Open Access Article
Open Access ArticleLysozyme coated copper nanoclusters for green fluorescence and their utility in cell imaging
Atul Gajanan
Thawari
a,
Piyush
Kumar
b,
Rohit
Srivastava
 b and
Chebrolu Pulla
Rao
b and
Chebrolu Pulla
Rao
 *ac
*ac
aBioinorganic Laboratory, Department of Chemistry, Indian Institute of Technology Bombay, Powai, Mumbai – 400076, India
bDepartment of Bioscience and Bioengineering, Indian Institute of Technology Bombay, Powai, Mumbai – 400076, India
cDepartment of Chemistry, Indian Institute of Technology Tirupati, Settipalli post, Tirupati – 517 506, Andhra Pradesh, India. E-mail: cprao@iittp.ac.in; Fax: +91 0877 250 3004; Tel: +91 0877 250 3036
First published on 14th July 2020
Abstract
Fluorescent, pH dependent and water soluble copper nanoclusters (CuNCs) were synthesized using lysozyme (lyz) as the stabilizing agent to give lysozyme coated copper nanoclusters, viz., lyz-CuNCs. The lyz-CuNCs were 3–5 nm in size at neutral pH and exhibit green fluorescence (λem ∼ 510 nm) when excited at 490 nm and exhibited a maximum quantum yield of 18%. However, under basic conditions, aggregates of lyz-CuNCs were seen with a particle size of ∼100 nm. The emission of lyz-CuNCs observed at 510 nm complements that from the blue (λem = 450 nm) and the red (λem = 650 nm) nanoclusters and thus bridges the gap. On the other hand, under acidic conditions, these show a size of 5–10 nm but weakly fluoresce. The particle size and aggregations were monitored by TEM studies carried out using the samples prepared under both the acidic and the basic conditions. The lyz-CuNCs prepared at neutral pH show >90% cell viability and hence can be used as a probe for cellular imaging. The imaging was carried out with both healthy and cancer cell lines, viz., NIH3T3 cells (mouse embryonic fibroblast cell), MCF7 cells (human breast cancer cells) and MDA-MB-231 cells (human estrogen negative breast cancer cells). The Z-stack study suggested the presence of lyz-CuNCs in the cells in cytoplasm. Thus, the green fluorescent lyz-CuNCs can be an alternate to green fluorescent protein (GFP) that is used for cell imaging purposes, since the latter needs a tedious procedure to express, purify and to conjugate.
Introduction
Metal nanoparticles of 3–5 nm show new physical and chemical properties owing to their nano-metric size1 and hence can be used in a broad range of applications, including, cell labeling,2 ion sensing,3 catalysis4 and anti-cancer activity.5 The recent literature primarily deals with the luminescent nanoclusters of Au6,7 and Ag8 stabilized by a variety of small chemical species as well as large biological molecules. To date, no abundant information is available regarding the synthesis of copper nanoparticles (CuNPs) and nanoclusters (CuNCs) due to the problems arising from the stability and the size of such nanoparticles.9–11However, the CuNPs have recently attracted special attention because of their low cost and novel optical, electrical, mechanical and thermal properties.5 The CuNPs are used in textiles, paints, plastics and food industries3,4 due to their antimicrobial properties. The protein coated CuNPs are expected to exhibit anticancer activity owing to their redox ability.5 Recent studies showed that the fluorescent CuNPs are stabilized by proteins, such as BSA, which gives red fluorescence, and lysozyme, which gives both red and blue fluorescence.12–18 Therefore, the stabilization of fluorescent nanoclusters of copper emitting in the visible region is still a challenging task. Proteins have been widely used in directing the synthesis of functional nanomaterials owing to the coordinating ability of –NH2, –COOH, –CONH–, –OH and –SH groups present as side chains in proteins.18 The use of lysozyme in the present study derives its impetus from (i) high pH stability due to the presence of six Lys and eleven Arg (pKa = 10) residues, (ii) coordination ability towards metal ions through carboxylates (Asp and Glu) and imidazole (His), (iii) ability to reduce metal ions such as Cu2+, Ag+, and Au+ using Tyr side chains19 and (iv) its well-known antibacterial property.20 Hence, lysozyme has been widely used for the synthesis of metal nanoclusters, such as copper,13 gold21,22 and silver23 nanoclusters. Herein, we report the lysozyme (lyz) directed synthesis of stable, water-soluble and green fluorescent copper nanoclusters lyz-CuNCs, with excitation (λex) at 490 nm and emission (λem) at 510 nm and demonstrated their utility in cell imaging.
