Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Fluorescence imaging of a potential diagnostic biomarker for breast cancer cells using a peptide-functionalized fluorogenic 2D material†
Wei-Tao
Dou‡
a,
Li-Fang
Liu‡
ab,
Jie
Gao
ab,
Yi
Zang
*b,
Guo-Rong
Chen
a,
Robert A.
Field
 c,
Tony D.
James
c,
Tony D.
James
 d,
Jia
Li
*b and
Xiao-Peng
He
d,
Jia
Li
*b and
Xiao-Peng
He
 *a
*a
aKey Laboratory for Advanced Materials, Feringa Nobel Prize Scientist Joint Research Center, School of Chemistry and Molecular Engineering, East China University of Science and Technology, 130 Meilong Rd, Shanghai 200237, China. E-mail: xphe@ecust.edu.cn
bNational Center for Drug Screening, State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, 189 Guo Shoujing Rd, Shanghai 201203, P. R. China. E-mail: yzang@simm.ac.cn; jli@simm.ac.cn
cDepartment of Biological Chemistry, John Innes Centre, Norwich Research Park, Norwich NR4 7UH, UK
dDepartment of Chemistry, University of Bath, Bath, BA2 7AY, UK
First published on 2nd October 2019
Abstract
Protein C receptor (PROCR) is a recently discovered transmembrane biomarker for several tissue stem cells and is highly expressed in triple-negative breast cancer (TNBC) patient-derived xenografts. Herein, to enrich the toolbox for the biochemical evaluation of PROCR, we have developed a peptide-functionalized fluorogenic 2D material based on the self-assembly between a fluorescent peptide probe and thin-layer molybdenum disulfide. The material developed was suitable for the sensitive detection of PROCR recombinant protein in buffer solution and the fluorescence imaging of TNBC cells that express high levels of PROCR.
Breast cancer (BC) is the leading cause of cancer-related deaths among women in the world.1,2 BC is a highly heterogeneous disease3 with different biological features and clinical behaviours,4,5 which poses great challenges for the treatment of BC. Triple-negative breast cancer (TNBC) is the most aggressive form of BC, and it accounts for a much higher recurrence and metastasis rate due to the limited expression of progesterone receptor (PR), estrogen receptor (ER), and human epidermal growth factor receptor 2 (HER2).6 TNBC accounts for 15–20% of all breast carcinomas.7 Moreover, TNBC is not sensitive to existing hormone inhibitory drugs and hormone suppression methods.8 Clinically, there is a lack of effective treatment to treat breast cancer at an early stage, to complement tumor removal, which runs the risk of metastasis of tumour cells, however. The currently applied clinical methods for BC diagnosis, such as X-ray photography and immunohistochemistry (IHC),9 may cause harmful side effects. Therefore, it is important to develop new diagnostic means for early-stage TNBC diagnosis.
Recent studies have shown that BC is initiated by mammary stem cells (MaSCs) or progenitor cell lesions.10 Therefore, breast stem cells have become a research focus by oncologists. Recently, Zeng and colleagues discovered a new group of breast stem cells and showed that MaSCs specifically express protein C receptor (PROCR), which plays an important role in the initiation as well as epithelial-to-mesenchymal transition-driven metastasis of BC. Wang et al. reported that PROCR is differentially expressed in breast cancer cell lines but more prevalently in TNBC cells. Therefore, PROCR is important for the in-depth exploration of breast cancer development and tumorigenesis mechanisms, and would be potentially useful for the treatment of TNBC which lacks effective transmembrane biomarkers for targeted diagnosis and therapy.11 PROCR is a Type I transmembrane glycoprotein,12 which mediates cell signalling, promotes migration, invasion and angiogenesis of cancer cells, and inhibits cancer cell apoptosis.13 In addition, PROCR is highly expressed in extremely invasive basal BC subtype cells.14 On the basis of these pathological properties, Shipitsin and colleagues suggested PROCR as one of the most promising biomarkers for breast cancer.15
Two-dimensional (2D) nanomaterials (2D nanomaterials) have attracted significant research interest due to their highly diverse physical, chemical and optical properties. In recent years, with the in-depth exploration and application of graphene, a number of other 2D materials such as layered double hydroxides (LDHs), transition-metal dichalcogenides (TMDs) and transition metal oxides (TMOs) have been synthesized.16,17 In particular 2D materials have been widely employed as the support onto which biomolecules can be modified in order to enhance biocompatibility for biomedical applications including drug delivery, in vitro and in vivo imaging and disease theranostics.18,19 Among the 2D materials reported, 2D molybdenum disulfide (MoS2) has been extensively used by researchers because of its low cytotoxicity, good water dispersibility and high biocompatibility, as proven by in vivo studies.20
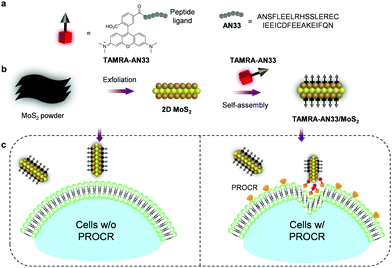
Here, to expand the chemical toolbox for the biochemical investigation of PROCR, we constructed a fluorogenic 2D material based on the self-assembly between a synthetic peptide ligand (Fig. 1a) that is labelled with a fluorophore and 2D MoS2 (Fig. 1b). Using this composite, we established a fluorogenic assay for the sensitive detection of PROCR recombinant protein as well as the imaging of PROCR-expressed TNBC cells (Fig. 1c). Importantly, using this simple 2D material, we have demonstrated that PROCR could be a potential surface biomarker for the targeted diagnosis and therapy of TNBC.
