Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
An evolutionary perspective on zinc uptake by human fungal pathogens†
Duncan
Wilson
Aberdeen Fungal Group, School of Medical Sciences, Institute of Medical Sciences, University of Aberdeen, Foresterhill, Aberdeen, AB25 2ZD, UK. E-mail: Duncan.Wilson@abdn.ac.uk
First published on 26th January 2015
Abstract
The mammalian immune system has evolved sophisticated mechanisms to withhold essential micronutrients from invading pathogens. These processes, collectively known as nutritional immunity serve to limit microbial proliferation and bolster killing of the invader. Successful pathogens, therefore, have developed strategies to counteract nutritional immunity and acquire essential micronutrients in the restrictive environment of the infected host. Here I take advantage of the now large number of sequenced fungal genomes to explore the zinc acquisition strategies of human fungal pathogens and reflect on the evolutionary context of these uptake pathways.
Fungal pathogens of humans
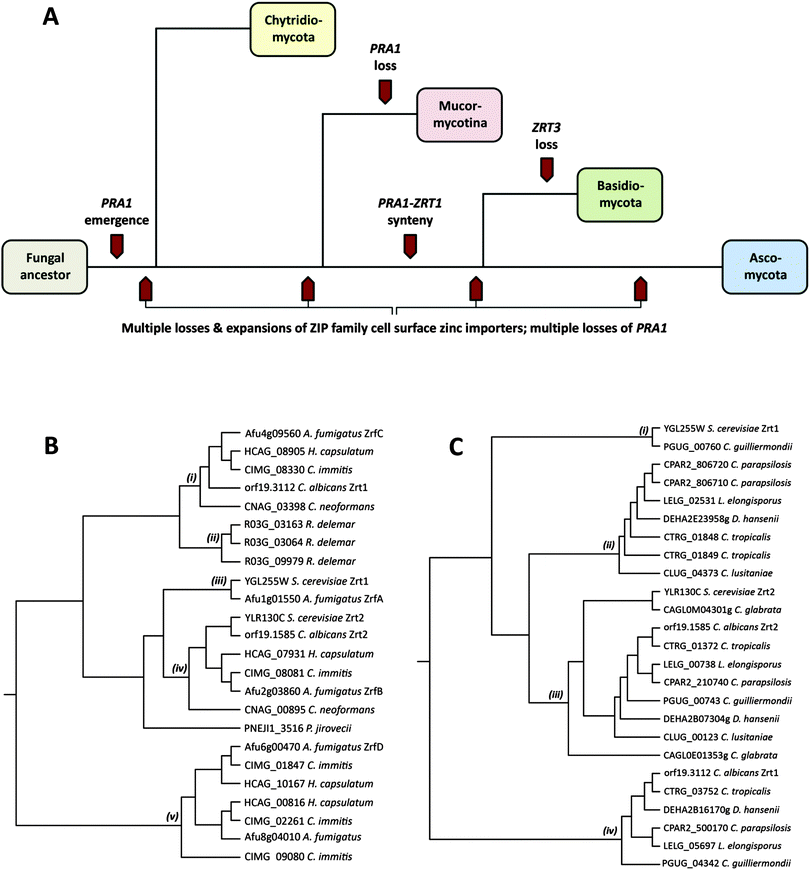
Pathogenic fungi represent an enormous, but under-appreciated burden on human health, with invasive mycoses killing approximately one and a half million people each year.1 The most common invasive fungal diseases of humans, and the dominant aetiological agents are: aspergillosis (Aspergillus fumigatus), candidiasis (Candida albicans), cryptococcosis (Cryptococcus neoformans), mucormycosis (Rhizopus oryzae/delemar), pneumocystis (Pneumocystis jirovecii), and the endemic mycoses histoplasmosis (Histoplasma capsulatum), coccidioidomycosis (Coccidioides immitis), blastomycosis (Blastomyces dermatitidis), paracoccidioidomycosis (Paracoccidioides brasiliensis) and penicilliosis (Penicillium marneffei).1These major fungal pathogens are highly diverse from both evolutionary and ecological perspectives.2 Pathogenic species are distributed throughout the Fungal Kingdom and found in three of the major fungal phyla: the Mucormycotina (Rhizopus), the Basidiomycota (Cryptococcus) and the Ascomycota (all other major invasive pathogenic species). For orientation, Fig. 1A shows a simplified overview of the Fungal Kingdom, with highlights of key events in the evolution of zinc acquisition pathways, which are discussed below. The Chytridiomycota represent the most basal fungal lineage shown in Fig. 1A. The next to diverge, the Mucormycotina, encompasses several important human pathogenic species, including Rhizopus oryzae/delemar. Within the Basidiomycota, the only major invasive pathogen of humans are the Cryptococcal species, C. neoformans and C. gattii, however C. neoformans is a major killer, responsible for more than one million life-threatening infections per year, predominantly in HIV infected individuals.1 Both Rhizopus and Cryptococci exist in environmental niches such as soil or bird guano. Finally, the Ascomycota encompasses the largest number of human pathogenic species. However, even within this phylum, the major pathogenic species are evolutionarily diverse. The moulds, such as A. fumigatus, and the endemic fungal pathogens (e.g. C. immitis and H. capsulatum) both inhabit environmental niches such as compost or soil and cause disease in susceptible individuals, typically upon inhalation of spores or hyphal fragments (hyphae are the long, thin filamentous structures formed by fungi). In contrast, Candida species, such as C. albicans, are not found in the environment, but are commensal members of the normal mucosal microbial flora of humans. P. jirovecii is also intimately associated with humans and cannot be cultured in vitro.
