Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Bottom-up supramolecular assembly in two dimensions
Ignacio
Insua
 ,
Julian
Bergueiro
,
Julian
Bergueiro
 ,
Alejandro
Méndez-Ardoy
,
Alejandro
Méndez-Ardoy
 ,
Irene
Lostalé-Seijo
,
Irene
Lostalé-Seijo
 and
Javier
Montenegro
and
Javier
Montenegro
 *
*
Centro Singular de Investigación en Química Biolóxica e Materiais Moleculares (CiQUS), Departamento de Química Orgánica, Universidade de Santiago de Compostela, 15705, Spain. E-mail: javier.montenegro@usc.es
First published on 19th January 2022
Abstract
The self-assembly of molecules in two dimensions (2D) is gathering attention from all disciplines across the chemical sciences. Attracted by the interesting properties of two-dimensional inorganic analogues, monomers of different chemical natures are being explored for the assembly of dynamic 2D systems. Although many important discoveries have been already achieved, great challenges are still to be addressed in this field. Hierarchical multicomponent assembly, directional non-covalent growth and internal structural control are a just a few of the examples that will be discussed in this perspective about the exciting present and the bright future of two-dimensional supramolecular assemblies.
1. Introduction
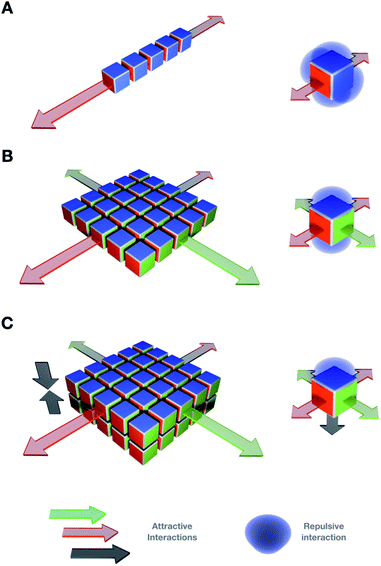
The discovery of graphene in 2004 sparked interest in 2D materials with broad cross-disciplinary resonance.1 In the two following decades, other covalent 2D architectures2 and inorganic allotropes were developed, such as elemental 2D films3 and metal–organic frameworks,4 amongst others.5 The intriguing properties of different 2D molecular systems are currently attracting great attention towards tackling important societal challenges such as green energy sourcing,6 new-generation catalysts7 and biomedical technologies.8 Alternatively to static covalent and inorganic nanomaterials, supramolecular 2D assemblies allow controlled structural and functional reconfiguration.9 The design of small self-assembling monomers bearing orthogonal and directional binding motifs has led to a plethora of supramolecular 2D nanomaterials.8,10 The non-covalent foundation of these systems allows the dynamic exchange of monomers and structural remodelling in response to external conditions (e.g. temperature, pH, and ionic strength),11 which endows the resulting materials with new functional properties such as self-healing,12 polymorphism,13 or molecular capture and sensing.14–16 Despite the continuous development of new supramolecular 2D materials, there is still a know-how gap in predicting the supramolecular outcome of monomers at the molecular design stage. The establishment of precise assembly guidelines remains elusive, so each new monomer must undergo its own structural optimisation. Since a single monomer can in many cases self-organise into several supramolecular ensembles, many efforts currently focus on controlling the assembly pathway towards isolating a single supramolecular product (see Section 2).17,18 To control competing assembly pathways, external conditions such as heating–cooling ramps, solvent exchange or time-dependent structural interconversion must be screened.19Simple molecules with few non-covalent binding domains, which would not be localised or directionally restricted, can display high rotational freedom and different potential assembly pathways. However, under specific external conditions (e.g. physiological), the highest degree of structural accuracy and assembly reproducibility is achieved by complex monomers with a high number of precisely positioned non-covalent binding points (e.g. tubulin). In this framework, a two-dimensional assembly is achieved by controlling the orientation of non-covalent interactions on the suitable synthetic monomers (Fig. 1), whereas high internal order would be required to control material properties such as surface functionalisation and morphology; static crystalline assemblies display limited stimuli-responsiveness due to a high structural stability. Hence, a key challenge towards new-generation 2D supramolecular materials lies in understanding the geometrical constraints required for monomers to assemble highly ordered molecular lattices and avoid competing assembling pathways, while maintaining an adaptive stimuli-responsive behaviour.
In this perspective, we will discuss concepts in supramolecular polymerisation and monomer design distilled from the literature on aqueous 2D assemblies.8 The ideas presented here navigate the structure-assembly relationships of monomers, aiming to balance assembly complexity and ordered monomer packing in 2D. Firstly, supramolecular polymerisation strategies will be presented with a focus on controlling polydispersity, aspect ratio and other key structural features of 2D nanomaterials. Secondly, monomer design will be discussed from a geometrical perspective, addressing the orientation of supramolecular interactions to promote internal order in 2D ensembles. Finally, a discrete collection of edge applications in chemistry, materials science and biology is presented to highlight the current and potential uses of these advanced nanomaterials.
2. Assembly mechanisms and strategies
Control over the assembly mechanisms is key to direct precise supramolecular architectures with dimensional order. A combination of at least two orthogonal non-covalent forces is required to elongate monomers in 2D (Fig. 1).However, the inherent complexity of non-covalent interactions still hinders a precise framework for multidimensional assembly prediction.20,21 Recent discoveries contribute to the potential prediction of supramolecular multiscale materials by molecular design, both generally and specifically in 2D. Different concepts and mechanisms important for two-dimensional supramolecular polymerisation will be discussed in this section.
2.1. Energy landscape
Contrary to covalent systems, supramolecular polymers are frequently under thermodynamic control, particularly under dilute conditions. Nonetheless, supramolecular assemblies have also shown kinetic control in solution.22 This scenario evolves into the definition of pathway complexity, which addresses the variety of supramolecular products that can originate in solution from a single monomer.18 Small changes in the monomer structure can produce radical changes in the supra-polymerisation mechanism, transiting from thermodynamic to kinetic control and even generating different metastable states.23 Even small amounts of co-solvent in the ppm range can also alter the system's energy and the expected assembly pathway.24 Solution properties such as ionic strength may also impact the shape and internal order of the assembly.25 Environmental factors such as mechanical stress produced at moving air–water interfaces can play an important role in the energetic assembly pathway.13 The control of the assembly pathway can also have a drastic effect on the size and number of defects of supramolecular 2D assemblies.26,27 Therefore, all factors affecting the assembly pathway have to be considered when designing a potential monomer for 2D self-assembly.282.2. Suprapolymerisation mechanism
Two main general mechanisms can be considered: isodesmic, where monomer association constants do not vary along the polymerisation process (such as covalent step-growth polymerisation), and cooperative (chain-growth), in which monomer association constants vary with polymer length.29 First reports of living chain-growth supramolecular polymerisation gave hints on the basic molecular design principles needed to achieve control over block copolymerisation.30 This strategy also allowed a certain degree of control in the suprapolymer's length and molecular weight.31,32 Monomer design has been employed to control the chain-growth polymerisation mechanism to afford bidimensional materials.33 For generating 2D structures, it could be desirable to ensure higher polymerisation degrees in the sheet's plane. Here, a cooperative mechanism would ensure that the polymer weight fraction is skewed towards a higher degree of polymerisation.29 However, the precise translation of the mechanistic aspects of supramolecular polymerisation for the construction of 2D assemblies is not always a trivial task.2.3. Supramolecular copolymerisation
The combined polymerisation of different supramolecular monomers allows access to hybrid materials with new assembly capacities and functionalities.21 However, the dynamics of non-covalent forces can hinder the control of the internal microstructure of supramolecular copolymers compared to covalent analogues.34 Block and alternating isodesmic supramolecular copolymerisations have been achieved, mostly for 1D assemblies, taking advantage of the directional interaction of H-bonding donor–acceptor groups.35 Temperature-controlled copolymerisations were reported to be assembled into block tubular36 and fibrillar37 nanostructures. Specific host–guest interactions can also be employed in dynamic self-sorting of sequence-controlled 1D polymers.38 Fluorescently labelled monomers have recently allowed the structural characterisation of multiblock tubular polymers with low polydispersity.39,40 More challenging is block segregation in 2D, which has been achieved by thermodynamic assembly of flexible monomers into lamellar phase-segregated blocks.27 However, imposing sequence restrictions on supramolecular polymerisation from monomer design – solely based on structural complexity and geometry – for the construction of 2D materials currently seems to be a difficult, yet stimulating challenge for supramolecular chemists.412.4. Structural control
A variety of physicochemical processes can be used to control the polymerisation energy pathway and thus select a potential 2D supramolecular structure:Withal, supramolecular chemists aspire to master all the mentioned factors to finely control the fate of the polymer's morphology and potential responsiveness. However, the molecular complexity required to control the dynamic nature of non-covalent bonds by design could be very high, which challenges the synthetic chemist in exploring new monomer possibilities.
