70 and 35 μg mL−1 of the Ag@S-GQDs nanocomposite are sufficient to avert the growth of P. aeruginosa and S. aureus, respectively. Comparatively, the corresponding MIC values of S-GQDs and AgNPs were found to be much higher and more than 750 and 230 μg mL−1, respectively. This significant enhancement in the MIC values enables us to conclude that the Ag@S-GQDs nanocomposite has superior antibacterial behavior over S-GQDs and AgNPs. Next, to understand any possible synergy between S-GQDs and AgNPs in the Ag@S-GQDs nanocomposite, FIC index values were also calculated using the MIC values of 750 and 230 μg mL−1 as the lower bound for S-GQDs and AgNPs, respectively. The calculated upper bound of FIC index values (0.35 for P. aeruginosa and 0.20 for S. aureus) was found below 0.5, thereby signifying the presence of a synergistic effect between S-GQDs and AgNPs.21,37 A comparison between previously reported antibacterial nanomaterials and our synthesized nanocomposite is demonstrated in Table 2. The MIC results shown in Table 2 suggest that our prepared Ag@S-GQDs nanocomposite exhibited superiority over many previously reported antibacterial agents and appears comparable with the best performing ones. Hence, it could be inferred that the Ag@S-GQDs nanocomposite may serve as a next-generation antibacterial agent for various biomedical applications.| Samples tested | MIC (μg mL−1) | |
|---|---|---|
| S. aureus | P. aeruginosa | |
| S-GQDs | >750 | >750 |
| AgNPs | >230 | >230 |
| Ag@S-GQDs | 35 | 70 |
Furthermore, the agar well diffusion method (Fig. S2, ESI†) was also used to assess the antibacterial effectiveness of the Ag@S-GQDs nanocomposite and to validate the results of the macrodilution method. The measured zone of inhibition diameter shown by the diffusion of S-GQDs, AgNPs and the Ag@S-GQDs nanocomposite at the same concentration (70 μg mL−1) is summarized in Table 3. It can be observed from Table 3 that only the Ag@S-GQDs nanocomposite hindered the growth of both Gram-positive and Gram-negative bacteria, while no inhibition was shown by S-GQDs and AgNPs at a concentration (70 μg mL−1) lower than their MIC values. Since no inhibition zone was observed in the case of AgNPs and SGQDs, statistical analysis is not needed to be performed for these two compounds. For the case of Ag@S-GQDs, the statistical analysis of the inhibition zone obtained for both bacteria is performed using Student's t-test. The p-value between S. aureus and P. aeruginosa was estimated to be 0.0046, which is <0.01, revealing a significant difference in efficacy between the two bacterial strains. These results confirm that the Ag@S-GQDs nanocomposite may serve as a better antibacterial agent as compared to S-GQDs and AgNPs due to the existing synergistic effect and this conforms with the observations of the macrodilution method.
| Samples tested | Inhibition zone (mm) | |
|---|---|---|
| S. aureus | P. aeruginosa | |
**![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Represents the p-value (0.0046) <0.01. Represents the p-value (0.0046) <0.01. |
||
| S-GQDs | — | — |
| AgNPs | — | — |
| Ag@S-GQDs | 19.4 ± 0.635 | 15.1 ± 0.404** |
3.3 Possible antibacterial mechanism
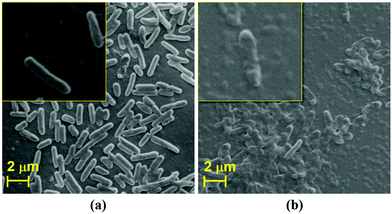
It is clearly evident from the above discussed comparison and FIC index results that the antibacterial response of the as-synthesized Ag@S-GQDs nanocomposite is stronger than that of S-GQDs and AgNPs due to the existing synergistic effect of the S-GQDs and AgNPs. In order to comprehend the existence of a synergistic effect, it is quite necessary to discuss the feasible interaction among Ag@S-GQDs and bacteria. Although the exact mechanism of antimicrobial activity of silver–carbon nanocomposites is not fully elucidated, several possible interactions between bacteria and silver–carbon nanocomposites (including Ag–graphene, Ag–GO, Ag–CNTs and Ag–GQDs) have been reported to describe the inhibition mechanism.11,12,21,24 It has been suggested that the binding of Ag+ ions of Ag–CNT nanocomposites on the cellular surface generates holes and permits the composite to penetrate the bacterial cell wall.24 It has been reported that damage to the bacterial cell wall might also be possible due to the irregularities generated by silver–carbon nanocomposites in some physiological processes associated with the outer membrane of bacteria.62 Some reports suggest that this damage of the bacterial cell wall may be attributed to the oxidative stress caused by oxygen radicals or the sharp edge of graphene, GO, CNTs, rGO and GQD sheets present