Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Hydrogels containing water soluble conjugates of silver(I) ions with amino acids, metabolites or natural products for non infectious contact lenses†
C. N.
Banti
 *a,
M.
Kapetana
a,
C.
Papachristodoulou
b,
C. P.
Raptopoulou
c,
V.
Psycharis
*a,
M.
Kapetana
a,
C.
Papachristodoulou
b,
C. P.
Raptopoulou
c,
V.
Psycharis
 c,
P.
Zoumpoulakis
d,
T.
Mavromoustakos
e and
S. K.
Hadjikakou
c,
P.
Zoumpoulakis
d,
T.
Mavromoustakos
e and
S. K.
Hadjikakou
 *af
*af
aInorganic and Analytical Chemistry, Department of Chemistry, University of Ioannina, 45110 Ioannina, Greece. E-mail: cbanti@uoi.gr; shadjika@uoi.gr
bDepartment of Physics, University of Ioannina, Greece
cNCSR “Demokritos”, Institute of Nanoscience and Nanotechnology, A. Paraskevi, Attikis, Greece
dLaboratory of Chemistry, Analysis & Design of Food Processes, Department of Food Science and Technology, University of West Attica, Greece
eOrganic Chemistry Laboratory, Department of Chemistry, University of Athens, Greece
fUniversity Research Center of Ioannina (URCI), Institute of Materials Science and Computing, Ioannina, Greece
First published on 11th August 2021
Abstract
The poor handling and hygiene practices of contact lenses are the key reasons for their frequent contamination, and are responsible for developing ocular complications, such as microbial keratitis (MK). Thus there is a strong demand for the development of biomaterials of which contact lenses are made, combined with antimicrobial agents. For this purpose, the known water soluble silver(I) covalent polymers of glycine (GlyH), urea (U) and the salicylic acid (SalH2) of formulae [Ag3(Gly)2NO3]n (AGGLY), [Ag(U)NO3]n (AGU), and dimeric [Ag(salH)]2 (AGSAL) were used. Water solutions of AGGLY, AGU and AGSAL were dispersed in polymeric hydrogels using hydroxyethyl-methacrylate (HEMA) to form the biomaterials pHEMA@AGGLY-2, pHEMA@AGU-2, and pHEMA@AGSAL-2. The biomaterials were characterized by X-ray fluorescence (XRF) spectroscopy, thermogravimetric differential thermal analysis (TG-DTA), differential scanning calorimetry (DTG/DSC), attenuated total reflection spectroscopy (FT-IR-ATR) and single crystal diffraction analysis. The antibacterial activity of AGGLY, AGU, AGSAL, pHEMA@AGGLY-2, pHEMA@AGU-2 and pHEMA@AGSAL-2 was evaluated against the Gram negative species Pseudomonas aeruginosa (P. aeruginosa) and Gram positive ones Staphylococcus epidermidis (S. epidermidis) and Staphylococcus aureus (S. aureus), which mainly colonize in contact lenses. The in vitro toxicity of the biomaterials and their ingredients was evaluated against normal human corneal epithelial cells (HCECs) whereas the in vitro genotoxicity was evaluated by the micronucleus (MN) assay in HCECs. The Artemia salina and Allium cepa models were applied for the evaluation of in vivo toxicity and genotoxicity of the materials. Following our studies, the new biomaterials pHEMA@AGGLY-2, pHEMA@AGU-2, and pHEMA@AGSAL-2 are suggested as efficient candidates for the development of antimicrobial contact lenses.
Introduction
The use of soft contact lenses is a popular method for correcting eye refractive errors.1 Their poor handling and hygiene practices are the key reasons for their frequent contamination, which can lead to microbial keratitis (MK).2 MK is an infection of the cornea, which occurs at a rate of approximately 3 per 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 wearers per year.3,4 Despite the rare occurrences of MK development, the high number of soft contact lens wearers (45 million in the United States) leads to thousands of cases, annually.5,6 A strategy for manufacturing next generation soft contact lenses involves the use of novel active biomaterials which control microbial colonisation and thus the incidence of MK.3 Attempts towards this direction have been made either by loading an antimicrobial agent in the hydrogels, of which contact lenses are made, or by functionalizing the surface of contact lenses with an antimicrobial component.2
000 wearers per year.3,4 Despite the rare occurrences of MK development, the high number of soft contact lens wearers (45 million in the United States) leads to thousands of cases, annually.5,6 A strategy for manufacturing next generation soft contact lenses involves the use of novel active biomaterials which control microbial colonisation and thus the incidence of MK.3 Attempts towards this direction have been made either by loading an antimicrobial agent in the hydrogels, of which contact lenses are made, or by functionalizing the surface of contact lenses with an antimicrobial component.2
Silver(I), on the other hand, exhibits a broad spectrum of antimicrobial activity against both Gram-positive and Gram-negative bacteria.1 Currently, silver nitrate solution (1–2%) is extensively used for curing neonatal conjunctivitis.4 Moreover, silver-impregnated contact lens cases, (MicroBlock®) have been on the market in the United Kingdom since 2004, and were approved for use in the United States in 2005.1 Upon adding the contact lens solution to the MicroBlock® (or Pro-Guard) case, silver ions at concentrations of approximately 10 μg L−1 are released into the solution for about one month protecting the lenses from microbial colonaization.4 Generally, a slow release of silver ions maintains the antimicrobial activity in a macromolecular environment by inhibiting the growth of microorganisms.7 Therefore, the development of biomaterials of which contact lenses are made combined with silver based antimicrobial agents is a research, technological and financial issue. For this purpose, two strategies have been developed by our group. One involves the use of silver nanoparticles using extracts from natural products as combined reducing and capping agents.8 The second one involves the use of small molecules which act as antimicrobial agents as well.5,8–10 However, it is of interest to study whether small molecules which are natural product ingredients could lead to biocompatible, non-toxic agent formation and thus to the formation of effective antimicrobial materials. In the course of our studies on the development of new antimicrobial agents5,8,9,11–21 and their non-infectious contact lenses the novel biomaterials pHEMA@AGGLY-2, pHEMA@AGU-2, and pHEMA@AGSAL-2 were synthesised by dispersion in polymeric poly(2-hydroxyethyl methacrylate) (pHEMA) of the known water soluble silver(I) covalent polymers [Ag3(Gly)2NO3]n (AGGLY), [Ag(U)NO3]n (AGU), and dimeric [Ag(salH)]2 (AGSAL) (GlyH = glycine, U = urea and SalH2 = salicylic acid). The biomaterials were characterized by XRF, TG-DTA, DTG/DSC and FT-IR-ATR analytical techniques. The prepared materials were evaluated for their antibacterial activity against the Gram negative species P. aeruginosa and Gram positive ones S. epidermidis and S. aureus which are abundant in microbial keratitis. The in vitro and in vivo toxicity of the biomaterials was tested against HCECs, by the MN assay, using Artemia salina and Allium cepa models.
Results and discussion
General aspects
The known silver(I) covalent polymers AGGLY, AGU and dimeric AGSAL were readily obtained by reacting AgNO3 with GlyH, U or SalH2 (Scheme 1).22–24 Briefly, AgNO3 reacts with the potassium salts of GlyH or salH2 in dd water in 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 (AGGLY) and 1
3 (AGGLY) and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (AGSAL) molar ratios (Scheme 1). The white precipitates were crystallized from DMSO solutions to form colourless crystals of the named compounds of formulae [Ag3(Gly)2NO3]n (AGGLY) or [Ag(salH)]2 (AGSAL).13,22–24 Colourless crystals of [Ag(U)NO3]n (AGU) were grown from the methanol/acetonitrile solution of AgNO3 and U (1
1 (AGSAL) molar ratios (Scheme 1). The white precipitates were crystallized from DMSO solutions to form colourless crystals of the named compounds of formulae [Ag3(Gly)2NO3]n (AGGLY) or [Ag(salH)]2 (AGSAL).13,22–24 Colourless crystals of [Ag(U)NO3]n (AGU) were grown from the methanol/acetonitrile solution of AgNO3 and U (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio). AGGLY, AGU and AGSAL are soluble in H2O and DMSO.