Experimental
Preparation of copper nanoclusters coated with lysozyme (lyz-CuNCs)
Thirty mg of lysozyme was added to 3 ml of water. To this 100 μl of 100 mM CuCl2 was added. The solution was stirred for 30 min and to this 100 μl of 1 M NaOH was added in 10 μl portions each time ten times over a period of 10 min and stirred for another 30 min. Then 100 μl of hydrazine was similarly added over a period of 10 min and stirred for one day. The pH of the resultant solution was 12. The pH of the solution was brought down to 7, 5 and 3 in different vials by adding requisite volume of dil HCl and the final volume was maintained at 10 ml and the solutions were stirred for one day. At pH = 7, the lyz-CuNC solutions are stable for more than a month as confirmed by fluorescence spectral measurements. In order to remove the un-reacted small molecular components, the resultant solutions were dialyzed against water for 24 h by replacing the medium three times intermittently. The dialyzed samples showed similar characteristics to the un-dialyzed ones as proven by fluorescence and TEM studies. The dialyzed lyz-CuNCs were used for cell imaging studies.Quantum yield (QY) measurement
The fluorescence quantum yields of lyz-CuNCs were determined using the equation,24| Φx = (ΦST × AST × FX × ηX2)/(AX × FST × ηST2), |
Sample preparation and studies
Fluorescence studies were performed on a PerkinElmer LS55 Fluorescence Spectrometer. The lyz-CuNC solutions were used for the fluorescence studies by exciting these at 490 nm and measuring the spectra from 500 to 700 nm using a 5 nm slit width for the excitation and emission gates at a scan speed of 300 nm min−1. UV-visible absorption studies were performed on a Shimadzu UV-2101 PC. One ml solutions were used for collecting the spectrum using a faster scan speed and slit width of 5 nm in the range of 200–800 nm. Far-UV CD spectra were collected using a Jasco J-815 CD spectrometer. The path length of the cuvette was 0.1 cm and the bandwidth of the light used was 1 nm. Three scans were recorded with a scan speed of 100 nm min−1 and the scans were averaged.Spectroscopy and microscopy studies
The matrix assisted laser desorption ionization (MALDI) studies were performed on AutoflexIII TOF/TOF (MALDI-TOF mass spectrometer, Bruker Daltonics Co.) using sinapinic acid as a matrix and by using a 355 nm laser as an ionization source. The FTIR studies were performed using a PerkinElmer FT-IR spectrometer. One mg of lyophilised sample of lyz-Cu NCs was ground with KBr and a pellet was made from this. The spectrum for only lysozyme was also recorded for comparison. In order to carry out field emission gun transmission electron microscopy (FEG-TEM) or just TEM microscopy studies, samples were spread over carbon coated copper grids having 200 mesh and dried and analyzed by using JEOL JEM-2100F (FEG-TEM) operating at 200 kV. X-ray photoelectron spectroscopy (XPS) was performed using Mg Kα excitation (1253.6 eV) with a VG ESCALAB MKII spectrometer. Calibration for the binding energy was done based on carbon 1s being at 284.6 eV.Results and discussion
Lysozyme coated copper nanoclusters (lyz-CuNCs)
The lyz-CuNCs were initially prepared from a solution of lysozyme and copper(II) chloride at pH = 12 upon treating with a mild reducing agent, such as hydrazine, and the solution was adjusted to the requisite pH using HCl. The lyz-CuNCs were characterized by UV-Vis absorption, fluorescence, CD and XPS spectroscopy, and also by TEM.Emission and absorption studies
