Open Access Article
Open Access ArticleMolecular crowding induces primer extension by RNA polymerase through base stacking beyond Watson–Crick rules†
Shuntaro Takahashi a,
Hiromichi Okuraa,
Pallavi Chilkaa,
Saptarshi Ghosha and
Naoki Sugimoto
a,
Hiromichi Okuraa,
Pallavi Chilkaa,
Saptarshi Ghosha and
Naoki Sugimoto *ab
*ab
aFrontier Institute for Biomolecular Engineering Research (FIBER), Konan University, 7-1-20 Minatojima-minamimachi, Kobe 650-0047, Japan
bGraduate School of Frontiers of Innovative Research in Science and Technology (FIRST), Konan University, 7-1-20 Minatojima-minamimachi, Kobe 650-0047, Japan. E-mail: sugimoto@konan-u.ac.jp; Fax: +81-78-303-1495; Tel: +81-78-303-1416
First published on 7th September 2020
Abstract
The polymerisation of nucleic acids is essential for copying genetic information correctly to the next generations, whereas mispolymerisation could promote genetic diversity. It is possible that in the prebiotic era, polymerases might have used mispolymerisation to accelerate the diversification of genetic information. Even in the current era, polymerases of RNA viruses frequently cause mutations. In this study, primer extension under different molecular crowding conditions was measured using T7 RNA polymerase as a model for the reaction in the prebiotic world. Interestingly, molecular crowding using 20 wt% poly(ethylene glycol) 2000 preferentially promoted the primer extensions with ATP and GTP by T7 RNA polymerase, regardless of Watson–Crick base-pairing rules. This indicates that molecular crowding decreases the dielectric constants in solution, resulting in enhancement of stacking interactions between the primer and an incorporated nucleotide. These findings suggest that molecular crowding could accelerate genetic diversity in the prebiotic world and may promote transcription error of RNA viruses in the current era.
Introduction
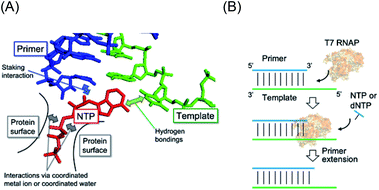
High fidelity oligonucleotide synthesis is beneficial for maintaining genetic information over many generations and preventing mutations that can initiate and promote diseases. For accurate oligonucleotide synthesis, polymerases must distinguish the correct nucleotide that pairs with the complementary base in the template strand based on Watson–Crick (WC) hydrogen bonding (Fig. 1A).1 On the contrary, oligonucleotide synthesis with low fidelity is beneficial for the evolution of species,2,3 for enhanced genetic diversity leading to increased survival of viruses and microbes when subjected to environmental change,4 and for the development of an immune system.5 Mutation rates tend to be minimised and approach the lower limits imposed by the efficiency of selection or the physiological costs of replication fidelity.6 Among all species, RNA viruses such as human immunodeficiency virus (HIV-1), influenza virus, and the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) constitute a major group of pathogens characterised by their extremely high rates of spontaneous mutation. For example, the mutation rate of HIV-1 in vivo is 4.1 × 10−3 per base per cell.7 This rate is orders of magnitude higher than those of DNA-based organisms and DNA viruses.8 However, DNA-dependent RNA polymerases in higher species operate with much higher fidelity due to their proofreading activity.9 According to established evolutionary hypotheses, the precursors of enzymes could have been less specific than current enzymes, which evolved to possess higher specificities.10 Therefore, virus polymerases should be the closest to those in the prebiotic world owing to their small molecular size and low polymerisation fidelity.The low fidelity of such ancestral and viral polymerases implies that the mismatched substrate is also allowed to be incorporated. The polymerisation reaction occurs when the incorporating substrate stably pairs a base on the template in the catalytic centre of the polymerase. The efficiency of polymerisation is determined by the stability of the intermediate of the base pair. As a mismatch base pair sometimes stabilises the duplex structure of nucleic acids, the incorporation of a non-cognate substrate can occur.11 When considering the stability of nucleic acid duplexes, not only hydrogen bonding between base pairs but also stacking interactions between the nearest base pairs are important. Interestingly, the energies of hydrogen bonding and stacking interactions have a compensatory relationship.12 This finding suggests that the incorporation of rUTP and rATP, which form fewer hydrogen bonds with the cognate counterparts of dATP and dTTP than those of rGTP and rCTP, efficiently occurs due to energetic compensation via stacking interactions at the active site of RNA polymerases. Therefore, manipulation of the energies of hydrogen bonding and stacking interactions should enable changes in the fidelity of polymerisation.