in the respective silver–carbon nanocomposite.11,12,21,31,63 In our case, shape distortion and physical damage to P. aeruginosa bacteria were observed when treated with Ag@S-GQDs, as shown in Fig. 4. It can be observed from the SEM image that the surface of untreated bacteria (Fig. 4a and inset) was smooth and intact like a native cell, whereas Fig. 4b and the inset depict the shape distortion and fragmentation of bacteria after treatment with the Ag@S-GQDs nanocomposite. Recently, Habiba et al. also reached a similar conclusion and reported that fragmentation and deformation processes can physically damage the bacteria when treated with Ag–GQDs. Akhavan et al. also conveyed that the direct contact of sharp edge materials could damage the bacterial cell membrane. Another report suggests that silver–carbon nanocomposites effectively stabilize the AgNPs and avert them from aggregation, which highly affects the antibacterial activity of AgNPs.64 Hence, the above discussed mechanisms individually or jointly may take place and result in an increased antibacterial activity of the Ag@S-GQDs nanocomposite.3.4 Cytotoxicity assay
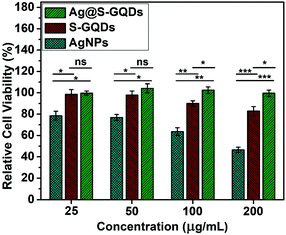
Biocompatibility plays an important role in evaluating the potential of new materials for their usage in various biomedical applications. Therefore, the biocompatibility assay of all compounds (S-GQDs, AgNPs and Ag@S-GQDs) was performed on HEK 293 cell lines, as shown in Fig. 5. Different concentrations (25, 50, 100 and 200 μg mL−1) of SGQDs, AgNPs and the Ag@S-GQDs nanocomposite were supplemented in the wells of 96-well plates containing (5 × 103 cells) HEK 293 cells and the effect was observed after 24 hours. It was found that the viability of cells reduced by about 21.4, 1.54 and 0.35% in response to the lowest concentration (25 μg mL−1) of AgNPs, S-GQDs and Ag@S-GQDs nanocomposite, respectively. Interestingly, the viability of cells treated with the Ag@S-GQDs nanocomposite was found to be almost 100% indicating no cell death while both AgNPs and S-GQDs showed significant cell death albeit with varying potencies (p < 0.05). Therefore, it can be concluded that AgNPs exhibited high toxicity to HEK 293 cells as compared to the Ag@S-GQDs nanocomposite even at lower concentrations after 24 hours of incubation. This improvement in the cell viability of the Ag@S-GQD nanocomposite could be ascribed to almost no availability of toxic NaBH4 in comparison to that in AgNPs and S-GQDs. Such a pattern of cytotoxicity along with antibacterial activities suggests that the Ag@S-GQDs nanocomposite can be an effective next-generation antibacterial agent as compared to AgNPs.4. Conclusions
In summary, a new biocompatible Ag@S-GQDs nanocomposite consisting of AgNPs uniformly decorated by S-GQDs with superior antibacterial properties has been synthesized successfully. The characterization results demonstrated that the uniform surface decoration of AgNPs with S-GQDs ensured the stability and dispersibility of the Ag@S-GQDs nanocomposite. The as-prepared Ag@S-GQDs nanocomposite exhibited enhanced antibacterial activity as compared to AgNPs and S-GQDs with MIC values as low as 70 and 35 μg mL−1 to inhibit the growth of P. aeruginosa and S. aureus model bacterial strains, respectively. The FIC index result of less than 0.5 demonstrated that the surface passivation of AgNPs with S-GQDs encourages a synergistic effect between AgNPs and S-GQDs in inhibiting bacterial growth. In addition, the Ag@S-GQDs nanocomposite revealed superior cell viability for HEK 293 cell lines in comparison to bare AgNPs and S-GQDs. These enhanced antibacterial and cell viability results co-jointly allow us to conclude that the Ag@S-GQDs nanocomposite can serve as a next-generation antibacterial agent for various biomedical applications like the fabrication of wound dressing textiles, antibacterial coatings, disinfectants and practical health care products. Additional studies are still required in order to elucidate the exact mechanism behind the bacterial cytotoxicity of the Ag@S-GQDs nanocomposite.Conflicts of interest
The author declares no conflicts of interest.Acknowledgements
The first author gratefully acknowledges the Ministry of Human Resources and Development (MHRD), Govt. of India, for providing the fellowship. The authors duly acknowledge the staff of the Institute Instrumentation Centre (IIC) and the Centre of Nanotechnology, IIT Roorkee for providing the facilities for characterization.References
- W. Shao and Q. Zhao, Colloids Surf., B DOI:10.1016/j.colsurfb.2009.10.018.
