Open Access Article
Open Access ArticleA microphysiological system for handling graphene related materials under flow conditions†
Alodia
Lacueva-Aparicio
ab,
Viviana Jehová
González
 b,
Ana Rosa
Remacha
a,
Daniel
Woods
a,
Eduardo
Prado
b,
Ignacio
Ochoa
b,
Ana Rosa
Remacha
a,
Daniel
Woods
a,
Eduardo
Prado
b,
Ignacio
Ochoa
 ac,
Sara
Oliván
ac,
Sara
Oliván
 *a and
Ester
Vázquez
*a and
Ester
Vázquez
 *bd
*bd
aTissue Microenvironment (TME) Lab, I3A _ IIS Aragón, University of Zaragoza, 50018 Zaragoza, Spain
bInstituto Regional de Investigación Científica Aplicada (IRICA), University of Castilla-La Mancha, Ciudad Real 13071, Spain. E-mail: ester.vazquez@uclm.es
cCIBER in Bioengineering, Biomaterials and Nanomedicine (CIBER-BBN), Madrid, Spain
dFaculty of Chemical Science and Technology, University of Castilla-La Mancha, Ciudad Real 13071, Spain
First published on 5th April 2024
Abstract
The field of nanotechnology has developed rapidly in recent decades due to its broad applications in many industrial and biomedical fields. Notably, 2D materials such as graphene-related materials (GRMs) have been extensively explored and, as such, their safety needs to be assessed. However, GRMs tend to deposit quickly, present low stability in aqueous solutions, and adsorb to plastic materials. Consequently, traditional approaches based on static assays facilitate their deposition and adsorption and fail to recreate human physiological conditions. Organ-on-a-chip (OOC) technology could, however, solve these drawbacks and lead to the development of microphysiological systems (MPSs) that mimic the microenvironment present in human tissues. In light of the above, in the present study a microfluidic system under flow conditions has been optimised to minimise graphene oxide (GO) and few-layer graphene (FLG) adsorption and deposition. For that purpose, a kidney-on-a-chip was developed and optimised to evaluate the effects of exposure to GO and FLG flakes at a sublethal dose under fluid flow conditions. In summary, MPSs are an innovative and precise tool for evaluating the effects of exposure to GRMs and other type of nanomaterials.
New concepts2D materials are becoming increasingly common in commercial or near-market products. This has made it necessary to analyse their safety using standardized studies, such as the OECD guidelines, which are generally designed for chemicals that are mostly soluble in aqueous dispersions. Are these traditional static test methods useful for analysing 2D materials? These materials, unlike other nanomaterials, exhibit poor stability in aqueous solutions, adsorb to plastic structures and tend to deposit rapidly. As a result, traditional approaches are often not reproducible and lead to errors in biological assays. Our study has optimized for the first time a microfluidic system under flow conditions to minimize the adsorption of graphene materials onto components such as tubes and devices and their deposition on cells. Using this design, we have compared the response of a cell model under static and flow conditions, verifying the viability of our proposal, and we have designed a kidney-on-a-chip system that emulates the shear stress experienced by cells in this type of tissues. This work paves the way for the preparation of other microphysiological systems that recreate different tissues whose functions can be altered by interaction with 2D materials. |
Introduction
The discovery of graphene and, subsequently, the broader family of 2D materials has triggered new lines of research in a multitude of disciplines. Numerous research groups have worked on the synthesis and property characterisation of these new structures, which has led to the development of many applications in different fields.1–3 Despite the expectation created, experience has shown us that translation to the real world takes time and that, for this to happen, it is also necessary to assess the safety of the material and its interaction with the environment and living organisms.4,5 For this reason, especially in the last decade, great efforts have been made to determine the reliability of these materials, their interaction with biological barriers such as skin, the lungs, the placenta, the cardiovascular system, the gastrointestinal system, the immune systems and the central nervous system, as well as their compatibility with various organisms from different ecosystems.6As is the case for other nanoparticles, in general, most studies have been conducted following the same models that have been used for decades to evaluate chemicals that dissolve perfectly in the cell culture medium. However, in the case of 2D materials, agglomeration and aggregation must also be considered as diffusion is not the main process that takes place.7 Even in those materials containing hydrophilic functional groups, such as graphene oxide (GO), visible sedimentation occurs on culture plates. Moreover, during prolonged treatments, the material often remains anchored to the cell wall despite washing. Therefore, it is questionable, for example, whether the initial concentration of the material accurately represents the dose at which the studies are conducted, or what would happen if the experimental conditions were such that precipitation could be avoided.8 To solve these problems, some protocols recommend centrifuging the materials until stable dispersions are achieved. However, this solution does not seem appropriate either, as it eliminates those materials that are less stable in such media due to their physicochemical characteristics, thus meaning that the toxicity assessment is biased. Other solutions, such as the use of surfactants or chemical modification of materials, may also lead to results that are not representative of the material itself.9,10
The observation that traditional cell culture methods are not the optimal choice for assessing the biological behavior of nanoparticles has already been demonstrated by a variety of studies. In fact, some authors note that this could be a major problem when testing these nanoparticles in clinical studies since, in general, the results observed in vitro differ significantly from those in vivo. For this reason, many efforts have been made to develop more realistic in vitro studies that are able to better simulate tissue physiology.11,12
To date, studies on the interaction of graphene-related materials (GRMs) with different tissues or cell types, and their cytotoxic effect, have mainly been performed in two ways. The first uses animal models, which arguably fail to recreate human physiology and have bioethical implications,13,14 whereas the second uses static in vitro models, in which a GRM dispersion is deposited on the cells, although this fails to recreate the fluidic or mechanical properties present in the human body. This point is particularly relevant in several tissues, such as the renal or vascular system, where cells live in a dynamic environment and are exposed to forces related to fluid movement.15 Furthermore, non-fluidic assays could alter the findings concerning the effects of GRMs since, as already mentioned, these materials tend to deposit quickly on the cells and form a film, in some cases promoting an artificial interaction with them.
A newer type of in vitro model that has arisen in recent years and could solve the problems mentioned above, involves microphysiological systems (MPSs), which are also referred to as organs-on-a-chip (OOC).16 This technology integrates complex cell-culture techniques (hydrogels, organoids, or even co-cultures) and mechano-physiological conditions to recreate the tissue microenvironment.17 Furthermore, these systems allow cellular interaction with the extracellular matrix and incorporate electrical and mechanical stimuli.18 As OOCs can reproduce the physiology of the tissue, they could also reproduce the response to different drugs and toxins.17 When using OOC technology, multiple advantages, such as minimising the materials and reagents needed, better control of critical parameters through integrated sensors and minimising or even replacing animal models, can also be found.19–21 Thus, OOC appears to be an excellent instrument to avoid nanoparticle precipitation and to develop specific models recreating specific conditions.22,23
Several studies have been carried out using organ-on-a-chip technology for the evaluation of different NPs such as gold nanoparticles in endothelial cells20,24 but none of them have been previously performed analysing the cell response to GO or FLG. The novelty of the present work relies on the fact that there are no studies with GRMs under fluid flow conditions applied to cell culture. Establishing microphysiological systems capable of circulating graphene-based nanomaterials through microfluidic devices is crucial to mimic physiological conditions and explore the effects of these particles on various tissues within the human body.
In the present work, a microfluidic model under fluid flow conditions was developed to evaluate GRM interactions. Although this system could apparently help to solve GRM deposition by maintaining a stable nanoparticle dispersion and could therefore be a good alternative for performing biological studies, it was necessary to assess the interaction between the aggregates and the system compounds (mainly tubing and devices). Graphene tends to adsorb to plastic materials commonly used in experimental procedures or biomedical applications.24 As such, the loss of nanoparticles throughout the microfluidic system could influence the GRM concentration, altering the effects of particles and, therefore, the results. For that reason, the use of different materials in the microfluidic system setup was tested in order to reduce GRM adsorption.