Actually, the stability of base pairs is influenced by environmental factors. One of the important factors in cellular conditions is molecular crowding. Molecular crowding critically affects structures, affinities, and reactions of biomolecules by altering properties of the solution, including decreases in the dielectric constant13 and water activity,14–16 and increases in viscosity17 and excluded volume.18 The base pairs of a DNA duplex, RNA duplex, and DNA/RNA hybrid are destabilised by molecular crowding, but the magnitude of destabilisation is different between them.19–21 Furthermore, the instability caused by mismatched base pairs can become very slight compared to that by matched pairs.22 These altered properties can affect forces for substrate recognition by polymerases.23 The in vivo mutation rate in viruses is two orders of magnitude higher than that predicted by in vitro studies;7 thus, it is possible that the chemical environment of polymerase reactions influences polymerisation fidelity. Recently, we investigated primer extension along RNA templates as a model of prebiotic gene replication, which is different from the canonical transcription reaction requiring the promoter sequence (Fig. 1B).24 We used single-subunit RNA polymerase, T7 RNA polymerase (T7 RNAP), as a model of the ancestral RNAP25 and found that T7 RNAP preferentially incorporated dNTPs over NTPs under specific crowding conditions.24 Moreover, the fidelity of NTP incorporation was decreased in the crowding condition. These results suggest that primer extension by T7 RNAP enables non-canonical polymerisation of RNA and DNA, which is highly responsive to molecular crowding. However, primer extension along a DNA template is also an interesting reaction because transcription is a reaction to polymerise RNA along the DNA sequence. It is possible that, in the prebiotic world, reactions using RNA and DNA templates have been distinguished. However, primer extension using a DNA template as a model of transcription has not been addressed.
In this study, we investigated the effect of molecular crowding on substrate selectivity of primer extension along a DNA template by T7 RNAP. Regardless of the matching of the 3′ terminus of the primer with the template DNA, primer extension by NTP incorporation occurred. Interestingly, the fidelity of polymerisation using the mismatched primer was better than that of the matched primer. Furthermore, in the crowding condition with polyethylene glycol 2000 (PEG2000, average molecular weight 2000), ATP and GTP were favoured as substrates, thus lowering the fidelity of polymerisation. These results suggest that the crowding condition induced substrate selection via stacking interactions over Watson–Crick base-pairings due to a decrease in the dielectric constant of the solution.