A previously reported21 peptide ligand (ANSFLEELRHSSLERECIEEICDFEEAKEIFQN – AN33) known to bind PROCR was used (PDB: 1LQV). TAMRA (tetramethyl-rhodamine) was used as a fluorescent label to covalently couple with the peptide, producing the peptide probe TAMRA-AN33. Subsequently, self-assembly of the peptide probe with 2D MoS2 synthesized by a known exfoliation method22 was conducted in phosphate-buffered saline (PBS, 0.01 M, pH 7.4) as solvent to form the TAMRA-AN33/MoS2 composite.
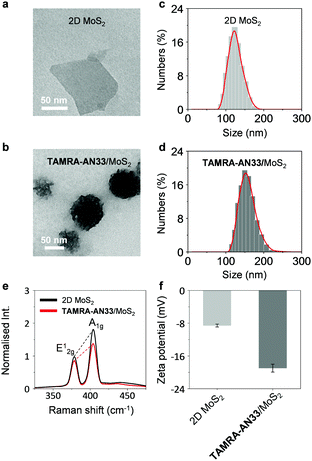
To study the morphology of the peptide-functionalized 2D material, we used high-resolution transmission electron microscopy (HRTEM). 2D MoS2 alone was observed to be a thin layered nanosheet structure (Fig. 2a). After assembling with TAMRA-AN33, peptide aggregates were seen on the surface of the 2D MoS2 nanosheet producing an entangled fibril architecture (Fig. 2b). The size distribution of the 2D MoS2 and TAMRA-AN33/MoS2 was measured by dynamic light scattering (DLS). The diameter of the 2D MoS2 sheets ranged from 90 nm to 180 nm, and the average particle size was determined to be ca. 125 nm (Fig. 2c). After assembly with the peptide, the particle size increased to 110–250 nm with an average particle size of 152 nm (Fig. 2d). Subsequent element distribution mapping using energy dispersive X-ray spectrometry indicated a universal distribution of the peptide ligand on the surface of 2D MoS2 (Fig. S1, ESI†). To measure the thickness increase of 2D MoS2 after assembly with the peptide, atomic force microscopy (AFM) was used. In the representative AFM images of TAMRA-AN33/MoS2, we observed particle-like aggregates on the surface of 2D MoS2, resulting in a height increase of ∼2 nm with respect to 2D MoS2 alone (Fig. S2, ESI†). These results suggest the attachment of TAMRA-AN33 to the surface of the 2D MoS2.
