Open Access Article
Open Access ArticleDevelopment of an immunochromatographic lateral flow strip for the simultaneous detection of aminoglycoside residues in milk
Yaning Sun†
a,
Jifei Yang†a,
Suzhen Yang† a,
Qingbo Sangd,
Man Tenga,
Qingmei Lia,
Ruiguang Denga,
Lili Fengb,
Xiaofei Hu*a and
Gaiping Zhang*ac
a,
Qingbo Sangd,
Man Tenga,
Qingmei Lia,
Ruiguang Denga,
Lili Fengb,
Xiaofei Hu*a and
Gaiping Zhang*ac
aKey Laboratory of Animal Immunology of the Ministry of Agriculture, Henan Provincial Key Laboratory of Animal Immunology, Henan Academy of Agricultural Sciences, Zhengzhou, 450002, China. E-mail: huxf1972@126.com
bInstitute of Agricultural Economics and Information, Henan Academy of Agriculture Sciences, Zhengzhou, 450002, China
cCollege of Animal Science and Veterinary Medicine, Henan Agricultural University, No. 63 Nongye Road, Zhengzhou, 450002, China. E-mail: zhanggaiping2003@163.com
dZhengzhou Ruilong Food Science and Technology Limited Company, Zhengzhou, 450064, China
First published on 5th March 2018
Abstract
A colloidal gold-based immunochromatographic strip with a competitive format has been developed for the rapid, simultaneous, semi-quantitative and quantitative detection of several aminoglycoside residues in milk, including gentamicin sulfate (GM), neomycin sulfate (NEO) and kanamycin sulfate (KN). Three monoclonal antibodies against the three corresponding aminoglycosides were conjugated to colloidal gold particles and applied to the conjugate pads of the strip. The competitors [GM-bovine serum albumin (GM-BSA), NEO-BSA and KN-BSA conjugates] of GM, NEO and KN were immobilized onto a nitrocellulose (NC) membrane at three detection zones, T1, T2, and T3, respectively. The minimal cut-off values of the strip were 10 ng mL−1 for GM, and 100 ng mL−1 for NEO and KN, which are lower than the maximum residue levels (MRLs) established for aminoglycosides. The IC50 values of the strip were 0.737 ng mL−1, 8.971 ng mL−1 and 11.110 ng mL−1 for GM, NEO and KN respectively. In conclusion, the immunochromatographic lateral flow strip could be used for rapid, simultaneous, semi-quantitative and quantitative detection of GM, NEO and KN residues in milk.
Introduction
Aminoglycosides are a class of antibiotics characterised by two or more amino-sugars linked by glycosidic bonds to an aminocyclitol scaffold. They have been used extensively for their broad-spectrum antibacterial effectiveness in both human and veterinary medicine to treat various bacterial infections.1–3 They act directly by interfering with bacterial protein synthesis. These drugs are usually administered via intramuscular injection or intramammary infusion, and are generally excreted through the urinary tract. However, residues have also been found in milk and muscle because of their inappropriate overuse.1 The residual of aminoglycosides in the foodstuff may lead to the development of antibiotic resistance in pathogenic bacterial strains and cause directly ototoxicity, nephrotoxicity and neuromuscular blockade, and subsequently endanger the consumer.4–6 So monitoring for aminoglycoside residues in food of animal origin is mandatory in many countries to ensure compliance with veterinary good practice and food security. Currently, maximum residual limits (MRLs) have been established by the EU and the FDA, for the aminoglycosides residues in milk, edible tissues and other food stuffs (Table 1).3,7| Active ingredient | Species or product | Market commodity (μg kg−1) | ||
|---|---|---|---|---|
| Kidney | Fat, liver, muscle | Milk | ||
| Dihydrostreptomycin | Cattle | 1000 | 600 | 200 |
| Pig | 1000 | 600 | ||
| Sheep | 200 | |||