3. Directing monomer self-assembly in 2D
It is key to note the importance of directionality in non-covalent interactions to ensure structural order and pathway simplicity in 2D self-assembly. For example, directional interactions such as H-bonding and π–π stacking can guide the ordered elongation of monomers across multiple dimensions (Fig. 1).62 However, non-directional forces (e.g. hydrophobic effects) are valuable orthogonal motifs that require the assistance of directional interactions to facilitate supramolecular polymerisation in 2D. Pre-folded scaffolds, such as α-helical peptide backbones63,64 or self-assembled nanotubes,65 can also direct monomer elongation by segregating supramolecular motifs into domains with defined geometries (e.g. Leu zippers (Fig. 2C) or polar contacts (Fig. 4B)). With these ideas in mind, four important design concepts that provide directionality and internal order to the assembly of 2D supramolecular systems will be discussed in this section.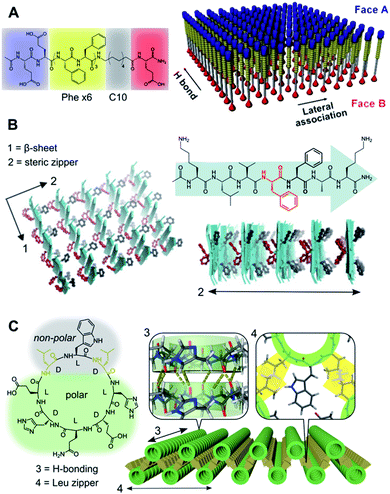 | ||
| Fig. 2 Hydrophobic packing of triblock (A and B) and diblock (C) peptide amphiphiles as mono- and bilayers, respectively. (A) Janus nanosheets obtained from peptides with segregated hydrophobic blocks (Phe6vs. C10), which favour parallel monomer alignment by Phe H-bonding and π–π stacking in two perpendicular axes. Reprinted with permission from ref. 70. Copyright 2017 American Chemical Society. (B) Steric zipping of complementary hydrophobic interfaces between adjacent β-sheets. Reprinted with permission from ref. 71. (C) Amphiphilic cyclic peptide with alternating (D/L) chirality with 2D self-assembly via inter-backbone axial H-bonding (3) and lateral Leu zippers between nanotubes (4). Adapted with permission from ref. 65. Copyright 2019 American Chemical Society. | ||
3.1. Engineering directionality in hydrophobic packing
Ubiquitous for amphiphilic monomers in water, the hydrophobic effect constitutes a primary driving force in aqueous self-assembly.66,67 A variety of amphiphilic monomers have been assembled in water across two dimensions, all of which capitalise on burying different hydrophobic segments in the core of a supramolecular ensemble. Alternatively, liquid–liquid interfaces can template 2D self-assembly based on the partial solubility of different monomer domains.68 Structure-wise, these amphiphilic monomers can be split into two broad categories: diblock and triblock amphiphiles (Fig. 2). Whereas diblock monomers lead to bilayers, triblock amphiphiles present a polar–hydrophobic–polar structure that allows the assembly of monolayers.69 Importantly, triblock monomers can be arranged in parallel or antiparallel orientation based on the directionality of their non-covalent binding domains. Stevens et al. developed parallel triblock amphiphilic assemblies by implementing mixed aliphatic and Phe cores, so H-bonding and π–π stacking between Phe units drive face separation into Janus nanosheets (Fig. 2A).70 Alternatively, an antiparallel order was achieved by hydrophobic ‘steric zippers’ in cationic triblock amyloid-forming peptides, where the antiparallel β-sheet stacking of monomers creates complementary open/closed Phe interfaces (Fig. 2B).71 It should be noted that the presence of ionic terminal moieties normally requires the adjustment of pH and/or ionic strength to externally control 2D self-assembly. These examples illustrate the two perpendicular axes of growth of peptides in the β-sheet configuration: backbone H-bonding versus side-chain interactions,72 directing the monomer orientation in a 2D lattice from the hydrophobic block (Fig. 2). Beyond peptide monomers, triblock bolaamphiphiles capitalise on the incorporation of mesogenic core units (e.g. biphenyl73 or pyrene74) and hydrophilic/ionic termini that prearrange their hydrophobic packing.In contrast to triblock monomers, diblock amphiphiles can self-organise into 2D bilayers by both lateral and axial hydrophobic packing. Rigid diblock monomers can also incorporate ionic domains to control directional growth, which allows the ordered assembly of 2D bilayers from flat amphiphilic peptoids75 and cylindrical coiled-coils.57 Our group recently reported cyclic D/L-alternating peptides that grow 1D nanotubes via backbone H-bonding,76 whose hydrophobic face induces their subsequent assembly into 2D bilayers by inter-tubular hydrophobic interactions (Fig. 2C).65 In this design, interlayer Leu–Leu contacts at specific angles afforded staggered nanotubular bilayers, providing an excellent monomer scaffold for further functionalisation in all three dimensions.77,78
The examples in Fig. 2 showcase the ability of directional interactions, such as H-bonding, and nanotube interfaces to guide the hydrophobic packing of monomers, which becomes evident in the perfect alignment of monomers throughout the entire internal structure of these 2D assemblies.
3.2. π–π stacking of rigid molecular blocks
Aromatic rings and polycycles have been widely explored in 2D self-assembly capitalising on three particular features: (a) the oriented disposition of substituents at specific angles in the same plane (e.g. n·60° in benzene derivatives; Fig. 3A and B); (b) the rigidity of aromatic units allows the geometrical design of polycyclic shapes and interfaces for complementary stacking in a variety of 2D arrays; and (c) the directional interaction between π electron clouds (δ−) and electron-deficient atoms (δ+) directs their hydrophobic packing in a dipole–dipole fashion. Aromatic geometry can range from simple flat pyrene interfaces,26 to perpendicular aromatic diblocks,79 seesaw-shaped scaffolds,80 dual planar ‘attractor’ and non-planar ‘repeller’ surfaces,81 curved monomers82 and boat-shaped building blocks,83 amongst others (Fig. 3C–F).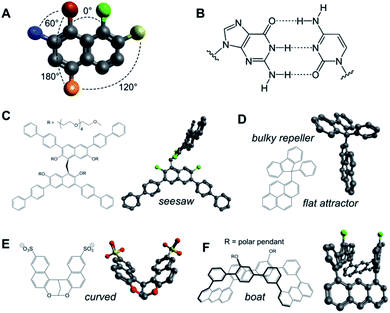 | ||
| Fig. 3 (A and B) Aromatic scaffolds allow precise control over the geometrical disposition of their substituents (A) and the establishment of complementary multivalent interfaces (e.g. nucleobases, B). (C–F) Examples of rigid aromatic monomers capable of 2D self-assembly and their minimised 3D structures (Avogadro v.1.2.0): seesaw (C),80 attractor–repeller (D),81 curved (E)82 and boat-shaped (F);83 green substituents represent polar pendants. | ||
Porous nanosheets have been also assembled from aromatic macrocycles using either covalent macrocyclic tiles of fixed geometry (Fig. 5A)14 or fully supramolecular tiles based on the cation–π stacking of flat aromatic monomers.84 Overall, these designs demonstrate the importance of the geometrical precision at which aromatic interfaces can be synthesised, posing the challenge of incorporating more complex aromatic monomers such as helicenes85 and porous aromatic frameworks86 into 2D self-assemblies.