Among the studies carried out at pH = 12, 10, 7, 5 and 3, the one that is studied at the neutral pH, i.e., a mixture of lyz and copper salt solution (details given in the experimental section) resulted in dark yellow colour after one day of incubation and the fluorescence emission suggested the formation of CuNCs. Photographs of the vials of lyz-CuNCs in aqueous solution at all pH values under visible and UV light are given in Fig. 1. At pH = 12 and 10, the solutions were transparent and exhibited no green fluorescence when observed under UV light. The green fluorescence was stronger at neutral pH. However, there was only a minimal amount of green fluorescence at pH = 5 and 3 due to the precipitation and/or agglomeration. In the literature, lysozyme capped CuNCs were synthesized at pH 11–12 and a temperature of 40–45 °C, but using two different ratios of protein to the copper salt to result in blue emission (λex = 360 & λem = 450 nm) in one case13 and red emission (λex = 365 & λem = 600 nm) in the other.14 Therefore, the literature reports of lyz-CuNCs sharply differ from our case where our lyz-CuNCs were synthesized at neutral pH and RT and these show green fluorescence with λem = 510 nm, and not the blue or the red emission. While the excitation is in the UV region (360–365 nm), in the present case the excitation is in the visible region at 490 nm. Thus the lyz-CuNCs reported in this paper exhibited a shift of 60 nm in their fluorescence emission when compared to the blue lyz-CuNCs reported in the literature.13 Thus, our green fluorescent lyz-CuNCs complement the blue13 and red14 nanoclusters reported in the literature by completing the range of emission of 450 to 600 nm via bridging the gap with its emission centered at 510 nm.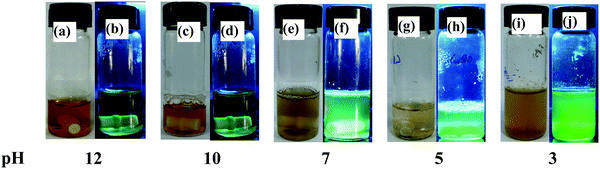 | ||
| Fig. 1 Photographs of lyz-CuNCs in aqueous solution under visible (left side vial) and UV (right side vial) light in each pair at the indicated pH. | ||
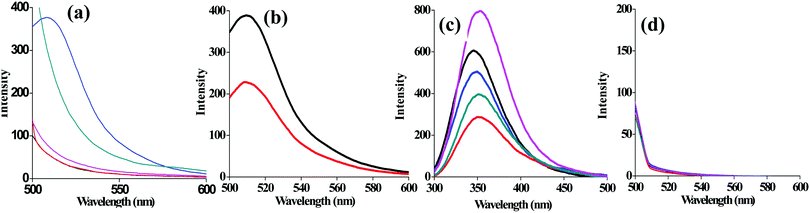
As compared to other pH conditions studied, at pH = 7, the agglomeration is prevented due to the capping of lyz that results in exhibiting significant fluorescence emission intensity at 510 nm (Fig. 2a). Minimal fluorescence emission intensity was noticed for the solution of lyz-CuNCs at pH = 5 and no fluorescence intensity was noticed for that studied at pH = 12, 10 and 3. Solution of lyz-CuNCs was dialyzed for removing unbound copper ions as well as other small molecular components. Even after the dialysis, the solutions exhibited fluorescence emission for the study carried out at pH = 7 (Fig. 2b). The quantum yields of the lyz-CuNCs determined as given in the experimental section are 0.014, 0.016, 0.18, 0.037 and 0.0093 at pH values of 3, 5, 7, 10 and 12 respectively, with respect to quinine hemisulfate, thus showing the highest quantum yield at pH = 7. The protein lyz alone shows maximum emission at ∼350 nm upon excitation at 280 nm at all the pH values studied due to the presence of aromatic amino acids, viz., Trp and Tyr as can be noticed from Fig. 2c. When excited at 490 nm, lyz alone doesn’t show any fluorescence (Fig. 2d) which suggested that the green fluorescence is emitted only by the protein coated CuNPs, viz., lyz-CuNCs, but not simply by the protein.
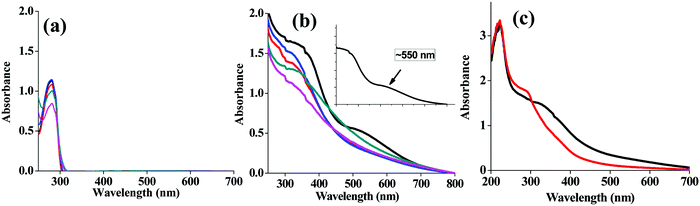
The absorption spectrum of lysozyme alone shows a typical protein absorption spectrum with a maximum peak at 280 nm and without any SPR band at all the pH studied (Fig. 3a). However, the lyz-CuNCs show two peaks, one broad peak at ∼300 nm and a sharp peak at ∼220 nm (Fig. 3b). In the absorption spectra at pH = 7, no SPR band was noticed at 550 nm due to smaller size of the copper nanoparticles as measured based on TEM (3–5 nm) and these are referred to as nanoclusters (NCs) (Fig. 3b and c). Thus we find a significant increase in the absorbance of the SPR band observed at ∼550 nm in the case of pH = 12 which distinguishes this from the remaining pH values and this supports the nanoparticle formation as can be noted from the inset of Fig. 3b. Thus, the SPR band almost diminishes as the pH of the solution decreases and turns to acidic (Fig. 3b). The spectrum of lyz alone is totally different from the absorption spectra obtained for lyz-CuNCs and the latter is characteristic of the copper bound lyz (Fig. 3a and b).