| Neomycin | Cattle | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
500 | 1500 |
| Sheep | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
500 | 1500 | |
| Pig | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
500 | ||
| Turkey | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
500 | ||
| Spectinomycin | Cattle | 1000 | 600 | |
| Chicken | 5000 | 2000 (fat, liver) | ||
| 500 (muscle) | ||||
| Streptomycin | Chicken | 1000 | 600 | |
| Pig | 1000 | 600 | ||
| Gentamicin | Cattle | 5000 | 100 (muscle, fat) | 200 |
| 2000 (liver) | ||||
| Pig | 5000 | 100 (muscle, fat) | ||
| 2000 (liver) | ||||
| Avilamycin | Turkey | 200 | 200 (muscle, fat) | |
| 300 (liver) | ||||
| Chicken | 200 | 200 (muscle, fat) | ||
| 300 (liver) | ||||
| Pig | 200 | 200 (muscle, fat) | ||
| 300 (liver) | ||||
| Rabbit | 200 | 200 (muscle, fat) | ||
| 300 (liver) | ||||
To monitor aminoglycoside levels, a number of analytical methods have been developed, such as high-performance liquid chromatography (HPLC),2,8,9 capillary electrophoresis,10 and surface plasmon resonance.11 Although these methods are accurate and very sensitive, the required instruments are expensive and the techniques are time-consuming and require skilled technical support.12 These methods typically require sample purification with organic solvents making them unsuitable for screening large numbers of samples or real-time detection.
In the past decade, immunochemical methods have become increasingly important in antimicrobial drug monitoring, in particular the immunochromatographic lateral flow strip test (IL-FST). This methodology provides the advantages of sensitivity, specificity, rapidity, simplicity, low cost and high throughput and with a grayscale scanner it can provide quantitative as well as qualitative answers.13–16
IL-FST have been reported on the detection of aminoglycosides residues, but these have been limited to the detection of a single analyte.17–19 Simultaneous detection of multi-aminoglycosides with an IL-FST represents a considerable challenge because of the structural similarity of these compounds and the difficulty of producing sufficiently specific monoclonal antibodies. In this study we present a novel format of an immunochromatographic lateral flow strip for rapid, simultaneous, semi-quantitative and quantitative detection of GM, NEO, and KN in milk samples based on the use of three different highly specific monoclonal antibodies.
Experimental section
Reagents and apparatus
Gentamicin sulfate (GM) standard is a mixture of gentamicin C, C1a and C2, neomycin sulfate (NEO), kanamycin sulfate (KN), apramycin (AP) and streptomycin (SM) were purchased from Dr Ehrenstorfer (Augsburg, Germany). Gold chloride trihydrate was purchased from Sigma Aldrich (St Louis, MO, USA). PVP-10 was purchased from Prospec (Rehovot, Israel). Tween-20 was purchased from Merck & Co. Inc. Bovine serum albumin (BSA) and ovalbumin (OVA) were purchased from BDH (VWR International Ltd.). Trehalose was purchased from Sigma Wako (Osaka, Japan). Nitrocellulose membrane (HF13502S25, 30 × 2 cm2), glass fiber and absorbent pad were purchased from Millipore (Bedford, MA). GM-BSA, NEO-BSA and KN-BSA conjugate, anti-mouse IgG as well as three monoclonal antibodies against GM, NEO and KN were prepared in our laboratory. The other reagents and solvents were of analytical grade or higher.Ultrapure water purification system was purchased from Millipore (Bedford, MA). Zetasizer Nano ZS90 was purchased from Malvern (Worcestershire, UK). An XYZ Biostrip Dispenser, CM 4000 Cutter, and TSR3000 membrane strip reader were all purchased from Bio-Dot (Richmond, CA).