To elaborate the driving force by which TAMRA-AN33 and 2D MoS2 self-assembles, we used Raman spectroscopy and zeta potentiometry to characterize the scattering signals and potential of the materials, respectively. Typical Raman peaks centred at 378 cm−1 and 404 cm−1, which correspond to the in-plane vibration (E12g) of Mo and S atoms and the reverse interlaminar vibration of two S atoms (A1g), respectively, were observed for both 2D MoS2 and the composite (Fig. 2e).23 The ratio of E12g and A1g was used to characterize the change in vibration level and thickness of the MoS2 sheets. In the presence of TAMRA-AN33, the E12g/A1g ratio of the 2D MoS2 increased from 0.55 to 0.63, suggesting an increased degree of the in-plane Mo–S vibration and a decreased interlaminar vibration of the two S atoms. This implies that the coating of TAMRA-AN33 to the surface of 2D MoS2 increased the degree of conjugation and the thickness of the sheet. The zeta potential of 2D MoS2 decreased from −8.6 mV to −19.6 mV (Fig. 2f), suggesting an electrostatic attraction between the peptide and the 2D material. In addition, the increase of the absolute potential value suggests an enhanced stability of TAMRA-AN33/MoS2 in the aqueous phase with respect to 2D MoS2.24
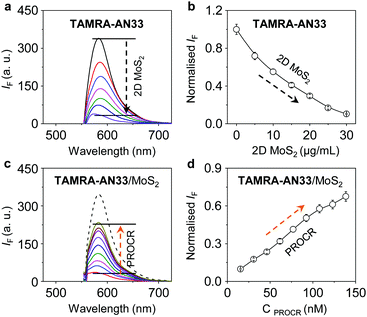
2D MoS2 features a wide spectral absorption band, which can absorb the photon energy of adjacent fluorophores (less than 10 nm), thus quenching the fluorescence emission. As a result, we tested the fluorescence quenching of TAMRA-AN33 with 2D MoS2 using graphene oxide (GO) as a control. We determined that with the continuous addition of 2D MoS2 (Fig. 3a) and GO (Fig. S3a, ESI†), the fluorescence of TAMRA-AN33 decreased in a concentration-dependent manner (Fig. 3b and Fig. S3b, ESI† for 2D MoS2 and GO, respectively). Subsequently, the addition of recombinant PROCR to a solution of TAMRA-AN33/MoS2 (Fig. 3c) and TAMRA-AN33/GO (Fig. S3c, ESI†) led to a concentration-dependent recovery of the fluorescence (Fig. 3d and Fig. S3d, ESI† for 2D MoS2 and GO, respectively). The limit of detection of TAMRA-AN33/MoS2 (0.76 nM) for PROCR was determined to be lower than that of TAMRA-AN33/GO (1.26 nM) (Fig. S4, ESI†). These results suggest that the peptide probe could be removed from the surface of the 2D materials through binding to PROCR, leading to fluorescence recovery. This is in accordance with our previous observations.25
To confirm that the fluorescence enhancement originates from the association between TAMRA-AN33 and PROCR, AN33 without being modified with TAMRA was used as a competing agent (Fig. S5, ESI†). We found that pre-incubation of bare AN33 with PROCR significantly reduced the fluorescence enhancement of TAMRA-AN33/MoS2, suggesting binding between the peptide ligand and the protein. To further test the binding selectivity of the material composite, we used a variety of other proteins, including soybean agglutinin (SBA), concanavalin A (Con A), pepsin (PEP), bovine serum albumin (BSA), wheat germ agglutinin (WGA) and peanut agglutinin (PNA) (Fig. S6, ESI†). With the same protein concentration, we observed that the fluorescence intensity of TAMRA-AN33/MoS2 with PROCR is significantly higher than that with the other proteins, implying that TAMRA-AN33/MoS2 has good selectivity for PROCR detection. These results provide the basis for developing a new in vitro fluorescence assay to sensitively detect, as well as screen new binding agents for, PROCR.
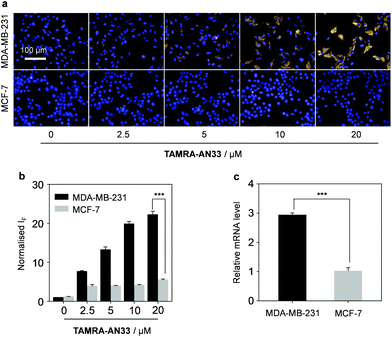
Next, we tested the capacity of the material composite for the fluorescence imaging of TNBC that highly expresses PROCR (Fig. 4). A human TNBC cell line (MDA-MB-231) known to highly express the protein26 and another breast cancer cell line (MCF-7) with a lower PROCR expression level27 were used. We first determined that with an increasing concentration of TAMRA-AN33, the fluorescence produced in MDA-MB-231 cells was gradually enhanced (Fig. 4a and b). Whereas, the fluorescence in MCF-7 cells did not significantly change. This agrees with the PROCR expression level of the two cells, as measured by real-time quantitative polymerase chain reaction (RT qPCR) (Fig. 4c).