CD and XPS studies
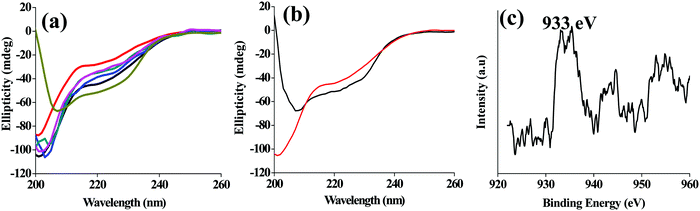
While the simple lyz is predominantly present in the α-helical (222 nm band in CD) structure, the lyz-CuNCs exhibit mostly a random coil (200 nm band in CD) like structure under all the pH conditions studied (Fig. 4a). Even after the dialysis, at pH = 7, the random coil structure was still seen to be predominant (Fig. 4b). This suggested the partial denaturation of the protein in lyz-CuNCs which helps in opening an α-helix as well as in reducing Cu2+ to Cu0 and the protein, lyz is coated onto the CuNCs. XPS analysis was performed with lyz-CuNCs obtained after the dialysis. The XPS spectra were characteristic of the reduction of Cu2+ to Cu0 both based on the main peak as well as the satellite peaks. While the Cu2+ shows two strong satellites of which one is at 962 eV, the Cu0 shows two weak satellites with the main peak being centered around 933 eV. All these features can be clearly seen from the XPS spectra given in Fig. 4c and the spectra correspond to the Cu0 species.Characterization by MALDI and FTIR studies
The lysozyme, in its native state, showed a m/z peak around 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 Da (for z = 1) and 7145 Da (for z = 2). Upon addition of 20 equivalents of Cu2+ at pH 10, the peak shifts in the molecular weight corresponding to the presence of higher copper clusters, such as Cu1, Cu2, Cu4 and Cu6 as noticed from the m/2 peak (Fig. 5a). Under these conditions, the peak corresponding to the protein alone almost completely diminished and the copper bound ones were noticed as marked in the spectra. Upon addition of hydrazine, the Cu2+ ion is reduced to Cu0 and results in a lysozyme bound nanocluster which was then dialyzed. As the desorption of the dialysed nanocluster is not abundant, the corresponding MALDI spectra resulted in greater noise. In the case of blue fluorescent lyz-CuNCs, 2 to 9 copper bound nanoclusters (Cu2, Cu4 and Cu9) were reported,13 while no MALDI studies were reported in the case of red lyz-CuNCs.14 An FTIR spectrum of simple protein shows an amide peak at ∼1600 cm−1 along with a shoulder at ∼1700 cm−1. In the case of lyz-CuNCs, only the shoulder disappears and the amide band is retained, suggesting the capping of CuNCs by the protein, lysozyme. Thus, the protein acts as a capping as well as a stabilizing agent in these CuNCs (Fig. 5b).
300 Da (for z = 1) and 7145 Da (for z = 2). Upon addition of 20 equivalents of Cu2+ at pH 10, the peak shifts in the molecular weight corresponding to the presence of higher copper clusters, such as Cu1, Cu2, Cu4 and Cu6 as noticed from the m/2 peak (Fig. 5a). Under these conditions, the peak corresponding to the protein alone almost completely diminished and the copper bound ones were noticed as marked in the spectra. Upon addition of hydrazine, the Cu2+ ion is reduced to Cu0 and results in a lysozyme bound nanocluster which was then dialyzed. As the desorption of the dialysed nanocluster is not abundant, the corresponding MALDI spectra resulted in greater noise. In the case of blue fluorescent lyz-CuNCs, 2 to 9 copper bound nanoclusters (Cu2, Cu4 and Cu9) were reported,13 while no MALDI studies were reported in the case of red lyz-CuNCs.14 An FTIR spectrum of simple protein shows an amide peak at ∼1600 cm−1 along with a shoulder at ∼1700 cm−1. In the case of lyz-CuNCs, only the shoulder disappears and the amide band is retained, suggesting the capping of CuNCs by the protein, lysozyme. Thus, the protein acts as a capping as well as a stabilizing agent in these CuNCs (Fig. 5b).