Despite these evolutionary and ecological differences, these species are devastating human pathogens and must therefore be able to effectively utilise zinc from their host environment during infection.3
Evolutionary dynamics of fungal zinc transporters
Fungi import zinc via members of the ZIP (ZRT/IRT protein) family of zinc transporters. Members of the ZIP family transport their substrate from the extracellular environment, or from subcellular compartments, into the cytoplasm, and thus facilitate cellular utilisation of zinc. To date, fungal ZIP family transporters have been functionally characterised in C. albicans, A. fumigatus and the model yeast, Saccharomyces cerevisiae.3I will begin with a comparison of cellular zinc importers in these three characterised species. S. cerevisiae, encodes two cellular zinc importers: the high affinity Zrt1 and the low affinity Zrt2.4 Like S. cerevisiae, the pathogenic yeast C. albicans also encodes two zinc importers (also named Zrt1 and Zrt2); however, as illustrated in Fig. 1B, whilst the ZRT2 genes of both species are homologous (iv), C. albicans Zrt1 and S. cerevisiae Zrt1 are not directly related to each other, aligning to distinct branches of the tree (i), (iii). Interestingly, the more distantly related pathogenic mould, A. fumigatus, encodes homologues of S. cerevisiae Zrt1 (ZrfA), C. albicans Zrt1 (ZrfC) and a third transporter (ZrfB), which shares similarity with Zrt2 from both yeast species.
From a functional perspective, whilst the S. cerevisiae Zrt1 and Zrt2 are optimised for high- and low-affinity zinc import, respectively, the A. fumigatus zinc importers are pH-specialists: AfZrfC operates at neutral-alkaline pH, whilst ZrfA and ZrfB are required for zinc utilisation under acidic pH.5
Despite the closer evolutionary relationship between the yeasts C. albicans and S. cerevisiae,2C. albicans zinc import appears to be functionally analogous to the more distantly related mould, A. fumigatus. For example, CaZrt1 (the AfZrfC orthologue) is upregulated and functional under neutral-alkaline pH whilst CaZrt2 is expressed at acidic pH.6,7A. fumigatus and C. albicans are not closely related and the two species inhabit very different ecological niches in nature (soil/compost and human mucosal surfaces, respectively). Therefore, it would appear that pH-dependent regulation of zinc import may be conserved in fungi, at least within the major Ascomycota phylum (Fig. 1A). There are several possible explanations for the pH-optimisation of fungal zinc transporters. For example, zinc typically becomes much less soluble and bioavailable as pH increases. Moreover, histidine residues of zinc transporters have been implicated in metal binding and import.8 As the protonation state of histidine (and thus its chemical properties) is pH-dependent, it is possible that the arrangement of histidines has been optimised for function at acidic and alkaline pH in fungal zinc importers.