The present study represents a pioneering effort in validating the components of a microfluidic system tailored specifically for the manipulation of 2D materials, particularly graphene-based nanomaterials (GRMs), with precise control over their adsorption and precipitation processes. Furthermore, the current kidney-on-a-chip model allows a physiological shear stress to be applied under fluid flow conditions, thereby mimicking the human renal proximal tubule. Thus, MPS are postulated as a more realistic model for the evaluation of the possible cytotoxic effects of GRMs and other nanomaterials.
Results and discussion
Characterisation of GO and FLG prior to cell culture exposure
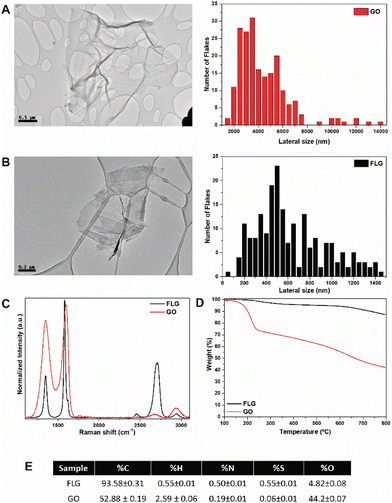
HRTEM imaging was used to assess the morphology of GO (Fig. 1A) and FLG (Fig. 1B), revealing graphene aggregates with average sizes of 602.30 ± 320.20 nm for FLG and 4143.98 ± 2130.77 nm for GO. As shown in Fig. 1C, Raman spectroscopy was employed to analyse the structural properties of the nanomaterials, focusing on key graphene bands: D at 1350 cm−1 (associated with an amorphous phase in the carbon rings of graphene), G at 1580 cm−1 (representing sp2 carbon bonds in the hexagonal structure), and 2D at 2700 cm−1 (indicative of carbon ring quality and used to determine the number of layers (NG) in the graphene structure).25–27 FLG exhibited an average of four layers (FWHM of 64.25 cm−1) calculated according to Paton et al.,25 whereas GO lacked the 2D band, thus indicating a high degree of structural defects in its graphene layers.3 This observation was further supported by the intensity ratio of the D and G bands (ID/IG), which is associated with the defect density of graphene,28 with GO displaying a value of 1.36 and FLG a value of 0.36, thus suggesting fewer defects in FLG compared to GO. Thermogravimetric analysis of GO and FLG demonstrated the thermal stability of the nanomaterials (Fig. 1D). Thus, under a nitrogen atmosphere, GO and FLG exhibited a weight loss of 44.3% and 5.7% respectively, thereby indicating a higher density of defects and oxygen content in GO, the latter of which is attributed to residual oxygen-containing groups at the edges of graphene. Generally, the primary mass loss occurred in the temperature range of 100–300 °C due to the presence of functional groups such as –OH, –COOH, –C–O–C, esters, etc., within the structure of the nanomaterials.3,29 Finally, Fig. 1E presents the elemental analysis of FLG and GO, revealing an oxygen percentage of 44.2 wt% for GO and 4.8 wt% for FLG, thereby corroborating the values observed in the TGA analysis.Optimised microphysiological system for handling GRM nanomaterials
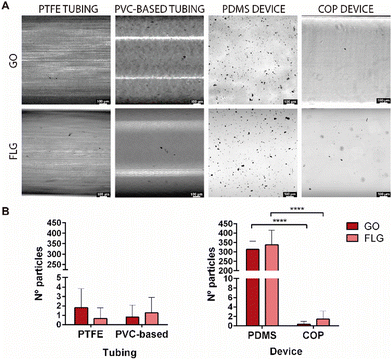
Two components are relevant when designing an MPS: tubing and microfluidic devices. The nature of their composition is critical to avoid the possibility of retaining graphene aggregates on their surface and thus to ensure the concentration of nanomaterial to which the cells will be exposed. The primary materials employed for tubing are PTFE and PVC, therefore these materials were chosen for comparison. Similarly, in the case of devices, PDMS and COP have been studied. These materials are highly hydrophobic, resistant to organic chemicals, autoclavable, chemically inert towards most reagents used in cellular studies, and have high mechanical resistance.After perfusing the GO and FLG suspension at a 10 μg mL−1 concentration in distilled water, the tubing and devices analysed presented some black particles adsorbed on the surfaces. Distilled water was used to avoid possible artefacts from the culture medium or PBS containing salts or other components that could lead to an error in counting particle aggregates. Bright-field microscopy images showed that, in general, these particles have a high heterogeneity and were more abundant in the PDMS device than in the other materials analysed (Fig. 2). The results from counting the particles per area (300 μm2) of the tubing system showed that GO and FLG particles do not tend to adhere to the surface of both PTFE and PVC-based tubing. With regard to microfluidic devices, the results showed that graphene aggregates deposited preferentially on PDMS devices rather than COP devices, with the number of particles per area found on the surface of PDMS being several times higher than for COP. This different adhesion could be related to surface treatments or the chemical composition, which could facilitate preferential adhesion towards PDMS devices. Moreover, commercial COP-based devices are typically manufactured by injection moulding and subsequent polishing, with solvent vapour or chemicals, to achieve optical transparency. These data show that the adsorption of graphene depends more on the material employed to make the device than on the type of GRM used. However, the different aggregation degree of GO and FLG on different material surfaces, can be rationalised by analysing their intrinsic physicochemical characteristics. GO, with its considerably larger flake size and substantial oxygen functionalities, exhibits enhanced surface interaction capabilities. These oxygen-containing groups not only increase the hydrophilicity of GO but also provide numerous active sites for binding, facilitating more pronounced aggregation on material surfaces. In contrast, FLG, characterised by its smaller flake size and lower defect and oxygen content, is less susceptible to such interactions. The minimal presence of functional groups on FLG reduces its surface activity, leading to lower aggregation compared to GO.
Nevertheless, we wanted to confirm that the black particles on the surface of the tubing and the device were GO and FLG aggregates and not defects in the processing of the device and tubing materials, which could result in imperfections or aggregates within the channels of the device or the tubing part of the system. To that end, surface sections were visualised by HRSEM microscopy, to observe their morphology (Supplementary 1, ESI†). The results showed that the morphology of these particles resembled GO and FLG agglomerates, as described in the literature.30 A compositional EDS analysis was also conducted (Supplementary 2, ESI†). As noted by the manufacturer, the data showed that PTFE and PDMS materials contain mainly fluorine and silicon, respectively, in their composition, in addition to carbon and oxygen. In the case of the found particles, only carbon and oxygen were present, with carbon being the main component. Thus, these data corroborate that the black particles observed correspond to GRM aggregates (Supplementary 2, ESI†). The quantity of particles deposited on the COP surface was too low to allow HRSEM images to be recorded and compositional analysis to be carried out.
According to the results, PTFE and PVC-based in tubing and COP in devices should be the materials of choice when working under flow conditions with microfluidic platforms since they better maintain the initial graphene concentration.