Results and discussion
Extension of RNA primer by T7 RNAP under molecular crowding conditions
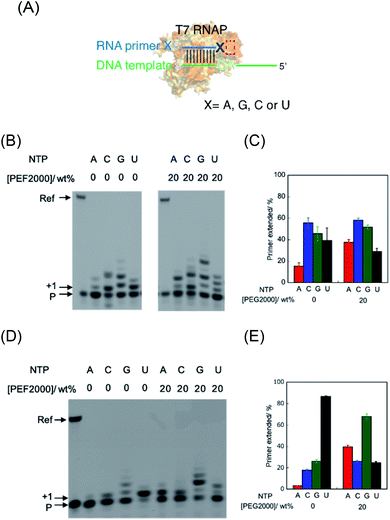
The T7 RNAP was used as a model of RNAPs to understand the effect of molecular crowding on primer extension mimicking a prebiotic reaction.24 T7 RNAP is known to first bind the DNA promoter region and initiate RNA polymerisation from downstream of the promoter region by forming an elongation complex with the template DNA and the nascent RNA strand. However, T7 RNAP can also form elongation complexes only through incubation with an RNA oligonucleotide as a primer and a DNA oligonucleotide as a template, which resembles the RNA elongation step (Fig. 2A).26 Thus, T7 RNAP can start the polymerisation of nucleotides without an initiation step.24,27 Therefore, after forming the elongation complex, the fidelity of the base elongated at the primer terminus can be estimated.First, we examined polymerisation using RNA primer G, which matches to form a WC base pair with cytosine at the 3′ end of the template DNA. As a result, all four NTPs were polymerised (Fig. 2B), although UTP should have been specifically incorporated against adenine in the template DNA (Fig. 2A), as per transcriptional rules. Moreover, there are extra elongated primers comprising more than a single base extension with each NTP. To quantitatively analyse the extension, the percentage of primers extended was calculated as the fluorescence intensity (LAU mm−2) of all bands of the extended primer (products showing larger molecular weight than the primer) divided by the summed fluorescence intensities (LAU mm−2) of all detectable bands, including those of the primer. After performing the reaction for 1 hour, 38% of the primer was extended with UTP as a substrate, whereas the primer was more extended with CTP (55%) and GTP (43%) (Fig. 2C). We previously reported that primer extension by T7 RNAP using RNA primer G and RNA template favours the polymerisation of matched UTP, although CTP and GTP also react slightly.24 A DNA/RNA duplex forms an A-like helix but shows a slightly longer helical rise and smaller helical twist per base pair than that in an RNA/RNA duplex that forms an A-type helix.28 Therefore, it is possible that the helical structure geometry of the RNA primer and the DNA template did not match in the active centre of T7 RNAP to catalyse polymerisation based on the formation of the WC base pair. Instead of WC formation, another interaction, such as mismatch base pairing or a stacking interaction, might drive the promotion of the substrate geometry to catalyse the T7 RNAP.
Next, we used RNA primer A, which does not match with cytosine to form a WC base pair of the counterpart base on the template DNA (Fig. 2A). Interestingly, the cognate substrate, UTP, was preferentially polymerised more than the other NTPs. Specifically, 86% of the primer was extended with UTP as a substrate (Fig. 2D and E). In the cases of ATP, CTP, and GTP, only 4.1%, 17%, and 26% of the primers were extended, respectively. This result indicates that T7 RNAP preferentially extends UTP, which forms WC base pairing with an adenine base in the template. The different reactivity between these primers suggests that the mismatch base pair between rA and dC provided different geometry of the primer terminus, which promoted the reaction in the catalytic centre via WC base-pair formation, as previously observed in the reaction using an RNA primer and RNA template.24 We also tested mismatched RNA primer C and RNA primer U. As observed in the case of RNA primer A, UTP was preferentially incorporated compared to that in the case using matched RNA primer G (Fig. S1†), although GTP was well-incorporated in the case of RNA primer C, which suggests that polymerisation based on a non-WC rule also occurred. Therefore, in the primer extension by T7 RNAP using the RNA primer and DNA template, the reaction can be driven based on the WC rule or non-WC rule according to the environment of the catalytic centre.