Aromatic scaffolds also serve as rigid units to segregate polar and hydrophobic substituents at specific angles, allowing the 2D self-assembly of diblock87 and triblock88 amphiphiles (Section 3.1) that capitalise on π–π interactions to order their hydrophobic cores. Importantly, variations in the pendant structure allow the tuning of the nanosheet aspect ratio.89 Connection of aromatic cores to stimuli-responsive polar pendants has also been recently employed for the assembly of reversible aqueous supramolecular polymers.80,83 Similar to supramolecular assemblies, dynamic covalent chemistry offers alternative access to highly crystalline 2D networks from aromatic monomers with potential structural reconfiguration through covalent bond exchange.90
3.3. Host–guest chemistry
Supramolecular host–guest interactions involve the binding between converging (host) and diverging (guest) binding sites based on non-covalent interactions and hydrophobic effects.91 Integration of host–guest pairs as ‘mortar’ to specifically bind cross-linking ‘bricks’ has been widely exploited to build 2D supramolecular networks.92 A structural requirement for the host–guest complex is to preserve the molecular directionality of the building blocks to prevent cross-linking in the perpendicular axis of the nanosheet. A variety of cavitands have been explored in the assembly of supramolecular 2D nanostructures, as reviewed in the literature.93 Inspection of the reported systems reveals a few recurrent structural features. One is the use of planar building blocks, which in some cases introduce π–π stacking as the driving force into the formation of the host–guest complex within the cavitand. Additionally, the inclusion of moieties that prevent 3D growth is frequently required. A remarkable example combining both of these features is the formation of 2D honeycombs with CB[8]-based supramolecular complexes.94 The overall assembly is defined by a multivalent guest scaffold and its cross-linking with CB[8] units (Fig. 4A). Importantly, suppression of interlayer stacking can be achieved by incorporation of hydrophilic pendants into the planar scaffold. Refinement of this architectural concept aimed to release molecular strain to yield larger nanosheets,95 provide stimuli-sensitivity96 and attach specific fluorophores.97 The use of semiflexible chains resulted in the generation of polymorphic 2D structures with lyotropic properties.98 In addition to cucurbituril cavitands, some other host scaffolds such as calixarenes and cyclodextrins have been integrated into 2D self-assemblies.93 Lacking the capabilities of forming ternary host–guest systems, these supramolecular connectors are less predictable in their final orientation geometry. Therefore, 2D host–guest designs must apply certain strategies to direct their planar elongation, such as amphiphilic coronas to prevent the stacking of the aromatic guest,99 or the inclusion of multivalent synthetic hosts/guests that control the cross-linking and branching of the supramolecular ensemble.100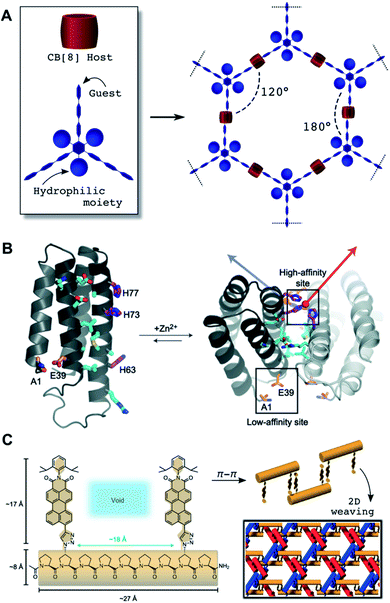 | ||
| Fig. 4 (A) Illustration of geometric control attainable from trigonal multivalent host monomers and cucurbituril CB[8] cavitands as porous hexagonal 2D networks. (B) 2D self-assembly topologically encoded on the outer surface of pre-folded protein four-helix bundles: Two high-affinity Zn2+ coordination sites allow for the elongation of this protein in helical 1D assemblies, while a low affinity site provides a perpendicular elongation point for 2D growth. Polar contacts at interfacing protein domains (cyan) contribute to the stabilisation of the assembly. Reprinted with permission from ref. 101. Copyright 2012 Springer Nature. (C) Woven 2D assembly obtained from oligoproline monomers elongated though π–π stacking of their aromatic pendants. Each individual 1D suprapolymer (red, blue and orange) interlocks by top-down alternation of monomers, thus leaving periodically spaced voids filled by the other 1D chains. Adapted with permission from ref. 123. Copyright 2017 Springer Nature. | ||
3.4. 2D macromolecular assemblies
Macromolecules can act as monomers by exploiting their pre-folding into rigid scaffolds to orient multivalent domains for 2D self-assembly. Biomacromolecules provide inspiration for synthetic designs based on their intrinsic ability to adopt rigid backbone folds (e.g. α-helices and duplexed DNA). For example, helical peptide and protein scaffolds have been assembled in 2D by controlling binding domains such as metal coordination groups (Fig. 4B),101 oppositely charged amphiphiles,102 hydrophobic zipping of coiled-coils64 and trigonal orientation of orthogonal domains.63 Protein interfaces have also produced porous 2D assemblies by geometrically restricted oligomerisation of covalent protein dimers103 and hydrophobic interfaces.104 Even allosteric control has been introduced in protein blocks for supramolecular 1D–2D switching.105 DNA nanotechnology has provided unparalleled design flexibility to access increasingly complex 2D suprastructures, from patterned DNA origami wireframes,106,107 to honeycomb DNA networks,108,109 DNA tile arrays110,111 and hybrid DNA–polymer nanoassemblies,112 amongst others.113 Thus, DNA hybridisation capitalises on directional inter-strand H-bonding and intra-strand π–π stacking between nucleobases114 to assemble helical blocks into a myriad of crystalline materials.115Unlike proteins and nucleic acids, most non-biogenic polymers are not folded into precise secondary structures, hence requiring alternative mechanisms to achieve 2D growth. Crystallisable blocks have been used to provide internal order to the hydrophobic core of the assembly.116–118 An important breakthrough was made by developing crystallisation-driven self-assembly methodologies (Section 2.4).119 Note that many of these examples use organic (co)solvents for self-assembly, posing the standing challenge of translating these supramolecular concepts into physiological environments. Woven 2D assemblies have been recently developed by metal coordination of polycyclic monomers into pre-assembled tiles for intertwined 1D covalent polymerisation.120,121 Other metal–organic suprapolymers can result in dynamic 2D assemblies, from woven to ordered complexes and randomly entangled networks.122 Oligoproline monomers provide a metal-free and fully supramolecular alternative for 2D weaving based on π–π stacking of aromatic pendants at triaxial crossing points (Fig. 4C).123 Woven 2D nanomaterials demonstrate the structural complexity that can be engineered at the molecular level from suitable monomers equipped with directional binding motifs, providing valuable inspiration for new designs to come.