In addition to these three characterised zinc transporters (ZrfA, B and C), A. fumigatus encodes a further two putative cellular zinc importers. Orthologues of these alternative transporters (Fig. 1B(v)) are not present in Mucormycotina or Basidiomycete species, or in the Saccharomycotina (e.g. Candida and Saccharomyces species), but rather appear to have arisen independently in the Pezizomycetes subphylum of filamentous fungi, which encompasses numerous environmental species, including the Aspergilli and the endemic fungal pathogens, H. capsulatum and C. immitis. An alignment of A. fumigatus zinc importer sequences is available in Fig. S1 (ESI†).
Rhizopus delemar, which belongs to the basal Mucormycotina lineage, encodes three cell surface zinc importers (Fig. 1B(ii)). However, these three proteins are more closely related to each other than to transporters in other species, suggesting that a single ancestral ZIP family zinc importer has undergone sequential duplication in Rhizopus since divergence of the Mucormycotina.
ZIP family gene expansion has also occurred in other fungal sub-phyla; let's consider the medically-important Candida genus in more detail. Although most cases of candidiasis are caused by C. albicans, several other Candida species are of medical relevance. The genus Candida is actually polyphyletic. However, with the exception of C. glabrata and C. krusei, all other medically relevant species belong to the “CUG clade”, a monophyletic group of yeasts which uniquely decode the CUG codon to serine instead of leucine.9
Fig. 1C(iii) shows that all Candida species encode orthologues of C. albicans Zrt2. All CUG species, with the exception of C. lusitaniae, encode orthologues of C. albicans Zrt1 (Fig. 1C(iv)), indicating that this single (sequenced) member of the CUG clade has lost this gene. On the other hand, only C. guilliermondii encodes an orthologue of S. cerevisiae Zrt1 (Fig. 1C(i)). This is interesting because, barring horizontal gene transfer, it implies that the ScZrt1 orthologue was present in the Candida ancestor, but has only been maintained by this single (sequenced) modern-day species. Moreover, because C. guilliermondii is unlikely to represent the most basal Candida species,9 it would appear that the ScZrt1 orthologue has been lost at least twice within the CUG clade.
In counterpoint to these gene loss events in the CUG clade, Candida tropicalis, parapsilosis, lusitaniae, D. hansenii and L. elongisporus all possess additional zinc importers, which, whilst most closely related to CaZrt2, form a distinct clade on the tree (Fig. 1C(ii)). As these transporters are more closely related to Candida Zrt2, than to other classes of transporters, this sub-set of Zrt2-relatives (Fig. 1C(ii)) must have arisen from gene duplication (of ZRT2) following divergence of the CUG clade. Since then, this sub-family has been independently lost by both C. albicans and C. guilliermondii. This conclusion can be drawn because C. albicans (together with C. tropicalis and C. parapsilosis) and C. guilliermondii (together with D. hansenii) belong to distinct sub-clades of the CUG clade.9
As well as being twice lost, this sub-family has also undergone recent expansion in C. tropicalis and C. parapsilosis. Indeed, in these two species, this recent expansion appears to have been via independent tandem gene duplication, as the encoding genes are syntenically arranged in the genome of C. tropicalis and C. parapsilosis. Therefore, even amongst these seven species of the CUG clade, the ZIP family of zinc importers has experienced a series of expansion and contraction events.
Outside the CUG clade, perhaps the most striking example of gene loss is for P. jirovecii, which encodes only a single ZIP family zinc importer (Fig. 1B). This fungus is obligately associated with its human host and cannot be cultured in vitro. Therefore reduction in zinc transporter copy number may reflect an adaptation to the specific environment of the human lung. In this context, it is worthwhile noting that Malassezia globosa, which also intimately associates with its human host, also encodes only one predicted zinc importer (not shown).