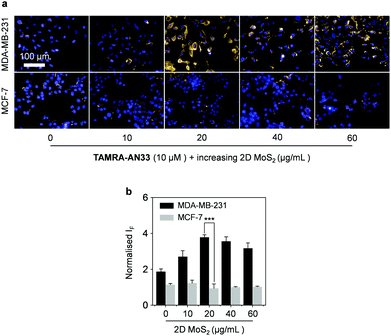
Then, we incubated TAMRA-AN33/MoS2 with cells using the peptide probe alone as a control. While no obvious fluorescence change was seen for MCF-7, a fluorescence enhancement was observed for MDA-MB-231 when the concentration of 2D MoS2 was increased from 0 to 20 μg mL−1 (Fig. 5a and b). However, a further increase of the material concentration led to a gradually suppressed fluorescence. This suggests that the presence of 2D MoS2 could enhance the binding of the peptide probe with membrane-bound PROCR probably through a clustering effect, as observed in our previous research.28 A subsequent cell proliferation assay indicated that the peptide probe was not toxic to MDA-MB-231 cells (Fig. S7, ESI†), suggesting its good compatibility for in-depth pharmacological investigations. To further demonstrate the targeting ability of the composite developed, we used a normal cell line (human hepatocyte – L02) for comparison (Fig. S8, ESI†). We observed that the imaging signature of the composite for the breast cancer cells was significantly stronger than that of the hepatocyte. This also agrees with their PROCR expression level measured by RT qPCR (Fig. S9, ESI†).
In summary, we have developed a fluorogenic 2D TAMRA-AN33/MoS2 composite through the self-assembly of a fluorophore-labelled PROCR ligand and 2D MoS2. The resulting composite offers scope to establish a concise fluorogenic assay for in vitro sensing and screening of binding agents of PROCR, as well as for the imaging of TNBC cells that highly express the protein biomarker. Considering the excellent therapeutic effect of 2D materials as demonstrated by numerous investigations,29 this composite could be extended to the fluorescence imaging and therapy of TNBC in vivo.
Prof. Yi Zeng is warmly thanked for her helpful discussion. This research was supported by the Natural Science Foundation of China (no. 21788102, 91853201, 21722801 and 21776078), the Shanghai Municipal Science and Technology Major Project (No. 2018SHZDZX03), the Shanghai Science and Technology Committee (No. 19410712600), and the National Postdoctoral Program for Innovative Talents (BX20190115). T. D. J. thanks the Royal Society for a Wolfson Research Merit Award. All cells were obtained from ATCC.
Conflicts of interest
There are no conflicts to declare.Notes and references
- Q. Yan, Z. Xiaorong, Z. Zhang, W. Bing, Y. Feng and B. Hong, Pathol., Res. Pract., 2017, 213, 1173 CrossRef CAS PubMed.
- R. Abdollahzadeh, A. Daraei, Y. Mansoori, M. Sepahvand, M. M. Amoli and J. Tavakkoly-Bazzaz, J. Cell. Physiol., 2019, 234, 10080 CrossRef CAS.
- B. Weigelt, F. C. Geyer and J. S. Reis-Filho, Mol. Oncol., 2010, 4, 192 CrossRef CAS PubMed.
- A. Bahrami, M. Khazaei, S. Shahidsales, S. M. Hassanian, M. Hasanzadeh, M. Maftouh, G. A. Ferns and A. Avan, J. Cell. Biochem., 2018, 1192, 213 CrossRef PubMed.
- S. Alimirzaie, M. Bagherzadeh and M. R. Akbari, Clin. Genet., 2019, 95, 643 CrossRef CAS PubMed.
- C. Belli, B. A. Duso, E. Ferraro and G. Curigliano, Breast, 2019, 45, 15 CrossRef PubMed.
- A. G. Waks and E. P. Winer, J. Am. Med. Assoc., 2019, 321, 288 CrossRef CAS PubMed.
- A. C. Garrido-Castro, N. U. Lin and K. Polyak, Cancer Discovery, 2019, 9, 176 CrossRef PubMed.
- I. A. Nemeir, J. Saab, W. Hleihel, A. Errachid, N. Jafferzic-Renault and N. Zine, Sensors, 2019, 19, 2373 CrossRef PubMed.
- P. Bonetti, M. Climent, F. Panebianco, C. Tordonato, A. Santoro, M. J. Marzi, P. G. Pelicci, A. Ventura and F. Nicassio, Oncogene, 2019, 38, 360 CrossRef CAS PubMed.
- B. Lloyd-Lewis, O. B. Harris, C. J. Watson and F. M. Davis, Trends Cell Biol., 2017, 27, 556 CrossRef PubMed.
- D. Wang, C. Cai, X. Dong, Q. Yu, X.-O. Zang, L. Yang and Y. Zeng, Nature, 2015, 517, 81 CrossRef CAS PubMed.
- K. Fukudome, X. Ye, N. Tsuneyoshi, O. Tokunaga, K. Sugawara, H. Mizokami and M. Kimoto, J. Exp. Med., 1998, 187, 1029 CrossRef CAS PubMed.
