Development of a label-free Raman imaging technique for differentiation of malaria parasite infected from non-infected tissue†
Laura
Frame
 a,
James
Brewer
b,
Rebecca
Lee
b,
Karen
Faulds
a and
Duncan
Graham
a,
James
Brewer
b,
Rebecca
Lee
b,
Karen
Faulds
a and
Duncan
Graham
 *a
*a
aCentre of Molecular Nanometrology, Department of Pure and Applied Chemistry, University of Strathclyde, Technology and Innovation Centre, 99 George Street, G1 1RD, UK. E-mail: Duncan.graham@strath.ac.uk
bInstitute for Infection, Immunity and Inflammation, University of Glasgow, G12 8QQ, UK
First published on 16th November 2017
Abstract
During malarial infection, the host uses the spleen to clear the malaria parasites, however, the parasites have evolved the ability to bind to endothelial receptors in blood vessels of tissues to avoid removal, known as sequestration, and this is largely responsible for the symptoms and severity of infection. So a technique which could non-invasively diagnose tissue burden could be utilised as an aid for localised malaria diagnosis within tissue. Raman spectroscopy is a label-free imaging technique and can provide unique and chemically specific Raman ‘fingerprint’ spectrum of biological samples such as tissue. Within this study, Raman imaging was used to observe the changes to the molecular composition of mice spleen tissue under malarial infection, compared with non-infected samples. From analysis of the Raman imaging data, both tissue types showed very similar spectral profiles, which highlighted that their biochemical compositions were closely linked. Principal component analysis showed very clear separation of the two sample groups, with an associated increase in concentration of heme-based Raman vibrations within the infected dataset. This was indicative of the presence of hemozoin, the malaria pigment, being detected within the infected spleen. Separation also showed that as the hemozoin content within the tissue increased, there was a corresponding change to hemoglobin and some lipid/nucleic acid vibrations. These results demonstrate that Raman spectroscopy can be used to easily discriminate the subtle changes in tissue burden upon malarial infection.
Introduction
Malaria is a mosquito-borne infectious disease, caused by the Plasmodium parasite1 and is a leading cause of morbidity and mortality in the developing world.2 Five species are known to cause malaria infection in humans, with Plasmodium falciparum (P. falciparum) being responsible for a large number of cases in Africa.1,3During the erythrocytic infection phase, the parasite catabolises up to 80%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 of the hosts’ hemoglobin. This occurs within the digestive vacuole of the parasite5 and produces free heme as a by-product (ferrous [FeII] protoporphyrin IX),6 which is toxic to the parasite.5 To detoxify this, the parasite oxidises it to an insoluble, crystalline material known as hemozoin or the malaria pigment.5,7 At the end of the erythrocytic cycle, red blood cells (RBCs) rupture, releasing hemozoin and parasites into the bloodstream, with the parasites going on to infect further RBCs and continue the cycle of infection. While hemozoin, under physiological conditions, is not degraded and circulates in the bloodstream until it is digested by macrophages present within specific organs.7 The spleen is the primary organ involved in the development of an immune response8 during the erythrocytic stage of infection along with the elimination of parasitised RBCs and hemozoin.9,10 However, P. falciparum proteins on the surface of infected erythrocytes give these cells the ability to bind to endothelial receptors in blood vessels of tissue, leading to sequestration2,11 of these infected cells and hence reduced clearance from the bloodstream.2 Sequestration of parasites into tissue is largely responsible for the severity and symptoms of infection,12,13 therefore, a technique which could non-invasively detect tissue burden would be a great step forward.