The major Basidiomycete pathogen of humans, C. neoformans, conservatively possesses only two zinc importers. One is related to the Ascomycete neutral/alkaline pH-optimised transporters (e.g. C. albicans Zrt1 and A. fumigatus ZrfC), whilst the second (CNAG_00895) clusters with the Zrt2 importers (Fig. 1B(i), (iv)). This suggest that the ancestor of the two major fungal phyla (the Basidiomycota and the Ascomycota – Fig. 1A) possessed (at least) two zinc importers and that these have been maintained by many modern Basidiomycete and Ascomycete species.
In the above section, I have examined cell surface zinc importers; however, in addition to zinc acquisition from the environment, fungi can utilise zinc from intracellular storage compartments. The most notable of these “zinc sinks” is the fungal vacuole, an organelle, which in S. cerevisiae, can hold up to 7 × 108 ions (100 mM) of zinc.4 This extraordinary capacity for storage can allow a single mother cell to generate up to 200 progenitor cells without utilisation of exogenous zinc. Yeast cells shuttle zinc from the vacuole for use in the cytoplasm via the vacuolar-associated ZIP transporter, Zrt3.4 Interestingly, within the Fungal Kingdom, only species from the Basidiomycota appear to lack an orthologue of ZRT3. This is a significant observation because the Mucormycotina (a more basal lineage than the Basidiomycota) do possess ZRT3 orthologues, indicating that the entire Basidiomycota lineage (or at least all Basidiomycetes which have been sequenced so far) may have lost the vacuolar zinc exporter. Therefore, the molecular mechanism employed by Basidiomycota for zinc shuttling out of the vacuole remains unknown.
Conserved transcriptional control of zinc uptake in the fungi
In spite of the dramatic mutability in transporter copy number discussed above, the transcriptional network governing the expression of zinc importers appears to be highly conserved in fungi. The transcription factor, Zap1 (Zinc-responsive Activator Protein), is the master regulator of zinc uptake and homeostasis.4 Since its discovery in S. cerevisiae, Zap1 orthologues have been characterised in the human fungal pathogens, C. albicans, A. fumigatus and Cryptococcus gattii.10–12 In all three of these distantly related species, Zap1 (also known as Csr1 in C. albicans and ZafA in A. fumigatus) positively regulates the expression of the cell surface zinc importers, facilitating growth under zinc-limiting conditions. Significantly, mutant strains of C. albicans, A. fumigatus and C. gattii lacking their respective ZAP1 orthologues have all been tested for virulence in relevant animal infection models and all three mutants exhibit reduced virulence in comparison to wild type fungi. These three independent studies, spanning at least half a billion years of fungal evolution, underline the essentiality of coordinated zinc assimilation by pathogens during infection.The dynamic history of the fungal zincophore
In addition to transporter mediated uptake, C. albicans has also been shown to scavenge zinc via a secreted “zincophore”.7C. albicans secretes a zinc binding protein (Pra1) which can sequester zinc and subsequently reassociate with the fungal cell. C. albicans was found to sequester zinc from host cells; whilst deletion of PRA1 blocked this zinc scavenging. PRA1 was also found to be essential for growth on and damage of host cells only in the absence of exogenous zinc, and recombinant Pra1 exhibited zinc-binding activity in vitro. Interestingly, reassociation of soluble Pra1 to the fungal cell appears to be mediated by the cell surface zinc transporter (Zrt1), because deletion of ZRT1 precluded cellular reassociation of soluble Pra1. The two genes of this system, PRA1 and ZRT1, are syntenic – encoded at the same genetic locus in C. albicans with a shared promoter. In line with its function in zinc scavenging from host cells, this locus is positively regulated by zinc starvation and by physiological (neutral/alkaline) pH.6,7Although zincophore activity has, as yet, only been demonstrated for C. albicans, the orthologous genes in A. fumigatus are also required for growth under zinc-depletion at neutral-alkaline pH13 (Fig. S2, ESI† displays an alignment of C. albicans Pra1 and it's A. fumigatus orthologue, AspF2). As A. fumigatus and C. albicans are not closely related species, this would suggest a conserved role for PRA1 orthologues in zinc scavenging, at least within the Ascomycota.
