Open Access Article
Open Access ArticleMining proteomes for zinc finger persulfidation†
Haoju
Li
,
Andrew T.
Stoltzfus
and
Sarah L. J.
Michel
 *
*
Department of Pharmaceutical Sciences, University of Maryland School of Pharmacy, Baltimore, MD 21201, USA. E-mail: smichel@rx.umaryland.edu
First published on 13th May 2024
Abstract
Hydrogen sulfide (H2S) is an endogenous gasotransmitter that signals via persulfidation. There is evidence that the cysteine residues of certain zinc finger (ZF) proteins, a common type of cysteine rich protein, are modified to persulfides by H2S. To determine how frequently ZF persulfidation occurs in cells and identify the types of ZFs that are persulfidated, persulfide specific proteomics data were evaluated. 22 datasets from 16 studies were analyzed via a meta-analysis approach. Persulfidated ZFs were identified in a range of eukaryotic species, including Homo sapiens, Mus musculus, Rattus norvegicus, Arabidopsis thaliana, and Emiliania huxley (single-celled phytoplankton). The types of ZFs identified for each species encompassed all three common ZF ligand sets (4-cysteine, 3-cysteine-1-histidine, and 2-cysteine-2-hisitidine), indicating that persulfidation of ZFs is broad. Overlap analysis between different species identified several common ZFs. GO and KEGG analysis identified pathway enrichment for ubiquitin-dependent protein catabolic process and viral carcinogenesis. These collective findings support ZF persulfidation as a wide-ranging PTM that impacts all classes of ZFs.
Introduction
Hydrogen sulfide (H2S) is a gaseous signalling molecule for which numerous biological roles have been identified. A role for H2S in signalling was first recognized in the 1990s, by Kimura and co-workers who discovered that the enzymatic production of H2S was an essential process for neuromodulation.1 Subsequent roles for H2S were then identified, including roles in cellular metabolism, cell survival, and oxidative stress.2,3 These collective findings led to the designation of H2S as an endogenous gasotransmitter, making it the third such molecule after carbon monoxide (CO) and nitric oxide (NO).4,5 H2S continues to garner significant attention, as roles in additional biological processes are identified. These include roles in the regulation of ER stress, inflammation, and cardiovascular function.6,7 If H2S levels in cells are not properly controlled, deleterious pathophysiological consequences are observed, including neurodegenerative diseases, cancer, and cardiovascular diseases.8,9Enzymes including cystathionine gamma-lyase (CSE), cystathionine-beta synthase (CBS), and 3-mercaptopyruvate sulfurtransferase (3-MST) are known to participate in hydrogen sulfide (H2S) metabolism.6 These enzymes are involved in the transsulfuration pathway and form various sulfur-containing metabolites. Specifically, they facilitate the homeostasis of various persulfide (SSH) and polysulfide (RS-Sn-H, where n > 1, and RS-Sn-R′, where n ≥ 1) moieties which are collectively referred to as the “sulfane sulfur pool”.10,11 These reactive sulfur species (RSS) exhibit crosstalk with reactive oxygen species (ROS) and reactive nitrogen species (RNS), thus connecting these molecules and their metabolic enzymes.12 RSS biomolecules exhibit antioxidant properties, thereby offering protection against oxidative stress, such as H2O2-induced cellular damage.10,13 Furthermore, persulfides and polysulfides affect cell redox signaling, mitochondrial membrane energetics, metalloproteins, and serve as a defense to pathogenic infection.14–17
One key function of H2S is signalling via the post-translational modification (PTM) of cysteine residues to persulfides (RSH → RSSH).10 Persulfidated cysteine residues are more reactive than their cysteine thiol counterparts and are thought to be critical to the ‘signaling’ function of H2S.18 We do not have a broad understanding of the proteins that are persulfidated in cells nor the functional consequences of persulfidation. There is emerging evidence that zinc finger proteins (ZFs), which are cysteine rich, are targets for persulfidation by H2S.19–21
Zinc finger proteins (ZFs) require zinc as a structural co-factor to fold and function.22–24 ZFs share a common feature of modular domains with four cysteine (C) and histidine (H) residues that serve as ligands.25 Zinc binds to these ligands in a tetrahedral geometry which leads to a folded and functional domain (Fig. 1A).26,27 As increasing numbers of proteomics and genome sequencing data have become available, it has been observed that zinc-binding proteins, and Cys residues, are present at higher numbers in more complex organisms; today approximately 9% of all human proteins are predicted to bind Zn(II), with the majority being ZFs.19,21,28,29 In contrast, while the Ros/MucR family of CCHH ZFs has been characterized in Agrobacterium tumefaciens and other prokaryotic species, these domains feature several distinctions from the eukaryotic CCHH ZFs.30–32 Namely, the prokaryotic ZF domains are larger and only occur once in a protein sequence, as opposed to the well-established tandem arrangement of multiple eukaryotic ZF domains required for full protein function.31 Thus, prokaryotic ZFs are less abundant overall, and their biological importance is not well established. Nevertheless, eukaryotic ZFs regulate numerous biological processes, including immune response, inflammation, gene regulation, and cancer development.33–35
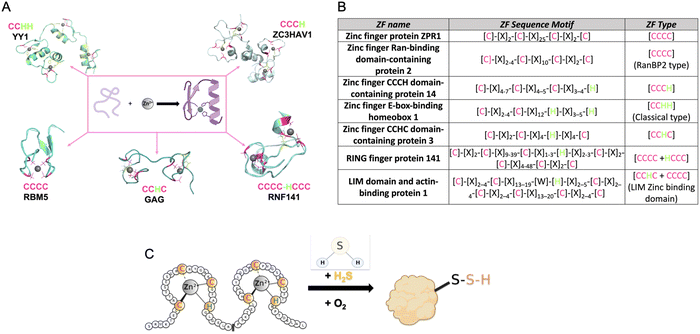 | ||
| Fig. 1 (A) Structures of common types of ZFs, (PDB number: YY1 = 1UBD, RNF141 = 5XEK, GAG = 1BJ6, RBM5 = 2LK0, ZC3HAV1 = 6UEI). (B) Table listing the consensus sequences for several commons ZF. (C) Cartoon diagram of the reaction of a ZF with H2S to form a ZF persulfide; this reaction requires zinc coordination and O2. | ||
The first ZF, Xenopus protein transcription factor IIIA (TFIIIA), was discovered in 1985.26,36,37 Each zinc centre in TFIIIA binds zinc with a CCHH ligand set, and the protein contains nine of these zinc binding domains.22 As more ZFs have been identified, efforts have been made to classify them. This includes categorizing ZFs based upon their amino acid sequence, their three-dimensional structures, or their functions. As an example, in PROSITE, ZFs are classified based upon cellular location, interactions with other proteins, amino acid sequence, and their participation in certain biological processes. Currently, there are sixty-nine different types of ZFs delineated in PROSITE.38 Another common way that ZFs are sorted is by the number of cysteine and histidine residues present within each ZF domain. This classification is often refined to a consensus sequence when it includes both the number of Cys and His resides, the specific spacing between these residues, and sometimes additional conserved amino acids. Once a ZF is defined, it can then be used to classify newly discovered ZFs.27,39,40 For example, all ZFs with a CCHH conserved ligand set and a spacing of [C]-[X]2–4-[C]-[X]12-[H]-[X]3–5-[H] are referred to as ‘classical zinc finger proteins’.33 Other examples of ZF classifications are shown in Fig. 1B.