4. Edge applications
Supramolecular materials can bring advantages and new properties such as fine tuning of chemical topology and shape, adaptation to their environment and reversible structural transitions. Capitalising on these interesting properties, a wide variety of applications have already been proposed for 2D supramolecular systems.124,125 A short collection of recent applications will be briefly discussed below to highlight the strong potential of 2D assemblies and the clear necessity of new monomers and conceptual designs.4.1. Chemical separation
2D nanosheets have potential use in several separation problems due to the occurrence of nanopores in these materials. A particularly remarkable example was reported by Sun et al. in the separation of racemic mixtures of L and D-tryptophan by macrocycle-based homochiral porous nanosheets (Fig. 5A).14 This system can close the pores on-demand by addition of ammonium acetate, thus, the selective binding of the D-stereoisomer can be prevented or expelled from the 2D matrix if already captured. The size of these pores can be adjusted at the monomer synthesis stage, suggesting the possibility of adapting this design for the stereo-selective capture of other molecular targets. Some other examples in this sense have been reported, with excellent enantiomeric separation (>99% ee).126 Additionally, highly porous 2D assemblies can be used for the size-selective filtration of nanoparticles,96,127 where the thermal expansion of the pores can tune the size separation cut-off according to the pore distribution achieved.96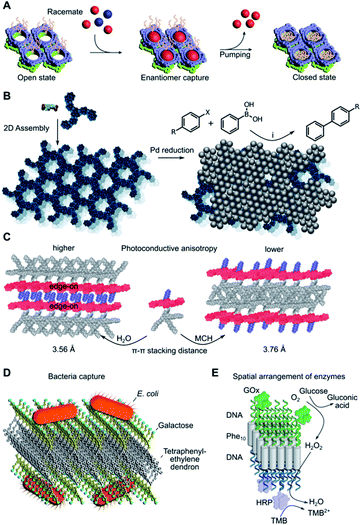 | ||
| Fig. 5 Edge applications of supramolecular 2D assemblies. (A) Enantiomeric molecular recognition and pumping of racemic guests on chiral 2D porous nanosheets. Adapted with permission from ref. 14. Copyright 2018 Springer Nature. (B) Immobilisation of Pd nanoparticles on 2D nanosheets and subsequent catalytic Suzuki–Miyaura coupling. X = I or Br, R = H, CH3, CH3O, NO2, CHO or CN, (i) K2CO3, ethanol, 65 °C. (C) Solvent-dependent self-assembly results in different supramolecular π-stacking and photoconductive anisotropy. MCH = methylcyclohexane. (D) Bacterial agglutination mediated by galactose-decorated nanosheets. Adapted from ref. 143. Copyright 2020 Wiley-VCH. (E) DNA-functionalised enzymes anchored selectively to each side of a DNA–peptide–DNA hybrid Janus nanosheet. GOx = glucose oxidase; HRP = horseradish peroxidase; TMB = 3,3′,5,5′-tetramethylbenzidine. Adapted from ref. 149. Copyright 2021 Wiley-VCH. | ||
4.2. Catalysis
Porous 2D nanosheets can incorporate a range of catalytically active nanoparticles and thus prevent their aggregation-induced loss in catalytic activity. Some examples include the catalysis of the Suzuki–Miyaura coupling reaction on the surface of PdNPs84 (Fig. 5B) or electrochemical nitrogen reduction mediated by PtNPs.128 A selective approach can take advantage of precisely defined molecular-sized pores to allow specific molecular transformations within nanosheet environments. This strategy must consider the molecular dimensions and chemical properties of the chosen reactant. Some illustrative examples include the macrocyclisation of linear pre-organised substrates within the pores of carefully designed 2D nanosheets via Suzuki–Miyaura coupling,129 or in-pore asymmetric catalysis of small substrates based on their enantio-specific fixation.130 Questions arise on the catalytic limitations of these nanopores in comparison to enzymatic binding pockets, where active groups are precisely positioned to manipulate the substrate conformation and its catalytic transformation.131,1324.3. Charge transport
Supramolecular chemists have also designed bottom-up 2D materials with optical and electronic properties. The precise orientation and spacing of monomers in the assembled state provide an opportunity to modulate charge transport processes. While this phenomenon is well known in organic semiconductors,133 less attention has been focused on 2D nanosheets. New developments in this field include the assembly of aromatic amphiphiles as 2D nanosheets with high anisotropic charge-carrier mobility, which results from the defined orientation of monomers in the 2D lattice (Fig. 5C).134,135 Other molecular designs have shown similar properties,88,136 supporting the correlation between precise monomer packing parameters and the specific charge transport capabilities of the material. However, it is still unclear whether proton transport could benefit from a similar lattice effect in 2D systems.137,138 Future efforts to measure and understand charge transport processes in new 2D supramolecular materials are necessary for their implementation in next-generation fuel cell membranes, photovoltaic devices and sensors.4.4. Biological applications
2D assemblies are present in many biological structures, so it is not surprising that many supramolecular nanosheets are suitable materials for biological and biomedical applications.8,139 High surface multivalency in 2D is useful for the development of sensitive biosensors,15,16,140,141 or to capture microorganisms such as bacteria142,143 or viruses (Fig. 5D).71 The stimuli-responsiveness of some nanosheets can be used for the controlled release of drugs, such as light-triggered doxorubicin release,144 or as bacteria sensors with fluorescence output after enzymatic degradation of the assembly.145 Incorporation of fluorophores into the nanosheets opens access to photodynamic therapy applications146 and sensitive biocompatible probes.147,148 Fine control over monomer packing allows for the assembly of Janus nanosheets with distinct enzymatic activities on either leaflet (Fig. 5E).149 Other structures may be of interest for future tissue regeneration applications, such as 2D sheets that serve as templates for mineralisation by calcium carbonate deposition,150 or dynamic nanosheets with self-healing properties.12,151 Even though this is still a nascent field, these examples highlight the vast potential of supramolecular nanosheets as advanced responsive materials in biology and biomedicine.5. Outlook
New supramolecular strategies are emerging for the assembly of two-dimensional materials that could combine molecular order, function and adaptive behaviour. These bottom-up approaches improve the synthetic access to 2D materials, as well as surmount the additional heterogeneous modification steps that could be required in classical top-down strategies. In this perspective, we have highlighted selected examples of current strategies and potential future challenges in the assembly of 2D supramolecular materials. From the design standpoint, combining orthogonal supramolecular interactions and preventing layer stacking have been proved critical to obtaining controlled bidimensional growth. A careful selection of monomer geometry and directional non-covalent forces is needed to obtain smooth free-energy pathways that will lead to discrete 2D nanostructures. Regarding reversibility, multi-monomer systems should avoid potential kinetic traps to allow monomer exchange and structural reshuffle along the whole assembly pathway. Sensitivity to dilution could result in different degrees of polymerisation, which may require post-assembly cross-linking at the expense of potential dynamic network reconfiguration. Beyond stimuli responsive behaviour, the potential self-assembly of multicomponent 2D functional systems, with a monoatomic thickness, also remains elusive. Overall, the successful development of effective 2D nanomaterials and their future impact as a reliable technology will depend on efficient and predictable strategies at the lowest possible synthetic cost. Here, the supramolecular approach is yet to show its real potential to achieve control of properties and functions, which will not be accessible by static covalent strategies. The development of new monomers and the accurate understanding and prediction of their hierarchical 2D self-assembly at the molecular level, constitute a fundamental, yet not trivial exercise in this research field. The growing number of different chemical concepts and structural designs applied to the assembly of 2D supramolecular systems indicates that an important effort is still needed and currently ongoing in these directions.Note from the authors
In the interest of conciseness, the examples included in this perspective have been limited to a representative collection of published structures and mechanistic studies. The authors apologise for any insightful reports that might have been left out in this manuscriptt.Author contributions
I. I. drafted the first version of the manuscript with contributions from all authors. All authors reviewed and contributed to the final version of the paper.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was partially supported by the Spanish Agencia Estatal de Investigación (AEI) (SAF2017-89890-R, PCI2019-103400, and PID2020-117143RB-I00), Xunta de Galicia (ED431C 2017/25, Centro singular de investigación de Galicia accreditation 2019–2022, ED431G 2019/03) and the European Commission (ERDF). I. I. thanks the European Commission (EC) and AEI for MSCA-IF (2018-843332) and Juan de la Cierva (FJCI-2017-31795) fellowships, respectively. J. M. thanks the ERC-STG (DYNAP, 677786), Instituto de Salud Carlos III (COV20/00297), ERC-POC (TraffikGene, 838002), Xunta de Galicia (Oportunius Program) and Human Frontier Science Programme Young Investigator Grant (RGY0066/2017) for funding.References
- K. S. Novoselov, A. K. Geim, S. V. Morozov, D. Jiang, Y. Zhang, S. V. Dubonos, I. V. Grigorieva and A. A. Firsov, Science, 2004, 306, 666–669 CrossRef CAS PubMed.