4 of the hosts’ hemoglobin. This occurs within the digestive vacuole of the parasite5 and produces free heme as a by-product (ferrous [FeII] protoporphyrin IX),6 which is toxic to the parasite.5 To detoxify this, the parasite oxidises it to an insoluble, crystalline material known as hemozoin or the malaria pigment.5,7 At the end of the erythrocytic cycle, red blood cells (RBCs) rupture, releasing hemozoin and parasites into the bloodstream, with the parasites going on to infect further RBCs and continue the cycle of infection. While hemozoin, under physiological conditions, is not degraded and circulates in the bloodstream until it is digested by macrophages present within specific organs.7 The spleen is the primary organ involved in the development of an immune response8 during the erythrocytic stage of infection along with the elimination of parasitised RBCs and hemozoin.9,10 However, P. falciparum proteins on the surface of infected erythrocytes give these cells the ability to bind to endothelial receptors in blood vessels of tissue, leading to sequestration2,11 of these infected cells and hence reduced clearance from the bloodstream.2 Sequestration of parasites into tissue is largely responsible for the severity and symptoms of infection,12,13 therefore, a technique which could non-invasively detect tissue burden would be a great step forward.
Raman spectroscopy has been extensively used for the identification of malaria infection most commonly via the detection of hemozoin. It has been widely reported by groups including Frosch et al.4 and Wood et al.14 that the choice of Raman excitation wavelength is key to gaining selective enhancement of hemoglobin and hemozoin bands over other non-resonant cellular components. Wood et al.15 reported that by exploiting the resonance Raman characteristics of hemozoin, coupled with partial dark field microscopy, there was potential to detect low-pigmented phases of the malaria parasite's life-cycle within infected erythrocytes. More recently, Hobro et al.16 applied resonance Raman imaging to study the progression of malaria infection via blood and plasma samples. They showed that within infected blood samples, changes associated with a loss of hemoglobin and an increase in hemozoin could be detected after four days of infection. However, hemozoin detection limits were shown to be reduced to only one day, following Plasmodium infection, when plasma samples were analysed due to a reduced heme background. Resonance Raman spectroscopy has also been used within a multimodal imaging approach to study the disease mechanisms of murine cerebral malaria.17 This work by Hackett et al.17 showed strong evidence that during cerebral malaria infection iron-mediated protein oxidation occurs within the tissue.
These studies highlight how valuable Raman spectroscopic techniques are for the study of biological matrices within malarial infection. Because Raman spectroscopy is a powerful, label-free technique, it can provide a unique and chemically specific Raman ‘fingerprint’ spectrum of biological samples.18–20 It has a number of advantages for studying these types of materials including requiring minimal to no sample preparation;18 being a non-destructive technique;21 and allowing specific chemical information to be gained without the use of stains.22 During the pathogenesis of disease, biological alterations occur, which lead to changes in the biochemical composition of affected tissues. These changes in structure, concentration and composition of biomolecules can be reflected in the Raman spectra of tissue samples. Indeed, Raman spectroscopy has been used to study the subtle molecular changes in tissue samples associated with other diseases including, lung23 cancer; breast20,24 cancer; and diabetes.18
In this work, we used Raman spectroscopic imaging to study the changes in the biochemical composition of mouse spleen tissue sections following infection with the rodent parasite, Plasmodium berghei (P. berghei). We compared the Raman spectra of infected with non-infected samples to detect changes in the Raman spectral signature associated with infection. The multivariate chemometric analysis technique, principal component analysis (PCA) was applied to decompose the spectral data into components and attempt to identify subtle changes to biological components within tissue sections which could differentiate infected tissue burden from non-infected samples.
Materials and methods
Infections and tissue preparation
Procedures on mice were approved following local ethical review by the University of Glasgow Animal Procedures and Ethics Committee and were performed in strict accordance with the UK Home Office Animals (Scientific Procedures) Act 1986 (project licence no. P2F28B003). Two female, BALB/c mice (Harlan, Bicester, UK; 6–8 weeks old) were infected with asynchronous P. berghei ANKA, and euthanised when peripheral blood parasitemia reached 5%. At this level, although high, the infection is asymptomatic in BALB/c mice. Spleens were removed from these and from two uninfected control mice and embedded in tissue freezing medium (Tissue-Tek O.C.T.).25 This freezing medium was used to preserve the tissue structure. Frozen tissue blocks were then positioned in a cryostat chamber at −22 °C, cut into sections with a thickness of 10 μm and placed on poly-L-lysine glass slides with Tissue-Tek O.C.T. Ten tissue sections were taken from the infected spleens, along with a further ten from the uninfected spleens. Tissue sections on glass slides were stored at −20 °C prior to analysis. Spleen tissue was chosen as it is a key tissue for the sequestration of the parasite.Reference spectra
Synthetic hemozoin was prepared using methods reported by Egan et al.26,27 (full synthetic method is given in ESI S1†) and a 2.5 mg mL−1 solution prepared with PBS (10 mM, pH 7.2). Hemoglobin was purchased from Sigma-Aldrich and a 10 mg mL−1 solution prepared with PBS. 100 μL of each solution were spotted on glass coverslips and allowed to dry overnight before analysis.Raman spectroscopy
Tissue sections were left to thaw at room temperature, washed once with PBS (10 mM, pH 7.2) and finally rinsed in d.H2O. Raman mapping was performed using a Renishaw inVia Raman spectrometer equipped with 532 nm laser excitation (5% laser power, 1 mW) and an 1800 l mm−1 grating. Raman images of uninfected and infected malaria tissue were recorded using a ×50 objective (NA = 0.75) with an integration time of 3 s per spectrum, a step size of 1 μm, and in the spectral range of 680–2330 cm−1.Hemoglobin and hemozoin reference spectra were collected using a ×20 objective, 30 s acquisition time per spectra and a 532 nm laser excitation (1% laser power, 0.1 mW).
Data pre-processing
In all datasets, a cosmic ray removal algorithm was first applied within WiRE 4.2 software. This algorithm sets values for width and height of features to detect potential cosmic rays, which are then accepted or rejected by the programmer. Noise filtering was then applied within the same software, to reduce the level of random noise within the data whilst maintaining the important Raman spectral features. This pre-processing method applies a form of PCA to the dataset, which subsequently provides scores and loadings for each principal component (PC) to determine whether components primarily relate to real Raman signals or noise. The optimum number of PCs, which describe all the real Raman signal with minimal noise interference, can then be selected for each Raman map. Raman spectra were baseline corrected using an asymmetric least square smoothing algorithm28 operating in Matlab R2013a software (The MathWorks, Natick, MA, USA). Finally, all datasets were truncated so only the fingerprint region of interest was studied (700–1700 cm−1).The reference spectra shown are the average of five spectra, that were baselined using the same asymmetric least square smoothing algorithm as above.
Empirical analysis
In this study, the empirical analysis focused on the ratio of the Raman peak intensity at ∼745 cm−1 (v15 hemoglobin),29,30 to the peak intensity at ∼1307 cm−1 (v21 hemoglobin & lipids)18,29 for both tissue types. An unpaired student's t-test was used to assess whether the difference in the Raman intensity ratio (I745/I1307) between non-infected and infected tissues was statistically significant.18,19 To manage the data for analysis, all infected Raman maps were combined to give one large data matrix containing thousands of Raman spectra, the same process was carried out with the uninfected data. These were then averaged to leave 43 final infected Raman spectra and 43 control spectra (n = 43). These data were subsequently used for empirical and multivariate analysis. The intensity ratios for all samples were plotted to observe sample distribution, along with the average intensity ratio plot for both the control and infected tissue. This analysis was performed on the same data set, used for the PCA model.Multivariate analysis
PCA was applied to highlight any spectral separation between the control and infected data sets using Matlab software version R2013a (The MathWorks, Natick, MA, USA). Multiple maps for both tissue types were combined and averaged to give 43 final spectra for both the control and infected tissue data sets. The dataset was then normalised and mean centred prior to PCA being applied. Principal component one (PC1) was then plotted against principal component two (PC2). The loadings plots were also plotted to determine correlation. All analysis within Matlab software was performed using custom scripts.Results and discussion
Average spectra of non-infected and malaria parasite infected tissue
In this study two approaches were used to analyse the large spectral data sets. Firstly, spectral features were studied and changes in relative peak intensity investigated using intensity ratio maps and empirical analysis. The second approach used PCA to determine the primary spectral features resulting in correlations or anti-correlation between the infected and non-infected tissue, by creating principal components to explain the data set variance in an unsupervised manner.Fig. 1A shows the averaged, normalised Raman spectra obtained from a non-infected (control) tissue map and a P. berghei infected mouse spleen tissue map. Both tissue types showed a very similar spectral profile, highlighting that the biochemical compositions were closely linked. Tentative peak assignments of the Raman bands are listed in Table 1. However, differences in the relative Raman peak intensities for infected vs. control tissue were observed. These were highlighted through subtraction of the average, normalised control spectra from the infected (Fig. 1B). This emphasised some peaks that appeared to increase (∼745, 1130, 1170 and 1225 cm−1) and decrease (∼1087 and 1439 cm−1) in Raman signal intensity within the P. berghei infected tissue compared with the control. This change in relative peak intensity suggests an alteration in the biological composition of the spleen at this level of infection, characterised by changes in level and/or composition of the biological species assigned to these Raman signals.