ZF proteins have long been considered inert due to their metal centre, Zn(II), which has a d10 electron configuration, making it redox-inactive.41 As such, their biological roles have been considered solely structural. The ZF is folded in a manner that allows for a binding interaction with a target DNA or RNA, leading to regulation of transcription or translation.42 There is some recent evidence for persulfidation of the ZFs SIRT1, androgen receptor (AR), SP-1, and PHD2 from cell-based studies.43–47 In addition, there are biochemical data from our laboratory for persulfidation of the ZF tristetraprolin (TTP). We discovered that persulfidation by H2S occurs for TTP, in a reaction that requires Zn(II) and O2. Utilizing cryogenic electrospray ionization mass spectrometry (Cryo-ESI-MS) and orthogonal spectroscopic methods, we obtained evidence for TTP persulfidation, radical intermediate species, and disulfide bond formation with concomitant zinc ejection. This process abrogated TTP's ability to bind RNA in solution.48 From these studies, a mechanism was proposed in which Zn(II) serves as a conduit for electron transfer between H2S and O2 (Fig. 1C). These findings led us to hypothesize that persulfidation of ZF proteins may be a general post-translational modification. To test this hypothesis, we applied a persulfide-specific proteomics screen (dimedone switch method) to M. musculus embryonic fibroblast (MEF) cells. Analysis of the proteomics data led to the confirmation of our hypothesis: ZFs were broadly persulfidated.49 We sorted the ZFs into three groups: CCCC, CCCH/CCHC and CCHH, and observed persulfidation in all cases. We then analysed a published data set in which a different persulfidation selective proteomics screen was applied to different cell types (five human cell types). Here, too, persulfidation was broadly observed in ZFs of different classes.49
Our finding of ZF persulfidation in six different types of mammalian cells led to the question: how broad is ZF persulfidation across different species? Herein we answer via a meta-analysis approach whereby we analysed all published proteomics work that focussed on identifying protein persulfidation for ZF proteins.
Methods
Data sources and search strategy
To identify the publications for which persulfide specific proteomics assays were reported, a search on PubMed was performed using the keywords “Persulfidation” and “Proteomics.” The search included all publications until February 2023. The preferred reporting items for systematic review and meta-analyses (PRISMA) guidelines were used to verify these studies.50 Only studies written in English were included (Fig. S1, ESI†).Study selection
The first publication that involved persulfide specific proteomics was reported in 2010, therefore the studies utilized for the analysis reported here span 2010–2023. All data included in the analyses presented here were from studies that involved a method to enrich persulfidated proteins from cell lysates. In these studies, the proteins were either detected by mass spectrometry or extracted from existing proteomics data. All proteomics data analysed included identifiable protein accession IDs such as Uniprot accession ID, or NCBI gene IDs that could be searched against protein databases. Individual proteins that had been isolated and analysed for persulfidation were not included in this study. We placed no restriction on the persulfidation enrichment, or the mass spectrometry method used to detect the proteins. Reviews, book chapters, and secondary data were excluded.Data extraction and assessment
The following data were collected from each publication: citation (author/year), sample preparation methods for persulfidated protein enrichment and mass spectrometry analysis, and persulfidated protein list (Uniprot accession ID, NCBI Geninfo Identifier).Synthesis method to extract persulfidated zinc finger proteins
To identify the ZFs present in each dataset, care was taken to use comparable metrics across studies. First, all detected proteins were converted to UniProt accession names. Second, duplicates were removed if the protein appeared multiple times in the proteomics data. To extract the ZFs that had been detected in the persulfidated specific proteomics studies, the full protein list for each study was parsed for the presence of annotated ZF proteins (using UniProt database with the term “zinc finger protein”). Only reviewed ZF proteins (Swiss-Prot) were used. The ZF classification was retrieved for each identified protein by selecting “zinc finger” under the family and domain option. Each ZF was then grouped manually based upon the cysteine and histidine content of the ZF domains. Four main ZF classification types were applied in this study: CCCC, CCCH, CCHC, and CCHH, with C = cysteine and H = histidine. If more than one domain type was present in a single ZF, each domain was included in the analysis. Once identified, the ZFs were grouped based upon the species the data came from (Mus musculus, Rattus norvegicus, Homo sapiens, Arabidopsis thaliana). All the ZFs sorted were confirmed to be annotated as “Zinc Finger Protein” in Uniprot. ZFs that were annotated as “putative” were not included in this work.Gene ontology (GO) and kyoto encyclopedia of genes and genomes (KEGG) enrichment analysis of proteomics data
The data were processed using Microsoft Excel (Version 16.70). GO enrichment and KEGG pathway were analyzed using the database for annotation, visualization, and integrated discovery (DAVID) at P value < 0.05. Results of enriched terms and KEGG pathways were visualized using GraphPad Prism.Identification of persulfidated ZFs that are present in multiple species
To identify persulfidated ZFs common to multiple species, UniProt accession names were converted into Uniprot entry names. Data was processed on RStudio (Version 2022.12.0 + 353) using package VennDiagram (Version 1.7.3).Results and discussion
Study selections and characteristics
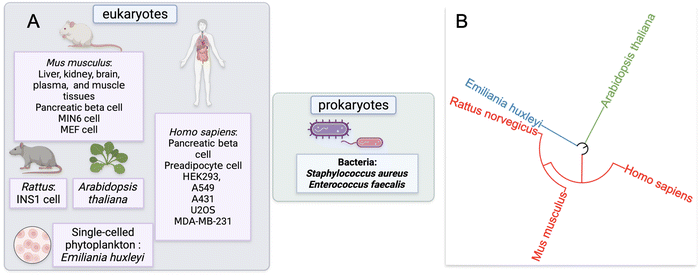