Additionally, COP devices can provide certain advantages for cellular studies compared with PDMS-based devices, although both materials have been shown to be biocompatible and adequate for cell culture and have been used to produce microfluidic devices. Firstly, COP does not retain small molecules due to its low porosity and it also exhibits lower autofluorescence, which makes it useful for microscopy based on immunofluorescence or when performing experiments with fluorescent molecules. Moreover, COP is impermeable to gases, thus allowing the possibility to perform hypoxia assays, amongst others.31–33
The next step was to choose the best propulsion system and microfluidic configuration for conducting the assays in the presence of GRMs. The main tools for liquid manipulation on a small scale are microfluidic flow control systems based on pressure or vacuum control, peristaltic and syringe pumps. Peristaltic pumps have many advantages when compared with other perfusion systems, such as pressure or syringe pumps. For example, they allow cell culture media to be recirculated through the system. This is paramount when performing experiments with expensive cell-culture media or in long-term experiments: recirculation minimises the handling, whereas unidirectional experiments require the reservoir to be refilled every so often. Fluid flow can also be controlled since peristaltic pumps can perfuse a constant fluid flow, thus resulting in an absence of changes that might affect cell culture. Moreover, fluid flow conditions provide fresh nutrients to the cell culture and a low level of waste products. Differently, under static conditions the same volume is maintained throughout the whole experiment, leading to cell culture media containing the metabolic cell products and a decreasing level of nutrients over time.
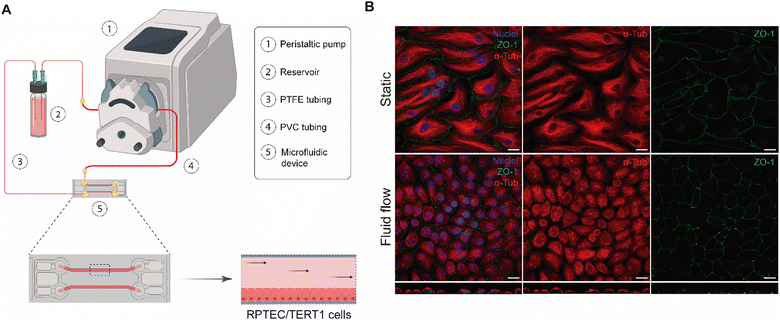
Consequently, after testing different materials and configurations, the optimised MPS is composed of a peristaltic pump, PTFE and PVC tubing, a COP microfluidic device, and a glass reservoir with a magnetic stirrer (Fig. 3A).
Validation of a kidney-on-a-chip model
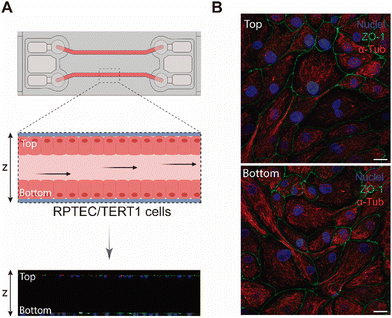
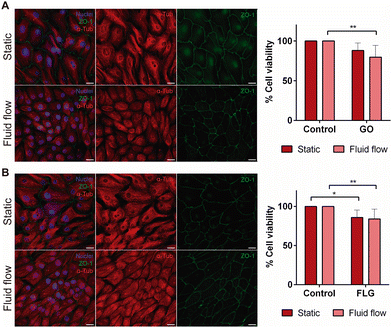
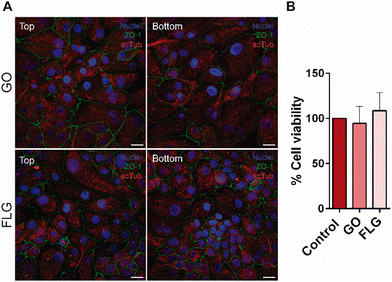
The aim of our study was to validate a MPS for handling graphene under physiological fluid flow conditions. To that end, a kidney-on-a-chip model based on OOC technology was optimised to evaluate and compare the effect of GO and FLG particles on cells under static and flow conditions. This model should make it possible to recreate physiological conditions and to maintain the integrity of the monolayer and cell viability, thus validating the model for cytotoxicity assays. An established cell line from human renal proximal tubular cells (RPTEC/TERT1) was used for developing our model because the proximal tubule plays a key role in xenobiotic transport. That is the reason why the segment of the nephron is more often used for toxicity assays.34Firstly, the monolayer integrity, cytoskeleton organisation and cell morphology were evaluated after applying a physiological shear stress by perfusing cell culture medium through the system in comparison with static conditions (Fig. 3B). With that purpose, the identification of Zonula occludens 1 (ZO-1), also known as Tight Junction Protein-1, which is crucial for establishing a functional barrier-forming layer to visualise membrane integrity, was employed. Meanwhile, the expression of α-tubulin protein was used to assess cytoskeleton organisation and the presence of primary cilia. The differences in morphology observed between the static and flow conditions are explained by the fact that cells were seeded on cover slides (static) and the microfluidic device (flow). Under static conditions, RPTEC/TERT1 cells exhibited greater growth and expansion due to the larger surface area available for proliferation compared to those seeded within the microfluidic channels. It is noteworthy that substrate stiffness can influence cell morphology, potentially contributing to differences between cells on coverslips in multi-well plates and those within the microfluidic device. Furthermore, experiments under fluid flow conditions involved a continuous circulation of 5 mL of culture medium, providing more nutrients, whereas under the static condition, the smaller volume resulted in lower nutrient availability, potentially leading to reduced proliferation rates. After perfusing cell culture medium for 72 hours whilst applying physiological shear stress (0.2 dyne cm−2), the RPTEC/TERT1 cells maintained the cell–cell connections, the presence of primary cilia and cytoskeleton organisation observed as seen in previous studies where proximal tubular epithelial cells were exposed to shear stress.15,35 In order to recreate the renal proximal tubule present in vivo, a tubule-like structure was reproduced in the microfluidic device (Video 1, ESI†). To that end, the next step was to seed RPTEC/TERT1 cells both at the bottom and top of each channel (Fig. 4A). After applying a higher fluid flow rate corresponding to a shear stress of 0.48 dyne cm−2 for 72 hours, the monolayer remained intact both at the bottom and at the top of the microdevice channels (Fig. 4B). Both fluid flow regimes exposed the cell culture to physiological levels of shear stress in the kidney, as described in the literature.15,36,37 Hence, both developed models have been validated for use in GRM assays.
A key characteristic of the microfluidic technology is the presence of fluid dynamics in the systems mimicking the constant native fluid flow, which presents varying velocities (slower flow rates in the interstitium and the fastest inside blood vessels).
Flow applies shear stress, which impacts cellular function and viability and can result in cellular adhesion, activation, differentiation, extravasation, long-term survival, and even how cells uptake different nanoparticles, thus altering the cellular response to these materials, as well as an increased cell-to-cell adhesion.38,39 As such, the mechanical forces related to the flow generated must be considered in this model.
