Open Access Article
Open Access ArticleCytotoxicity of Pd(II) and Pt(II) complexes of 2′,6′-di(thiazol-2-yl)-2,4′-bipyridine: insights into the mode of cell death and cell cycle arrest†
Ahmed M. Mansour‡
 *ab,
Krzysztof Radacki
*ab,
Krzysztof Radacki c,
Ola R. Shehab
c,
Ola R. Shehab *b,
Gamal A. E. Mostafad,
Essam A. Alid and
Mahmoud T. Abo-Elfadl‡
*b,
Gamal A. E. Mostafad,
Essam A. Alid and
Mahmoud T. Abo-Elfadl‡ ef
ef
aDepartment of Chemistry, United Arab Emirates University, Al-Ain, United Arab Emirates. E-mail: Mansour_am@uaeu.ac.ae; mansour@sci.cu.edu.eg; inorganic_am@yahoo.com
bDepartment of Chemistry, Faculty of Science, Cairo University, Gamma Street, Giza, Cairo 12613, Egypt. E-mail: Olashehab@sci.cu.edu.eg
cInstitut für Anorganische Chemie, Julius-Maximilians-Universität Würzburg, Am Hubland, D-97074 Würzburg, Germany
dDepartment of Pharmaceutical Chemistry, College of Pharmacy, King Saud University, Riyadh 11451, Saudi Arabia
eCancer Biology and Genetics Laboratory, Centre of Excellence for Advanced Sciences, National Research Centre, Dokki, Giza 12622, Egypt
fBiochemistry Department, Biotechnology Research Institute, National Research Centre, Dokki, Giza 12622, Egypt
First published on 16th April 2025
Abstract
Square-planar complexes were synthesized by the reaction of 2′,6′-di(thiazol-2-yl)-2,4′-bipyridine with either Na2[PdCl4] or K2[PtCl4], and these were thoroughly structurally characterized using some analytical and spectroscopic techniques. Density functional theory computations, including natural bond orbital analysis, were used to complement the experimental work to gain insight into the natural charge and electronic arrangement of the metal ion, as well as the strength of the metal–ligand bonds. The Pd(II) complex exhibited exceptional cytotoxicity against the A549 and HCT-116 cell lines with IC50 values of 60.1 ± 3.45 and 23.8 ± 1.48 μM, respectively. Unfortunately, the Pd(II) complex was harmful to the Vero normal cell line with an IC50 value of 24.5 ± 2.13 μM. The Pt(II) complex is unstable and has a high likelihood of exchanging the chlorido ligand for solvent molecules such as DMSO. The fluorescent-stain photos of the treated HCT-116 cells with the Pd(II) complex showed increased apoptotic bodies, indicating both early (18%) and late apoptosis (32%), as well as a necrosis ratio of about 10%. Flow cytometric analysis demonstrated that a cell arrest was induced by the Pd(II) complex on HCT-116 cells in the G2/M phase.
1 Introduction
The metal-based anticancer agent cisplatin is one of the most widely used clinical medications for treating a variety of tumours despite its adverse effects, which include vomiting, neurotoxicity, nephrotoxicity, and ototoxicity. Even now, some tumours become resistant to cisplatin, which lessens its effectiveness.1,2 Therefore, the development of more effective cancer therapy treatments is therefore necessary.3 There are two main approaches to develop new anticancer drugs based on the cisplatin structure and its next generations. One way to deal with the side effects of cisplatin is to synthesize analogues by substituting the labile chlorido or inert ammine ligands.4–7 The second approach to counteract the negative side effects of cisplatin includes using metal ions other than Pt(II), such as Pd(II), Au(III), Ru(II), and Pt(IV). One strategy to improve the effectiveness of cisplatin against the resistant cell lines is co-treatment. Given the structural similarities between the d8 metal ions, the closely related Pd(II) complexes have garnered lots of interest in this area.6,8–12 Since Pd(II) complexes exchange ligands 105 times more quickly than their Pt(II) analogues and are more likely to interact with sulfur-donor biomolecules and other cell components, they are indeed labile. The quick exchange and/or hydrolysis of Pd(II) complexes was resolved with the aid of certain chelating ligands that offered high thermodynamic stability and a kinetically inert nature.One of the intriguing ligands that is widely utilized to create biologically active Pd(II) and Pt(II) complexes for addressing the high lability of Pd(II) complexes is 2,2′:6′,2′′-terpyridine.13,14 Due to its exceptional complexing ability towards a variety of metal ions, 2,2′:6′,2′′-terpyridine has been widely used as a tridentate motif among the well-known ligands used as building blocks for the synthesis of diverse complexes containing nitrogen donor ligands.15–17 When applied to the Cellosaurus cell line (Bel-7402), the Pd(II) complex of 4′-(4-(2-(piperidin-1-yl)ethoxy)phenyl)-2,2′:6′,2′′-terpyridine (Fig. 1a) is seven times more cytotoxic (IC50 = 1.01 ± 0.13 μM) than cisplatin (IC50 = 8.12 ± 0.20 μM).18 It has shown that the Pd(II) complex of 2,2′:6′,2′′-terpyridine (Fig. 1b) is more cytotoxic (IC50 = 4.0 ± 0.5 μM) towards MCF7 cell line than the corresponding two Pd(II) complexes of 2,6-bis(N-pyrazol-2-yl)pyridine derivatives (IC50 = 13.0–27.0 μM).19 Yet, the three complexes demonstrated harm to the normal Vero cell line with CC50 < 50 μM. The microscopic images of the treated MCF7, with the three complexes, revealed indications of cell blebbing, early-stage apoptosis induction, and necrotic cell death. The Pd(II) terpyridine complexes (Fig. 1c), decorated with either R![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) OH20 or I exhibited the highest level of effectiveness in inhibiting the growth of the Bel-7402 cell line within the series.21 However, the complex decorated with OH was less cytotoxic than cisplatin against MCF-7, A549, K562, HT-29, and HepG2.20 Flow cytometry on the Bel-7402 cell line shows that the complex, with R
OH20 or I exhibited the highest level of effectiveness in inhibiting the growth of the Bel-7402 cell line within the series.21 However, the complex decorated with OH was less cytotoxic than cisplatin against MCF-7, A549, K562, HT-29, and HepG2.20 Flow cytometry on the Bel-7402 cell line shows that the complex, with R![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) OH, prevents cell proliferation during the G0/G1 phase, causing most apoptotic cells to be late apoptotic.