Our approach to identify and analyze ZF proteins that are persulfidated in different cell types began with a PubMed search. Two keywords: “persulfidation” and “proteomics” were queried leading to the retrieval of sixty-five publications. The abstracts and text of these publications were then assessed. Fifteen of these publications were reviews or book chapters, so were excluded from data analysis; likewise, thirty-six studies did not provide persulfide specific proteomics data, so were also excluded from data analysis.This yielded sixteen studies that met the criteria for meta-analysis, and these studies were subsequently analyzed for the presence of persulfidated ZFs.51–65 The proteomics data from these sixteen studies were from seven distinct species: H. sapiens, M. musculus, R. norvegicus, A. thaliana, Emiliania huxleyi (a single-celled phytoplankton), Staphylococcus aureus, and Enterococcus faecalis. Two of the species were bacteria, three were mammals, one was plant, and one was phytoplankton (Fig. 2A). The seven distinct species are displayed with regards to their evolutionary relationships using a phylogenetic tree in Fig. 2B. The methods employed to enrich for persulfidated proteins for mass spectrometry analysis varied across the publications. Overall, there were ten distinct methods reported for persulfide enrichment and detection of proteins.18,53,56,57,59–62,64,66
Protein persulfide enrichment and detection methods
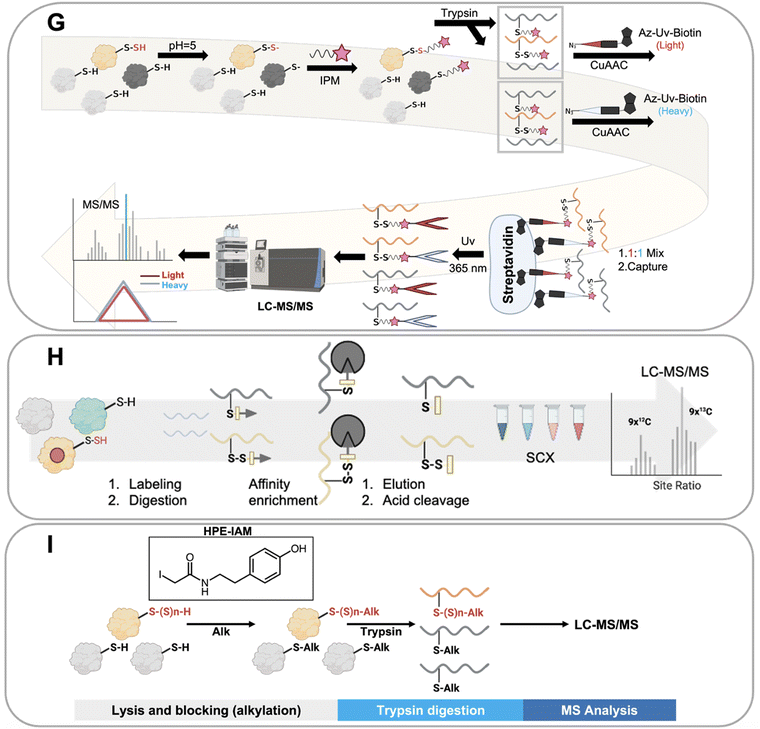
Ten distinct proteomics methods that selectively identify persulfidated proteins were utilized to obtain the collective data that we have analyzed in this work. These methods, which are shown in Fig. 3, include: tag switch, dimedone switch, quantitative persulfide site identification (qPerS-SID), ‘b-IAM-Avidin enrichment followed by TCEP elution and IAM incubation,’ an unnamed method developed by the Giedroc lab (Giedroc's method), biotin thiol assay (BTA), tandem mass tag(TMT)-BTA, site-specific quantification of persulfidome (SSQPer), HPE-IAM blocking, low-pH quantitative thiol reactivity profiling (QTRP), and a software-assisted search approach that identified persulfidated proteins using existing proteomics data.18,53,56,57,59–62,64,66 Some of these methods employ similar chemistries to enrich for persulfidated proteins, while others rely on mass spectrometry data analysis post sample to search for PTMs that have a persulfide-specific mass-to-charge ratio. These methods are described in more detail here.The tag switch and dimedone switch methods share similar chemistries but have different labeling and switching reagents. In the tag switch method, methylsulfonyl benzothiazole (MSBT) is used to universally label all thiol and persulfide groups on proteins (Fig. 3A). In the dimedone switch method, the blocking reagent is 4-chloro-7-nitrobenzofurazan (NBF-Cl) which blocks all thiols, persulfides, sulfenic acids, and amines (Fig. 3B).18,66 Both labeling methods result in the formation of an activated disulfide bond for the labeled persulfide moiety, which is then modified by the addition of a second label that selectively tags the persulfidated species because the outer sulfur of the persulfide is more electrophilic and amenable to “switching”.67 The switching molecule for the tag switch method is biotin-linked cyanoacetate (CN-biotin), and for the dimedone switch method is biotin-linked dimedone. Once the tag switch has occurred, the persulfidated proteins are isolated via a biotin–avidin interaction and analyzed.18,66
Another method for persulfide enrichment is the biotin thiol assay (BTA) (Fig. 3C),56 for which there are several related approaches: TMT-BTA (Fig. 3D), qPerS-SID (Fig. 3E), and Giedroc's method (Fig. 3F). BTA and TMT-BTA use a maleimide biotin to label thiols (–SH) and persulfides (–SSH) in proteins. Labelled proteins are then further enriched via a biotin–avidin interaction. The thiol reaction forms a thioether while the persulfide forms a disulfide bond. The disulfide bond is more reactive than the thioether and can be reduced using either tris(2-carboxyethyl) phosphine (TCEP) or dithiothreitol (DTT), which releases the persulfidated peptide from the avidin beads. The peptides can then be detected and identified by LC-MS/MS. TMT-BTA is a modification of the BTA method in which peptides are labelled with iodo-TMT 6 plex after the release of the persulfidated peptide, and then detected via mass spectrometry. qPerS-SID utilizes trichloroacetic acid (TCA) to denature proteins and protonate all thiol and persulfide groups to prevent oxidation. After this, a thiol and persulfide specific reagent is then added, iodoacetyl-PEG2-Biotin (IAM-Bio), and the proteins are isolated via the biotin–avidin interaction. After elution with TCEP, iodoacetamide (IAM) is added to block peptides for specific mass spectrometry detection. Giedroc's method is similar to qPerS-SID, except that it omits TCA for protein denaturation. Collectively, these strategies are based upon the BTA method, but include modifications to improve the sensitivity of persulfidated protein detection by mass spectrometry.60,61,64
The low-pH quantitative thiol reactivity profiling (QTRP) is another method used to detect persulfides (Fig. 3G). This approach is the only currently published method that allows for direct detection of thiols and persulfides on proteins. Using the differences in pKa of cysteine persulfide and thiol (pKa ∼4.3 and ∼8.3, respectively), the pH is lowered to 5, and almost all thiols are protonated but persulfides are mostly deprotonated. In this case, persulfides maintain a higher reactivity (more RSS- is available than RS-) and are more likely to react with carbon electrophiles, such as iodo-N-(prop-2-yn-1-yl) acetamide (IPM). This minimizes the click chemistry reaction between thiols and IPM, thus enhancing the signal of IPM labeled persulfides. Detection involves enrichment by click-chemistry and cleavage through UV-biotinylated probes. No reduction step is involved in this method. Thiols and persulfides can be differentiated by comparing mass-to-charge ratios using mass spectrometry.62