Preparation of monoclonal antibody-based colloidal gold probe
Gold chloride was reduced with 1% trisodium citrate to produce colloidal gold. To 100 mL of boiling 0.01% (w/v) gold chloride solution 1.6 mL of a 1% solution of trisodium citrate was added. The resulting solution initially had a gray color, which changed to lavender and with continued boiling, after 1 to 3 minutes, developed a red hue. The solution was boiled for another 5 min, and then cooled to room temperature.20 The quality and the size of the gold nanoparticles were evaluated using a Malvern Zetasizer Nano ZS90. The colloidal gold solution was stored at 4 °C until use.The quality and the size of gold nanoparticles were evaluated using a Malvern Zetasizer Nano ZS90. In this study, 24.1 ± 4.3 nm particles were prepared by reducing 100 mL of 0.01% gold chloride with 1.6 mL of sodium citrate. The zeta potential of the gold nanoparticles was −45 mV.
The monoclonal antibodies (mAb) were purified from mouse ascitic fluid using a caprylic acid and ammonium sulfate method, and the final concentration was adjusted to 20 mg mL−1 with PBS.
The pH value of the colloidal gold solution was adjusted to pH 8.2 with 0.2 mol L−1 K2CO3. The concentration of mAb was adjusted to 2 mg mL−1 with Ultrapure water (UPW). To determine the most appropriate protein concentration for conjugation 10 μL of mAb solution was 2-fold serially diluted in UPW, before mixing with 125 μL of colloidal gold solution, following incubation for 5 min at room temperature, 125 μL of 10% NaCl solution was added. The color of the mixtures changed from brilliant red to blue as the concentration of mAb decreased. The optimum concentration of mAb for colloidal gold labeling was the lowest concentration of mAb solution that did not produce a change in color.21
Briefly, 1 mg of the purified mAb (anti-NEO, anti-KN or anti-GM) diluted in 1 mL of UPW, was added gently into 100 mL of gold colloid solution previously adjusted to pH 8.2 with 0.2 mol L−1 K2CO3. Following incubation for 70 min at room temperature, 10 mL of 10% BSA (w/v) was added, 10 min later the mixture was centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4 °C for 30 min and the supernatant discarded. The final pellets were suspended in 40 mL of gold suspension buffer (20 mmol L−1 borate buffer, 10 g L−1 BSA, 30 g L−1 trehalose, and 0.3 g L−1 sodium azide, pH 8.5) and stored at 4 °C until use.
000 rpm at 4 °C for 30 min and the supernatant discarded. The final pellets were suspended in 40 mL of gold suspension buffer (20 mmol L−1 borate buffer, 10 g L−1 BSA, 30 g L−1 trehalose, and 0.3 g L−1 sodium azide, pH 8.5) and stored at 4 °C until use.
Preparation of the strip
The immunochromatographic lateral flow strip was based on the competitive immunoassay principle, and was prepared from three pads (sample pads, 30 × 1.5 cm; conjugate pads, 30 × 0.8 cm; absorbent pads, 30 × 1.8 cm) and a nitrocellulose (NC) membrane (HF13502S25, Millipore, 30 × 2 cm) all assembled on a semirigid polyethylene sheet. Sample pads were soaked with sample pads treatment buffer, and were dried at 42 °C for 4 h. The conjugate pads were coated with a mixture of gold-antibody conjugates [anti-GM-mAb-gold, anti-NEO-mAb-gold and anti-KN-mAb-gold in a proportion of 1/1.5/1 (v/v/v)] using a XYZ Biostrip Dispenser at jetting rate of 8 μL cm−1 and were dried at 42 °C for 1 h. The test lines for GM, NEO and KN, were GM-BSA (test lines1/T1), NEO-BSA (test lines2/T2) and KN-BSA (test lines3/T3) conjugates respectively and the control lines was anti-mouse IgG (control line/C), diluted with normal saline (NS) and spotted onto the NC membrane using a XYZ Biostrip Dispenser at jetting rate of 0.9 μL cm−1. The distance between the lines was 0.3 cm. After spotting, the membrane was dried at 42 °C for 4 h. All the materials were vacuum packaged in a plastic bag containing silica as moisture absorbent.The immunochromatographic lateral flow strip was assembled as shown in Fig. 1A. The NC membrane was pasted at the center of a semi-rigid polyethylene sheet, and the conjugate pad and the absorbent pad were attached with overlaps of 2 mm respectively with the NC membrane. The sample pad was pasted with 2 mm overlap on the conjugate pad. Finally the assembled sheet was cut into 2.8 mm strips.22