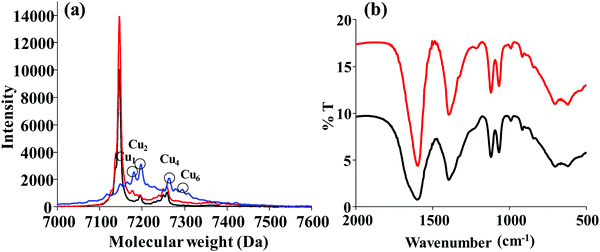 | ||
Fig. 5 (a) MALDI-TOF spectra of simple lyz (—) and the lyz treated with Cu2+ in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 ratio at pH 7 ( 20 ratio at pH 7 ( ) and at pH 10 ( ) and at pH 10 ( ). (b) FTIR spectra of simple lyz (—) and lyz-CuNCs ( ). (b) FTIR spectra of simple lyz (—) and lyz-CuNCs ( ). ). | ||
TEM studies
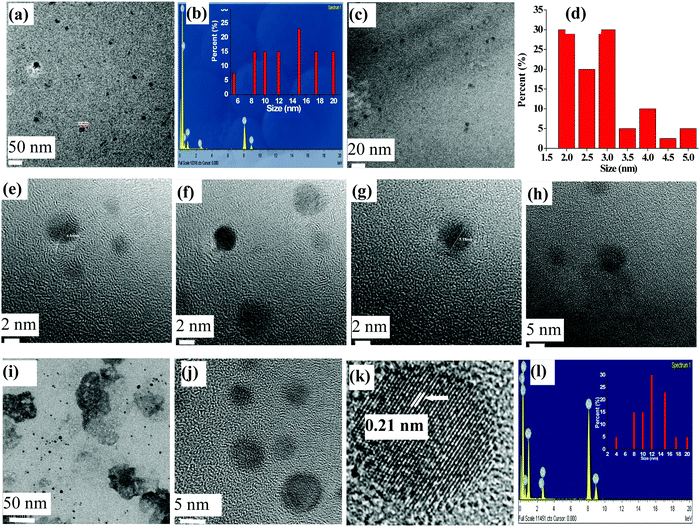
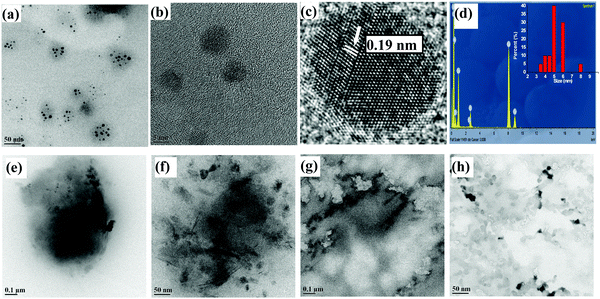
The TEM micrographs were measured for lyz-CuNCs at pH = 7 both before and after the dialysis. Before the dialysis, the lyz-CuNCs exhibited particle sizes in the range of 5–20 nm (Fig. 6a, e and f). The protein coating in lyz-CuNCs was proven by EDAX, which shows peaks for copper along with N, C, O, and S (Fig. 6b) and particle size distribution is shown in the inset. Since some larger size particle of 10–20 nm was noticed in this sample, TEM was performed again with the dialyzed and centrifuged solution for establishing the size distribution of lyz-CuNCs. After the dialysis, the particle size distribution was restricted to 3–5 nm (Fig. 6c, d, g and h). The particles in the size distribution (Fig. 6d) of 3–5 nm of lyz-CuNCs were expected to show fluorescence emission and these indeed showed green fluorescence as presented in this paper.At acidic pH, the size of lyz-CuNCs was in the range of 5–10 nm but these nanoparticles were precipitated out from the solution and hence weak fluorescence was noticed (Fig. 6i, j and 7a, b). The micrograph indicates a clear-cut aggregation of the small nanoparticles into clusters of 50–100 nm size. Even the EDAX measurement (Fig. 6l and 7d) carried out with the samples at pH = 5 and 3 clearly showed the protein coating by exhibiting the peaks for C, N, O, and S along with copper. On going from pH = 5 to 3, spherical aggregates are more prevalent. Thus, TEM clearly supports the fact that 3–5 nm size lyz-CuNCs are formed mainly at pH = 7 and these give rise to green emission, while the particles observed at all other pH values are of larger size or aggregate leading to minimal or no fluorescence. At pH = 5 and 3, the HR-TEM image shows an interplanar distance of 0.21 and 0.19 nm respectively that corresponds to the (111) lattice planes of metallic copper, supporting the reduction of Cu2+ to Cu0 (Fig. 6k and 7c). The presence of Cu0 was already shown by XPS. The interplanar distance observed in the present case agrees well with that reported in the literature.25 At pH = 12 and 10, larger aggregates of the small nanoclusters were noticed clearly showing the agglomeration at these pH values to result in fiber like structures (Fig. 7e–h).