Previous studies have demonstrated that GRMs can exert different cytotoxic effects dependent on concentration and/or particle size and oxidation degree. Toxic effects in vitro include ROS generation, DNA damage, reduced cell viability, and/or membrane damage.40–45 Meanwhile, in vivo studies performed with animal models have shown that GRMs tend to accumulate throughout the organism, mainly in the liver, lungs, spleen and kidney, thus resulting in varying degrees of damage to these organs.46–52
In the specific case of renal cell studies, cells have shown that their viability presents a dose-dependent cytotoxic effect when human embryonic kidney cells were seeded using traditional cell culture and were kept under static conditions after GO exposure.53 This cell line showed no significant toxicity when cell cultures were exposed to GO at sublethal concentrations of up to 10 μg mL−1 under static conditions.54 Graphene toxicity assays have also been performed in animal models such as mice. One such study showed that small-GO (s-GO) and large-GO (l-GO) were eliminated by glomerular filtration and tubular secretion, respectively. To confirm these results, a renal proximal tubular cell line (HK-2) was used and the uptake of l-GO was found to be higher than s-GO after 4 and 24 hours of graphene exposure for HK-2 cells in traditional cell culture.49
As mentioned previously, graphene toxicity studies in renal tissue have been performed in vivo and in vitro although they have some limitations. Studies performed in cell culture do not recreate the microenvironment of the kidney since static conditions do not mimic the physiological properties of the proximal tubule. With regard to animal models, their use is expensive, time-consuming and raises ethical concerns. As such, a kidney-on-a-chip model could be extremely useful for assessing nanomaterial toxicity. The validated kidney-on-a-chip model recreates the biomechanical properties of the renal proximal tubule since fluid flow allows us to apply a physiological shear stress to the cell culture in the microfluidic device.37
GO and FLG exposure assays
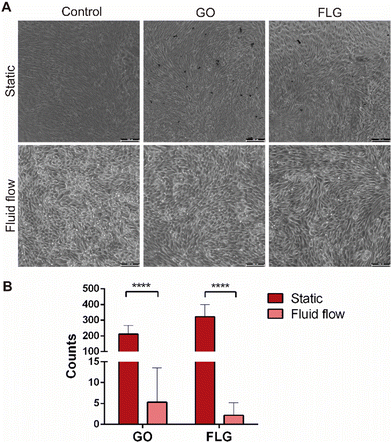
All experiments, both under static and fluid flow conditions, were performed at a sublethal dose of GO and FLG (10 μg mL−1). After 72 hours of exposure, GO and FLG aggregates were visible under the static condition because they deposit on top of the cell culture (Fig. 5A). However, such aggregates were not present within the channels of the microfluidic devices under low fluid flow conditions since GO and FLG circulate through the system, which mimics what would happen under physiological conditions (Fig. 5A). Aggregates were counted to compare both static and fluid flow conditions, and the analysis showed that the number of GO and FLG particles was significantly much higher under static conditions than under fluid flow conditions.In the case of GO, the flow rate reduced the presence of aggregates significantly when compared with the control (5.30 ± 8.24 GO aggregates for fluid flow vs. 211.70% ± 55.40 GO aggregates for static; p < 0.0001), whereas the reduction for FLG was significantly greater (2.2 ± 2.27 FLG aggregates for fluid flow vs. 321.9 ± 76.41 FLG aggregates for static; p > 0.0001) (Fig. 5B).
It is known that static conditions can result in the deposition of other nanoparticles (NP), thus leading to substantial modifications in both the interactions between NP and cells and the kinetics of their transportation, thereby affecting the cellular uptake process. On the other hand, under fluid flow conditions a tangential force known as shear stress is exerted on the surface of the cell culture, thus leading to changes in the interactions between cells and NPs. These changes include the uptake of NPs by cells and the viability of the cell culture after NP exposition.22
Cell integrity and morphology were monitored after GO and FLG exposure to determine whether the presence GO and FLG aggregates affected the plasma membrane integrity or cytoskeleton organisation. The monolayer remained intact after GO and FLG exposure in both static and fluid flow models, thus allowing us to conclude that, at low concentrations, there appears to be no mechanical damage under fluid flow conditions (Fig. 6A and B). A similar finding was obtained for the model in which cells recreated a tubule-like structure under fluid flow conditions, applying a higher shear stress than in the model containing only a monolayer at the bottom of the channels (Fig. 7A).
Finally, a cell viability assay was carried out to determine whether GO and FLG exposure affected cell culture in kidney-on-a-chip model when compared with the static model. Cell viability was lower when cells were exposed to GO in both the static and kidney-on-a-chip models, being significantly lower for the fluid flow model when compared with its control (79.79 ± 14.70% vs. 100%; *p < 0.1) (Fig. 6A). Similar results were obtained in terms of cell viability when cells were exposed to FLG (static 85.87 ± 9.66% vs. fluid flow 84.12 ± 12.65%; p* < 0.1, ** < 0.01), being significantly lower in both models when compared with the control with no FLG exposure (Fig. 6B). According to the results obtained with the kidney-on-a-chip model, the preliminary data show that GO and FLG materials decrease cell viability, with this effect being statistically significant under flow conditions. No significant decrease in cell viability was observed when cells were exposed to GO under static conditions at a non-toxic sublethal dose (10 μg mL−1) (Fig. 6B), and this effect has already been described by other authors.54 Although the integrity of the monolayer remains intact and microscopic images do not show cell deterioration a priori, our results provide evidence that cell viability under flow could be compromised. As such, the impairment of cellular metabolic activity could be related to the mechanical effect of the flakes on cells. This finding indicates that graphene flakes could compromise the physiological state of cells, even when non-toxic doses are applied. Furthermore, the chemistry of the nanomaterials also plays an important role since the higher the oxygen concentration the higher the oxidative stress exerted on the cells. GO, which is highly oxidized, generates a higher increase in ROS.55 This effect could explain the lower viability obtained in the assays where cells were exposed to GO in comparison to FLG assays.
The more advanced kidney-on-a-chip model was also used for cell viability assays after exposing RPTEC/TERT1 to GO and FLG for 72 hours at a higher fluid flow rate (Fig. 7B). The presence of both 2D materials altered cell viability when compared to its control under fluid flow conditions, showing that GO and FLG are affecting the cell culture, making this model suitable for toxicity assays.
Many types of cell culture media used for in vitro studies contain fetal bovine serum (FBS) to enhance cell proliferation even though it is known that the proteins present in FBS can interfere in the interaction of NPs with the cell culture medium, thus generating the protein corona.9,56 Cell culture media used for all experiments performed in the present work did not contain FBS so the differences in cell viability between conditions are not related to interferences between FBS and the 2D materials used. However, regarding the interaction of GRMs with other components of the cell culture media, it is known that they have the capability to adsorb and deplete nutrients from the cell culture media at low graphene doses. This effect could lead to a lower proliferation of the cell culture as described by Creighton et al.57 Nutrient depletion could explain the lower viability shown in the assays where basal cell culture medium was used in the first set of experiments where cells were exposed to GO and FLG under static and fluid flow conditions (Fig. 6). However, this effect could not have been observed in the optimised kidney-on-a-chip model since the cell culture medium (ProxUp) was enriched with more nutrients than in the previous experiments (Fig. 7). The reason why the cell culture medium was enriched with nutrients such as glucose was because the cell culture was consuming more nutrients since there were cells at the top and the bottom of the channels, not only at the bottom as in our first model. These results show how important the selection of the cell culture media is for exposing cells to GRMs.
Finally, two main advantages of using this new MPS can be discussed. On the one hand, 2D materials have the ability to aggregate over time. Some components of the cell culture media such as ions and biomolecules can play an essential role in mediating their agglomeration and deposition. For instance, ions can interact with the negatively charged functional groups on 2D materials, influencing their dispersion behaviour. In some cases, biomolecules like proteins can be used to stabilize the dispersions because they can be adsorbed onto the surface of the GRM, creating a steric barrier that hinders aggregation. Therefore, the biological response of these 2D materials is highly dependent on these interactions.58 The use of these MPSs in which 2D materials are in constant movement throughout the system avoids the problem of aggregation and deposition on the cells and the physiological response could be more accurate compared to traditional cell culture under static conditions.
Another important point to consider is that, due to the increasing use of 2D materials in real world applications, they must undergo standardised testing such as OECD guidelines in order to determine their safety. These test guidelines are described in general for chemicals and the aggregation of 2D materials under the test conditions may lead to problems in the application of these guidelines. For example, this is the case of a widely used technique for studying genotoxicity (OECD TG 487).59 Thus, the application of fluid flow conditions could be a solution for the appropriate evaluation of the toxicity of the 2D materials in in vitro studies.