An alternative approach used to detect persulfides is to differentiate protein persulfides from other sulfur moieties by analyzing proteomics data that detects both persulfides and thiols. This approach involves applying software analysis after the mass spectrometry analysis of the sample to identify the persulfidated proteins. The SSQPer method uses a cleavable isotope-coded affinity tag (c-ICAT) to label thiols and persulfides on proteins. Samples are further enriched by streptavidin agarose beads. Then the tagged proteins are cleaved by TFA and followed by strong cation exchange (SCX) based fractionation which enriches for thiols and persulfides. The identity of the modification (persulfide vs. thiol) is determined by evaluating mass differences in the proteomics software (Fig. 3H). The HPE-IAM method follows a similar strategy but foregoes the enrichment step. Instead, β-(4-hydroxyphenyl)ethyl iodoacetamide (HPE-IAM) is used to tag all reactive thiols, persulfides, and hydropolysulfides followed by trypsin digestion and LC-MS/MS. The persulfidated proteins are then extracted and identified via anaylsis of the mass spectrometry data (Fig. 3I).57,59
A final approach that has been reported to identify persulfidated proteins involves the trans-proteomic pipeline (Comet and PeptideProphet) which is data analysis software used to search for protein PTMs based on the MS/MS spectra of proteomics data obtained from a traditional proteomics screen. Persulfidation is reported with proteins with Cys (31.9721 Da) modification (Fig. 3J).53
Mining the persulfide specifics proteomics datasets for persulfidated ZFs
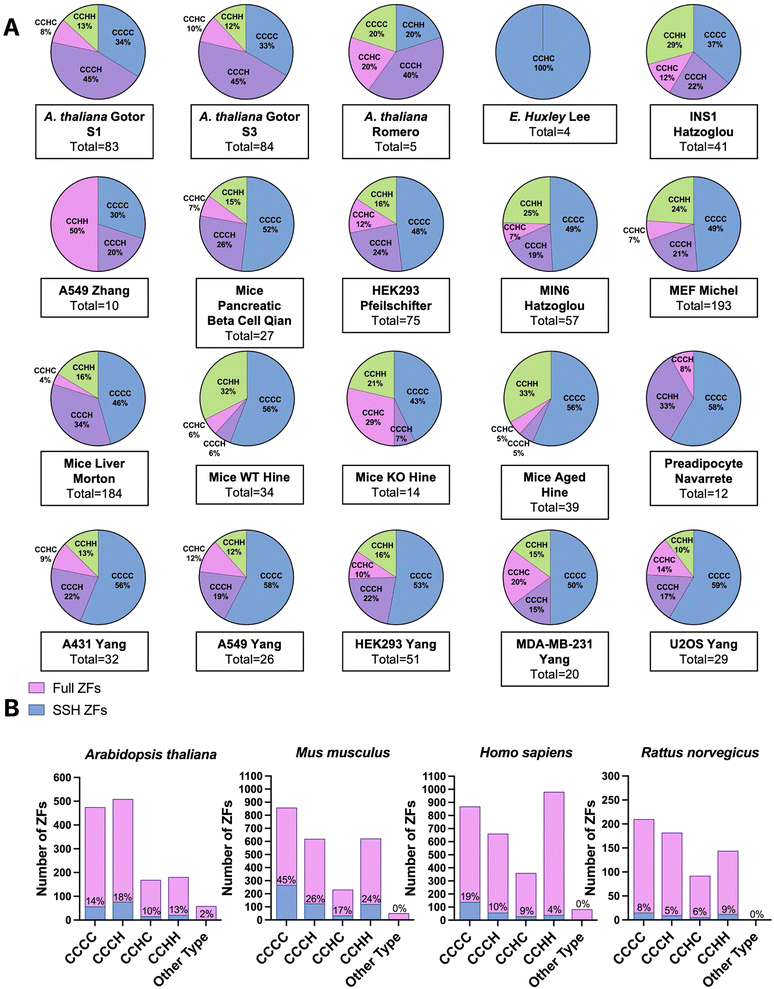
We extracted the proteomics data from the sixteen studies and five species identified in our initial query. With these data in hand, we searched for the presence of ZFs. Depending on the proteomics strategy utilized, between 4 to 193 total ZF proteins were identified as persulfidated. Remarkably, in all but one eukaryotic data set analyzed, multiple ZFs were identified as persulfidated. The data set for which no ZFs were observed was significantly smaller than other published data (i.e., only 70 total proteins were identified as persulfidated in this data), and the dataset may not be complete enough for ZFs to be identified.61 In the two bacterial data sets, no ZFs were identified as persulfidated.64,65 There are only a handful of ZFs that have been identified in prokaryotes, suggesting that the paucity of ZFs in bacteria explains the lack of persulfidated ZFs observed. There were six reported proteomics data sets that involved M. musculus tissue samples. These included liver, kidney, brain, plasma, muscle, and heart. There were fourteen cell lines total that contained persulfidated ZFs. Seven were human cell lines including A549, A431, HEK293, U2OS, MDA-MB-231, EndoC-BH3, and human preadipocyte cell obtained from participants. A549, A431, U2OS, and MDA-MB-231 are cancer cell lines. HEK293 is a human embryonic kidney cell line; preadipocyte cell and EndoC-BH3 derive from human adipose tissue and pancreatic islets, respectively. In addition, three of the cell lines are from M. musculus: MIN6, MEF, and pancreatic beta cells. MIN6 and pancreatic beta cells both originate from the M. musculus pancreas and the MEF cell is an embryonic fibroblast cell line. One cell line from R. norvegicus was included in this study, INS1, which is an insulinoma cell line.68 In total, 548 persulfidated ZFs were found in M. musculus, 267 persulfidated ZFs were found in H. sapiens, 23 in R. norvegicus, and 172 in A. thaliana; duplicates were removed when consolidating ZFs acrossstudies. We note that the studies which reported a larger persulfidated proteome resulted in detection of more persulfidated ZFs. In all cases, the ratio of ZFs/total protein tracked between 3–10%, which is consistent with what is known about the abundance of zinc binding proteins in cells.19Classification of persulfidated zinc finger proteins
To classify the ZFs present in the persulfide specific proteomics data, we first generated a list of the total ZFs, using UniProt and the annotation “Zinc Finger” domain. Only proteins for which there is experimental evidence that the protein is a bona fide ZF were included; putative ZFs were excluded. ZF lists for each species were constructed (H. sapiens, M. musculus, R. norvegicus, A. thaliana, E. huxleyi).We have previously reported that the frequency of persulfidation in ZFs is related to the number of cysteines present in the proteins’ ZF domain(s). This finding came from analysis of persulfide specific proteomics data using MEF cells and several human cancer cell lines.41 We postulated that this relationship between the number of cysteine residues and the frequency of persulfidation would be observed generally in cells and tested this hypothesis by evaluating the 20 different datasets analyzed here (Fig. 4A). The ZFs were sorted into categories based on their ligand set compositions: CCCC, CCCH, CCHC, and CCHH type. Datasets from S. aureus and E. faecalis, both prokaryotes, were excluded from this study because no ZFs were detected in the persulfide specific proteomes. In the A. thaliana datasets, the most frequently persulfidated ZFs had the CCCH ligand set, followed by CCCC, CCHH, and CCHC. Only ZFs with the CCHC ligand set were found in the single-celled phytoplankton E. huxleyi. The classification of persulfidated ZFs in mammals (H. sapiens, M. musculus, and R. norvegicus) follows the same trend as we observed in our earlier work. The most frequent ligand set of the persulfidated ZFs was the CCCC ligand set, followed by CCCH/CCHC, and finally CCHH. We then grouped these data based upon species, and we compared the number of each type of ZF persulfidated to the total ZFs present in each species. As Fig. 4B shows, for, A. thaliana (total 172 ZFs) the relative frequencies of ZF persulfidation tracked as CCCH and CCHC > CCCC > CCHH. M. musculus showed the frequencies as: CCCC > CCCH and CCHC > CCHH. H. sapiens showed the frequencies as: CCCH/CCHC > CCCC > CCHH. R. norvegicus showed the frequencies as: CCHH > CCCC > CCCH and CCHC. We note that for R. norvegicus, the sample size is small (23 total persulfidated proteins detected), potentially making the analysis of this species less comprehensive. The ligand type CCHH is the least persulfidated among M. musculus, Homo sapiens and A. thaliana (Fig. 4B).