- G. Bian, J. Yin and J. Zhu, Small, 2021, 17, 2006043 CrossRef CAS PubMed.
- N. R. Glavin, R. Rao, V. Varshney, E. Bianco, A. Apte, A. Roy, E. Ringe and P. M. Ajayan, Adv. Mater., 2020, 32, 1904302 CrossRef CAS PubMed.
- G. Chakraborty, I.-H. Park, R. Medishetty and J. J. Vittal, Chem. Rev., 2021, 121, 3751–3891 CrossRef CAS PubMed.
- C. Tan, X. Cao, X.-J. Wu, Q. He, J. Yang, X. Zhang, J. Chen, W. Zhao, S. Han, G.-H. Nam, M. Sindoro and H. Zhang, Chem. Rev., 2017, 117, 6225–6331 CrossRef CAS PubMed.
- H. Shao, N. H. Ladi, H. Pan, X. L. Zhang, Y. Shen and M. Wang, Sol. RRL, 2021, 5, 2000566 CrossRef CAS.
- X. Jin, T.-H. Gu, K.-G. Lee, M. J. Kim, Md. S. Islam and S.-J. Hwang, Coord. Chem. Rev., 2020, 415, 213280 CrossRef CAS.
- T. Kim, J. Y. Park, J. Hwang, G. Seo and Y. Kim, Adv. Mater., 2020, 32, 2002405 CrossRef CAS PubMed.
- S. Cai, W. Zhang, R. N. Zuckermann, Z. Li, X. Zhao and Y. Liu, Adv. Mater., 2015, 27, 5762–5770 CrossRef CAS PubMed.
- W. Zhang and P. Yang, Adv. Compos. Hybrid Mater., 2019, 2, 201–213 CrossRef CAS.
- I. Insua and J. Montenegro, Chem, 2020, 6, 1652–1682 CAS.
- H. Jin, F. Jiao, M. D. Daily, Y. Chen, F. Yan, Y.-H. Ding, X. Zhang, E. J. Robertson, M. D. Baer and C.-L. Chen, Nat. Commun., 2016, 7, 12252 CrossRef CAS PubMed.
- J. Zhou, L. Venturelli, L. Keiser, S. K. Sekatskii, F. Gallaire, S. Kasas, G. Longo, T. P. J. Knowles, F. S. Ruggeri and G. Dietler, ACS Nano, 2021, 15, 944–953 CrossRef CAS PubMed.
- B. Sun, Y. Kim, Y. Wang, H. Wang, J. Kim, X. Liu and M. Lee, Nat. Mater., 2018, 17, 599–604 CrossRef CAS PubMed.
- J. H. Kim, S. C. Kim, M. A. Kline, E. M. Grzincic, B. W. Tresca, J. Cardiel, M. Karbaschi, D. C. Dehigaspitiya, Y. Chen, V. Udumula, T. Jian, D. J. Murray, L. Yun, M. D. Connolly, J. Liu, G. Ren, C.-L. Chen, K. Kirshenbaum, A. R. Abate and R. N. Zuckermann, ACS Nano, 2020, 14, 185–195 CrossRef CAS PubMed.
- A. Battigelli, J. H. Kim, D. C. Dehigaspitiya, C. Proulx, E. J. Robertson, D. J. Murray, B. Rad, K. Kirshenbaum and R. N. Zuckermann, ACS Nano, 2018, 12, 2455–2465 CrossRef CAS PubMed.
- E. Mattia and S. Otto, Nat. Nanotechnol., 2015, 10, 111–119 CrossRef CAS PubMed.
- P. A. Korevaar, T. F. A. de Greef and E. W. Meijer, Chem. Mater., 2014, 26, 576–586 CrossRef CAS.
- J. Matern, Y. Dorca, L. Sánchez and G. Fernández, Angew. Chem., Int. Ed., 2019, 58, 16730–16740 CrossRef CAS PubMed.
- P. K. Hashim, J. Bergueiro, E. W. Meijer and T. Aida, Prog. Polym. Sci., 2020, 105, 101250 CrossRef CAS.
- B. Adelizzi, N. J. V. Zee, L. N. J. de Windt, A. R. A. Palmans and E. W. Meijer, J. Am. Chem. Soc., 2019, 141, 6110–6121 CrossRef CAS PubMed.
- M. Wehner and F. Würthner, Nat. Rev. Chem., 2020, 4, 38–53 CrossRef CAS.
- P. A. Korevaar, S. J. George, A. J. Markvoort, M. M. J. Smulders, P. A. J. Hilbers, A. P. H. J. Schenning, T. F. A. D. Greef and E. W. Meijer, Nature, 2012, 481, 492–496 CrossRef CAS PubMed.
- N. J. V. Zee, B. Adelizzi, M. F. J. Mabesoone, X. Meng, A. Aloi, R. H. Zha, M. Lutz, I. A. W. Filot, A. R. A. Palmans and E. W. Meijer, Nature, 2018, 558, 100–103 CrossRef PubMed.
- B. Dai, S. Kang, T. Huynh, H. Lei, M. Castelli, J. Hu, Y. Zhang and R. Zhou, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 8543–8548 CrossRef CAS PubMed.
- M. Vybornyi, A. Rudnev and R. Häner, Chem. Mater., 2015, 27, 1426–1431 CrossRef CAS.
- B. A. G. Lamers, R. Graf, B. F. M. de Waal, G. Vantomme, A. R. A. Palmans and E. W. Meijer, J. Am. Chem. Soc., 2019, 141, 15456–15463 CrossRef CAS PubMed.
- T. Fukui, S. Kawai, S. Fujinuma, Y. Matsushita, T. Yasuda, T. Sakurai, S. Seki, M. Takeuchi and K. Sugiyasu, Nat. Chem., 2017, 9, 493–499 CrossRef CAS PubMed.
- M. M. J. Smulders, M. M. L. Nieuwenhuizen, T. F. A. deGreef, P. van der Schoot, A. P. H. J. Schenning and E. W. Meijer, Chem.–Eur. J., 2010, 16, 362–367 CrossRef CAS PubMed.
- J. Kang, D. Miyajima, T. Mori, Y. Inoue, Y. Itoh and T. Aida, Science, 2015, 347, 646–651 CrossRef CAS PubMed.
- R. D. Mukhopadhyay and A. Ajayaghosh, Science, 2015, 349, 241–242 CrossRef CAS PubMed.
- S. Ogi, K. Sugiyasu, S. Manna, S. Samitsu and M. Takeuchi, Nat. Chem., 2014, 6, 188–195 CrossRef CAS PubMed.
- A. Chakraborty, G. Ghosh, D. S. Pal, S. Varghese and S. Ghosh, Chem. Sci., 2019, 10, 7345–7351 RSC.
- L. Albertazzi, D. van der Zwaag, C. M. A. Leenders, R. Fitzner, R. W. van der Hofstad and E. W. Meijer, Science, 2014, 344, 491–495 CrossRef CAS PubMed.
- H. Frisch, J. P. Unsleber, D. Lüdeker, M. Peterlechner, G. Brunklaus, M. Waller and P. Besenius, Angew. Chem., Int. Ed., 2013, 52, 10097–10101 CrossRef CAS PubMed.
- W. Zhang, W. Jin, T. Fukushima, A. Saeki, S. Seki and T. Aida, Science, 2011, 334, 340–343 CrossRef CAS PubMed.
- B. Adelizzi, A. Aloi, A. J. Markvoort, H. M. M. T. Eikelder, I. K. Voets, A. R. A. Palmans and E. W. Meijer, J. Am. Chem. Soc., 2018, 140, 7168–7175 CrossRef CAS PubMed.