A. fumigatus aspF2 (the orthologue of C. albicans PRA1) also shares its promoter with zrfC (the orthologue of C. albicans ZRT1), and deeper phylogenetic analysis revealed that this syntenic arrangement of secreted zinc binding protein- and zinc transporter-encoding genes is conserved in numerous fungal species.7 Indeed, conserved synteny is present in both Ascomycete and Basidiomycete species – the two major fungal phyla, which diverged at least half a billion years ago. Although a PRA1 orthologue was identified in the Chytrid, Spizellomyces punctatus, this gene does not share synteny with a ZRT1 orthologue. Therefore, whilst the PRA1 ancestor gene arose in an ancestral fungal lineage (PRA1 orthologues are not found outside the Fungal Kingdom), synteny with the ZRT1 ancestor probably occurred after divergence of the Chytridiomycota from higher phyla (Fig. 1A). However, it should be noted that only a small number of Chytridiomycota species have been sequenced, and the PRA1 orthologue was only identified in one (S. punctatus). Although present in the basal Chytrid lineage, PRA1 appears to have undergone wholesale loss from the Mucormycotina phylum, as the orthologue is not present in any sequenced Mucor species. However, again, it should be noted that drawing firm conclusions on gene loss and maintenance in the Chytridiomycota and Mucormycotina is problematic due to the relative scarcity of sequenced species.
On the other hand, a large number of genome sequences are available for fungi of the Basidiomycota and the Ascomycota. This has revealed perhaps an even more interesting phenomenon: PRA1 has been lost a remarkable number of times throughout fungal evolution. Within the Candida CUG clade alone, PRA1 is encoded by C. albicans, C. dubliniensis, C. tropicalis C. guilliermondii and D. hansenii and has been lost by C. lusitaniae, C. orthopsilosis, C. parapsilosis and L. elongisporus. This pattern is noteworthy, because it means that PRA1 has been maintained and lost by species in both the haploid (e.g. C. guilliermondii and C. lusitaniae) and the diploid (e.g. C. albicans and C. parapsilosis) sub-clades of the CUG clade,9 and indicates that Pra1 is under dynamic selective pressure.
Outside the CUG clade, Pra1 has been lost independently from multiple fungal lineages, including the major human pathogens C. neoformans, H. capsulatum and C. glabrata. Is there any relationship between zincophore status and fungal physiology? In C. albicans and A. fumigatus (the two species in which Pra1/AspF2 has been characterised), this system is involved in zinc assimilation at neutral-alkaline pH and tightly repressed under acidic conditions. It is therefore conceivable that, in species which inhabit acidic ecological niches, Pra1 is not under positive selective pressure.
C. immitis and H. capsulatum are both endemic fungal pathogens of North America which have maintained and lost the PRA1 orthologue, respectively. Although these two species are quite closely related, they exhibit discrete geographical localisation. C. immitis is endemic to the south-western United States, whilst H. capsulatum is found in central eastern areas. Direct comparison of the soil pH with endemic areas shows that C. immitis is limited to regions of neutral-alkaline soil (Fig. 2). This correlation is most clearly evident in California, where a finger-like projection of C. immitis endemicity stretches up the central region of this state, enclosed on both sides by areas of acidic soil (Fig. 2).