OH, prevents cell proliferation during the G0/G1 phase, causing most apoptotic cells to be late apoptotic.
 | ||
| Fig. 1 Previously reported cytotoxic Pd(II)- and Pt(II)-terpyridine complexes and closely related structures, functionalized with tridentate ligands. | ||
Due to a combination of autophagic and apoptotic cell death pathways, the Pd(II) complexes of 2,6-bis(thiazol-2-yl)pyridine (Fig. 1d), adorned with 9-phenantryl and 1-pyrenyl, exhibited potency against the A2780 cell line, comparable to cisplatin.22 Reactive oxygen species were produced by both complexes, and they could be associated to the apoptotic pathway trigger. The IC50 values (22−26 μM) of Pt(II) complex of 2,6-bis(2-benzimidazolyl) pyridine (Fig. 1e) against HepG2 and HT29 demonstrated that it had significantly more cytotoxic activity than cisplatin. Using flow cytometric analysis and fluorescence microscopy, it was shown that this complex caused apoptotic cell death, as shown by morphological alterations, loss of mitochondrial trans-membrane potential, and a significant increase of cells in the sub G0/G1 phase of the cell cycle.23 The Pd(II) complex of the mentioned ligand demonstrated selectivity and cytotoxic potency against HeLa cells (IC50 = 16.3 ± 4.9 μM) that were at par with cisplatin (IC50 = 11.4 ± 3.5 μM).24 In contrast to cisplatin, this Pd(II) complex did not significantly trigger caspase-dependent apoptosis.
Herein, we report the synthesis and characterization of 2′,6′-di(thiazol-2-yl)-2,4′-bipyridine (1) and its Pd(II) and Pt(II) complexes ([MCl(1)]·PF6 (M = Pd(II) 2 and Pt(II)) 3) (Scheme 1) using a variety of analytical and spectroscopic tools. The stability of the complexes was examined before researching their antiproliferative properties against five cancerous and one normal cell lines. The Pt(II) complex is unfortunately unstable and has a high likelihood of exchanging the chlorido ligand for DMSO. Therefore, the Pt(II) complex was not further investigated for its cytotoxic properties because it showed low and poor stability in a media appropriate for biological applications. A singlet oxygen trap named 1,3-diphenyliso-benzofuran was utilized to track the production of the reactive oxygen species. The effect of 2 on the cell cycle was also investigated by the flow cytometry technique. The acridine orange/ethidium bromide stain (AO/EtBr) is used to regulate the manner of cell death that occurs when the HCT-116 cell line (for example) is treated with complex 2.
 | ||
| Scheme 1 Synthesis of 2′,6′-di(thiazol-2-yl)-2,4′-bipyridine (1), and its Pd(II) (2) and Pt(II) (3) complexes. | ||
2 Results and discussion
2.1. Synthesis and characterization
Scheme 1 illustrates how 2-pyridinecarboxaldehyde and two equivalents of 2-acetyl thiazole reacted in strongly basic media (potassium tert-butoxide and ammonia) to produce 2′,6′-di(thiazol-2-yl)-2,4′-bipyridine (1).25,26 Ligand 1 was structurally characterized by IR (Fig. S1†), 1H NMR (Fig. S2†), 13C NMR (Fig. S3†), {1H, 1H} COS90 (Fig. S4†), and {1H, 13C} HSQC (Fig. S5†). In CDCl3, the 1H NMR spectrum (Fig. S2†) of 1 is characterized by a singlet signal at δ 8.84 ppm assigned to the protons of the central ring, as well as two doublets, and two triplets, at δ 8.77, 7.99, 7.82 and 7.35 ppm, due to the terminal pyridine ring. For thiazole ring, two doublets are observed at δ 7.96 and 7.48 ppm in the 1H NMR spectrum of 1. After being treated with a saturated solution of NH4PF6, the reaction between 1 and sodium tetrachloropalladate produced 2. As 1, the Pd(II) complex 2 was fully characterized by IR (Fig. S6†), NMR (Fig. S7–S12†) (1H, 13C, {1H, 1H} COS90, {1H, 13C} HSQC, 19F and 31P), electrospray ionization mass spectrometry (Fig. S13†) and elemental analysis. In the positive mode, the ESI MS spectrum (Fig. S13†) of 2 is characterized by a fragment at m/z 464.9059 (calcd 464.9065) due to {M–PF6}+ (M is a molecular formula of 2). A review of the literature revealed that terpyridine ligands and their analogues typically coordinate to metal ions in one of three ways: either in a tridentate fashion via all three nitrogen donor atoms,27–29 in a bidentate к2-N1,1′ mode where one of the two outer heterocyclic rings is bent out of the mean plane of the other two coordinated centers,30–32 or in a few instances, in a monodentate к1-N1 binding mode.33 A singlet 1H NMR signal for the central pyridine-H3/H5 indicates that the ligand 1 is in the tridentate chelation mode. The PF6− counterion was traced by observing the septet 31P NMR signal (Fig. S11†) and doublet 19F NMR signal (Fig. S12†) at δ −144.2 (1JPF = 711 Hz) and −70.1 ppm (1JPF = 711 Hz), respectively. After 1 and K2[PtCl4] reacted together, NH4PF6 was added, yielding 3. Unfortunately, complex 3 decomposes in DMSO (Fig. S14†) because of the quick exchange of the chlorido ligand with DMSO. Besides, complex 3 has low solubility in most of the organic solvents. This has been noted for some Pt(II) terpyridine complexes.28,34 Therefore, complex 3 was structurally characterized by IR (Fig. S15†), solid-state NMR {13C, 15N, 19F, and 31P} (Fig. S16−S19†), ESI MS (Fig. S20†), and elemental analysis. The ESI MS spectrum (Fig. S20†) of 3, in the positive mode, displays a unique fragment at m/z 552.9657 (calcd 552.9679) due to {M−PF6}+ (M is a molecular formula). In 31P (Fig. S17†) and 19F (Fig. S18†) NMR spectra of 3, the PF6− signals are detected at δ = −143.9 and −68.3 ppm, respectively. The 15N NMR spectrum (Fig. S19†) of 3 is characterized by a signal at δ −168.6 ppm, which is typical for both the Pt central atom and the nitrogen trans-to Cl.34–362.2. Density functional theory calculations