Conclusions
We have developed and optimised the first MPS to evaluate the effects of GO and FLG aggregates on cells under flow conditions. This system allows control over the deposition of aggregates by keeping GRMs in continuous movement. Moreover, as a result of the materials used in tubing and devices, the loss of GRMs due to adherence to them is minimised. As such, MPS under fluid flow conditions provide a more accurate system for evaluating the cytotoxic effects of 2D materials in suspension, not only GRMs, while helping to preserve their intrinsic properties. Our results not only showed that at a non-lethal dose of these GRMs they were not more toxic under fluid flow conditions than under static conditions, but also that fluid flow conditions enhance the solubility of GO and FLG when compared to static conditions. In this sense, these innovative models emerge as a more realistic alternative to conventional in vitro assays, representing a paradigm shift in experimental approaches. Apart from the kidney-on-a-chip model, some other OOCs recreating different tissues, such as the liver, lungs or blood vessels, whose functions may be altered by interaction with GRMs, could also be optimised.Materials and methods
Synthesis of GO and FLG nanomaterials
GO synthesised by Grupo Antolin (https://www.antolin.com) was prepared using helical-ribbon carbon nanofibers (GANF®) as the starting material. A modified Hummer's method was employed utilising oxidising agents such as a KMnO4/H2SO4 mixture and sodium nitrate at 0 °C. The resulting GO was washed multiple times with Milli-Q water to a pH of approximately 5 in order to eliminate any residual acid traces. The washing process involved re-dispersion and centrifugation at 4000 rpm for 30 minutes. Finally, the GO was obtained in powder form by lyophilization at –80 °C and a pressure of 0.005 bar.FLG was synthesised using mechanochemical treatment following the methodology described in ref. 60 and 61. In this process, 7.5 mg of graphite (from Bay Carbon, Inc.) and 22.5 mg of melamine (Sigma-Aldrich) were mixed in a 25 mL stainless steel jar containing 10 stainless steel balls with a diameter of 1 cm. The mixture was milled in a Retsch PM 100 planetary mill, at 100 rpm for 30 minutes. The resulting powder was dispersed in 20 mL of Milli-Q water and dialyzed at 70 °C with five solution changes. After five days of sedimentation, FLG was obtained by extracting the supernatant. To obtain FLG in powder form, the supernatant was further lyophilized at –80 °C and a pressure of 0.005 bar.
Characterisation of nanomaterials
Different techniques were used to characterise the nanomaterials. An InVia Renishaw microspectrometer with a 532 nm laser was used to record Raman spectra. To avoid the effects of laser heating, the power density was kept below 1 mW μm−2. Raman analyses were conducted on solid samples under normal conditions, and multiple random locations on each sample (at least 30–40) were analysed. Lorentzian-shaped bands were fitted to the D, G, and 2D peaks of the spectra to determine their positions, widths, and intensities. A GeminiSEM 500 field emission instrument manufactured by Zeiss was utilized to conduct SEM imaging, elemental mapping, and EDS analyses.For TEM analyses, stable graphene dispersions were prepared, diluted as needed and dip-casted onto Lacey carbon supported copper grids (3.00 mm, 200 mesh) coated with carbon film. The samples were then dried under vacuum. High-Resolution transmission electron microscopy (HRTEM) was conducted using a JEOL 2100 instrument with an accelerating voltage of 100 kV.
Thermogravimetric analyses (TGA) were carried out using a TGA Q50 instrument (TA Instruments). Analyses were performed under a nitrogen flow, starting from 100 °C and increasing at a rate of 10 °C min−1 up to 800 °C.
High-resolution scanning electron microscopy (HRSEM) and elemental analysis
Scanning electron microscopy was performed to verify that the particles observed during the microscopic analysis were indeed GO and not any other kind of particles that could have entered the microfluidic system during the experiment. HRSEM was performed with a GeminiSEM 500 (Zeiss). The samples had been previously treated with a gold coating to improve visualisation. Different images were taken at random points of the devices used for GO and FLG experiments, and unused materials/devices through which no flow had been passed were used as controls. An atomic compositional analysis was performed using an 80 mm2 energy dispersive spectroscopy (EDS) detector (Oxford) at 10 kV to further verify the nature of the deposited particles and the surface composition of the device.Microfluidic system
The microfluidic flow system includes the tubing to carry the medium, the microfluidic device, the glass reservoir containing the graphene solution or the medium, and a peristaltic pump to move the liquid through the system (Watson–Marlow 323d; Fig. 4A). Two different types of tubing were evaluated: one based on a fluorocarbon polymer (Polytetrafluoroethylene (PTFE), 1608-20, Darwin Microfluidics) and the other based on polyvinyl chloride (PVC) materials (Tygon S3 E-3603, Saint-Gobain). Depending on the microfluidic device, two materials were assessed: polydimethylsiloxane (PDMS, Sylgard 184) and a thermoplastic cyclic olefin polymer (COP). PDMS microfluidic devices were fabricated using Sylgard® 184, mixing with a curing agent at a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. A mould was filled with the PDMS and degassed using a vacuum chamber. After this process, it was cured for at least an hour in an oven at 85 °C. PDMS devices were then removed from the mould and fixed on top of a crystal coverslip using oxygen plasma. The PDMS-based device consists of three parallel channels, each channel being 2 mm wide, 2 mm high, and 35 mm long. Meanwhile, COP microdevices are commercial devices (BE-Flow, BeOnChip). All channels of both PDMS and COP devices had a rectangular section.
1 ratio. A mould was filled with the PDMS and degassed using a vacuum chamber. After this process, it was cured for at least an hour in an oven at 85 °C. PDMS devices were then removed from the mould and fixed on top of a crystal coverslip using oxygen plasma. The PDMS-based device consists of three parallel channels, each channel being 2 mm wide, 2 mm high, and 35 mm long. Meanwhile, COP microdevices are commercial devices (BE-Flow, BeOnChip). All channels of both PDMS and COP devices had a rectangular section.
Fluid shear stress (T) calculations were performed using the Hagen–Poiseuille equation, where η is the viscosity of the medium, Q is the flow rate, w is the channel width and h is the channel height:
| T = 6ηQ/wh2 |
PDMS and COP devices were used for these experiments combining them with PTFE and PVC-based tubing for perfusing GO and FLG aggregates through the system. The assays were performed at 1 dyne cm−2 for 6 hours at room temperature, and the microfluidic systems were washed with distilled water for 30 minutes to remove non-adhered nanoparticles. After washing, 20 pictures for each condition were taken using a bright-field microscope for aggregate counting. Raw tubing and devices were employed as controls for all experiments.
GO and FLG suspension and microscopy visualisation
GO and FLG solutions were prepared by suspending GO and FLG powder in distilled water at a concentration of 10 μg mL−1 followed by ultrasonication (DK Sonic Ultrasonic Cleaner) for one minute. This concentration represents a sublethal dose, so cytotoxic effects must not be present.62Bright-field microscopy was performed to observe particle adhesion to the tubing and the microfluidic devices (Microscope DMi-8, Leica). Particles deposited on the surface of tubing and devices were counted manually. Counting was performed at random areas on the surface of each material, each measuring 300 μm2 (n = 20 for each condition).