1 molar ratio). AGGLY, AGU and AGSAL are soluble in H2O and DMSO.
The formation of AGGLY, AGU and AGSAL was verified from their physical constants (m.p. and ATR-FT-IR).13,22–24 The vibrational spectra of AGGLY and AGSAL (Fig. S1 and S2†) are identical to those of the published ones.13,22–24 The ATR-FT-IR spectrum of AGU shows vibration bands at 1674 and 3354 cm−1, which are attributed to ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) and ν(N–H), respectively (Fig. S3†). The corresponding vibration bands in the IR spectrum of urea are observed at 1690 and 3355 cm−1 (Fig. S4†).25,26 The ν(C–N) vibration band is observed at 1465 cm−1 in the spectrum of AGU.25 The vibration band at 1384 cm−1 is assigned to the NO3 group.25
O) and ν(N–H), respectively (Fig. S3†). The corresponding vibration bands in the IR spectrum of urea are observed at 1690 and 3355 cm−1 (Fig. S4†).25,26 The ν(C–N) vibration band is observed at 1465 cm−1 in the spectrum of AGU.25 The vibration band at 1384 cm−1 is assigned to the NO3 group.25
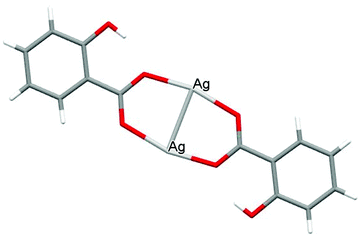
The crystal structures of both AGGLY and AGSAL are already refined by single crystal X-ray crystallography and they are reported elsewhere (Fig. 1 and 2).13,22–24 Their structures are briefly described here: AGGLY shows a polymeric 3D structure (Fig. 1).22 There are three symmetry independent Ag(I) cations, two symmetry independent deprotonated glycine ligands, one nitrate anion and one water solvent molecule in a lattice site with half occupancy in the asymmetric unit of the unit cell. Thus, the charge balanced stoichiometric formula of AGGLY is {[Ag3(Gly)2NO3]·0.5H2O} (Fig. 1).22 The architecture of the structure is based on the centrosymmetric Ag6(NO3)24− unit presented in Fig. 1.22AGSAL is dimeric (Fig. 2). Two silver atoms are bridged together by two carboxylic groups from two SalH2 ligands (Fig. 2).13,23 Strong Ag⋯Ag interactions (2.856 Å) are established due to argentophilicity, which leads to metalloaromaticity in the 5-member rings. This adds further stabilization to the assembly.
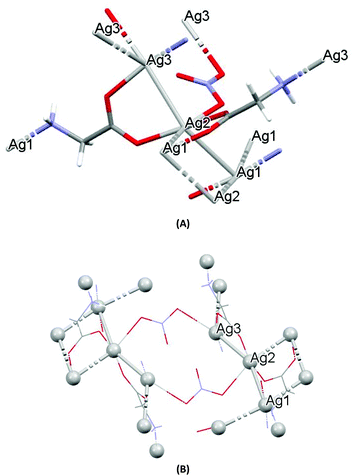 | ||
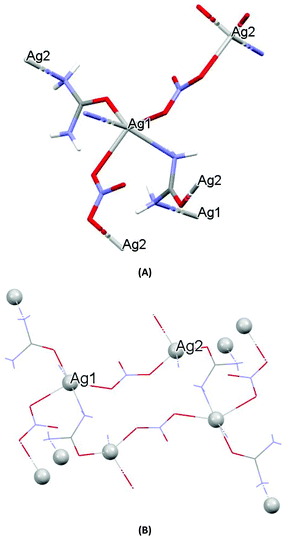
| Fig. 1 (A) Molecular diagrams of AGGLY. (B) Covalent Ag–O and Ag–N intra-molecular interactions lead to supramolecular assembly. | ||
The structure of AGU has been studied previously at room temperature and in the present discussion an emphasis is given in the packing and the hydrogen bond interactions.24AGU crystallized in P21/n with a = 6.2509(1), b = 16.8667(4), c = 10.2467(3) Å, 102.191(1)°, V = 1055.97(4) Å3, Z = 4, R = 2.31%, while the reported one crystallized in P21/n with a = 6.314(4), b = 16.886(4), c = 10.270(8) Å, β = 104.80(5)°, V = 1068(l) A3, Z = 4, R = 7.7% (ref. 24). Selected bond distances and angles are listed in Fig. 3, while extensive quoting is listed in Table AGU-1, ESI.† A detailed description of the supramolecular assembly is provided in the ESI (see the ESI†). The building block of the entire AGU is shown in Fig. 3A. The compound is polymeric. The chains consist of [–Ag(U)–NO3–]n units. The secondary N(urea)–Ag bonds link different chains (Fig. 3). The geometry around silver is distorted trigonal bi-pyramidal. Three oxygen atoms from two NO3− groups and one from urea coordinate to the metal centre, while the coordination sphere is completed by two N atoms of amide groups originating from two different ureas.