Regardless of the method utilized to detect persulfidated proteins, persulfidated ZF proteins always appear. At the evolutionary scale, the cells that come from higher order organisms (i.e., mammals) appear to prefer CCCC type ZF persulfidation, whereas plant cells have a preference for CCCH, and the lower order organism (E. huxleyi) has a preference for CCHC. A. thaliana is known to have multiple CCCH type ZFs for dealing with environmental stress and regulation of growth, and the persulfidation may be related to the frequency of the specific ZFs in each organism.69–71
Commonly present zinc finger proteins across species
To identify commonly persulfidated ZFs across different species we performed a Venn diagram analysis using RStudio with the VennDiagram package to visualize common proteins (Fig. 5).72 The persulfidation datasets of ZFs from M. musculus, R. norvegicus, H. sapiens, and A. thaliana were compared after removing duplicates. E. huxleyi data were excluded due to the absence of common persulfidated ZF proteins with the other species. (ESI† Table S9) Evolutionarily, the phytoplankton are distant from the other organisms studied, perhaps explaining the lack of common persulfidated ZF proteins. Intersect analysis, which is designed to identify common proteins, revealed a single protein present across all four species: the DNA replication licensing factor MCM2. This protein, which contains a CCCC zinc finger domain in its sequence, is located in the nucleus where it is part of a larger protein complex called MCM2-7 that associates with chromosomes when activated. As a part of the MCM2-7 complex, MCM2 plays a pivotal role in DNA initiation and elongation in eukaryotic cells.73 When we limited our analysis to M. musculus, H. sapiens, and R. norvegicus, reflecting their evolutionary similarities within the animal kingdom, we identified eight ZFs that are common to all three species. These included CCHC-type zinc finger nucleic acid binding protein, transcription intermediary factor 1-beta, E3 ubiquitin-protein ligase UBR4, ATPase WRNIP1, DNA (cytosine-5)-methyltransferase 1, activity-dependent neuroprotector homeobox protein (Adnp), poly [ADP-ribose] polymerase 1 (parp1), and chromodomain-helicase-DNA-binding protein 5 (CHD5). The intersect analysis of H. sapiens, M. musculus, and A. thaliana showed a significantly lower number of common ZFs, with only MCM2 being present in all three species. This may be reflective of the evolutionary distance between H. sapiens and M. musculus, when compared to A. thaliana. Further analysis of overlapping ZFs between M. musculus and H. sapiens, M. musculus and R. norvegicus, and H. sapiens and R. norvegicus yielded additional insights. We found 51 ZFs common to M. musculus and H. sapiens, constituting 37% and 20% of the total persulfidated ZFs in H. sapiens and M. musculus, respectively. Conversely, only 17 proteins overlapped between M. musculus and R. norvegicus, and 9 proteins between H. sapiens and R. norvegicus. This smaller overlap may be attributed to the limited persulfide-specific sample size for R. norvegicus. Specifically, only 22 unique ZFs were detected in the INS-1 cell line generated from R. norvegicus, whereas multiple datasets, each of which contains more proteins (e.g., 20-fold more) were available for the other species. One limitation to this Venn diagram approach is that the proteomics data compared were obtained using different persulfide-specific proteomics methods.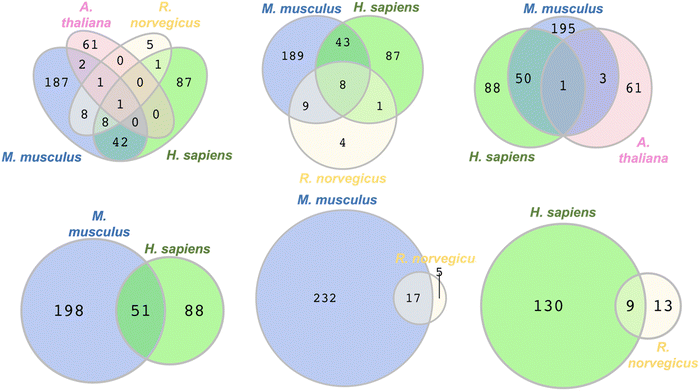 | ||
| Fig. 5 Venn diagrams depicting the overlap in the types of persulfidated ZFs from different species, M. musculus, R. norvegicus, H. sapiens, and A. thaliana, (see Table S9 for protein names, ESI†). | ||
To improve the Venn diagram approach, we also generated a Venn diagram that was generated from the data from the three publications that utilized same persulfide-specifics proteomics approach. These publications by the groups of Hatzoglou, Morton, and Hine used the BTA method for sample preparation and analysis. The Hatzoglou group worked with M. musculus MIN6 cells, the Morton group with liver tissues of M. musculus, and the Hine group with tissue samples from the heart, brain, kidney, and liver of M. musculus.54–56 (Fig. S2, ESI†) The Venn diagram generated identified two proteins across the three datasets: RAN binding protein 2 (RANBP2) and Ubiquitin carboxyl-terminal hydrolase 5 (UBP5). Furthermore, nine proteins were found to overlap between the MIN6 cells and liver tissue datasets, while eight proteins were common between the liver tissues studied by Morton and Hine's group (encompassing heart, brain, kidney, and liver). The lack of overlap in proteins identified across the analyses may be due to differences in sample size and the organ-specific nature of the proteins.
These collective data show that limitations of the Venn diagram approach include the persulfide-specific proteomic sample workflows, which differ between publications, and the limited numbers of proteins identified in certain published data (e.g., the data from the BTA methods had as few as nineteen proteins identified).
GO and KEGG enrichment analysis of proteome data
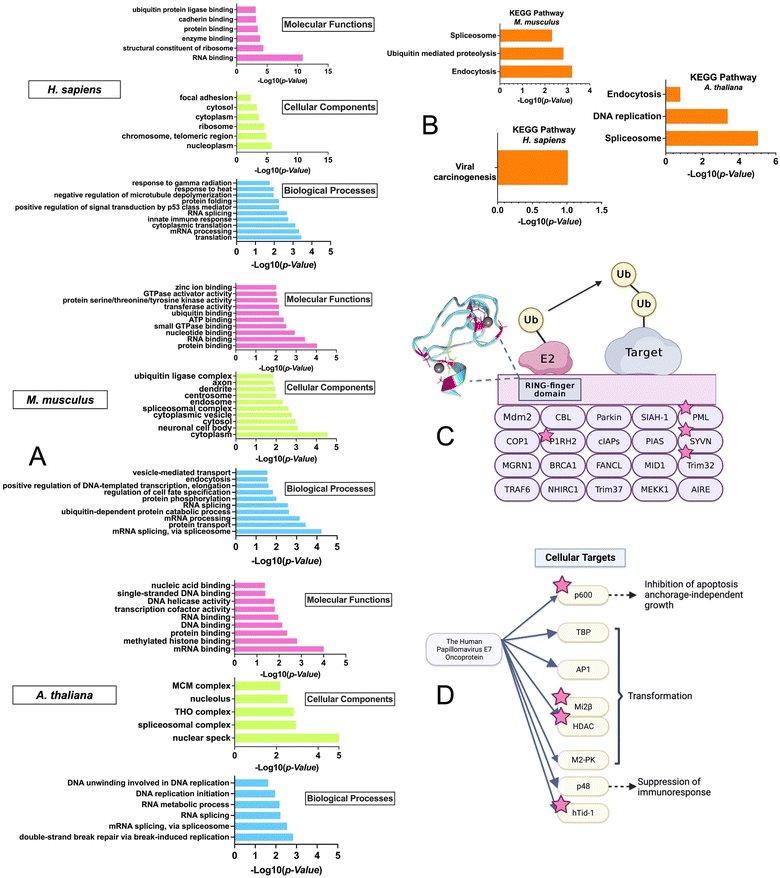
To gain insight into the biological pathways and molecular functions that were enriched in the ZFs across species, GO enrichment analysis was performed using the DAVID bioinformatics tool with the M. musculus, H. sapiens, and A. thaliana datasets (Fig. 6A).74–76 The R. norvegicus dataset was excluded from the GO and KEGG enrichment analysis due to the limited number of ZF proteins identified (i.e., 22) and the enrichment results do not meet the P-value cutoff of 0.05 for this study. GO enrichment analysis was performed using two approaches: the first utilized the complete list of ZFs from each species for the background. (Fig. 6A) while the second was conducted using the entire proteome as the background. (Fig. S3, ESI†) As shown in Fig. 6A, a variety of molecular functions were determined to be enriched by the GO analysis using the ZF proteome as the background. These include predicted functions for ZFs, such as translation (9 protein hits) and mRNA processing (36 protein hits). Examples include the CCHC-type zinc finger nucleic acid binding protein, which contains multiple CCHC zinc finger domains that can interact with specific single stranded DNA and RNA sequences to regulate transcription and translation.77,78 In addition, ubiquitin-dependent protein catabolic process, a biological process that is not common to all ZFs, was also found to be enriched, especially in M. musculus. The GO analysis was also conducted using the entire proteome as a background. The results showed a similar trend, with ubiquitination-related functions being enriched in H. sapiens and M. musculus. (Fig. S3, ESI†)This finding was further confirmed by the enrichment results from the KEGG analysis, which focuses on pathways; here too, “ubiquitin-mediated proteolysis” was a major enriched pathway. This finding suggests that persulfidation may be linked to ZFs that participate in protein ubiquitination (Fig. 6B).
We note that there is evidence from cell-based studies for persulfidation of a ZF called Parkin, which is involved in ubiquitination.79,80 Parkin contains 4 ZF domains (3 of which are RING-type, CCCC–CCHC) and is part of the RING-finger type E3 ligase family (Fig. S4, ESI†). This family facilitates the transfer of ubiquitin from the E2 conjugating enzyme to the target protein by acting as a scaffold to bring E2 close to the target protein.81 The activation of Parkin and its recruitment to the mitochondrial membrane by PTEN induced kinase 1 (PINK1) have been well studied.82,83 Phosphorylation of Parkin at S65 in the Ubiquitin-like domain (UBL) by PINK1 and binding of phospho-Ubiquitin (pUb) to the RING-type 0 ZF domain are required for full activation of Parkin's E3 ligase activity. These PTMs induce a change to Parkin's typical auto-inhibitory conformation in the cytoplasm, where the multiple ZF domains self-associate and obscure the catalytic C431, to an open conformation where the RING-type 1 ZF domain can bind the E2 enzyme in ubiquitin transfer. C431 of the RING-type 2 ZF domain is accessible for ubiquitin binding and transfer to Parkin substrates.84–86
In addition to this Parkin activation via phosphorylation, a recent study has shown that persulfidation of Parkin, predominantly in the UBL and RING-type 0 domains, can increase the activity of the enzyme, thus enhancing the ubiquitination and degradation of target proteins (Fig. S4, ESI†).80 The cystine rich nature of Parkin may contribute to the likelihood for it to become persulfidated.
Interestingly, in the M. musculus KEGG pathway: “ubiquitin-mediated proteolysis” enrichment results, four additional single RING-finger type E3 proteins (P1RH2, PML, SYVN, Trim32) were identified in the persulfide-specific proteomics screen (Fig. 6), suggesting these types of ZF proteins may be a common target for persulfidation (Fig. 6C).
The KEGG pathway enrichment analysis showed that the most enriched pathway for H. sapiens is “viral carcinogenesis”. The human papillomavirus (HPV) E7 oncoprotein has four cellular targets that appeared in the ZFs persulfide screen (p600, Mi2β, HDAC, and hTid-1). HPV E7 plays a key role in the development of cervical cancer, and it can perturb many cellular processes by interacting with different protein targets, immortalizing, and transforming cells, which eventually leads to malignant growth (Fig. 6D).87 The most enriched KEGG pathway in A. thaliana is ‘spliceosome’. It has been shown that ZFs are involved in spliceosome activities.88 These collective findings that ZFs from different biological pathways can be persulfidated supports persulfidation as a common PTM for ZFs.
Conclusions
In the work presented here, we sought to determine whether ZF persulfidation is a general PTM in different types of cells and organisms. By analysing all of the published persulfide specific proteomics data for the presence of persulfidated ZFs, we found that persulfidation of ZFs is common in eukaryotes and occurs in a range of species. The number of persulfidated ZFs that were identified varied between datasets, in part because the proteomics methods used to identify persulfidated proteins for each dataset yielded a range of total proteome sizes (i.e., 193 total proteins identified in MEF cells by Michel et al., to 4 total proteins identified in Emiliania huxleyi (single-celled phytoplankton) by Lee et al.). Despite the differences in dataset size, some commonalities between data sets were observed. One commonality was seen in the types of persulfidated ZFs in the mammalian species when the data were evaluated via Venn diagram analysis. Another commonality was in biological functions, for example persulfidation of ZFs involved in ubiquitination was found in human and mouse data via GO enrichment and KEGG analysis. Our analysis was limited by the availability of persulfide specific proteomics data, making our findings mammalian centric, as most data were from these types of cells. Another limitation is that the current literature does not provide information on which cysteine is persulfidated in the proteomics datasets. As more persulfide specific proteomics data becomes available from different organisms, we will be able to expand our understanding of ZFs and persulfidation on the species level and learn more about the delineation of ZF type and persulfidation. Together, these data reveal that ZF persulfidation is a common PTM and opens the door for biochemical and cellular studies to understand functional consequences.Author contributions
Conceptualization: SLJM, HL, AS; methodology: SLJM, HL; investigation: SLJM, HL; visualization: HL; funding acquisition: SLJM; project administration: SLJM; supervision: SLJM; writing – original draft: SLJM, HL, AS; writing – review and editing: SLJM, HL, AS.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are grateful to the NIH (R01GM139854) for support. Figures are created using Biorender.com and iTOL (https://itol.embl.de).References
- R. Hosoki, N. Matsuki and H. Kimura, Biochem. Biophys. Res. Commun., 1997, 237, 527–531 CrossRef CAS PubMed.
- G. Yang, Expert Rev. Clin. Pharmacol., 2011, 4, 33–47 CrossRef CAS PubMed.
- G. Cirino, C. Szabo and A. Papapetropoulos, Physiol. Rev., 2023, 103, 31–276 CrossRef CAS.
- M. R. Filipovic, J. Zivanovic, B. Alvarez and R. Banerjee, Chem. Rev., 2018, 118, 1253–1337 CrossRef CAS PubMed.
- B. D. Paul and S. H. Snyder, Nat. Rev. Mol. Cell Biol., 2012, 13, 499–507 CrossRef CAS PubMed.
- J. I. Sbodio, S. H. Snyder and B. D. Paul, Br. J. Pharmacol., 2019, 176, 583–593 CrossRef CAS.
- B. He, Z. Zhang, Z. Huang, X. Duan, Y. Wang, J. Cao, L. Li, K. He, E. C. Nice, W. He, W. Gao and Z. Shen, Biochem. Pharmacol., 2023, 209, 115444 CrossRef CAS PubMed.
- E. Donnarumma, R. K. Trivedi and D. J. Lefer, Compr. Physiol., 2011, 7, 583–602 Search PubMed.
- H. Kimura, Antioxid. Redox Signaling, 2015, 22, 362–376 CrossRef CAS PubMed.
- T. Ida, T. Sawa, H. Ihara, Y. Tsuchiya, Y. Watanabe, Y. Kumagai, M. Suematsu, H. Motohashi, S. Fujii, T. Matsunaga, M. Yamamoto, K. Ono, N. O. Devarie-Baez, M. Xian, J. M. Fukuto and T. Akaike, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 7606–7611 CrossRef CAS PubMed.
- M. Iciek, A. Bilska-Wilkosz and M. Górny, Acta Biochim. Pol., 2019, 66, 533–544 CAS.
- N. Lau and M. D. Pluth, Curr. Opin. Chem. Biol., 2019, 49, 1–8 CrossRef CAS.
- J. M. Fukuto, L. J. Ignarro, P. Nagy, D. A. Wink, C. G. Kevil, M. Feelisch, M. M. Cortese-Krott, C. L. Bianco, Y. Kumagai and A. J. Hobbs, FEBS Lett., 2018, 592, 2140–2152 CrossRef CAS PubMed.
- B. D. Paul, S. H. Snyder and K. Kashfi, Redox. Biol., 2021, 38, 101772 CrossRef CAS PubMed.
- R. Kumar and R. Banerjee, Crit. Rev. Biochem. Mol. Biol., 2021, 56, 221–235 CrossRef CAS PubMed.
- D. P. Giedroc, G. T. Antelo, J. N. Fakhoury and D. A. Capdevila, Curr. Opin. Chem. Biol., 2023, 76, 102358 CrossRef CAS PubMed.
- A. Domán, É. Dóka, D. Garai, V. Bogdándi, G. Balla, J. Balla and P. Nagy, Redox. Biol., 2023, 60, 102617 CrossRef PubMed.