- US Department of Health and Human Services, Centers for Disease Control and Prevention, Antibiotic Resistance Threats in the United States, 2013.
- P. Madhavan, P. Y. Hong, R. Sougrat and S. P. Nunes, ACS Appl. Mater. Interfaces DOI:10.1021/am505594c.
- M. C. Wu, A. R. Deokar, J. H. Liao, P. Y. Shih and Y. C. Ling, ACS Nano DOI:10.1021/nn304782d.
- S. Agnihotri, S. Mukherji and S. Mukherji, RSC Adv. 10.1039/c3ra44507k.
- N. V. Ayala-Núñez, H. H. Lara Villegas, L. Del Carmen Ixtepan Turrent and C. Rodríguez Padilla, Nanobiotechnology DOI:10.1007/s12030-009-9029-1.
- M. Guzman, J. Dille and S. Godet, Nanomedicine, 2012, 37–45, DOI:10.1016/j.nano.2011.05.007.
- M. Rai, A. Yadav and A. Gade, Biotechnol. Adv., 2009, 76–83, DOI:10.1016/j.biotechadv.2008.09.002.
- A. Azam, A. S. Ahmed, M. Oves, M. S. Khan, S. S. Habib and A. Memic, Int. J. Nanomed. DOI:10.2147/IJN.S35347.
- S. Kang, M. Herzberg, D. F. Rodrigues and M. Elimelech, Langmuir DOI:10.1021/la800951v.
- O. Akhavan and E. Ghaderi, ACS Nano DOI:10.1021/nn101390x.
- S. Liu, T. H. Zeng, M. Hofmann, E. Burcombe, J. Wei, R. Jiang, J. Kong and Y. Chen, ACS Nano DOI:10.1021/nn202451x.
- B. Z. Ristic, M. M. Milenkovic, I. R. Dakic, B. M. Todorovic-Markovic, M. S. Milosavljevic, M. D. Budimir, V. G. Paunovic, M. D. Dramicanin, Z. M. Markovic and V. S. Trajkovic, Biomaterials DOI:10.1016/j.biomaterials.2014.02.014.
- H. Sun, N. Gao, K. Dong, J. Ren and X. Qu, ACS Nano DOI:10.1021/nn501640q.
- S. Jaworski, M. Wierzbicki, E. Sawosz, A. Jung, G. Gielerak, J. Biernat, H. Jaremek, W. Łojkowski, B. Woźniak, J. Wojnarowicz, L. Stobiński, A. Małolepszy, M. Mazurkiewicz-Pawlicka, M. Łojkowski, N. Kurantowicz and A. Chwalibog, Nanoscale Res. Lett. DOI:10.1186/s11671-018-2533-2.
- V. H. Nguyen, B. K. Kim, Y. L. Jo and J. J. Shim, J. Supercrit. Fluids DOI:10.1016/j.supflu.2012.08.005.
- T. C. Dakal, A. Kumar, R. S. Majumdar and V. Yadav, Front. Microbiol. DOI:10.3389/fmicb.2016.01831.
- M. Yamanaka, K. Hara and J. Kudo, Appl. Environ. Microbiol. DOI:10.1128/AEM.71.11.7589-7593.2005.
- Y. Matsumura, K. Yoshikata, S.-I. Kunisaki and T. Tsuchido, Appl. Environ. Microbiol. DOI:10.1128/AEM.69.7.4278-4281.2003.
- C. N. Lok, C. M. Ho, R. Chen, Q. Y. He, W. Y. Yu, H. Sun, P. K. H. Tam, J. F. Chiu and C. M. Che, J. Proteome Res. DOI:10.1021/pr0504079.