Cell culture
The RPTEC/TERT1 cell line (CHT-003-0002, Evercyte) was developed from human proximal tubular epithelial cells and was cultured according to the supplier's guidelines. Cells were seeded and maintained in culture flasks at 37 °C with DMEM/F12 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (PAN-Biotech, P04-41154), 10 mM HEPES-buffer (Sigma-Aldrich, H0887), 10 ng mL−1 hEGF (Sigma-Aldrich, E9644), 5 pM 3,3′,5-triiodo-L-thyronine sodium salt (Sigma-Aldrich, T6397), 3.5 μg mL−1L-ascorbic acid (Sigma-Aldrich, A4544), 5 μg mL−1 transferrin holo (Merck Millipore, 616424), 25 ng mL−1 prostaglandin E1 (Sigma-Aldrich, P8908), 25 ng mL−1 hydrocortisone (Sigma-Aldrich, H0396), 8.65 ng mL−1 sodium-selenite (Sigma-Aldrich, S5261), 100 μg mL−1 G418 (InvivoGen, ant-gn-5) and 5 μg mL−1 insulin (Sigma-Aldrich, I9278).
1) (PAN-Biotech, P04-41154), 10 mM HEPES-buffer (Sigma-Aldrich, H0887), 10 ng mL−1 hEGF (Sigma-Aldrich, E9644), 5 pM 3,3′,5-triiodo-L-thyronine sodium salt (Sigma-Aldrich, T6397), 3.5 μg mL−1L-ascorbic acid (Sigma-Aldrich, A4544), 5 μg mL−1 transferrin holo (Merck Millipore, 616424), 25 ng mL−1 prostaglandin E1 (Sigma-Aldrich, P8908), 25 ng mL−1 hydrocortisone (Sigma-Aldrich, H0396), 8.65 ng mL−1 sodium-selenite (Sigma-Aldrich, S5261), 100 μg mL−1 G418 (InvivoGen, ant-gn-5) and 5 μg mL−1 insulin (Sigma-Aldrich, I9278).
Before seeding cells in a 96-well plate for the static model, a collagen type I coating (Corning, 354236) at a final concentration of 371 μg mL−1 in 0.02 N acetic acid was applied for one hour at 37 °C in an incubator with 5% CO2 and washed with PBS (Lonza, BE17-516F). Immediately after this step, RPTEC/TERT1 cells were seeded in the 96-well plate 3.75 × 105 cells per mL. Once cells had formed a complete monolayer after 72 hours, they were exposed to GO and FLG at a concentration of 10 μg mL−1 in DMEM-F12 (Biowest, L0090-500; Biowest, L0091-500) 5 mM glucose culture media. After keeping the cell culture exposed to GO and FLG during 72 hours with no cell culture media refreshing during the experiment, different assays were performed and bright-field pictures were obtained using the Thunder Leica Microscope.
Kidney-on-a-chip model
A commercial microfluidic device was used for the kidney-on-a-chip model. After applying the collagen I coating described previously in the static model, RPTEC/TERT1 cells were seeded at the bottom or both the bottom and top of the channels of the microfluidic device at 2.5 × 106 cells per mL. After 72 hours, the cells reached confluency in the channels of the device. At this point, RPTEC/TERT1 cells were exposed to a physiological shear stress of 0.25 dyne cm−2 with a fluid flow of 75 μL min−1 using a peristaltic pump (Reglo Digital Pump, 4-Channel 12-Roller, Masterflex Ismatec) when cells were only seeded at the bottom of the channels. For experiments in which RPTEC/TERT1 cells were seeded at the bottom and top of the channels, the fluid flow was 140 μL min−1, applying a physiological shear stress of 0.48 dyne cm−2. The microfluidic system was set up as follows. A glass reservoir was connected with Sterican Safety needles 20 G × 1½′′ (Braun, 4670050S-01) to PVC-based tubing (VWR, Tygon LMT 55, VERN070534 06 ND) via luer connectors. The PVC-based tubing was then connected to the microfluidic device using PTFE tubing (Darwin microfluidics, LVF-KTU-15). Once the microfluidic system had been assembled, the reservoir was filled with 3.5 mL of the cell culture medium DMEM-F12 5 mM glucose for experiments under low fluid flow (75 μL min−1) or ProxUp cell-culture medium for the experiments under high fluid flow (140 μL min−1). For GRM exposure, GO and FLG were added to the glass reservoir at a final concentration of 10 μg mL−1. Experiments were performed in an incubator at 37 °C. To keep GO and FLG particles in suspension in the cell-culture media, a PTFE Micro Stirrer Bar (VWR, 442-0594) was introduced within the glass reservoir and a magnetic stirrer (VELP Scientifica, AGE Magnetic stirrer) was used during the experiment under fluid flow conditions.Cell viability assays
The viability of RPTEC/TERT1 cells after 72 hours of GO and FLG exposure at a low fluid flow rate was assessed using an MTT-based kit (In vitro toxicology assay kit; Sigma-Aldrich, TOX1) following the supplier's instructions. Briefly, cells under static and flow conditions were washed once with PBS and MTT was added in an amount equal to 10% of the culture medium volume. After an incubation period of 2 hours at 37 °C in the incubator, the resulting formazan crystals were dissolved by adding an amount of MTT solubilization solution equal to the original culture medium volume. In the case of the MTT assay in the device, the final volume was pipetted up and down several times until solutions of cell culture medium with formazan crystals from each channel of the device were transferred to a 96-well plate. Absorbance was measured at a wavelength of 570 nm in a well-plate reader (Biotek, Synergy HT).RPTEC/TERT1 cell viability at high fluid flow was assessed using a Cell Counting Kit-8 (Dojindo, CK04-11) following the supplier's instructions. This cell viability assay was used in order to minimise sample handling compared to the previously used MTT kit. Cells were washed with PBS and Cell Counting Kit-8 (CCK8) solution was diluted at 10% concentration in ProxUp. After washing, CCK8 10% was added in the devices and incubated for 1 hour at 37 °C. The cell culture medium with the resultant water-soluble WST-8 formazan was transferred from the channels of the device to a 96-well plate. Absorbance was measured at a wavelength of 450 nm in the well-plate reader.
Immunofluorescence
Cells were fixed in 4% paraformaldehyde for 20 minutes at room temperature (RT). After that, they were permeabilized with 0.1% Triton X-100 diluted in PBS for 10 minutes at room temperature. Samples were then incubated with a blocking solution containing 3% bovine serum albumin in PBS for 2 hours at room temperature. Cells were treated with the following antibodies overnight at 4 °C: rabbit monoclonal anti-ZO-1 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, ThermoFisher, 40-2200), mouse monoclonal anti-α-Tubulin (1
200, ThermoFisher, 40-2200), mouse monoclonal anti-α-Tubulin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, SantaCruz, sc-23948). After incubating with the primary antibodies, the cells were incubated with secondary antibodies conjugated to AlexaFluor 488 (1
200, SantaCruz, sc-23948). After incubating with the primary antibodies, the cells were incubated with secondary antibodies conjugated to AlexaFluor 488 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, Life Technologies, A11001) and AlexaFluor 555 (1
500, Life Technologies, A11001) and AlexaFluor 555 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, Invitrogen, A2148) for 1.5 hours at room temperature. A Hoechst 33342 was used to visualise DNA by confocal microscopy (Nikon, Ti Eclipse EZ-C1).
500, Invitrogen, A2148) for 1.5 hours at room temperature. A Hoechst 33342 was used to visualise DNA by confocal microscopy (Nikon, Ti Eclipse EZ-C1).