Agent loading in pHEMA
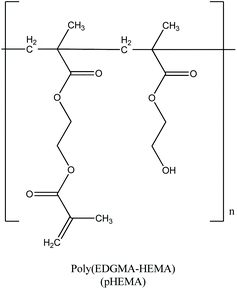
Aqua solutions of AGGLY, AGU and AGSAL (2 mM) were dispersed in pHEMA (Scheme 2) during the polymerization procedure towards the formation of biomaterials pHEMA@AGGLY-2, pHEMA@AGU-2, and pHEMA@AGSAL-2. The dispersion of AGGLY, AGU and AGSAL in pHEMA was qualitatively verified by XRF, UV solid, TG-DTA and DTG/DSC analyses.X-ray fluorescence spectroscopy
The XRF spectra of pHEMA@AGGLY-2, pHEMA@AGU-2 and pHEMA@AGSAL-2 confirm the presence of Ag. Moreover, the Ag Kα X-ray emission was used for the quantitative determination of Ag. The content of silver in pHEMA@AGGLY-2, pHEMA@AGU-2 and pHEMA@AGSAL-2 was determined to be 0.19 ± 0.05, 0.16 ± 0.04 and 0.23 ± 0.05% w/w, while the calculated one is 0.11 (pHEMA@AGGLY-2), 0.04 (pHEMA@AGU-2) and 0.08 (pHEMA@AGSAL-2) % respectively. The variations between the experimental and calculated values are due to its low quantity, which is close to the limits of the method.X-ray powder diffraction analysis (XRPD)
Powders of dry pHEMA@AGGLY-2, pHEMA@AGU-2 and pHEMA@AGSAL-2 discs were used for XRPD spectra recording (Fig. S5–S7†). The absence of diffraction in the XRPD spectra of pHEMA@AGGLY-2, pHEMA@AGU-2 and pHEMA@AGSAL-2, such as those observed in the XRPD diagrams of their corresponding ingredients AGGLY, AGU and AGSAL shows either the effective phase transition from the crystalline to amorphous phase during confinement into pHEMA or their low concentrations in the disc.Solid state UV-vis spectra
The dispersion of AGGLY, AGU and AGSAL in pHEMA within pHEMA@AGGLY-2, pHEMA@AGU-2 and pHEMA@AGSAL-2 was verified by solid state UV-Vis spectroscopy (Fig. S8†). The characteristic bands in the solid state UV-Vis spectra of pHEMA (240 nm) and free ingredients AGGLY (280 nm), AGU and AGSAL (280, 295 and 295 nm respectively) undergo a hypsochromic shift in the cases of pHEMA@AGGLY-2, pHEMA@AGU-2 and pHEMA@AGSAL-2 (230, 210 and 240 nm respectively) suggesting the incorporation of the silver complexes in pHEMA.Thermo-gravimetric analysis of biomaterials pHEMA@AGGLY-2, pHEMA@AGU-2 and pHEMA@AGSAL-2
Antibacterial activity
| P. aeuroginosa | S. epidermidis | S. aureus | Ref. | |
|---|---|---|---|---|
| a In this work; HCIP·HCl = ciprofloxacin hydrochloride and HCIP = ciprofloxacin. | ||||
| MIC (μM) | ||||
| AGGLY | 21.4 ± 4.6 | 49.6 ± 6.6 | 51.3 ± 5.1 | |
| AGU | 94.6 ± 3.9 | 90.3 ± 10.7 | 92.0 ± 10.4 | |
| AGSAL | 28.0 | 50.0 | 42.0 | 13 |
| AgNO3 | 60.0 | 122.0 | 95.0 | 13 and 14 |
| MBC (μM) | ||||
| AGGLY | 28.0 ± 4.5 | 55.0 ± 9.0 | 75 ± 0 | |
| AGU | 120 ± 14 | 132 ± 16 | 140 ± 23 | |
| AGSAL | 33 | 90 | 50 | 13 |
| AgNO3 | 153.3 ± 13.1 | 140 | 135.0 ± 6.7 | 13 and 14 |
| MBC/MIC | ||||
| AGGLY | 1.31 | 1.11 | 1.46 | |
| AGU | 1.27 | 1.46 | 1.52 | |
| AGSAL | 1.18 | 1.80 | 1.19 | 13 |
| AgNO3 | 2.56 | 1.15 | 1.42 | 13 and 14 |
| Bacterial viability (%) | ||||
| pHEMA@AGGLY-2 | 1.2 | 5.2 | 3.3 | |
| pHEMA@AGU-2 | 0.8 | 4.9 | 2.4 | |
| pHEMA@AGSAL-2 | 1.1 | 2.1 | 3.1 | |
| IZ (mm) | ||||
| AGGLY (1 mM) | 11.7 ± 1.8 | 13.0 ± 1.9 | 12.5 ± 0.9 | |
| AGGLY (2 mM) | 13.9 ± 1.6 | 16.4 ± 1.6 | 15.3 ± 1.7 | |
| AGU (1 mM) | 12.5 ± 0.9 | 12.3 ± 1.7 | 14.0 ± 1.7 | |
| AGU (2 mM) | 14.3 ± 1.5 | 14.5 ± 1.7 | 16.1 ± 1.3 | |
| AGSAL (1 mM) | 13.0 | 13.0 | 12.0 | 13 |
| AGSAL (2 mM) | 13.4 ± 1.5 | 14.7 ± 1.3 | 13.8 ± 1.4 | |
| pHEMA@AGGLY-2 | 12.4 ± 1.0 | 12.2 ± 1.1 | 12.5 ± 1.0 | |
| pHEMA@AGU-2 | 14.5 ± 1.0 | 14.5 ± 1.0 | 13.3 ± 1.8 | |
| pHEMA@AGSAL-2 | 14 ± 1.9 | 15.2 ± 0.3 | 14.3 ± 2.4 | |
| BEC (μM) | ||||
| AGGLY | 150 ± 22 | — | 294 ± 4 | |
| AGU | 694 ± 139 | — | 687 ± 38 | |
| AGSAL | 875 | 648 | 13 | |
| HCIP·HCl | 670 | 952 | 27 | |
| HCIP | 2140 | 2463 | 27 | |
| IC 50 (μM) towards HCECs | ||||
| AGGLY | 2.87 ± 0.13 | |||
| AGU | 8.69 ± 0.65 | |||
| AGSAL | 10.22 ± 0.33 | |||
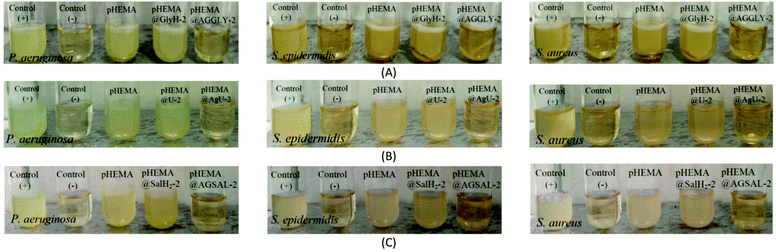
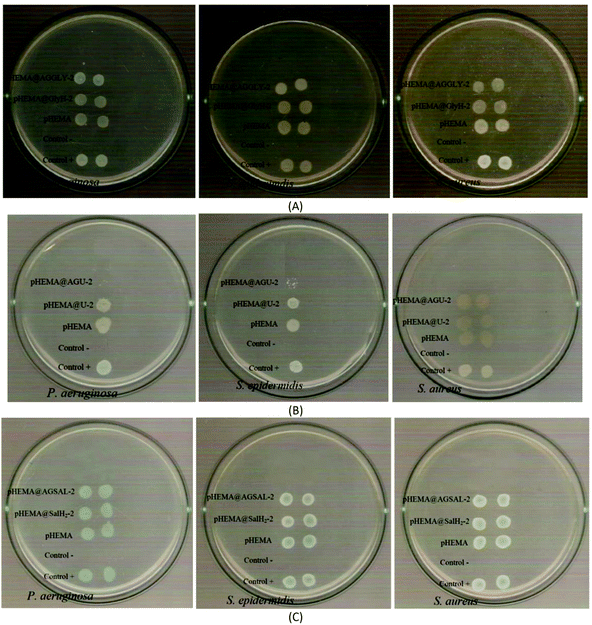
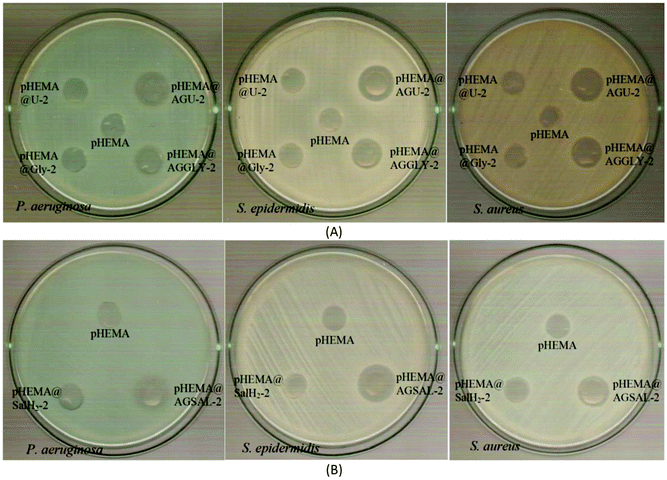
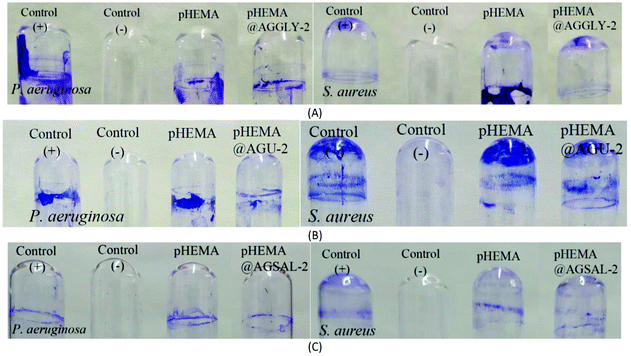
The calculated % bacterial viability of P. aeruginosa, S. epidermidis and S. aureus upon their incubation with the new biomaterials pHEMA@AGGLY-2, pHEMA@AGU-2, and pHEMA@AGSAL-2 is extremely low in the range of 0.8–1.2%, 2.1–5.2% and 2.4–3.3% against P. aeruginosa, S. epidermidis and S. aureus, respectively (Table 1). The P. aeruginosa and S. aureus are eliminated by the discs of pHEMA@AGU-2, up to 99.2 and 97.6%, respectively, while the bacteria of S. epidermidis are eliminated by the disc of pHEMA@AGSAL-2 up to 97.6%. No influence on the bacterial viability was observed upon their treatment with the discs of pure pHEMA or pHEMA@Gly-2, pHEMA@U-2 and pHEMA@SalH2-2 (Fig. 5). However, bacterial colonies of P. aeruginosa, S. epidermidis and S. aureus are grown in agar plates when 4 μL of the supernatants of the solutions were used, which were initially treated with the discs of pHEMA and pHEMA@AGGLY-2, pHEMA@AGU-2, and pHEMA@AGSAL-2 (Fig. 6). Therefore, the biomaterials prohibit microbial growth only when they are present in the microbe cultures (Fig. 8).