- K. Habiba, D. P. Bracho-Rincon, J. A. Gonzalez-Feliciano, J. C. Villalobos-Santos, V. I. Makarov, D. Ortiz, J. A. Avalos, C. I. Gonzalez, B. R. Weiner and G. Morell, Appl. Mater. Today DOI:10.1016/j.apmt.2015.10.001.
- Y. Wei, X. Zuo, X. Li, S. Song, L. Chen, J. Shen, Y. Meng, Y. Zhao and S. Fang, Mater. Res. Bull. DOI:10.1016/j.materresbull.2014.02.015.
- X. D. Youngmi Koo, J. Nanomed. Nanotechnol. DOI:10.4172/2157-7439.1000279.
- S. J. Kazmi, M. A. Shehzad, S. Mehmood, M. Yasar, A. Naeem and A. S. Bhatti, Sens. Actuators, A DOI:10.1016/j.sna.2014.06.002.
- J. Tang, Q. Chen, L. Xu, S. Zhang, L. Feng, L. Cheng, H. Xu, Z. Liu and R. Peng, ACS Appl. Mater. Interfaces DOI:10.1021/am4005495.
- D. A. Dinh, K. S. Hui, K. N. Hui, Y. R. Cho, W. Zhou, X. Hong and H. H. Chun, Appl. Surf. Sci. DOI:10.1016/j.apsusc.2014.01.101.
- Z. Xu, H. He, S. Zhang, B. Wang, J. Jin, C. Li, X. Chen, B. Jiang and Y. Liu, Environ. Sci.: Nano 10.1039/c8en01090k.
- R. Foldbjerg, E. S. Irving, J. Wang, K. Thorsen, D. S. Sutherland, H. Autrup and C. Beer, Toxicol. Res. 10.1039/c3tx50099c.
- Y. Chong, Y. Ma, H. Shen, X. Tu, X. Zhou, J. Xu, J. Dai, S. Fan and Z. Zhang, Biomaterials DOI:10.1016/j.biomaterials.2014.03.021.
- M. Nurunnabi, Z. Khatun, K. M. Huh, S. Y. Park, D. Y. Lee, K. J. Cho and Y. K. Lee, ACS Nano DOI:10.1021/nn402043c.
- C. Wang, C. Wu, X. Zhou, T. Han, X. Xin, J. Wu, J. Zhang and S. Guo, Sci. Rep. DOI:10.1038/srep02852.
- L. Li, G. Wu, G. Yang, J. Peng, J. Zhao and J. J. Zhu, Nanoscale, 2013, 4015–4039, 10.1039/C3NR33849E.
- Y. Tian, F. Wang, Y. Liu, F. Pang and X. Zhang, Electrochim. Acta DOI:10.1016/j.electacta.2014.08.133.
- S. Liu, B. Yu and T. Zhang, RSC Adv. 10.1039/c3ra44492a.
- S. Kadian, G. Manik, A. Kalkal, M. Singh and R. P. Chauhan, Nanotechnology DOI:10.1088/1361-6528/ab3566.
- S. S. Suner, M. Sahiner, A. Akcali and N. Sahiner, J. Appl. Polym. Sci. DOI:10.1002/app.48352.
- S. Ruden, K. Hilpert, M. Berditsch, P. Wadhwani and A. S. Ulrich, Antimicrob. Agents Chemother. DOI:10.1128/AAC.01106-08.
- Sulmiyati, N. Said, D. U. Fahrodi, R. Malaka and F. Maruddin, IOP Conference Series: Earth and Environmental Science, 2019.
- K. Nikhil, S. Sharan, A. Chakraborty and P. Roy, PLoS One DOI:10.1371/journal.pone.0093335.
- J. Wu, P. Wang, F. Wang and Y. Fang, Nanomaterials DOI:10.3390/nano8100864.
- M. Amjadi, T. Hallaj, H. Asadollahi, Z. Song, M. de Frutos and N. Hildebrandt, Sens. Actuators, B DOI:10.1016/j.snb.2017.01.003.
- N. T. N. Anh and R. Doong, ACS Appl. Nano Mater. DOI:10.1021/acsanm.8b00210.