The tridentate coordination mode of 1 towards Pd(II) and Pt(II) ions was shown by the spectroscopic and analytical data in the previous section. The geometries around 2 and 3 could be illustrated as square-planar, defined by a tridentate N,N,N-ligand, and chlorido ligand. The local minimum structures of 2 and 3 (Fig. S21 and Fig. S22†), in the ground state, were attained by optimizing the models representing their molecular structures, using the Becke 3-parameter (exchange) Lee–Yang–Parr functional37,38 and LANL2DZ basis set39,40 and by validating as a minimum on the potential energy surface by computing the vibrational modes at the same computational level of theory. Tables S1 and S2† contain the atomic coordinates for the local minimum structures of 2 and 3. Selected bond lengths and angles of the local minimum structures of 2 and 3 are given in Table 1. In both complexes, the thiazole trans- M–N bonds are the same, but the ones to the Pt(II) ion are 0.020 Å shorter than the ones to the Pd(II) ion. The reason for this is that Pd(II) and Pt(II) ions differ in size. Similarly, Pt(II) is 0.016 Å closer to the central pyridine nitrogen than the Pd(II) ion. In contrast, the Pt–Cl is 0.034 Å longer than the Pd–Cl, which could be the cause of the complex's instability in DMSO due to the quick exchange between Cl and the solvent molecules. In both complexes, the corresponding cis- and trans-angles around the metal center are equal. This means that the shift of the metal center from Pd(II) to Pt(II) ion does not cause any change in the bond angles. Since the bond angles deviate from the linearity, the geometries of 2 and 3 should be regarded as distorted square planar.| 2 | 3 | ||
|---|---|---|---|
| Pd–N(py) | 1.990 Å | Pt–N(py) | 1.974 Å |
| Pd–N(thiazole) | 2.055 Å | Pt–N(thiazole) | 2.035 Å |
| Pd–Cl | 2.348 Å | Pt–Cl | 2.382 Å |
| N(py)–Pd–N(thiazole) | 79.9° | N(py)–Pt–N(thiazole) | 80.1° |
| N(thiazole)–Pd–Cl | 100.2° | N(thiazole)–Pt–Cl | 99.7° |
| N(thiazole)–Pd–Cl | 100.0° | N(thiazole)–Pt–Cl | 99.9° |
| N(py)–Pd–Cl | 179.8° | N(py)–Pt–Cl | 179.9° |
| N(thiazole)–Pd–N(thiazole) | 159.7° | N(thiazole)–Pt–N(thiazole) | 160.3° |
The natural charge of the metal center, population of the d-orbitals, type of bonding, and strength of the M–N bonds (pyridine vs. triazole) in 2 and 3 could be ascertained using the natural bond orbital (NBO) analysis of Weinhold and co-workers41 and the second order perturbation theory analysis. Moreover, the NBO approach has been extensively used to assess the intra- and intermolecular contacts and offers a good basis for researching conjugative interactions or charge transfer in several molecular systems. To ascertain the contributions of atomic orbitals to σ or π natural bond orbital, for bonded atom pairs, NBO analysis was performed. In general, the types of NBOs are bonding, core (CR), and lone pair (LP). The ideal Lewis structure is composed of Lewis σ-type (donor) NBOs and non-Lewis σ-type (acceptor) NBOs. Filled NBOs define covalency in the molecules, while anti-bonds represent vacant valence-shell capacity. Weak valence anti-bond occupancies are a sign of an intricate departure from a localized Lewis picture, or actual “delocalization effects.” According to NBO analysis, the electronic configuration of the Pd(II) ion in 2 is [Kr]5s0.344d8.835p0.305d0.01. Thus, 35.99 core electrons, 9.46 valence electrons, and 0.01 Rydberg electrons give 45.46 electrons. Since the surrounding ligands donate an electron density to the metal center, the calculated natural charge on the Pd ion (0.534e) is significantly less than the formal charge (+2) (Table 2). The occupancies of 4d orbitals are as follows: dxy1.97701dxz1.93809dyz1.95669dx2−y21.01202dz21.94578. The four bonds that form between Pd and the N or Cl-atoms can be explained by NBO analysis as the donation of electron density from each nitrogen atom's lone pair (LP) orbital to palladium molecular orbitals. Alternatively, the electronic arrangement of Pt(II) ion in 3 is [Xe]6s0.515d8.626p0.316d0.01. Thus, 67.98 core electrons, 9.43 valence electrons, and 0.01 Rydberg electrons give the 67.98 electrons. The computed Pt ion natural charge (0.560e) is comparable to the Pd natural charge. The occupancies of 5d orbitals are as follows: dxy1.96248dxz1.88053dyz1.91837dx2−y20.96476dz21.88930. Similar to 2, the four bonds in 3 might be accounted for as an electron donation to the Pt ion from the coordination sites, with the exception of the bond of the central pyridine ring, which has a slightly covalent charter. This may be seen in the trans-effect, the elongation of the Pt–Cl bond, and thus the quick solvent–Cl exchange. For 3, the σ(Pd–N) bond (N is the pyridine nitrogen) is formed from a sp2.76 hybrid on N atom, which is a mixture of 26.63% s and 73.37% p, and sp0.36d5.81 hybrid orbital on Pt atom (a mixture of 14.60% s, 5.24% p, and 5.49% d). NBO analysis implied that the σ(Pt–N) bond is strongly polarized towards the N atoms with about 74.85% of electron density concentrated on the N atom.
| Complex | Electronic arrangement of Pd atom | dxy | dxz | dyz | dx2−y2 | dz2 | Natural charge |
|---|---|---|---|---|---|---|---|
| 2 | [Kr]5 s0.344d8.835p0.305d0.01 | 1.97701 | 1.93809 | 1.95669 | 1.01202 | 1.94578 | 0.534e |
| 3 | [Xe]6 s0.515d8.626p0.316d0.01 | 1.96248 | 1.88053 | 1.91837 | 0.96476 | 1.88930 | 0.560e |
Hyperconjugation is the stabilizing effect caused by the overlap of two orbitals, one of which is occupied and the other is electron deficient. The second-order perturbation theory and the Fock Matrix in NBO basis were used to determine the hyper-conjugative interaction energy between the acceptor and donor orbitals. The strength of the electronic interactions between the donors such as the coordination sites of 1 and acceptors e.g., Pd(II) ion is indicated by the second-order interaction energy (E2) (Table 3). The E2 values of LP(1)N(triazole) → RY*(4)Pd are equal, 0.74 kcal−1 mol−1. However, these two interactions are weaker than the corresponding one from the pyridine ring, 1.96 kcal−1 mol−1. The interaction LP(4)Cl → RY*(4)Pd has E2 value of 0.27 kcal−1 mol−1. In the case of 3, the LP(1)N(triazole) → RY*(9)Pt, σ(Pt–N(pyridine)) → RY*(9)Pt and LP(4)Cl → RY*(9)Pt have E2 values of 1.48, 0.56 and 1.32 kcal−1 mol−1. Once more, both triazole ligands have equal and greater interactions than the chlorido ligand.