- I. Antón, E. Molina, D. Luis-Ravelo, C. Zandueta, K. Valencia, C. Ormazabal, S. Martínez-Canarias, N. Perurena, M. J. Pajares, J. Aqorreta, L. M. Montuenqa, V. Segura, I. I. Wistuba, J. D. L. Rivas, J. Hermida and F. Lecanda, Am. J. Respir. Crit. Care Med., 2012, 186, 96 CrossRef PubMed.
- S. Y. Park, H. E. Lee, H. Li, M. Shipitsin, R. Gelman and K. Polyak, Clin. Cancer Res., 2010, 16, 876 CrossRef CAS PubMed.
- M. Shipitsin, L. L. Campbell, P. Argani, S. Weremowicz, N. Bloushtain-Qimron, J. Yao, T. Nikolskaya, T. Serebryiskaya, R. Beroukhim, M. Hu, M. K. Halushka, S. Sukumar, L. M. Parker, K. S. Anderson, L. N. Harris, J. E. Garber, A. L. Richardson, S. J. Schnitt, Y. Nikolsky, R. S. Gelman and K. Polyak, Cancer Cell, 2007, 11, 259 CrossRef CAS PubMed.
- C. Tan, X. Cao, X.-J. Wu, Q. He, J. Yang, X. Zhang, J. Chen, W. Zhao, S. Han, G.-H. Nam, M. Sindoro and H. Zhang, Chem. Rev., 2017, 117, 6225 CrossRef CAS PubMed.
- G. R. Bhimanapati, Z. Lin, V. Meunier, Y. Jung, J. Cha, S. Das, D. Xiao, Y. Son, M. S. Strano, V. R. Cooper, L. Liang, S. G. Louie, E. Ringe, W. Zhou, S. S. Kim, R. R. Naik, B. G. Sumpter, H. Terrones, F. Xia, Y. Wang, J. Zhu, D. Akinwande, N. Alem, J. A. Schuller, R. E. Schaak, M. Terrones and J. A. Robinson, ACS Nano, 2015, 9, 11509 CrossRef CAS PubMed.
- A. Gnach, T. Lipinski, A. Bednarkiewicz, J. Rybka and J. A. Capobianco, Chem. Soc. Rev., 2015, 44, 1561 RSC.
- T. Liu and Z. Liu, Adv. Healthcare Mater., 2018, 7, 1701158 CrossRef PubMed.
- J. Hao, G. Song, T. Liu, X. Yi, K. Yang, L. Cheng and Z. Liu, Adv. Sci., 2017, 4, 1600160 CrossRef PubMed.
- V. Oganesyan, N. Oganesyan, S. Terzyan, D. Qu, Z. Dauter, N. L. Esmon and C. T. Esmon, J. Biol. Chem., 2002, 277, 24851 CrossRef CAS PubMed.
- J. N. Coleman, M. Lotya, A. O'Neill, S. D. Bergin, P. J. King, K. Young, A. Gaucher, S. De, R. J. Smith, I. V. Shvets, S. K. Arora, G. Stanton, H.-Y. Kim, K. Lee, G. T. Kim, G. S. Duesberg, T. Hallam, J. J. Boland, J. J. Wang, J. F. Donegan, J. C. Grunlan, G. Moriarty, K. Theuwissen, D. W. McComb, P. D. Nellist and V. Nicolosi, Science, 2011, 331, 568 CrossRef CAS PubMed.
- F. Bonaccorso, P. H. Tan and A. C. Ferrari, ACS Nano, 2013, 7, 1838 CrossRef CAS PubMed.
- D.-K. Ji, Y. Zhang, Y. Zang, J. Li, G.-R. Chen, X.-P. He and H. Tian, Adv. Mater., 2016, 28, 9356 CrossRef CAS PubMed.
- Y.-H. Ma, W.-T. Dou, Y.-F. Pan, L.-W. Dong, Y.-X. Tan, X.-P. He, H. Tian and H.-Y. Wang, Adv. Mater., 2017, 29, 1604253 CrossRef PubMed.
- D. Wang, C. Liu, J. Wang, Y. Jia, X. Hu, H. Jiang, Z.-M. Shao and Y. A. Zeng, J. Biol. Chem., 2018, 293, 1413 CrossRef CAS PubMed.
- J. Eisermann, A. Kerth and D. Hinderberger, RSC Adv., 2019, 9, 18627 RSC.
- W.-T. Dou, Y. Kong, X.-P. He, G.-R. Chen, Y. Zang, J. Li and H. Tian, ACS Appl. Mater. Interfaces, 2017, 9, 14709 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Additional figures and experimental section. See DOI: 10.1039/c9cc06399d |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2019 |