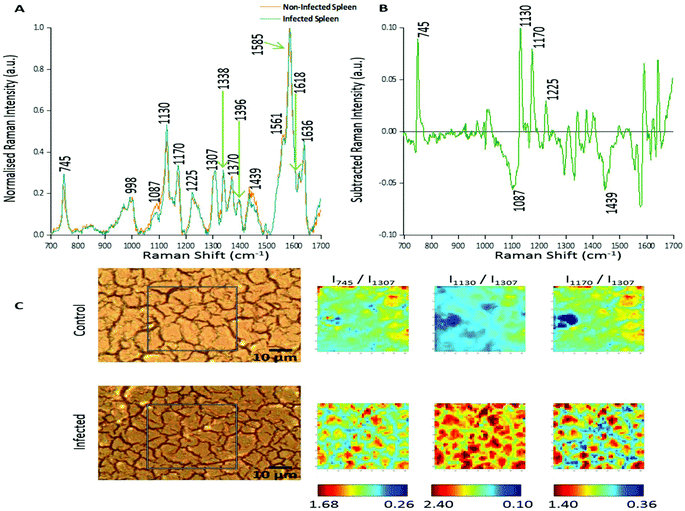 | ||
| Fig. 1 Analysis of Raman peak intensity for control and P. berghei-infected tissue sections. (A) Averaged, normalised spectra from a mapped area of control and infected tissue. Spectra were then normalised against the peak at 1585 cm−1. Tentative peak assignments are also given in Table 1. (B) Subtracted Raman spectrum. The average, normalised control spectrum was subtracted from the average, normalised infected spectrum, both shown in (A), to highlight any potential biological components that were being altered within the malaria infected tissue. (C) Image shows white light images of both tissue sections, with the grey box indicating the area that was mapped, along with peak intensity ratio maps for selected Raman peaks. Intensity ratio maps were created using the peak intensities for: 745 cm−1, 1130 cm−1, 1170 cm−1, and 1307 cm−1 (I745/I1307; I1130/I1307; I1170/I1307). These ratio maps visually show relative changes in the intensity of these Raman peaks within the area of P. berghei-infected tissue that was mapped compared to the uninfected area. These maps suggest an increase or alteration to some key biological components within the infected tissue including, multiple hemoglobin vibrations,29 lipids,31 and proteins.30 | ||
| Raman shift (cm−1) | Assignmenta |
|---|---|
| a Abbreviations: v – stretch vibration; δ – in-plane bending vibration; sym – symmetric; asym – asymmetric; Hb – hemoglobin; Phe – phenylalanine; Tyr – tyrosine. Assignments from ref. 16, 18 and 29–31. | |
| 745 | v 15 Hb [v(pyr breathing)] |
| 998 | Proteins (Phe), v45 Hb [v(CC)asym] |
| 1087 | DNA/nucleic acids [vs(PO2−), v(CC)], Hb |
| 1130 | Lipids, v5 Hb [δ(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2)4] CH2)4] |
| 1170 | Proteins (Tyr), v30 Hb [v(pyr half-ring)asym] |
| 1225 | Proteins (amide III), v13/v42 Hb [δ(CH)] |
| 1307 | Lipids, v21 Hb [δ(CH)] |
| 1338 | Nucleic acid modes, v41 Hb [v(pyr half-ring)sym] |
| 1370 | v 4 Hb [v(pyr half-ring)sym] |
| 1396 | CH2 deformation, v20 Hb [v(pyr quarter-ring)] |
| 1439 | Lipids [δ(CH2)] |
| 1561 | v 11 Hb [v(CC)] |
| 1585 | v 37 Hb [v(CC)asym] |
| 1618 | v 19 Hb [v(CC)asym] |
| 1636 | Proteins (amide I), v10 Hb [v(CC)asym] |