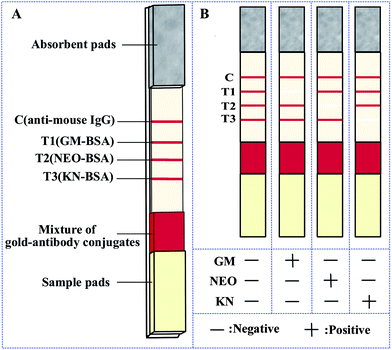 | ||
| Fig. 1 (A) Schematic illustration of the strip. (B) Schematic illustration of the result determination. Note: C, control line; T1, test line 1; T2, test line 2; T3, test line 3. | ||
Principle and procedure of the strip
The immunochromatographic lateral flow strip for the simultaneous detection of three analytes was based on competition for the mAb-gold conjugate mixture between the free analytes in the sample and the hapten-protein conjugates bound to the NC membrane.23 A diagram of the results using standards are shown in Fig. 1B. When a sample is placed on the sample pad, the mAb-gold conjugates are dissolved and then trained along by the liquid through capillary action. In the absence of analyte in the sample (negative sample), the mAb-gold conjugate (anti-GM mAb-gold, anti-NEO mAb-gold and anti-KN mAb-gold) would be trapped by the capture reagents (GM-BSA, NEO-BSA and KN-BSA) on the NC membrane. The test results in four vivid red lines (T1, T2, T3 and C lines). On the other hand if the sample contains a sufficient quantity of a target analyte no red line is observed in the corresponding location (Fig. 1B). The control line is always colored red, indicating that the strip and the test procedure are correct. If no color appears at the control line it would mean that either the test procedure is improperly conducted or the strip is invalid and the sample should be tested again using a new strip.Sensitivity and specificity of the strip
The sensitivity of the strip was determined with normal saline (NS) containing different concentrations of the three analytes (GM 10 ng mL−1, NEO 100 ng mL−1 and KN 100 ng mL−1 were 2-fold serially diluted in normal saline and used as the standards). The cut-off values were defined as the lowest concentration failing to produce color at the test lines by direct visual observation.24With a TSR3000 membrane strip reader provides a more quantitative detection.24 When targets existed in the sample, they would compete with the immobilized capture reagents for the limited amount of mAb-gold conjugate. Hence, the more targets in the sample, the more shallow the test line colored. The TSR3000 membrane strip reader gives a relative optical density (ROD) at the test line. A standard curve is constructed by plotting the (G/D) × A (mean density of the scanned points times sampling window area) values obtained from standard samples against the log[concentration]. The IC50 values were calculated from the calculated regression. Linearity was assessed by the coefficient of determination (R2).25
Specificity was evaluated by investigating cross-reactivity between three test lines of the strip with GM, NEO, KN and other aminoglycosides (apramycin and streptomycin) standard solution. The cross-reactivities were expressed as the percentage of the concentration at cut-off values of GM, NEO or KN compared to other aminoglycosides.
Sample preparation
Ten milliliters of each milk sample was weighed into a 25 mL plastic centrifuge tube. After centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min at 4 °C, the lipid layer was removed. The supernatant was then diluted with normal saline, and tested using the strip.
000 rpm for 5 min at 4 °C, the lipid layer was removed. The supernatant was then diluted with normal saline, and tested using the strip.
The matrix effects in milk
Negative milk samples (without GM, NEO, KN previously confirmed by HPLC) were treated according to 2.7, then diluted with normal saline at 1/2, 1/3, 1/4 and 1/8. The diluted samples were tested using the strip and the result read by unaided visual assessment and the TSR3000 membrane strip reader.GM, NEO and KN standard solutions were added to different dilutions (1/2, 1/4, 1/8) of milk samples at concentrations of 5, 2.5, 1.25, 0.625, 0.3125 ng mL−1, 10, 5, 2.5, 1.25, 0.625 ng mL−1 and 10, 5, 2.5, 1.25, 0.625 ng mL−1, respectively. The sensitivity of the strip was determined with the above spiked solutions in different dilutions of milk.