- X. Shi, X. Zhang, X.-L. Ni, H. Zhang, P. Wei, J. Liu, H. Xing, H.-Q. Peng, J. W. Y. Lam, P. Zhang, Z. Wang, H. Hao and B. Z. Tang, Macromolecules, 2019, 52, 8814–8825 CrossRef CAS.
- Z. M. Hudson, D. J. Lunn, M. A. Winnik and I. Manners, Nat. Commun., 2014, 5, 3372 CrossRef PubMed.
- J. Y. Rho, H. Cox, E. D. H. Mansfield, S. H. Ellacott, R. Peltier, J. C. Brendel, M. Hartlieb, T. A. Waigh and S. Perrier, Nat. Commun., 2019, 10, 4708 CrossRef PubMed.
- D. vanderZwaag, T. F. A. de Greef and E. W. Meijer, Angew. Chem., Int. Ed., 2015, 54, 8334–8336 CrossRef CAS PubMed.
- L. R. MacFarlane, H. Shaikh, J. D. Garcia-Hernandez, M. Vespa, T. Fukui and I. Manners, Nat. Rev. Mater., 2021, 6, 7–26 CrossRef CAS.
- J. R. Finnegan, X. He, S. T. G. Street, J. D. Garcia-Hernandez, D. W. Hayward, R. L. Harniman, R. M. Richardson, G. R. Whittell and I. Manners, J. Am. Chem. Soc., 2018, 140, 17127–17140 CrossRef CAS PubMed.
- H. Qiu, Y. Gao, C. E. Boott, O. E. C. Gould, R. L. Harniman, M. J. Miles, S. E. D. Webb, M. A. Winnik and I. Manners, Science, 2016, 352, 697–701 CrossRef CAS PubMed.
- S. Yang, S.-Y. Kang and T.-L. Choi, Nat. Commun., 2021, 12, 2602 CrossRef CAS PubMed.
- Z. M. Hudson, C. E. Boott, M. E. Robinson, P. A. Rupar, M. A. Winnik and I. Manners, Nat. Chem., 2014, 6, 893–898 CrossRef CAS PubMed.
- X. He, M.-S. Hsiao, C. E. Boott, R. L. Harniman, A. Nazemi, X. Li, M. A. Winnik and I. Manners, Nat. Mater., 2017, 16, 481–488 CrossRef CAS PubMed.
- X. He, Y. He, M.-S. Hsiao, R. L. Harniman, S. Pearce, M. A. Winnik and I. Manners, J. Am. Chem. Soc., 2017, 139, 9221–9228 CrossRef CAS PubMed.
- H. Li, L. Han, Y. Zhu, P. Fernández-Trillo and F. He, Macromolecules, 2021, 54, 5278–5285 CrossRef CAS.
- J. Boekhoven, W. E. Hendriksen, G. J. M. Koper, R. Eelkema and J. H. van Esch, Science, 2015, 349, 1075–1079 CrossRef CAS PubMed.
- S. A. P. van Rossum, M. Tena-Solsona, J. H. van Esch, R. Eelkema and J. Boekhoven, Chem. Soc. Rev., 2017, 46, 5519–5535 RSC.
- R. Booth, I. Insua, S. Ahmed, A. Rioboo and J. Montenegro, Nat. Commun., 2021, 12, 6421 CrossRef CAS PubMed.
- R. Booth, I. Insua, G. Bhak and J. Montenegro, Org. Biomol. Chem., 2019, 17, 1984–1991 RSC.
- S. Dhiman, R. Ghosh, S. Sarkar and S. J. George, Chem. Sci., 2020, 11, 12701–12709 RSC.
- D. Bochicchio, S. Kwangmettatam, T. Kudernac and G. M. Pavan, ACS Nano, 2019, 13, 4322–4334 CrossRef CAS PubMed.
- T. Jiang, E. L. Magnotti and V. P. Conticello, Interface Focus, 2017, 7, 20160141 CrossRef PubMed.
- A. D. Merg, G. Touponse, E. van Genderen, T. B. Blum, X. Zuo, A. Bazrafshan, H. M. H. Siaw, A. McCanna, R. B. Dyer, K. Salaita, J. P. Abrahams and V. P. Conticello, J. Am. Chem. Soc., 2020, 142, 199956–199968 CrossRef PubMed.
- H. Qiu, A. M. Oliver, J. Gwyther, J. Cai, R. L. Harniman, D. W. Hayward and I. Manners, Macromolecules, 2019, 52, 113–120 CrossRef CAS.
- M. M. J. Smulders, M. M. L. Nieuwenhuizen, M. Grossman, I. A. W. Filot, C. C. Lee, T. F. A. de Greef, A. P. H. J. Schenning, A. R. A. Palmans and E. W. Meijer, Macromolecules, 2011, 44, 6581–6587 CrossRef CAS.
- S. Sim, T. Niwa, H. Taguchi and T. Aida, J. Am. Chem. Soc., 2016, 138, 11152–11155 CrossRef CAS PubMed.
- S. C. Karunakaran, B. J. Cafferty, K. S. Jain, G. B. Schuster and N. V. Hud, ACS Omega, 2020, 5, 344–349 CrossRef CAS PubMed.
- A. C. Mendes, E. T. Baran, R. L. Reis and H. S. Azevedo, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2013, 5, 582–612 CAS.
- E. L. Magnotti, S. A. Hughes, R. S. Dillard, S. Wang, L. Hough, A. Karumbamkandathil, T. Lian, J. S. Wall, X. Zuo, E. R. Wright and V. P. Conticello, J. Am. Chem. Soc., 2016, 138, 16274–16282 CrossRef CAS PubMed.
- J. M. Galloway, H. E. V. Bray, D. K. Shoemark, L. R. Hodgson, J. Coombs, J. M. Mantell, R. S. Rose, J. F. Ross, C. Morris, R. L. Harniman, C. W. Wood, C. Arthur, P. Verkade and D. N. Woolfson, Small, 2021, 17, 2100472 CrossRef CAS PubMed.
- I. Insua and J. Montenegro, J. Am. Chem. Soc., 2020, 142, 300–307 CrossRef CAS PubMed.
- D. Chandler, Nature, 2002, 417, 491 CrossRef CAS PubMed.
- E. Krieg, M. M. C. Bastings, P. Besenius and B. Rybtchinski, Chem. Rev., 2016, 116, 2414–2477 CrossRef CAS PubMed.
- P. Wei, Q. Luo, K. J. Edgehouse, C. M. Hemmingsen, B. J. Rodier and E. B. Pentzer, ACS Appl. Mater. Interfaces, 2018, 10, 21765–21781 CrossRef CAS PubMed.
- L. Liu, L. H. Klausen and M. Dong, Nano Today, 2018, 23, 40–58 CrossRef CAS.
- Y. Lin, M. R. Thomas, A. Gelmi, V. Leonardo, E. T. Pashuck, S. A. Maynard, Y. Wang and M. M. Stevens, J. Am. Chem. Soc., 2017, 139, 13592–13595 CrossRef CAS PubMed.
- B. Dai, D. Li, W. Xi, F. Luo, X. Zhang, M. Zou, M. Cao, J. Hu, W. Wang, G. Wei, Y. Zhang and C. Liu, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 2996–3001 CrossRef CAS PubMed.
- F. Sheehan, D. Sementa, A. Jain, M. Kumar, M. Tayarani-Najjaran, D. Kroiss and R. V. Ulijn, Chem. Rev., 2021, 121, 13869–13914 CrossRef CAS PubMed.
- Z. Li and Z. Lin, ACS Nano, 2021, 15, 3152–3160 CrossRef CAS PubMed.
- J. Huang, S. Wang, G. Wu, L. Yan, L. Dong, X. Lai, S. Yin and B. Song, Soft Matter, 2013, 10, 1018–1023 RSC.