- S. Kadian, B. D. Arya, S. Kumar, S. N. Sharma, R. P. Chauhan, A. Srivastava, P. Chandra and S. P. Singh, Electroanalysis DOI:10.1002/elan.201800207.
- W. Shao, X. Liu, H. Min, G. Dong, Q. Feng and S. Zuo, ACS Appl. Mater. Interfaces DOI:10.1021/acsami.5b00937.
- D. Qu, M. Zheng, P. Du, Y. Zhou, L. Zhang, D. Li, H. Tan, Z. Zhao, Z. Xie and Z. Sun, Nanoscale 10.1039/c3nr04402e.
- S. Kadian, Luminescence, 2020, 1–10 Search PubMed.
- L. Scudiero, H. Wei and H. Eilers, ACS Appl. Mater. Interfaces DOI:10.1021/am900582w.
- B. Shamieh, S. Obuchovsky and G. L. Frey, J. Mater. Chem. C 10.1039/c5tc04141d.
- S. Bian, C. Shen, Y. Qian, J. Liu, F. Xi and X. Dong, Sens. Actuators, B DOI:10.1016/j.snb.2016.11.044.
- C. Wu, Y. Yuan, Q. He and R. Song, Nanotechnology DOI:10.1088/0957-4484/27/48/48LT02.
- P. Prieto, V. Nistor, K. Nouneh, M. Oyama, M. Abd-Lefdil and R. Díaz, Appl. Surf. Sci. DOI:10.1016/j.apsusc.2012.05.095.
- R. Dietsche, D. C. Lim, M. Bubek, I. Lopez-Salido, G. Ganteför and Y. D. Kim, Appl. Phys. A: Mater. Sci. Process. DOI:10.1007/s00339-007-4344-7.
- V. Kumar, R. K. Gupta, R. K. Gundampati, D. K. Singh, S. Mohan, S. H. Hasan and M. Malviya, RSC Adv. 10.1039/c7ra11466d.
- G. B. Hoflund, Z. F. Hazos and G. N. Salaita, Phys. Rev. B: Condens. Matter Mater. Phys. DOI:10.1103/PhysRevB.62.11126.
- N. Maiti, S. Thomas, A. Debnath and S. Kapoor, RSC Adv. 10.1039/c6ra09569k.
- N. J. Firet, M. A. Blommaert, T. Burdyny, A. Venugopal, D. Bohra, A. Longo and W. A. Smith, J. Mater. Chem. A 10.1039/c8ta10412c.
- A. Biswas, P. Khandelwal, R. Das, G. Salunke, A. Alam, S. Ghorai, S. Chattopadhyay and P. Poddar, J. Mater. Chem. B 10.1039/c6tb02446g.
- I. Ocsoy, M. Temiz, C. Celik, B. Altinsoy, V. Yilmaz and F. Duman, J. Mol. Liq. DOI:10.1016/j.molliq.2016.12.015.
- M. R. Das, R. K. Sarma, S. C. Borah, R. Kumari, R. Saikia, A. B. Deshmukh, M. V. Shelke, P. Sengupta, S. Szunerits and R. Boukherroub, Colloids Surf., B DOI:10.1016/j.colsurfb.2012.12.033.
- W. P. Xu, L. C. Zhang, J. P. Li, Y. Lu, H. H. Li, Y. N. Ma, W. Di Wang and S. H. Yu, J. Mater. Chem. 10.1039/c0jm03376f.
- L. Hui, J. Huang, G. Chen, Y. Zhu and L. Yang, ACS Appl. Mater. Interfaces DOI:10.1021/acsami.5b10132.
- A. A. Chaudhari, S. L. Jasper, E. Dosunmu, M. E. Miller, R. D. Arnold, S. R. Singh and S. Pillai, J. Nanobiotechnol. DOI:10.1186/s12951-015-0085-5.
- H. Yun, J. D. Kim, H. C. Choi and C. W. Lee, Bull. Korean Chem. Soc. DOI:10.5012/bkcs.2013.34.11.3261.
- S. Kumar-Krishnan, E. Prokhorov, M. Hernández-Iturriaga, J. D. Mota-Morales, M. Vázquez-Lepe, Y. Kovalenko, I. C. Sanchez and G. Luna-Bárcenas, Eur. Polym. J. DOI:10.1016/j.eurpolymj.2015.03.066.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9tb02823d |
| This journal is © The Royal Society of Chemistry 2020 |