Recoveries from spiked samples
Negative milk samples were spiked with GM, NEO and KN at concentrations 5, 10, 20 ng mL−1, 50, 100, 200 ng mL−1 and 50, 100, 200 ng mL−1, respectively. The analyses were performed on ten replicates for each concentration (n = 10). This test was used to evaluate the accuracy of the strip.Results and discussion
Optimization of the strip
In general, the analytical performance of the strip is affected by a number of parameters, including the type and pore size of the membrane, buffer and the amount of the immune reagents.Three different mAbs and conjugates were used. The sample pad treatment buffer was evaluated for possible effects on the different mAbs and analytes. 20 mmol L−1 borate buffer (pH 9.0) containing 10 g L−1 BSA, 1 mL L−1 tween-20, 2 g L−1 PVP-10 and 0.3 g L−1 sodium azide was chosen as the optimal sample pads treatment buffer for the sample pads to achieve the required visibility and the best sensitivity.
In order to further improve the strip system, the size of gold nanoparticles, the optimal concentrations of the capture reagents (GM-BSA/NEO-BSA/KN-BSA), gold-antibody conjugates, and the volume of samples (60–150 μL) were optimized. Under optimized conditions, it was found that the gold particles around 24 nm gave satisfactory performance and the optimal amounts of GM-BSA, NEO-BSA, KN-BSA, anti-mouse IgG and gold-antibody conjugates were 5.8 ng cm−1, 24 ng cm−1, 11.3 ng cm−1, 900 ng cm−1 and 6 μL cm−1, separately and 100 μL of sample provided effective dissolution of the gold-antibody conjugates from the conjugate pads.
Sensitivity of the strip
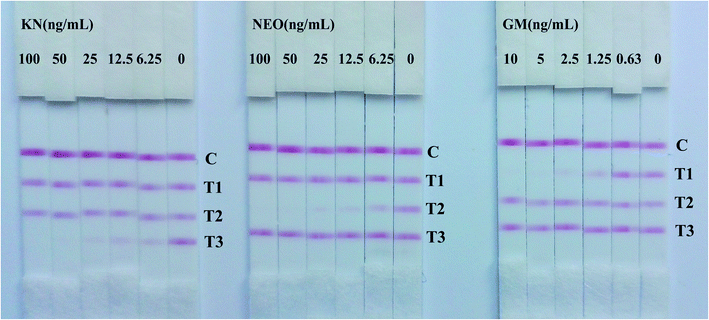
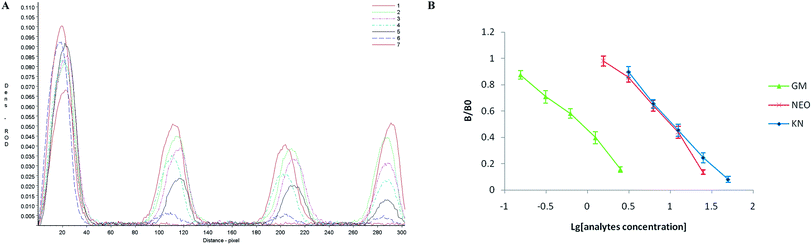
In semi-quantitative detection, the sensitivity of the strip is evaluated by testing standard solutions of the three analytes. The results can be seen in Fig. 2. When standard solutions were applied to the test strip, the color of the T1, T2, T3 zones disappeared at 10 ng mL−1, 100 ng mL−1 and 100 ng mL−1 respectively. Therefore, the cut-off values was 10 ng mL−1 for GM, and 100 ng mL−1 for NEO and KN.Following scanning with a Bio-Dot TSR3000 Membrane Strip Reader gave standard curves of good linearity (Fig. 3B). The regression equations were y = −0.580x + 0.423 (R2 = 0.986), y = −0.700x + 1.167 (R2 = 0.979) and y = −0.678x + 1.209 (R2 = 0.996) for GM, NEO and KN respectively and the calculated IC50 values of GM, NEO and KN were 0.737 ng mL−1, 8.971 ng mL−1 and 11.110 ng mL−1, respectively.