- K. T. Nam, S. A. Shelby, P. H. Choi, A. B. Marciel, R. Chen, L. Tan, T. K. Chu, R. A. Mesch, B.-C. Lee, M. D. Connolly, C. Kisielowski and R. N. Zuckermann, Nat. Mater., 2010, 9, 454–460 CrossRef CAS PubMed.
- A. Fuertes, M. Juanes, J. R. Granja and J. Montenegro, Chem. Commun., 2017, 53, 7861–7871 RSC.
- A. Mendez-Ardoy, A. Bayón-Fernández, Z. Yu, C. Abell, J. R. Granja and J. Montenegro, Angew. Chem., Int. Ed., 2020, 132, 6969–6975 CrossRef.
- S. Díaz, I. Insua, G. Bhak and J. Montenegro, Chem.–Eur. J., 2020, 26, 14765–14770 CrossRef PubMed.
- T. Nishikawa, H. Narita, S. Ogi, Y. Sato and S. Yamaguchi, Chem. Commun., 2019, 55, 14950–14953 RSC.
- H. Sun, L. Dong, Y. Kim and M. Lee, Chem.–Asian J., 2020, 15, 2470–2474 CrossRef CAS PubMed.
- Y. Li, X. Dong, M. Yu, W. Liu, Y. Nie, J. Zhang, L. Xie, C. Xu, J. Liu and W. Huang, Small, 2021, 17, 2102060 CrossRef CAS PubMed.
- H. Yao, L.-P. Yang, X.-Y. Pang, J.-R. Li and W. Jiang, Chem. Commun., 2018, 54, 10847–10850 RSC.
- Y. Wang, Y. Kim and M. Lee, Angew. Chem., Int. Ed., 2016, 55, 13122–13126 CrossRef CAS PubMed.
- X. Xiao, H. Chen, X. Dong, D. Ren, Q. Deng, D. Wang and W. Tian, Angew. Chem., Int. Ed., 2020, 59, 9534–9541 CrossRef CAS PubMed.
- T. Mori, Chem. Rev., 2021, 121, 2373–2412 CrossRef CAS PubMed.
- Y. Tian and G. Zhu, Chem. Rev., 2020, 120, 8934–8986 CrossRef CAS PubMed.
- L. Han, M. Wang, X. Jia, W. Chen, H. Qian and F. He, Nat. Commun., 2018, 9, 865 CrossRef PubMed.
- A. Mukherjee, T. Sakurai, S. Seki and S. Ghosh, Langmuir, 2020, 36, 13096–13103 CrossRef CAS PubMed.
- N. Sasaki, J. Yuan, T. Fukui, M. Takeuchi and K. Sugiyasu, Chem.–Eur. J., 2020, 26, 7840–7846 CrossRef CAS PubMed.
- A. M. Evans, L. R. Parent, N. C. Flanders, R. P. Bisbey, E. Vitaku, M. S. Kirschner, R. D. Schaller, L. X. Chen, N. C. Gianneschi and W. R. Dichtel, Science, 2018, 361, 52–57 CrossRef CAS PubMed.
- J. W. Steed and J. L. Atwood, Supramolecular Chemistry, Wiley, UK, 2nd edn, 2009 Search PubMed.
- J. Tian, L. Chen, D.-W. Zhang, Y. Liu and Z.-T. Li, Chem. Commun., 2016, 52, 6351–6362 RSC.
- N. Cheng and Y. Liu, Handbook of Macrocyclic Supramolecular Assembly, Springer, Singapore, 1st edn, 2020 Search PubMed.
- K.-D. Zhang, J. Tian, D. Hanifi, Y. Zhang, A. C.-H. Sue, T.-Y. Zhou, L. Zhang, X. Zhao, Y. Liu and Z.-T. Li, J. Am. Chem. Soc., 2013, 135, 17913–17918 CrossRef CAS PubMed.
- M. Pfeffermann, R. Dong, R. Graf, W. Zajaczkowski, T. Gorelik, W. Pisula, A. Narita, K. Müllen and X. Feng, J. Am. Chem. Soc., 2015, 137, 14525–14532 CrossRef CAS PubMed.
- Q. Zhang, R.-J. Xing, W.-Z. Wang, Y.-X. Deng, D.-H. Qu and H. Tian, iScience, 2019, 19, 14–24 CrossRef PubMed.
- Q. Zhao and Y. Liu, Chem. Commun., 2018, 54, 6068–6071 RSC.
- J. del Barrio, J. Liu, R. A. Brady, C. S. Y. Tan, S. Chiodini, M. Ricci, R. Fernández-Leiro, C.-J. Tsai, P. Vasileiadi, L. D. Michele, D. Lairez, C. Toprakcioglu and O. A. Scherman, J. Am. Chem. Soc., 2019, 141, 14021–14025 CrossRef PubMed.
- Y. Yi, S. Fa, W. Cao, L. Zeng, M. Wang, H. Xu and X. Zhang, Chem. Commun., 2012, 48, 7495–7497 RSC.
- H. Sun, Y. Chen, X. Han and Y. Liu, Angew. Chem., Int. Ed., 2017, 56, 7062–7065 CrossRef CAS PubMed.
- J. D. Brodin, X. I. Ambroggio, C. Tang, K. N. Parent, T. S. Baker and F. A. Tezcan, Nat. Chem., 2012, 4, 375–382 CrossRef CAS PubMed.
- A. D. Merg, G. Touponse, E. van Genderen, X. Zuo, A. Bazrafshan, T. Blum, S. Hughes, K. Salaita, J. P. Abrahams and V. P. Conticello, Angew. Chem., Int. Ed., 2019, 58, 13507–13512 CrossRef CAS PubMed.
- J. F. Matthaei, F. DiMaio, J. J. Richards, L. D. Pozzo, D. Baker and F. Baneyx, Nano Lett., 2015, 15, 5235–5239 CrossRef CAS PubMed.
- A. J. Ben-Sasson, J. L. Watson, W. Sheffler, M. C. Johnson, A. Bittleston, L. Somasundaram, J. Decarreau, F. Jiao, J. Chen, I. Mela, A. A. Drabek, S. M. Jarrett, S. C. Blacklow, C. F. Kaminski, G. L. Hura, J. J. D. Yoreo, J. M. Kollman, H. Ruohola-Baker, E. Derivery and D. Baker, Nature, 2021, 589, 468–473 CrossRef CAS PubMed.
- M. Xu, R. Zeng, J. Xiang and Q. Yan, J. Am. Chem. Soc., 2019, 141, 13724–13728 CrossRef CAS PubMed.
- H. Jun, F. Zhang, T. Shepherd, S. Ratanalert, X. Qi, H. Yan and M. Bathe, Sci. Adv., 2019, 5, eaav0655 CrossRef PubMed.
- H. Jun, X. Wang, W. P. Bricker and M. Bathe, Nat. Commun., 2019, 10, 5419 CrossRef PubMed.
- P. Wang, S. Gaitanaros, S. Lee, M. Bathe, W. M. Shih and Y. Ke, J. Am. Chem. Soc., 2016, 138, 7733–7740 CrossRef CAS PubMed.
- H. Yu, D. T. L. Alexander, U. Aschauer and R. Häner, Angew. Chem., Int. Ed., 2017, 56, 5040–5044 CrossRef CAS PubMed.
- F. Hong, S. Jiang, X. Lan, R. P. Narayanan, P. Šulc, F. Zhang, Y. Liu and H. Yan, J. Am. Chem. Soc., 2018, 140, 14670–14676 CrossRef CAS PubMed.
- M. Ali, N. Afshan, C. Jiang, H. Zheng and S.-J. Xiao, Nanoscale, 2019, 11, 22216–22221 RSC.
- N. Hannewald, P. Winterwerber, S. Zechel, D. Y. W. Ng, M. D. Hager, T. Weil and U. S. Schubert, Angew. Chem., Int. Ed., 2021, 60, 6218–6229 CrossRef CAS PubMed.
- P. Wang, T. A. Meyer, V. Pan, P. K. Dutta and Y. Ke, Chem, 2017, 2, 359–382 CAS.