Cell viability of the fluorescent lyz-CuNCs prepared at pH = 7
The MTT assay was carried out to determine the cell viability of lyz-CuNCs upon treating the NIH3T3 cells with these nanoclusters and incubating these for a period of 24 h. The cell viability was studied as a function of the concentration of lyz-CuNCs as shown in Fig. 8. At up to 20 μg ml−1 concentration of lyz-CuNCs, the cell viability is ∼100%. Even at very high concentration of lyz-CuNCs (i.e., 80 μg ml−1), ∼90% cell viability was noticed, supporting that these nanoclusters are non-toxic to cells and hence are well suited for cellular imaging work.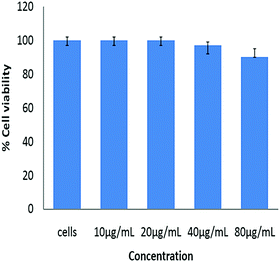 | ||
| Fig. 8 Percent viability of NIH3T3 cells after the treatment with various concentrations of lyz-CuNCs. | ||
Cell imaging by the fluorescent lyz-CuNCs prepared at pH = 7
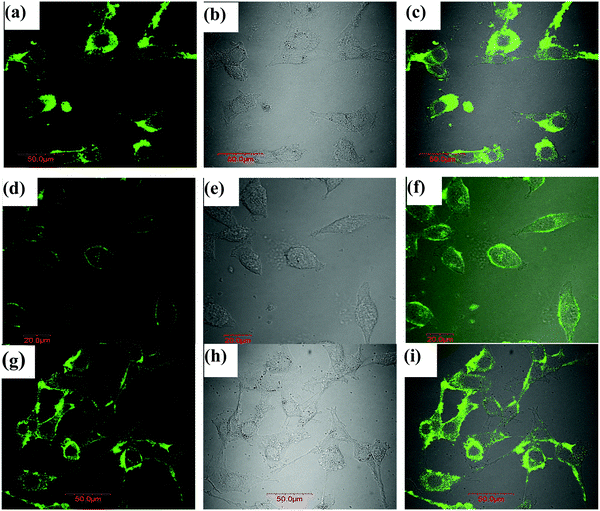
The utility of lyz-CuNCs in cellular imaging was successfully demonstrated using three cell lines, namely, NIH3T3 cells (mouse embryonic fibroblast cell), MCF7 cells (human breast cancer cells) and MDA-MB-231 cells (human estrogen negative breast cancer cells) using fluorescence confocal microscopy. The fluorescence image collected under 490 nm excitation showed bright green fluorescence in cells (Fig. 9). In all the three types of cells, green fluorescence was mostly located in the periphery, though some fluorescence intensity was seen inside the cells particularly in the case of MCF7 cells.Evidence for the cellular uptake of lyz-CuNCs
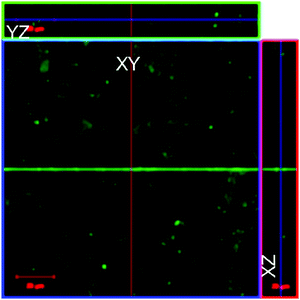
Initially, the cellular uptake of lyz-CuNCs was studied using the same three cell lines, viz., NIH3T3, MCF7 and MDA-MB-231 using fluorescence confocal microscopy. The localization of lyz-CuNCs was determined in L929 cells by the Z-stack experiment and it is noted that lyz-CuNCs were present in the cells predominantly in the perinuclear cytoplasm (Fig. 10). The fluorescence in blue lines in YZ and XZ axes clearly indicate that lyz-CuNCs entered the cells and mostly localized in the cytoplasm.Conclusions and comparisons
Lysozyme was chosen as the model protein to stabilize the copper nanoclusters to give lyz-CuNCs. In the literature, lyz was used for the synthesis of gold nanoparticles within a single crystal of lysozyme.26,27 In our paper, we have shown that the protein, lyz, stabilizes the copper nanoparticles due to its high pKa over a wide pH range. Lysozyme is a monomer containing 129 amino acid residues with 4 disulfide bridges and has a molecular weight of 14.3 kDa.26 The presence of 14% basic amino acids, such as Lys and Arg, makes this protein well suited for binding to copper depending on the pH and this has been clearly demonstrated from the studies reported in this paper by carrying out these at different pH and varying protein to copper mole ratios.The fluorescence intensity of lyz-CuNCs observed at pH = 7 was high with a quantum yield of 0.18 with respect to quinine hemisulfate. The small size (3–5 nm) of lyz-CuNCs results in the formation of fluorescent nanoclusters. As compared to other pH values studied, at pH = 7, the TEM image showed relatively homogeneous distribution of 3–5 nm particles without any significant aggregation of proteins resulting in green fluorescence. However, at both lower and higher pH, aggregated species of different types were observed in TEM, supporting that at neutral pH the binding of copper dominates resulting in smaller size nanoclusters to give green fluorescence emission. A schematic representation of the nanoparticle formation under different pH conditions is shown in Fig. 11a–c.