Intensity ratio maps (Fig. 1C) were created to visually show these relative peak changes, using the 1307 cm−1 peak as a constant background. These heat intensity maps showed a clear overall increase in the peak intensity ratios within the infected mapped areas compared to the control samples. This suggests alterations to biological components including hemoglobin29,32 (∼745, 1130 and 1170 cm−1), lipids30 (∼1130 cm−1) and other proteins30 (∼1170 cm−1) at this level of P. berghei infection. Ratio maps were also created for different samples to show this overall increase was common throughout the infected tissue (ESI Fig. S2†). Within these maps, for both uninfected and infected samples, particularly intense regions are present (Fig. 1C and ESI Fig. S2†). This can be explained through enhanced Raman intensity from resonance contributions of the biological components within these areas of tissue. These ratio maps visually show relative changes in biological peaks in a simple manner and can be used alongside empirical analysis and PCA to aid in classification of tissue types from Raman spectroscopy data.
Empirical analysis of Raman spectra
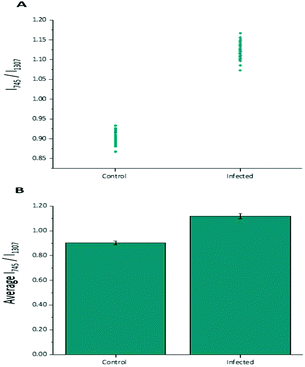
Diagnostic algorithms based on peak intensity ratios have been widely employed in the literature for tissue classification using Raman spectroscopy.18,19,23 These have been shown to successfully correlate variations in tissue spectra with tissue pathology19 in a simple manner.An empirical analysis based on the peak intensity ratio of two Raman bands was used for tissue classification within this study. Fig. 2A shows the scatter plot of the ratio of Raman peak intensity at 745 cm−1 to 1307 cm−1 (I745/I1307) for the 43 averaged P. berghei infected samples and 43 uninfected samples. A clear separation was observed in the scatter plot for the two data sets, with replicate data within each tissue type being grouped closely together, indicating little intra-sample variation in the intensity ratios. The mean ratio values (mean ± std. dev.) for both control (0.903 ± 0.015) and infected (1.119 ± 0.021) data sets were also plotted (Fig. 2B). It was shown that the differences between the mean ratios for each tissue type were statistically relevant (unpaired student's t-test, p < 0.0001). Infected tissue showed a higher ratio value compared with control, this could be explained by the increase in heme-based Raman vibrations (∼745 cm−1 assigned to v15 Hb29) due to the presence of hemozoin within the infected samples.
From these results, this intensity ratio is a potential simple and reliable marker to indicate the presence of infection. It has also been shown that other peak ratios can also be used to classify the two tissue types with good results (ESI Fig. S3†). However, this method does not consider the entire Raman spectrum. To help improve tissue analysis and classification, PCA was employed to highlight the most diagnostically significant Raman peaks.