- P. Yakovchuk, E. Protozanova and M. D. Frank-Kamenetskii, Nucleic Acids Res., Spec. Publ., 2006, 34, 564–574 CrossRef CAS PubMed.
- A. Heuer-Jungemann and T. Liedl, Trends Chem, 2019, 1, 799–814 CrossRef CAS.
- F. Ishiwari, G. Okabe, H. Ogiwara, T. Kajitani, M. Tokita, M. Takata and T. Fukushima, J. Am. Chem. Soc., 2018, 140, 13497–13502 CrossRef CAS PubMed.
- R. Qi, Y. Zhu, L. Han, M. Wang and F. He, Macromolecules, 2020, 53, 6555–6565 CrossRef CAS.
- S. Ganda, M. Dulle, M. Drechsler, B. Förster, S. Förster and M. H. Stenzel, Macromolecules, 2017, 50, 8544–8553 CrossRef CAS.
- L. MacFarlane, C. Zhao, J. Cai, H. Qiu and I. Manners, Chem. Sci., 2021, 12, 4661–4682 RSC.
- D. P. August, R. A. W. Dryfe, S. J. Haigh, P. R. C. Kent, D. A. Leigh, J.-F. Lemonnier, Z. Li, C. A. Muryn, L. I. Palmer, Y. Song, G. F. S. Whitehead and R. J. Young, Nature, 2020, 588, 429–435 CrossRef CAS PubMed.
- Z. Wang, A. Błaszczyk, O. Fuhr, S. Heissler, C. Wöll and M. Mayor, Nat. Commun., 2017, 8, 14442 CrossRef CAS PubMed.
- K. C. Bentz and S. M. Cohen, Angew. Chem., Int. Ed., 2018, 57, 14992–15001 CrossRef CAS PubMed.
- U. Lewandowska, W. Zajaczkowski, S. Corra, J. Tanabe, R. Borrmann, E. M. Benetti, S. Stappert, K. Watanabe, N. A. K. Ochs, R. Schaeublin, C. Li, E. Yashima, W. Pisula, K. Müllen and H. Wennemers, Nat. Chem., 2017, 9, 1068–1072 CrossRef CAS PubMed.
- B. Shen, Y. Kim and M. Lee, Adv. Mater., 2020, 1905669 CrossRef CAS PubMed.
- A. Ciesielski, C. Palma, M. Bonini and P. Samorì, Adv. Mater., 2010, 22, 3506–3520 CrossRef CAS PubMed.
- X. Feng, B. Shen, B. Sun, J. Kim, X. Liu and M. Lee, Angew. Chem., Int. Ed., 2020, 59, 11355–11359 CrossRef CAS PubMed.
- Y. Zhou, G. Zhang, B. Li and L. Wu, ACS Appl. Mater. Interfaces, 2020, 12, 30761–30769 CrossRef CAS PubMed.
- C.-C. Zhang, X. Liu, Y.-P. Liu and Y. Liu, Chem. Mater., 2020, 32, 8724–8732 CrossRef CAS.
- X. Liu, X. Zhou, B. Shen, Y. Kim, H. Wang, W. Pan, J. Kim and M. Lee, J. Am. Chem. Soc., 2020, 142, 1904–1910 CrossRef CAS PubMed.
- B. Sun, B. Shen, A. Urushima, X. Liu, X. Feng, E. Yashima and M. Lee, Angew. Chem., Int. Ed., 2020, 59, 22690–22696 CrossRef CAS PubMed.
- F. Wang, O. Gnewou, S. Wang, T. Osinski, X. Zuo, E. H. Egelman and V. P. Conticello, Matter, 2021, 4, 3217–3231 CrossRef CAS PubMed.
- F. M. Koehler, N. A. Luechinger, D. Ziegler, E. K. Athanassiou, R. N. Grass, A. Rossi, C. Hierold, A. Stemmer and W. J. Stark, Angew. Chem., Int. Ed., 2008, 48, 224–227 CrossRef PubMed.
- V. Coropceanu, J. Cornil, D. A. d. S. Filho, Y. Olivier, R. Silbey and J.-L. Brédas, Chem. Rev., 2007, 107, 926–952 CrossRef CAS PubMed.
- S. Ghosh, D. S. Philips, A. Saeki and A. Ajayaghosh, Adv. Mater., 2017, 29, 1605408 CrossRef PubMed.
- B. Narayan, S. P. Senanayak, A. Jain, K. S. Narayan and S. J. George, Adv. Funct. Mater., 2013, 23, 3053–3060 CrossRef CAS.
- Z. Yao, Y. Zheng, Q. Li, T. Lei, S. Zhang, L. Zou, H. Liu, J. Dou, Y. Lu, J. Wang, X. Gu and J. Pei, Adv. Mater., 2019, 31, 1806747 CrossRef PubMed.
- M. Kumar, N. L. Ing, V. Narang, N. K. Wijerathne, A. I. Hochbaum and R. V. Ulijn, Nat. Chem., 2018, 10, 696–703 CrossRef CAS PubMed.
- D. D. Ordinario, L. Phan, W. G. W. IV, J.-M. Jocson, E. Karshalev, N. Hüsken and A. A. Gorodetsky, Nat. Chem., 2014, 6, 596–602 CrossRef CAS PubMed.
- X. Zhang, C. Gong, O. U. Akakuru, Z. Su, A. Wu and G. Wei, Chem. Soc. Rev., 2019, 48, 5564–5595 RSC.
- G. K. Olivier, A. Cho, B. Sanii, M. D. Connolly, H. Tran and R. N. Zuckermann, ACS Nano, 2013, 7, 9276–9286 CrossRef CAS PubMed.
- L. Zhu, Z. Zhao, P. Cheng, Z. He, Z. Cheng, J. Peng, H. Wang, C. Wang, Y. Yang and Z. Hu, Adv. Mater., 2017, 29, 1700057 CrossRef PubMed.
- J. H. Kim, E. M. Grzincic, L. Yun, R. K. Spencer, M. A. Kline and R. N. Zuckermann, Soft Matter, 2019, 16, 907–913 RSC.
- N. Krishnan, D. Perumal, S. Atchimnaidu, K. S. Harikrishnan, M. Golla, N. M. Kumar, J. Kalathil, J. Krishna, D. K. Vijayan and R. Varghese, Chem.–Eur. J., 2020, 26, 1037–1041 CrossRef CAS PubMed.
- T. Zhang and C.-H. Zhang, J. Mater. Chem. B, 2019, 7, 7736–7743 RSC.
- H. Zhang, F. Liang and Y. Yang, Chem.–Eur. J., 2020, 26, 198–205 CrossRef CAS PubMed.
- D. Perumal, M. Golla, K. S. Pillai, G. Raj, A. Krishna and R. Varghese, Org. Biomol. Chem., 2021, 19, 2804–2810 RSC.
- M. Wang, Y. Song, P. Mu, X. Cai, Y. Lin and C.-L. Chen, ACS Appl. Bio Mater., 2020, 3, 6039–6048 CrossRef CAS PubMed.
- Y. Song, M. Wang, S. Akkineni, W. Yang, J. J. Hettige, H. Jin, Z. Liao, P. Mu, F. Yan, M. Baer, J. J. D. Yoreo, D. Du, Y. Lin and C.-L. Chen, ACS Mater. Lett., 2021, 3, 420–427 CrossRef CAS.
- S. K. Albert, S. Lee, P. Durai, X. Hu, B. Jeong, K. Park and S. Park, Small, 2021, 17, 2006110 CrossRef CAS PubMed.
- J. M. V. Jun, M. V. P. Altoe, S. Aloni and R. N. Zuckermann, Chem. Commun., 2015, 51, 10218–10221 RSC.
- F. Jiao, Y. Chen, H. Jin, P. He, C.-L. Chen and J. J. D. Yoreo, Adv. Funct. Mater., 2016, 26, 8960–8967 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2022 |