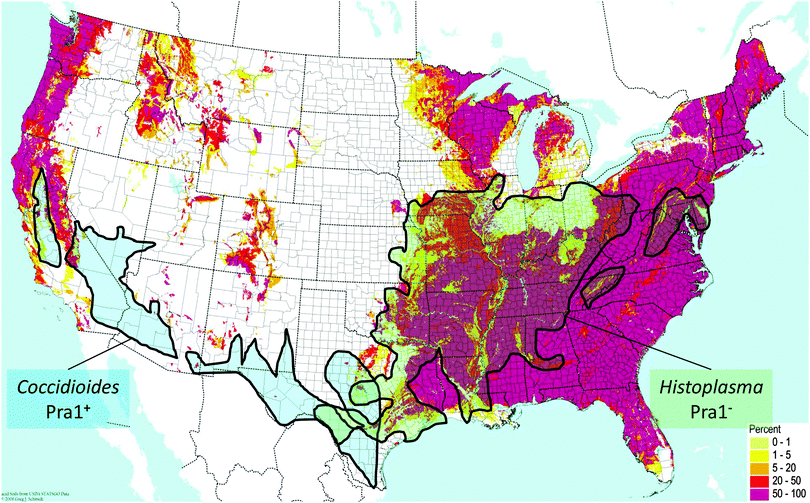 | ||
| Fig. 2 Distribution of endemic fungal pathogens relative to zincophore status and environmental pH. Map of soil acidity in the contiguous USA is from the BONAP website (http://www.bonap.org/), reproduced with permission from Greg Schmidt, 2008, and includes data from the USDA Natural Resource Conservation Service. The pink colouring on the map indicates areas with high percentages (50–100%) of acidic soil (pH < 6). Superimposed is endemicity data for C. immitis and H. capsulatum from ref. 19. | ||
In contrast, regions of H. capsulatum endemicity map clearly with areas of acidic soil. In addition to soil, H. capsulatum is frequently associated with bird guano, which is also an acidic environment. Therefore, at least for these two endemic species, their ecological pH correlates with the presence or absence of the zincophore system. Obviously, pH is only one environmental factor among many which have driven the speciation of C. immitis and H. capsulatum; however, persistent occupation of acidic ecological niches may have driven the loss of Pra1, as a neutral-alkaline pH-adapted zincophore would not be under selective evolutionary pressure in such environments. This is supported by the fact that, in the distantly related species, C. neoformans, which inhabits similar environmental niches as H. capsulatum (e.g. bird guano), the Pra1 orthologue has also been lost.7
Zincophore loss may have more far-reaching implications than an evolutionary adaptation to acidic environments: in addition to its function in zinc acquisition, the secreted factor (Pra1/AspF2) is also highly immunogenic in the context of human infections. The Aspergillus orthologue, AspF2, cross-reacts with the sera of over 80% of individuals suffering from aspergilloma or allergic bronchopulmonary aspergillosis and serves as a major allergen in humans. Moreover, Pra1 from C. albicans has several immune-modulatory roles.14 In addition to a well-established role in modulating the human complement system, CaPra1 serves as a major ligand for the neutrophil αMβ2 receptor, and C. albicans cells can be killed by neutrophils via Pra1-mediated recognition. Indeed, genetic deletion of PRA1 in C. albicans renders the fungus more resistant to neutrophil killing both in vitro and in vivo.15
This raises the question of whether wild type fungal pathogen zincophore status impacts pathogenesis? As noted above, Pra1 serves to recruit neutrophils during systemic candidiasis. In this context, it is noteworthy that both characterised zincophore+ species, C. albicans and A. fumigatus, exhibit aggressive invasive infections associated with a high degree of inflammation. In contrast, C. neoformans, H. capsulatum and C. glabrata, three distantly related species, each of which has independently lost the zincophore gene, are all facultative intracellular parasites of macrophages. Notably, the macrophage phagolysosome is an acidic environment. It is conceivable that the long-term ecological adaptation of species such as C. neoformans and H. capsulatum to environments of acidic soil and bird guano has prepared these fungi for life within the macrophage phagosome. Finally, it is tempting to speculate that, like the genetically manipulated C. albicans PRA1-deletion strain,15 these natural “pra1Δ mutants” may benefit by avoiding aggressive immune responses during human infections. However, future studies, perhaps utilising ectopic zincophore expression in these natural pra1Δ null mutants, will be required to confirm this.