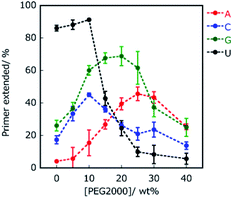
The environment-dependent manner of this reaction suggests variability of polymerisation substrate selection, which is mediated by environmental change. Thus, we investigated the effect of molecular crowding on substrate selection of the primer extension using an RNA primer and DNA template. As a crowding reagent, PEG2000 was utilised because it is one of the well-studied crowding reagents.29 For the reaction using RNA primer G (matched), the addition of 20 wt% PEG2000 promoted the extension of ATP (37%) but suppressed the extension of UTP (29%), compared to the same reaction in the absence of PEG2000 (15% and 39%, respectively as shown in Fig. 2B and C). However, the extensions of CTP (58%) and GTP (51%) were only slightly promoted by the addition of PEG2000. More drastic changes were observed in the reaction using RNA primer A (mismatched) (Fig. 2C and D). Interestingly, at 20 wt% PEG2000, T7 RNAP preferentially incorporated GTP (67%), ATP (41%), and CTP (26%) over UTP (23%), indicating that the molecular environment enabled T7 RNAP to change substrate specificity. These different tendencies of GTP between primer G and primer A could be caused by the stronger influences of dipole interactions of GTP with primer G than with primer A. Furthermore, we changed the concentration of PEG2000 to study the trend in substrate specificity with respect to crowding magnitude (Fig. 3). When PEG2000 was added at a concentration of 10 wt%, the amount of primer extended with each NTP increased. Further increasing the concentration of PEG2000 from 10 wt% to 20 wt% promoted the extensions of ATP and GTP but suppressed the extensions of CTP and UTP. At 20 wt% PEG2000, T7 RNAP preferentially incorporated GTP and ATP over UTP. At concentrations of PEG2000 greater than 20 wt%, the extended primer levels decreased with each NTP. From these results, it was revealed that the peak of effective extension for each NTP correlated with specific peak concentrations of PEG2000. The peak concentrations of PEG2000 for purines (25 wt% for ATP and 20 wt% for GTP) were higher than those for pyrimidines (10 wt% for CTP or UTP). The stacking interaction is stronger between purine bases than between pyrimidine bases.30 Thus, substrate selectivity of T7 RNAP could be differentiated according to the stacking interaction beyond WC base-pairing.
Effect of dielectric constants on the extension of RNA primer
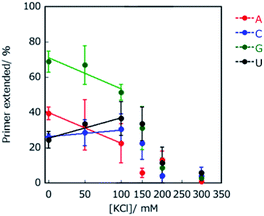
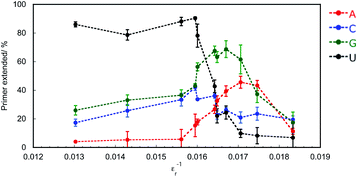
To investigate the contribution of electrostatic interactions between ions, dipole moments, and/or induced dipole moments for substrate-dependent effects of molecular crowding on primer extension, the primer A was extended in solutions containing 20 wt% PEG2000 and various concentrations of KCl, which were 0, 50, 100, 150, 200, and 300 mM. Extended primer amounts showed linear correlations with KCl concentration, although these correlations were lost at concentrations higher than 150 mM KCl (Fig. 4). The lower KCl concentrations (<150 mM) reflect the different kinds of electrostatic interactions, whereas KCl concentrations higher than 150 mM would be sufficient to inhibit binding of T7 RNAP to the template and the primer to catalyse the reaction. At lower KCl concentrations, with an increase in KCl concentration, the amounts of extended primer with ATP and GTP decreased, but those with CTP and UTP increased. The difference in the trends with KCl is linked to the identity of nucleotide bases on NTPs, that is, purines or pyrimidines. These trends are also consistent with the extension trends earlier described here. Thus, differences in electrostatic interactions of NTPs at the incorporation can cause differences in the extension efficiencies of purine NTPs and pyrimidine NTPs in the solutions containing PEG2000.Crowding reagents, including PEGs, change the solution properties such as water