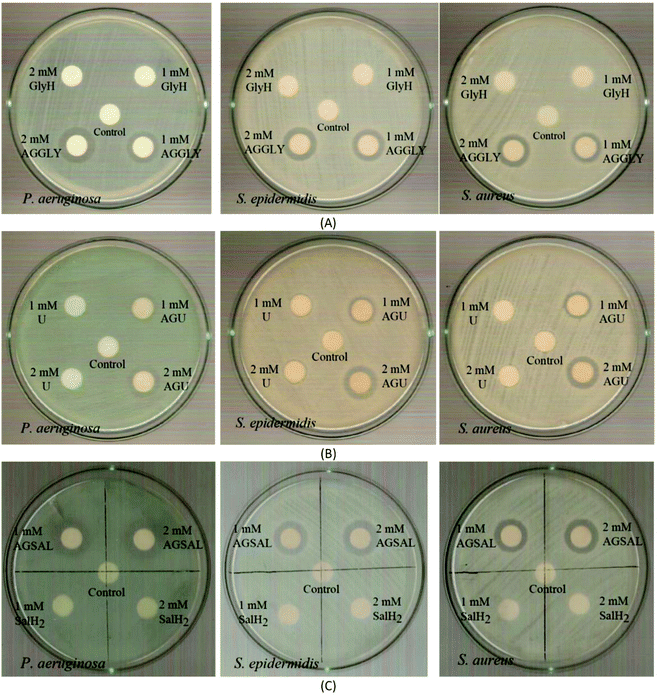 | ||
| Fig. 7 IZs which are developed in agar plates of P. aeruginosa, S. epidermidis and S. aureus by the AGGLY, GlyH (A), AGU, urea (B) and AGSAL, SalH2 (C) at 1 and 2 mM. | ||
The bacteria tested show higher sensitivity to the concentration of 2 mM than 1 mM for the biomaterial ingredients AGGLY, AGU and AGSAL. Therefore, these strains exhibit dose dependent response on the agents. No inhibition zones were developed when the microbes were incubated with glycine, urea and salicylic acid at 1 or 2 mM. The Gram positive bacteria possess higher sensitivity to the agents than the corresponding negative ones. Particularly, AGGLY and AGSAL develop greater IZ towards S. epidermidis, (IZ 16.4 ± 1.6 and 14.7 ± 1.3 mm, respectively) while AGU exhibits the highest activity against S. aureus with an IZ value of 16.1 ± 1.3 mm. The microbial strains are classified into three categories according to the size of IZ, caused by an antimicrobial agent in their agar dilution culture: (i) strains, where the agent causes IZ ≥ 17 mm, are susceptible, (ii) those where an agent creates IZ between 13 and 16 mm (13 ≤ IZ ≤ 16 mm) are intermediate, while (iii) those where the agent causes IZ ≤ 12 mm are considered as resistant ones.5,8,9,13–15,27 Therefore, the response of P. aeruginosa, S. epidermidis and S. aureus can be considered as an intermediate against AGGLY, AGU and AGSAL at both concentrations used (1 or 2 mM).
The inhibition zones which were developed in agar plates of P. aeruginosa, E. coli, S. epidermidis and S. aureus microbes around the pHEMA@AGGLY-2, pHEMA@AGU-2 and pHEMA@AGSAL-2 discs suggest mild antimicrobial activity (Table 1 and Fig. 8). Moreover, no inhibition zones were developed when pHEMA or pHEMA@Gly-2, pHEMA@U-2 and pHEMA@SalH2-2 were used against the bacterial strains (Table 1 and Fig. 8).
The IZs of pHEMA@AGU-2 and pHEMA@AGSAL-2 hydrogels are similar to those of the soaked paper discs with the ingredient solutions (2 mM). In the case of the pHEMA@AGGLY-2 discs, however, the IZs developed against P. aeruginosa, E. coli, S. epidermidis and S. aureus (12.4 ± 1.0, 12.2 ± 1.1 and 12.5 ± 1.0 mm) are shorter than the soaked paper discs with AGGLY (2 mM) (13.9 ± 1.6, 16.4 ± 1.6 and 15.3 ± 1.7 mm), respectively. This suggests the negligible release of the disc ingredient.
The biofilm formation on the surface of contact lenses increases the survival of the bacteria and their replication on the lens surface.28In vitro studies have demonstrated that the bacteria can adhere to all types of contact lenses.28 The percent of removal of the preformed biofilm was assessed using the crystal violet assay.5 The discs of pHEMA@AGGLY-2, pHEMA@AGU-2 and pHEMA@AGSAL-2 eliminate the biofilm of P. aeruginosa by 40.3, 27.5 and 39.7%, respectively while they eliminate the biofilm of S. aureus by 42.0, 24.1 and 23.2%, respectively; no inhibitory activity is observed against the biofilm of S. aureus (Fig. 9).
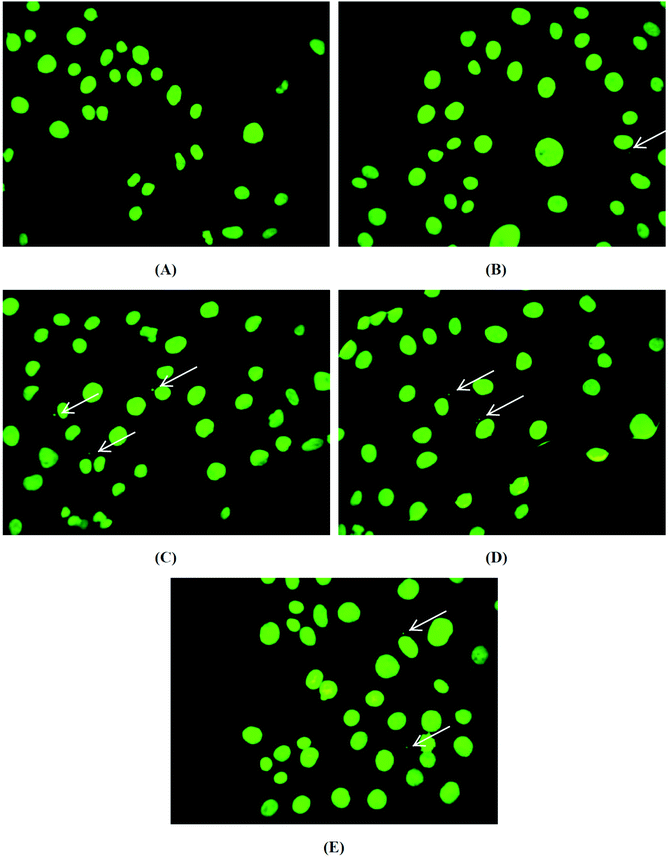
After the incubation of brine shrimp larvae with the biomaterials, pHEMA, pHEMA@AGGLY-2, pHEMA@AGU-2 and pHEMA@AGSAL-2, for a period of 24 h, no mortality level was found. Thus a non-toxic behaviour of the biomaterial discs is suggested.