Statistical analysis
Data are presented as mean ± standard deviation. Statistical analysis was performed using the GraphPad Prism 6 software (GraphPad, San Diego, CA, USA). The D’Agostino–Pearson omnibus test was performed to analyse normality. A Kruskal–Wallis test was performed for nonparametric comparisons, followed by the Mann–Whitney U-test. Statistical significance was set at p < 0.05. A two-way ANOVA was performed to analyse the number of GO and FLG particles and for cell-viability assays, followed by Tukey's multiple comparisons test.Author contributions
Conceptualization: A. L., E. V. and S. O. Methodology: A. L. and D. W. Validation: A. L. and S. O. Forman analysis: A. L., V. G and E. P. Investigation: A. L., V. G and E. P. Writing – original draft: A. L., E. V. and S. O. Writing – review and editing: A. L., E. V. and S. O. Visualization: A. L., E. V. and S. O. Resources: A. R. Supervision: I. O., E. V., and S. O. Funding acquisition: E. V. Project administration: S. O.Conflicts of interest
I. O. and S. O. are promoters for BeOnChip S. L. (Zaragoza, Spain). BeOnChip S. L. does not benefit or take part in any economic decisions of this work. A. L., V. G., A. R., E. P., D. W. and E. V. have no conflicts to declare.Acknowledgements
The authors acknowledge financial support from the European Flagship project GA881603 Graphene Core 3 and the Spanish government (project PID2020-113080RB-I00). This study forms part of the Advanced Materials programme and was supported by MCIN with funding from the European Union NextGenerationEU (PRTR-C17.I1) and the Junta de Comunidades de Castilla-La Mancha.References
- H. Zhang, H. M. Cheng and P. Ye, Chem. Soc. Rev., 2018, 47, 6009–6012 RSC.
- A. Qadir, T. K. Le, M. Malik, K. A. Amedome Min-Dianey, I. Saeed, Y. Yu, J. R. Choi and P. V. Pham, RSC Adv., 2021, 11, 23860–23880 RSC.
- Y. Chen, Z. Fan, Z. Zhang, W. Niu, C. Li, N. Yang, B. Chen and H. Zhang, Chem. Rev., 2018, 118, 6409–6455 CrossRef CAS PubMed.
- A. N. Ghulam, O. A. L. Dos Santos, L. Hazeem, B. P. Backx, M. Bououdina and S. Bellucci, J. Funct. Biomater., 2022, 13, 1–28 Search PubMed.
- P. Braylé, E. Pinelli, L. Gauthier, F. Mouchet and M. Barret, Environ. Sci.: Nano, 2022, 9, 3725–3741 RSC.
- B. Fadeel, C. Bussy, S. Merino, E. Vázquez, E. Flahaut, F. Mouchet, L. Evariste, L. Gauthier, A. J. Koivisto, U. Vogel, C. Martín, L. G. Delogu, T. Buerki-Thurnherr, P. Wick, D. Beloin-Saint-Pierre, R. Hischier, M. Pelin, F. Candotto Carniel, M. Tretiach, F. Cesca, F. Benfenati, D. Scaini, L. Ballerini, K. Kostarelos, M. Prato and A. Bianco, ACS Nano, 2018, 12, 10582–10620 CrossRef CAS PubMed.
- E. C. Cho, Q. Zhang and Y. Xia, Nat. Nanotechnol., 2011, 6, 385–391 CrossRef CAS PubMed.
- J. G. Teeguarden, P. M. Hinderliter, G. Orr, B. D. Thrall and J. G. Pounds, Toxicol. Sci., 2007, 95, 300–312 CrossRef CAS PubMed.
- L. S. Franqui, M. A. De Farias, R. V. Portugal, C. A. R. Costa, R. R. Domingues, A. G. Souza Filho, V. R. Coluci, A. F. P. Leme and D. S. T. Martinez, Mater. Sci. Eng., C, 2019, 100, 363–377 CrossRef CAS PubMed.
- P. Bihari, M. Vippola, S. Schultes, M. Praetner, A. G. Khandoga, C. A. Reichel, C. Coester, T. Tuomi, M. Rehberg and F. Krombach, Part. Fibre Toxicol., 2008, 5, 1–14 CrossRef PubMed.
- F. Joris, B. B. Manshian, K. Peynshaert, S. C. De Smedt, K. Braeckmans and S. J. Soenen, Chem. Soc. Rev., 2013, 42, 8339–8359 RSC.
- E. J. Guggenheim, S. Milani, P. J. F. Röttgermann, M. Dusinska, C. Saout, A. Salvati, J. O. Rädler and I. Lynch, NanoImpact, 2018, 10, 121–142 CrossRef.
- M. Jucker, Nat. Med., 2010, 16, 1210–1214 CrossRef CAS PubMed.
- M. von Scheidt, Y. Zhao, Z. Kurt, C. Pan, L. Zeng, X. Yang, H. Schunkert and A. J. Lusis, Cell Metab., 2017, 25, 248–261 CrossRef CAS PubMed.
- K. J. Jang, A. P. Mehr, G. A. Hamilton, L. A. McPartlin, S. Chung, K. Y. Suh and D. E. Ingber, Integr. Biol., 2013, 5, 1119–1129 CrossRef CAS PubMed.
- S. Peel and M. Jackman, Am. J. Physiol.: Cell Physiol., 2021, 320, C669–C680 CrossRef CAS PubMed.
- D. E. Ingber, Nat. Rev. Genet., 2022, 23, 467–491 CrossRef CAS PubMed.
- B. Zhang, A. Korolj, B. F. L. Lai and M. Radisic, Nat. Rev. Mater., 2018, 3, 257–278 CrossRef.
- Y. S. J. Li, J. H. Haga and S. Chien, J. Biomech., 2005, 38, 1949–1971 CrossRef PubMed.
- Y. Y. Chen, A. M. Syed, P. MacMillan, J. V. Rocheleau and W. C. W. Chan, Adv. Mater., 2020, 32, 1906274 CrossRef CAS PubMed.
- M. J. Gomez-Garcia, A. L. Doiron, R. R. M. Steele, H. I. Labouta, B. Vafadar, R. D. Shepherd, I. D. Gates, D. T. Cramb, S. J. Childs and K. D. Rinker, Nanoscale, 2018, 10, 15249–15261 RSC.
- M. Abdelkarim, L. Perez-Davalos, Y. Abdelkader, A. Abostait and H. I. Labouta, Expert Opin. Drug Deliveryt, 2023, 20, 13–30 CrossRef CAS PubMed.
- S. Kang, S. E. Park and D. D. Huh, Nano Converg., 2021, 8, 1–15 CrossRef PubMed.
- H. Klingberg, S. Loft, L. B. Oddershede and P. Møller, Nanoscale, 2015, 7, 11409–11419 RSC.
- K. R. Paton, E. Varrla, C. Backes, R. J. Smith, U. Khan, A. O’Neill, C. Boland, M. Lotya, O. M. Istrate, P. King, T. Higgins, S. Barwich, P. May, P. Puczkarski, I. Ahmed, M. Moebius, H. Pettersson, E. Long, J. Coelho, S. E. O’Brien, E. K. McGuire, B. M. Sanchez, G. S. Duesberg, N. McEvoy, T. J. Pennycook, C. Downing, A. Crossley, V. Nicolosi and J. N. Coleman, Nat. Mater., 2014, 13, 624–630 CrossRef CAS PubMed.
- S. Some, Y. Kim, Y. Yoon, H. Yoo, S. Lee, Y. Park and H. Lee, Sci. Rep., 2013, 3, 1–5 Search PubMed.
- V. González, J. Frontiñan-Rubio, M. V. Gomez, T. Montini, M. Durán-Prado, P. Fornasiero, M. Prato and E. Vázquez, ACS Nano, 2023, 17, 606–620 CrossRef PubMed.
- F. Torrisi, T. Hasan, W. Wu, Z. Sun, A. Lombardo, T. S. Kulmala, G. W. Hsieh, S. Jung, F. Bonaccorso, P. J. Paul, D. Chu and A. C. Ferrari, ACS Nano, 2012, 6, 2992–3006 CrossRef CAS PubMed.
- S. Watcharotone, D. A. Diking Sasha Stankovich, R. Pinery, I. Jung, G. H. B. Dommett, G. Evmenenko, S. E. Wu, S. F. Chen, C. P. Liu, S. T. Nguyen and R. S. Ruoff, Nano Lett., 2007, 7, 1888–1892 CrossRef CAS PubMed.
- L. Di Cristo, B. Grimaldi, T. Catelani, E. Vázquez, P. P. Pompa and S. Sabella, Mater. Today Bio, 2020, 6, 100050 CrossRef CAS PubMed.
- R. K. Jena and C. Y. Yue, Biomicrofluidics, 2012, 6, 012822 CrossRef.
- P. S. Nunes, P. D. Ohlsson, O. Ordeig and J. P. Kutter, Microfluid. Nanofluid., 2010, 9, 145–161 CrossRef CAS.
- E. Gencturk, S. Mutlu and K. O. Ulgen, Biomicrofluidics, 2017, 11, 1–41 CrossRef PubMed.
- P. F. Secker, N. Schlichenmaier, M. Beilmann, U. Deschl and D. R. Dietrich, Arch. Toxicol., 2019, 93, 1965–1978 CrossRef CAS PubMed.
- Y. Duan, N. Gotoh, Q. Yan, Z. Du, A. M. Weinstein, T. Wang and S. Weinbaum, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 11418–11423 CrossRef CAS PubMed.
- S. J. Kunnen, T. B. Malas, C. M. Semeins, A. D. Bakker and D. J. M. Peters, J. Cell. Physiol., 2018, 233, 3615–3628 CrossRef CAS PubMed.
- E. J. Ross, E. R. Gordon, H. Sothers, R. Darji, O. Baron, D. Haithcock, B. Prabhakarpandian, K. Pant, R. M. Myers, S. J. Cooper and N. J. Cox, Sci. Rep., 2021, 11, 1–14 CrossRef PubMed.
- U. Holzwarth, U. Cossío, J. Llop and W. G. Kreyling, Front. Pharmacol., 2019, 10, 1293 CrossRef CAS PubMed.
- P. M. Van Midwoud, A. Janse, M. T. Merema, G. M. M. Groothuis and E. Verpoorte, Anal. Chem., 2012, 84, 3938–3944 CrossRef CAS PubMed.
- A. Zuchowska, M. Chudy, A. Dybko and Z. Brzozka, Sens. Actuators, B, 2017, 243, 152–165 CrossRef CAS.
- G. Cibecchini, M. Veronesi, T. Catelani, T. Bandiera, D. Guarnieri and P. P. Pompa, ACS Appl. Mater. Interfaces, 2020, 12, 22507–22518 CrossRef CAS PubMed.
- J. Frontiñán-Rubio, M. Victoria Gómez, C. Martín, J. M. González-Domínguez, M. Durán-Prado and E. Vázquez, Nanoscale, 2018, 10, 11604–11615 RSC.
- Y. Li, Y. Liu, Y. Fu, T. Wei, L. Le Guyader, G. Gao, R. S. Liu, Y. Z. Chang and C. Chen, Biomaterials, 2012, 33, 402–411 CrossRef CAS PubMed.
- K. H. Liao, Y. S. Lin, C. W. MacOsko and C. L. Haynes, ACS Appl. Mater. Interfaces, 2011, 3, 2607–2615 CrossRef CAS PubMed.
- M. Pelin, H. Lin, A. Gazzi, S. Sosa, C. Ponti, A. Ortega, A. Zurutuza, E. Vázquez, M. Prato, A. Tubaro and A. Bianco, Nanomaterials, 2020, 10, 1–14 CrossRef PubMed.
- K. P. Wen, Y. C. Chen, C. H. Chuang, H. Y. Chang, C. Y. Lee and N. H. Tai, J. Appl. Toxicol., 2015, 35, 1211–1218 CrossRef CAS PubMed.
- B. Strojny, N. Kurantowicz, E. Sawosz, M. Grodzik, S. Jaworski, M. Kutwin, M. Wierzbicki, A. Hotowy, L. Lipińska and A. Chwalibog, PLoS One, 2015, 10, 1–18 CrossRef PubMed.
- A. Schinwald, F. Murphy, A. Askounis, V. Koutsos, K. Sefiane, K. Donaldson and C. J. Campbell, Nanotoxicology, 2014, 8, 824–832 CrossRef CAS PubMed.
- W. Chen, B. Wang, S. Liang, M. Wang, L. Zheng, S. Xu, J. Wang, H. Fang, P. Yang and W. Feng, J. Nanobiotechnol., 2023, 21, 1–18 CrossRef PubMed.
- A. F. Rodrigues, L. Newman, D. Jasim, S. P. Mukherjee, J. Wang, I. A. Vacchi, C. Ménard-Moyon, A. Bianco, B. Fadeel, K. Kostarelos and C. Bussy, Adv. Sci., 2020, 7, 1–17 Search PubMed.
- S. P. Mukherjee, G. Gupta, K. Klöditz, J. Wang, A. F. Rodrigues, K. Kostarelos and B. Fadeel, Small, 2020, 16, 1–15 Search PubMed.
- L. Newman, D. A. Jasim, E. Prestat, N. Lozano, I. De Lazaro, Y. Nam, B. M. Assas, J. Pennock, S. J. Haigh, C. Bussy and K. Kostarelos, ACS Nano, 2020, 14, 10168–10186 CrossRef CAS PubMed.
- S. Gurunathan, M. A. Iqbal, M. Qasim, C. H. Park, H. Yoo, J. H. Hwang, S. J. Uhm, H. Song, C. Park, J. T. Do, Y. Choi, J. H. Kim and K. Hong, Nanomaterials, 2019, 9, 1–22 Search PubMed.
- Y. Sun, H. Dai, S. Chen, M. Xu, X. Wang, Y. Zhang, S. Xu, A. Xu, J. Weng, S. Liu and L. Wu, Nanotoxicology, 2018, 12, 117–137 CrossRef CAS PubMed.
- V. C. Sanchez, A. Jachak, R. H. Hurt and A. B. Kane, Chem. Res. Toxicol., 2012, 25, 15–34 Search PubMed.
- A. Lesniak, F. Fenaroli, M. P. Monopoli, C. Åberg, K. A. Dawson and A. Salvati, ACS Nano, 2012, 6, 5845–5857 CrossRef CAS PubMed.
- M. A. Creighton, J. R. Rangel-Mendez, J. Huang, A. B. Kane and R. H. Hurt, Small, 2013, 9, 1921 CrossRef CAS PubMed.
- S. H. Chen, D. R. Bell and B. Luan, Adv. Drug Delivery Rev., 2022, 186, 114336 CrossRef CAS PubMed.
- S. García-Carpintero, V. Jehová González, J. Frontiñán-Rubio, A. Esteban-Arranz, E. Vázquez and M. Durán-Prado, Carbon, 2023, 215, 118426 CrossRef.
- V. León, A. M. Rodriguez, P. Prieto, M. Prato and E. Vázquez, ACS Nano, 2014, 8, 563–571 CrossRef PubMed.
- J. M. González-Domínguez, V. León, M. I. Lucío, M. Prato and E. Vázquez, Nat. Protoc., 2018, 13, 495–506 CrossRef PubMed.
- J. Frontiñan-Rubio, V. J. González, E. Vázquez and M. Durán-Prado, Sci. Rep., 2022, 12, 7664 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nh00064a |
| This journal is © The Royal Society of Chemistry 2024 |