The matrix effects in milk
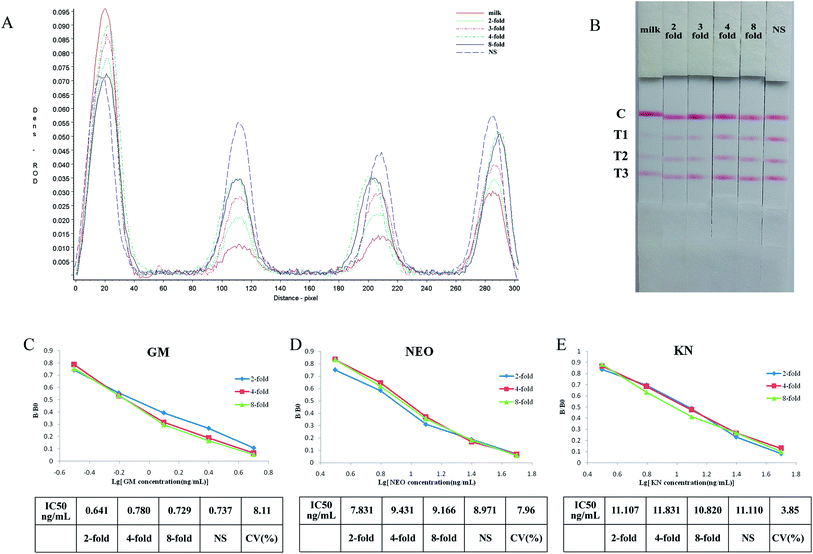
Standards of the aminoglycosides prepared in the supernatant of negative milk samples, which had been treated as in 2.7 above and further diluted 1/2, 1/3, 1/4 and 1/8 with normal saline then tested with the strip. The results indicated that the three test lines were weaker when tested using negative milk extract rather than normal saline as solvent, however this effect was progressively removed on dilution of the milk extract (Fig. 4A and B).The sensitivity of the test strip in milk extract can be seen in Fig. 4C–E. When different dilutions of milk extract were used as solvent the standard curves show good linearity, and the coefficients of variation (CV) of IC50's for the individual drugs were below 10%.
Although the test lines of the strip were less clear when using milk extract, they were clear when the samples were diluted 2-fold or higher and there was no significant difference between the IC50's. So the strip can be used for the semi-quantitative or quantitative detection of GM, NEO and KN in milk if it is diluted at least 2-fold. The required dilution of milk samples is in part determined by the specific MRL so for example, if the MRL standard for GM was 200 ng mL−1 milk samples should be diluted 20-fold.