| MI | CA | |
|---|---|---|
| Untreated cells | 7.3 ± 1.0 | 0.9 ± 0.3 |
| pHEMA | 6.8 ± 1.7 | 0.9 ± 0.4 |
| pHEMA@AGGLY-2 | 7.2 ± 1.3 | 1.1 ± 0.5 |
| pHEMA@AGU-2 | 6.5 ± 1.0 | 1.0 ± 0.1 |
| pHEMA@AGSAL-2 | 6.4 ± 0.5 | 0.4 ± 0.1 |
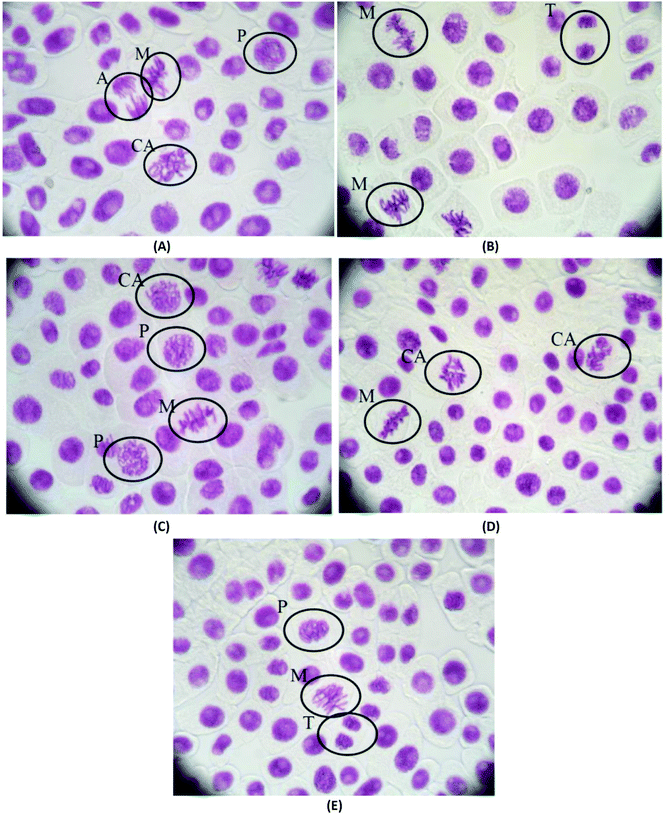
The mitotic index and the chromosomal aberrations of the cell division of Allium cepa are not affected when they are treated with pHEMA@AGGLY-2, pHEMA@AGU-2 and pHEMA@AGSAL-2 for 48 h, indicating that the biomaterials are not mutagenic or genotoxic (Fig. 11). The mitotic indexes of the treated Allium cepa meristematic root cells with pHEMA@AGGLY-2, pHEMA@AGU-2 and pHEMA@AGSAL-2 are 7.2 ± 1.3, 6.5 ± 1.0 and 6.4 ± 0.5%, respectively. These values are in accordance with those of the untreated group or the group treated with pHEMA (7.3 ± 1.0 and 6.8 ± 1.7%, respectively). Therefore no in vivo genotoxic effect can be concluded for the new biomaterials.
Conclusion
Since the contact lens wear can be implicated with the occurrence of microbial keratitis (MK), novel active biomaterials pHEMA@AGGLY-2, pHEMA@AGU-2, and pHEMA@AGSAL-2 were synthesised by the dispersion in pHEMA of the known water soluble silver(I) covalent polymers AGGLY, AGU and AGSAL of the natural products GlyH, U and SalH2.The silver(I) covalent polymers were tested for the antibacterial efficiency against microbes which colonise in contact lenses. AGGLY and AGSAL exhibit higher antimicrobial activity than silver nitrate, which increases up to 2.8-, 1.2- and 1.4-fold against P. aeruginosa, S. epidermidis and S. aureus, respectively (Table 1). In contrast, AGU exhibits moderate activity against the tested microbes. The MBC/MIC values of AGGLY, AGU and AGSAL classified them as bactericidal ones (Table 1). AGGLY, AGU and AGSAL were dispersed in pHEMA aiming for the development of new non infected soft contact lens materials. The biomaterials can eliminate up to 94.8–99.2% of the bacteria after their incubation with the hydrogels. This activity is superior to those exhibited by pHEMA@(SLS@[Zn3(CitH)2]) (CitH4 = citric acid; SLS = sodium lauryl sulphate) which were found to lie between 6.5 and 29.0% respectively, pHEMA@ORLE_2 (ORLE = oregano leaf extract) and pHEMA@AgNPs(ORLE)_2 (AgNPs(ORLE) = silver nanoparticles of oregano leaf extract) which were evaluated at 59.6–88.3% against P. aeruginosa, S. epidermidis and S. aureus which lie between 6.5 and 29.0% respectively.8,10 However, the activity of pHEMA@AGGLY-2, pHEMA@AGU-2, and pHEMA@AGSAL-2 is comparable to that of the corresponding pHEMA@AGMNA-1 (AGMNA = {[Ag6(μ3-HMNA)4(μ3-MNA)2]2−·[(Et3NH)+]2·(DMSO)2·(H2O)}) which was found to eliminate bacterial strains such as P. aeruginosa, S. epidermidis and S. aureus by 92.3–99.6% respectively.5 Moreover, the biomaterials pHEMA@AGGLY-2, pHEMA@AGU-2, and pHEMA@AGSAL-2 exhibit a low in vitro genotoxic effect against HCECs and negligible in vivo toxicity against Artemia salina and Allium cepa making them important candidates for the development of innovative non toxic biomaterials for sterilised contact lens preparation with high resistance against microbes which are involved in microbial keratitis.