| Complex | M–N and M–Cl interactions | ||
|---|---|---|---|
| 2 | LP(1)N(triazole) → RY*(4)Pd = 0.74 | LP(1)N(pyridine) → RY*(4)Pd = 1.96 | LP(4)Cl → RY*(4)Pd = 0.27 |
| 3 | LP(1)N(triazole) → RY*(9)Pt = 1.48 | σ(Pt–N (pyridine)) → RY*(9)Pt = 0.56 | LP(4)Cl → RY*(9)Pt = 1.32 |
2.3. Time-dependent density functional calculations
Time-dependent density functional theory (TDDFT) calculations were carried out in the singlet state using a Becke 3-parameter (exchange) Lee–Yang–Parr functional37,38 with LANL2DZ basis set39,40 and polarizable continuum solvation model (DMSO),42 to gain an insight into the nature of the electronic transitions expected to be observed in the electronic spectra of 2 and 3. The computed electronic spectra electronic spectra (Fig. S23†) were modelled using the Gauss-Sum software. A Gaussian convolution was utilized for each excited state, with a full width at a maximum of 3000 cm−1. The calculated electronic transitions, their energies, and assignments are tabulated in Table S3.† The computed excitation state energies with transition strength (f > 0.05) were considered. The electronic spectrum of 2 shows two bands at 273 and 321 nm as well as a shoulder at 403 nm due to HOMO → LUMO+3, HOMO−1 → LUMO+1 and HOMO → LUMO, in that order. As shown in Fig. 2, HOMO and HOMO−1 are mostly d(Pd)/p(Cl), while the orbitals of the LUMOs stated in the above transitions are obtained from π(ligand). Therefore, the three main transitions featuring the electronic spectrum of 2 are mainly MLCT in nature. On the other hand, the calculated electronic absorption spectrum of 3 is characterized by three main bands at 295, 350, and 454 nm corresponding to HOMO−6 → LUMO+1, HOMO−5 → LUMO and HOMO → LUMO, respectively. Whereas LUMO is mostly made up of the ligand π-framework with a minor contribution from d(Pt), HOMO orbitals are mostly contained upon d(Pt)/p(Cl). Thus, the lowest energy transition at 454 nm is a combination of MLCT/d–d. Similarly, the transition at 350 nm is MLCT/d–d. The transition at 295 nm is mainly LLCT in nature.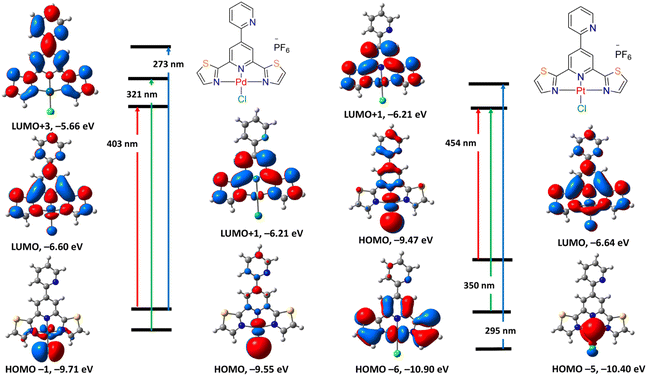 | ||
| Fig. 2 Selected frontier molecular orbitals of 2 (left) and 3 (right) obtained by B3LYP/LANL2DZ method. The electronic transitions were obtained at the same level of theory. | ||
2.4. Cell viability assay
The cytotoxicity of 2 was assessed against Human epithelial mammary gland breast adenocarcinoma (MCF7), human epithelial hepatocellular carcinoma (HepG2), human epithelial-like lung carcinoma (A549), human epithelial large intestine colorectal carcinoma (HCT-116), and normal epithelial kidney of an African green monkey, Vero cell line, using the MTT assay. All cell lines were obtained from the American Type Cell Culture Bank (ATCC, Manassas, USA). Because complex 3 demonstrated low stability in DMSO and poor solubility in a medium suitable for biological applications, its cytotoxic properties were not further studied. For a duration of 24 h, the normal and malignant cultures were co-incubated with 2 at concentrations ranging from 5.13 to 164 μM. The concentration (IC50 value ± SD) (Table 4) required to reduce the cell growth by 50% was determined for 2 (Fig. S24†) and cisplatin (Fig. S25†). The viability of the examined cells is not significantly impacted by the medium. The compound exhibited varying degrees of cytotoxicity against the distinct cell lines that were being studied. Complex 2 demonstrated a strong effect on A549 and HCT-116 cell lines, with IC50 values of 60.1 ± 3.45 and 23.8 ± 1.48 μM, respectively, while remaining safe on the MCF7 and HepG2 cell lines. Unfortunately, the Pd(II) complex was harmful to the Vero normal cell line with an IC50 value of 24.5 ± 2.13 μM. As can be shown from Table 4, complex 2 exhibits cytotoxicity against HCT-111 and Vero cell lines that is comparable to that of cisplatin.| Complex | MCF7 | HepG2 | A549 | HCT-116 | Vero |
|---|---|---|---|---|---|
| 2 | — | — | 60.1 ± 3.45 | 23.8 ± 1.48 | 24.5 ± 2.13 |
| Cisplatin | 110.58 ± 12.5 | 27.95 ± 2.5 | 8.71 ± 0.5 | 14.61 ± 1.1 | 17.09 ± 1.4 |
2.5. Mode of cell death
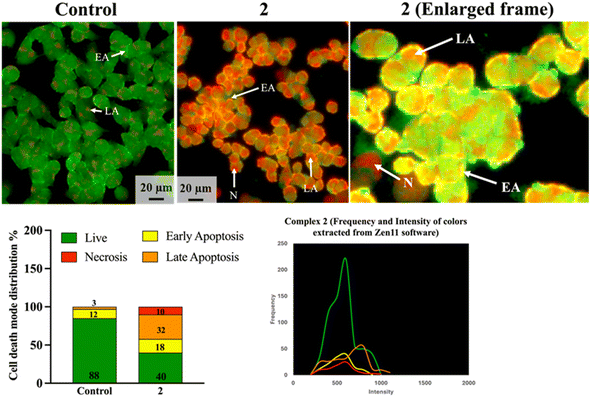
The acridine orange/ethidium bromide stain (AO/EtBr) is used to regulate the manner of cell death that occurs when the HCT-116 cell line (for example) is treated with 2. The necrotic cells as well as the early and late apoptotic cells are shown when the exclusion dye, ethidium bromide, diffuses through the ruined cell membranes and attaches itself to the DNA. The different colors of orange to red, yellowish green, and yellowish orange are utilized for distinguishing between necrotic, late, and early apoptotic cell types. The photos and cell death distribution are presented in Fig. 3. By using the fluorescent dye of AO/EtBr, the HCT-116 cells treated with 2 at a sub-toxic dose (11.5 μM) for 24 h showed the largest late apoptotic alterations (32%) compared to only 3% in the control cells. The cell death mechanism also displays early apoptosis, with ratios of 18% and 12% in cells for 2 and control, respectively. The cells treated with 2 showed a necrosis ratio of roughly 10%, while the control cells showed no necrosis.2.6. Cell cycle