activity, dielectric constant, and the excluded volume effect. As structure and stability of DNA and RNA can be affected by the changes in water activity and dielectric constant,29 we hypothesised that either of these changes or both are the main cause of the difference in extension efficiencies of purine NTPs and pyrimidine NTPs in the solutions containing PEG2000. To examine the effects of water activity and dielectric constant on primer extension with each NTP, crowding reagents with different molecular weights were next examined. Water activity was determined by the osmotic stress method using vapour phase osmometry (Table S1†).31 Dielectric constants were determined by a blue shift in the emission maximum of the fluorescent probe, 1-anilino-8-naphthalene sulfonate.32 As the average molecular weights increased from 68 (ethylene glycol; EG) to 2000 (PEG2000), the values of water activity, aw, increased and those of dielectric constant, εr, decreased, respectively, in the presence of 20 wt% of each crowding reagent (Table S2†). Although the amounts of extended primers with NTPs did not continuously change with the values of aw (Fig. S2†), the amounts did continuously change with the values of εr−1 (Fig. 5). In the case of 20 wt% EG (molecular weight is 68), εr was 70.0 (εr−1 = 0.0143). The extended primer amount with each NTP was slightly changed (5.1% for ATP, 23% for CTP, 31% for GTP, and 81% for UTP), compared to that in the absence of crowding reagents (ερ = 76.8, εr−1 = 0.0130). In the case of εr−1 increasing from 0.016 to 0.017, the extended primer amounts with purine NTPs increased, and those with pyrimidine NTPs decreased. Although the relationship between εr−1 and the amount of primer extended with each NTP was not linear, larger εr−1 values were associated with larger extended amounts with purine NTPs and smaller extended amounts with pyrimidine NTPs. The potential energy of the interactions between induced dipole moments, which is the main force in base stacking, and the electric static forces have a ratio of the inverse square of εr. Therefore, this result suggests that interactions between induced dipole moments cause the substrate-dependent effects of molecular crowding on primer extension. Additionally, purine NTPs have wider stacking areas than pyrimidine NTPs. Importantly the trends in primer extension mediated by changing the dielectric constant (Fig. 5) resembled those of the crowding reagent concentration as observed in Fig. 3. Regarding the excluded volume effect, the volumetric effect of the incorporating NTP stacked on the end of the primer should be considered. In the case of DNA, it has been reported that the order of volumetric contribution to the formation of the nearest neighbour base pair was AC < AG < AT < AA.33 This order indicates how the excluded volume effect destabilises base stacking. Thus, conversely, the order of AA < AT < AG < AC is the expected order for substrate selection of the primer extension because the reaction having smaller volumetric changes is stabilized by the excluded volume effect. However, the primer extension in the presence of 20 wt% PEG2000 was in the order of AG < AA < AC < AU, suggesting that the contribution of the excluded volume effect was not substantial for the substrate selection of the primer extension. Therefore, decreasing dielectric constants by molecular crowding is the main physical factor that changes the substrate specificity and promotes the incorporation of purine NTPs by enhancing stacking interactions with purine bases.
Proposed mechanism of RNA primer extension regulated by molecular crowding conditions