Experimental
Materials and instruments
All solvents used were of reagent grade and were used with no further purification. Glycine, urea and salicylic acid were purchased from Sigma-Aldrich. Melting points were measured in open tubes with Stuart Scientific apparatus and are uncorrected. Mid infrared spectra (4000–400 cm−1) were obtained on a Cary 670 FTIR spectrometer (Agilent Technologies). 2-Hydroxyethyl-methacrylate (pHEMA), ethylene-glycole-dimethacrylate (EGDMA, Merck), diphenyl(2,4,6-trimethylbenzoyl)phosphine oxide (TPO 97%, Sigma Aldrich) as well as sodium chloride (NaCl, Merck) and hydrochloric acid (HCl 37%, Merck) were used. X-ray fluorescence XRF measurements were carried out using an Am-241 radioisotopic source (exciting radiation 59.5 keV). For the detection of X-ray fluorescence, a Si (Li) detector was used. The measuring time was chosen to collect ∼2000 data on the weaker Kα peak. The tryptone tryptophan medium, beef extract powder, bacteriological peptone, and soy peptone were purchased from Biolife. Agar and yeast extract were purchased from Fluka Analytical. Sodium chloride, D(+)-glucose, and di-potassium hydrogen phosphate trihydrate were purchased from Merck. Dulbecco's modified Eagle's medium, (DMEM), fetal bovine serum, glutamine and trypsin were purchased from Gibco, Glasgow, UK. Phosphate buffer saline (PBS) was purchased from Sigma-Aldrich. Dimethyl sulfoxide was purchased from Riedel-de Haën. For the toxicity experiments, Brine Shrimp Eggs (Artemia salina) were purchased from Ocean Nutrition.Synthesis and crystallization of AGGLY, AGU and AGSAL
AGGLY and AGSAL were synthesized as described in a previous work.13,22,23 For the synthesis of AGU, a solution of 0.5 mmol AgNO3 (0.085 g) and 1 mmol urea (0.060 g) were stirred in 10 mL of methanol and 10 mL of acetonitrile. Crystals of pure AGU were grown from the slow evaporation of the solution.
AGGLY: Colorless crystal, melting point: 147–149 °C; Elemental analysis found: C: 9.30; H: 1.42, N: 7.70% calculated for C4H8Ag3N3O7: C: 9.00; H: 1.51, N: 7.87%. IR (cm−1): 3104w, 2426w, 2360w, 1764m, 1594s, 1496s, 1384vs, 1127m, 1043m, 930m, 891m, 826s, 667m, 506s; 1H-NMR (ppm) in D2O: 3.332 (s, C[H]; UV-vis (dd H2O): λ = 194 nm (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε = 4.86).
ε = 4.86).
AGU: Colorless crystal, melting point: 90–100 °C; Elemental analysis found: C: 5.08; H: 1.90, N: 18.70%; calculated for CH4AgN3O4: C: 5.22; H: 1.75, N: 18.28%; IR (cm−1): 3437 (m), 2353 (m), 2170 (m), 1381 (vs), 1151 (m), 839 (m), 825 (m), 557 (w).
AGSAL: White-gray crystal, melting point: >280 °C; Elemental analysis found: C = 34.34, H = 2.11%; calc: C = 34.32, H = 2.06%; IR (cm−1): 1588 (w), 1556 (s), 1455 (s), 1383 (m), 1334 (m), 1301 (m), 1257 (m), 1213 (w), 1141 (m), 1020 (w), 879 (w), 860 (s), 809 (s), 779 (s), 745 (w), 708 (vs), 667 (s), 546 (s), 475 (w), 423 (w), 407 (s), 394 (s), 377 (s); 1H NMR (ppm) in D2O: 7.77–7.75 (d, H[dC], 7.41–7.37 (t, H [fOH]), 6.92–6.87 (m, H[e,gC]; UV–Vis (H2O): λ = 202 nm (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε = 2.5), λ = 228 nm (log
ε = 2.5), λ = 228 nm (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε = 1.82), λ = 295 nm (log
ε = 1.82), λ = 295 nm (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε = 1.38).
ε = 1.38).
X-ray crystal structure determination
A crystal of AGU, (0.07 × 0.32 × 0.40 mm) was taken from the mother liquor and immediately cooled to −93 °C. Diffraction measurements for AGU were performed on a Rigaku R-AXIS SPIDER Image Plate diffractometer using graphite monochromated Mo Kα radiation.39 Data collection (ω-scans) and processing (cell refinement, data reduction and empirical absorption correction) were performed using the CrystalClear program package. Important crystallographic data are listed in Table S3.† The structures were solved by direct methods using SHELXS v.2013/1 and refined by full-matrix least-squares techniques on F2 with SHELXL ver. 2014/6.40,41 Further experimental crystallographic details for AGU: 2θmax = 54.0°; reflections collected/unique/used, 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 437/2294 [Rint = 0.0321]/2294; 195 parameters refined; (Δ/σ)max = 0.003; (Δρ)max/(Δρ)min = 0.913/−0.704 e Å−3; R1/wR2 (for all data), 0.0231/0.0475. All hydrogen atoms were located from difference Fourier maps and were refined freely. All non-hydrogen atoms were refined anisotropically. Plots of the structures were drawn using the Diamond 3 program package.42
437/2294 [Rint = 0.0321]/2294; 195 parameters refined; (Δ/σ)max = 0.003; (Δρ)max/(Δρ)min = 0.913/−0.704 e Å−3; R1/wR2 (for all data), 0.0231/0.0475. All hydrogen atoms were located from difference Fourier maps and were refined freely. All non-hydrogen atoms were refined anisotropically. Plots of the structures were drawn using the Diamond 3 program package.42
Crystallographic data (excluding structure factors) for AGU reported in this paper have been deposited with the Cambridge Crystallographic Data Centre as supplementary publication no. CCDC 2089744.†
Synthesis of pHEMA@AGGLY-2, pHEMA@AGU-2 and pHEMA@AGSAL-2
Hydrogels of pHEMA incorporated with AGGLY, AGU and AGSAL were obtained as follows: 2.7 mL of HEMA was mixed with 2 mL of double distilled water (ddw), which contains AGGLY, AGU and AGSAL (2 mM) and 10 μL of EGDMA. The solution was then degassed by bubbling with nitrogen for 15 minutes. The TPO initiator (6 mg) was added to the solution and mixed for 5 min at 800 rpm. The solution was poured into the mold and was then placed under a UV mercury lamp (λmax = 280 nm), 15 watt, where photopolymerization occurred, for 40 minutes. Unreacted monomers were removed, by immersing the gel in boiling water for 15 min. Discs with 10 mm diameter were cut, and they were washed by immersion in water, NaCl 0.9%, HCl 0.1 M, and again in water. The discs were then dried at 40 °C until no weight change would occur.X-ray powder diffraction (XRPD)
The study of the samples by using X-ray powder diffraction was accomplished by using a diffraction-meter D8 Advance Bruker, Department of Physics, University of Ioannina. Radiation CuKα (40 kV, 40 mA, λKα) and the monochromator system of diffracted beam were used. The X-ray powder diffraction patterns were measured in the area of 2θ angles between 2° and 80° at a rotation step of 0.02° and time of 2 s per step. All samples measured with the above diffraction-meter were in the fine-grained powdered form.Thermogravimetric differential thermal analysis (TG-DTA) and differential scanning calorimetry (DTG/DSC)
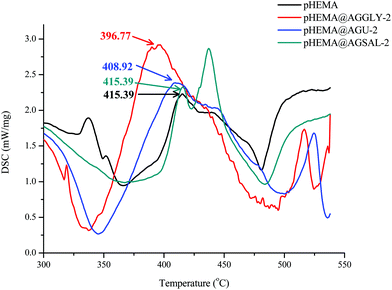
The measurements were performed on a DTG/TG NETZSCH STA 449C. For the measurements, 9.8, 9.2 and 14.6 mg of pHEMA@AGGLY-2, pHEMA@AGU-2 and pHEMA@AGSAL-2, respectively were placed inside a platinum capsule with alumina as the reference sample. The temperature increase rate was 10 °C min−1 in the range of 25–500 °C and the measurements were performed in an air atmosphere. The samples measured were in the fine-grained powdered form.Biological tests
In order to evaluate the viability of the microbes on the pHEMA, pHEMA@AGGLY-2, pHEMA@AGU-2 and pHEMA@AGSAL-2 discs, the materials were placed in the test tubes which contain 5 × 105 cfu mL−1 of the P. aeruginosa, S. epidermidis and S. aureus microbes.5,8,9,13–15,27 The optical densities of the supernatant solutions were then measured to give the % viability of the microbes after incubation for 18–24 h.5,8,9,13–15,27
For the evaluation of IZ, AGGLY, glycine, AGU, urea at 1 mM and 2 mM, and the disks of pHEMA, pHEMA@AGGLY-2, pHEMA@AGU-2 and pHEMA@AGSAL-2 were placed on the agar surface and the Petri plates were incubated for 20 h.5,8,9,13–15,27
Conflicts of interest
There are no conflicts to declare by the authors.Acknowledgements
This research has been co-financed by the European Union and Greek national funds through the Operational Program Competitiveness, Entrepreneurship and Innovation, under the call RESEARCH – CREATE – INNOVATE (project code: T1EDK-02990). Professor T. Vaimakis is acknowledged for the TG-DTA/DSC measurements. The COST Action CA15114 “Anti-Microbial Coating Innovations to prevent infectious diseases (AMICI)” is acknowledged for the stimulating discussions. This work was carried out in fulfilment of the requirements for the Master thesis of Ms M. K. according to the curriculum of the International Graduate Program in “Biological Inorganic Chemistry”, which operates at the University of Ioannina within the collaboration of the Departments of Chemistry of the Universities of Ioannina, Athens, Thessaloniki, Patras, Crete and the Department of Chemistry of the University of Cyprus (http://bic.chem.uoi.gr/BIC-En/index-en.html) under the supervision of Prof. S. K. H.References
- A. Xiao, C. Dhand, C. M. Leung, R. W. Beuerman, S. Ramakrishna and R. Lakshminarayanan, Strategies to design antimicrobial contact lenses and contact lens cases, J. Mater. Chem. B, 2018, 6, 2171–2186 RSC.