One of the main factors in the development and spread of cancer is the dysregulation of cell cycle progression, which results in unchecked cell growth halt and excessive cell proliferation.43 The first sign that complex 2 is interfering with the cellular process is a change in the distribution of cells across the phases of mitotic division when compared to samples that were not treated. Though there may be many different processes causing cells to accumulate during a given stage of the cell cycle, these findings offer justification for choosing which direction to focus future research. Based on the inhibitory effect on cell proliferation, the effect of 2 on the cell cycle of HCT-116 cells was examined. The cell cycle distribution of HCT-116 cells treated with a sub-toxic dose of 2 (11.5 μM) for 24 h was examined by flow cytometric analysis of the DNA content. The distribution of cell populations in each phase of the cell cycle is shown in Fig. 4. The results demonstrated that the G0/G1 phase was significantly reduced in the cells treated with 2 as compared to the control cells (84.90% for the control and 62.12% for 2, p < 0.001). By comparing the S phases of the control and the cells treated with 2, there was no significant difference among the control cells (14.00%) and 2 (18.18%, p > 0.05). On the other hand, there was significant cell arrest in the G2/M phase in cells treated with 2 (19.3%) compared to the control cells (1.1%) with a high significant value p < 0.001. These results indicated that the antiproliferative mechanism induced by 2 on HCT-116 cells was cell arrest in the G2/M phase.2.7. Reactive oxygen species
It is well recognized that the aerobic cells' mitochondrial oxidative metabolism results in the production of reactive oxygen species (ROS). When properly controlled, ROS can act as ubiquitous mediators of post-translational signalling and the control of gene expression.44 Nonetheless, several diseases linked to aging and metabolism have one thing in common: chronically high ROS production is very detrimental to cellular constituents.45 Interestingly, in pathological conditions, mitochondria are the main source of ROS in cells because electrons that have accumulated in the electron transport chain as a result of disrupted metabolism can mostly escape from complex I by reverse electron flow or straight from complex III.44 Superoxide and other strong oxidants like hydrogen peroxide and hydroxyl radical are created when these electrons are then carried directly to O2. As shown in Fig. 5, the quantity of ROS generated from the treated HCT-116 was ambiguous. Although it was as high as the positive control cells (treated with the strong ROS-releasing agent; tert-butyl hydroperoxide) with no significant difference between them (p = 0.32), the ROS level was still not significantly different from the untreated negative control cells (p = 0.11).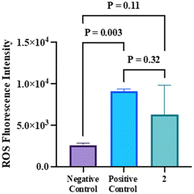 | ||
| Fig. 5 The ROS fluorescence intensity of 2 in the HCT-116 cell compared to the positive and negative controls. The data are represented as mean ± Sd. | ||
3 Conclusions
In the present contribution, we reported the synthesis, and structural characterization of chlorido Pd(II) and Pt(II) complexes of the N,N,N-tridentate ligand, 2′,6′-di(thiazol-2-yl)-2,4′-bipyridine. The experimental studies were complemented with density functional theory calculations. The Pt(II) complex disintegrated in DMSO and displayed poor solubility in the rest of the organic solvents. It is highly likely that the chlorido ligand of the Pt(II) complex was exchanged for DMSO. Therefore, Pt(II) was not investigated for its cytotoxic properties. This has been seen for some Pt(II) complexes that are structurally related.28 It is evident from the local minimum structures of the Pd(II) and Pt(II) complexes that the Pt–Cl is 0.034 Å longer than the Pd–Cl. This could be the reason for the complex's instability in DMSO because of the rapid exchange between Cl and the solvent molecules. This has been confirmed by natural bond orbital analysis. On the other hand, the corresponding Pd(II) complex showed greater stability in DMSO and as a result, the complex was tested for possible cytotoxicity against normal cell types and cancer. The cell viability assay showed that the Pd(II) complex was potent to A549 and HCT-116 cell lines, with IC50 values of 60.1 ± 3.45 and 23.8 ± 1.48 μM, respectively, while remaining non-cytotoxic to MCF7 and HepG2 cell lines. In fact, complex 2 demonstrated greater or comparable cytotoxicity against HCT-1116 in comparison to cisplatin and certain Pd(II) complexes that have been previously described; nevertheless, the other complexes might be less detrimental to normal cell lines.46–48 Unfortunately, the Pd(II) complex was harmful to the Vero normal cell line with an IC50 value of 24.5 ± 2.13 μM. Treating cells with the Pd(II) complex released ROS not high enough to be significantly different from the untreated control cells. In the mode of cell death study, the fluorescent images of the treated HCT-116 cells with the Pd(II) complex, displayed more apoptotic bodies, indicating both early (18%) and late (32%), as well as a necrosis ratio of roughly 10%. Flow cytometric analysis revealed that the antiproliferative mechanism induced by the Pd(II) complex on HCT-116 cells was a cell arrest in the G2/M phase.4 Experimental
4.1. Materials and instruments
Organic solvents, potassium tetrachloroplatinate, and sodium tetrachloropalladate were bought and utilized exactly as supplied. 2′,6′-di(thiazol-2-yl)-2,4′-bipyridine25,26 was prepared by following the previously published methods. 1H, 13C, 19F and 31P NMR spectra were recorded with Brucker-Avance 500 (1H, 500.13 MHz; 13C{1H}, 125.77 MHz; 19F, 470.59 MHz; 31P, 202.46 MHz) and Bruker-Avance 400 (1H, 400.40 MHz; 13C{1H}, 100.68 MHz; 31P, 162.08 MHz) spectrometers. Assignments of NMR signals were performed with the aid of two-dimensional NMR methods, {1H, 1H} COS90 and {1H, 13C} HSQC. A ThermoFisher Exactive Plus device equipped with an Orbitrap mass analyser was used to record electrospray mass spectra at a solvent flow rate of 50 μl min−1 and a resolution of R = 70.000. Elemental micro-analysis was achieved with a Vario Micro Cube analyser of Elementar Analysensysteme or an EA 3000 elemental analyzer from HEKtech. A Nicolet 380 FT-IR spectrometer equipped with a smart iFTR adapter was used to gather vibrational spectra in the solid state. The Specord 210 Plus spectrophotometer was used to record electronic absorption spectra.4.2. Synthesis
1: (ref. 25 and 26) To a flat-bottom flask charged with 2-acetylthiazole (1 g; 7 mmol), ammonia solution (20 mL), potassium tert-butoxide, and ethanol (20 mL), was added 2-pyridine carboxaldehyde (0.42 g; 0.35 mmol). Directly, a dark brown colour developed. For 24 h, the reaction mixture was agitated at room temperature. As time passes, the dark-grown colour fades, and a white precipitate forms. Filtration was used to collect the precipitate, which was then repeatedly cleaned with water and diethyl ether. IR (ATR): ν = 3055, 1586, 1503, 1430, 1251, 1067, 991, 783 cm−1. 1H NMR (CDCl3, 500.13 MHz): δ = 8.84 (s, 2H, py-H3′/H5′), 8.77 (d, 3JH,H = 4.8 Hz, 1H, py-H6), 7.99 (d, 3JH,H = 7.9 Hz, 1H, py-H3), 7.96 (d, 3JH,H = 3.22 Hz, 2H, thiazole-H4), 7.82 (td, 3JH,H = 7.89 Hz, 4JH,H = 1.81 Hz, 1H, py-H4), 7.48 (d, 3JH,H = 3.15 Hz, 2H, thiazole-H5), 7.35 (m, 1H, py-H5) ppm. 13C NMR (CDCl3, 125.77 MHz): δ = 168.8, 153.9, 151.9, 150.2, 149.1, 144.3, 137.2, 124.0, 122.0, 121.2, 117.8 ppm. C16H10N4S2: C 59.61, H 3.13, N 17.38, S 19.89; found C 59.75, H 3.12, N 17.54, S 19.60.
2: Methanol (25 mL) was added to a round-bottom flask that was charged with 1 (0.5 mmol; 161 mg) and Na2PdCl4 (0.55 mmol; 147 mg), and then the reaction mixture was heated to reflux for 12 h. Yellow precipitate was collected by filtration, washed with methanol (3 × 5 mL), and chloroform (3 × 5 mL). The complex was treated with NH4PF6 (1.0 mmol; 163 mg) in 75% (acetone/water) mixture. Stirring was done overnight. Yellow precipitate was collected by filtration, washed with water (5 × 5 mL), diethyl ether, and dried in vacuo. IR (ATR): ν = 3065 (br, CH), 1611, 1472, 1330, 1252, 840 and 779 cm−1.1H NMR (DMSO-d6, 500.13 MHz): δ = 9.14 (s, 2H, py-H3′/H5′), 8.90 (m, 1H, py-H6), 8.51 (m, 1H, py-H3), 8.40 (m, 2H, thiazole-H4), 8.18 (m, 1H, py-H4), 7.96 (m, 2H, thiazole-H5), 7.71 (m, 1H, py-H5) ppm. 13C-NMR (DMSO-d6, 125.77 MHz): δ = 167.8, 152.5, 150.4, 150.3, 141.5, 138.1, 128.3, 126.4, 125.1, 123.5, 123.0 and 120.6 ppm. 31P NMR (DMSO-d6, 202.46 MHz): δ = −144.2 (sept, 1JPF = 711 Hz, PF6−). 19F NMR (DMSO-d6, 470.59 MHz): δ = −70.1 (doublet, 1JPF = 711 Hz, PF6−) ppm. ESI-MS (positive mode, DMSO/methanol): m/z = 464.9058 {M–PF6−}+ (M: molecular formula). C16H10ClF6N4PPdS2·H2O: C 30.64, H 1.93, N 8.93, S 10.22; found C 30.77, H 1.59, N 9.25, S 10.11.
3: To a flat-bottom flask charged with 1 (0.5 mmol; 161 mg) and methanol (30 mL), was added 5 mL aqueous solution of K2PtCl4 (0.5 mmol; 205 mg). The reaction mixture was heated to reflux for 12 h, whereupon brown precipitate formed. The complex was collected by filtration, washed with chloroform (3 × 5 mL), and water (3 × 5 mL). Next, the complex was suspended in 75% (acetone/water) solution that included NH4PF6 (1.0 mmol; 163 mg). Stirring was done overnight. Brown precipitate was collected by filtration, washed with water, diethyl ether, and dried in vacuo. IR (ATR): ν = 3065, 1611, 1457, 1252, 1208, 840, 779 cm−1. The complex decomposed in DMSO-d6 (see Fig. S15†) and had poor solubility in DMF-d7. Solid-state NMR analyses were done. 13C NMR (solid-state, 100.68 MHz): δ = 168.9, 166.9, 165.8, 160.4, 159.3, 149.9, 141.3, 138.1, 127.4, 126.3, 122.1, 120.0 and 116.7 ppm. 31P NMR (solid-state, 161.94 MHz): δ = −143.9 ppm (sept, 1JPF = 710 Hz, PF6−). 19F NMR (solid-state, 396.5 MHz): δ = −68.3 (br, PF6−) ppm. ESI-MS (positive mode, DMSO/methanol): m/z = 552.9658 {M–PF6−}+ (M: molecular formula). 15N NMR (solid-state, 40.56 MHz): δ = −168.6 ppm. C16H10ClF6N4PPtS2·3H2O: C 25.56, H 2.14, N 7.45, S 8.53; found C 25.41, H 2.09, N 7.39, S 8.46.
4.3. Density functional theory calculations
Ground-state geometry optimizations of 2 and 3, were executed using Gaussian03,49 with a Becke 3-parameter (exchange) Lee–Yang–Parr functional37,38 and LANL2DZ basis set.39,40 The local minimum structure of the complex was verified to be the lowest on the potential energy surface by computing their vibrational modes. No imaginary vibrations were detected. Using the computed approach utilized in the optimization procedure, natural bond orbital (NBO) analysis of Weinhold and co-workers was carried.41 Time-dependent density functional theory (TDDFT) calculations were done also by B3LYP/LANL2DZ method, including the polarizable continuum model (PCM).42 Visualization of the electronic spectra and frontier molecular orbitals was achieved using Gaussview03.504.4. Cell viability assay
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in PBS
1 in PBS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethidium bromide (100 μg mL−1) and acridine orange (100 μg mL−1). Ten minutes later, the slides were washed with PBS and viewed on an AxioImager Z2 fluorescent microscope using Zen 11 software (Zeiss, Jena, Germany).
ethidium bromide (100 μg mL−1) and acridine orange (100 μg mL−1). Ten minutes later, the slides were washed with PBS and viewed on an AxioImager Z2 fluorescent microscope using Zen 11 software (Zeiss, Jena, Germany).Data availability
Data is available in main text and ESI.†Author contributions
A. Mansour: conceptualization, investigation, validation, resources, formal analysis, software, writing – original draft, writing – review & editing. K. Radacki: methodology, investigation, resources. O. Shehab: investigation, writing – review & editing. G. Mostafa, writing – review & editing, resources. E. Ali, writing – review & editing, resources. M. Abo-Elfadl: conceptualization, methodology, investigation, validation, resources, formal analysis, writing – original draft, writing – review & editing.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
The authors extend their appreciation to the researchers Supporting Project Number (RSPD2025R1000) at King Saud University Riyadh Saudi Arabia, for funding this work. The authors wish to thank United Arab Emirates University, Al Ain, UAE, for research support. A. Mansour thanks Dr Rüdiger Bertermann, Julius-Maximilians-Universität Würzburg, for solid-state NMR analysis.Notes and references
- A. Rennicke, W. Voigt, T. Mueller, A. Fruehauf, H.-J. Schmoll, C. Beyer and W. Dempke, Anticancer Res., 2005, 25, 1147–1155 CAS.
- H. Timmer-Bosscha, N. Mulder and E. De Vries, Br. J. Cancer, 1992, 66, 227–238 CrossRef CAS PubMed.
- M. Tanaka, H. Kataoka, S. Yano, H. Ohi, K. Kawamoto, T. Shibahara, T. Mizoshita, Y. Mori, S. Tanida and T. Kamiya, BMC Cancer, 2013, 13, 1–9 CrossRef PubMed.
- A. M. Mansour and O. R. Shehab, Dalton Trans., 2018, 47, 3459–3468 RSC.
- N. T. Abdel-Ghani and A. M. Mansour, Eur. J. Med. Chem., 2012, 47, 399–411 CrossRef CAS PubMed.
- A. M. Mansour and N. T. Abdel-Ghani, Inorg. Chim. Acta, 2015, 438, 76–84 CrossRef CAS.
- A. M. Mansour, Inorg. Chim. Acta, 2016, 453, 697–703 CrossRef CAS.
- T. Lazarević, A. Rilak and Ž. D. Bugarčić, Eur. J. Med. Chem., 2017, 142, 8–31 CrossRef PubMed.
- E. Gao, C. Liu, M. Zhu, H. Lin, Q. Wu and L. Liu, Anti-Cancer Agents Med. Chem., 2009, 9, 356–368 CrossRef CAS PubMed.
- A. G. Quiroga and C. N. Ranninger, Coord. Chem. Rev., 2004, 248, 119–133 CrossRef CAS.
- A. M. Mansour, R. M. Khaled, K. Radacki, M. A. Abo-Zeid, O. R. Shehab, G. A. Mostafa, E. A. Ali and M. T. Abo-Elfadl, Dalton Trans., 2024, 53, 5073–5083 RSC.
- A. M. Mansour, R. M. Khaled, K. Radacki, O. R. Shehab, G. A. Mostafa, E. A. Ali and M. T. Abo-Elfadl, Chem. Biodiversity, 2024, 21, e202400363 CrossRef CAS PubMed.
- R. R. Panicker and A. Sivaramakrishna, Coord. Chem. Rev., 2022, 459, 214426 CrossRef CAS.
- R. Abhijnakrishna, K. Magesh, A. Ayushi and S. Velmathi, Coord. Chem. Rev., 2023, 496, 215380 CrossRef CAS.
- B. Z. Momeni, N. Davarzani, J. Janczak, N. Ma and A. S. Abd-El-Aziz, Coord. Chem. Rev., 2024, 506, 215619 CrossRef CAS.
- R. R. Panicker and A. Sivaramakrishna, Coord. Chem. Rev., 2022, 459, 214426 CrossRef CAS.
- C. Wei, Y. He, X. Shi and Z. Song, Coord. Chem. Rev., 2019, 385, 1–19 CrossRef CAS PubMed.
- W. Chu, Y. Wang, S. Liu, X. Yang, S. Wang, S. Li, G. Zhou, X. Qin, C. Zhou and J. Zhang, Bioorg. Med. Chem. Lett., 2013, 23, 5187–5191 CrossRef CAS PubMed.
- D. O. Onunga, R. Bellam, G. K. Mutua, M. Sitati, M. D. BalaKumaran, D. Jaganyi and A. Mambanda, J. Inorg. Biochem., 2020, 213, 111261 CrossRef CAS PubMed.
- F. Darabi, H. Hadadzadeh, J. Simpson and A. Shahpiri, New J. Chem., 2016, 40, 9081–9097 RSC.
- Z. Wang, J. Li, R. Liu, X. Jia, H. Liu, T. Xie, H. Chen, L. Pan and Z. Ma, J. Inorg. Biochem., 2023, 244, 112219 CrossRef CAS PubMed.
- K. Choroba, B. Machura, L. R. Raposo, J. G. Małecki, S. Kula, M. Pająk, K. Erfurt, A. M. Maroń and A. R. Fernandes, Dalton Trans., 2019, 48, 13081–13093 RSC.
- S. Rubino, P. Portanova, F. Giammalva, M. Girasolo, S. Orecchio, G. Calvaruso and G. Stocco, Inorg. Chim. Acta, 2011, 370, 207–214 CrossRef CAS.
- R. O. Omondi, R. Bellam, S. O. Ojwach, D. Jaganyi and A. A. Fatokun, J. Inorg. Biochem., 2020, 210, 111156 CrossRef CAS PubMed.
- T. Klemens, K. Czerwińska, A. Szlapa-Kula, S. Kula, A. Świtlicka, S. Kotowicz, M. Siwy, K. Bednarczyk, S. Krompiec and K. Smolarek, Dalton Trans., 2017, 46, 9605–9620 RSC.
- A. Maroń, S. Kula, A. Szlapa-Kula, A. Świtlicka, B. Machura, S. Krompiec, J. G. Małecki, R. Kruszyński, A. Chrobok and E. Schab-Balcerzak, Eur. J. Org Chem., 2017, 2017, 2730–2745 CrossRef.
- A. M. Mansour, J. Mol. Struct., 2021, 1242, 130737 CrossRef CAS.
- A. M. Mansour, RSC Adv., 2021, 11, 39748–39757 RSC.
- A. M. Mansour, K. Radacki, G. A. Mostafa, E. A. Ali and O. R. Shehab, Bioorg. Chem., 2024, 146, 107262 CrossRef CAS PubMed.
- A. M. Mansour and K. Radacki, Inorg. Chim. Acta, 2020, 511, 119806 CrossRef CAS.
- O. S. Taniya, D. S. Kopchuk, A. F. Khasanov, I. S. Kovalev, S. Santra, G. V. Zyryanov, A. Majee, V. N. Charushin and O. N. Chupakhin, Coord. Chem. Rev., 2021, 442, 213980 CrossRef CAS.
- A. M. Mansour, K. Radacki and O. R. Shehab, Dalton Trans., 2021, 50, 1197–1201 RSC.
- A. Hildebrandt, N. Wetzold, P. Ecorchard, B. Walfort, T. Rüffer and H. Lang, Eur. J. Inorg. Chem., 2010, 2010, 3615–3627 CrossRef.
- A. M. Mansour, K. Radacki, G. A. Mostafa, E. A. Ali and O. R. Shehab, RSC Adv., 2023, 13, 34826–34835 RSC.
- L. Pazderski, in Annual Reports on NMR Spectroscopy, Elsevier, 2013, vol. 80, pp. 33–179 Search PubMed.
- W. P. Ozimiński, P. Garnuszek, E. Bednarek and J. C. Dobrowolski, Inorg. Chim. Acta, 2007, 360, 1902–1914 CrossRef.
- A. Becke, Chem. Phys., 1993, 98, 5648 CAS.
- A. D. Becke, Phys. Rev. A, 1988, 38, 3098 CrossRef CAS PubMed.
- P. J. Hay and W. R. Wadt, J. Chem. Phys., 1985, 82, 299–310 CrossRef CAS.
- P. J. Hay and W. R. Wadt, J. Chem. Phys., 1985, 82, 270–283 CrossRef CAS.
- A. E. Reed, L. A. Curtiss and F. Weinhold, Chem. Rev., 1988, 88, 899–926 CrossRef CAS.
- M. Cossi, V. Barone, R. Cammi and J. Tomasi, Chem. Phys. Lett., 1996, 255, 327–335 CrossRef CAS.
- R. Farghadani, J. Rajarajeswaran, N. B. M. Hashim, M. A. Abdulla and S. Muniandy, RSC Adv., 2017, 7, 24387–24398 RSC.
- D. C. Wallace, W. Fan and V. Procaccio, Annu. Rev. Pathol.: Mech. Dis., 2010, 5, 297–348 CrossRef CAS PubMed.
- S. Raha and B. H. Robinson, Trends Biochem. Sci., 2000, 25, 502–508 CrossRef CAS PubMed.
- M. Ž. Mijajlović, M. V. Nikolić, V. V. Jevtić, Z. R. Ratković, B. S. Marković, V. Volarević, N. N. Arsenijević, S. B. Novaković, G. A. Bogdanović and S. R. Trifunović, Polyhedron, 2015, 90, 34–40 CrossRef.
- A. Savić, T. Marzo, F. Scaletti, L. Massai, G. Bartoli, R. Hoogenboom, L. Messori, R. Van Deun and K. Van Hecke, BioMetals, 2019, 32, 33–47 CrossRef PubMed.
- M. N. Alam and F. Huq, Coord. Chem. Rev., 2016, 316, 36–67 CrossRef CAS.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, V. G. Zakrzewski, J. A. Montgomery, J. C. Burant, R. E. Stratmann, S. Dapprich, J. M. Millam, A. D. Daniels, K. N. Kudin, M. C. Strain, O. Farkas, J. Tomasi, V. Barone, M. Cossi, R. Cammi, B. Mennucci, C. Pomelli, C. Adamo, S. Clifford, J. Ochterski, G. A. Petersson, P. Y. Ayala, Q. Cui, K. Morokuma, D. K. Malick, A. D. Rabuck, K. Raghavachari, J. B. Foresman, J. Cioslowski, J. V. Ortiz, A. G. Baboul, B. B. Stefanov, A. L. G. Liu, I. K. P. Piskorz, R. Gomperts, R. L. Martin, D. J. Fox, T. Keith, M. A. Al-Laham, C. Y. Peng, A. Nanayakkara, C. Gonzalez, M. Challacombe, P. M. W. Gill, B. G. Johnson, W. Chen, M. W. Wong, J. L. Andres, M. Head-Gordon, E. S. Replogle and J. A. Pople, Gaussian 03 (Revision A.9), Gaussian, Inc., Pittsburgh, 2003 Search PubMed.
- A. Frisch, A. B. Nielson and A. J. Holder, Gaussian, Inc., Pittsburgh, PA, 2000.
- M. B. Hansen, S. E. Nielsen and K. Berg, J. Immunol. Methods, 1989, 119, 203–210 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5ra00647c |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2025 |