Specificity of the strip
To ensure the cross reactions of the strip from the three analytes and common aminoglycosides, the specificity of the strip was evaluated by testing analytes separately to exclude false-positive results. The results were summarized in Fig. 5, the monoclonal antibodies of GM and NEO present good specificity, there were no any reaction with other aminoglycosides (T1 and T2). However, the signal of KN (T3) reduced in different degree when testing sample contain 1 μg mL−1 GM or 1 μg mL−1 AP (Fig. 5A), and the signal of KN (T3) vanished when the amount of GM and AP reach to 2.5 μg mL−1 and 10 μg mL−1 severally (Fig. 5B and C). The cross-reactivity between KN with GM and AP were 4% and 1% respectively. Due to the drug incompatibility of aminoglycosides, they can not be used simultaneously or continuously, two and three positive results on one strip are non-existent. When T1 and T3 vanished concurrently indicated there has more than 2.5 μg mL−1 GM in the tested samples.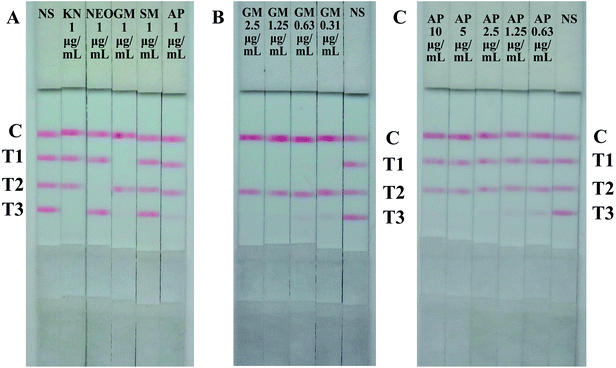 | ||
| Fig. 5 Results of specificity evaluation of the strip. (A) Cross-reactivity of aminoglycosides by the strip. (B) Cross-reactivity of GM by the strip. (C) Cross-reactivity of AP by the strip. | ||
Accuracy of the strip
GM, NEO and KN standard solutions were spiked into the blank milk samples with concentrations of 5, 10, 20 ng mL−1, 50, 100, 200 ng mL−1 and 50, 100, 200 ng mL−1, respectively. The results of recovery and repeatability for GM, NEO and KN in milk samples are shown in Table 2. Each sample was tested 10 times with a single batch of test strips. The average recoveries and their CVs were calculated. For GM, recoveries were from 91.80 to 97.03% with the highest CV at 11.70%. For NEO, recoveries were from 88.90 to 100.72% with the highest CV at 11.09%. For KN, recoveries were from 82.01 to 98.42% with the highest CV at 11.60%. The results indicated recoveries better than 82% of theoretical values and coefficients of variation below 15%, and were acceptable.| Spiked (ng mL−1) | Strip (ng mL−1) | Recovery (%) | CV (%) |
|---|---|---|---|
| GM/NEO/KN | GM/NEO/KN | GM/NEO/KN | GM/NEO/KN |
| 5/50/50 | 4.80/47.11/47.15 | 96.04/94.22/94.30 | 11.70/8.93/6.46 |
| 10/100/100 | 9.70/100.72/82.01 | 97.03/100.72/82.01 | 5.50/6.62/7.23 |
| 20/200/200 | 18.36/177.80/196.85 | 91.80/88.90/98.42 | 9.62/11.09/11.60 |
Conclusions
We have developed a colloidal gold-based immunochromatographic strip test for the rapid and simultaneous, semi-quantitative and quantitative detection of GM, NEO and KN. The three mAbs used in the preparation of the strip have a high specificity for their respective haptens and show little or no cross-reactivity with the tested antibiotics. The major advantages of the test strip is the results can be obtained within 5–10 min and all needed reagents are included in the strip, it can be used to simultaneous screen for the aminoglycosides GM, NEO and KN residues. Results of the strips obtained by naked eyes showed that the cut-off values of the strip were 10 ng mL−1 for GM, 100 ng mL−1 for NEO and KN separately. The TSR3000 membrane strip reader scanned results indicated that the IC50 were 0.737 ng mL−1, 8.971 ng mL−1 and 11.110 ng mL−1 for GM, NEO and KN respectively. The sample matrix effects on strip performance was evaluated using milk samples, although 2-fold diluted milk affected the color intensity of the test lines, the IC50 was no clear affected. The results also showed good recoveries of added analytes and good reproducibility of the developed strip.This study suggests that this immunological test strip would provide a feasible on site screen for the residues of three important antibiotics in animal products.
Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported by the National Science & Technology Pillar Program of ‘12th Five-Year Plan’ (2014BAD13B05) and China Agriculture Research System (CARS-36).References
- S. B. Turnipseed, S. B. Clark, C. M. Karbiwnyk, W. C. Andersen, K. E. Millerand and M. R. Madson, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2009, 877, 1487–1493 CrossRef CAS PubMed.
- J. B. Arsand, L. ì. S. Jank, M. T. Martins, R. B. Hoff, F. Barreto, T. N. M. Pizzolato and C. Sirtori, Talanta, 2016, 154, 38–45 CrossRef CAS PubMed.