- C. Dhand, C. Y. Ong, N. Dwivedi, J. Varadarajan, M. H. Periayah, E. J. Lim, V. Mayandi, E. T. Leng Goh, R. P. Najjar, L. W. Chan, R. W. Beuerman, L. L. Foo, X. J. Loh and R. Lakshminarayanan, Mussel-Inspired Durable Antimicrobial Contact Lenses: The Role of Covalent and Noncovalent Attachment of Antimicrobials, ACS Biomater, Sci. Eng., 2020, 6, 3162–3173 CAS.
- D. Kharaghani, D. Dutta, P. Gitigard, Y. Tamada, A. Katagiri, D.-N. Phan, M. D. P. Willcox and I. Soo Kim, Development of antibacterial contact lenses containing metallic nanoparticles, Polym. Test., 2019, 79, 106034 CrossRef CAS.
- S. Ahmad Khan and C.-S. Lee, Recent progress and strategies to develop antimicrobial contact lenses and lens cases for different types of microbial keratitis, Acta Biomater., 2020, 113, 101–118 CrossRef.
- A. K. Rossos, C. N. Banti, A. Kalampounias, C. Papachristodoulou, K. Kordatos, P. Zoumpoulakis, T. Mavromoustakos, N. Kourkoumelis and S. K. Hadjikakou, pHEMA@AGMNA-1: A novel material for the development of antibacterial contact lens, Mater. Sci. Eng., C, 2020, 111, 110770 CrossRef CAS PubMed.
- https://www.cdc.gov/contactlenses/fast-facts.html .
- D. Dutta and M. D. P. Willcox, Antimicrobial Contact Lenses and Lens Cases, Eye Contact Lens, 2014, 40, 312–324 CrossRef PubMed.
- A. Meretoudi, C. N. Banti, P. K. Raptis, C. Papachristodoulou, N. Kourkoumelis, A. A. Ikiades, P. Zoumpoulakis, T. Mavromoustakos and S. K. Hadjikakou, Silver Nanoparticles from Oregano Leaves’ Extracts as Antimicrobial Components for Non-Infected Hydrogel Contact Lenses, Int. J. Mol. Sci., 2021, 22, 3539 CrossRef CAS PubMed.
- I. Milionis, C. N. Banti, I. Sainis, C. P. Raptopoulou, V. Psycharis, N. Kourkoumelis and S. K. Hadjikakou, Silver ciprofloxacin (CIPAG): a successful combination of chemically modified antibiotic in inorganic–organic hybrid, J. Biol. Inorg. Chem., 2018, 23, 705–723 CrossRef CAS PubMed.
- V. A. Karetsi, C. N. Banti, N. Kourkoumelis, C. Papachristodoulou, C. D. Stalikas, C. P. Raptopoulou, V. Psycharis, P. Zoumpoulakis, T. Mavromoustakos, I. Sainis and S. K. Hadjikakou, An Efficient Disinfectant, Composite Material {SLS@[Zn3(CitH)2]} as Ingredient for Development of Sterilized and Non Infectious Contact Lens, Antibiotics, 2019, 8, 213 CrossRef CAS PubMed.
- L. Kyros, C. N. Banti, N. Kourkoumelis, M. Kubicki, I. Sainis and S. K. Hadjikakou, Synthesis, characterization, and binding properties towards CT-DNA and lipoxygenase of mixed-ligand silver(I) complexes with 2-mercaptothiazole and its derivatives and triphenylphosphine, J. Biol. Inorg. Chem., 2014, 19, 449–464 CrossRef CAS PubMed.
- I. Sainis, C. N. Banti, A. M. Owczarzak, L. Kyros, N. Kourkoumelis, M. Kubicki and S. K. Hadjikakou, New antibacterial, non-genotoxic materials, derived from the functionalization of the anti-thyroid drug methimazole with silver ions, J. Inorg. Biochem., 2016, 160, 114–124 CrossRef CAS PubMed.
- M.-E. K. Stathopoulou, C. N. Banti, N. Kourkoumelis, A. G. Hatzidimitriou, A. G. Kalampounias and S. K. Hadjikakou, Silver complex of salicylic acid and its hydrogel-cream in wound healing chemotherapy, J. Inorg. Biochem., 2018, 181, 41–55 CrossRef CAS PubMed.
- M. P. Chrysouli, C. N. Banti, I. Milionis, D. Koumasi, C. P. Raptopoulou, V. Psycharis, I. Sainis and S. K. Hadjikakou, A water-soluble silver(I) formulation as an effective disinfectant of contact lenses cases, Mater. Sci. Eng. C, 2018, 93, 902–910 CrossRef CAS PubMed.
- I. Ketikidis, C. N. Banti, N. Kourkoumelis, C. G. Tsiafoulis, C. Papachristodoulou, A. G. Kalampounias and S. K. Hadjikakou, Conjugation of Penicillin-G with Silver(I) Ions Expands Its Antimicrobial Activity against Gram negative Bacteria, Antibiotics, 2020, 9, 25 CrossRef CAS.
- P. J. Cox, P. Aslanidis, P. Karagiannidis and S. Hadjikakou, Silver(I) complexes with heterocyclic thiones and tertiary phosphines as ligands. Part 4. Dinuclear complexes of silver(I) bromide: the crystal structure of bis[bromo-(pyrimidine-2-thione)(triphenylphosphine)silver(I), Inorg. Chim. Acta, 2000, 310, 268–272 CrossRef CAS.
- S. Zartilas, N. Kourkoumelis, S. K. Hadjikakou, N. Hadjiliadis, P. Zachariadis, M. Kubicki, A. Y. Denisov and I. S. Butler, A new silver(I) aggregate having an octagonal Ag4S4 core where μ3-S bonding interactions lead to a nano-tube assembly which exhibits quasi-aromaticity, Eur J. Inorg. Chem., 2007, 1219–1224 CrossRef CAS.