RNA extension progresses in a series of steps. Fig. 6 shows the proposed mechanism of primer extension mediated by T7 RNAP in crowding and non-crowding conditions. In the non-crowding condition, T7 RNAP initially captures the NTPs on the template stand via WC base-pairing at the 3′ terminus, as the T7 RNAP is at the “initial binding position”, which is far from the 3′ terminus of the primer where the NTPs attach (Fig. 6 left and see also Fig. S3†).26 The ionic interactions between the phosphate group of NTPs and the basic amino acids (Arg 627 and Lys 631) of T7 RNAP leads to conformational changes. The conformational change orients NTPs for the polymerase reaction at the “Mg2+ binding position”, which is nearer to the 3′ terminus of the primer than the initial binding position and is coordinated via Mg2+ ion between phosphate groups of NTPs and Asp 537 and Asp 812 (see Fig. S3†).27 Considering the substrate-binding pocket of T7 RNAP, the decreasing dielectric constant caused by molecular crowding would promote the direct incorporation of NTPs at the Mg2+ binding position by enhancing stacking interactions between the purine bases of NTPs and the 3′ terminal base of the primer (Fig. 6 right). This kind of direct incorporation of NTPs is feasible because this kind of complex structure was reported in de novo RNA priming.34,35 These findings suggest that not just T7 RNAP, but any DNA-dependent RNA polymerases in viruses that orient at the 3′ base of the primer onto the solvent-accessible surface, could extend purine NTPs at high concentrations of crowding reagents by decreasing the dielectric constant.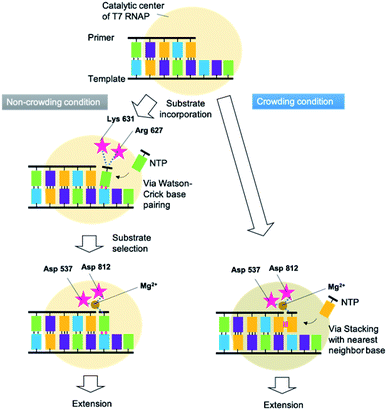 | ||
| Fig. 6 Proposed mechanisms of primer extension mediated by T7 RNAP and regulated by crowding and non-crowding conditions. | ||
T7 RNAP is one of the smallest polymerases. A phylogenetic study revealed that these T7-like RNAPs share a common ancestor and that DNA polymerases and reverse transcriptases evolved from this polymerase.36,37 These backgrounds of T7 RNAP suggest that in the prebiotic world, the DNA and RNA polymerases might have increased their mutation rate in response to the changes in the dielectric constant of the solution. Even in the current era, changes in the substrate preference induced by molecular crowding might generally occur with respect to the RNA polymerase of RNA viruses. Therefore, molecular crowding, depending on the environment of the host cell, might enable RNA viruses to promote genetic diversity at the transcription level. This study suggests that it could be important to consider the dielectric constant in the cells of host organisms, which would cause an inflated mutation rate of HIV in vivo and pandemic-related RNA viruses through mutations in their genomes.
Conclusions
In conclusion, molecular crowding using concentrated PEG2000 shifted preferable substrates of T7 RNA polymerase from substrates which formed WC base pairs with template bases to purine NTPs. The purine NTPs were incorporated by stacking interactions between a purine base and the 3′ terminal base of the primer beyond the rule of WC formation. In the prebiotic world, the polymerase might need to both catalyse correct polymerisation to transfer genetic information to the next generation and incorrect polymerisation to induce evolution. Our findings suggest that molecular crowding emphasises stacking interactions more than WC base-pairing, which results in the switching of substrate specificity based on the simple chemistry between nucleotides. This mechanism could have accelerated genetic diversity in the prebiotic world. The structural similarity between T7 RNAP and other RNA polymerases in viruses implies that even in the current era, the transcriptional mutation rate might be enhanced, as found based on the inflated mutation rate of HIV in vivo and the spread of infection mediated by RNA viruses that are triggered by genomic mutations.Experimental
Materials
NTP solutions were purchased from Thermo Scientific Japan, EG and PEG200 were purchased from Wako Pure Chemicals Japan. PEG2000 was purchased from Sigma-Aldrich. FITC-labelled RNA primers and DNA templates were purchased from Japan Bio Service. These were used without further purification. T7 RNAP was purchased from Takara Bio. Other reagents were purchased from Wako Pure Chemicals.Primer extension assay