Nevertheless, based on the observations made in C. albicans and A. fumigatus, it is evident that our immune system has learned to recognise this fungal zinc uptake system. This seems to be a recurrent immunological theme, as several bacterial zinc uptake systems are effectively recognised by our immune systems and are now being considered as vaccine targets – e.g.ref. 16. From the perspective of nutritional immunity, this is perhaps not surprising: during infection, zinc is a generally limiting factor, and thus, in this setting, pathogens must express high affinity uptake systems in order to proliferate. Indeed, it is possible that our immune system has developed this positive feedback loop for the recognition and killing of microbial invaders.17 By creating a circuit whereby zinc deficiency is enforced via nutritional immunity and microbial high-affinity zinc uptake systems are targeted, our immune systems force the invader to reveal itself.
In summary, zinc uptake pathways have experienced a dynamic evolutionary history in the Fungal Kingdom. The environmental Ascomycete species, A. fumigatus, H. capsulatum and C. immitis (along with other members of the Pezizomycetes) have experienced expansion in zinc importer copy number, resulting in an entirely new sub family (Fig. 1B(v)). The CUG clade have also generated new zinc importers, most likely via the duplication of Zrt2 (Fig. 1C(ii)) and these have been subsequently lost by important pathogenic species, including C. albicans. The basal Mucormycotina, R. delemar has generated three importers, again via lineage-specific duplication (Fig. 1B(ii)). Finally, the obligately host-associated P. jirovecii has maintained only one zinc importer. Paralleling zinc transporter family expansion and contraction, the zincophore encoding gene (PRA1) has experienced a staggering number of independent losses from modern fungal genomes. Together, these events have coincided with the emergence of a diverse group of modern-day pathogenic species (Fig. 1A), capable of assimilating sufficient zinc from their host environment to proliferate and cause disease. It is becoming increasingly apparent that efficient zinc acquisition represents a critical component of microbial pathogenicity. This is because the mammalian host actively attempts to withhold essential trace nutrients (such as zinc).18 Therefore, the challenge now will be to understand the mechanisms of fungal metal acquisition from host tissues, and how these processes contribute to the pathogenesis of the devastating diseases caused by fungi.
Acknowledgements
DW is supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (Grant Number 102549/Z/13/Z).References
- G. D. Brown, D. W. Denning, N. A. Gow, S. M. Levitz, M. G. Netea and T. C. White, Hidden killers: human fungal infections, Sci. Transl. Med., 2012, 4, 165rv113 CrossRef PubMed.
- D. A. Fitzpatrick, M. E. Logue, J. E. Stajich and G. Butler, A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis, BMC Evol. Biol., 2006, 6, 99 CrossRef PubMed.
- D. Wilson, F. Citiulo and B. Hube, Zinc exploitation by pathogenic fungi, PLoS Pathog., 2012, 8, e1003034 CAS.
- D. J. Eide, Zinc transporters and the cellular trafficking of zinc, Biochim. Biophys. Acta, 2006, 1763, 711–722 CrossRef CAS PubMed.
- J. Amich and J. A. Calera, Zinc Acquisition: A Key Aspect in Aspergillus fumigatus Virulence, Mycopathologia, 2014, 178, 379–385 CrossRef PubMed.
- E. S. Bensen, S. J. Martin, M. Li, J. Berman and D. A. Davis, Transcriptional profiling in Candida albicans reveals new adaptive responses to extracellular pH and functions for Rim101p, Mol. Microbiol., 2004, 54, 1335–1351 CrossRef CAS PubMed.
- F. Citiulo, I. D. Jacobsen, P. Miramon, L. Schild, S. Brunke, P. Zipfel, M. Brock, B. Hube and D. Wilson, Candida albicans scavenges host zinc via Pra1 during endothelial invasion, PLoS Pathog., 2012, 8, e1002777 CAS.
- B. Milon, Q. Wu, J. Zou, L. C. Costello and R. B. Franklin, Histidine residues in the region between transmembrane domains III and IV of hZip1 are required for zinc transport across the plasma membrane in PC-3 cells, Biochim. Biophys. Acta, 2006, 1758, 1696–1701 CrossRef CAS PubMed.