Principal component analysis
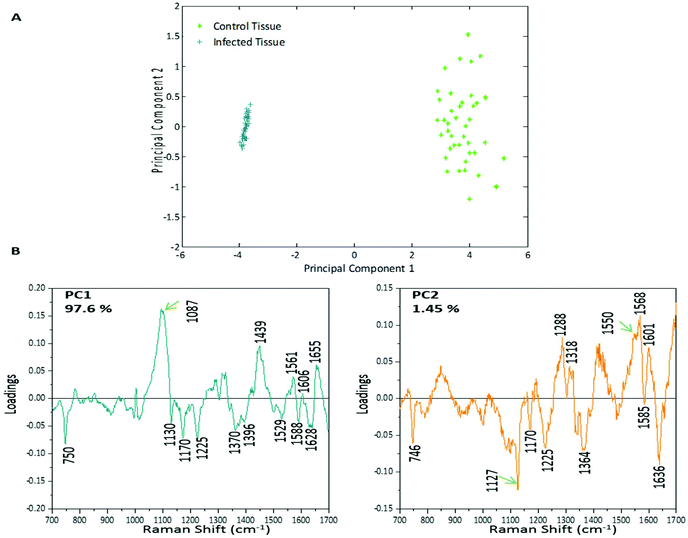
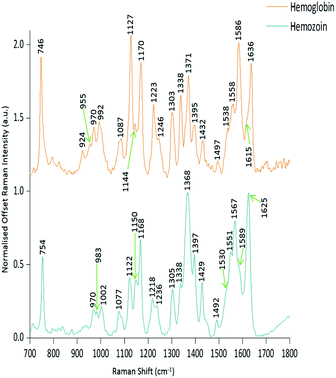
PCA was used to analyse this data set to better distinguish the spectral differences between the non-infected and infected spleen tissue. Fig. 3 shows the scores plot for PC1 (97.6% variance) and PC2 (1.45% variance), along with the corresponding loadings vectors. As can be seen from the scores plot (Fig. 3A) PC1 was primarily responsible for the separation of the two tissue types. The loadings plot (Fig. 3B) for PC1 indicated that the peaks in the negative half of the plot were spectrally important for describing the separation of the infected tissue (∼750, 1130, 1170, 1225, 1370, 1529, 1588 and 1628 cm−1), with the latter few being more indicative of hemozoin than hemoglobin. The negative peaks at ∼1529, 1588 and 1628 cm−1 matched closely with that of the hemozoin reference spectra in Fig. 4 (core structures of hemoglobin and hemozoin shown in ESI S4† to highlight structural similarities) and therefore suggested detection of the insoluble pigment by Raman. The negative ∼1370 cm−1 peak was present in both reference spectra; however, it was much more prominent within hemozoin and so suggested that hemozoin was contributing to this peaks appearance within the PC1 loadings plot. It is well known that hemozoin accumulates within the spleen8,9 (presence of hemozoin within the infected spleen can be seen in stained tissue images, ESI S5†), therefore it follows that there will be an associated increase in concentration of heme-based Raman vibrations. This is clearly shown in this PCA model as many of these peaks separating the infected tissue from the uninfected samples can be assigned to heme-vibrations, including hemozoin. Although other proteins and lipids will be present and exhibit Raman bands in similar positions to those of hemoglobin (∼1130, 1170, 1225 & 1396 cm−1), resonance effects would mean hemoglobin will tend to dominate the spectra (excitation in the regions of Q-absorption bands),29,32,33 with the other proteins and lipids having only small contributions to the observed spectra.Within the positive peaks in the PC1 loadings plot, the relatively broad ∼1087 cm−1 peak is likely to have a significant contribution from hemoglobin (Fig. 4), however, its broadness suggested contributions from other components. Raman bands within this region can also be assigned to C–C and O–P–O vibrations of nucleic acids.18,30 The other prominent positive peak at ∼1439 cm−1 can mainly be assigned to Raman vibrations from lipids,18,31 although it may also have slight hemoglobin contributions (∼1432 cm−1 peak in reference spectra). Analysis of the PC1 loadings plot indicated that at this level of P. berghei malaria infection Raman spectroscopy can clearly detect hemozoin accumulation within the spleen, with the positive peaks (∼1087 and 1439 cm−1) indicating that as the amount of hemozoin increases within the spleen there is a corresponding change to hemoglobin and some lipids/nucleic acid vibrations.
PC2 describes little separation between the control and infected samples. From the PC2 loadings plot (Fig. 3B) the positive peaks (∼1550, 1568, 1601 cm−1) originate mainly from hemoglobin vibrations, whereas the negative peaks have contributions from hemoglobin as well as from other proteins18,30 (∼1170, 1225 and 1636 cm−1) and lipids31 (∼1127 cm−1). This PCA analysis indicates that Raman imaging can easily distinguish between P. berghei infected spleen tissue from uninfected samples, through identification of hemozoin accumulation and alterations to other key biological components.