T7 RNAP (0.5 μM) was incubated with fluorescein isothiocyanate (FITC)-labelled 10-mer RNA primer G (5′- fluorescein-CUGCCAACCG-3′), RNA primer A (5′-fluorescein-CUGCCAACCA-3′), RNA primer C (5′-fluorescein-CUGCCAACCC-3′), or RNA primer U (5′-fluorescein-CUGCCAACCU-3′) (0.5 μM in each case), 39-mer DNA template (5′-GUCAAUGACACGCUUCGCACGGUUGGCAGAAAAAAAAAA-3′) (0.5 μM), and each NTP (100 μM) in 50 mM Tris–HCl (pH 8.0) and 10 mM MgCl2 at 25 °C for 1 hour. RNA primer A, C, and U were designed for mispolymerisation, while primer G was for correct continuous elongation. Reactions were performed in 0–40 wt% crowding reagents. The complementary strand of the DNA template was used as a reference for electrophoresis. Reactants and products of the polymerase reaction were separated by denaturing polyacrylamide gel electrophoresis (PAGE) as reported previously.24 The gel images were captured using a Fujifilm FLA-5100 fluorescent imager. Bands corresponding to extended primers and unreacted primers were quantified by measuring fluorescence intensity as follows: the percentage of primers extended was calculated as the fluorescence intensity (LAU mm−2) of the bands of the extended primers divided by the summed fluorescence intensities (LAU mm−2) of all detectable bands. Data are averages of three samples. Errors are standard deviations.Measurements of solution properties
Solution properties were examined using solutions containing 50 mM Tris–HCl (pH 8.0), 2 mM MgCl2, and 0–40 wt% co-solute at 25 °C. Water activity was determined by the osmotic stress method using vapour phase osmometry (Wescor pressure osmometer, 5520XR). For determining the dielectric constant, the fluorescent probe 1,8-ANS (1-anilino-8-naphthalene sulfonate) was employed in sample solutions. The probe responds to changes in the dielectric property of a solution by a shift in the blue emission maximum, particularly in media of low dielectric constants. Fluorescence emission spectra, with excitation at 356 nm, were measured using a fluorometer (JASCO, F-6500). The dielectric constant was calculated using a standard curve of several organic solvents with known values.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), the Japan Society for the Promotion of Science (JSPS); a Grant-in-Aid for Scientific Research on Innovative Areas “Chemistry for Multimolecular Crowding Biosystems” (JSPS KAKENHI Grant No. JP17H06351), a JSPS International Research Fellowship to S. G. (19F19337), the Hirao Taro Foundation of Konan Gakuen for Academic Research, the Chubei Itoh Foundation and the Nakatani Foundation for the advancement of measuring in biomedical engineering.Notes and references
- E. T. Kool, Annu. Rev. Biophys. Biomol. Struct., 2001, 30, 1–22 CrossRef CAS.
- D. Metzgar and C. Wills, Cell, 2000, 101, 581–584 CrossRef CAS.
- O. J. Rando and K. J. Verstrepen, Cell, 2007, 128, 655–668 CrossRef CAS.
- R. Moxon, C. Bayliss and D. Hood, Annu. Rev. Genet., 2006, 40, 307–333 CrossRef CAS.
- S. B. Lloyd, S. J. Kent and W. R. Winnall, AIDS Res. Hum. Retroviruses, 2014, 30, 8–16 CrossRef.
- M. Lynch, Trends Genet., 2010, 26, 345–352 CrossRef CAS.
- J. M. Cuevas, R. Geller, R. Garijo, J. López-Aldeguer and R. Sanjuán, PLoS Biol., 2015, 13, e1002251 CrossRef.
- S. Duffy, L. A. Shackelton and E. C. Holmes, Nat. Rev. Genet., 2008, 9, 267–276 CrossRef CAS.
- J. F. Sydow and P. Cramer, Curr. Opin. Struct. Biol., 2009, 19, 732–739 CrossRef CAS.
- R. A. Jensen, Annu. Rev. Microbiol., 1976, 30, 409–425 CrossRef CAS.
- N. Sugimoto, M. Nakano and S. Nakano, Biochemistry, 2000, 39, 11270–11281 CrossRef CAS.
- D. H. Turner, N. Sugimoto, R. Kierzek and S. D. Dreiker, J. Am. Chem. Soc., 1987, 109, 3783–3785 CrossRef CAS.
- R. Harada, Y. Sugita and M. Feig, J. Am. Chem. Soc., 2012, 134, 4842–4849 CrossRef CAS.