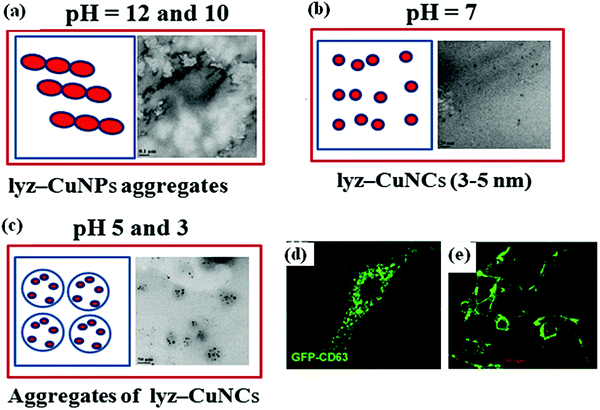 | ||
| Fig. 11 (a–c) The schematic representation of the formation of lyz-CuNPs and lyz-CuNCs under the indicated pH conditions. Confocal microscopy images: (d) BT-CD63 cells labeled with GFP-CD63 exosomes30 and (e) MCF7 cells labeled with lyz-CuNCs at pH 7. | ||
To our knowledge, this is the first example of the formation of protein mediated green fluorescent copper nanoparticles. Blue and red fluorescent CuNCs stabilized with protein, DNA and ascorbic acid were reported in the literature.12–18 However, in the present study, the emission is around ∼510 nm owing to its green fluorescence. The green fluorescence reported in our paper emerges from the clusters of size 3–5 nm when the study was carried out at neutral pH.
The report of our green fluorescent lyz-CuNCs complements the blue (λex = 360 & λem = 450 nm)13 and red (λex = 365 & λem = 600 nm)14 nanoclusters reported in the literature by completing the range of emission of 450 to 600 nm through bridging the gap with its emission centered at 510 nm. The green fluorescence property of the lyz-CuNCs has been proven to be useful in the cell imaging of normal (NIH3T3 cells) as well as cancerous cells (MCF7 and MDA-MB-231 cells). As reported in the literature, the lyz-CuNCs were used for imaging only the HeLa cell lines.13 Our study reports the imaging of one healthy cell line (NIH3T3 cells) and two cancerous cells (MCF7 and MDA-MB-231 cells) along with proving its presence inside the cell by Z-stacking experiment. The Z-stacking experiment showed that lyz-CuNCs go inside the cell and are localized in the cytoplasm. Thus, our lyz-CuNCs provide broader applicability for cell or tissue imaging. Our attention was attracted towards green fluorescent CuNPs due to their similarity to the green fluorescent protein (GFP) reported in the literature.25 Similar to the present lyz-CuNCs, the GFP from A. victoria has its excitation peak at 395 nm and the emission peak at 509 nm.28 GFP is widely used for cell labeling and as a marker for gene expression.29–31 The lyz-CuNC material can also be used in labeling various types of cells similar to that where GFP is being used. Comparison of these two in the case of cancer cell labeling can be viewed from Fig. 11d and e. In our case the excitation wavelength is clearly in the visible region unlike those reported in the literature regarding the blue and the red copper nanoclusters wherein the excitation wavelength was in the UV region. BT-CD63 cells labeled with GFP-CD63 exosomes exhibit bright green fluorescence.30 Similar green fluorescence was also noticed in our studies of lyz-CuNCs with MCF-7 cells. Hence, green fluorescent lyz-CuNCs can be used for cancer diagnosis and treatment when appropriately tuned and conjugated with a drug molecule. Therefore, the green fluorescent lyz-CuNCs can be an alternate choice for GFP, since the GFP needs a tedious procedure to express, purify and conjugate and is also expensive to procure. Hence these lysozyme coated copper nanocluster (lyz-CuNC) materials may find real time applications in biology, medical diagnosis and even in therapy when appropriately tuned.
Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
CPR acknowledges the Department of Science and Technology for the JC Bose National Fellowship (SB/S2/JCB/066/2015) and IIT Tirupati for providing an MHRD Professorship. CPR expresses his gratitude to IIT Bombay for fostering his academic and research activities for over three decades. We thank the central facilities, such as TEM, XPS, MALDI-TOF and confocal microscopy of IIT Bombay, for providing services.References
- B. S. González, M. J. Rodríguez, C. Blanco, J. Rivas, M. A. L’opez-Quintela and J. M. G. Martinho, Nano Lett., 2010, 10, 4217–4221 CrossRef PubMed.
- S. I. Tanaka, J. Miyazaki, D. K. Tiwari, T. Jin and Y. Inouye, Angew. Chem., Int. Ed., 2011, 50, 431–435 CrossRef CAS PubMed.
- C. C. Huang, Z. Yang, K. H. Lee and H. T. Chang, Angew. Chem., Int. Ed., 2007, 46, 6824–6828 CrossRef CAS PubMed.
- M. Hyotanishi, Y. Isomura, H. Yamamoto, H. Kawasaki and Y. Obora, Chem. Commun., 2011, 47, 5750–5752 RSC.
- G. P. Jose, S. Santra, S. K. Mandal and T. K. Sengupta, J. Nanobiotechnol., 2011, 9, 9–15 CrossRef CAS.