Conclusions
In conclusion, a combination of Raman imaging with multivariate analysis resulted in very clear discrimination of P. berghei infected tissue from non-infected samples. Separation highlighted an overall increase in heme-based Raman vibrations within the infected samples, which correlates with the accumulation of hemozoin within the spleen contributing to this. Hemozoin detection was clear with the identification of key peaks occurring ∼1370, 1529, 1588, and 1628 cm−1 which matched closely with the reference spectra. Chemometric analysis also showed that at the level of P. berghei infection reported here, changes to hemoglobin and some other lipid/nucleic acid vibrations corresponded with an increase in hemozoin content within the spleen. Other proteins (tyrosine ∼1130 cm−1, and amide III ∼1225 cm−1) and lipids (∼1130 cm−1) were also contributing to the separation of infected tissue from control. However, due to the resonance effects of heme-based compounds the separation contribution from these biological components would have been small.The use of peak ratio maps and empirical analysis also highlighted changes to the biological components as you move towards an infected tissue state. These two simple data analysis methods could be used as tools in combination with the PCA to help improve efficiency of Raman tissue analysis and classification.
As the Raman imaging technique used in this study had a short penetration depth, it would not be suitable for in vivo diagnostics. However, with the development of spatially offset Raman spectroscopy, which allows accurate analysis through millimetres of material when obscuring barriers such as skin are present,34,35 this may allow for biochemical information from tissue to be detected in vivo in the near future. Overall, this approach demonstrates the potential use of Raman spectroscopy to provide detailed chemical information from both malaria parasite infected and control tissue and when coupled with PCA, discrimination due to infection burden is achievable between the two sample groups.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the University of Strathclyde. Data associated with research published in this paper is accessible at: http://dx.doi.org/10.15129/89483183-40b2-4ebb-8b10-43a4615e40db and http://dx.doi.org/10.15129/b069f380-61ae-494d-9419-dcdf28a5b160.Notes and references
- World Health Organisation, World Malaria Report 2015, 2015 Search PubMed.
- W. C. Aird, L. O. Mosnier and R. M. Fairhurst, Blood, 2014, 123, 163–168 CrossRef CAS PubMed.
- CDC: malaria parasites, https://www.cdc.gov/malaria/about/biology/parasites.html (accessed Sep 22, 2017).
- T. Frosch, S. Koncarevic, K. Becker and J. Popp, Analyst, 2009, 134, 1126–1132 RSC.
- L. M. Coronado, C. T. Nadovich and C. Spadafora, Biochim. Biophys. Acta, 2014, 1840, 2032–2041 CrossRef CAS PubMed.
- T. Frosch, S. Koncarevic, L. Zedler, M. Schmitt, K. Schenzel, K. Becker and J. Popp, J. Phys. Chem. B, 2007, 111, 11047–11056 CrossRef CAS PubMed.
- M. Boura, R. Frita, A. Góis, T. Carvalho and T. Hänscheid, Trends Parasitol., 2013, 29, 469–476 CrossRef CAS PubMed.
- M. Ferrer, L. Martin-Jaular, M. De Niz, S. M. Khan, C. J. Janse, M. Calvo, V. Heussler and H. A. Del Portillo, Parasitol. Int., 2014, 63, 195–205 CrossRef PubMed.
- H. A. DelPortillo, M. Ferrer, T. Brugat, L. Martin-Jaular, J. Langhorne and M. V. G. Lacerda, Cell. Microbiol., 2012, 14, 343–355 CrossRef CAS PubMed.
- C. R. Engwerda, L. Beattie and F. H. Amante, Trends Parasitol., 2005, 21, 75–80 CrossRef PubMed.
- B. Franke-Fayard, J. Fonager, A. Braks, S. M. Khan and C. J. Janse, PLoS Pathog., 2010, 6, 1–10 Search PubMed.
- B. Autino, Y. Corbett, F. Castelli and D. Taramelli, Mediterr. J. Hematol. Infect. Dis., 2012, 4, 1–12 Search PubMed.
- D. S. Khoury, D. Cromer, S. E. Best, K. R. James, P. S. Kim, C. R. Engwerda, A. Haque and M. P. Davenport, Infect. Immun., 2014, 82, 212–220 CrossRef PubMed.