- S. Nakano, H. Karimata, T. Ohmichi, J. Kawakami and N. Sugimoto, J. Am. Chem. Soc., 2004, 126, 14330–14331 CrossRef CAS.
- M. J. Blandamer, J. B. Engberts, P. T. Gleeson and J. C. Reis, Chem. Soc. Rev., 2005, 34, 440–458 RSC.
- D. Miyoshi, H. Karimata and N. Sugimoto, J. Am. Chem. Soc., 2006, 128, 7957–7963 CrossRef CAS.
- N. Kozer, Y. Y. Kuttner, G. Haran and G. Schreiber, Biophys. J., 2007, 92, 2139–2149 CrossRef CAS.
- A. P. Minton, J. Biol. Chem., 2001, 276, 10577–10580 CrossRef CAS.
- S. Pramanik, S. Nagatoishi, S. Saxena, J. Bhattacharyya and N. Sugimoto, J. Phys. Chem. B, 2011, 115, 13862–13872 CrossRef CAS.
- S. Ghosh, S. Takahashi, T. Ohyama, T. Endoh, H. Tateishi-Karimata and N. Sugimoto, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 14194–14201 CrossRef CAS.
- M. S. Adams and B. M. Znosko, Nucleic Acids Res., 2019, 47, 3658–3666 CrossRef CAS.
- Y. Teng, S. Pramanik, H. Tateishi-Karimata, T. Ohyama and N. Sugimoto, Biochem. Biophys. Res. Commun., 2018, 496, 601–607 CrossRef CAS.
- Y. Sasaki, D. Miyoshi and N. Sugimoto, Biotechnol. J., 2006, 1, 440–446 CrossRef CAS.
- S. Takahashi, H. Okura and N. Sugimoto, Biochemistry, 2019, 58, 1081–1093 CrossRef CAS.
- D. Forrest, K. James, Y. Yuzenkova and N. Zenkin, Nat. Commun., 2017, 8, 15774 CrossRef CAS.
- D. Temiakov, V. Patlan, M. Anikin, W. T. McAllister, S. Yokoyama and D. G. Vassylyev, Cell, 2004, 116, 381–391 CrossRef CAS.
- R. Sousa and R. Padilla, EMBO J., 1995, 14, 4609–4621 CrossRef CAS.
- G. L. Conn, T. Brown and G. A. Leonard, Nucleic Acids Res., 1999, 27, 555–561 CrossRef CAS.
- S. Nakano, D. Miyoshi and N. Sugimoto, Chem. Rev., 2014, 114, 2733–2758 CrossRef CAS.
- K. H. Scheller, F. Hofstetter, P. R. Mitchell, B. Prijs and H. Sigel, J. Am. Chem. Soc., 1981, 103, 247–260 CrossRef CAS.
- R. Goobes, N. Kahana, O. Cohen and A. Minsky, Biochemistry, 2003, 42, 2431–2440 CrossRef CAS.
- S. Nakano, H. Hirayama, D. Miyoshi and N. Sugimoto, J. Phys. Chem. B, 2012, 116, 7406–7415 CrossRef CAS.
- D. N. Dubins and R. B. Macgregor, Biopolymers, 2004, 73, 242–257 CrossRef CAS.
- W. P. Kennedy, J. R. Momand and Y. W. Yin, J. Mol. Biol., 2007, 370, 256–268 CrossRef CAS.
- R. S. Basu, B. A. Warner, V. Molodtsov, D. Pupov, D. Esyunina, C. Fernandez-Tornero, A. Kulbachinskiy and K. S. Murakami, J. Biol. Chem., 2014, 289, 24549–24559 CrossRef CAS.
- N. Glansdorff, Y. Xu and B. Labedan, Biol. Direct, 2008, 3, 29 CrossRef.
- F. Werner and D. Grohmann, Nat. Rev. Microbiol., 2011, 9, 85–98 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra06502a |
| This journal is © The Royal Society of Chemistry 2020 |