- G. Butler, M. D. Rasmussen, M. F. Lin, M. A. Santos, S. Sakthikumar, C. A. Munro, E. Rheinbay, M. Grabherr, A. Forche, J. L. Reedy, I. Agrafioti, M. B. Arnaud, S. Bates, A. J. Brown, S. Brunke, M. C. Costanzo, D. A. Fitzpatrick, P. W. de Groot, D. Harris, L. L. Hoyer, B. Hube, F. M. Klis, C. Kodira, N. Lennard, M. E. Logue, R. Martin, A. M. Neiman, E. Nikolaou, M. A. Quail, J. Quinn, M. C. Santos, F. F. Schmitzberger, G. Sherlock, P. Shah, K. A. Silverstein, M. S. Skrzypek, D. Soll, R. Staggs, I. Stansfield, M. P. Stumpf, P. E. Sudbery, T. Srikantha, Q. Zeng, J. Berman, M. Berriman, J. Heitman, N. A. Gow, M. C. Lorenz, B. W. Birren, M. Kellis and C. A. Cuomo, Evolution of pathogenicity and sexual reproduction in eight Candida genomes, Nature, 2009, 459, 657–662 CrossRef CAS PubMed.
- M. A. Moreno, O. Ibrahim-Granet, R. Vicentefranqueira, J. Amich, P. Ave, F. Leal, J. P. Latge and J. A. Calera, The regulation of zinc homeostasis by the ZafA transcriptional activator is essential for Aspergillus fumigatus virulence, Mol. Microbiol., 2007, 64, 1182–1197 CrossRef CAS PubMed.
- C. J. Nobile, J. E. Nett, A. D. Hernday, O. R. Homann, J. S. Deneault, A. Nantel, D. R. Andes, A. D. Johnson and A. P. Mitchell, Biofilm matrix regulation by Candida albicans Zap1, PLoS Biol., 2009, 7, e1000133 Search PubMed.
- O. Schneider Rde, S. Fogaca Nde, L. Kmetzsch, A. Schrank, M. H. Vainstein and C. C. Staats, Zap1 regulates zinc homeostasis and modulates virulence in Cryptococcus gattii, PLoS One, 2012, 7, e43773 Search PubMed.
- J. Amich, R. Vicentefranqueira, F. Leal and J. A. Calera, Aspergillus fumigatus survival in alkaline and extreme zinc-limiting environments relies on the induction of a zinc homeostasis system encoded by the zrfC and aspf2 genes, Eukaryotic Cell, 2010, 9, 424–437 CrossRef CAS PubMed.
- P. F. Zipfel, C. Skerka, D. Kupka and S. Luo, Immune escape of the human facultative pathogenic yeast Candida albicans: the many faces of the Candida Pra1 protein, Int. J. Med. Microbiol., 2011, 301, 423–430 CrossRef CAS PubMed.
- D. A. Soloviev, W. A. Fonzi, R. Sentandreu, E. Pluskota, C. B. Forsyth, S. Yadav and E. F. Plow, Identification of pH-regulated antigen 1 released from Candida albicans as the major ligand for leukocyte integrin alphaMbeta2, J. Immunol., 2007, 178, 2038–2046 CrossRef CAS.
- M. Stork, M. P. Bos, I. Jongerius, N. de Kok, I. Schilders, V. E. Weynants, J. T. Poolman and J. Tommassen, An outer membrane receptor of Neisseria meningitidis involved in zinc acquisition with vaccine potential, PLoS Pathog., 2010, 6, e1000969 Search PubMed.
- E. K. LeGrand and J. Alcock, Turning up the heat: immune brinksmanship in the acute-phase response, Q. Rev. Biol., 2012, 87, 3–18 CrossRef.
- K. Subramanian Vignesh, J. A. Landero Figueroa, A. Porollo, J. A. Caruso and G. S. Deepe Jr., Zinc sequestration: arming phagocyte defense against fungal attack, PLoS Pathog., 2013, 9, e1003815 Search PubMed.
- S. Meyerson and D. A. Harpole, in General Thoracic Surgery, ed. T. W. Shields, J. LoCicero, C. E. Reed and R. H. Feins, 7th edn, 2011, ch. 92, pp. 1159–1178 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4mt00331d |
| This journal is © The Royal Society of Chemistry 2015 |