- J. Zivanovic, E. Kouroussis, J. B. Kohl, B. Adhikari, B. Bursac, S. Schott-Roux, D. Petrovic, J. L. Miljkovic, D. Thomas-Lopez, Y. Jung, M. Miler, S. Mitchell, V. Milosevic, J. E. Gomes, M. Benhar, B. Gonzalez-Zorn, I. Ivanovic-Burmazovic, R. Torregrossa, J. R. Mitchell, M. Whiteman, G. Schwarz, S. H. Snyder, B. D. Paul, K. S. Carroll and M. R. Filipovic, Cell Metab., 2019, 30, 1152–1170 CrossRef CAS.
- C. Andreini, L. Banci, I. Bertini and A. Rosato, J. Proteome Res., 2006, 5, 3173–3178 CrossRef CAS PubMed.
- K. Ok, M. R. Filipovic and S. L. Michel, Eur. J. Inorg. Chem., 2021, 3795–3805 CrossRef CAS.
- C. Andreini, L. Banci, I. Bertini and A. Rosato, J. Proteome Res., 2006, 5, 196–201 CrossRef CAS.
- A. Klug, Annu. Rev. Biochem., 2010, 79, 213–231 CrossRef CAS PubMed.
- M. Padjasek, A. Kocyła, K. Kluska, O. Kerber, J. B. Tran and A. Krężel, J. Inorg. Biochem., 2020, 204, 110955 CrossRef CAS.
- S. Negi, M. Imanishi, M. Hamori, Y. Kawahara-Nakagawa, W. Nomura, K. Kishi, N. Shibata and Y. Sugiura, J. Biol. Inorg. Chem., 2023, 28, 249–261 CrossRef CAS PubMed.
- J. L. Michalek, A. N. Besold and S. L. Michel, Dalton Trans., 2011, 40, 12619–12632 RSC.
- J. M. Berg, Proc. Natl. Acad. Sci. U. S. A., 1988, 85, 99–102 CrossRef CAS PubMed.
- R. Gamsjaeger, C. K. Liew, F. E. Loughlin, M. Crossley and J. P. Mackay, Trends Biochem. Sci., 2007, 32, 63–70 CrossRef CAS PubMed.
- I. Bertini, L. Decaria and A. Rosato, J. Biol. Inorg. Chem., 2010, 15, 1071–1078 CrossRef CAS PubMed.
- C. Wiedemann, A. Kumar, A. Lang and O. Ohlenschläger, Front. Chem., 2020, 8, 280 CrossRef CAS.
- A. Y. Chou, J. Archdeacon and C. I. Kado, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 5293–5298 CrossRef CAS.
- G. Malgieri, M. Palmieri, L. Russo, R. Fattorusso, P. V. Pedone and C. Isernia, FEBS J., 2015, 282, 4480–4496 CrossRef CAS.
- P. A. Bertram-Drogatz, I. Quester, A. Becker and A. Pühler, MGG, 1998, 257, 433–441 CAS.
- S. J. Lee and S. L. Michel, Acc. Chem. Res., 2014, 47, 2643–2650 CrossRef CAS PubMed.
- J. Jen and Y.-C. Wang, J. Biomed. Sci., 2016, 23, 53 CrossRef PubMed.
- G. Rakhra and G. Rakhra, Mol. Biol. Rep., 2021, 48, 5735–5743 CrossRef CAS PubMed.
- J. Miller, A. McLachlan and A. Klug, EMBO J., 1985, 4, 1609–1614 CrossRef CAS PubMed.
- A. D. Frankel, J. M. Berg and C. O. Pabo, Proc. Natl. Acad. Sci. U. S. A., 1987, 84, 4841–4845 CrossRef CAS.
- C. J. A. Sigrist, E. De Castro, P. S. Langendijk-Genevaux, V. Le Saux, A. Bairoch and N. Hulo, Bioinformatics, 2005, 21, 4060–4066 CrossRef CAS PubMed.
- L. S. Johnson, S. R. Eddy and E. Portugaly, BMC Bioinform., 2010, 11, 1–8 CrossRef PubMed.
- C. Sathyaseelan, L. P. P. Patro and T. Rathinavelan, Pattern Recognit., 2023, 135, 109134 CrossRef.
- N. J. Pace and E. Weerapana, Biomolecules, 2014, 4, 419–434 CrossRef PubMed.
- W. Maret, Int. J. Mol. Sci., 2017, 18, 2285 CrossRef PubMed.
- B. Cui, Q. Pan, D. Clarke, M. O. Villarreal, S. Umbreen, B. Yuan, W. Shan, J. Jiang and G. J. Loake, Nat. Commun., 2018, 9, 4226 CrossRef PubMed.
- A. Dey, S. Prabhudesai, Y. Zhang, G. Rao, K. Thirugnanam, M. N. Hossen, S. K. D. Dwivedi, R. Ramchandran, P. Mukherjee and R. Bhattacharya, Sci. Adv., 2020, 6, eaaz8534 CrossRef CAS PubMed.
- Y. Yuan, L. Zhu, L. Li, J. Liu, Y. Chen, J. Cheng, T. Peng and Y. Lu, Antioxid. Redox Signaling, 2019, 31, 1302–1319 CrossRef CAS PubMed.
- K. Zhao, S. Li, L. Wu, C. Lai and G. Yang, J. Biol. Chem., 2014, 289, 20824–20835 CrossRef CAS PubMed.
- C. Du, X. Lin, W. Xu, F. Zheng, J. Cai, J. Yang, Q. Cui, C. Tang, J. Cai and G. Xu, Antioxid. Redox Signaling, 2019, 30, 184–197 CrossRef CAS PubMed.
- M. Lange, K. Ok, G. D. Shimberg, B. Bursac, L. Markó, I. Ivanović-Burmazović, S. L. Michel and M. R. Filipovic, Angew. Chem., 2019, 131, 8081–8085 CrossRef.
- A. T. Stoltzfus, J. G. Ballot, T. Vignane, H. Li, M. M. Worth, L. Muller, M. A. Siegler, M. A. Kane, M. R. Filipovic, D. P. Goldberg and S. L. J. Michel, Angew. Chem., 2024 Search PubMed , accepted for publication.
- M. J. Page, J. E. McKenzie, P. M. Bossuyt, I. Boutron, T. C. Hoffmann, C. D. Mulrow, L. Shamseer, J. M. Tetzlaff, E. A. Akl, S. E. Brennan, R. Chou, J. Glanville, J. M. Grimshaw, A. Hróbjartsson, M. M. Lalu, T. Li, E. W. Loder, E. Mayo-Wilson, S. McDonald, L. A. McGuinness, L. A. Stewart, J. Thomas, A. C. Tricco, V. A. Welch, P. Whiting and D. Moher, BMJ, 2021, 372, n71 CrossRef PubMed.
- A. Aroca, J. M. Benito, C. Gotor and L. C. Romero, J. Exp. Bot., 2017, 68, 4915–4927 CrossRef CAS PubMed.
- A. Jurado-Flores, L. C. Romero and C. Gotor, Antioxidants, 2021, 10, 508 CrossRef CAS PubMed.
- V.-A. Duong, O. Nam, E. Jin, J.-M. Park and H. Lee, Molecules, 2021, 26, 2027 CrossRef CAS PubMed.
- N. Bithi, C. Link, Y. O. Henderson, S. Kim, J. Yang, L. Li, R. Wang, B. Willard and C. Hine, Nat. Commun., 2021, 12, 1745 CrossRef CAS PubMed.