- B. R. Wood, S. J. Langford, B. M. Cooke, F. K. Glenister, J. Lim and D. McNaughton, FEBS Lett., 2003, 554, 247–252 CrossRef CAS PubMed.
- B. R. Wood, A. Hermelink, P. Lasch, K. R. Bambery, G. T. Webster, M. A. Khiavi, B. M. Cooke, S. Deed, D. Naumann and D. McNaughton, Analyst, 2009, 134, 1119–1125 RSC.
- A. J. Hobro, A. Konishi, C. Coban and N. I. Smith, Analyst, 2013, 138, 3927–3933 RSC.
- M. J. Hackett, J. B. Aitken, F. El-Assaad, J. a. McQuillan, E. a. Carter, H. J. Ball, M. J. Tobin, D. Paterson, M. D. de Jonge, R. Siegele, D. D. Cohen, S. Vogt, G. E. Grau, N. H. Hunt and P. A. Lay, Sci. Adv., 2015, 1, 1–13 Search PubMed.
- K. Kochan, K. M. Marzec, K. Chruszcz-Lipska, A. Jasztal, E. Maslak, H. Musiolik, S. Chłopicki and M. Baranska, Analyst, 2013, 138, 3885–3890 RSC.
- S. K. Teh, W. Zheng, K. Y. Ho, M. Teh, K. G. Yeoh and Z. Huang, Br. J. Cancer, 2008, 98, 457–465 CrossRef CAS PubMed.
- C. J. Frank, R. L. McCreery and D. C. Redd, Anal. Chem., 1995, 67, 777–783 CrossRef CAS PubMed.
- T. Huser and J. Chan, Adv. Drug Delivery Rev., 2015, 89, 57–70 CrossRef CAS PubMed.
- A. J. Hobro, N. Pavillon, K. Fujita, M. Ozkan, C. Coban and N. I. Smith, Analyst, 2015, 140, 2350–2359 RSC.
- Z. Huang, A. McWilliams, H. Lui, D. I. McLean, S. Lam and H. Zeng, Int. J. Cancer, 2003, 107, 1047–1052 CrossRef CAS PubMed.
- A. S. Haka, K. E. Shafer-Peltier, M. Fitzmaurice, J. Crowe, R. R. Dasari and M. S. Feld, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 12371–12376 CrossRef CAS.
- Tissue-Tek O.C.T Compound and Cryomolds, http://www.sakura.eu/Our-products/item/11/Cryotomy/48/Tissue-Tek-OCT-Compound-and-Cryomolds, (accessed Sep 15, 2017).
- T. J. Egan, D. C. Rossa and P. A. Adamsb, FEBS Lett., 1994, 352, 54–57 CrossRef CAS PubMed.
- T. J. Egan, W. W. Mavuso and K. K. Ncokazi, Biochemistry, 2001, 40, 204–213 CrossRef CAS PubMed.
- P. H. C. Eilers and H. F. M. Boelens, Life Sci., 2005, 1–26 Search PubMed.
- G. Rusciano, A. C. De Luca, G. Pesce and A. Sasso, Sensors, 2008, 8, 7818–7832 CrossRef CAS PubMed.
- Z. Movasaghi, S. Rehman and I. U. Rehman, Appl. Spectrosc. Rev., 2007, 42, 493–541 CrossRef CAS.
- C. Krafft, D. Codrich, G. Pelizzo and V. Sergo, Analyst, 2008, 133, 361–371 RSC.
- M. Polakovs, N. Mironova-Ulmane, N. Kurjane, E. Reinholds and M. Grube, Proc. SPIE, 2008, 7142, 1–8 CrossRef.
- T. G. Spiro and T. C. Strekas, J. Am. Chem. Soc., 1974, 96, 338–345 CrossRef CAS PubMed.
- N. Stone and P. Matousek, Cancer Res., 2008, 68, 4424–4430 CrossRef CAS PubMed.
- M. D. Keller, S. K. Majumder and A. Mahadevan-Jansen, Opt. Lett., 2009, 34, 926–928 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7an01760j |
| This journal is © The Royal Society of Chemistry 2018 |