- H. Zhang, T. Watanabe, M. Okumura, M. Haruta and N. Toshima, Nat. Mater., 2012, 11, 49–52 CrossRef.
- Z. Wu, M. Wang, J. Yang, X. Zheng, W. Cai, G. Meng, H. Qian, H. Wang and R. Jin, Small, 2012, 8, 2028–2035 CrossRef CAS PubMed.
- W. J. Cho, Y. Kim and J. K. Kim, ACS Nano, 2012, 6, 249–255 CrossRef CAS PubMed.
- N. Vilar-Vidal, M. C. Blanco, M. A. Lopez-Quintela, J. Rivas and C. Serra, J. Phys. Chem. C, 2010, 114, 15924–15930 CrossRef CAS.
- H. Kawasaki, Y. Kosaka, Y. Myoujin, T. Narushima, T. Yonezawa and R. Arakawa, Chem. Commun., 2011, 47, 7740–7742 RSC.
- W. Wei, Y. Lu, W. Chen and S. Chen, J. Am. Chem. Soc., 2011, 133, 2060–2063 CrossRef CAS PubMed.
- N. Goswami, A. Giri, M. S. Bootharaju, P. L. Xavier, T. Pradeep and S. K. Pal, Anal. Chem., 2011, 83, 9676–9680 CrossRef CAS PubMed.
- R. Ghosh, A. K. Sahoo, S. S. Ghosh, A. Paul and A. Chattopadhyay, ACS Appl. Mater. Interfaces, 2014, 6, 3822–3828 CrossRef CAS PubMed.
- C. Wang, S. Shu, Y. Yao and Q. Song, RSC Adv., 2015, 5, 101599–101606 RSC.
- Y. Guo, F. Cao, X. Lei, L. Mang, S. Cheng and J. Song, Nanoscale, 2016, 8, 4852–4863 RSC.
- S. Kim, J. H. Kim, W. Y. Kwon, S. H. Hwang, B. S. Cha, J. M. Kim, S. S. Oh and K. S. Park, Microchim. Acta, 2019, 186, 479–484 CrossRef PubMed.
- S. Momeni, R. Ahmadi, A. Safavi and I. Nabipour, Talanta, 2017, 175, 514–521 CrossRef CAS.
- C. Wang, C. Wang, L. Xu, H. Cheng, Q. Lin and C. Zhang, Nanoscale, 2014, 6, 1775–1781 RSC.
- H. Kawasaki, K. Hamaguchi, I. Osaka and R. Arakawa, Adv. Funct. Mater., 2011, 21, 3508–3515 CrossRef CAS.
- H. Schwalbe, S. B. Grimshaw, A. Spencer, M. Buck, J. Boyd, C. M. Dobson, C. Redfield and L. J. Smith, Protein Sci., 2001, 10, 677–688 CrossRef CAS PubMed.
- S. Chandirasekar, J. G. You, J. H. Xue and W. L. Tseng, J. Mater. Chem. B, 2019, 10, 3876–3883 RSC.
- B. A. Russell, B. Jachimska, P. Komorek, P. A. Mulheran and Y. Chen, Phys. Chem. Chem. Phys., 2017, 19, 7228–7235 RSC.
- S. Pang and S. Liu, Anal. Methods, 2017, 9, 6713–6718 RSC.
- X. Zhou, R. Lai, H. Li and C. I. Stains, Anal. Chem., 2015, 87, 4081–4086 CrossRef CAS PubMed.
- A. Chahadih, H. E. Hamzaoui, O. Cristini, L. Bigot, R. Bernard, C. Kinowski, M. Bouazaoui and B. Capoen, Nanoscale Res. Lett., 2012, 7, 1–6 CrossRef.
- M. Liang, L. Wang, X. Liu, W. Qi, R. Su, R. Huang, Y. Yu and Z. He, Nanotechnology, 2013, 24, 245601 CrossRef PubMed.
- H. Wei and Y. Lu, Chem. – Asian J., 2012, 7, 680–683 CrossRef CAS PubMed.
- M. Ormö, A. B. Cubitt, K. Kallio, L. A. Gross, R. Y. Tsien and S. J. Remington, Science, 1996, 273, 1392–1395 CrossRef PubMed.
- M. Chalfie, Y. Tu, G. Euskirchen, W. W. Ward and D. C. Prasher, Science, 1994, 263, 802–805 CrossRef CAS.
- R. B. Koumangoye, A. M. Sakwe, J. S. Goodwin, T. Patel and J. Ochieng, PLoS One, 2011, 6, e24234 CrossRef CAS PubMed.
- M. H. Robert, Lab. Invest., 2015, 95, 432–452 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2020 |