- R. N. Carter, M. T. G. Gibbins, M. E. Barrios-Llerena, S. E. Wilkie, P. L. Freddolino, M. Libiad, V. Vitvitsky, B. Emerson, T. Le Bihan, M. Brice, H. Su, S. G. Denham, N. Z. M. Homer, C. Mc Fadden, A. Tailleux, N. Faresse, T. Sulpice, F. Briand, T. Gillingwater, K. H. Ahn, S. Singha, C. McMaster, R. C. Hartley, B. Staels, G. A. Gray, A. J. Finch, C. Selman, R. Banerjee and N. M. Morton, Cell Rep., 2021, 37, 109958 CrossRef CAS PubMed.
- X.-H. Gao, D. Krokowski, B.-J. Guan, I. Bederman, M. Majumder, M. Parisien, L. Diatchenko, O. Kabil, B. Willard, R. Banerjee, B. Wang, G. Bebek, C. R. Evans, P. L. Fox, S. L. Gerson, C. L. Hoppel, M. Liu, P. Arvan and M. Hatzoglou, eLife, 2015, 4, e10067 CrossRef PubMed.
- Q. Wu, B. Zhao, Y. Weng, Y. Shan, X. Li, Y. Hu, Z. Liang, H. Yuan, L. Zhang and Y. Zhang, Anal. Chem., 2019, 91, 14860–14864 CrossRef CAS PubMed.
- J. L. F. Comas, F. Ortega, M. A. Rodríguez, M. Kern, A. Lluch, W. Ricart, M. Blüher, C. Gotor, L. C. Romero, J. M. Fernández-Real and J. M. Moreno-Navarrete, Antioxid. Redox Signaling, 2021, 35, 319–340 CrossRef PubMed.
- X. Li, N. J. Day, S. Feng, M. J. Gaffrey, T.-D. Lin, V. L. Paurus, M. E. Monroe, R. J. Moore, B. Yang, M. Xian and W.-J. Qian, Redox. Biol., 2021, 46, 102111 CrossRef CAS PubMed.
- X.-H. Gao, L. Li, M. Parisien, J. Wu, I. Bederman, Z. Gao, D. Krokowski, S. M. Chirieleison, D. Abbott, B. Wang, P. Arvan, M. Cameron, M. Chance, B. Willard and M. Hatzoglou, Mol. Cell. Proteomics, 2020, 19, 852–870 CrossRef CAS PubMed.
- S. Longen, F. Richter, Y. Köhler, I. Wittig, K.-F. Beck and J. Pfeilschifter, Sci. Rep., 2016, 6, 29808 CrossRef CAS PubMed.
- L. Fu, K. Liu, J. He, C. Tian, X. Yu and J. Yang, Antioxid. Redox Signaling, 2020, 33, 1061–1076 CrossRef CAS PubMed.
- B. Pedre, D. Talwar, U. Barayeu, D. Schilling, M. Luzarowski, M. Sokolowski, S. Glatt and T. P. Dick, Nat. Chem. Biol., 2023, 19, 507–517 CrossRef CAS PubMed.
- H. Peng, Y. Zhang, L. D. Palmer, T. E. Kehl-Fie, E. P. Skaar, J. C. Trinidad and D. P. Giedroc, ACS Infect. Dis., 2017, 3, 744–755 CrossRef CAS PubMed.
- B. J. C. Walsh, S. S. Costa, K. A. Edmonds, J. C. Trinidad, F. M. Issoglio, J. A. Brito and D. P. Giedroc, Antioxidants, 2022, 11, 1607 CrossRef CAS PubMed.
- D. Zhang, I. Macinkovic, N. O. Devarie-Baez, J. Pan, C.-M. Park, K. S. Carroll, M. R. Filipovic and M. Xian, Angew. Chem., Int. Ed., 2014, 53, 575–581 CrossRef CAS PubMed.
- J. Pan and M. Xian, Chem. Commun., 2010, 47, 352–354 RSC.
- W. L. Chick, S. Warren, R. N. Chute, A. A. Like, V. Lauris and K. C. Kitchen, Proc. Natl. Acad. Sci. U. S. A., 1977, 74, 628–632 CrossRef CAS PubMed.
- Z. Li and T. L. Thomas, Plant Cell, 1998, 10, 383–398 CrossRef CAS PubMed.
- J. Li, D. Jia and X. Chen, Plant Cell, 2001, 13, 2269–2281 CrossRef CAS PubMed.
- G. Han, M. Wang, F. Yuan, N. Sui, J. Song and B. Wang, Plant Mol. Biol., 2014, 86, 237–253 CrossRef CAS PubMed.
- H. Chen and P. C. Boutros, BMC Bioinform., 2011, 12, 35 CrossRef PubMed.
- N. J. Rzechorzek, S. W. Hardwick, V. A. Jatikusumo, D. Y. Chirgadze and L. Pellegrini, Nucleic Acids Res., 2020, 48, 6980–6995 CrossRef CAS PubMed.
- D. W. Huang, B. T. Sherman and R. A. Lempicki, Nat. Protoc., 2009, 4, 44–57 CrossRef CAS PubMed.
- W. Huang da, B. T. Sherman and R. A. Lempicki, Nucleic Acids Res., 2009, 37, 1–13 CrossRef PubMed.
- H. Ogata, S. Goto, K. Sato, W. Fujibuchi, H. Bono and M. Kanehisa, Nucleic Acids Res., 1999, 27, 29–34 CrossRef CAS PubMed.
- T. B. Rajavashisth, A. K. Taylor, A. Andalibi, K. L. Svenson and A. J. Lusis, Science, 1989, 245, 640–643 CrossRef CAS PubMed.
- D. Benhalevy, S. K. Gupta, C. H. Danan, S. Ghosal, H. W. Sun, H. G. Kazemier, K. Paeschke, M. Hafner and S. A. Juranek, Cell Rep., 2017, 18, 2979–2990 CrossRef CAS PubMed.
- D. Petrovic, E. Kouroussis, T. Vignane and M. R. Filipovic, Front. Aging Neurosci., 2021, 13 Search PubMed.
- M. S. Vandiver, B. D. Paul, R. Xu, S. Karuppagounder, F. Rao, A. M. Snowman, H. Seok Ko, Y. Il Lee, V. L. Dawson, T. M. Dawson, N. Sen and S. H. Snyder, Nat. Commun., 2013, 4, 1626 CrossRef PubMed.
- R. J. Deshaies and C. A. P. Joazeiro, Annu. Rev. Biochem., 2009, 78, 399–434 CrossRef CAS PubMed.
- L. Barazzuol, F. Giamogante, M. Brini and T. Calì, Int. J. Mol. Sci., 2020, 21 Search PubMed.
- A. Kazlauskaite, R. J. Martínez-Torres, S. Wilkie, A. Kumar, J. Peltier, A. Gonzalez, C. Johnson, J. Zhang, A. G. Hope, M. Peggie, M. Trost, D. M. van Aalten, D. R. Alessi, A. R. Prescott, A. Knebel, H. Walden and M. M. Muqit, EMBO Rep., 2015, 16, 939–954 CrossRef CAS PubMed.
- M. Gundogdu, R. Tadayon, G. Salzano, G. S. Shaw and H. Walden, Biochim. Biophys. Acta, Gen. Subj., 2021, 1865, 129894 CrossRef CAS PubMed.
- T. Wauer, M. Simicek, A. Schubert and D. Komander, Nature, 2015, 524, 370–374 CrossRef CAS PubMed.
- C. Gladkova, S. L. Maslen, J. M. Skehel and D. Komander, Nature, 2018, 559, 410–414 CrossRef CAS PubMed.
- M. E. McLaughlin-Drubin and K. Münger, Virology, 2009, 384, 335–344 CrossRef CAS PubMed.
- J. Font and J. P. Mackay, Engineered Zinc Finger Proteins: Methods and Protocols, 2010, pp. 479–491 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3cb00106g |
| This journal is © The Royal Society of Chemistry 2024 |