- S. K. Hadjikakou, I. I. Ozturk, M. N. Xanthopoulou, P. C. Zachariadis, S. Zartilas and N. Hadjiliadis, Synthesis, Structural Characterization and Biological Study of New Organotin(IV), Silver(I) and Antimony(III) Complexes with Thioamides, J. Inorg. Biochem., 2008, 102, 1007–1015 CrossRef CAS.
- S. Zartilas, S. K. Hadjikakou, N. Hadjiliadis, N. Kourkoumelis, L. Kyros, M. Kubicki, M. Baril, I. S. Butler, S. Karkabounas and J. Balzarini, Tetrameric 1:1 and monomeric 1:3 complexes of silver(I) halides with tri(p-tolyl)-phosphine: A structural and biological study, Inorg. Chim. Acta, 2009, 362, 1003–1010 CrossRef CAS.
- P. C. Zachariadis, S. K. Hadjikakou, N. Hadjiliadis, A. Michaelides, S. Skoulika, Y. Ming and Y. Xiaolin, Synthesis, study and structural characterisation of a new water soluble hexanuclear silver(I) cluster with the 2-mercapto-nicotinic acid with possible antiviral activity, Inorg. Chim. Acta, 2003, 343, 361–365 CrossRef CAS.
- P. C. Zachariadis, S. K. Hadjikakou, N. Hadjiliadis, A. Michaelides, S. Skoulika, J. Balzarini and E. D. Clercq, Synthesis, characterization and in vitro study of the cytostatic and antiviral activity of new polymeric silver(I) complexes with ribbon structures, derived from the conjugated heterocyclic thioamide 2-mercapto-3,4,5,6-tetrahydro-pyrimidine, Eur. J. Inorg. Chem., 2004, 1420–1426 CrossRef CAS.
- C. N. Banti, C. P. Raptopoulou, V. Psycharis and S. K. Hadjikakou, Novel silver glycinate conjugate with 3D polymeric intermolecular self-assembly architecture; an antiproliferative agent which induces apoptosis on human breast cancer cells, J. Inorg. Biochem., 2021, 216, 111351 CrossRef CAS PubMed.
- M. Poyraz, C. N. Banti, N. Kourkoumelis, V. Dokorou, M. J. Manos, M. Simčič, S. Golič-Grdadolnik, T. Mavromoustakos, A. D. Giannoulis, I. I. Verginadis, K. Charalabopoulos and S. K. Hadjikakou, Synthesis, structural characterization and biological studies of novel mixed ligand Ag(I) complexes with triphenylphosphine and aspirin or salicylic acid, Inorg. Chim. Acta, 2011, 375, 114–121 CrossRef CAS.
- D. S. Sagatys, R. C. Bott, G. Smith, K. A. Byriel and C. H. L. Kennard, The preparation and crystal structure of a polymeric (1:1)-silver nitrate urea complex, [(AgNO3)2(CH4N2O)2]n, Polyhedron, 1992, 11, 49–52 CrossRef CAS.
- F. J. Carmona, G. D. Sasso, G. B. Ramírez-Rodríguez, Y. Pii, J. M. Delgado-López, A. Guagliardi and N. Masciocchi, Urea-functionalized amorphous calcium phosphate nanofertilizers: optimizing the synthetic strategy towards environmental sustainability and manufacturing costs, Sci. Rep., 2021, 11, 3419 CrossRef CAS PubMed.
- M. A. C. de Oliveira, P. P. M. Pico, W. da Silva Freitas, A. D'Epifanio and B. Mecheri, Iron-Based Electrocatalysts for Energy Conversion: Effect of Ball Milling on Oxygen Reduction Activity, Appl. Sci., 2020, 10, 5278 CrossRef.
- M. P. Chrysouli, C. N. Banti, N. Kourkoumelis, E. E. Moushi, A. J. Tasiopoulos, A. Douvalis, C. Papachristodoulou, A. G. Hatzidimitriou, T. Bakas and S. K. Hadjikakou, Ciprofloxacin conjugated to diphenyltin(IV): a novel formulation with enhanced antimicrobial activity, Dalton Trans., 2020, 49, 11522–11535 RSC.
- M. Elder, F. Stapleton, E. Evans and J. K. G. Dart, Biofilm-related infections in ophthalmology, Eye, 1995, 9, 102–109 CrossRef PubMed.
- H. Hintzsche, G. Montag and H. Stopper, Induction of micronuclei by four cytostatic compounds in human hematopoietic stem cells and human lymphoblastoid TK6 cells, Sci. Rep., 2018, 8, 3371 CrossRef.
- M. Fenech, Cytokinesis-block micronucleus cytome assay, Nat. Protoc., 2007, 2, 1084–1104 CrossRef CAS PubMed.
- C. N. Banti and S. K. Hadjikakou, Evaluation of Genotoxicity by Micronucleus Assay in vitro and by Allium cepa Test in vivo, Bio-Protoc., 2019, 9, e3311 Search PubMed.
- C. N. Banti and S. K. Hadjikakou, Evaluation of Toxicity with Brine Shrimp Assay, Bio-Protoc., 2021, 11, e3895 Search PubMed.
- A. F. Trompeta, I. Preiss, F. Ben-Ami, Y. Benayahu and C. A. Charitidis, Toxicity testing of MWCNTs to aquatic organisms, RSC Adv., 2019, 9, 36707–36716 RSC.
- M. Ates, J. Daniels, Z. Arslan, I. O. Farah and H. F. Rivera, Comparative evaluation of impact of Zn and ZnO nanoparticles on brine shrimp (Artemia salina) larvae: effects of particle size and solubility on toxicity, Environ. Sci.: Processes Impacts, 2013, 15225–15233 Search PubMed.
- B. Zhu, S. Zhu, J. Li, X. Hui and G.-X. Wang, The developmental toxicity, bioaccumulation and distribution of oxidized single walled carbon nanotubes in Artemia salina, Toxicol. Res., 2018, 7897–7906 Search PubMed.
- C. N. Banti, N. Kourkoumelis, A. G. Hatzidimitriou, I. Antoniadou, A. Dimou, M. Rallis, A. Hoffmann, M. Schmidtke, K. McGuire, D. Busath, A. Kolocouris and S. K. Hadjikakou, Amantadine copper(II) chloride conjugate with possible implementation in influenza virus inhibition, Polyhedron, 2020, 185, 114590 CrossRef CAS.
- M. Ç. Akgündüz, K. Çavuşoğlu and E. Yalçın, The Potential Risk Assessment of Phenoxyethanol with a Versatile Model System, Sci. Rep., 2020, 10, 1209 CrossRef.
- S. A. S. Mercado and J. D. Q. Caleño, Cytotoxic evaluation of glyphosate, using Allium cepa L. as bioindicator, Sci. Total Environ., 2020, 700, 134452 CrossRef CAS PubMed.
- Rigaku/MSC, CrystalClear, Rigaku/MSC Inc., The Woodlands, Texas, USA, 2005 Search PubMed.
- G. M. Sheldrick, A short history of SHELX, Acta Crystallogr., Sect. A: Found. Crystallogr., 2008, 64, 112–122 CrossRef CAS PubMed.
- G. M. Sheldrick, Crystal structure refinement with SHELXL, Acta Crystallogr., Sect. C: Struct. Chem., 2015, 71, 3–8 Search PubMed.
- DIAMOND-Crystal and Molecular Structure Visualization, Ver. 3.2, Crystal Impact, Rathausgasse 30, 53111, Bonn, Germany Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 2089744. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/d1dt02158c |
| This journal is © The Royal Society of Chemistry 2021 |