Open Access Article
Open Access ArticleChemical tools for study and modulation of biomolecular phase transitions
Raymond F.
Berkeley
 and
Galia T.
Debelouchina
and
Galia T.
Debelouchina
 *
*
Department of Chemistry and Biochemistry, University of California San Diego, La Jolla, CA, USA. E-mail: gdebelouchina@ucsd.edu
First published on 21st November 2022
Abstract
Biomolecular phase transitions play an important role in organizing cellular processes in space and time. Methods and tools for studying these transitions, and the intrinsically disordered proteins (IDPs) that often drive them, are typically less developed than tools for studying their folded protein counterparts. In this perspective, we assess the current landscape of chemical tools for studying IDPs, with a specific focus on protein liquid–liquid phase separation (LLPS). We highlight methodologies that enable imaging and spectroscopic studies of these systems, including site-specific labeling with small molecules and the diverse range of capabilities offered by inteins and protein semisynthesis. We discuss strategies for introducing post-translational modifications that are central to IDP and LLPS function and regulation. We also investigate the nascent field of noncovalent small-molecule modulators of LLPS. We hope that this review of the state-of-the-art in chemical tools for interrogating IDPs and LLPS, along with an associated perspective on areas of unmet need, can serve as a valuable and timely resource for these rapidly expanding fields of study.
Introduction
Intrinsically disordered proteins and intrinsically disordered protein regions (henceforth collectively referred to as IDPs) are common in the proteome.1,2 All IDPs share a relatively flat folding energy landscape, and often completely lack a detectable folded low-energy state.3 Unlike proteins constructed from random amino acid sequences, which are also often intrinsically disordered, proteomic IDPs typically exhibit low sequence complexity with primary sequences enriched in a small number of amino acids.4–6 Despite their unfolded or conditionally folded nature, IDPs can act as important regulators of cellular functions, often through low-affinity interactions that are driven by the bulk chemical properties of residues in the IDP.2 Many IDPs can also undergo a process called liquid–liquid phase separation (LLPS) that allows them to concentrate into dynamic and spatially resolved condensates.7 This capability allows IDPs to rapidly and selectively recruit other biomolecules and entrenches their role as regulators of intracellular organization, with many common IDPs such as p53, FUS, and α-synuclein exhibiting pleiotropic regulatory behaviors that are essential for cell viability.8,9IDPs present unique challenges to structural biologists – their conformational plasticity and the range of dynamics that they can exhibit are difficult to characterize with tools that have been optimized for folded proteins. While early structural studies of IDPs often focused on proteins that fold upon binding, this behavior is not universal. For example, some IDPs engage in high affinity interactions while retaining their disorder, while others engage in nonspecific low affinity interactions that drive biomolecular LLPS and are dependent on their intrinsically disordered nature.10–12 In addition to the conceptual challenges brought on by the unstructured nature of these proteins, the sensitivity of IDPs to environmental conditions requires special consideration when working with them in vitro. Characterizing the structure–function relationship of IDPs, therefore, necessitates a shift in the way that we think about protein structure and a coincident shift in the methods and tools that we use to interrogate it, especially in the context of LLPS.13,14
In this perspective, we discuss how chemical biology can aid structural studies of IDPs, with an emphasis on chemical tools that are compatible with LLPS. This will include strategies for the recombinant preparation of IDPs, tools that enable the efficient and site-selective introduction of chemical probes and isotopic labels, and chemical modulators of IDPs and LLPS. We take an application-centric approach to highlight real use cases that are enabled by the current state-of-the-art in IDP chemical biology. We hope to provide insight into best practices for handling and studying these systems, along with a call for the development of minimally perturbative small-molecule chemical tools to aid their analysis and functional manipulation.
IDPs present distinct challenges for biophysical investigations in vitro
When working with IDPs in vitro, their intrinsic disorder brings considerations that are not present for folded proteins. IDPs are often less soluble than their globular counterparts near their isoelectric points due to the lack of a distinct fold, and they are often prone to LLPS or aggregation under these conditions due to nonspecific interactions between hydrophobic residues.15 On the other hand, the lack of a need to maintain a fold can facilitate IDP survival at extreme pH levels and temperatures that would lead to denaturation and aggregation of folded proteins, which can enable unique purification and manipulation strategies. The conditions under which IDPs are handled must therefore be chosen with these considerations in mind. In this section, we will discuss challenges associated with working with IDPs in vitro, along with approaches for purification that are distinct from those of typically folded proteins. We will highlight examples of IDP preparations that strategically employ pH, temperature, and other abiotic factors in ways that would not be applicable for folded proteins. This section will also provide some context for the challenges that need to be overcome when developing chemical tools to aid the structural and functional analysis of IDPs. For a more comprehensive overview of strategies and recommendations for purifying IDPs, the interested reader is referred to two excellent resources by Graether and Alberti et al.16,17Whether expressed in E. coli or other recombinant expression systems, one of the challenges in handling IDPs is avoiding undesirable aggregation or LLPS during the purification process. Fusing solubility tags such as MBP or GST to an IDP of interest can be a useful strategy for maintaining its solubility.18,19 As these solubility tags are large and may interfere with downstream functional and biophysical assays, they are typically removed at the end of the purification process through a suitably engineered cleavable site. This requires an additional purification step to remove the cleaved tag. Cleavage of a solubility tag after purification can be a facile approach for performing LLPS studies.20 In a compelling recent example of this approach, Morin et al. use an MBP-Klf4 fusion protein to construct a model describing the role of prewetting in the sequence-specific surface condensation of the transcription factor Klf4, which forms small LLPS condensates on DNA. In this work, the MBP-Klf4 fusion is capable of adsorbing onto DNA, but does not undergo LLPS. After adsorption, the MBP tag is removed and Klf4 condenses into droplets around sequences known to promote Klf4 binding.21 This strategy allows for the disambiguation of Klf4 adsorption and LLPS. In this case, a solubility tag is used to enable an LLPS study in an environment that would be sensitive to other means of LLPS initiation, such as a pH jump or changing the IDP or salt concentration.
Since IDPs are often enriched in hydrophilic and charged residues, tuning the pH can be a straightforward way to improve the performance of a purification protocol. Although there are reports that leverage pH in order to purposefully precipitate the IDP of interest,22 in most cases the selection of a buffer that optimizes solubility is preferred. Buffers that maintain unusually high or low pH conditions are often advantageous, and the disordered nature of the IDP means that pH-induced denaturation is not an issue. As an example, some purification approaches for the low-complexity domain of Fused in Sarcoma (FUS LC), which undergoes LLPS and/or aggregation at neutral pH, involve the extensive use of N-cyclohexyl-3-aminopropanesulfonic acid (CAPS), a buffer that maintains a pH of 10–11. At such high pH, the tyrosine residues on FUS are deprotonated and the protein is highly charged, which promotes solubility and enables the subsequent purification of the LC domain by size-exclusion chromatography without the need for a denaturant.18,23–25 On the other hand, low-pH conditions are often used to purify α-synuclein.26 This first step in the purification process precipitates many undesired cellular proteins, thereby leveraging α-synuclein's resistance to pH-dependent denaturation. A potential disadvantage of pH-based purification protocols is that they may not be compatible with chemical biology approaches for protein labeling or modification, as the reactions are typically sensitive to pH.
Temperature is another practical consideration that may require a different approach than that taken for a typical globular protein. Since there is no fold to maintain, some IDPs are resistant to high temperatures, a unique property that can be exploited with high-temperature protein purification approaches. Tau and α-synuclein, for example, are often purified by boiling crude cell lysate, which denatures and precipitates most cellular proteins and leaves a soluble fraction that is highly enriched in the desired protein.26 On the other hand, many IDPs undergo thermoresponsive phase transitions which may require the use of mild temperatures around 25 °C throughout the protein preparation,7 a characteristic that is counter-intuitive to those who are used to maintaining temperatures closer to 4 °C throughout the purification of a globular protein. Tolerance to higher temperatures may be beneficial for chemical labeling approaches of IDPs as it can speed up the relevant reactions.
For IDPs that are especially aggregation-prone, chemical denaturants are often necessary to achieve reasonable yields from a recombinant protein preparation. In many IDP preparations, chaotropes such as urea or guanidinium hydrochloride can be used to redissolve aggregated protein or to maintain solubility at a pH or temperature that would otherwise induce aggregation or LLPS.27 Chemical denaturants may also be required to keep IDPs soluble during size-exclusion or ion-exchange chromatography purification steps. In some cases, the IDP will be sequestered into inclusion bodies during expression, and urea or guanidinium hydrochloride may be required for extracting proteins from the inclusion body and for mitigating the risk of aggregation in subsequent steps.28 Chemical denaturants are often compatible with cysteine chemistry and even intein-based segmental labeling approaches, and can therefore be useful in the preparation of modified and labeled IDPs.29
Introduction of chemical probes for imaging and spectroscopic studies
Understanding the structure, dynamics, interactions, and functions of IDPs often relies on fluorescence-based approaches such as fluorescence recovery after photobleaching (FRAP) and fluorescence resonance energy transfer (FRET), and/or spectroscopic studies by nuclear magnetic resonance (NMR) and electron paramagnetic resonance (EPR).30–32 A common requirement for these techniques is the site-specific installation of chemical probes that report on the properties of the IDP or its environment. A wide range of technologies for site-specific labeling have been demonstrated, and many of these approaches are applicable to IDPs.33–35 Here we review the applications of cysteine chemistry, unnatural amino acid incorporation through genetic means (amber suppression), inteins, and sortase, with a particular emphasis on the unique challenges presented by IDPs and LLPS. We focus primarily on the installation of small chemical probes such as fluorescent labels, rather than the use of large fusion fluorescent proteins, as these approaches may be less familiar to the reader and may have distinct advantages in certain applications.Cysteine is the chemical handle of choice for facile labeling of IDPs
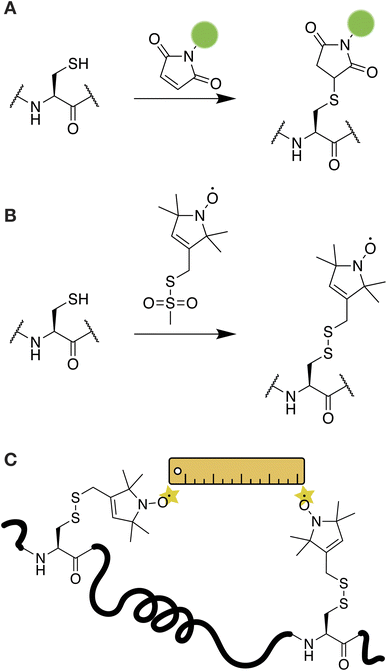
A wide range of small-molecule chemical tools are available for the site-selective modification of proteins under aqueous conditions.36,37 Of these, the most robust and practical reactions take place at nucleophilic cysteine residues. Cysteines are particularly rare in IDPs and are therefore valuable reactive handles that can be targeted selectively if present or added into the recombinantly-produced proteins if needed. The potent nucleophilicity of the cysteine thiol allows for chemoselectivity even in the presence of other nucleophiles such as the primary amines on lysines and protein N-termini.Covalent labeling at cysteine residues is a popular strategy for introducing small-molecule fluorescent probes in proteins including IDPs (Fig. 1A). This is most often achieved through maleimide or iodoacetamide functionalized dyes and there are many commercially available options covering a wide range of absorbance and emission properties, including cyanine-based probes, the Alexa Fluor® series, and the BODIPY family of dyes.38 Labelling at a cysteine residue with a maleimide-based probe is often as simple as incubating the dye with the protein for less than an hour and subsequently removing the unreacted dye via gel filtration or reverse-phase chromatography.39–41 In the case of IDPs, it may be beneficial to perform the labeling step while the protein is still fused to the solubility tag or under denaturing conditions to avoid issues with aggregation or premature LLPS. The straightforward and robust nature of cysteine chemistry make this the preferred method of choice for labeling IDPs, especially if the purification protocol is challenging and delivers relatively low yields. Sometimes, it may also be possible to use the primary amines of lysine residues to attach fluorescent probes through N-hydroxysuccinimide ester chemistry.42 However, there are typically many more lysine residues in a protein compared to cysteine and it is difficult to control the specificity of these reactions, especially in the context of IDPs where all side-chains are solvent exposed and accessible. The unique reactivity of the α-amine of a protein's N-terminus can also be exploited to attach imaging or spectroscopic probes through chemical or enzymatic means. Interested readers are referred to the comprehensive review of these methods by Rosen & Francis.43
In addition to fluorescent labeling, cysteine is a useful handle for introducing spin labels for NMR- and EPR-based methods. A common NMR approach for IDP studies is the paramagnetic relaxation enhancement (PRE) experiment.44 PREs are particularly powerful in detecting weak intra- and inter-molecular interactions at residue-specific resolution in dynamic biological systems and are often applied to characterize the molecular basis of LLPS.18,23,24,45,46 In a PRE experiment, the protein of interest is labeled with a paramagnetic relaxation enhancement probe, typically a nitroxide-based stable radical moiety such as S-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate (MTSL).44 The addition of MTSL results in an oxidation reaction and the formation of a disulfide bond with the targeted cysteine residue (Fig. 1B). The nitroxide moiety on the probe can induce distance dependent relaxation effects that reduce the peak intensity for residues within a 10–25 Å radius.13 This information can be used to construct a map of the residues that participate in intra- or intermolecular interactions for IDPs. For example, PRE-based NMR experiments have been used to describe the transient interactions formed by the low complexity domains of FUS, TDP-43, hnRNPA1, and hnRNPA2 in LLPS environments.23,24,45,47,48
The MTSL probe can also enable the characterization of IDP behavior by EPR. For example, MTSL-labeled tau was used to characterize tau dynamics within liquid–liquid droplets and to report on tau–water interactions.42 In this case, the native cysteine residues of tau were mutated to serine, and a new cysteine was introduced at a different position in the protein sequence for labeling so that the relevant interactions could be captured. A commonly used EPR experiment is double electron–electron resonance (DEER), which measures up to 10 nm distances between two electron spin probes.49 DEER is conceptually similar to FRET experiments and can be used to build a structural model of the protein of interest and to characterize protein–protein interactions (Fig. 1C).50,51 If intramolecular DEER is performed, then two spin labels need to be introduced in the protein. As DEER can probe distances between two identical spin probes, labeling can be easily achieved by introducing two cysteine residues at the relevant positions in the IDP. For example, in a recent study, DEER of a doubly-labeled construct of the FUS LC domain was used to interrogate the dynamics and conformational distributions of the protein within a phase separated sample.52 EPR and DEER can also be performed with Gd3+-based spin probes attached through cysteine chemistry.51,53
Despite their popularity, some important considerations need to be taken into account when working with cysteine-based labeling approaches. When oxidation-based reactions are used for labeling (e.g. with MTSL), the protein cysteines need to be reduced and available before the labeling reaction takes place.54 Once the disulfide bond between the protein and the probe is formed, care must be taken to avoid reducing conditions or agents in the sample buffer. Even the more chemically resilient maleimide labeling reactions can be susceptible to hydrolysis and thiol exchange, processes that can be exacerbated by higher pH and long storage in aqueous solutions.55 Cysteine-based reactions are also not bioorthogonal and are therefore not suitable for in-cell applications. Finally, using cysteine labeling approaches, it is difficult to introduce two different probes on the same IDP as may be required for intramolecular FRET experiments. In this case, cysteine chemistry may be combined with unnatural amino acid incorporation by genetic means and bioorthogonal labeling reactions, as discussed below.
Unnatural amino acids provide flexibility for specific and multiple labeling
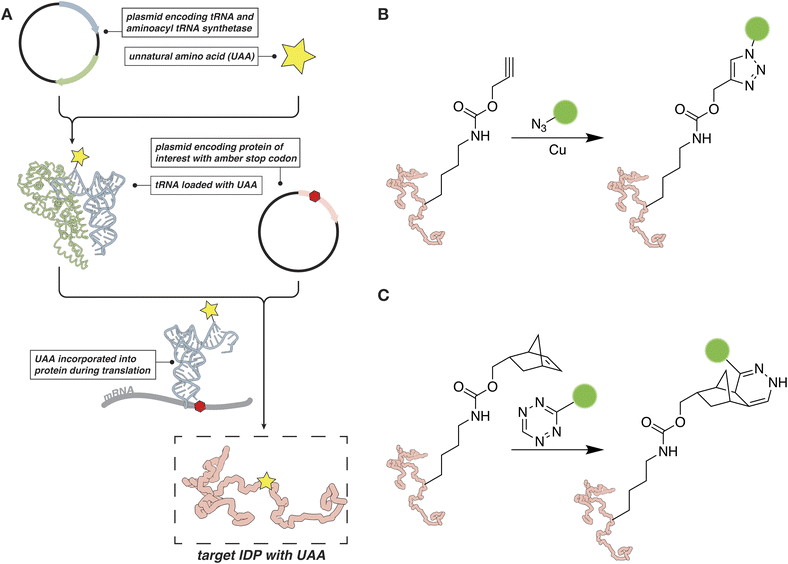
In cases where labeling at a cysteine is not an option, or a second distinct chemical probe needs to be site-specifically introduced to an IDP of interest, the incorporation of an unnatural amino acid (UAA) can help expand the reactive scope of the target protein.56,57 The introduction of an entirely unique chemistry into the IDP with a UAA precludes any cross-reactivity with other nucleophilic residues or issues with multiple labeling that come along with the classical thiol-reactive chemical probes described in the previous section. Unnatural amino acids can be introduced by genetic means through a technique often referred to as amber suppression (Fig. 2A).56,58,59 In this case, the amber stop codon, UAG/TAG, is assigned to the UAA and cloned at the desired position in the protein sequence. At the same time, the cells are transformed or transfected with a second plasmid that encodes an engineered tRNA/tRNA-synthetase pair. The tRNA synthetase can recognize the UAA and load it onto the cognate tRNA. The loaded tRNA, in turn, recognizes the amber stop codon and delivers the UAA to the ribosome for incorporation into the growing protein sequence.Amber suppression is often used to introduce a chemical handle for bioorthogonal chemical reactions, such as an azide, tetrazine, strained alkene or alkyne.60–65 These chemical moieties can then be targeted with a suitable “warhead” carrying the fluorescent or spectroscopic probe of interest. The bioorthogonal reactions can be performed both in vitro on the purified protein and in cells. For example, copper-catalyzed [3 + 2] azide–alkyne cycloaddition (CuAAC) reactions have been used to attach EPR probes to proteins in mammalian cells, while tetrazine-based reagents have been used to tether fluorescent probes (Fig. 2B and C).62,65 If a suitable engineered tRNA/tRNA-synthetase pair has been developed, fluorescent probes, spin labels, or other suitable moieties, can be introduced directly as the UAA.56–58,66 For example, several reports from Schmidt et al., describe the evolution and subsequent application of a tRNAPyl/pyrrolysyl-tRNA-synthetase pair capable of installing a spin-labeled nitroxide-based amino acid.67,68 This allows for the direct introduction of a probe without the need for performing chemistry on the target protein after expression, a capability that could prove especially useful for IDPs that are sensitive to abiotic conditions.
From a conceptual point of view, amber suppression is relatively straightforward to implement as it requires only the addition of two plasmids and the UAA before protein expression. In practice, it can often severely reduce the yield of the desired protein as truncation products are very common. This problem is even more pronounced if two or more UAAs need to be installed.69 Despite these shortcomings, amber suppression has great potential for the biophysical studies of IDPs. For example, it can be used in combination with cysteine chemistry or alone to install two distinct fluorescent and spectroscopic probes on the same protein. More importantly, as these reactions can be performed in cells, bioorthogonal chemistry and amber suppression can provide an alternative to fluorescent proteins for LLPS studies in the cellular milieu. In the context of spectroscopic probes, these strategies can enable the structural characterization of IDPs by EPR or PRE NMR in a native environment.65,70 While there are currently few examples in the literature of successful applications to IDP and LLPS studies,71 we expect that continuing developments to improve efficiency of UAA incorporation and speed of bioorthogonal labeling will make these approaches a reliable and useful option for biophysical analysis both in vitro and in cells.72,73
Inteins and sortase are versatile tools for segmental labeling of IDPs
While IDPs are often studied by NMR, their repetitive sequences and low chemical shift dispersion can make resonance assignments challenging.74 In such cases, it is helpful to segmentally label the protein, so that only a portion of the sequence is visible by NMR while the properties of the full-length polypeptide are preserved.75,76 Segmental labeling is often performed with split inteins, protein engineering tools that can connect two separate protein segments through a native peptide bond in a process called trans-splicing (Fig. 3A).77 To perform segmental labeling, the protein of interest (i.e. the extein) is divided into two fragments called the N- and C-exteins, respectively. Each fragment is fused with the corresponding N- or C-intein and the fusions are expressed and purified separately, so that each fusion construct can be labeled as desired (e.g.15N and natural abundance, or 15N and 13C respectively). After purification, the two constructs are mixed and the N- and C-inteins come together through non-covalent electrostatic interactions and adopt the functional intein horseshoe-like fold.78 Upon folding, the assembled intein performs trans-splicing of the extein fragments and releases the newly ligated native protein of interest. Since most optimized split inteins use cysteine chemistry to carry out the trans-splicing reaction, segmental labeling requires that the extein is split at a native cysteine residue or that a cysteine is introduced at the desired location. Some inteins may also require a few additional residues beyond the junction cysteine, although the most efficient engineered intein so far, Cfa GEP, is quite tolerant to variations in the extein sequence.29,79,80 Some inteins can also carry out splicing reactions at serine or threonine junctions, although their full extein dependency is less known.81,82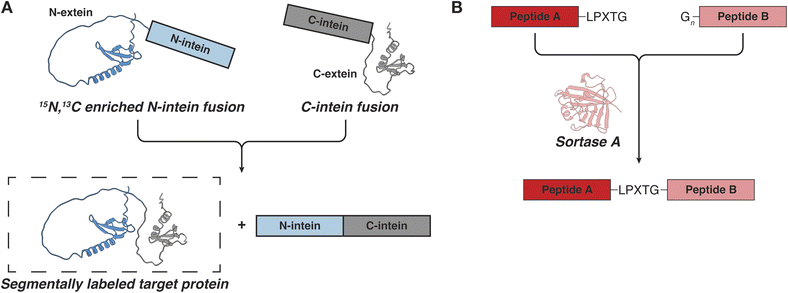 | ||
| Fig. 3 Approaches for segmental labeling of IDPs for NMR studies. (A) Intein mediated protein trans-splicing. (B) Ligation of recombinant or synthetic polypeptides by sortase. | ||
While inteins are gaining traction as segmental labeling tools for a variety of proteins, the biggest challenge for their application to most IDPs is the requirement that the trans-splicing reaction is performed under denaturing conditions to keep the reactants and final product soluble. The presence of urea or guanidinium can interfere with the folding of the intein and severely reduce the efficiency of splicing. The best intein to use in such cases is the Cfa GEP intein, which was engineered to withstand harsh conditions and can carry out splicing in buffers that contain up to 4 M guanidinium hydrochloride and 6 M urea.29,79 There are also inteins that are tolerant to very high salt concentrations and can in principle be used for splicing in such conditions that often prevent phase separation of the protein of interest.83 Extein–intein fusions may also have increased expression levels and higher solubility when compared to the full-length IDP construct, although this may be highly protein dependent. In cases where the solubility and purification of the extein–intein needs to be improved, solubility or affinity tags such as MBP or His6 can be fused on the intein-side of the construct.29,84 At this position, the tags do not interfere with the splicing reaction and are automatically removed from the extein when splicing takes place.
In addition to segmental labeling, inteins have other useful applications. For example, they can be adapted to attach C-terminal small molecule fluorophores to proteins of interest.77,79,85 In this case, an engineered or native contiguous intein is fused to the C-terminal side of the extein. The fused intein can be cleaved in the presence of a thiol which generates a C-terminal thioester on the extein. The thioester can then be reacted with a small peptide bearing the desired fluorophore, thus generating the labeled protein. This strategy may be useful when fluorescent proteins such as GFP are not compatible with LLPS studies (i.e. if they interfere with LLPS) and when cysteine labeling is not an option. In a different adaptation of this strategy, the intein can be hydrolyzed from the extein at slightly basic pH.86–89 If a suitable affinity tag is attached on the intein side, e.g. a His6 or chitin, then the intein can be hydrolyzed directly during affinity purification. In this case, the intein and the tag remain on the column while the pure protein of interest is released in solution. This strategy circumvents the need for a protease cleavage step to remove the affinity tag and may be a useful option when such steps are problematic in the IDP purification protocols.
An alternative tool for segmental labeling and protein modification is the transpeptidase sortase (Fig. 3B). This protein can stitch together two polypeptides in trans, provided that they carry an appropriate signal peptide.90–92 The recognition signal typically consists of the LPXTG sequence (where X is any amino acid) on one peptide, and one to five glycine residues on the other peptide.90 The two peptides are mixed and sortase is added in trans to carry out the ligation reaction, resulting in a minimum six residue “scar” in the newly formed protein. If one of the peptides is prepared in minimal isotopically labeled media, the resulting full-length protein will be segmentally labeled. In the context of IDPs and LLPS, there are several important considerations that need to be kept in mind.93,94 First, to carry out the ligation reaction, sortase needs to be folded, which precludes the use of high concentrations of denaturants. Second, the reaction requires the presence of Ca2+ ions in the buffer, which may be incompatible with LLPS studies for some proteins.92 And third, as the product polypeptide also contains the LPXTG recognition motif, the sortase reaction may be reversible and care must be taken to remove the product quickly and to minimize the generation of undesired products.93 Nevertheless, the sortase-based ligations are conceptually elegant and may provide a useful alternative when the protein of interest is incompatible with intein splicing (e.g. if the protein charge interferes with the binding and folding of the intein fragments which proceeds through electrostatic interactions). Sortase has also been used extensively to attach proteins to surfaces and to add IDRs to the folded regions of proteins.95,96 Interestingly, sortase has an IDR which undergoes a disorder-to-order transition upon binding of the signal peptide and Ca2+ ions.97
Introduction of post-translational modifications
Innate biological LLPS is often controlled by post-translational modifications (PTMs) including phosphorylation, acetylation, methylation, and ubiquitination.98,99 Faithfully recapitulating PTMs in vitro is therefore crucial for constructing experiments that interrogate LLPS. While there are a variety of chemical and biochemical tools for installing or modelling PTMs after a protein has been recombinantly purified, IDPs present unique challenges for the applications of these tools.98 Here, we review enzymatic approaches, bioisostere substitutions, cysteine chemistry and native chemical ligation, with the goal to give a range of options to biophysicists and biochemists interested in understanding the role of PTMs in IDPs and LLPS mechanisms and interactions.The most common strategy to introduce PTMs in recombinantly produced IDPs is enzymatic modification. This approach is flexible as it can generally be applied to any substrate protein if the appropriate enzyme can be purchased or expressed and purified in house. Enzymes also introduce the same chemistries that are seen in cells, avoiding issues that may arise with PTM isosteres or PTM mimetics that result from other methods for PTM installation. The chemical accuracy and ease of use of this approach has made it the method of choice for many studies involving a variety of combinations of IDPs and PTMs, including phosphorylation of tau,100 acetylation of FUS,101 and mono-, di-, and trimethylation of histone tails102 (among many others). The major drawback with enzymatic PTMs is that the activity of the enzyme can be difficult to control, and the resulting protein can be either overmodified (with the PTM present at multiple sites on the protein) or undermodified where the desired PTM is not installed efficiently. Controlling the installation site is also an issue, especially in unfolded IDPs whose residues are entirely solvent exposed. The proteins that result from mixtures of sites and distributions of PTMs that are generated by enzymes are often hard to separate from each other. These factors limit the enzymatic approach to cases where an appropriately active enzyme is available and there is either only one specific substrate residue in the protein or a distribution of modifications is desired, similar to the cases illustrated above.
A second straightforward strategy to incorporate PTMs is to genetically encode a bioisostere into the protein of interest. Bioisosteres, commonly used in medicinal chemistry and chemical biology, are atoms or functional groups with similar chemical and physical properties.103 In the context of PTMs, perhaps the best known example is the substitution of phosphorylated serine or threonine residues with glutamic acid. Similar to enzymatic PTM incorporation, ease of application is a major benefit with this approach: bioisosteric residues can be encoded in the protein with cloning, and no further modification is required after protein purification. The drawback is that few good bioisosteres for PTMs exist among the canonical amino acids. Despite this limitation, this strategy has been commonly used in the literature, especially in the context of phosphorylation.104 This includes a number of studies by the Fawzi group that reveal the influence of site-specific phosphorylation on the structural distributions and LLPS propensities of both FUS and TDP-43.23,46 As more tRNA/tRNA synthetase pairs that encode pre-modified amino acids are developed, we expect that the incorporation of chemically accurate PTMs via genetic encoding by amber suppression will become a valuable strategy for incorporating PTMs into IDPs. It is already possible to encode phosphotyrosine and acetyllysine, for example, and new tRNA/tRNA synthetases are actively being developed.105,106
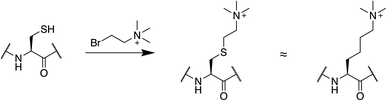
Cysteine chemistry presents a convenient way to introduce PTM mimics that are inaccessible with genetically encoded bioisosteres. Mimics of lysine methylation are common targets for this approach (Fig. 4). To use this strategy, a lysine residue is substituted with a cysteine, which is then alkylated with appropriate reagents to generate a mono-, di-, or trimethyllysine as desired.107 This produces a sidechain that is the same length as lysine but bearing a sulfur instead of a carbon atom at the γ position. The reaction is compatible with denaturing conditions and is very popular in chromatin studies as it presents a relatively straightforward way to generate methylated histones.107–110 It is important to note that in binding studies, alkylated cysteines display slightly higher Kd values compared to native methylated lysine residues.109 They are, however, a good option when large amounts of methylated protein are needed, and are especially useful in the context of isotopic labeling for NMR studies.108 Similarly, cysteine-based chemistries have been leveraged to introduce acetyl-lysine and methyl-arginine mimics into histone tails after cysteine mutations at endogenous lysine or arginine residues.111–113 Asymmetric disulfide linkages can also be used to attach ubiquitin at well-defined positions in a protein sequence.114,115
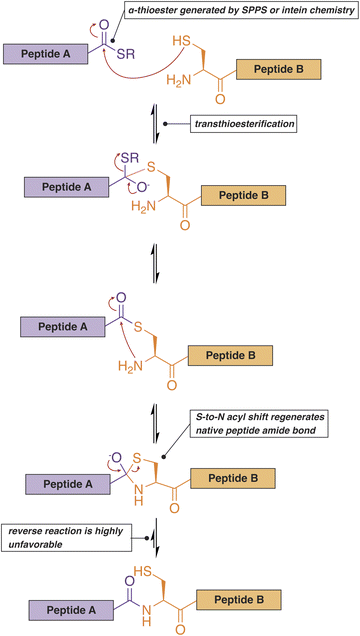
Native chemical ligation (NCL) represents another versatile option for the site-selective introduction of PTMs in IDPs (Fig. 5).116,117 Unlike most of the methods discussed above, this technique can generate proteins with multiple well-defined PTMs, enabling the construction of highly accurate models of post-translationally modified systems in vitro. In this case, a synthetic peptide bearing the necessary modifications is prepared through Fmoc- or Boc-mediated solid-phase peptide synthesis. The peptide is then ligated to another synthetic or recombinant fragment to build the full-length protein of interest. To perform NCL, the N-terminal fragment must contain a C-terminal α-thioester, while the C-terminal fragment needs to start with a nucleophilic amino acid such as cysteine. The thioester can be generated synthetically (native chemical ligation) or via the use of an intein that is fused to a recombinant protein or peptide fragment (expressed protein ligation, EPL).116,118 The α-thioester serves as a reactive handle for the formation of a native amide protein backbone via a trans-thioesterification reaction with the cysteine sidechain of the second peptide fragment. Since the development of NCL, however, the range of chemistries that can be formed at this ligation site has extended far beyond cysteine to include many other natural and unnatural amino acids,119 with recent methodologies expanding the scope to extremely challenging residues such as proline.120 Another notable improvement in the NCL methodology is the development of the C-terminal hydrazide as a more stable and flexible replacement for the C-terminal α-thioester.121 It should also be noted that the ligation reaction can proceed in the presence of urea or guanidinium hydrochloride and is therefore compatible with the production of IDPs that are prone to aggregation or premature LLPS.122
The primary advantage of using NCL or EPL to introduce PTMs into IDPs is that these techniques offer the capability to site-specifically introduce multiple distinct PTMs if desired. An excellent illustration of the value that this capability can offer to IDP research is provided by recent work by Ge et al., in which a semisynthetic construct of the partially disordered JARID2 protein was produced using NCL.123 In this report, multiple NCL reactions were utilized to generate a protein construct containing two distinct post translational modifications on individual residues in a site-specific manner. NCL and EPL have also been used extensively to produce α-synuclein and tau bearing a wide variety of PTMs, sometimes in combination with segmental isotopic labeling for NMR studies.124–128
Noncovalent small-molecule chemical tools for modulating LLPS and IDP behavior
One of the most exciting frontiers for IDP chemical biology is the development of noncovalent modulators of IDPs and LLPS. Disordered proteins do not offer hydrophobic pockets or other defined structural features that can serve as targets for small molecules. IDPs are therefore often avoided in high-throughput screens for small-molecule effectors due to the high potential for failure. Despite this, successful screens for high-value protein targets such as Aβ, Myc, and α-synuclein are present in the literature,129–134 suggesting that screening is a viable strategy for the discovery of chemical tools and drug leads that target IDPs directly.134 Rational structure-based design has also been challenging although there is now at least one example of a successful structure–activity relationship (SAR)-based campaign against IDPs in the literature.135 Given the central roles of IDPs and LLPS in cell biology and disease, the dearth of small-molecules capable of selectively engaging IDPs for use as chemical probes or therapeutic leads is an important problem in the field and makes this area of research especially exciting.136 In this section, we will outline the precedent and explore the prospects for the discovery and development of such small-molecule modulators of IDPs and LLPS.The current chemical landscape of LLPS modulators is sparse and lacks specificity
The chemical tool most often used to study LLPS is 1,6-hexanediol, a general disruptor of this process. Akyl alcohols including 1,6-hexanediol were first used to probe interactions between the FG-nucleoporins that gate the nuclear pore complex, which are IDPs that exhibit behavior consistent with LLPS.137–139 Since its demonstration as a modulator of FG-nucleoporins, 1,6-hexanediol has been used as a convenient probe for assaying the properties of droplet-like structures in cells, for the rapid characterization of LLPS in vitro, or for differentiating between LLPS droplets and solids.140–143 Despite its widespread use, 1,6-hexanediol is typically added at high concentrations of 5–10% w/v. Such high concentrations can interfere with the analysis of in vitro LLPS systems that are sensitive to abiotic conditions as well as with studies done in cells where 1,6-hexanediol disrupts a broad spectrum of cellular processes and can be cytotoxic.140,144,145 Additionally, 1,6-hexanediol works primarily by disrupting hydrophobic interactions, a mechanism that does not address electrostatic, dipolar, and cation-π interactions that are also known to drive LLPS.5 This suggests that the efficacy of 1,6-hexanediol in disrupting LLPS droplets may vary.5,146,147 Conclusions drawn from experiments that rely on 1,6-hexanediol must therefore be considered with care and with the limitations of this tool in mind.In addition to 1,6-hexanediol, a number of alkyl alcohols have been used to characterize LLPS. As a general trend, less hydrophobic alkyl alcohols (2,5-hexanediol and 1,2,3-hexanetriol, for example) are less effective at disrupting LLPS droplets, which is consistent with the proposed mechanism of action.141 This difference in efficacy can be used as a tool for differentiating between phase-separated structures with different susceptibilities to disruption by alkyl alcohols.143
On the other end of the spectrum from 1,6-hexanediol and related disruptors lie small-molecule promoters of LLPS. Promoters include physiologically-relevant small molecule hydrotopes such as ATP, which can enhance the propensity of FUS to undergo LLPS in a concentration-dependent manner.148,149 A synthetic example of a hydrotope capable of enhancing LLPS is 4,4′-dianilino-1,1′-binaphthyl-5,5′-disulfonic acid (bis-ANS). This molecule, along with a handful of similar but less effective naphthalene sulfonate derivatives, has been shown to promote LLPS for a number of common LLPS-prone model proteins such as TDP-43 low complexity domain, tau, and FUS LC.150 The study that introduces bis-ANS further demonstrates that Congo Red, a small molecule probe that is used as a reporter for amyloid, is capable of promoting LLPS in a similar manner to bis-ANS. This work suggests that bivalent, negatively charged compounds with hydrophobic cores can serve as hydrotopic drivers for LLPS. Some biologically important small molecules have also been shown to be capable of maintaining LLPS and modulating the onset of the liquid-to-solid transition of LLPS droplets. In one of the few systematic studies of its kind, Jonchhe et al. found that hydrophobic moieties in small molecules delay the onset of a liquid-to-solid phase transition in tau LLPS droplets.151 In the same work, TMAO is shown to be an especially potent inhibitor of the liquid-to-solid transition, which is consistent with the observation that amphiphilic compounds like bis-ANS and ATP can drive LLPS.
Although they do not necessarily modulate LLPS directly, LLPS-sensitive probes that fluoresce or otherwise report on LLPS, are valuable tools for characterizing this process and may prove superior to disruptive reporters such as 1,6-hexanediol in many experimental contexts. Molecular rotors such as thioflavin T are sensitive to viscosity and can be used to probe LLPS while also serving as a tool for identifying and visualizing protein aggregation and fibrilization.152 Recently, a novel aggregation-induced emission fluorogen, sodium 1,2-bis[4-(3-sulfonatopropoxyl)phenyl]-1,2-diphenylethene (BSPOTPE), was demonstrated to be capable of reporting on LLPS by partitioning into droplets in vitro and fluorescing in a viscosity-dependent manner.153 The continued development of chemical tools capable of reporting on LLPS without the need for toxic or otherwise disruptive concentrations of 1,6-hexanediol will allow for less perturbative in vitro and in-cell assays. Molecules designed to non-perturbatively probe LLPS characteristics may also be useful for providing SAR data for chemistries capable of selectively partitioning into LLPS droplets. Just like biological LLPS condensates, whose composition is tuned by the chemistry of the client and scaffold proteins, it is feasible that small molecules could be tuned to partition into LLPS droplets with a particular set of chemical properties.5,154 For example, recently, a handful of cancer therapeutics were shown to selectively partition into biomolecular LLPS condensates both in vitro and in cells.155 It is our hope that further development of selective small-molecule LLPS modulators will lead to better chemical tools for studying LLPS and better therapeutics for addressing clinically-relevant LLPS dysregulation.
Rethinking the physicochemical basis of binding to IDPs
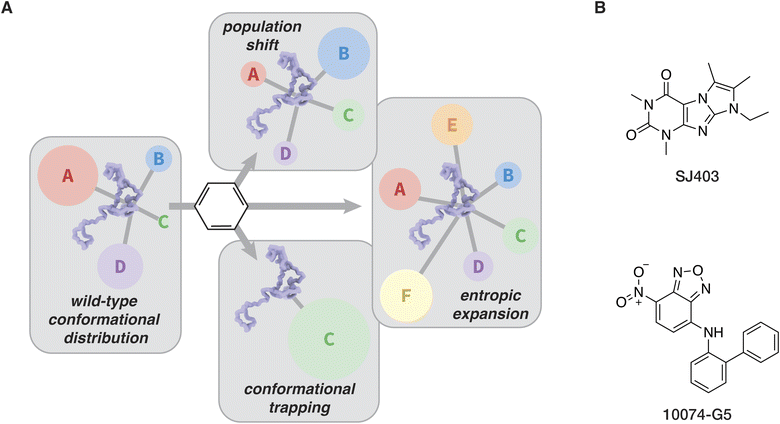
In order to develop more effective small molecule modulators of IDPs and LLPS, it is important to first consider the properties that would make a modulator effective in the first place. In general, three outcomes are available for small molecule binding to an IDP (Fig. 6A). First, the conformational distribution of an IDP may be changed or the conformational space available to an IDP may be reduced, a mechanism referred to as a population shift or conformational restriction.156,157 Alternatively, the conformational states available to an IDP may increase, a mechanism referred to as entropic expansion.158,159 Lastly, a small-molecule IDP modulator may bind to and induce a single conformation of an IDP, which we refer to as conformational trapping.The population shift mechanism relies on transient enthalpy-driven binding of a small molecule to an IDP. Many IDPs adopt transient secondary structure, either due to a predominant low-energy conformation or as the result of a transient interaction with another protein (in the case of LLPS, this interaction is often with another copy of the same protein). As an example, adoption of transient secondary structure by an IDP has been demonstrated by Conicella et al., who showed that transient α-helices in the C-terminal domain of TDP-43 exist both in the dispersed and LLPS phase, and that mutations that enhance the propensity for the formation of α-helices also enhance the propensity for LLPS.47,160 Molecular dynamics and NMR are particularly powerful for providing insight into the role of transient secondary structure in the behavior of IDPs and IDP interactors. In a recent study, Zhu et al. integrated solution NMR data with all-atom molecular dynamics to provide a structure-based explanation for SAR differences between a family of bisphenol A-based modulators of the intrinsically disordered transactivation domain of the androgen receptor.161 Rational design of IDPs that target transient structure has been achieved by clustering snapshots from molecular dynamics simulations and using favored structural ensembles as targets for docking experiments.162
The discovery and optimization of a molecule that confers a population shift in an IDP can also be performed in the absence of a full structural ensemble, as illustrated in work described by the Shelat, Zuo, and Kriwacki groups that shows the discovery and optimization of a small molecule capable of engaging the IDP p27Kip1. An initial fragment-based NMR screen against p27Kip1 yielded SJ403 (Fig. 6B), which can bind transient clusters of hydrophobic residues.163 SJ403 is shown to induce a population shift through this binding mode, and further SAR work on this molecule generated a compound capable of sequestering p27Kip1 into small soluble oligomers via the hydrophobic binding mechanism.135,156 This effort provides an elegant example of the implementation of well-established drug discovery and optimization approaches to discover a small-molecule modulator of an IDP.
The entropic expansion mechanism introduces a much stronger focus on the entropic contributions to interactions between a small-molecule modulator and IDPs.158 In the conformational trapping mechanism, and to a lesser degree the population shift mechanism, binding can be optimized through rational design principles against a favored IDP conformation. The strategy mirrors that of lead optimization for a folded protein, where the main goal is to introduce changes to a small molecule binder in order to decrease the enthalpy of binding.164 In the entropic expansion approach, on the other hand, the entropic benefit of introducing more diversity into the conformational ensemble of a population of IDPs is considered. Enthalpic contributions provide a degree of target specificity through transient, weak interactions with structural or sequence motifs in IDPs, while the entropic contribution favors interactions with the IDPs and effectively reduces unwanted LLPS or aggregation by expanding the range of conformations that the proteins can adopt.158 The only example to date of a molecule that works through the entropic expansion mechanism is 10074-G5 (Fig. 6B), a small molecule inhibitor of amyloid-β42 aggregation discovered by the Knowles, Dobson, and Vendruscolo groups.165 Since many IDPs undergo pathological conformational collapse into aggregates and fibrils, it is clear that molecules that exploit the entropic expansion mechanism could be viable therapeutic leads. It is possible that other small molecules that protect against aggregation of IDPs while preserving their wild-type function work through this mechanism.166,167 Further efforts exploring the entropic expansion mechanism will be valuable for the development of better chemical tools for studies of IDPs and LLPS.
For both the population shift and entropic expansion mechanisms, a framework for initial small-molecule binding and selectivity must exist. For enthalpically driven binding, selectivity can be explained in some cases by the existence of transient binding pockets. Recently, Robustelli et al. provided an alternative mechanism for selectivity in small molecule binders of IDPs. Using all-atom molecular dynamics simulations and solution NMR studies of a fragment of α-synuclein and fasudil, a known ligand for α-synuclein, Robustelli and coworkers demonstrated that the spacing of low-affinity binding sites for fasudil along the α-synuclein sequence can compensate for the lack of a single well-conserved or high-affinity binding site. This mechanism for selectivity, called “dynamic shuttling” provides a compelling explanation for small-molecule selectivity towards specific IDP sequences that is compatible with both the population shift and entropic expansion paradigms for small-molecule-IDP interactions.168
A final mechanism for IDP interactions with small molecules is conformational trapping. Due to the large entropic cost associated with restricting an IDP to a single conformation, this mode of interaction is challenging to achieve with a small molecule. An example of conformational trapping can be found in the biotinylated 5-aryl-isoxazole-3-carboxyamide molecule (b-isox) described by Kato et al. In this case, the microcrystalline form of b-isox is the active modality. Grooves on the crystal surface provide binding sites capable of overcoming the entropic penalty associated with conformational trapping of IDPs, and multiple IDPs associated with RNA granules can be trapped and subsequently isolated using b-isox as a chemical tool.169
Future directions
A quarter of a century of IDP research has revealed that unstructured polypeptides carry out a vast number of important and precise functions in the cell.10,170,171 Despite these advances, there is still much to learn regarding how cells translate a multitude of non-specific interactions into specific biological outcomes, and how the cellular machinery perceives functional and pathological states of IDPs. Further developments in both structural biology and protein chemistry will be crucial to these efforts.Here, we have reviewed the current state of the art in chemical and biochemical approaches that allow the preparation of IDPs for imaging and structural studies and enable the investigation of PTMs in physiologically relevant contexts (summarized in Table 1). While these tools have become much more efficient and versatile over time, the handling requirements and aggregation propensity of many IDPs still present a tremendous challenge. The development of the Cfa GEP intein, for example, enabled trans-splicing and segmental labeling of many new proteins, including IDPs such as FUS.29,79,172 Yet, more can be done to improve the efficiency of inteins in high concentrations of guanidinium hydrochloride and urea, conditions that are essential for the handling of many aggregation and LLPS prone IDPs. The development of cysteine alkylation approaches to mimic lysine methylation, on the other hand, has allowed NMR spectroscopists to study the role of this modification using specifically methylated, biologically relevant samples.107,108,110 The design of similarly robust and easy to implement methodologies to create acetylation and phosphorylation mimics or modifications on recombinantly produced templates, would be highly beneficial.
| Goal | Strategy | Advantages | Disadvantages | References |
|---|---|---|---|---|
| Purification of IDPs | Purification under denaturing conditions | • In principle, compatible with all IDP/IDRs | • Denaturant must be removed | 27, 28 and 172 |
| • Refolding required for constructs that contain both folded and IDR domains | ||||
| Purification at extreme pH | • Straightforward recovery of target protein | • Applicable to only a small subset of proteins | 18 and 22–26 | |
| Purification at extreme temperature | • Straightforward recovery of target protein | • Applicable to only a small subset of proteins | 26 | |
| Labeling with spectroscopic or imaging probes | Maleimide or iodoacetamide-based probes | • Site-specific at cysteine residues | • Not applicable in cells | 36–41 |
| • Compatible with isotopic labeling for NMR | • Potential labeling at multiple sites | |||
| • Probes may perturb IDP function and LLPS | ||||
| N-Hydroxysuccininimide ester-based chemistry | • Site-specific at lysine residues | • Not applicable in cells | 42 | |
| • Compatible with isotopic labeling for NMR | • Potential labeling at multiple sites | |||
| • Probes may perturb IDP function and LLPS | ||||
| Fusion protein-based approaches | • Genetically encoded | • Large fusion proteins may perturb IDP function and LLPS | See ref. 41 and 183 for comparison of the effects on LLPS of small molecule labeling vs. fusion protein labeling | |
| • Applicable in cells | ||||
| Bioorthogonal chemical approaches and amber suppression | • Genetically encoded | • UAA incorporation may be inefficient and lead to low protein yields | 56, 61–67, 70, 184 and 185 | |
| • Applicable in cells | • Probes may perturb IDP function and LLPS | |||
| • Site-specific labeling | ||||
| • May be compatible with isotopic labeling for NMR | ||||
| Native chemical ligation and expressed protein ligation | • Site-specific labeling | • Segments must be accessible synthetically | 77, 79, 116 and 118 | |
| • Multiple probes can be introduced at the same time | • Difficult to label sites away from the N- or C-terminus | Also see relevant references for introducing PTMs through NCL and EPL below | ||
| • Access to a wide range of chemical probes | • Final protein yields may be low | |||
| • May be compatible with isotopic labeling for NMR | • Not applicable in cells | |||
| • Probes may perturb IDP function and LLPS | ||||
| Segmental labeling for NMR spectroscopy | Cfa GEP | • Trans-splicing can be achieved under denaturing conditions | • Splicing reaction must take place at a cysteine residue | 29, 79 and 172 |
| • High efficiency | ||||
| • Robust to a range of extein sequences | ||||
| • May be applied in cells | ||||
| MCM2 | • Salt-inducible splicing can be used to control intein activity | • Splicing cannot be performed under denaturing conditions | 83 | |
| • Robust reactivity under high-salt conditions | • May not be applicable in cells | |||
| • Serine-based reaction mechanism provides versatility | • Splicing rate and extein sequence compatibility are poor compared to Cfa | |||
| Sortase | • A possible alternative to intein-based methods | • Requires the insertion of a five/six amino acid scar into the target protein | 90–96 and 186 | |
| • May be used to attach proteins to cell surfaces | • Continued reaction between reactants and desired product may reduce yield | |||
| Introduction of post-translational modifications | Enzymatic methods | • Straightforward introduction of native PTMs | • Lack of control over stoichiometry and location of PTM installation | 100–102 |
| Genetic encoding of a bioisostere | • Straightforward introduction of PTM mimics | • Scope of PTMs is limited to those that can be mimicked effectively by a bioisostere | 23, 46 and 104 | |
| • Applicable in cells | ||||
| Amber suppression | • Introduction of native PTMs or PTM mimics | • Scope of PTMs is limited to those that have a tRNA synthetase available | 105 and 106 | |
| • Applicable in cells | • UAA incorporation may be inefficient and lead to low protein yields | |||
| Cysteine alkylation | • Efficient and specific introduction of methyl-lysine mimics | • Methyl-lysine mimic may not faithfully reproduce the function of the native PTM | 41, 107, 109 and 110 | |
| • Reactions are compatible with denaturing conditions | • Not applicable in cells | |||
| Native chemical ligation and expressed protein ligation | • Site-specific introduction of PTMs | • Segments must be accessible synthetically | 125–128, 187 and 188 | |
| • Multiple PTMs can be introduced at the same time | • Difficult to introduce PTMs away from the N- or C-terminus | |||
| • Access to a wide range of modifications | • Final protein yields may be low | |||
| • Not applicable in cells |
Going forward, we expect that protein engineering efforts on IDPs will shift more and more to the cellular environment. This reflects a recent increase in efforts from imaging, NMR and EPR spectroscopy to understand how cells shape the conformational ensembles, interactions, and dynamics of IDPs and LLPS-based compartments.51,173,174 While most imaging efforts so far have relied on fusion fluorescent proteins such as GFP, the significant bulk of these tags may not be compatible with all IDPs or LLPS studies.41 In these situations, alternative labeling approaches with small molecule fluorescent probes are needed. Here, we expect that unnatural amino acid incorporation through amber suppression and bioorthogonal labeling methodologies would be particularly valuable. In addition to the delivery of fluorescent probes in cells, this technology is also gaining traction for EPR and sensitivity-enhanced NMR spectroscopy in cells.65,70 Recent developments have focused on improving the efficiency of UAA incorporation, and in particular, the installation of two or three UAAs in mammalian cells.66,72,73 It should also be mentioned that the intein approach can also be adapted to studies in cells, where it can be used to control IDP function and for the installation of PTMs.175,176
Perhaps the greatest challenge facing the IDP chemical biology field is the development of specific small molecule modulators that allow regulation of IDP and LLPS functions in cells. These efforts invariably tie back to more detailed understanding of the structural ensemble, dynamics, and interactions of the IDP of interest. These developments will no doubt require close collaborations between synthetic chemists, structural biologists, and computational chemists. A number of NMR studies, often complemented with all atom molecular dynamics simulations, have already provided valuable information regarding the interactions between small molecules and IDPs.135,156,158,163,165,168 There have also been exciting developments in the coarse-grained simulations of LLPS177–180 and we are looking forward to the extension of these studies to all atom simulations and the incorporation of small molecule modulators.181,182
While IDPs present significant challenges to chemical and structural biologists, they also put forward valuable opportunities to hone existing chemical and biophysical methodologies and to develop more efficient and precise tools. Such developments will have far reaching implications for the chemical and biological fields and will undoubtedly enrich our understanding of protein function in health and disease.
Author contributions
R. F. B. and G. T. D. conceived the idea and wrote the manuscript. R. F. B. prepared the figures.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This work was supported by a Research Education Component associated with NIH Grant P30 AG062429, an R21 AG069064 award to G. T. D., and a T32 GM112584 fellowship to R. F. B.References
- R. van der Lee, M. Buljan, B. Lang, R. J. Weatheritt, G. W. Daughdrill, A. K. Dunker, M. Fuxreiter, J. Gough, J. Gsponer, D. T. Jones, P. M. Kim, R. W. Kriwacki, C. J. Oldfield, R. V. Pappu, P. Tompa, V. N. Uversky, P. E. Wright and M. M. Babu, Classification of intrinsically disordered regions and proteins, Chem. Rev., 2014, 114, 6589–6631 CrossRef CAS PubMed.
- P. E. Wright and H. J. Dyson, Intrinsically disordered proteins in cellular signalling and regulation, Nat. Rev. Mol. Cell Biol., 2015, 16, 18–29 CrossRef CAS PubMed.
- J. N. Onuchic, Z. Luthey-Schulten and P. G. Wolynes, Theory of protein folding: the energy landscape perspective, Annu. Rev. Phys. Chem., 1997, 48, 545–600 CrossRef CAS PubMed.
- R. Neme, C. Amador, B. Yildirim, E. McConnell and D. Tautz, Random sequences are an abundant source of bioactive RNAs or peptides, Nat. Ecol. Evol., 2017, 1, 0217 Search PubMed.
- J. Wang, J. M. Choi, A. S. Holehouse, H. O. Lee, X. Zhang, M. Jahnel, S. Maharana, R. Lemaitre, A. Pozniakovsky, D. Drechsel, I. Poser, R. V. Pappu, S. Alberti and A. A. Hyman, A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins, Cell, 2018, 174, 688–699 CrossRef CAS PubMed.
- I. Pritisanac, R. M. Vernon, A. M. Moses and J. D. Forman Kay, Entropy and Information within Intrinsically Disordered Protein Regions, Entropy (Basel), 2019, p. 21 Search PubMed.
- S. F. Banani, H. O. Lee, A. A. Hyman and M. K. Rosen, Biomolecular condensates: organizers of cellular biochemistry, Nat. Rev. Mol. Cell Biol., 2017, 18, 285–298 CrossRef CAS PubMed.
- M. Ashburner, C. A. Ball, J. A. Blake, D. Botstein, H. Butler, J. M. Cherry, A. P. Davis, K. Dolinski, S. S. Dwight, J. T. Eppig, M. A. Harris, D. P. Hill, L. Issel-Tarver, A. Kasarskis, S. Lewis, J. C. Matese, J. E. Richardson, M. Ringwald, G. M. Rubin and G. Sherlock, Gene ontology: tool for the unification of biology. The Gene Ontology Consortium, Nat. Genet., 2000, 25, 25–29 CrossRef CAS PubMed.
- Gene Ontology Consortium, The Gene Ontology resource: enriching a GOld mine, Nucleic Acids Res., 2021, 49, D325–D334 Search PubMed.
- H. J. Dyson and P. E. Wright, Coupling of folding and binding for unstructured proteins, Curr. Opin. Struct. Biol., 2002, 12, 54–60 CrossRef CAS PubMed.
- A. Borgia, M. B. Borgia, K. Bugge, V. M. Kissling, P. O. Heidarsson, C. B. Fernandes, A. Sottini, A. Soranno, K. J. Buholzer, D. Nettels, B. B. Kragelund, R. B. Best and B. Schuler, Extreme disorder in an ultrahigh-affinity protein complex, Nature, 2018, 555, 61–66 CrossRef CAS PubMed.
- M. K. Hazra and Y. Levy, Affinity of disordered protein complexes is modulated by entropy-energy reinforcement, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2120456119 CrossRef CAS PubMed.
- A. C. Murthy and N. L. Fawzi, The (un)structural biology of biomolecular liquid-liquid phase separation using NMR spectroscopy, J. Biol. Chem., 2020, 295, 2375–2384 CrossRef CAS PubMed.
- L. Emmanouilidis, L. Esteban-Hofer, G. Jeschke and F. H. Allain, Structural biology of RNA-binding proteins in the context of phase separation: What NMR and EPR can bring?, Curr. Opin. Struct. Biol., 2021, 70, 132–138 CrossRef CAS PubMed.
- G. Tedeschi, M. Mangiagalli, S. Chmielewska, M. Lotti, A. Natalello and S. Brocca, Aggregation properties of a disordered protein are tunable by pH and depend on its net charge per residue, Biochim. Biophys. Acta, Gen. Subj., 2017, 1861, 2543–2550 CrossRef CAS PubMed.
- S. P. Graether, Troubleshooting Guide to Expressing Intrinsically Disordered Proteins for Use in NMR Experiments, Front. Mol. Biosci., 2018, 5, 118 CrossRef CAS PubMed.
- S. Alberti, S. Saha, J. B. Woodruff, T. M. Franzmann, J. Wang and A. A. Hyman, A User's Guide for Phase Separation Assays with Purified Proteins, J. Mol. Biol., 2018, 430, 4806–4820 CrossRef CAS PubMed.
- K. A. Burke, A. M. Janke, C. L. Rhine and N. L. Fawzi, Residue-by-Residue View of In Vitro FUS Granules that Bind the C-Terminal Domain of RNA Polymerase II, Mol. Cell, 2015, 60, 231–241 CrossRef CAS PubMed.
- M. Vivoli Vega, A. Nigro, S. Luti, C. Capitini, G. Fani, L. Gonnelli, F. Boscaro and F. Chiti, Isolation and characterization of soluble human full-length TDP-43 associated with neurodegeneration, FASEB J., 2019, 33, 10780–10793 CrossRef PubMed.
- J. Van Lindt, A. Bratek-Skicki, P. N. Nguyen, D. Pakravan, L. F. Duran-Armenta, A. Tantos, R. Pancsa, L. Van Den Bosch, D. Maes and P. Tompa, A generic approach to study the kinetics of liquid-liquid phase separation under near-native conditions, Commun. Biol., 2021, 4, 77 CrossRef CAS PubMed.
- J. A. Morin, S. Wittmann, S. Choubey, A. Klosin, S. Golfier, A. A. Hyman, F. Julicher and S. W. Grill, Sequence-dependent surface condensation of a pioneer transcription factor on DNA, Nat. Phys., 2022, 18, 271–276 Search PubMed.
- C. P. Pedersen, P. Seiffert, I. Brakti and K. Bugge, Production of Intrinsically Disordered Proteins for Biophysical Studies: Tips and Tricks, Methods Mol. Biol., 2020, 2141, 195–209 CrossRef CAS PubMed.
- Z. Monahan, V. H. Ryan, A. M. Janke, K. A. Burke, S. N. Rhoads, G. H. Zerze, R. O'Meally, G. L. Dignon, A. E. Conicella, W. Zheng, R. B. Best, R. N. Cole, J. Mittal, F. Shewmaker and N. L. Fawzi, Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity, EMBO J., 2017, 36, 2951–2967 CrossRef CAS PubMed.
- A. C. Murthy, G. L. Dignon, Y. Kan, G. H. Zerze, S. H. Parekh, J. Mittal and N. L. Fawzi, Molecular interactions underlying liquid-liquid phase separation of the FUS low-complexity domain, Nat. Struct. Mol. Biol., 2019, 26, 637–648 CrossRef CAS PubMed.
- R. F. Berkeley, M. Kashefi and G. T. Debelouchina, Real-time observation of structure and dynamics during the liquid-to-solid transition of FUS LC, Biophys. J., 2021, 120, 1276–1287 CrossRef CAS PubMed.
- A. D. Stephens, D. Matak-Vinkovic, A. Fernandez-Villegas and G. S. Kaminski Schierle, Purification of Recombinant alpha-synuclein: A Comparison of Commonly Used Protocols, Biochemistry, 2020, 59, 4563–4572 CrossRef CAS PubMed.
- M. Pesarrodona, I. Latorre and X. Salvatella, Intrinsically Disordered Proteins (IDP): Purification Under Denaturing Conditions, Methods Mol. Biol., 2022, 2406, 359–370 CrossRef PubMed.
- B. D. Fonda, K. M. Jami, N. R. Boulos and D. T. Murray, Identification of the Rigid Core for Aged Liquid Droplets of an RNA-Binding Protein Low Complexity Domain, J. Am. Chem. Soc., 2021, 143, 6657–6668 CrossRef CAS PubMed.
- A. J. Stevens, Z. Z. Brown, N. H. Shah, G. Sekar, D. Cowburn and T. W. Muir, Design of a Split Intein with Exceptional Protein Splicing Activity, J. Am. Chem. Soc., 2016, 138, 2162–2165 CrossRef CAS PubMed.
- N. O. Taylor, M. T. Wei, H. A. Stone and C. P. Brangwynne, Quantifying Dynamics in Phase-Separated Condensates Using Fluorescence Recovery after Photobleaching, Biophys. J., 2019, 117, 1285–1300 CrossRef CAS PubMed.
- L. A. Metskas and E. Rhoades, Single-Molecule FRET of Intrinsically Disordered Proteins, Annu. Rev. Phys. Chem., 2020, 71, 391–414 CrossRef CAS PubMed.
- S. Weickert, J. Cattani and M. Drescher, in Electron Paramagnetic Resonance, ed. V. Chechik and D. M. Murphy, 2018, vol. 26, pp. 1–37 Search PubMed.
- E. A. Hoyt, P. M. S. D. Cal, B. L. Oliveira and G. J. L. Bernardes, Contemporary approaches to site-selective protein modification, Nat. Rev. Chem., 2019, 3, 147–171 CrossRef CAS.
- G. T. Debelouchina and T. W. Muir, A molecular engineering toolbox for the structural biologist, Q. Rev. Biophys., 2017, 50, e7 CrossRef PubMed.
- E. T. Clark, E. E. Sievers and G. T. Debelouchina, A Chemical Biology Primer for NMR Spectroscopists, J. Magn. Reson. Open, 2022, 10–11 Search PubMed.
- J. N. deGruyter, L. R. Malins and P. S. Baran, Residue-Specific Peptide Modification: A Chemist's Guide, Biochemistry, 2017, 56, 3863–3873 CrossRef CAS PubMed.
- O. Boutureira and G. J. Bernardes, Advances in chemical protein modification, Chem. Rev., 2015, 115, 2174–2195 CrossRef CAS PubMed.
- Y. Fu and N. S. Finney, Small-molecule fluorescent probes and their design, RSC Adv., 2018, 8, 29051–29061 RSC.
- K. Rhine, M. A. Makurath, J. Liu, S. Skanchy, C. Lopez, K. F. Catalan, Y. Ma, C. M. Fare, J. Shorter, T. Ha, Y. R. Chemla and S. Myong, ALS/FTLD-Linked Mutations in FUS Glycine Residues Cause Accelerated Gelation and Reduced Interactions with Wild-Type FUS, Mol. Cell, 2020, 80, 1139 CrossRef CAS PubMed.
- Z. R. Grese, A. C. Bastos, L. D. Mamede, R. L. French, T. M. Miller and Y. M. Ayala, Specific RNA interactions promote TDP-43 multivalent phase separation and maintain liquid properties, EMBO Rep., 2021, 22, e53632 CrossRef CAS PubMed.
- A. G. Larson, D. Elnatan, M. M. Keenen, M. J. Trnka, J. B. Johnston, A. L. Burlingame, D. A. Agard, S. Redding and G. J. Narlikar, Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin, Nature, 2017, 547, 236–240 CrossRef CAS PubMed.
- Y. Lin, Y. Fichou, A. P. Longhini, L. C. Llanes, P. Yin, G. C. Bazan, K. S. Kosik and S. Han, Liquid-Liquid Phase Separation of Tau Driven by Hydrophobic Interaction Facilitates Fibrillization of Tau, J. Mol. Biol., 2021, 433, 166731 CrossRef CAS PubMed.
- C. B. Rosen and M. B. Francis, Targeting the N terminus for site-selective protein modification, Nat. Chem. Biol., 2017, 13, 697–705 CrossRef CAS PubMed.
- G. M. Clore, Practical Aspects of Paramagnetic Relaxation Enhancement in Biological Macromolecules, Methods Enzymol., 2015, 564, 485–497 CAS.
- V. H. Ryan, G. L. Dignon, G. H. Zerze, C. V. Chabata, R. Silva, A. E. Conicella, J. Amaya, K. A. Burke, J. Mittal and N. L. Fawzi, Mechanistic View of hnRNPA2 Low-Complexity Domain Structure, Interactions, and Phase Separation Altered by Mutation and Arginine Methylation, Mol. Cell, 2018, 69, 465–479 CrossRef CAS PubMed.
- A. Wang, A. E. Conicella, H. B. Schmidt, E. W. Martin, S. N. Rhoads, A. N. Reeb, A. Nourse, D. Ramirez Montero, V. H. Ryan, R. Rohatgi, F. Shewmaker, M. T. Naik, T. Mittag, Y. M. Ayala and N. L. Fawzi, A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing, EMBO J., 2018, 37 CAS.
- A. E. Conicella, G. H. Zerze, J. Mittal and N. L. Fawzi, ALS Mutations Disrupt Phase Separation Mediated by alpha-Helical Structure in the TDP-43 Low-Complexity C-Terminal Domain, Structure, 2016, 24, 1537–1549 CrossRef CAS PubMed.
- I. Ritsch, E. Lehmann, L. Emmanouilidis, M. Yulikov, F. Allain and G. Jeschke, Phase Separation of Heterogeneous Nuclear Ribonucleoprotein A1 upon Specific RNA-Binding Observed by Magnetic Resonance, Angew. Chem., Int. Ed., 2022, 61, e202204311 CrossRef CAS PubMed.
- G. Jeschke, DEER distance measurements on proteins, Annu. Rev. Phys. Chem., 2012, 63, 419–446 CrossRef CAS PubMed.
- N. A. Eschmann, E. R. Georgieva, P. Ganguly, P. P. Borbat, M. D. Rappaport, Y. Akdogan, J. H. Freed, J. E. Shea and S. Han, Signature of an aggregation-prone conformation of tau, Sci. Rep., 2017, 7, 44739 CrossRef PubMed.
- F. X. Theillet, A. Binolfi, B. Bekei, A. Martorana, H. M. Rose, M. Stuiver, S. Verzini, D. Lorenz, M. van Rossum, D. Goldfarb and P. Selenko, Structural disorder of monomeric alpha-synuclein persists in mammalian cells, Nature, 2016, 530, 45–50 CrossRef CAS PubMed.
- L. Emmanouilidis, L. Esteban-Hofer, F. F. Damberger, T. de Vries, C. K. X. Nguyen, L. F. Ibanez, S. Mergenthal, E. Klotzsch, M. Yulikov, G. Jeschke and F. H. Allain, NMR and EPR reveal a compaction of the RNA-binding protein FUS upon droplet formation, Nat. Chem. Biol., 2021, 17, 608–614 CrossRef CAS PubMed.
- E. H. Yardeni, T. Bahrenberg, R. A. Stein, S. Mishra, E. Zomot, B. Graham, K. L. Tuck, T. Huber, E. Bibi, H. S. McHaourab and D. Goldfarb, Probing the solution structure of the E. coli multidrug transporter MdfA using DEER distance measurements with nitroxide and Gd(III) spin labels, Sci. Rep., 2019, 9, 12528 CrossRef PubMed.
- V. Venditti and N. L. Fawzi, Probing the Atomic Structure of Transient Protein Contacts by Paramagnetic Relaxation Enhancement Solution NMR, Methods Mol. Biol., 2018, 1688, 243–255 CrossRef CAS PubMed.
- S. D. Fontaine, R. Reid, L. Robinson, G. W. Ashley and D. V. Santi, Long-term stabilization of maleimide-thiol conjugates, Bioconjugate Chem., 2015, 26, 145–152 CrossRef CAS PubMed.
- L. Wang, A. Brock, B. Herberich and P. G. Schultz, Expanding the genetic code of Escherichia coli, Science, 2001, 292, 498–500 CrossRef CAS PubMed.
- L. Wang and P. G. Schultz, Expanding the genetic code, Angew. Chem., Int. Ed., 2004, 44, 34–66 CrossRef PubMed.
- C. C. Liu and P. G. Schultz, Adding new chemistries to the genetic code, Annu. Rev. Biochem., 2010, 79, 413–444 CrossRef CAS PubMed.
- J. W. Chin, Expanding and reprogramming the genetic code, Nature, 2017, 550, 53–60 CrossRef CAS PubMed.
- K. Lang and J. W. Chin, Cellular incorporation of unnatural amino acids and bioorthogonal labeling of proteins, Chem. Rev., 2014, 114, 4764–4806 CrossRef CAS PubMed.
- K. Lang, L. Davis, J. Torres-Kolbus, C. J. Chou, A. Deiters and J. W. Chin, Genetically encoded norbornene directs site-specific cellular protein labelling via a rapid bioorthogonal reaction, Nat. Chem., 2012, 4, 298–304 CrossRef CAS PubMed.
- K. Lang, L. Davis, S. Wallace, M. Mahesh, D. J. Cox, M. L. Blackman, J. M. Fox and J. W. Chin, Genetic Encoding of Bicyclononynes and trans-Cyclooctenes for Site-Specific Protein Labeling in Vitro and in Live Mammalian Cells via Rapid Fluorogenic Diels-Alder Reactions, J. Am. Chem. Soc., 2012, 134, 10317–10320 CrossRef CAS PubMed.
- T. Plass, S. Milles, C. Koehler, J. Szymanski, R. Mueller, M. Wiessler, C. Schultz and E. A. Lemke, Amino acids for Diels-Alder reactions in living cells, Angew. Chem., Int. Ed., 2012, 51, 4166–4170 CrossRef CAS PubMed.
- J. W. Chin, S. W. Santoro, A. B. Martin, D. S. King, L. Wang and P. G. Schultz, Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli, J. Am. Chem. Soc., 2002, 124, 9026–9027 CrossRef CAS PubMed.
- P. Widder, J. Schuck, D. Summerer and M. Drescher, Combining site-directed spin labeling in vivo and in-cell EPR distance determination, Phys. Chem. Chem. Phys., 2020, 22, 4875–4879 RSC.
- D. de la Torre and J. W. Chin, Reprogramming the genetic code, Nat. Rev. Genet., 2021, 22, 169–184 CrossRef CAS PubMed.
- M. J. Schmidt, J. Borbas, M. Drescher and D. Summerer, A genetically encoded spin label for electron paramagnetic resonance distance measurements, J. Am. Chem. Soc., 2014, 136, 1238–1241 CrossRef CAS PubMed.
- M. J. Schmidt, A. Fedoseev, D. Bucker, J. Borbas, C. Peter, M. Drescher and D. Summerer, EPR Distance Measurements in Native Proteins with Genetically Encoded Spin Labels, ACS Chem. Biol., 2015, 10, 2764–2771 CrossRef CAS PubMed.
- A. Chatterjee, S. B. Sun, J. L. Furman, H. Xiao and P. G. Schultz, A versatile platform for single- and multiple-unnatural amino acid mutagenesis in Escherichia coli, Biochemistry, 2013, 52, 1828–1837 CrossRef CAS PubMed.
- B. J. Lim, B. E. Ackermann and G. T. Debelouchina, Targetable Tetrazine-Based Dynamic Nuclear Polarization Agents for Biological Systems, Chembiochem, 2020, 21, 1315–1319 CrossRef CAS PubMed.
- C. M. Haney, R. F. Wissner, J. B. Warner, Y. X. J. Wang, J. J. Ferrie, D. J. Covell, R. J. Karpowicz, V. M. Y. Lee and E. J. Petersson, Comparison of strategies for non-perturbing labeling of alpha-synuclein to study amyloidogenesis, Org. Biomol. Chem., 2016, 14, 1584–1592 RSC.
- W. E. Robertson, L. F. H. Funke, D. de la Torre, J. Fredens, T. S. Elliott, M. Spinck, Y. Christova, D. Cervettini, F. L. Boge, K. C. Liu, S. Buse, S. Maslen, G. P. C. Salmond and J. W. Chin, Sense codon reassignment enables viral resistance and encoded polymer synthesis, Science, 2021, 372, 1057–1062 CrossRef CAS PubMed.
- E. D. Ficaretta, C. J. J. Wrobel, S. J. S. Roy, S. B. Erickson, J. S. Italia and A. Chatterjee, A Robust Platform for Unnatural Amino Acid Mutagenesis in E. coli Using the Bacterial Tryptophanyl-tRNA synthetase/tRNA pair, J. Mol. Biol., 2022, 434, 167304 CrossRef CAS PubMed.
- J. Yao, H. J. Dyson and P. E. Wright, Chemical shift dispersion and secondary structure prediction in unfolded and partly folded proteins, FEBS Lett., 1997, 419, 285–289 CrossRef PubMed.
- T. Yamazaki, T. Otomo, N. Oda, Y. Kyogoku, K. Uegaki, N. Ito, Y. Ishino and H. Nakamura, Segmental isotope labeling for protein NMR using peptide splicing, J. Am. Chem. Soc., 1998, 120, 5591–5592 CrossRef CAS.
- R. Xu, B. Ayers, D. Cowburn and T. W. Muir, Chemical ligation of folded recombinant proteins: segmental isotopic labeling of domains for NMR studies, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 388–393 CrossRef CAS PubMed.
- N. H. Shah, G. P. Dann, M. Vila-Perello, Z. H. Liu and T. W. Muir, Ultrafast Protein Splicing is Common among Cyanobacterial Split Inteins: Implications for Protein Engineering, J. Am. Chem. Soc., 2012, 134, 11338–11341 CrossRef CAS PubMed.
- N. H. Shah, E. Eryilmaz, D. Cowburn and T. W. Muir, Naturally split inteins assemble through a “capture and collapse” mechanism, J. Am. Chem. Soc., 2013, 135, 18673–18681 CrossRef CAS PubMed.
- A. J. Stevens, G. Sekar, N. H. Shah, A. Z. Mostafavi, D. Cowburn and T. W. Muir, A promiscuous split intein with expanded protein engineering applications, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 8538–8543 CrossRef CAS PubMed.
- N. H. Shah, E. Eryilmaz, D. Cowburn and T. W. Muir, Extein residues play an intimate role in the rate-limiting step of protein trans-splicing, J. Am. Chem. Soc., 2013, 135, 5839–5847 CrossRef CAS PubMed.
- P. Carvajal-Vallejos, R. Pallisse, H. D. Mootz and S. R. Schmidt, Unprecedented Rates and Efficiencies Revealed for New Natural Split Inteins from Metagenomic Sources, J. Biol. Chem., 2012, 287, 28686–28696 CrossRef CAS PubMed.
- M. Bhagawati, T. M. E. Terhorst, F. Fusser, S. Hoffmann, T. Pasch, S. Pietrokovski and H. D. Mootz, A mesophilic cysteine-less split intein for protein trans-splicing applications under oxidizing conditions, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 22164–22172 CrossRef CAS PubMed.
- A. Ciragan, S. M. Backlund, K. M. Mikula, H. M. Beyer, O. H. Samuli Ollila and H. Iwai, NMR Structure and Dynamics of TonB Investigated by Scar-Less Segmental Isotopic Labeling Using a Salt-Inducible Split Intein, Front. Chem., 2020, 8, 136 CrossRef CAS PubMed.
- K. K. Frederick, V. K. Michaelis, M. A. Caporini, L. B. Andreas, G. T. Debelouchina, R. G. Griffin and S. Lindquist, Combining DNP NMR with segmental and specific labeling to study a yeast prion protein strain that is not parallel in-register, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 3642–3647 CrossRef CAS PubMed.
- N. H. Shah and T. W. Muir, Inteins: nature's gift to protein chemists, Chem. Sci., 2014, 5, 446–461 RSC.
- B. J. Lim, R. F. Berkeley and G. T. Debelouchina, Fused Split Inteins: Tools for Introducing Multiple Protein Modifications, Methods Mol. Biol., 2020, 2133, 163–181 CrossRef CAS PubMed.
- S. Batjargal, C. R. Walters and E. J. Petersson, Inteins as Traceless Purification Tags for Unnatural Amino Acid Proteins, J. Am. Chem. Soc., 2015, 137, 1734–1737 CrossRef CAS PubMed.
- S. R. Chong, F. B. Mersha, D. G. Comb, M. E. Scott, D. Landry, L. M. Vence, F. B. Perler, J. Benner, R. B. Kucera, C. A. Hirvonen, J. J. Pelletier, H. Paulus and M. Q. Xu, Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element, Gene, 1997, 192, 271–281 CrossRef CAS PubMed.
- M. W. Southworth, K. Amaya, T. C. Evans, M. Q. Xu and F. B. Perler, Purification of proteins fused to either the amino or carboxy terminus of the Mycobacterium xenopi gyrase A intein, Biotechniques, 1999, 27, 110–120 CrossRef CAS PubMed.
- H. Ton-That, G. Liu, S. K. Mazmanian, K. F. Faull and O. Schneewind, Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 12424–12429 CrossRef CAS PubMed.
- S. K. Mazmanian, G. Liu, H. Ton-That and O. Schneewind, Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall, Science, 1999, 285, 760–763 CrossRef CAS PubMed.
- A. W. Jacobitz, M. D. Kattke, J. Wereszczynski and R. T. Clubb, Sortase Transpeptidases: Structural Biology and Catalytic Mechanism, Adv. Protein Chem. Struct. Biol., 2017, 109, 223–264 CAS.
- H. Morgan, W. BruceTurnbull and M. Webb, Challenges in the use of sortase and other peptide ligases for site-specific protein modification, Chem. Soc. Rev., 2022, 51, 4121–4145 RSC.
- K. V. Boyko, E. A. Rosenkranz, D. M. Smith, H. L. Miears, M. Oueld Es Cheikh, M. Z. Lund, J. C. Young, P. N. Reardon, M. Okon, S. L. Smirnov and J. M. Antos, Sortase-mediated segmental labeling: a method for segmental assignment of intrinsically disordered regions in proteins, PLoS One, 2021, 16, e0258531 CrossRef CAS PubMed.
- L. Freiburger, M. Sonntag, J. Hennig, J. Li, P. J. Zou and M. Sattler, Efficient segmental isotope labeling of multi-domain proteins using Sortase A, J. Biomol. NMR, 2015, 63, 1–8 CrossRef CAS PubMed.
- C. P. Guimaraes, M. D. Witte, C. S. Theile, G. Bozkurt, L. Kundrat, A. E. Blom and H. L. Ploegh, Site-specific C-terminal and internal loop labeling of proteins using sortase-mediated reactions, Nat. Protoc., 2013, 8, 1787–1799 CrossRef PubMed.
- K. Moritsugu, T. Terada and A. Kidera, Disorder-to-order transition of an intrinsically disordered region of sortase revealed by multiscale enhanced sampling, J. Am. Chem. Soc., 2012, 134, 7094–7101 CrossRef CAS PubMed.
- A. Bah and J. D. Forman-Kay, Modulation of Intrinsically Disordered Protein Function by Post-translational Modifications, J. Biol. Chem., 2016, 291, 6696–6705 CrossRef CAS PubMed.
- M. Hofweber and D. Dormann, Friend or foe-Post-translational modifications as regulators of phase separation and RNP granule dynamics, J. Biol. Chem., 2019, 294, 7137–7150 CrossRef CAS PubMed.
- A. Savastano, D. Flores, H. Kadavath, J. Biernat, E. Mandelkow and M. Zweckstetter, Disease-Associated Tau Phosphorylation Hinders Tubulin Assembly within Tau Condensates, Angew. Chem., Int. Ed., 2021, 60, 726–730 CrossRef CAS PubMed.
- M. Saito, D. Hess, J. Eglinger, A. W. Fritsch, M. Kreysing, B. T. Weinert, C. Choudhary and P. Matthias, Acetylation of intrinsically disordered regions regulates phase separation, Nat. Chem. Biol., 2019, 15, 51–61 CrossRef CAS PubMed.
- E. T. Usher, K. E. W. Namitz, M. S. Cosgrove and S. A. Showalter, Probing multiple enzymatic methylation events in real time with NMR spectroscopy, Biophys. J., 2021, 120, 4710–4721 CrossRef CAS PubMed.
- A. I. Green and G. M. Burslem, Focused Libraries for Epigenetic Drug Discovery: The Importance of Isosteres, J. Med. Chem., 2021, 64, 7231–7240 CrossRef CAS PubMed.
- L. A. Gruijs da Silva, F. Simonetti, S. Hutten, H. Riemenschneider, E. L. Sternburg, L. M. Pietrek, J. Gebel, V. Dotsch, D. Edbauer, G. Hummer, L. S. Stelzl and D. Dormann, Disease-linked TDP-43 hyperphosphorylation suppresses TDP-43 condensation and aggregation, EMBO J., 2022, 41, e108443 CrossRef CAS PubMed.
- X. Luo, G. Fu, R. E. Wang, X. Zhu, C. Zambaldo, R. Liu, T. Liu, X. Lyu, J. Du, W. Xuan, A. Yao, S. A. Reed, M. Kang, Y. Zhang, H. Guo, C. Huang, P. Y. Yang, I. A. Wilson, P. G. Schultz and F. Wang, Genetically encoding phosphotyrosine and its nonhydrolyzable analog in bacteria, Nat. Chem. Biol., 2017, 13, 845–849 CrossRef CAS PubMed.
- S. J. Elsasser, R. J. Ernst, O. S. Walker and J. W. Chin, Genetic code expansion in stable cell lines enables encoded chromatin modification, Nat. Methods, 2016, 13, 158–164 CrossRef CAS PubMed.
- M. D. Simon, F. Chu, L. R. Racki, C. C. de la Cruz, A. L. Burlingame, B. Panning, G. J. Narlikar and K. M. Shokat, The site-specific installation of methyl-lysine analogs into recombinant histones, Cell, 2007, 128, 1003–1012 CrossRef CAS PubMed.
- M. Shoaib, Q. Chen, X. Shi, N. Nair, C. Prasanna, R. Yang, D. Walter, K. S. Frederiksen, H. Einarsson, J. P. Svensson, C. F. Liu, K. Ekwall, M. Lerdrup, L. Nordenskiold and C. S. Sorensen, Histone H4 lysine 20 mono-methylation directly facilitates chromatin openness and promotes transcription of housekeeping genes, Nat. Commun., 2021, 12, 4800 CrossRef CAS PubMed.
- K. Hiragami-Hamada, S. Soeroes, M. Nikolov, B. Wilkins, S. Kreuz, C. Chen, I. A. De La Rosa-Velazquez, H. M. Zenn, N. Kost, W. Pohl, A. Chernev, D. Schwarzer, T. Jenuwein, M. Lorincz, B. Zimmermann, P. J. Walla, H. Neumann, T. Baubec, H. Urlaub and W. Fischle, Dynamic and flexible H3K9me3 bridging via HP1beta dimerization establishes a plastic state of condensed chromatin, Nat. Commun., 2016, 7, 11310 CrossRef CAS PubMed.
- B. E. Ackermann and G. T. Debelouchina, Heterochromatin Protein HP1alpha Gelation Dynamics Revealed by Solid-State NMR Spectroscopy, Angew. Chem., Int. Ed., 2019, 58, 6300–6305 CrossRef CAS PubMed.
- R. Huang, M. A. Holbert, M. K. Tarrant, S. Curtet, D. R. Colquhoun, B. M. Dancy, B. C. Dancy, Y. Hwang, Y. Tang, K. Meeth, R. Marmorstein, R. N. Cole, S. Khochbin and P. A. Cole, Site-specific introduction of an acetyl-lysine mimic into peptides and proteins by cysteine alkylation, J. Am. Chem. Soc., 2010, 132, 9986–9987 CrossRef CAS PubMed.
- F. Li, A. Allahverdi, R. Yang, G. B. Lua, X. Zhang, Y. Cao, N. Korolev, L. Nordenskiold and C. F. Liu, A direct method for site-specific protein acetylation, Angew. Chem., Int. Ed., 2011, 50, 9611–9614 CrossRef CAS PubMed.
- D. D. Le, A. T. Cortesi, S. A. Myers, A. L. Burlingame and D. G. Fujimori, Site-specific and regiospecific installation of methylarginine analogues into recombinant histones and insights into effector protein binding, J. Am. Chem. Soc., 2013, 135, 2879–2882 CrossRef CAS PubMed.
- C. Chatterjee, R. K. McGinty, B. Fierz and T. W. Muir, Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation, Nat. Chem. Biol., 2010, 6, 267–269 CrossRef CAS PubMed.
- G. T. Debelouchina, K. Gerecht and T. W. Muir, Ubiquitin utilizes an acidic surface patch to alter chromatin structure, Nat. Chem. Biol., 2017, 13, 105–110 CrossRef CAS PubMed.
- P. E. Dawson, T. W. Muir, I. Clark-Lewis and S. B. Kent, Synthesis of proteins by native chemical ligation, Science, 1994, 266, 776–779 CrossRef CAS PubMed.
- R. E. Thompson and T. W. Muir, Chemoenzymatic Semisynthesis of Proteins, Chem. Rev., 2020, 120, 3051–3126 CrossRef CAS PubMed.
- T. W. Muir, D. Sondhi and P. A. Cole, Expressed protein ligation: a general method for protein engineering, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 6705–6710 CrossRef CAS PubMed.
- V. Agouridas, O. El Mahdi, V. Diemer, M. Cargoet, J. M. Monbaliu and O. Melnyk, Native Chemical Ligation and Extended Methods: Mechanisms, Catalysis, Scope, and Limitations, Chem. Rev., 2019, 119, 7328–7443 CrossRef CAS PubMed.
- O. Fuchs, S. Trunschke, H. Hanebrink, M. Reimann and O. Seitz, Enabling Cysteine-Free Native Chemical Ligation at Challenging Junctions with a Ligation Auxiliary Capable of Base Catalysis, Angew. Chem., Int. Ed., 2021, 60, 19483–19490 CrossRef CAS PubMed.
- G. M. Fang, Y. M. Li, F. Shen, Y. C. Huang, J. B. Li, Y. Lin, H. K. Cui and L. Liu, Protein chemical synthesis by ligation of peptide hydrazides, Angew. Chem., Int. Ed., 2011, 50, 7645–7649 CrossRef CAS PubMed.
- J. C. Shimko, C. J. Howard, M. G. Poirier and J. J. Ottesen, Preparing semisynthetic and fully synthetic histones h3 and h4 to modify the nucleosome core, Methods Mol. Biol., 2013, 981, 177–192 CrossRef CAS PubMed.
- E. J. Ge, K. S. Jani, K. L. Diehl, M. M. Muller and T. W. Muir, Nucleation and Propagation of Heterochromatin by the Histone Methyltransferase PRC2: Geometric Constraints and Impact of the Regulatory Subunit JARID2, J. Am. Chem. Soc., 2019, 141, 15029–15039 CrossRef CAS PubMed.
- M. R. Pratt, T. Abeywardana and N. P. Marotta, Synthetic Proteins and Peptides for the Direct Interrogation of alpha-Synuclein Posttranslational Modifications, Biomolecules, 2015, 5, 1210–1227 CrossRef CAS PubMed.
- M. Hejjaoui, S. Butterfield, B. Fauvet, F. Vercruysse, J. Cui, I. Dikiy, M. Prudent, D. Olschewski, Y. Zhang, D. Eliezer and H. A. Lashuel, Elucidating the role of C-terminal post-translational modifications using protein semisynthesis strategies: alpha-synuclein phosphorylation at tyrosine 125, J. Am. Chem. Soc., 2012, 134, 5196–5210 CrossRef CAS PubMed.
- D. Ellmer, M. Brehs, M. Haj-Yahya, H. A. Lashuel and C. F. W. Becker, Single Posttranslational Modifications in the Central Repeat Domains of Tau4 Impact its Aggregation and Tubulin Binding, Angew. Chem., Int. Ed., 2019, 58, 1616–1620 CrossRef CAS PubMed.
- M. Haj-Yahya and H. A. Lashuel, Protein Semisynthesis Provides Access to Tau Disease-Associated Post-translational Modifications (PTMs) and Paves the Way to Deciphering the Tau PTM Code in Health and Diseased States, J. Am. Chem. Soc., 2018, 140, 6611–6621 CrossRef CAS PubMed.
- B. Pan, J. H. Park, T. Ramlall, D. Eliezer, E. Rhoades and E. J. Petersson, Chemoenzymatic Semi-synthesis Enables Efficient Production of Isotopically Labeled alpha-Synuclein with Site-Specific Tyrosine Phosphorylation, ChemBioChem, 2021, 22, 1440–1447 CrossRef CAS PubMed.
- H. V. Erkizan, Y. Kong, M. Merchant, S. Schlottmann, J. S. Barber-Rotenberg, L. Yuan, O. D. Abaan, T. H. Chou, S. Dakshanamurthy, M. L. Brown, A. Uren and J. A. Toretsky, A small molecule blocking oncogenic protein EWS-FLI1 interaction with RNA helicase A inhibits growth of Ewing's sarcoma, Nat. Med., 2009, 15, 750–756 CrossRef CAS PubMed.
- N. Krishnan, D. Koveal, D. H. Miller, B. Xue, S. D. Akshinthala, J. Kragelj, M. R. Jensen, C. M. Gauss, R. Page, M. Blackledge, S. K. Muthuswamy, W. Peti and N. K. Tonks, Targeting the disordered C terminus of PTP1B with an allosteric inhibitor, Nat. Chem. Biol., 2014, 10, 558–566 CrossRef CAS PubMed.
- X. Yin, C. Giap, J. S. Lazo and E. V. Prochownik, Low molecular weight inhibitors of Myc-Max interaction and function, Oncogene, 2003, 22, 6151–6159 CrossRef CAS PubMed.
- C. Yu, X. Niu, F. Jin, Z. Liu, C. Jin and L. Lai, Structure-based Inhibitor Design for the Intrinsically Disordered Protein c-Myc, Sci. Rep., 2016, 6, 22298 CrossRef CAS PubMed.
- M. Convertino, A. Vitalis and A. Caflisch, Disordered binding of small molecules to Abeta(12-28), J. Biol. Chem., 2011, 286, 41578–41588 CrossRef CAS PubMed.
- G. Toth, T. Neumann, A. Berthet, E. Masliah, B. Spencer, J. Tao, M. F. Jobling, S. J. Gardai, C. W. Bertoncini, N. Cremades, M. Bova, S. Ballaron, X. H. Chen, W. Mao, P. Nguyen, M. C. Tabios, M. A. Tambe, J. C. Rochet, H. D. Junker, D. Schwizer, R. Sekul, I. Ott, J. P. Anderson, B. Szoke, W. Hoffman, J. Christodoulou, T. Yednock, D. A. Agard, D. Schenk and L. McConlogue, Novel Small Molecules Targeting the Intrinsically Disordered Structural Ensemble of alpha-Synuclein Protect Against Diverse alpha-Synuclein Mediated Dysfunctions, Sci. Rep., 2019, 9, 16947 CrossRef PubMed.
- L. I. Iconaru, S. Das, A. Nourse, A. A. Shelat, J. Zuo and R. W. Kriwacki, Small Molecule Sequestration of the Intrinsically Disordered Protein, p27(Kip1), Within Soluble Oligomers, J. Mol. Biol., 2021, 433, 167120 CrossRef CAS PubMed.
- Y. Cheng, T. LeGall, C. J. Oldfield, J. P. Mueller, Y. Y. Van, P. Romero, M. S. Cortese, V. N. Uversky and A. K. Dunker, Rational drug design via intrinsically disordered protein, Trends Biotechnol., 2006, 24, 435–442 CrossRef CAS PubMed.
- K. Ribbeck and D. Gorlich, The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion, EMBO J., 2002, 21, 2664–2671 CrossRef CAS PubMed.
- N. Nag, S. Sasidharan, V. N. Uversky, P. Saudagar and T. Tripathi, Phase separation of FG-nucleoporins in nuclear pore complexes, Biochim. Biophys. Acta, Mol. Cell Res., 2022, 1869, 119205 CrossRef CAS PubMed.
- N. Shulga and D. S. Goldfarb, Binding dynamics of structural nucleoporins govern nuclear pore complex permeability and may mediate channel gating, Mol. Cell. Biol., 2003, 23, 534–542 CrossRef CAS PubMed.
- S. Kroschwald, S. Maharana and A. Simon, Hexanediol: a chemical probe to investigate the material properties of membrane-less compartments, Matters, 2017, 3, e201702000010 Search PubMed.
- Y. Lin, E. Mori, M. Kato, S. Xiang, L. Wu, I. Kwon and S. L. McKnight, Toxic PR Poly-Dipeptides Encoded by the C9orf72 Repeat Expansion Target LC Domain Polymers, Cell, 2016, 167, 789–802 CrossRef CAS PubMed.
- J. R. Wheeler, T. Matheny, S. Jain, R. Abrisch and R. Parker, Distinct stages in stress granule assembly and disassembly, Elife, 2016, 5, e18413 CrossRef PubMed.
- M. Shi, K. You, T. Chen, C. Hou, Z. Liang, M. Liu, J. Wang, T. Wei, J. Qin, Y. Chen, M. Q. Zhang and T. Li, Quantifying the phase separation property of chromatin-associated proteins under physiological conditions using an anti-1,6-hexanediol index, Genome Biol., 2021, 22, 229 CrossRef CAS PubMed.
- R. Duster, I. H. Kaltheuner, M. Schmitz and M. Geyer, 1,6-Hexanediol, commonly used to dissolve liquid-liquid phase separated condensates, directly impairs kinase and phosphatase activities, J. Biol. Chem., 2021, 296, 100260 CrossRef PubMed.
- R. Meduri, L. S. Rubio, S. Mohajan and D. S. Gross, Phase-separation antagonists potently inhibit transcription and broadly increase nucleosome density, J. Biol. Chem., 2022, 102365, DOI:10.1016/j.jbc.2022.102365.
- S. Alberti, A. Gladfelter and T. Mittag, Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates, Cell, 2019, 176, 419–434 CrossRef CAS PubMed.
- S. S. Patel, B. J. Belmont, J. M. Sante and M. F. Rexach, Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex, Cell, 2007, 129, 83–96 CrossRef CAS PubMed.
- A. Patel, L. Malinovska, S. Saha, J. Wang, S. Alberti, Y. Krishnan and A. A. Hyman, ATP as a biological hydrotrope, Science, 2017, 356, 753–756 CrossRef CAS PubMed.
- J. Kang, L. Lim and J. Song, ATP enhances at low concentrations but dissolves at high concentrations liquid-liquid phase separation (LLPS) of ALS/FTD-causing FUS, Biochem. Biophys. Res. Commun., 2018, 504, 545–551 CrossRef CAS PubMed.
- W. M. Babinchak, B. K. Dumm, S. Venus, S. Boyko, A. A. Putnam, E. Jankowsky and W. K. Surewicz, Small molecules as potent biphasic modulators of protein liquid-liquid phase separation, Nat. Commun., 2020, 11, 5574 CrossRef CAS PubMed.
- S. Jonchhe, W. Pan, P. Pokhrel and H. Mao, Small Molecules Modulate Liquid-to-Solid Transitions in Phase-Separated Tau Condensates, Angew. Chem., Int. Ed., 2022, 61, e202113156 CrossRef CAS PubMed.
- V. I. Stsiapura, A. A. Maskevich, V. A. Kuzmitsky, V. N. Uversky, I. M. Kuznetsova and K. K. Turoverov, Thioflavin T as a molecular rotor: fluorescent properties of thioflavin T in solvents with different viscosity, J. Phys. Chem. B, 2008, 112, 15893–15902 CrossRef CAS PubMed.
- C. Q. Liang, L. Wang, Y. Y. Luo, Q. Q. Li and Y. M. Li, Capturing protein droplets: label-free visualization and detection of protein liquid-liquid phase separation with an aggregation-induced emission fluorogen, Chem. Commun., 2021, 57, 3805–3808 RSC.
- S. F. Banani, A. M. Rice, W. B. Peeples, Y. Lin, S. Jain, R. Parker and M. K. Rosen, Compositional Control of Phase-Separated Cellular Bodies, Cell, 2016, 166, 651–663 CrossRef CAS PubMed.
- I. A. Klein, A. Boija, L. K. Afeyan, S. W. Hawken, M. Fan, A. Dall'Agnese, O. Oksuz, J. E. Henninger, K. Shrinivas, B. R. Sabari, I. Sagi, V. E. Clark, J. M. Platt, M. Kar, P. M. McCall, A. V. Zamudio, J. C. Manteiga, E. L. Coffey, C. H. Li, N. M. Hannett, Y. E. Guo, T. M. Decker, T. I. Lee, T. Zhang, J. K. Weng, D. J. Taatjes, A. Chakraborty, P. A. Sharp, Y. T. Chang, A. A. Hyman, N. S. Gray and R. A. Young, Partitioning of cancer therapeutics in nuclear condensates, Science, 2020, 368, 1386–1392 CrossRef CAS PubMed.
- D. Ban, L. I. Iconaru, A. Ramanathan, J. Zuo and R. W. Kriwacki, A Small Molecule Causes a Population Shift in the Conformational Landscape of an Intrinsically Disordered Protein, J. Am. Chem. Soc., 2017, 139, 13692–13700 CrossRef CAS PubMed.
- P. Herrera-Nieto, A. Perez and G. De Fabritiis, Small Molecule Modulation of Intrinsically Disordered Proteins Using Molecular Dynamics Simulations, J. Chem. Inf. Model., 2020, 60, 5003–5010 CrossRef CAS PubMed.
- G. T. Heller, P. Sormanni and M. Vendruscolo, Targeting disordered proteins with small molecules using entropy, Trends Biochem. Sci., 2015, 40, 491–496 CrossRef CAS PubMed.
- G. T. Heller, M. Bonomi and M. Vendruscolo, Structural Ensemble Modulation upon Small-Molecule Binding to Disordered Proteins, J. Mol. Biol., 2018, 430, 2288–2292 CrossRef CAS PubMed.
- A. E. Conicella, G. L. Dignon, G. H. Zerze, H. B. Schmidt, A. M. D'Ordine, Y. C. Kim, R. Rohatgi, Y. M. Ayala, J. Mittal and N. L. Fawzi, TDP-43 alpha-helical structure tunes liquid-liquid phase separation and function, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 5883–5894 CrossRef CAS PubMed.
- J. Zhu, X. Salvatella and P. Robustelli, Small molecules targeting the disordered transactivation domain of the androgen receptor induce the formation of collapsed helical states, Nat. Commun., 2022, 13, 6390 CrossRef CAS PubMed.
- G. Toth, S. J. Gardai, W. Zago, C. W. Bertoncini, N. Cremades, S. L. Roy, M. A. Tambe, J. C. Rochet, C. Galvagnion, G. Skibinski, S. Finkbeiner, M. Bova, K. Regnstrom, S. S. Chiou, J. Johnston, K. Callaway, J. P. Anderson, M. F. Jobling, A. K. Buell, T. A. Yednock, T. P. Knowles, M. Vendruscolo, J. Christodoulou, C. M. Dobson, D. Schenk and L. McConlogue, Targeting the intrinsically disordered structural ensemble of alpha-synuclein by small molecules as a potential therapeutic strategy for Parkinson's disease, PLoS One, 2014, 9, e87133 CrossRef PubMed.
- L. I. Iconaru, D. Ban, K. Bharatham, A. Ramanathan, W. Zhang, A. A. Shelat, J. Zuo and R. W. Kriwacki, Discovery of Small Molecules that Inhibit the Disordered Protein, p27(Kip1), Sci. Rep., 2015, 5, 15686 CrossRef CAS PubMed.
- G. Klebe, Applying thermodynamic profiling in lead finding and optimization, Nat. Rev. Drug Discovery, 2015, 14, 95–110 CrossRef CAS PubMed.
- G. T. Heller, F. A. Aprile, T. C. T. Michaels, R. Limbocker, M. Perni, F. S. Ruggeri, B. Mannini, T. Lohr, M. Bonomi, C. Camilloni, A. De Simone, I. C. Felli, R. Pierattelli, T. P. J. Knowles, C. M. Dobson and M. Vendruscolo, Small-molecule sequestration of amyloid-beta as a drug discovery strategy for Alzheimer’s disease, Sci. Adv., 2020, 6(45), eabb5924 CrossRef CAS PubMed.
- J. Pujols, S. Pena-Diaz, D. F. Lazaro, F. Peccati, F. Pinheiro, D. Gonzalez, A. Carija, S. Navarro, M. Conde-Gimenez, J. Garcia, S. Guardiola, E. Giralt, X. Salvatella, J. Sancho, M. Sodupe, T. F. Outeiro, E. Dalfo and S. Ventura, Small molecule inhibits alpha-synuclein aggregation, disrupts amyloid fibrils, and prevents degeneration of dopaminergic neurons, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 10481–10486 CrossRef CAS PubMed.
- B. Xu, X. Mo, J. Chen, H. Yu and Y. Liu, Myricetin Inhibits alpha-Synuclein Amyloid Aggregation by Delaying the Liquid-to-Solid Phase Transition, Chembiochem, 2022, 23, e202200216 CAS.
- P. Robustelli, A. Ibanez-de-Opakua, C. Campbell-Bezat, F. Giordanetto, S. Becker, M. Zweckstetter, A. C. Pan and D. E. Shaw, Molecular Basis of Small-Molecule Binding to alpha-Synuclein, J. Am. Chem. Soc., 2022, 144, 2501–2510 CrossRef CAS PubMed.
- M. Kato, T. W. Han, S. Xie, K. Shi, X. Du, L. C. Wu, H. Mirzaei, E. J. Goldsmith, J. Longgood, J. Pei, N. V. Grishin, D. E. Frantz, J. W. Schneider, S. Chen, L. Li, M. R. Sawaya, D. Eisenberg, R. Tycko and S. L. McKnight, Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels, Cell, 2012, 149, 753–767 CrossRef CAS PubMed.
- P. Romero, Z. Obradovic, C. R. Kissinger, J. E. Villafranca, E. Garner, S. Guilliot and A. K. Dunker, Thousands of proteins likely to have long disordered regions, Pac. Symp. Biocomput., 1998, 437–448 CAS.
- H. J. Dyson and P. E. Wright, NMR illuminates intrinsic disorder, Curr. Opin. Struct. Biol., 2021, 70, 44–52 CrossRef CAS PubMed.
- D. T. Murray, M. Kato, Y. Lin, K. R. Thurber, I. Hung, S. L. McKnight and R. Tycko, Structure of FUS Protein Fibrils and Its Relevance to Self-Assembly and Phase Separation of Low-Complexity Domains, Cell, 2017, 171, 615–627 CrossRef CAS PubMed.
- J. M. Plitzko, B. Schuler and P. Selenko, Structural Biology outside the box-inside the cell, Curr. Opin. Struct. Biol., 2017, 46, 110–121 CrossRef CAS PubMed.
- E. Luchinat and L. Banci, In-cell NMR: From target structure and dynamics to drug screening, Curr. Opin. Struct. Biol., 2022, 74, 102374 CrossRef CAS PubMed.
- Y. David, M. Vila-Perello, S. Verma and T. W. Muir, Chemical tagging and customizing of cellular chromatin states using ultrafast trans-splicing inteins, Nat. Chem., 2015, 7, 394–402 CrossRef CAS PubMed.
- J. A. Gramespacher, A. J. Stevens, D. P. Nguyen, J. W. Chin and T. W. Muir, Intein Zymogens: Conditional Assembly and Splicing of Split Inteins via Targeted Proteolysis, J. Am. Chem. Soc., 2017, 139, 8074–8077 CrossRef CAS PubMed.
- G. L. Dignon, W. Zheng, Y. C. Kim, R. B. Best and J. Mittal, Sequence determinants of protein phase behavior from a coarse-grained model, PLoS Comput. Biol., 2018, 14, e1005941 CrossRef PubMed.
- Z. Benayad, S. von Bulow, L. S. Stelzl and G. Hummer, Simulation of FUS Protein Condensates with an Adapted Coarse-Grained Model, J. Chem. Theory Comput., 2021, 17, 525–537 CrossRef CAS PubMed.
- A. P. Latham and B. Zhang, Unifying coarse-grained force fields for folded and disordered proteins, Curr. Opin. Struct. Biol., 2022, 72, 63–70 CrossRef CAS PubMed.
- J. A. Joseph, A. Reinhardt, A. Aguirre, P. Y. Chew, K. O. Russell, J. R. Espinosa, A. Garaizar and R. Collepardo-Guevara, Physics-driven coarse-grained model for biomolecular phase separation with near-quantitative accuracy, Nat. Comput. Sci., 2021, 1, 732–743 CrossRef PubMed.
- M. Paloni, R. Bailly, L. Ciandrini and A. Barducci, Unraveling Molecular Interactions in Liquid-Liquid Phase Separation of Disordered Proteins by Atomistic Simulations, J. Phys. Chem. B, 2020, 124, 9009–9016 CrossRef CAS PubMed.
- W. Zheng, G. L. Dignon, N. Jovic, X. Xu, R. M. Regy, N. L. Fawzi, Y. C. Kim, R. B. Best and J. Mittal, Molecular Details of Protein Condensates Probed by Microsecond Long Atomistic Simulations, J. Phys. Chem. B, 2020, 124, 11671–11679 CrossRef CAS PubMed.
- N. M. Kanaan, C. Hamel, T. Grabinski and B. Combs, Liquid-liquid phase separation induces pathogenic tau conformations in vitro, Nat. Commun., 2020, 11, 2809 CrossRef CAS PubMed.
- J. E. Hoffmann, T. Plass, I. Nikic, I. V. Aramburu, C. Koehler, H. Gillandt, E. A. Lemke and C. Schultz, Highly Stable trans-Cyclooctene Amino Acids for Live-Cell Labeling, Chem.–Eur. J., 2015, 21, 12266–12270 CrossRef CAS PubMed.
- P. Widder, F. Berner, D. Summerer and M. Drescher, Double Nitroxide Labeling by Copper-Catalyzed Azide-Alkyne Cycloadditions with Noncanonical Amino Acids for Electron Paramagnetic Resonance Spectroscopy, ACS Chem. Biol., 2019, 14, 839–844 CrossRef CAS PubMed.
- Y. Ge, L. Chen, S. Liu, J. Zhao, H. Zhang and P. R. Chen, Enzyme-Mediated Intercellular Proximity Labeling for Detecting Cell-Cell Interactions, J. Am. Chem. Soc., 2019, 141, 1833–1837 CrossRef CAS PubMed.
- B. Fierz, C. Chatterjee, R. K. McGinty, M. Bar-Dagan, D. P. Raleigh and T. W. Muir, Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction, Nat. Chem. Biol., 2011, 7, 113–119 CrossRef CAS PubMed.
- B. Pan, E. Rhoades and E. J. Petersson, Chemoenzymatic Semisynthesis of Phosphorylated alpha-Synuclein Enables Identification of a Bidirectional Effect on Fibril Formation, ACS Chem. Biol., 2020, 15, 640–645 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |