Open Access Article
Open Access ArticleSynaptotagmin-1 C2B domains cooperatively stabilize the fusion stalk via a master-servant mechanism†
Ary Lautaro
Di Bartolo
 ab and
Diego
Masone
ab and
Diego
Masone
 *ac
*ac
aInstituto de Histología y Embriología de Mendoza (IHEM) - Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Universidad Nacional de Cuyo (UNCuyo), 5500, Mendoza, Argentina. E-mail: diego.masone@ingenieria.uncuyo.edu.ar
bFacultad de Ciencias Exactas y Naturales, Universidad Nacional de Cuyo (UNCuyo), 5500, Mendoza, Argentina
cFacultad de Ingeniería, Universidad Nacional de Cuyo (UNCuyo), 5500, Mendoza, Argentina
First published on 23rd February 2022
Abstract
Synaptotagmin-1 is a low-affinity Ca2+ sensor that triggers synchronous vesicle fusion. It contains two similar C2 domains (C2A and C2B) that cooperate in membrane binding, being the C2B domain mainly responsible for the membrane fusion process due to its polybasic patch KRLKKKKTTIKK (321–332). In this work, a master-servant mechanism between two identical C2B domains is shown to control the formation of the fusion stalk in a calcium-independent manner. Two regions in C2B are essential for the process, the well-known polybasic patch and a recently described pair of arginines (398![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 399). The master domain shows strong PIP2 interactions with its polybasic patch and its pair of arginines. At the same time, the servant analogously cooperates with the master to reduce the total work to form the fusion stalk. The strategic mutation (T328E, T329E) in both master and servant domains disrupts the cooperative mechanism, drastically increasing the free energy needed to induce the fusion stalk, however, with negligible effects on the master domain interactions with PIP2. These data point to a difference in the behavior of the servant domain, which is unable to sustain its PIP2 interactions neither through its polybasic patch nor through its pair of arginines, and in the end, losing its ability to assist the master in the formation of the fusion stalk.
399). The master domain shows strong PIP2 interactions with its polybasic patch and its pair of arginines. At the same time, the servant analogously cooperates with the master to reduce the total work to form the fusion stalk. The strategic mutation (T328E, T329E) in both master and servant domains disrupts the cooperative mechanism, drastically increasing the free energy needed to induce the fusion stalk, however, with negligible effects on the master domain interactions with PIP2. These data point to a difference in the behavior of the servant domain, which is unable to sustain its PIP2 interactions neither through its polybasic patch nor through its pair of arginines, and in the end, losing its ability to assist the master in the formation of the fusion stalk.
1 Introduction
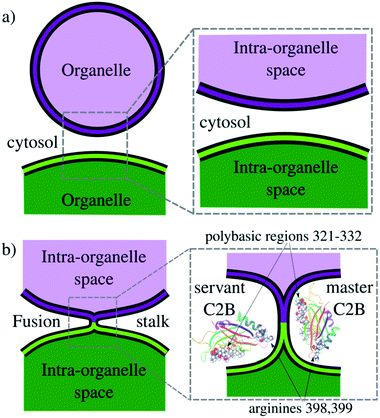
Exocytosis is an important process used by eukaryotic cells to release biological compounds and transport lipids and proteins through the plasma membrane. Specialized secretory cells experience regulated exocytosis as a response to physiological signals.1–3 Mainly, sperm exocytosis (or acrosome reaction) is a regulated secretion needed to fertilize the egg that requires large membrane remodeling, membrane bending and fusion.2,4–7 While this collective process develops, multiple fusion pores spontaneously form between the acrosomal and plasma membranes, connecting the acrosomal lumen to the extracellular milieu. Consequently, the fusion pore works as a remarkable mechanism to connect intracellular organelles and release vesicle contents during exocytosis. Fig. 1 schematically shows membrane remodeling and fusion stalk formation between organelles, which is the first step in the formation of a fusion pore.Synaptotagmin-1 (Syt1) is a vesicle-anchored protein known as a phospholipid binding machine.8 Syt1 has been related to synaptic vesicle fusion,9 fusion pore opening,10 stabilization5,11 and expansion.12,13 Syt1 contains two C2 domains (C2A and C2B) with Ca2+ binding loops, the latter (C2B) with a polybasic region KRLKKKKTTIKK (positions 321–332 in PDB ID: 1K5W8) that easily binds to negatively charged membrane patches, such as clusters of phosphatidylinositol-4,5-bisphosphate lipids (PI(4,5)P2, or simply PIP2), independently of Ca2+.14–18 Therefore, the Syt1–C2B domain has been identified as the main energetic driver during membrane fusion and evoked neurotransmitter release.19 Importantly, Cafiso and collaborators13 showed that two different regions of the C2B domain make unique contributions to the fusion process, namely, the polybasic region and a pair of arginines (R398, R399). However, the molecular mechanism by which Syt1 drives membrane fusion is yet not completely understood.13,20
In the present work, we use enhanced molecular dynamics with an ad hoc collective variable to induce a fusion stalk between lipid bilayers. We demonstrate that Syt1–C2B domains cooperatively facilitate the formation of the fusion stalk, significantly reducing its total thermodynamic work. We observe a master-servant mechanism between identical C2B domains, mainly driven by the polybasic regions 321–332 and arginines 398![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 399 while interacting with PIP2 lipids (see panel 1b). We show that mutations T328E and T329E in the polybasic region disrupt this mechanism of cooperation, drastically increasing the free energy needed to form the stalk.
399 while interacting with PIP2 lipids (see panel 1b). We show that mutations T328E and T329E in the polybasic region disrupt this mechanism of cooperation, drastically increasing the free energy needed to form the stalk.
2 Results and discussion
To induce the formation of the fusion stalk between two initially flat and planar bilayers surrounded by water molecules, we have followed a methodology originally developed by Hub and collaborators21,22 (Prof. Hub generously shared his GROMACS source code with us through personal communications). Using the MARTINI 3 coarse-grained model, we have prepared ternary lipid bilayers containing 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS) and the recently developed model for phosphatidylinositol-4-5-bisphosphate lipids (PIP2). This arrangement follows an experimental membrane composition proposed by Jahn and collaborators23 to trap Syt1 to the plasma membrane in the presence of calcium. Accordingly, lipid concentrations were set to POPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) POPS
POPS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PIP2 (87.5
PIP2 (87.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5), see Fig. S5 in the ESI.†
2.5), see Fig. S5 in the ESI.†
The collective variable designed by Hub and collaborators (ξ) induces a hydrophilic trans-membrane pore in a single lipid bilayer, using a membrane spanning cylinder that is decomposed into slices along the membrane normal.21 They also demonstrated that the same collective variable is capable of fusing bilayers at different hydration levels.22 Accordingly, in the present work, we have used ξ to fuse two bilayers and to study the effects of the Syt1–C2B domain on the process. To facilitate the repeatability of the results and to increase the versatility at the user level (and also for our own convenience), we have ported ξ into PLUMED24 as a collective variable (labeled ξf). The source code implementation for the PLUMED environment (including input file parameters and examples) is freely available at https://github.com/lautarodibartolo/MemFusion. See the ESI† for example input files and technical details on the collective variable set of parameters.
Practically, we have used eqn (1) to fuse membranes and form the stalk. The process starts with a pair of flat and parallel independent bilayers (ξf ∼ 0.2). Membrane fusion occurs in the interval 0.2 < ξf < 0.85 where the bilayers connect themselves forming the first stalk at ξf ∼ 0.58. The collective variable pulls from tail beads (C4A, C4B and C5A, for bead labeling see Fig. S14 in the ESI†) to fill a cylinder with Nsf = 85 slices, of thickness dsf = 0.1 nm, of radius Rcylf = 1.75 nm and with an occupation factor ζf = 0.5.
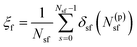 | (1) |
In eqn (1), N(p)sf accounts for the number of tail beads within the slice s inside the cylinder. δsf is a continuous function in the interval [0 1] (δsf = 0 for no beads in the slice s, and δsf = 1 for 1 or more beads in the slice s). For mathematical details, see the ESI† and the original article.21
2.1 Hysteresis-free sampling of the fusion stalk
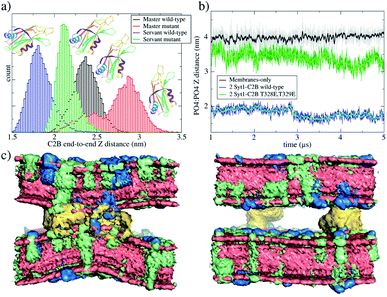
Here, we have equilibrated the inter-membrane distance between bilayers with the necessary amount of cytosolic water molecules to fit one and two C2B domains. Therefore, the PO4:PO4 inter-membrane distance was set to ∼3.9 nm, which requires ∼10 × 103 cytosolic water molecules, imposing a high hydration regime (∼34 cytosolic water beads per nm2). Phosphate PO4 beads in all lipid species were used for inter-membrane measurements along this work. For MARTINI bead labeling in lipid molecules, see Fig. S14 in the ESI.†The PO4:PO4 inter-membrane distance was measured along the Z axis from the average plane defined by all PO4 beads belonging to the upper leaflet of the lower bilayer to the average plane defined by all PO4 beads belonging to the lower leaflet of the upper bilayer. Therefore, the PO4:PO4 inter-membrane distance is a measure of the separation in the cytosolic space between bilayers.
As studied before,22,25 different amounts of water molecules between the bilayers result in different equilibrium inter-membrane distances, with significant effects on the free energy landscape for membrane fusion.
To avoid any sampling problems due to the high amount of water molecules we have included in the cytosolic space, we have verified the stalk fusion formation to be hysteresis-free. The free energy cost to evolve from different thermodynamic states (i.e. from parallel to fused bilayers) must be independent of the direction of the collective variable.26 Therefore, the forward and backward paths from parallel bilayers to the fusion stalk must be identical in the free energy profile. Any differences between them would suggest hysteresis problems, inadequate sampling and poor convergence.27
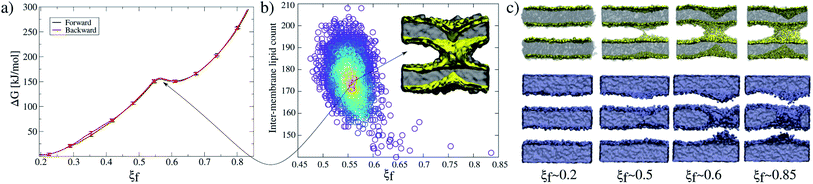
Accordingly, Fig. 2a shows PMF calculations in both directions of the collective variable: (i) forwards (black line) from ξf ∼0.2 (planar and parallel bilayers) to ξf ∼0.85 (formed fusion stalk) and (ii) backwards (red line) from ξf ∼0.85 to ξf ∼0.2. In this way, the collective variable space is sampled from initial configurations coming from different paths: parallel bilayers that fuse to form the stalk (forwards), and a formed stalk that shrinks and disconnects to recover parallel and planar bilayers (backwards). Initial configurations in both cases were taken from a slow-growth path in each direction, as originally suggested by Pearlman and Kollman.28 These profiles show no significant hysteresis.
Along membrane fusion, the first stalk is formed at ξf ∼0.58 with an energy cost of ∼150 kJ mol−1. From that state, the collective variable requires another ∼150 kJ mol−1 to reach the final state at ξf ∼0.85 (with a total cost of ∼300 kJ mol−1, see Fig. 2a). Importantly, ξf revealed an energy barrier for the fusion stalk (ξf ∼0.55) and a local minimum for a metastable stalk (ξf ∼0.6). See Fig. 2a, in agreement with minimum free energy path (MFEP) dynamics by Smirnova et al.29
Besides, μs-length unbiased molecular dynamics starting from a well-defined stalk (ξf ∼0.85) verified the existence of the free energy local minimum at 0.5 < ξf < 0.6 and hence the metastable stalk, see Fig. 2b. Panel 2c shows molecular dynamics snapshots of the fusion stalk at different stages for lipids only and waters only, separately.
PMF calculations with the new MARTINI 3 are in good agreement with previous results using the MARTINI 2.2 version,5 although a slight difference in the length of the pre-fusion region (ξf < 0.58) was observed. Under MARTINI 2.2, using the previous model of PIP2 lipids (POP2) and a different set of parameters, the first fusion stalk forms later in the collective variable space (0.65 < ξf < 0.7). This effect could also suggest that MARTINI 3 is prone to membrane fusion although less thermodynamic work would, in principle, be needed to fuse bilayers, as also pointed out by Hub and collaborators.22 Importantly, Vanni and collaborators have recently shown that MARTINI 3 is particularly suitable for characterizing mutagenesis experiments in peripheral proteins while binding to lipid bilayers.30 Such a characteristic is essential for studies (such as the present one) strongly based on relative comparisons between wild-type and mutant states.
2.2 One C2B domain has negligible effects on the fusion stalk free energy profile
Here, using ξf under the MARTINI 3 force-field, with two POPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) POPS
POPS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PIP2 (87.5
PIP2 (87.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5) lipid bilayers with ∼34 cytosolic water beads per nm2, we have shown that the necessary work to induce a fusion stalk between initially planar and parallel bilayers is ∼300 kJ mol−1 (see Fig. 2a), in good agreement with previous results of similar inter-membrane distances.5 Smirnova et al. demonstrated that increasing hydration levels, through larger inter-membrane distances, significantly increase the free energy cost for fusion stalk formation.25 Accordingly, Poojari et al. showed for MARTINI POPC bilayers that the energy cost for the fusion stalk is ∼175 kJ mol−1 for 18 water molecules per nm2.22
2.5) lipid bilayers with ∼34 cytosolic water beads per nm2, we have shown that the necessary work to induce a fusion stalk between initially planar and parallel bilayers is ∼300 kJ mol−1 (see Fig. 2a), in good agreement with previous results of similar inter-membrane distances.5 Smirnova et al. demonstrated that increasing hydration levels, through larger inter-membrane distances, significantly increase the free energy cost for fusion stalk formation.25 Accordingly, Poojari et al. showed for MARTINI POPC bilayers that the energy cost for the fusion stalk is ∼175 kJ mol−1 for 18 water molecules per nm2.22
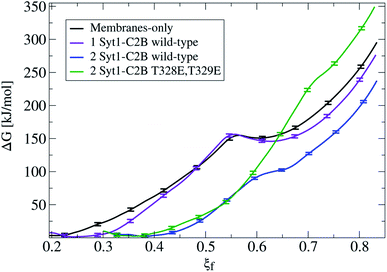
Fig. 3 shows that the introduction of one Syt1–C2B domain in the cytosolic space has little effects on the energy profile, in agreement with our previous study5 (a black line for membrane-only, and a violet line for membranes with one Syt1–C2B). The zero energy reference slightly displaces to the right (from ξf ∼0.2 to ξf ∼0.25) as a result of the inward protein pulling from the bilayers. The fusion stalk energy barrier at ξf ∼0.55 and the local minimum for the metastable stalk at ξf ∼0.6 are almost identical (between black and violet lines). The total cost for a fusion stalk at ξf ∼0.85 is slightly lower (∼275 kJ mol−1) due to the reduction of the inter-membrane distance (by ∼0.2 nm, as measured from unbiased simulations). This result is dependent on the initial parameters of the collective variable (see eqn (1)) that induces the stalk (adjusted here for the membrane-only system), i.e., the cylinder slices (Nsf), slice thickness (dsf), cylinder radius (Rcylf) and slice occupation factor (ζf). As pointed out previously, use of different sets of parameters defines different collective variables with different numerical predictions for the free energy.21
2.3 Two wild-type C2B domains reduce the free energy cost for the fusion stalk while two mutant C2B domains significantly increase it
Importantly, the presence of two Syt1–C2B domains in the cytosolic space drastically changes the free energy profile. Fig. 3 also shows the free energy curve to induce a fusion stalk with two wild-type C2B domains (blue line) and with two mutant T328E and T329E C2B domains (green curve), independently. More than a decade ago, Chapman and collaborators highlighted Syt1–C2B residue 328 as important for Ca2+ triggered fusion31 while carrying out scanning alanine mutagenesis to identify surfaces of Syt1 that may participate in regulated membrane fusion. Also, Mayorga and collaborators11,32 used a synthetic peptide RRLKKKKTTIKKNTL (res 411–425) from the synaptotagmin VI C2B domain to study acrosomal exocytosis. In both their studies,11,32 Thr418 and Thr419 in the polybasic region of the synaptotagmin VI C2B domain were changed to Glu (TE mutant) to mimic the negative charge introduced by phosphorylation. Analogously, we have mutated Thr328 and Thr329 of the Syt1–C2B domain with the same scope.For two wild-type domains (Fig. 3, blue line), it can be observed that both the stalk barrier at ξf ∼0.55 and the local minimum for the metastable stalk at ξf ∼0.6 have vanished. Besides, the zero reference is significantly displaced to the right (from ξf ∼0.2 to ξf ∼0.35), which lowers the total cost for a fusion stalk at ξf ∼0.85 to ΔG ∼240 kJ mol−1. To explore the displacement to the right of the zero-energy reference, we have performed μs-length unbiased molecular dynamics with two Syt1–C2B domains in the cytosolic space between two planar and parallel bilayers. Under these conditions, the measured equilibrium PO4:PO4 inter-membrane distance was ∼1.8 nm, a reduction of 2.1 nm from that of the membrane-only system (see Fig. 5b, blue line). In agreement with this observation, other studies have suggested that Syt1–C2AB domains are configured to bridge two membranes, driving opposed bilayers closer together,19,33 while the C2B domain has been proposed to cooperate with the SNARE complex in also bringing the two membranes together.9,34 Therefore, a slight displacement to the right in the formation of the first stalk is observed for two wild-type domains (see the inset of Fig. 4a).
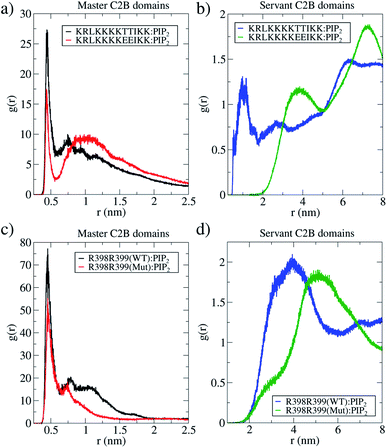
Measurements of PIP2 interactions with polybasic patches in wild-type C2B domains (both master and servant) show that these interactions are responsible for membranes being pulled together. Fig. 5c shows, under unbiased conditions and starting from initially flat and parallel bilayers, how 2 wild-type C2B effectively pull membranes together, inducing a local membrane curvature (see Fig. S6 and S9 in the ESI†). Strategic mutations T328E and T329E11,31,32 suffocate the effect in 2 mutant C2B domains. Accordingly, Fig. 6a and b quantify PIP2 interactions with polybasic patches in both C2B domains (master and servant) for wild-type and mutant cases when the fusion stalk has already formed (ξf ∼0.85). Although master domains (wild-type and mutant) show equivalent interactions between PIP2 and their polybasic patches (Fig. 6a), servant domains do not. Particularly, panel 6b shows increased interactions between PIP2 and wild-type servant C2B polybasic patch, with respect to its mutant counterpart (the green line significantly displaced to the right). Also, a similar effect is observed for PIP2 interactions with arginines 398 and 399 (equivalent interactions for the master domains in Fig. 6c, and the green line also displaced to the right in Fig. 6d).
For two mutant domains (Fig. 3, green line), the curve is similar to that of two wild-type C2B domains (blue line) until ξf ∼0.58. An equivalent displacement to the right of the zero-energy reference is observed, also without the fusion stalk energy barrier nor the metastable stalk. Remarkably, two C2B domains (either wild-type or mutants T328E and T329E) equivalently reduce the free energy in the interval (ξf < 0.58), right before the instant (in the ξf space) when the first stalk forms for the membrane-only system taken as the reference here). However, as observed in μs-length unbiased simulations, the inter-membrane distance is not reduced by two mutant C2B domains as much as it is by the two wild-type ones (see Fig. 5b and c). Consequently, the formation of the first stalk for two mutant domains is significantly delayed in the space of ξf, see Fig. 4a.
For ξf > 0.6, a change of regime takes place for two wild-type domains, and the free energy for two mutant domains significantly increases until reaching a total cost for a fusion stalk at ξf ∼0.85 of ΔG ∼350 kJ mol−1. This effect is due to the uncoordinated action between mutant master and servant C2B domains, as opposed to the cooperation between master and servant wild-type ones. While PIP2 lipids effectively interact with the now mutated polybasic patch KRLKKKKEEIKK in the master domain, as observed from the measurements in Fig. 6a (red line), the mutant servant (6b, green line) shows decreased interactions with respect to the wild-type servant (6b, blue line). This result is in agreement with the experimental description on how mutations lying in the polybasic path of C2B alter the fusion probability.13 Additionally, arginines 398 and 399 also seem to be essential in this free energy increase, describing a similar master-servant behavior, as shown in Fig. 6c (black and red lines), with equivalent master interactions and in 6d (green line) with significantly decreased ones in the mutant servant domain. See Sections 2.5 and 2.6 for more details on the specific PIP2 interactions with the polybasic patch and arginines 398 and 399 for master and servant C2B domains.
Also, we have conducted 10 μs of unbiased molecular dynamics initiated from the stalks at ξf ∼0.85 for systems with two Syt1–C2B domains (both wild-type and T328E and T329E mutants), see Fig. S11† in the ESI.† These simulations stabilize the stalk at ξf ∼0.58, which suggests that a metastable state still exists but is invisible for the collective variable. On the one hand, such a stabilized stalk systematically stretches due to its interactions with C2B domains (see Fig. S12†). On the other hand, this shape distortion is negligible when C2B domains are not present. As mentioned before, in the space of ξf, all stalks are either circular or square toroids (see Section 2.4). We hypothesize that the collective variable is unable to capture the energy minimum when the shape of the stalk is elongated, due to its intrinsic cylindrical geometry. Molecular dynamics snapshots of the stalk after 10 μs of unbiased simulations are provided in Fig. S12 in the ESI.†
2.4 Stalk ordering during membrane fusion: from a circular to a square toroid
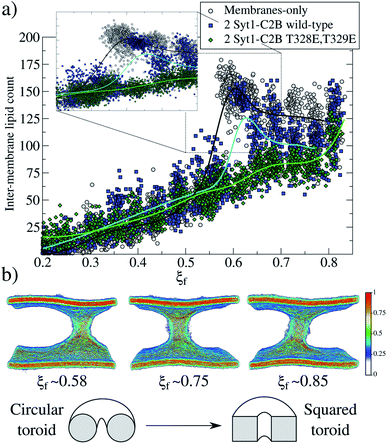
For the membrane-only system, after the first fusion stalk forms at ξf ∼0.58, the amount of lipids in the inter-membrane space has relatively small variations in the interval 0.58 < ξf < 0.85, see Fig. 4a (gray circles and a black line). This result indicates that the stalk does not widen significantly, although the evolution from the first stalk to ξf ∼0.85 is energetically demanding in all cases (see Fig. 3).The reason for almost doubling the free energy with apparently no evident effects is the following: at ξf ∼0.58, the collective variable has the majority of the necessary lipids to form the stalk already in the inter-membrane space, but they are disordered, and not all of them contribute to the collective variable by filling the cylinder slices. As ξf increases (until ξf ∼0.85), more tail beads from the lipid molecules already in the stalk, order themselves to fill the cylinder slices. Time-averaged densities for tail beads only (Fig. 4b) show how the geometry of the stalk changes from a circular-toroid at ξf ∼ 0.58 to a square-toroid at ξf ∼ 0.85.
Importantly, systems with membrane-only and containing 2 Syt1–C2B wild-type domains clearly describe the transition at the moment when the first stalk forms (ξf ∼0.58 for membrane-only, and ξ</i>f ∼0.6 for 2 Syt1–C2B wild-type domains). This event is characterized by an accelerated increase in the number of inter-membrane lipids, ξf ∼0.7 following an almost linear dependency with the amount of inter-membrane lipids, at least right before its final state at ξf ∼0.85. In the following two sections, we propose a master-servant mechanism of cooperation between C2B domains that explains these behaviors.
2.5 The master-servant mechanism (I): polybasic regions 321–332 and arginines 398![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) 399 function as molecular anchors for PIP2 lipids
399 function as molecular anchors for PIP2 lipids
The mechanism used by wild-type C2B domains to reduce the total work needed to induce a fusion stalk is revealed by specific aminoacid–lipid interactions. The polybasic region in Syt1–C2B domains (321–332) has extensively been studied and is thought to be responsible for crucial interactions with anionic PIP2 lipids,35–37 modulating the expansion rate12 and stabilizing the fusion pore5 through PIP2 micro-domains at the fusion sites.5,38 Moreover, PIP2 clusters have been reported to function as molecular beacons during vesicle recruitment.35,38 Recently, Cafiso and collaborators13 have shown that not only the polybasic patch in C2B domains is crucial for membrane fusion but also that arginines 398![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 399 are key during fusion pore expansion. In their study, they demonstrate how the C2B domain makes simultaneous membrane contact with arginines 398
399 are key during fusion pore expansion. In their study, they demonstrate how the C2B domain makes simultaneous membrane contact with arginines 398![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 399, the polybasic region and the Ca2+ binding loops.
399, the polybasic region and the Ca2+ binding loops.
Accordingly, we have performed μs-length unbiased molecular dynamics of 2 wild-type and 2 mutant C2B domains between initially flat and parallel bilayers, and we have analyzed C2B spatial orientations. To do so, we propose the Z-projection of the end-to-end maximum distance for each C2B domain as a measure of molecular alignment to the Z axis (perpendicular to the bilayers). This distance is calculated individually for a C2B domain by finding the maximum 3D distance between two beads belonging to this domain and projecting it to the Z axis. Fig. 5a shows histograms for the end-to-end distances along the Z axis of each C2B domain. Therefore, higher values of this distance indicate a C2B domain aligned with its normal axis to the bilayers, while lower values suggest a horizontally oriented domain (parallel to the bilayers). Fig. S10† shows the relation of this distance to an orientation angle for each C2B.
It can be observed that, in average, for both independent simulations, one domain (the master) orients itself parallel to the Z axis while the other (the servant) tends to be perpendicular to it. For the same unbiased trajectories, Fig. 5b shows the PO4:PO4 inter-membrane distance for the three systems: membrane-only (black line), 2 wild-type C2B domains (blue line) and 2 mutant C2B domains (green line). In the long run, 2 wild-type C2B domains significantly pull membranes together, reducing their PO4:PO4 inter-membrane distance. Importantly, this effect is negligible for 2 mutant C2B domains. Such membrane pulling is then due to interactions between the polybasic patch KRLKKKKTTIKK and PIP2 lipids, as shown here by the radial distribution function (RDF) measurements in Fig. 6 and in agreement with previous experimental studies.35–37 Also, supplementary Fig. S8† shows RDF calculations with POPS lipids, including the interaction with individual K326 and K327 residues in the polybasic patch and arginines 398 and 399. These data show that POPS interaction with key residues in C2B domains is also relevant, although of the second order with respect to the ones with PIP2. Therefore, these results also point to the interactions between PIP2 lipids and polybasic patches in wild-type domains as the key for membrane pulling.
Due to a saturation effect intrinsic to the geometry of the collective variable (see ESI, Fig. S13 and S15†), once the slices of the membrane spanning the cylinder are full (containing tail beads from opposed bilayers), further membrane pulling has no additional effects on ξf. Therefore, although inter-membrane distances are significantly different for systems with 2 wild-type and 2 mutant C2B domains (see Fig. 5b, blue and green lines), their free energy profiles show the same zero reference at ξf ∼0.35 (see Fig. 3).
Additionally, Fig. 5c shows averaged densities describing the induced curvature by 2 wild-type C2B domains (left) in contrast to almost planar bilayers with 2 mutant C2B domains (right). Noticeably, 2 Syt1–C2B wild-type domains locally bend the bilayers around the C2B location. This effect is not observable for 2 mutant domains nor for membrane-only systems. See supplementary Fig. S9† for a plot measuring the inter-membrane PO4:PO4Z distance as a function of the radial XY distance to the center of the defect, for all three systems under study.
Furthermore, we have measured the interactions between the polybasic region (321–332) with PIP2 lipids and of arginines 398![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 399 with the same PIP2 lipids, along two independent μs-length unbiased molecular dynamics simulations. One of these simulations contained 2 wild-type C2B domains and the other, 2 mutant C2B domains, both around a fusion stalk of ξf ∼ 0.85. Fig. 6 shows RDFs for all PIP2 lipids in the bilayers alternatively measured from the polybasic region 321–332 (panels 6a and b) and arginines 398
399 with the same PIP2 lipids, along two independent μs-length unbiased molecular dynamics simulations. One of these simulations contained 2 wild-type C2B domains and the other, 2 mutant C2B domains, both around a fusion stalk of ξf ∼ 0.85. Fig. 6 shows RDFs for all PIP2 lipids in the bilayers alternatively measured from the polybasic region 321–332 (panels 6a and b) and arginines 398![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 399 (panels 6c and d). For all wild-type domains, black lines represent the C2B domain that directly interacts with the stalk (the master), while blue lines represent the C2B domain that indirectly interacts with the stalk (the servant). See the ESI† for details on the calculation of RDFs. Analogously for all mutant domains, red lines represent the C2B domain that directly interacts with the stalk (the master), and green lines represent the C2B domain that indirectly interacts with the stalk (the servant).
399 (panels 6c and d). For all wild-type domains, black lines represent the C2B domain that directly interacts with the stalk (the master), while blue lines represent the C2B domain that indirectly interacts with the stalk (the servant). See the ESI† for details on the calculation of RDFs. Analogously for all mutant domains, red lines represent the C2B domain that directly interacts with the stalk (the master), and green lines represent the C2B domain that indirectly interacts with the stalk (the servant).
Black lines in panels 6a and c show that the wild-type master domain highly coordinates with PIP2 lipids (peaks at r ∼0.5 nm) through both its polybasic patch and its arginines 398![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 399. Simultaneously, blue lines in panels 6b and d show that the servant wild-type domain also coordinates well with PIP2 lipids through its polybasic patch (peak at r ∼ 1 nm) with a less frequent interaction of its arginines 398
399. Simultaneously, blue lines in panels 6b and d show that the servant wild-type domain also coordinates well with PIP2 lipids through its polybasic patch (peak at r ∼ 1 nm) with a less frequent interaction of its arginines 398![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 399 with PIP2 (peak at r ∼ 4 nm). Comparing servant domains (blue and green lines), wild-type ones show systematically better coordination with PIP2 lipids, both from their polybasic patches and their arginines 398
399 with PIP2 (peak at r ∼ 4 nm). Comparing servant domains (blue and green lines), wild-type ones show systematically better coordination with PIP2 lipids, both from their polybasic patches and their arginines 398![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 399.
399.
In the ESI†, two additional systems are described. The first one contains a tandem C2A–C2B of Syt1 domains (see Fig. S15†) using the crystal structure of the human synaptotagmin 1 C2A–C2B (PDB ID: 2R83).39 This system has shown to be the most effective in pulling membranes together, among the ones studied in this work (see Fig. S15b†). This result is in agreement with previous experimental studies, suggesting that Syt1 C2A–C2B domains drive opposed bilayers closer together.19,33 Accordingly, PMF calculations show a slight decrease in the free energy to induce the fusion stalk, with respect to the system containing two wild-type C2B domains (see Fig. S15a†). In the physiological context, more than one copy of a Syt1–C2 domain is required for catalyzing membrane fusion. Consequently, the presence of the C2A domain significantly lowers the free energy required to induce the fusion stalk, stabilizing the inter-membrane distance at the lowest value (∼1 nm) among the cases studied here. These results are in agreement with the experimental literature describing the behavior of Syt1–C2A–C2B with lipid bilayers19,33 and quantitatively supporting the simulation framework along this work for the fusion stalk.
The second system contains two C2B domains, one wild-type and one mutant (T328E and T329E), see Fig. S16.† The inter-membrane distance for this system (see Fig. S16b†) shows a slightly more effective membrane pulling than the system with 2 Syt1–C2B mutant domains, but a significant drawback when compared to the system with 2 Syt1–C2B wild-type domains. In agreement with the master-servant mechanism proposed here, replacing one wild-type C2B by a mutant T328E or T329E C2B weakens PIP2 interactions with the polybasic patches, hence, reducing the capability of the domains to pull membranes together. In agreement, PMF calculations (see Fig. S16a†) show that the free energy cost to induce a fusion stalk is slightly higher than the system with 2 Syt1–C2B wild-type domains, as expected for such an intermediate arrangement. Therefore, even under the wild-type/mutant scheme, these results support the mechanism proposed for a master-servant cooperation falling between the already described more extreme cases of 2 wild-type and 2 mutant C2B domains.
2.6 The master-servant mechanism (II): T328E and T329E mutations in C2B domains disrupt the cooperation
The same analysis was applied to 2 mutant C2B domains around an equivalent fusion stalk of ξf ∼ 0.85. Even with T328E and T329E mutations, the mutant master domain (red lines in panels 6a and c) shows good interactions with PIP2 lipids through both its polybasic patch and its arginines 398![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 399, although less frequent than those in the wild-type master domain (black lines). These results indicate that the main interactions between PIP2 and master C2B domains (either wild-type or mutant) are in the polybasic region 321–332, in line with previous studies describing PIP2 mediated membrane bending35,40–42 and fusion.41,43–45 Additionally, arginines 398
399, although less frequent than those in the wild-type master domain (black lines). These results indicate that the main interactions between PIP2 and master C2B domains (either wild-type or mutant) are in the polybasic region 321–332, in line with previous studies describing PIP2 mediated membrane bending35,40–42 and fusion.41,43–45 Additionally, arginines 398![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 399 appear to be key during C2B:PIP2 interactions for both wild-type and mutant master domains, in agreement with previous data as well.13
399 appear to be key during C2B:PIP2 interactions for both wild-type and mutant master domains, in agreement with previous data as well.13
However, servant domains exhibit a different behavior: while the wild-type servant keeps high interactions with PIP2 lipids through its polybasic patch, the mutant servant is unable to keep up (see panel 6). Also, a marginal reduction of arginines 398![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 399 with PIP2 interactions is observed between wild-type and mutant servants (see panel 6d).
399 with PIP2 interactions is observed between wild-type and mutant servants (see panel 6d).
The convenience of the RDF analysis becomes relevant when measurements are compared relatively between wild-type and mutant masters (and between wild-type and mutant servants). Therefore, panels 6a and c show respectively that master domains function equivalently in terms of their polybasic patches and interactions of arginines 398![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 399 with PIP2 lipids. However, panels 6b and d reveal a significantly different behavior of the servant domains, only due to mutations T328E and T329E. The evidence of the master-servant cooperation is then highlighted by an effective wild-type servant (blue line in panel 6b) with respect to an uninvolved mutant servant (green line in panel 6b), in terms of interactions with PIP2. A similar effect, although modest with respect to the polybasic patch, is observed in panel 6d for arginines 398 and 399.
399 with PIP2 lipids. However, panels 6b and d reveal a significantly different behavior of the servant domains, only due to mutations T328E and T329E. The evidence of the master-servant cooperation is then highlighted by an effective wild-type servant (blue line in panel 6b) with respect to an uninvolved mutant servant (green line in panel 6b), in terms of interactions with PIP2. A similar effect, although modest with respect to the polybasic patch, is observed in panel 6d for arginines 398 and 399.
The behavior showed by servant C2B domains when interacting with PIP2 reveals a cooperation mechanism with their respective masters, which we have been able to control with selected mutations identified in previous experimental studies.11,31,32 We expect this mechanism to function, in general terms, always in pairs of master-servant C2B domains. Therefore, simulations of larger membrane patches including more pairs of C2B domains could demonstrate the cumulative effect of the master-servant mechanism in controlling the fusion stalk.
Unfortunately, collective variable driven molecular dynamics for such systems are still prohibitive for the majority of researchers. A system containing just four C2B domains may require a membrane patch of ∼30 × 30 nm to avoid periodic interference. Two bilayers of such size would contain more than ∼8000 lipids and at least 120 × 103 water molecules. With the performance of the collective variable implemented in PLUMED in the order of ∼20% with respect to unbiased simulations, PMF calculations would require an uncomfortable amount of time, even for a modest supercomputer center.
3 Conclusions
Altogether, these data verify that C2B has two important regions that interact with anionic lipids, namely the well-known polybasic region KRLKKKKTTIKK (positions 321–332) and the recently described arginines 398![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 399.13 We observed a unique behavior of cooperation between identical C2B domains that facilitates the formation of the fusion stalk, as demonstrated by the free energy profile in Fig. 3 (blue line). While one domain (the master) binds to PIP2 lipids through its polybasic region and its arginines 398
399.13 We observed a unique behavior of cooperation between identical C2B domains that facilitates the formation of the fusion stalk, as demonstrated by the free energy profile in Fig. 3 (blue line). While one domain (the master) binds to PIP2 lipids through its polybasic region and its arginines 398![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 399 (see Fig. 6a and c, black lines), the other domain (the servant) successfully copies this behavior for its wild-type state (see Fig. 6b and d, blue lines). Remarkably, panels 6b and d show that wild-type servant domains are significantly more effective in their interactions with PIP2 than their mutant equivalents (comparing green and blue lines, either for polybasic patches in 6b or for arginines 398
399 (see Fig. 6a and c, black lines), the other domain (the servant) successfully copies this behavior for its wild-type state (see Fig. 6b and d, blue lines). Remarkably, panels 6b and d show that wild-type servant domains are significantly more effective in their interactions with PIP2 than their mutant equivalents (comparing green and blue lines, either for polybasic patches in 6b or for arginines 398![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 399 in 6d). Together, both wild-type C2B domains anchor PIP2 lipids from different regions to cooperatively reduce the free energy for the fusion stalk to form.
399 in 6d). Together, both wild-type C2B domains anchor PIP2 lipids from different regions to cooperatively reduce the free energy for the fusion stalk to form.
In silico mutagenesis (T328E and T329E) in both C2B domains not only terminates any cooperation to induce the fusion stalk but also increases the associated total work required, making the fusion event thermodynamically more difficult, with respect to the membrane-only system (see Fig. 3, black and green lines). Remarkably, in the presence of 2 Syt1–C2B mutant domains, the formation of the stalk takes place gradually, with a linear dependence on the lipid population of the stalk (see Fig. 4a, green line). This behavior contrasts with the drastic increase of the amount of lipids in the stalk when 2 Syt1–C2B wild-type domains control the process (blue line).
In terms of its interactions with PIP2 lipids, the mutant master domain suffers minor changes with respect to its wild-type counterpart (see Fig. 6a and c). It is the servant mutant domain who is unable to sustain PIP2 interactions with neither its polybasic patch nor its arginines 398![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 399 (see Fig. 6b and d). We propose that these reduced interactions are the reason for the whole disruption of the master-servant cooperation mechanism, ultimately responsible for the energetics of the fusion stalk.
399 (see Fig. 6b and d). We propose that these reduced interactions are the reason for the whole disruption of the master-servant cooperation mechanism, ultimately responsible for the energetics of the fusion stalk.
4 Computational methods
We have conducted all our simulations using Gromacs-2020.5,46–48 PLUMED-2.7.2 (ref. 24) and the Martini 3 coarse-grained model.49 Molecular dynamics simulations used the semi-isotropic NPT ensemble and a time step of 20 fs in all cases. The temperature was set to T = 303.15 K (ref. 6 and 50–53) and was controlled by a V-rescale thermostat54 with a coupling constant of 1 ps. The pressure was set at 1.0 bar with a compressibility equal to 3 × 10−4 bar−1, using a Parrinello–Rahman barostat55 with a 12 ps time constant. Neighbor search used a Verlet cut-off scheme with a buffer tolerance of 0.005 kJ mol−1 ps−1 and an update-frequency for the neighbor list equal to 25 steps. Periodic boundary conditions (PBCs) were used in all directions. Coulomb interactions used the reaction field method with a cut-off of 1.1 nm and a relative dielectric constant of 2.5. Van der Waals interactions followed a cut-off scheme set to 1.1 nm. All simulations in this work were conducted in a calcium-independent manner.In all cases, we have used a pair of lipid bilayers containing 1024 molecules each. These bilayer patches of ∼17 × 17 nm ensure negligible finite-size effects due to interactions between periodic images of the fusion pore.5,56 In all cases, the pair of bilayers were solvated in more than ∼30 × 103 W coarse-grained water molecules to fulfill the ample water conditions for MARTINI.57 The PO4:PO4 inter-membrane separation was adjusted to equilibrate at ∼3.9 nm to fit one and two Syt1–C2B domains. This inter-membrane distance results in ∼10 × 103 W water beads in the cytosolic space. PIP2 lipids for MARTINI 3 were modeled following a parametrization by Melo and collaborators58 (https://github.com/MeloLab/PhosphoinositideParameters).
Figures were created using Visual Molecular Dynamics (VMD),59 the academic version of Maestro,60 Grace (GRaphing, Advanced Computation and Exploration of data),61 Inkscape,62 GIMP (GNU Image Manipulation Program)63 and Gnuplot.64 Averaged densities from molecular dynamics simulations were generated using GROmaρs65 and PyMOL.60
4.1 PLUMED implementation of the collective variable
We have implemented the collective variable to induce membrane fusion as a modular C++ file compilable with PLUMED. The file is freely available at https://github.com/lautarodibartolo/MemFusion together with a README file and an example input system. Additionally, in the ESI†, we have included an example input file for PLUMED-2.7.2 to induce the fusion stalk using the same POPC:POPS:PIP2 bilayers described in this work. See the ESI† for more details on the collective variable (Fig. S13†).4.2 PMF calculations
Free energy profiles were computed with umbrella sampling66,67 in PLUMED24 and recovered using the Weighted Histogram Analysis Method (WHAM) using an implementation developed by Prof. Grossfield.68 Fusion stalk free energy profiles required between 16 and 18 windows to span ξf in the interval [0.2, 0.85] (depending on the simulation system, see the ESI† for details) using, in all cases, a force constant k = 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 kJ mol−1. All windows used to recover the free energy profile contained at least 100 ns in the steady-state regime, although the total simulation time required for each window varied (from 110 ns to 210 ns) depending on the region of the profile. See Fig. S1 to S4 in the ESI† for the convergence analysis on the free energy profiles and for technical details on umbrella sampling window distribution.
000 kJ mol−1. All windows used to recover the free energy profile contained at least 100 ns in the steady-state regime, although the total simulation time required for each window varied (from 110 ns to 210 ns) depending on the region of the profile. See Fig. S1 to S4 in the ESI† for the convergence analysis on the free energy profiles and for technical details on umbrella sampling window distribution.
Data availability
The following link provides access to computational data associated with this work: https://github.com/lautarodibartolo/MemFusion.Author contributions
DM conceived the idea and supervised the research. DM and ALDB designed the computational experiments and performed the numerical simulations. ALDB wrote the C++ PLUMED implementation of the code in consultation with DM. DM wrote the manuscript in consultation with ALDB. Both authors discussed the results and the conclusions.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The supercomputing time for this work was provided by CCAD (Centro de Computación de Alto Desempeño de la Universidad Nacional de Córdoba). Grants from CONICET (PIP-13CO01) and ANPCyT (PICT2017-1002) as well as GPU hardware granted by the NVIDIA Corporation are gratefully acknowledged. The authors thank Prof. Jochen Hub for generously sharing his GROMACS source code with us, Prof. Giovanni Bussi for his useful advice in PLUMED C++ coding and Dr Mariano Polo for critically reading the final manuscript.Notes and references
- M. F. Quevedo, M. A. Bustos, D. Masone, C. M. Roggero, D. M. Bustos and C. N. Tomes, Biochim. Biophys. Acta, Mol. Cell Res., 2019, 1866, 612–622 CrossRef CAS PubMed.
- A. T. Brunger, U. B. Choi, Y. Lai, J. Leitz and Q. Zhou, Annu. Rev. Biophys., 2018, 47, 469–497 CrossRef CAS PubMed.
- C.-W. Chang, C.-W. Chiang and M. B. Jackson, J. Gen. Physiol., 2017, 149, 301–322 CrossRef CAS PubMed.
- D. Das, H. Bao, K. C. Courtney, L. Wu and E. R. Chapman, Nat. Commun., 2020, 11, 231 CrossRef CAS PubMed.
- M. Caparotta, C. N. Tomes, L. S. Mayorga and D. Masone, J. Chem. Theory Comput., 2020, 16, 7840–7851 CrossRef CAS PubMed.
- D. Masone, M. Uhart and D. M. Bustos, J. Chem. Theory Comput., 2018, 14, 2240–2245 CrossRef CAS PubMed.
- E. Karatekin, FEBS Lett., 2018, 592, 3563–3585 CrossRef CAS PubMed.
- I. Fernandez, D. Arac, J. Ubach, S. H. Gerber, O.-h. Shin, Y. Gao, R. G. W. Anderson, T. C. Sudhof and J. Rizo, Neuron, 2001, 32, 1057–1069 CrossRef CAS PubMed.
- S. Wang, Y. Li and C. Ma, eLife, 2016, 5, e14211 CrossRef PubMed.
- Y. Lai, X. Lou, J. Diao and Y.-K. Shin, Sci. Rep., 2015, 5, 9267 CrossRef CAS PubMed.
- C. M. Roggero, G. A. De Blas, H. Dai, C. N. Tomes, J. Rizo and L. S. Mayorga, J. Biol. Chem., 2007, 282, 26335–26343 CrossRef CAS PubMed.
- M. Bendahmane, K. P. Bohannon, M. M. Bradberry, T. C. Rao, M. W. Schmidtke, P. S. Abbineni, N. L. Chon, S. Tran, H. Lin, E. R. Chapman, J. D. Knight and A. Anantharam, Mol. Biol. Cell, 2018, 29, 834–845 CrossRef CAS PubMed.
- S. B. Nyenhuis, N. Karandikar, V. Kiessling, A. J. B. Kreutzberger, A. Thapa, B. Liang, L. K. Tamm and D. S. Cafiso, Nat. Commun., 2021, 12, 761 CrossRef CAS PubMed.
- J. Bai, W. C. Tucker and E. R. Chapman, Nat. Struct. Mol. Biol., 2004, 11, 36–44 CrossRef CAS PubMed.
- L. Li, O.-H. Shin, J.-S. Rhee, D. Arac, J.-C. Rah, J. Rizo, T. Sudhof and C. Rosenmund, J. Biol. Chem., 2006, 281, 15845–15852 CrossRef CAS PubMed.
- A. Radhakrishnan, A. Stein, R. Jahn and D. Fasshauer, J. Biol. Chem., 2009, 284, 25749–25760 CrossRef CAS PubMed.
- W. Kuo, D. Z. Herrick, J. F. Ellena and D. S. Cafiso, J. Mol. Biol., 2009, 387, 284–294 CrossRef CAS PubMed.
- G. van den Bogaart, S. Thutupalli, J. H. Risselada, K. Meyenberg, M. Holt, D. Riedel, U. Diederichsen, S. Herminghaus, H. Grubmüller and R. Jahn, Nat. Struct. Mol. Biol., 2011, 18, 805–812 CrossRef CAS PubMed.
- C. Gruget, O. Bello, J. Coleman, S. S. Krishnakumar, E. Perez, J. E. Rothman, F. Pincet and S. H. Donaldson, Sci. Rep., 2020, 10, 18011 CrossRef CAS PubMed.
- Z. Wu, N. Dharan, Z. A. McDargh, S. Thiyagarajan, B. O'Shaughnessy and E. Karatekin, eLife, 2021, 10, e68215 CrossRef CAS PubMed.
- J. S. Hub and N. Awasthi, J. Chem. Theory Comput., 2017, 13, 2352–2366 CrossRef CAS PubMed.
- C. S. Poojari, K. C. Scherer and J. S. Hub, Nat. Commun., 2021, 12, 6594 CrossRef CAS PubMed.
- A. Perez-Lara, A. Thapa, S. B. Nyenhuis, D. A. Nyenhuis, P. Halder, M. Tietzel, K. Tittmann, D. S. Cafiso and R. Jahn, eLife, 2016, 5, e15886 CrossRef PubMed.
- G. Tribello, M. Bonomi, D. Branduardi, C. Camilloni and G. Bussi, Comput. Phys. Commun., 2014, 185, 604–613 CrossRef CAS.
- Y. G. Smirnova, H. J. Risselada and M. Müller, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 2571–2576 CrossRef CAS PubMed.
- C. Neale and R. Pomes, Biochim. Biophys. Acta, Biomembr., 2016, 1858, 2539–2548 CrossRef CAS PubMed.
- N. Awasthi and J. S. Hub, J. Chem. Theory Comput., 2016, 12, 3261–3269 CrossRef CAS PubMed.
- D. A. Pearlman and P. A. Kollman, J. Chem. Phys., 1989, 91, 7831–7839 CrossRef CAS.
- Y. G. Smirnova, M. Fuhrmans, I. A. B. Vidal and M. Müller, J. Phys. D: Appl. Phys., 2015, 48, 343001 CrossRef.
- S. Srinivasan, V. Zoni and S. Vanni, Faraday Discuss., 2021, 232, 131–148 RSC.
- J. D. Gaffaney, F. M. Dunning, Z. Wang, E. Hui and E. R. Chapman, J. Biol. Chem., 2008, 283, 31763–31775 CrossRef CAS PubMed.
- C. M. Roggero, C. N. Tomes, G. A. De Blas, J. Castillo, M. A. Michaut, M. Fukuda and L. S. Mayorga, Dev. Biol., 2005, 285, 422–435 CrossRef CAS PubMed.
- W. Kuo, D. Z. Herrick and D. S. Cafiso, Biochemistry, 2011, 50, 2633–2641 CrossRef CAS PubMed.
- K. D. Brewer, T. Bacaj, A. Cavalli, C. Camilloni, J. D. Swarbrick, J. Liu, A. Zhou, P. Zhou, N. Barlow, J. Xu, A. B. Seven, E. A. Prinslow, R. Voleti, D. Häussinger, A. M. J. J. Bonvin, D. R. Tomchick, M. Vendruscolo, B. Graham, T. C. Südhof and J. Rizo, Nat. Struct. Mol. Biol., 2015, 22, 555–564 CrossRef CAS PubMed.
- A. Honigmann, G. van den Bogaart, E. Iraheta, H. J. Risselada, D. Milovanovic, V. Mueller, S. Mullar, U. Diederichsen, D. Fasshauer, H. Grubmuller, S. W. Hell, C. Eggeling, K. Kuhnel and R. Jahn, Nat. Struct. Mol. Biol., 2013, 20, 679–686 CrossRef CAS PubMed.
- Y. Park, J. B. Seo, A. Fraind, A. Perez-Lara, H. Yavuz, K. Han, S.-R. Jung, I. Kattan, P. J. Walla, M. Choi, D. S. Cafiso, D.-S. Koh and R. Jahn, Nat. Struct. Mol. Biol., 2015, 22, 815–823 CrossRef CAS PubMed.
- Z. Wu, L. Ma, J. Zhu, N. Courtney, Y. Zhang, E. R. Chapman and E. Karatekin, Biophys. J., 2020, 118, 400a CrossRef.
- G. van den Bogaart, K. Meyenberg, H. J. Risselada, H. Amin, K. I. Willig, B. E. Hubrich, M. Dier, S. W. Hell, H. Grubmüller, U. Diederichsen and R. Jahn, Nature, 2011, 479, 552–555 CrossRef CAS PubMed.
- K. L. Fuson, M. Montes, J. J. Robert and R. B. Sutton, Biochemistry, 2007, 46, 13041–13048 CrossRef CAS PubMed.
- S. Shukla, R. Jin, J. Robustelli, Z. E. Zimmerman and T. Baumgart, Biophys. J., 2019, 117, 962–974 CrossRef CAS PubMed.
- H. T. McMahon, M. M. Kozlov and S. Martens, Cell, 2010, 140, 601–605 CrossRef CAS PubMed.
- K. L. Lynch, R. R. L. Gerona, D. M. Kielar, S. Martens, H. T. McMahon and T. F. J. Martin, Mol. Biol. Cell, 2008, 19, 5093–5103 CrossRef CAS PubMed.
- L. Mu, Z. Tu, L. Miao, H. Ruan, N. Kang, Y. Hei, J. Chen, W. Wei, F. Gong, B. Wang, Y. Du, G. Ma, M. W. Amerein, T. Xia and Y. Shi, Nat. Commun., 2018, 9, 4259 CrossRef PubMed.
- Y. Park and J.-K. Ryu, FEBS Lett., 2018, 592, 3480–3492 CrossRef CAS PubMed.
- D. J. James, C. Khodthong, J. A. Kowalchyk and T. F. J. Martin, J. Cell Biol., 2008, 182, 355–366 CrossRef CAS PubMed.
- D. van der Spoel, E. Lindahl, B. Hess, G. Groenhof, A. E. Mark and H. J. C. Berendsen, J. Comput. Chem., 2005, 26, 1701–1718 CrossRef CAS PubMed.
- S. Pronk, S. Páll, R. Schulz, P. Larsson, P. Bjelkmar, R. Apostolov, M. R. Shirts, J. C. Smith, P. M. Kasson, D. van der Spoel, B. Hess and E. Lindahl, Bioinformatics, 2013, 29, 845–854 CrossRef CAS PubMed.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess and E. Lindahl, SoftwareX, 2015, 1–2, 19–25 CrossRef.
- P. C. T. Souza, R. Alessandri, J. Barnoud, S. Thallmair, I. Faustino, F. Grünewald, I. Patmanidis, H. Abdizadeh, B. M. H. Bruininks, T. A. Wassenaar, P. C. Kroon, J. Melcr, V. Nieto, V. Corradi, H. M. Khan, J. Domański, M. Javanainen, H. Martinez-Seara, N. Reuter, R. B. Best, I. Vattulainen, L. Monticelli, X. Periole, D. P. Tieleman, A. H. de Vries and S. J. Marrink, Nat. Methods, 2021, 18, 382–388 CrossRef CAS PubMed.
- M. Caparotta, D. M. Bustos and D. Masone, Phys. Chem. Chem. Phys., 2020, 22, 5255–5263 RSC.
- S. Jo, J. B. Lim, J. B. Klauda and W. Im, Biophys. J., 2009, 97, 50–58 CrossRef CAS PubMed.
- H. Rui, K. I. Lee, R. W. Pastor and W. Im, Biophys. J., 2011, 100, 602–610 CrossRef CAS PubMed.
- S. Jo, T. Kim and W. Im, PLoS One, 2007, 2, e880 CrossRef PubMed.
- G. Bussi, D. Donadio and M. Parrinello, J. Chem. Phys., 2007, 126, 014101 CrossRef PubMed.
- M. Parrinello and A. Rahman, J. Appl. Phys., 1981, 52, 7182–7190 CrossRef CAS.
- M. Caparotta, M. Puiatti and D. Masone, Soft Matter, 2021, 17, 8314–8321 RSC.
- H. I. Ingolfsson, M. N. Melo, F. J. van Eerden, C. Arnarez, C. A. Lopez, T. A. Wassenaar, X. Periole, A. H. de Vries, D. P. Tieleman and S. J. Marrink, J. Am. Chem. Soc., 2014, 136, 14554–14559 CrossRef CAS PubMed.
- L. Borges-Araújo, P. C. T. Souza, F. Fernandes and M. N. Melo, J. Chem. Theory Comput., 2022, 18, 357–373 CrossRef PubMed.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS PubMed.
- Schrödinger LLC, New York, NY, 2015.
- Grace Development Team, GRACE: GRaphing, Advanced Computation and Exploration of data, https://plasma-gate.weizmann.ac.il/Grace/ Search PubMed.
- Inkscape Project, Inkscape, https://inkscape.org Search PubMed.
- The GIMP Development Team, GIMP, https://www.gimp.org Search PubMed.
- T. Williams, C. Kelley and et al., Gnuplot 4.6: an interactive plotting program, http://gnuplot.sourceforge.net/, 2013 Search PubMed.
- R. Briones, C. Blau, C. Kutzner, B. L. de Groot and C. Aponte-Santamaría, Biophys. J., 2019, 116, 4–11 CrossRef CAS PubMed.
- G. Torrie and J. Valleau, J. Comput. Phys., 1977, 23, 187–199 CrossRef.
- B. Roux, Comput. Phys. Commun., 1995, 91, 275–282 CrossRef CAS.
- A. Grossfield, WHAM: the weighted histogram analysis method, http://membrane.urmc.rochester.edu/wordpress/?pageid=126,%20version%202.0.9.1 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Umbrella sampling convergence analysis; time-averaged densities for stalk formation; molecular dynamics snapshots in the μs scale; lipid species density profiles; membrane curvature measurements; inter-membrane lipid count over unbiased simulations, radial distribution function calculation details, additional simulation systems (tandem C2A–C2B and wild-type/mutant C2B domains), collective variable physical interpretation (with mathematical details) and example input parameters for PLUMED. See DOI: 10.1039/d1sc06711g |
| This journal is © The Royal Society of Chemistry 2022 |