- F. Farouk, H. M. E. Azzazy and W. M. A. Niessen, Anal. Chim. Acta, 2015, 890, 21–43 CrossRef CAS PubMed.
- Y. Zhu, P. Chandra, K. Song, C. Ban and Y. Shim, Biosens. Bioelectron., 2012, 36, 29–34 CrossRef CAS PubMed.
- D. N. Heller, S. B. Clark and H. F. Righter, J. Mass Spectrom., 2000, 35, 39–49 CrossRef CAS.
- G. J. Kaloyanides and E. Pastoriza-Munoz, Kidney Int., 1980, 18, 571–582 CrossRef CAS PubMed.
- C. Alimentarius, CAC/MRL 2–2015.
- R. Oertel, V. Neumeister and W. Kirch, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2004, 1058, 197–201 CAS.
- A. Kaufmann and K. Maden, J. AOAC Int., 2005, 88, 1118–1125 CAS.
- Y. H. Long, M. Hernandez, E. Kaale, A. V. Schepdael, E. Roets, F. Borrull, M. Calull and J. Hoogmartens, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2003, 784, 255–264 CrossRef CAS.
- S. R. Raz, M. G. E. G. Bremer, W. Haasnoot and W. Norde, Anal. Chem., 2009, 81, 7743–7749 CrossRef PubMed.
- L. Guo, S. Song, L. Liu, J. Peng, K. Hua and C. Xu, Biomed. Chromatogr., 2015, 29, 1432–1439 CrossRef CAS PubMed.
- M. E. Jolley, S. D. Stroupe, C. H. Wang, H. N. Panas, C. L. Keegan, R. L. Schmidt and K. S. Schwenzer, Exp. Fluids, 1971, 60, 1559–1564 Search PubMed.
- Y. Chen, L. Guo, L. Liu, S. Song, H. Kuang and C. Xu, J. Agric. Food Chem., 2017, 65(37), 8248–8255 CrossRef CAS PubMed.
- J. Peng, L. Liu, L. Xu, S. Song, H. Kuang, G. Cui and C. Xu, Nano Res., 2017, 10, 108–120 CrossRef CAS.
- Y. Chen, L. Liu, L. Xu, S. Song, H. Kuang, G. Cui and C. Xu, Nano Res., 2017, 10, 2833–2844 CrossRef CAS.
- D. Mukunzi, J. Isanga, S. Suryoprabowo, L. Liu and H. Kuang, Food Agric. Immunol., 2017, 28, 1–13 CrossRef.
- J. J. C. H. Yong Jin, J. Vet. Sci., 2006, 7, 111–117 CrossRef.
- Y. Jin, J. W. Jang, C. H. Han and M. H. Lee, J. Agric. Food Chem., 2005, 53, 7639–7643 CrossRef CAS PubMed.
- G. Frens, Nature Phys. Sci., 1973, 241, 20–22 CrossRef CAS.
- Y. Zhao, G. Zhang, Q. Liu, M. Teng, J. Yang and J. Wang, J. Agric. Food Chem., 2008, 56, 12138–12142 CrossRef CAS PubMed.
- A. Y. Kolosova, S. De Saeger, L. Sibanda, R. Verheijen and C. Van Peteghem, Anal. Bioanal. Chem., 2007, 389, 2103–2107 CrossRef CAS PubMed.
- C. Xing, L. Liu, S. Song, F. Min, K. Hua and C. Xu, Biosens. Bioelectron., 2015, 66, 445–453 CrossRef CAS PubMed.
- C. Song, Q. Liu, A. Zhi, J. Yang, Y. Zhi, Q. Li, X. Hu, R. Deng, J. Casas and L. Tang, J. Agric. Food Chem., 2011, 59, 9319–9326 CrossRef CAS PubMed.
- Y. Sun, X. Hu, Y. Zhang, J. Yang, F. Wang, Y. Wang, R. Deng and G. Zhang, J. Agric. Food Chem., 2014, 62, 11116–11121 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |