Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Microfluidic methods for the diagnosis of acute respiratory tract infections
Kan-Zhi
Liu
a,
Ganghong
Tian
a,
Alex C.-T.
Ko
a,
Matthias
Geissler
 *b,
Lidija
Malic
*b,
Lidija
Malic
 b,
Byeong-Ui
Moon
b,
Liviu
Clime
b,
Byeong-Ui
Moon
b,
Liviu
Clime
 b and
Teodor
Veres
b
b and
Teodor
Veres
b
aLife Sciences Division, Medical Devices Research Centre, National Research Council of Canada, 435 Ellice Avenue, Winnipeg, MB R3B 1Y6, Canada
bLife Sciences Division, Medical Devices Research Centre, National Research Council of Canada, 75 de Mortagne Boulevard, Boucherville, QC J4B 6Y4, Canada. E-mail: matthias.geissler@cnrc-nrc.gc.ca
First published on 7th October 2024
Abstract
Acute respiratory tract infections (ARTIs) are caused by sporadic or pandemic outbreaks of viral or bacterial pathogens, and continue to be a considerable socioeconomic burden for both developing and industrialized countries alike. Diagnostic methods and technologies serving as the cornerstone for disease management, epidemiological tracking, and public health interventions are evolving continuously to keep up with the demand for higher sensitivity, specificity and analytical throughput. Microfluidics is becoming a key technology in these developments as it allows for integrating, miniaturizing and automating bioanalytical assays at an unprecedented scale, reducing sample and reagent consumption and improving diagnostic performance in terms of sensitivity, throughput and response time. In this article, we describe relevant ARTIs—pneumonia, influenza, severe acute respiratory syndrome, and coronavirus disease 2019—along with their pathogenesis. We provide a summary of established methods for disease diagnosis, involving nucleic acid amplification techniques, antigen detection, serological testing as well as microbial culture. This is followed by a short introduction to microfluidics and how flow is governed at low volume and reduced scale using centrifugation, pneumatic pumping, electrowetting, capillary action, and propagation in porous media through wicking, for each of these principles impacts the design, functioning and performance of diagnostic tools in a particular way. We briefly cover commercial instruments that employ microfluidics for use in both laboratory and point-of-care settings. The main part of the article is dedicated to emerging methods deriving from the use of miniaturized, microfluidic systems for ARTI diagnosis. Finally, we share our thoughts on future perspectives and the challenges associated with validation, approval, and adaptation of microfluidic-based systems.
1 Introduction
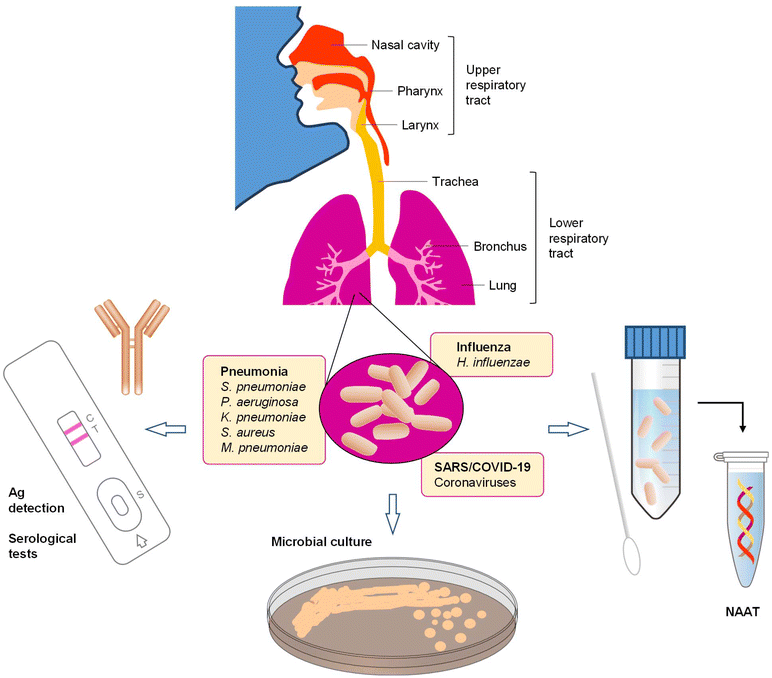
Acute respiratory tract infections (ARTIs) are affecting millions of people each year through seasonal or pandemic outbreaks. Especially infections of the lower respiratory tract are a leading cause of morbidity and mortality worldwide, with children and elderly people who suffer from chronic conditions being the most vulnerable population subgroups.1 According to estimates by the World Health Organization (WHO), 1.9 million children die from ARTIs each year,2 primarily in Africa and Asia.3 By the end of 2019 and the beginning of 2020, a new variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) had spread around the world within months, causing a global health crisis that stands at 702.9 million people infected and more than 6.9 million fatalities at the time of writing.4 The number of infections and the complications deriving from the disease have overwhelmed public health systems, resulting in substantial collateral damage and additional deaths due to insufficient capacities for treatment and care of other, non-respiratory diseases. Moreover, the socioeconomic consequences of the pandemic are far-reaching and have prompted governments around the world to assume trillions of dollars in relief and recovery programs.5The human respiratory tract (Fig. 1) is a primary toxicological target area for many microbial organisms as they enter the human body through inhalation. The upper respiratory tract, composed of nasopharyngeal and laryngeal tissues, allows for filtration, humidification and temperature adjustment of the inhaled air. It thus protects the more sensitive and fragile lower respiratory tract, which serves as the gas exchange medium in respiration cycles. ARTIs are mainly caused by virus or bacteria, but can also involve more than one pathogen from both categories. Symptoms of ARTIs are not pathogen-specific, thus examination of patients based on symptoms is usually insufficient for differentiating between infections since they can share very similar clinical characteristics. While discrimination between viral and bacterial infections is important, rapid and accurate identification of the causative pathogen is required to enable a targeted approach to administering medication and treat the disease.6 Diagnostic uncertainty commonly leads to over-prescription of antibiotics and extra diagnostic testing (along with costs) to rule out bacterial infections. Rapid detection of viral pathogens could overcome these disadvantages. Besides, prompt viral diagnosis may lead to rapid implementation of infection control measures, early administration of antiviral medication, if available, and shorter hospitalization, resulting in reduced healthcare costs.7,8 Therefore, the development of technology for rapid and accurate diagnosis of respiratory disease syndromes remains of paramount importance.6
Public health agencies rely on both diagnostic and surveillance tests in response to outbreaks of ARTIs. Diagnostic tests are typically employed for symptomatic individuals and require high specificity to avoid false positives. Surveillance tests, on the other hand, are aimed at identifying asymptomatic individuals in an effort to prevent the disease from spreading and thus necessitate high sensitivity to avoid false negatives. While traditional culture-based methods are usually accurate, processes related to isolation and growth of microbial organisms are lengthy. Delays in the diagnosis are often the reason for antibiotic misuse deriving from broad-spectrum antimicrobials administered by physicians to treat infected patients. The use of broad-spectrum antimicrobials, in turn, promotes antibiotic resistance, which can complicate the treatment of infected patients.6 Moreover, antimicrobial resistance (AMR) is considered to be one of the most pressing challenges for public health globally today.9 Timely detection and identification of microbial pathogens will enable clinicians to make decisions early with regard to patient management and treatment. The assessment of AMR profiles would further allow for adequate use of antibiotics, thus resulting in more effective treatment and reduced healthcare cost.
The development and widespread adoption of molecular and point-of-care (POC) testing methods over the past decade has led to significant improvements in the capacity to detect and identify respiratory pathogens. This article is intended to provide an overview of these developments, complementing previous reviews devoted to this field.6,10–15 First, we introduce selected ARTIs which have clinical and social impact today, especially during outbreaks. We then discuss established methods that are currently being used for the diagnosis of these diseases, mainly in clinical laboratories. This is followed by an introduction to microfluidics with an emphasis on operational principles for mediating flow at reduced scale. We subsequently provide a short overview of commercial instruments that employ these actuation principles. After that, we highlight emerging methods and recent developments in the area of miniaturized, microfluidic systems for detecting respiratory disease. We conclude this article with personal remark and an outlook on where developments might lead in the future.
2 Selected ARTIs
2.1 Pneumonia
Pneumonia is an alveolar infection that occurs when the innate immune system is unable to clear a pathogen from the lower airway and alveoli.16 It is a common infection of the lungs affecting approximately 450 million people per year in all parts of the world.17 Pneumonia was the 4th leading cause of death in the world in 2016, resulting in 3.0 million fatalities. There are over 30 microbial organisms that cause pneumonia, including bacteria, viruses, and fungi, each of which can induce different epidemiology, symptoms, and clinical course. Our innate defense systems, such as inflammatory factors and cytokines can cause additional harm to the lung parenchyma, which result in secondary symptoms such as fever, chills, and fatigue.18 Currently, the classification of pneumonia is based upon the causative microorganism, although pathologic demarcation exists between lobar and bronchopneumonia.19 Epidemiologic categories refer to nosocomial pneumonia and community-acquired pneumonia (CAP). Nosocomial pneumonia is commonly divided into two distinct groups: hospital-acquired pneumonia (HAP) and ventilator-associated pneumonia (VAP). HAP denotes an episode of pneumonia that occurs more than 48 h after hospital admission and was not in incubation at the time of the admission.20 VAP is defined as pneumonia that occurs more than 48 h after the initiation of invasive mechanical ventilation. Symptoms for nosocomial pneumonia may include cough, expectoration, rising body temperature, chest pain or dyspnea. Signs include fever, tachypnea, consolidations or crackles. CAP is defined as an acute lung infection that occurs without recent healthcare exposure of the patient;21 it is the most common type of bacterial pneumonia with Streptococcus pneumoniae dominating in nearly 50% of cases.17 Up to one third of all CAP cases are caused by Mycoplasma pneumoniae which is particularly challenging to diagnose.22,23 Patients with mycoplasmal infections often lack the typical CAP-associated symptoms as the disease has an insidious onset followed by several days or weeks of slowly worsening dry cough, fever, and malaise.222.2 Influenza
Influenza is caused by influenza viruses, which belong to the Orthomyxoviridae family and are divided into three types (A, B, and C).24 Influenza A and B viruses (IAV and IBV, respectively) cause seasonal epidemics, whereas influenza C viruses (ICV) generally cause mild disease.25 There are several subtypes of IAV based on the antigenic properties of their two surface glycoproteins, hemagglutinin and neuraminidase. Influenza viruses are enveloped particles that contain a single-stranded, segmented RNA genome.25 They mainly affect the upper respiratory tract, but other organs such as the heart, brain, and muscles can also be involved. The virus is transmitted from person to person with respiratory droplets produced upon coughing or sneezing. Influenza can occur globally, causing considerable morbidity and mortality with pandemic, epidemic, or seasonal patterns.24 The most severe pandemic (known as the Spanish Flu) occurred from 1918 till 1920 and caused an estimated 20–50 million (possibly up to 100 million) deaths worldwide.26,27 The presentation of seasonal influenza ranges from an asymptomatic infection to a fulminant illness, depending on the characteristics of both host and virus. Symptoms appear after an incubation period of 1–2 days and are characterized by various systemic features, including fever, chills, headache, myalgia, malaise, and anorexia, accompanied by respiratory symptoms, such as non-productive cough, nasal discharge, and sore throat. Individuals who contracted the virus usually recover after a few days, but it can give rise to severe complications in high-risk groups.2.3 SARS
SARS is caused by coronaviruses—a class of enveloped, positive-sense RNA viruses characterized by club-shaped spikes that project from their surface, an unusually large RNA genome, and a unique replication strategy.28 Compared to DNA viruses, RNA viruses are technically more virulent as they infect cells by injecting RNA which quickly transcribes and replicates viral proteins in the host cell. This also makes it extremely challenging to detect the virus at an early stage of the infection. Structurally, coronavirus particles contain four essential proteins—spike (S), membrane, envelope, and nucleocapsid (N)—all of which are encoded within the 3′ end of the viral genome.29,30 The virus binds to cells expressing the virus receptors, such as angiotensin-converting enzyme (ACE2), widely expressed in the respiratory tract on the epithelial cells of alveoli, trachea, bronchi, bronchial serous glands, as well as alveolar monocytes and macrophages. Furthermore, as a surface molecule, ACE2 is also diffusely localized on the endothelial cells of arteries and veins, the mucosal cells of the intestines, tubular epithelial cells of the kidneys, epithelial cells of the renal tubules, and cerebral neurons and immune cells, providing a variety of susceptible target locations for SARS-CoV.31 As a result, respiratory secretions as well as urine, stool, and sweat from SARS patients contain infective viral particles, which may be excreted into and contaminate the environment.Clinically, SARS is an emerging infectious disease with flu-like symptoms, including high fevers, myalgia, dry non-productive dyspnea, lymphopenia, and infiltrate on chest radiography. Transmission occurs through human contact and interaction, with international air travel facilitating the rapid dissemination of this virus on a global scale. In the 2003 SARS pandemic, over 8000 people were affected, with a fatality rate of 9.5%.32 Atypical pneumonia with rapid respiratory deterioration and failure can be induced by SARS-CoV infection because of increased levels of activated proinflammatory chemokines and cytokines.33,34 The outbreak of the original SARS epidemic was largely brought under control by simple public health measures. Testing people with symptoms (fever and respiratory problems), isolating and quarantining suspected cases, and restricting travel all had an effect.
2.4 Coronavirus disease 2019 (COVID-19)
COVID-19 is the clinical syndrome caused by SARS-CoV-2 infection.35 While some COVID-19 patients show no symptoms at all, most infected individuals experience one or several manifestations of the disease such as fever, cough, fatigue, sputum production, shortness of breath, sore throat, loss of taste, and headache. In severe cases, infections can cause interstitial pneumonia, acute respiratory distress syndrome (ARDS),36 kidney failure, and death. Comparative analysis of SARS-CoV-2 with other epidemic viral strains serves as background information on which researchers can build upon to create potential technologies to quickly address the demands of disease control in an emergency. COVID-19 has a fatality rate of 2.3%, which is lower than that of SARS and Middle East respiratory syndrome (MERS) which account for 9.5 and 34.4%, respectively.37 Analysis of receptor affinity shows that SARS-CoV-2 binds ACE2 human cell receptor more efficiently than the SARS-CoV strain of 2003.38 MERS-CoV, on the other hand, uses dipeptidyl peptidase 4 to enter host cells.38 Whether the higher receptor affinity of SARS-CoV-2 for ACE2 could lead to a more severe lung involvement in COVID-19 than in SARS requires further investigation.37 The basic reproduction number (R0) of COVID-19 is estimated at 2–5,39,40 while R0 amounts to 2–4 for SARS40 and 0.47 for MERS,41 suggesting that COVID-19 has a higher pandemic potential.423 Diagnostic methods
Diagnosis of ARTIs is challenging because symptoms can vary in severity and clinical signs of viral infections often overlap with those of bacterial infections.43,44 Initial assessment of pneumonia is typically performed through observation of physical manifestations resulting from lower respiratory tract infection, focal chest sounds, exclusion of other possible diagnosis and radiological evidences such as alveolar consolidation and pleural effusion.45,46 Clinicians are likely to diagnose influenza infection when fever and cough are part of the case definition, when influenza rates are high in the community, and when patients are severely ill or are at an increased risk of developing complications.47,48 Likewise, no pathognomonic signs or symptoms of SARS can be used to differentiate the disease from other causes of community- or hospital-acquired pneumonia. Signatures of COVID-19 infection which appear in computer-assisted tomography (CT) scans include areas of subpleural regions of ground glass opacification affecting the lower parts of either a single lobe or both lobes.49 In addition, data from COVID-19 patients have shown consolidation of fluids in the lungs. While CT scans are likely to remain one of the most important tools in the early diagnosis of COVID-19, the major challenge for the radiologists lies in the distinction of symptoms from other lung disorders or pneumonia-like symptoms which are not due to COVID-19 infections. In addition, CT scans are expensive and require advanced technical skills for operation and analysis. The technology therefore is considered only complementary for SARS-CoV-2 detection.Diagnostic methods comprise a triad of complementary approaches based on molecular and non-molecular methods for clinical use as well as surveillance of larger population groups at the point of need (Fig. 1). Molecular methods are becoming widely adopted and the progressive commercialization and clinical application is placing them at the forefront of etiological diagnosis.50–52 Molecular methods are based on nucleic acid (NA) amplification techniques (NAATs) where respiratory tract samples are being interrogated through amplification of genetic target strands specific to causative microbial pathogens. Non-molecular methods include antigen (Ag) detection, serological testing as well as microbial culture from respiratory, fecal, and tissue specimens. At present, NAATs are the only technical approach that can provide definite etiological diagnosis.
3.1 NAATs
Detection of pathogenic agents by NAAT involves different forms of polymerase chain reaction (PCR) which relies on an enzymatic amplification process and specific primers to produce multiple identical copies from a target sequence.53,54 For the detection of common respiratory pathogens, several NAAT assays are available commercially (Table 1).55–58 Rapid thermocycling instruments along with closed-tube assays and real-time detection using fluorescence technologies have enabled the widespread adoption of PCR in clinical microbiology laboratories, making it possible, in combination with rapid sample preparation processes, to detect microbes and their antibiotic resistance genes directly from clinical specimens.59 Standard PCR uses end-point detection based on gel electrophoresis or microarray hybridization where fluorescently-labelled PCR products are interrogated with a set of probes immobilized on a solid support to confirm or disprove the presence of a particular target gene. Real-time or quantitative PCR, on the other hand, records the entire reaction profile, making it possible to identify reactions that deviate in amplification efficiency. The concentration of NAs is thereby derived from amplification rates relative to a standard curve. Reverse transcription PCR (RT-PCR) has become the method of choice for detecting viral RNA in research and clinical laboratories with hydrolysis probes and molecular beacons being the most common probe formats. Loop-mediated amplification (LAMP) provides a reliable substitute for quick and accurate detection of low-copy-number NAs in the diagnosis of several viral diseases, including SARS and COVID-19. In LAMP, DNA is amplified under isothermal conditions.60,61 This method uses a set of specially designed primers, and a DNA polymerase with strand displacement activity. However, the main disadvantage of LAMP-based methods is the complexity of primer design to achieve the specificity of a PCR test. Multiplex PCR systems in which several PCR targets are sought after simultaneously in a single reaction have gained wider acceptance, particularly among commercial assays. The accuracy of these tests not only depends on the assay composition, but also on type, quantity and quality of the specimens collected. There are several different sample types commonly used for PCR, which include nasopharyngeal swab (NPS), nasal swab (NS), nasopharyngeal wash (NPW), nasal wash (NW), nasal aspirate and throat swab (TS). Many PCR assays require relatively clean sample solutions that are free of inhibitors, making it necessary to extract and purify microbial DNA or RNA from lysates. It should also be pointed out that the pathogenic viruses enter the human body through the mucus membrane cells in the bronchi and alveolar cells. The sample obtained by swab collection therefore may not contain virus, although individuals were infected by the pathogens. This could explain why some patients display typical COVID-19 symptoms in spite of a negative PCR test.| Test | Manufacturer | Technology | Targets | Sample type | Time-to-result |
|---|---|---|---|---|---|
| a Adapted in part from ref. 55. A more complete compilation of NAAT assays for the detection of respiratory pathogens is provided in ref. 56. For comprehensive overviews of SARS-CoV-2 detection assays, see ref. 57 and 58. | |||||
| Bio-Rad SARS-CoV-2 ddPCR kit | Bio-Rad | ddPCR | SARS-CoV-2 | NPS and others | 120 min |
| Alere i Influenza A & B | Alere Scarborough | Isothermal NA amplification | RSV | NPS | <15 min |
| ARIES R flu A/B & RSV assay | Luminex Corporation | Real-time PCR | IAV, IBV, RSV | NPS | <2 h |
| FilmArray Respiratory Panel 2.1 | BioFire Diagnostics | Nested multiplex RT-PCR | Multiplex panel of 22 targets, including M. pneumoniae, IAV, IBV, SARS-CoV-2 | NPS | ∼45 min |
| Cobas Influenza A/B Nucleic Acid Test for use on the Cobas Liat System | Roche Molecular Diagnostics | Real-time RT-PCR | IAV, IBV, RSV | NPS | ∼20 min |
| Illumigene Mycoplasma Direct DNA Amplification Assay | Meridian Bioscience | LAMP | M. pneumoniae | NPS, TS | <1 h |
| Panther fusion flu A/B/RSV | Hologic | Real-time RT-PCR | IAV, IBV, RSV | NPS | ∼2.5 h |
| Xpert Xpress Flu | Cepheid | Real-time RT-PCR | IAV, IBV, RSV | NPS, nasal aspirate, and NW | ∼30 min |
3.2 Ag testing
Direct fluorescent antibody (Ab) testing and rapid immunoassays (RIAs) have long been used for the diagnosis of ARTIs. They are aimed at detecting microbial Ag in body fluids.62 RIAs are typically performed as near-patient tests and can provide results in less than 30 min. There are four main formats for RIA tests: latex agglutination, lateral flow (LF) devices, horizontal flow devices and optical immunoassays. The LF immunoassay (LFIA) is the most versatile and widely used immunochromatographic method. LFIAs are relatively inexpensive and easy to perform. They are invaluable tools for primary care, emergency departments and low resource settings. The development of immunoassays is reliant on the identification of suitable Ag targets present in detectable quantities in clinical specimens. To date, commercial assays have been developed only for a limited range of pathogens. For instance, a newer generation immunochromatographic test that detects the C-polysaccharide cell wall Ag in urine has been an important advance in the diagnosis of pneumococcal disease.63 Viral Ag such as the S protein in circulating blood can be used for the prognosis of SARS-related viremia.64 RIAs have been commonly applied to the detection of IAV and IBV as well as respiratory syncytial virus (RSV). In the recent pandemic situation, LFIAs were routinely used for screening of coronavirus infections at the point of need. Other examples include the detection of Ag related to S. pneumoniae and Legionella pneumophila serogroups. These assays are more rapid than culture-based tests, however their sensitivity is generally inferior to that of culture. Also, they cannot provide information on susceptibility of microorganisms to antimicrobial drugs.3.3 Serological testing
Serological tests detect Abs produced in response to an infection. They allow for successfully identifying Abs for most respiratory pathogens, including SARS-CoV, adenovirus and Haemophilus influenzae. Pathogen-specific Abs usually appear about 10 days after exposure, which renders serological testing unsuitable for detection during the early stage of infection. Earlier SARS-CoV-related research and most recent studies on SARS-CoV-2 have shown that SARS-specific Abs, such as immunoglobulin G (IgG), immunoglobulin M (IgM), and immunoglobulin A (IgA) can be detected in serum as early as seven days after viral infection or 3–5 days after the symptoms appear and can last for several years after recovery.64,65 Detection of these antibodies could be extremely useful for surveillance of the population and also for plasma transfusion to combat the active virus.66 Standard serological procedures involve the use of microtiter plates such as the 96-, 192- or 384-well formats, where Abs are detected via protein–protein interactions that are visualized using a colorimetric or fluorescence assay assisted by enzymes involved in the reaction. For example, detection based on enzyme-linked immunosorbent assays (ELISA) has been shown for IgM and IgG antibodies raised to SARS-CoV-2 nucleoprotein Rp3 during the initial stages of COVID-19 disease.67 However, serological tests for detecting parainfluenza virus and adenovirus are considerably less sensitive than molecular methods such as PCR. Also, bacterial infections are difficult to detect using Ab-based testing, which limits the usefulness of serological methods for diagnostics in clinical settings.683.4 Culture-based methods
Observation of the cytopathic effect and hemadsorption in viral culture has been the ‘gold standard’ for the diagnosis of ARTIs for decades.69 Standard tube-based culture methods allow for growing a large number of viruses, but obtaining results can take 5–10 days. Over the years, turnaround times have been reduced to 24 h through the use of improved cell culture methods such as the centrifugation-enhanced shell vial method.70 However, several clinically relevant viruses such as rhinovirus (RV) and CoV are difficult to grow in culture.71 Also, subjecting samples to multiple freeze/thaw cycles prior to culture can affect viral titer and growth. Bacterial pathogens can be detected and identified through culture, which is often performed in conjunction with antimicrobial susceptibility testing.72 As for viral culture, bacterial culture requires experienced technologists, specialized media, and prolonged incubation periods (e.g., 48–96 h). Also, there are limitations to the diagnosis of several ARTIs such as mycoplasmal CAP by culture. For these reasons, culture-based methods are becoming less relevant for clinical diagnosis, patient care and antimicrobial therapy.72,734 Microfluidics
The embracement of microfluidics for diagnostic applications is largely driven by the need for automating complex analytical procedures and providing affordable solutions for POC testing.10,74–78 Microfluidics is the science and technology of manipulating fluids in channels with dimensions below 100 μm.79,80 As such, it provides a framework for reducing sample and reagent consumption by scaling the reaction volumes down to sub-microliter levels. Microfluidics is closely related to the concept of lab-on-a-chip (LOC), which broadly aims at the integration of analytical workflows into a compact, miniaturized format.76,81 LOC systems offer the prospect of shortening process times and improving sensitivity thanks to the unique characteristics deriving from the confinement of flow to small-scale dimensions, which include relatively high surface-to-volume ratios as well as faster rates of mass and heat transfer.From a technical perspective, there are two key components to microfluidics, which are intrinsically linked to one another: (i) a chip (or other suitable matrix) that hosts the fluidic pathway, and (ii) an actuation scheme (or pumping system) to mediate and control flow. The chip design accounts for the dimensions of both channels and reservoirs, the type of control elements that need to be present (for example, to achieve metering, mixing or aliquoting of assay components), and the potential need for implementing sensor and detection units on the chip.75 These criteria will also inform the selection of appropriate materials and the methods that can be employed for fabrication. We refer the reader to excellent review articles that cover these topics at length.82,83 The pumping system is central to the functioning and performance of the chip; it determines the flow rates that can be achieved as well as the overall volumes that can processed on a microfluidic device.75 The fluid actuation scheme must reflect the complexity of the assay that is to be performed, the level of control that needs to be exercised over the fluid manipulation steps involved, and the requirements associated with POC testing (e.g., portability, a low footprint, and ease of operation). Microfluidic flow can be generated using active and passive means of pumping (Fig. 2) which are described in more detail below.
 | ||
| Fig. 2 Principles for mediating flow at low volume and reduced scale along with microfluidic devices serving as illustrative examples for each category. Centrifugal microfluidics: structures on a microfluidic disk. Reprinted with permission from ref. 91. Copyright © 2010 The Royal Society of Chemistry. EWOD: optical microscope image showing manipulation of discrete droplets on a DMF device. Courtesy of Alphonsus Ng, Lisa Ngo and Prof. Aaron R. Wheeler (University of Toronto). Capillary microfluidics: photograph of a microfluidic chip comprising a programmable resistor array and a 15 μm-deep flow path. Reprinted with permission from ref. 144. Distributed under Creative Commons Attribution License 4.0. Lateral flow systems: photograph of a rapid COVID-19 Ag test device after use with a positive SARS-CoV-2 NPS sample. Pressure-driven microfluidics: photograph of a cell sorting chip. Copyright © 2022 Fluigent. | ||
In recent years, droplet microfluidics has emerged as a groundbreaking technology with the potential to transform clinical diagnostics. This innovative approach harnesses the power of controlled generation, manipulation, and merging of droplets in channels for high-throughput experimentation.84–86 The ability to compartmentalize reactions into discrete droplets (ranging from femto- to nanoliter volumes) leads to enhanced control over reaction conditions and increased throughput, while minimizing sample consumption and reducing reagent waste. Droplet formation is commonly achieved through extrusion and shearing of immiscible multiphase flows (e.g., water in oil) inside microchannels that can involve both active and passive fluid actuation mechanisms.87 Droplet digital PCR (ddPCR) is the most important application of droplet microfluidics in the context of disease diagnostics (ARTIs and others); it provides higher sensitivity and accuracy than real-time PCR, and allows for absolute quantification without the need for a standard curve.88 The partitioning afforded by ddPCR reduces interference from inhibitors present in the sample, which renders the technique advantageous for early diagnostics of ARTIs when the viral load is low.89
4.1 Centrifugal systems
Centrifugal microfluidics is closely related to the concept of lab-on-a-disk90 where microfluidic elements such as reservoirs, channels and valves are implemented into a compact disk-shaped format.91,92 Centrifugal microfluidics makes use of the centrifugal force as the main actuation method for fluidic operations—a force that can easily be generated by spinning the disk around its center axis with the help of an electrical rotor. For any particular operational protocol, microfluidic architectural elements are designed based on the interplay between centrifugal and capillary forces developed at the interface between liquids and solid surfaces inside the microfluidic conduit. A common dimensionless quantity used to evaluate the importance of body forces (such as gravitational or centrifugal) compared to surface tension is the Bond number Bo = ρacpL2/σ, where ρ is the density of the liquid, acp is the centripetal acceleration generated upon rotation, L is the characteristic length (e.g., the size of a microfluidic channel or the radius of curvature for a liquid meniscus) and σ is the surface tension coefficient (for water σ = 72.8 mN m−1 at 20 °C). At low angular velocity the centrifugal fields are weak and the Bond number therefore is small, suggesting that surface tension (capillary) effects remain dominant, which is different from macroscopic behaviour. Several flow control elements such as capillary valves93 and siphon valves94 are taking advantage of the interplay between centrifugal and capillary forces. The former is controlled by changing the angular velocity of the driving motor; the latter through the dimensions and surface properties of relevant microfluidic components. Combining these flow control elements with modules for heating and temperature adjustment at specific locations on the disk has proven suitable for integrating relatively complex assays, some of which were performed in an automated fashion.92,95Despite the level of maturity displayed by these developments, there are several limitations inherent to centrifugal systems. One is the so-called ‘footprint issue’: due to the unidirectional nature of the centrifugal force, serious drawbacks in placing microfluidic architectural elements along the fluidic path on the microfluidic cartridge are inconveniently increasing the size of the chip (radially) while decreasing the integration density (number of microfluidic features per square centimeter). Capillary forces at low Bond numbers further render the manipulation protocol sensitive to the physical parameters of the input sample (e.g., different viscosities, wetting properties, etc.). Finally, the practical use of microfluidics often remains hampered by the lack of adequate ‘world-to-chip’ interfaces96,97 capable of (i) properly transferring liquids to and from the chip, and (ii) providing connectivity for sensory elements.
Innovative solutions aiming at solving these problems at least partially were also proposed. For example, an elegant way of addressing limitations induced by the unidirectional nature of the centrifugal force consisted in considering additional step motors installed on the rotor which are capable of changing the orientation of the microfluidic chips in the centrifugal field landscape in a synchronized fashion through controlled rotations with respect to a secondary axis.98 Various active control99 and pumping100 methods directed at improving microfluidic control and integration density were also proposed. An important step towards advanced microfluidic functionality, high density microfluidic integration and world-to-chip interfacing capabilities has been made by mounting electronic pumps and programmable electromechanical valves on the rotor of a centrifugal platform and apply regulated air pressure at specific ports of the microfluidic circuit to conduct advanced unit operations such as valving, switching, reverse pumping, bubble mixing as well as on- and off-chip liquid transfer.101–105
4.2 Flow-controlled (pressure-driven) microfluidic systems
Pressure-driven microfluidics uses syringe pumps, pressure control systems as well as gravity to induce and mediate flow in microchannels. The flow generated in this way is typically laminar, well-defined and predictable. In the absence of convection, mixing of solution is mainly achieved by diffusion which is inherently slow, emphasizing the need for fluidic control elements such as valves, split and recombination units or topographic elements for passive mixing to be incorporated into the fluidic circuit. Flow manipulation with the help of syringe pumps is the most commonly used method in many research laboratories as it involves an experimental setup that is fast and easy to implement.106–109 The flow rate is controlled by mechanical actuation to push or pull liquid to and from a microfluidic device, which is interfaced by tubing. Inside the microfluidic channels, the flow generated by syringe pumps remains inherently pulsative and thus is prone to fluctuation.110Conversely, pneumatic systems offer stable and responsive flows as the pressure controllers are able to establish non-pulsatile movement of liquid in a short period of time.111,112 In general, the pneumatic unit (that can involve a compressed gas cylinder, for example) is interfaced with a tube to pressurize the sample so it can be transferred out of the container to a microfluidic chip. Unlike syringe pump-driven flow systems, the absence of actuating mechanical parts allows for driving the flows smoothly and enables flow rates to be changed without delay. When supported by an adequate manifold, pressure can be applied to multiple wells on demand.113,114
The use of hydrostatic, gravity-driven flow is a simple yet useful technique.115,116 For example, sample-containing pipette tips can be vertically inserted into the inlets of the microfluidic device. Based on the height difference of the fluid column, the hydrostatic pressure makes the fluid move along microchannels. The pressure can be calculated by P = ρgΔH, where ρ is the density of the fluid, g = 9.81 m s−2 is the gravitational constant, and ΔH is the height of the fluid column. This technique is beneficial when a weak pressure gradient and relatively low flow rates are envisaged.117,118
4.3 Digital microfluidics (DMF)
DMF relies on individual manipulation of discrete unit droplets to execute fluidic operations required for a particular assay.119 Actuation forces used to transport droplets on hydrophobic surfaces include electrowetting-on-dielectric (EWOD),119,120 surface acoustic waves,121 dielectrophoresis,122 thermocapillary forces,123 magnetic forces,124,125 and opto-electrowetting.126 Devices relying on EWOD present the most promising avenue for diagnostic applications.EWOD refers to an electrically induced modulation of interfacial tension between a polarizable and/or conductive droplet and a solid electrode coated with a hydrophobic dielectric film.119,127,128 When voltage is applied, the stored electric charge changes the free energy on the dielectric surface and reduces the interfacial tension, thus inducing a change in wettability on the surface. Switching the voltage “on” and “off” between a series of adjacent electrodes then results in an interfacial tension gradient that can be used to individually manipulate droplets on a solid substrate with no moving parts. As such, the advantage of EWOD over other actuation methods lies in the capability of these devices to execute complex analytical protocols using a finite set of elemental fluidic operations (e.g., droplet dispensing from a reservoir, transport, merging, mixing, and splitting).119,120
EWOD devices have been implemented using either a co-planar electrode scheme (open) with the ground electrode positioned in the same plane as the actuation electrodes, or a two-plane configuration (closed system) with the droplet being sandwiched between two parallel plates separated by a spacer. In a typical closed system, the bottom plate houses an array of equidistant actuation electrodes, while the top plate is reserved for the continuous ground electrode. This configuration allows for more precise control over droplet transport, dispensing and splitting in comparison to open systems and is the method of choice for implementation of automated biochemical assays.129 EWOD devices are typically fabricated using a silicon or glass substrate onto which patterned electrodes (e.g., using standard photolithography, physical deposition and lift-off techniques) are covered with a thin dielectric film (e.g., parylene, SU-8, SiO2, Si3N4 and others) and a hydrophobic coating (e.g., Teflon, SiC, etc.). In order to render these devices more amenable to industrial translation, low-cost printed circuit board EWOD devices have been demonstrated.130,131 Owing to these developments, several commercial products that rely on EWOD microfluidic technology are emerging for a number of bioanalytical applications, which include DNA library preparation, sample preparation, biosensing, and molecular diagnostics. Commercial potential of these devices has been outlined by Li and Kim,132 while several other reviews provide excellent overviews of technical and analytical developments.133–135
4.4 Capillary microfluidics
Capillary-driven microfluidics136–141 has become a rapidly growing field within the realm of microfluidics, focusing on the manipulation and control of fluids using capillary forces. Unlike conventional microfluidic systems that rely on external pumps or power sources, capillary microfluidics leverages the inherent properties of surface tension to mediate fluid flow in microchannels. As such, the approach offers significant advantages in terms of simplicity, cost effectiveness and portability. The fundamental principle of capillary microfluidics lies in the interplay between surface tension and channel geometry.142 Surface tension, a cohesive force resulting from the attraction between molecules at the liquid/gas interface, generates capillary forces that propel the fluid within channels. The fluid thereby flows from regions of higher surface tension to those of lower surface tension in a fully autonomous fashion, following the path of least resistance. The ability to further regulate liquid transport, such as the possibility of metering flow and engaging fluids sequentially, has been shown in conjunction with capillary pumps (CPs).143–145 CPs are typically implemented at the end of the flow path and can include arrays of posts providing a predetermined capillary pressure encoded in the microstructure.The fabrication of capillary-driven microfluidic devices requires precise engineering of microchannels and chambers to harness capillary forces effectively.144 Various fabrication techniques such as hot embossing, molding, micromachining and additive manufacturing can be employed to this end.82 These methods allow for the creation of intricate microstructures with tailored dimensions and surface properties in a variety of materials. The flow rate and the direction of flow can both be controlled with precision by adjusting channel dimensions and surface characteristics such as wettability. The advantages of capillary microfluidic systems are manifold. First, these devices are generally easy to use. Second, they are cost effective since they do not require expensive components for operation. Third, they are highly portable, making them suitable for on-site and POC applications. These characteristics render capillary microfluidics particularly attractive for resource-limited settings where affordability, simplicity and portability are essential.
4.5 LF and paper-based devices
LF and paper-based devices are related conceptually and share similar characteristics. LF devices are one of the most widely used POC diagnostic tools for the detection of infectious diseases; they are appealing due to their relative simplicity, easy operation, extended shelf life, short response time, and cost-effectiveness.146–149 The components of a typical LF device are nitrocellulose membrane, sample pad, conjugate pad, and absorbent pad with the required reagents prestored on the test strip. The sample pad is designed for loading sample fluid; the conjugate pad is impregnated with specific molecule-conjugated biolabels for binding to the analyte and generating signal; the nitrocellulose membrane supporting immobilized capture molecules is the core component for testing; and the absorbent pad provides capillarity to draw and collect excess sample fluid. Commonly, there are two formats of LF devices: sandwich and competitive. In a sandwich LF device, a small volume of sample is loaded to the sample pad, which then propagates across the conjugate pad, nitrocellulose membrane, and absorbent pad through wicking and capillary effects. The target in the fluid will specifically recognize reporter molecules with appropriate biolabels prestored on the conjugate pad to form a complex (or aggregate), which is then captured in the testing area. The signal generated on test and control lines provides contrast so that the analytical result can be visualized by the naked eye. Therefore, in a sandwich LF device, the target concentration is proportional to the test line signal. In a competitive LF device, the target analyte will compete with the capture molecules immobilized on the test zone to bind to the reporter molecules prestored on the conjugation pad. Thus, in the presence of target analyte, the test line will not display any signal, while in the absence of the analyte target, signals emerge for both the test and the control line.Microfluidic paper-based analytical devices (μPADs) are receiving a great deal of attention for POC diagnostic applications.150–153 As a substrate, paper is appealing for the fabrication of LF systems because it is porous, hydrophilic and readily available at low cost. In addition, paper is biocompatible, can be cut and folded, and disposed of easily. Depending on the fabrication methods employed, μPADs can be classified into 2D and 3D μPADs. 2D μPADs are commonly fabricated by patterning hydrophobic boundaries (that can be physical or chemical in nature) on the paper to produce liquid guiding structures. Photolithography, laser treatment, plotting, cutting, various forms of etching, wax printing, and screen printing have been attempted for producing 2D μPADs.151 The fabrication of 3D μPADs is more complicated than that of 2D μPADs and typically involves multiple layers of 2D microfluidic paper stacked on top of each other. Origami, slip techniques, and 3D-printing have also been shown for the fabrication of 3D μPADs.151,154 Over the past decade, there have been a large number of novel μPADs reported for the detection of infectious diseases, while the contribution of these devices to the prevention and control of disease spreading is likely to increase in the years to come.
5 Commercial diagnostic instruments using microfluidics
Commercial instruments for the detection of respiratory diseases have witnessed significant advancements in recent years, driven by technology development, automation, and miniaturization. These tools and devices (some of which are listed in Table 2) offer a range of options for rapid and accurate diagnosis, while also catering to diverse clinical and operational needs.155–163 During the COVID-19 pandemic, considerable efforts have been dedicated to the development (or adaptation) of instrumentation for the detection of SARS-CoV-2. Microfluidics plays an increasing role in many of these technologies as part of the overall diagnostic workflow, impacting pathogen detection and disease diagnosis in both clinical and POC settings.11,164| Instrument | Characteristics | Manufacturer | Pathogens | Time | Ref. |
|---|---|---|---|---|---|
| a Adapted in part from ref. 11. For a comprehensive overview, see also ref. 165. | |||||
| GeneXpert | Miniaturized PCR device | Cepheid | Tuberculosis | 120 min | 159 |
| FilmArray | Miniaturized PCR device | BioFire Diagnostics | Respiratory viruses and bacteria | 60 min | 157 |
| ML FluAB-RSV assay | Miniaturized PCR device | Enigma | IAV, IBV | 95 min | 158 |
| Alere™ i | LAMP device | Alere | IAV, IBV | 15 min | 156 |
| Revogene® | Centrifugal RT-PCR system | Meridian Bioscience | Respiratory viruses and bacteria | 70 min | 160 |
| Simplexa™ | Centrifugal microfluidic real-time PCR device | Focus Diagnostics | IAV, IBV, RSV | 150 min | 155 |
| QX200 Droplet Digital PCR System | ddPCR system | Bio-Rad | SARS-CoV-2 | 120 min | 163 |
| COVID-19 IgM/IgG™ | LF device | Truvian Sciences | SARS-CoV-2 | 10 min | 162 |
| Abbott Panbio™ COVID-19 IgG/IgM Rapid Test | LF device | Abbott Diagnostics | SARS-CoV-2 | 10–20 min | 161 |
LOC systems integrating NAATs enable multiplexed analysis of respiratory pathogens with higher throughput than conventional methods. Companies like BioFire Diagnostics and Cepheid have developed PCR-based LOC platforms capable of detecting a wide array of respiratory pathogens with high accuracy. POC devices have become an indispensable part of the healthcare industry for they bring diagnostic capabilities directly to the patient's bedside or clinic, eliminating the need for sample transportation and centralized laboratory facilities. Microfluidic-based POC devices for respiratory infections offer rapid and accurate results, facilitating prompt clinical decision-making. They also generally do not require skilled personnel or heavy equipment for operation. Notable examples are the Alere i Influenza A & B test and the Quidel Sofia® 2 Fluorescent Immunoassay Analyzer, both of which utilize microfluidic technology to detect influenza viruses within minutes. The Revogene® instrument, originally developed by GenePOC59,165 and now being marketed by Meridian Bioscience, is a fully automated centrifugal platform with single- and multiplex PCR testing capabilities. The QX200 Droplet Digital PCR System from Bio-Rad Laboratories and the Naica® system for Crystal Digital PCR from Stilla Technologies offer ddPCR-based solutions for respiratory infection diagnosis, enabling high-throughput screening of pathogens with minimal sample volumes and reagent consumption. These systems provide unparalleled flexibility and scalability in sample processing and analysis.
Rapid Ag tests have gained prominence during the COVID-19 pandemic for their ability to deliver results quickly. Commercially available rapid Ag tests often come in the form of LF assays, where NS or TS sample is applied to a test strip containing antibodies that bind to the viral Ag, if present. These tests are relatively inexpensive and can be performed without the need for specialized laboratory equipment, making them suitable for use in POC settings. Several types of LF test strips have been employed for the diagnosis of SARS-CoV-2, which include the Cellex qSARS-CoV-2 IgG/IgM Rapid Test, Wondfo SARS-CoV-2 Ab test, SARS-CoV-2 DETECTR (Mammoth Biosciences) and SGTi-flex COVID-19 IgM/IgG (Sugentech). These and other LF devices are discussed in more detail elsewhere.164,166,167 It should be noted, however, that the need for rapid testing has dropped drastically in the post-pandemic era which, in turn, has posed significant economic challenges to smaller and medium-sized companies and eventually has led to the discontinuation of several products in this market segment.
6 Emerging diagnostic methods using microfluidics
6.1 Centrifugal (lab-on-a-disk) systems
Stumpf et al. have demonstrated fully automated sample-to-answer detection of IAV H3N2 using a centrifugal LabDisk.168 Their system incorporates prestored reagents, making the initial supply of the sample the only manual handling step. The self-contained LabDisk performs the entire protocol for PCR-based pathogen detection, which includes lysis, NA extraction, aliquoting of the eluate into 8 reaction cavities, and RT-PCR. Prestored reagents comprise air dried specific primers and fluorescence probes, lyophilized RT-PCR master mix and stick-packaged liquid reagents for NA extraction. The authors demonstrated successful detection of the respiratory pathogen in a total of 18 LabDisk devices with sample concentrations down to 2.39 × 104 viral RNA copies per mL, which is in the range of clinical relevance. Rombach et al.169 presented the RespiDisk, enabling fully automated detection of up to 19 respiratory pathogens from a single sample in a multiplex format using RT-PCR. RespiDisk comprises a centrifugal microfluidic LabDisk platform and combines new and existing advanced unit operations for liquid control, thereby automating all assay steps through spinning frequency and temperature settings in combination with the use of a permanent magnet for in situ handing of magnetic particles used for NA extraction (Fig. 3A). The capabilities of the system were demonstrated with 36 tested samples mimicking clinical conditions (clinical and/or cultured material suspended in transport medium or synthetic bronchoalveolar lavage) from past external quality assessment panels covering 13 of the 19 integrated detection assays for respiratory infection. A strong feature of the platform is its universality since its components allow for simultaneous detection of a broad panel of bacteria and viruses in a single run, thereby enabling the differentiation between diseases that can be treated using antibiotics. | ||
| Fig. 3 Centrifugal microfluidic systems for respiratory pathogen detection. (A) Microfluidic layout of the RespiDisk. Reprinted with permission from ref. 169. Copyright © 2020 The Royal Society of Chemistry. (B) Centrifugal microfluidic POC testing platform for CRISPR-assisted RT-RAA detection of SARS-CoV-2. Reprinted with permission from ref. 173. Copyright © 2022 American Chemical Society. (C) Automated sample-to-answer SARS-CoV-2 detection performed on a centrifugal platform with pneumatic actuation. The microfluidic cartridge is prefilled with reagents for conducting RNA extraction, followed by amplification of E and N genes using RT-LAMP. Color change is detected by the optical imaging system of the platform. Reprinted with permission from ref. 175. Copyright © 2022 The Royal Society of Chemistry. (D) Implementation of an automated ddPCR-based sample-to-answer detection process integrated on a microfluidic cartridge that is operated on a centrifugal platform with active pneumatic pumping. Components depicted in the setup: (1) external tube heater for PCR thermal cycling; (2) world-to-chip interface tubing connecting the cartridge to the PCR tube inside the tube heater; (3) pneumatic manifold embedded on the floor of the rotating stage; (4) microfluidic cartridge; (5) rotating stage with frame to hold the cartridge in place; and (6) platform casing. Scale bars: 400 μm. Reprinted with permission from ref. 176. Copyright © 2024 The Royal Society of Chemistry. | ||
Huang et al. have designed an air-insulated microfluidic chip integrated with LAMP for automated distribution of samples to 24 test cells, enabling the parallel identification of multiple pathogens related to clinical pneumonia.170 All test and buffer cells were connected to an approximate sine-type channel pipe (0.2 mm). Six primers were designed to identify one pathogen which all were embedded at the bottom of the test cell using low melting point Sepharose CL-4B. The prepared DNA sample and isothermal NA amplification reactants were evenly mixed in a 1 mL microcentrifuge tube. Then, the mixture was injected into the microfluidic chip from the inlet hole using a pipette. Sepharose CL-4B dissolves when the device heats to 50 °C, allowing all primers to be released into the mixtures of DNA sample and LAMP reagents. EvaGreen fluorescence marker is used to bind to the amplified strands while NA sequences are being amplified at 65 °C, enabling detection in real-time. The authors demonstrated that this device can simultaneously detect 24 species, which is adequate for common pneumonia pathogen identification.
Tian et al.171 developed a fully automated centrifugal microfluidic system with sample-in-answer-out capability. Their setup consists of a microfluidic disk for housing samples and reagents and a signal-sensing instrument for rapid detection of SARS-CoV-2. The viral RNA released from oropharyngeal swab samples is amplified by reverse transcription LAMP (RT-LAMP) with fluorescence signal being detected automatically. The authors found that the limit of detection (LOD) for RNA particles is 2 copies per reaction with a throughput of 21 reactions per disk and a sample-to-result time of ∼70 min. RT-LAMP has also been used by de Oliveira et al. for molecular diagnosis of COVID-19 from RNA extracted samples.172 The authors employed a centrifugal microfluidic device made from polystyrene-toner which is manually controlled by a fidget spinner. The reaction was performed using a simple heating block and amplification results were obtained with on-chip visual detection using SYBR Green I intercalating dye, aided by a hand-held UV source and a smartphone for imaging.
Chen et al. have demonstrated reverse transcriptase recombinase-aided amplification (RT-RAA) of SARS-CoV-2 RNA extracts along with detection facilitated by clustered regularly interspaced short palindromic repeats (CRISPR) using a centrifugal microfluidic platform (Fig. 3B).173 The sensitivity of the assay was increased by segregating the amplification (RT-RAA) from the detection (CRISPR) using separate compartments of the device, while minimizing time-to-result, allowing the system to detect down to 1 copy per μL within 30 min. Using RNA extracts from 26 positive and 8 negative clinical samples, the centrifugal microfluidic device achieved 100% accuracy compared to RT-PCR.
Soares et al. have integrated a sample-to-answer LAMP assay for the detection of SARS-CoV-2 on a centrifugal platform using a simple optical setup for fluorescence readout and a smartphone camera.174 The reaction was performed directly using heat-inactivated NPS samples and included agarose beads for signal enhancement after NA amplification, exhibiting a sensitivity of 10–100 RNA copies per μL. We have developed an integrated SARS-CoV-2 sample-to-answer molecular diagnostic assay that combines RNA extraction from NPS specimens with RT-LAMP (Fig. 3C).175 The fully automated assay was performed on a centrifugal microfluidic platform with active pneumatic pumping using a disposable, polymer-based microfluidic cartridge and lyophilized reagents. The LOD of the sample-to-answer assay was 0.5 copies per μL, with 60 min time-to-result, showing promise for the practical implementation of rapid and sensitive COVID-19 diagnostics at the point-of-need. Using a cohort of 12 clinical samples, we demonstrated the capacity to distinguish positive and negative samples for SARS-CoV-2, thereby obtaining 100% agreement with RT-PCR results.
Using a more advanced version of this platform, we have recently shown a fully-integrated system seamlessly combining viral lysis, RNA extraction, emulsification, RT-ddPCR, and fluorescence readout in a sample-to-answer format (Fig. 3D).176 Highly monodisperse droplets (∼50 μm in diameter) were produced using centrifugal step emulsification and automatically transferred to an integrated heating module for target amplification. The platform had been equipped with a miniature fluorescence imaging system enabling on-chip readout of droplets after RT-ddPCR. Sample-to-answer detection was demonstrated for SARS-CoV-2 N and E genes, along with RNase P endogenous reference, using hydrolysis probes and multiplexed amplification within single droplets for concentrations as low as 0.1 copy per μL. We also tested 14 NPS specimens from patients and were able to distinguish positive and negative SARS-CoV-2 samples with 100% accuracy, surpassing results obtained by RT-PCR. Van Nguyen et al.177 demonstrated a prototype centrifugal microfluidic platform that automates the molecular diagnostic workflow for parallel detection of four respiratory pathogens, including IAV (H1N1, H3N2), IBV, and SARS-CoV-2, in a high-throughput manner. The microfluidic device implemented a zigzag aliquoting structure, enabling division of reagent solutions into 30 aliquots that can be processed simultaneously. The sample-to-answer reaction comprised RNA purification, RT-LAMP and the real-time fluorescence detection, and could be completed in 90 min, exhibiting an LOD of 100 copies per μL.
Lin et al.178 developed a POC microfluidic immunoassay system for simultaneously detecting IgG/IgM/Ag deriving from SARS-CoV-2 infection within 15 min. This centrifugal microfluidic platform is equipped with SARS-CoV-2 diagnostic microchips, multiple immunoassays and a home-made fluorescence analyzer. The microchip is composed of a sample loading chamber, a waste reservoir, and a fluorescence immunoassay fluid channel comprising a capture region (5 × 6 spots) and a test region (3 × 3 spots). The possibility to detect multiple biomarkers offers sensitive and accurate diagnosis of SARS-CoV-2. Liu et al. have integrated μPADs on a centrifugal microfluidic disk for detection of the SARS-CoV-2 N protein.179 The device employs a smartphone camera in concert with a colorimetric detection scheme using gold nanoparticles (NPs) to positively identify the protein in salivary samples with a sensitivity of 10 pg mL−1 and total assay time of 8 min.
6.2 Methods based on pressure-driven flow
Wang et al.180 developed a microfluidic chip platform for rapid detection of respiratory pathogens using LAMP. The platform comprises a polymer/paper microfluidic chip and a portable device for temperature control and remote visualization of the results. This chip, with rapid sample-in-product-out capability for respiratory clinical specimens, can be utilized for genomic DNA extraction, amplification, and readout. Genomic DNA of the pathogen is extracted with the help of magnetic particles. A chromatography paper disk is preloaded with LAMP primers and placed in each microchamber so that the chip can recognize specific NA fragments of S. pneumoniae and M. pneumoniae. Confirmation of these pathogens is achieved by visual assessment of green fluorescence using a portable device equipped with a UV light pen, a built-in heating module, a Wi-Fi camera, and a dedicated smartphone app interface to communicate and receive results. For quality control, these specimens were tested against PCR performed using bench-top instrumentation, yielding positive and negative predictive values for M. pneumoniae of 96.9% and 100%, respectively. Zhu et al.181 developed an LOC device integrating DNA extraction, solid-phase PCR and detection of human papillomavirus (HPV). With the microfluidic system, the investigators showed a linear correlation between the extracted DNA concentration and initial positive sample concentration, suggesting high efficacy of the device for NA extraction. They also demonstrated sufficient sensitivity for detecting five high-risk HPV genotypes with an LOD of 50 copies per reaction. It should be pointed out that all the procedures could be completed within 1 h, which is significantly shorter than the time required using commercially available methods.Zhang et al.182 conducted an exploratory study to detect and simultaneously subtype multiple influenza viruses (H1N1, H3N2, H9N2) by combining a microfluidic chip with magnetic NPs (MNPs) and quantum dots (QDs). This microfluidic chip incorporates channels for multiple detections, and is integrated with a controllable micromagnetic field to confine MNPs and a heating region modified with capture probes for recognizing the corresponding target cDNA sequences of IAV. Yeh et al. introduced a portable microfluidic platform comprising carbon nanotube arrays with differential filtration porosity designed for virus enrichment and optical virus identification.183 This platform effectively enriched RV, IAV, and parainfluenza virus, resulting in successful real-time virus identification directly from clinical samples with a virus specificity of 90%. Notably, viral detection was achieved with a 70-fold enrichment in just a few minutes.
Kim et al.184 developed an integrated microfluidic preconcentration and NA amplification system (μFPNAS) for highly sensitive detection of IAV H1N1. In this study, H1N1 virus particles were first preconcentrated using MNPs conjugated with an Ab specific to the virus. Their isolated RNA was then reversely transcribed and amplified to cDNAs through thermocycling in a trapezoidal chamber of the μFPNAS. LOD as low as 100 TCID50 (50% tissue culture infective dose) in saliva can be obtained within 2 h. Ganguli et al. demonstrated the rapid detection of SARS-CoV-2 virus from NPS transferred to viral transport medium (VTM), using a portable hand-held reader and an additively manufactured microfluidic cartridge (Fig. 4A).185 The POC microfluidic cartridge was inserted into the cradle of the portable instrument and operated by pressure-driven syringe flow. An isothermal RT-LAMP assay was performed for rapid and cost-effective detection of SARS-CoV-2, providing an LOD of 50 RNA copies per μL in VTM. A smartphone camera is used to monitor in real-time the fluorescence emission generated during amplification. Najjar et al. developed a 3D-printed microfluidic device capable of detecting SARS-CoV-2 RNA and anti-SARS-CoV-2 Abs in saliva and plasma via multiplexed electrochemical outputs based on horseradish peroxidase (HRP)-mediated conversion of 3,3′,5,5′-tetramethylbenzidine (TMB) (Fig. 4B).186 The fluid flow was controlled by a peristaltic pump, allowing for electrochemical assays, reagent and sample mixing, as well as LAMP and CRISPR reactions to be conducted sequentially according to the requirements of the analytical protocol. With their multiplexed assay, they were able to combine and perform serological and NA diagnostics simultaneously and validated multiplexed sensing with simultaneous detection of SARS-CoV-2 viral RNA and host Abs.
 | ||
| Fig. 4 Microfluidic systems operated using pressure-driven flow. (A) Disposable microfluidic device for rapid detection of SARS-CoV-2. RT-LAMP reagents and patient samples are injected using syringes connected to Luer lock inlet ports. Fluorescence images depict the result of SARS-CoV-2 analysis with patient samples including both positive and negative specimens. Reprinted from ref. 185. Distributed under Creative Commons Attribution License 4.0. (B) CRISPR-based electrochemical assay. Without viral RNA, the biotinylated ssDNA reporter probe (RP) is not cleaved; therefore, the polystreptavidin-HRP binds to the capture probe/biotin-DNA duplex when added to the sensor chip and consequently precipitates TMB, resulting in an increase in current. In contrast, the biotinylated ssDNA RP is hydrolysed in the presence of viral target RNA, cleaving the biotin group; thus, polystreptavidin-HRP does not bind to the chip surface, resulting in no TMB precipitation and no increase in current. The cyclic voltammogram shows the typical current peak signal achieved after incubation of samples from both SARS-CoV-2 negative and positive clinical specimens. Clinical samples that contained SARS-CoV-2 viral RNA had low signals and were clearly distinguishable from the high signals obtained for samples that did not contain viral RNA. Reprinted with permission from ref. 186. Copyright © 2022 Springer Nature. (C) Design of gravity-driven microfluidic cartridge incorporating sample lysis and real-time PCR amplification for viral RNA/DNA detection. The histogram shows the evaluation of analytical specificity for SARS-CoV-2 and influenza A/B in NA testing. Reprinted with permission from ref. 187. Copyright © 2022 The Royal Society of Chemistry. (D) Schematic illustration of the Evalution™ technology, which includes digitally barcoded microparticles, a microfluidic cartridge and an integrated instrument for fluid manipulation and readout. Scale bar: 20 μm. Reprinted with permission from ref. 188. Distributed under Creative Commons Attribution License 4.0. | ||
Zai et al. described a gravity-driven microfluidic cartridge for rapid detection of SARS-CoV-2, influenza A/B, and HPV16/18 (Fig. 4C).187 The microfluidic cartridge integrates sample lysis and direct quantitative PCR amplification for the detection of viral RNA/DNA. The resulting data showed high sensitivity and specificity in the detection of SARS-CoV-2, influenza A/B viral RNA, and HPV16/18 viral DNA, indicating an all-in-one sample-to-answer POC solution. Rutten et al.188 evaluated a fully integrated, commercial microfluidic platform (Evalution™ developed by MyCartis) in conjunction with digitally barcoded microparticles to detect disease-related biomarkers (Fig. 4D). Introduced by pneumatic pressure, the encoded microparticles serve as substrates for hybridization chain reaction amplification and subsequent detection of target marker genes. The system distinguished virus subtypes (human adenovirus type B and D) and antibiotic-resistant bacteria (S. pneumoniae) and exemplified specificity. Utilizing their highly simplified and enzyme-free isothermal amplification approach, the system demonstrated an LOD of 309 ± 80 fM for human adenovirus type B in NPS samples.
Yamaguchi et al. designed a microfluidic system for the detection of L. pneumophila in aquatic environments.189 The chip contained mainly two portions, one for mixing water samples with fluorescently-labelled Ab and another for detection and enumeration of bacterial cells. In this study, a home-built portable instrument, consisting of two syringe pumps, a CCD camera and a laptop was used for image acquisition and bacterial counting. The authors found that counts of the L. pneumophila cells in the standard samples measured with their microfluidic system were comparable to those determined with conventional fluorescence microscopy. Furthermore, they showed that L. pneumophila counts in cooling-tower water detected with the two methods were also comparable, showing a similar trend following disinfection and recirculation of the cooling tower. This study demonstrates that microfluidic techniques can be an effective approach for pathogen identification in disease management, environmental monitoring and public health surveillance. Singh et al. reported a microfluidic chip integrated with a reduced graphene oxide (RGO)-based electrochemical immunosensor for label-free detection of IAV H1N1.190 In this study, three microelectrodes were fabricated on a glass substrate using photolithography, and the working electrode was functionalized using RGO and monoclonal Abs specific to the virus. These chips were integrated with polydimethylsiloxane (PDMS) microchannels. Electrochemical studies revealed good selectivity and an enhanced detection limit of 0.5 plaque-forming units (pfu) per mL, where the chronoamperometric current increased linearly with H1N1 virus concentration within the range of 1 to 104 pfu mL−1. Yang et al. presented the generation of monoclonal Abs specific to the SARS-CoV-2 N protein and selected an optimal Ab pair for detecting the viral pathogen.191 Particularly, the produced Abs 1G6 and 3E10 exhibited higher reactivity and were shown to be superior candidates than others used for a sandwich ELISA platform to identify SARS-CoV-2. Using a single syringe pump-driven POC microfluidic platform, the authors showcased the detection of SARS-CoV-2 N protein and its variants as a protein-based diagnostic tool.
6.3 DMF platforms
Wulff-Burchfield et al.192 evaluated a microfluidic real-time PCR device (developed by Advanced Liquid Logic) for detection of M. pneumoniae that is rapid, portable, and fully automated. The Advanced Liquid Logic technology involves electrical fields to rapidly and precisely manipulate discrete nanoliter-sized droplets within an oil-filled chamber. Each droplet manipulation is directly controlled within a software program enabling complex, multi-step protocols to be implemented and easily reconfigured without requiring modifications to the chip design. In their study, the authors enrolled patients with CAP and extracted DNA from NPW specimens using a biotinylated capture probe and streptavidin-coupled magnetic beads. Each extract was tested for M. pneumoniae-specific DNA by real-time PCR in comparison to conventional PCR using a bench-top thermocycling instrument. Three out of 59 NPWs were positive, and agreement between the methods was 98%. Their data suggests that microfluidic real-time PCR and conventional PCR displayed comparable sensitivity and specificity. However, the former was easier to perform and at least three times faster, emphasizing the potential applicability of the microfluidic system to POC use for pneumonia patients. Prakash et al.193 have used DMF technology, integrated with suitably tailored resistive microheaters and temperature sensors, to achieve chip-based real-time PCR. The authors leveraged two actuation techniques, electrostatic/droplet dielectrophoresis and electrowetting, through tailored microelectrode architectures to facilitate the required on-chip droplet manipulation steps. The integrated device was used to analyze in vitro synthesized IAV and ICV RNA. The study demonstrates the utility of DMF for real-time PCR as it allows viral RNA to be quantified with a detection threshold of <5 copies. The capabilities of the device were further confirmed through detection and quantification of pathogenic samples comprised of IAV and ICV, where accuracy levels were found to be comparable to those obtained by bench-top PCR (efficiency ∼95%).Wang et al. have combined DMF with surface-enhanced Raman scattering (SERS) for the quantitative detection of avian IAV H5N1 in buffer and human serum (Fig. 5A).194 The SERS-based methodology shows excellent sensitivity (LOD of 74 pg mL−1) and selectivity for H5N1 detection with a relatively short assay time (<1 h) and low reagent consumption (∼30 μL) compared to standard ELISA. Therefore, the technology holds great potential for automated and sensitive diagnosis of a variety of infectious diseases.
 | ||
| Fig. 5 Respiratory disease diagnosis using DMF. (A) Schematic illustration of a SERS-based immunomagnetic assay. The droplet is sandwiched between top and bottom plates of the device. Quantitative analysis can be performed by measuring the Raman intensity of the SERS tags (e.g., at 1071 cm−1), which is determined by the target concentration. Reprinted with permission from ref. 194. Copyright © 2018 American Chemical Society. (B) Illustration of a DMF device that uses distance-based detection of LAMP amplicons. The assay starts with extracted viral RNA being loaded into the device along with the LAMP master mix. Following isothermal amplification, LAMP amplicons are cleaned up using Capto™ adhere beads and introduced into a distance-based detection substrate after mixing with SYBR® Safe intercalating dye. The fluorescence signal allows for semi-quantitative measurement of viral load in saliva samples. Reprinted with permission from ref. 197. Copyright © 2024 The Royal Society of Chemistry. (C) Design of the Virus Hunter microfluidic device that implements sample-to-answer detection of SARS-CoV-2 as well as influenza A/B using an RT-PCR assay. The device comprises microchannels for sample preparation and NA extraction and a DMF module for amplification. Amplification is performed using a standard TaqMan assay with fluorescence signal detected by the instrument. Prestored reagents allow for a sample-in-to-answer-out operation without manual intervention. Reprinted with permission from ref. 198. Copyright © 2024 The Royal Society of Chemistry. | ||
Zhang et al.195 have implemented a two-step CRISPR-assisted LAMP assay on a DMF platform comprising two inlets, two temperature control zones, and a fluorescence detection area. The reaction droplets were immersed in silicone oil to minimize formation of aerosols and prevent contamination between amplification and detection reactions. The automated assay was able to detect both wild and mutant SARS-CoV-2 and simultaneously monitor the T478K, D614G, P681H, and P681R mutations in samples comprising 100 copies per μL. Anderson et al.196 demonstrated successful implementation of a SARS-CoV-2 sample-to-answer assay on a thin-film transistor DMF (TFT-DMF) platform comprising a disposable microfluidic cartridge and control hardware with an embedded fluorescence detection system. The TFT-DMF device incorporates tenths of thousands of independently addressable electrodes with the capacity to perform multiple parallel reactions and implement complex analytical workflows by manipulating large numbers of droplets along any pathway. Using saliva samples spiked with SARS-CoV-2 RNA, the assay steps including magnetic bead-based NA extraction and RT-PCR were performed by the system with minimal user intervention.
Ho et al.197 have combined DMF with distance-based detection of LAMP products as a direct signal readout strategy to detect amplified SARS-CoV-2 RNA (Fig. 5B). Here, semi-quantitative detection of target analyte is reported based on the travelled length of a visual signal on a patterned paper reporter, such that identification can be performed by the naked eye. Using RNA extracts from saliva spiked with inactivated SARS-CoV-2 viral particles, the authors have demonstrated successful discrimination between low (10 copies per μL), and high viral load (105 copies per μL) as well as negative control samples. Future integration of sample preparation steps with the proposed approach could provide utility in portable applications outside of standard laboratory environments. Dong et al.198 have demonstrated a cost-effective sample-to-answer molecular analysis system termed Virus Hunter, which combines DMF technology with a continuous-flow microchannel-based subsystem for detection of SARS-CoV-2 and influenza A/B using RT-PCR (Fig. 5C). Using such a combinatorial approach, the authors demonstrated the capacity of the system to store reagents on the chip and process hundreds of microliters of sample volume to perform all assay steps in an automated manner, including bead-based NA extraction, purification, and amplification. The continuous-flow subsystem performed sample preparation steps before delivering purified RNA to the digital unit. Using EWOD, the sample was partitioned to eight independent droplets for RT-PCR, which were subsequently imaged using a 4-channel fluorescence reader. The sample-to-answer assay could be completed in 2 h exhibiting an LOD of 0.30 and 0.94 copies per μL for SARS-CoV-2 ORF1 and N genes, respectively, as well as 1.03 and 1.26 copies per μL for IAV and IBV, respectively. The platform was evaluated using clinical specimens, providing 100% agreement with standard (bench-top) RT-PCR.
6.4 Capillary-based microfluidic systems
Ramalingam et al.199 made use of isothermal helicase-dependent amplification (HDA) for viral detection using an integrated microfluidic (PDMS/glass) chip. Their system harbors open (unsealed) reactors in conjunction with a single-step capillary-based flow scheme for sequential distribution of the PCR mixture. The performance of HDA using their chip was evaluated in comparison to PCR on a PDMS/glass chip and HDA in a tube, and was found to be comparable in terms of NA amplification yield. The combination of autonomous, capillary-based liquid displacement and the specificity of HDA makes the system appealing for use in resource-limited settings, where expensive instruments are not available. Ma et al. have developed a capillary-driven microfluidic device for rapid detection of IAV H1N1 using RT-LAMP (Fig. 6A).200 The liquid flow is mediated via capillary forces and stopped using hydrophobic soft valves. Specifically, liquid flow from the hydrophobic area into the downstream chamber is initiated by pressing the hydrophobic soft valves. The device is fabricated in PDMS and integrates virus isolation via H1N1-specific aptamers conjugated to magnetic beads, lysis, isothermal NA amplification and colorimetric detection of the viral RNA. The entire procedure can be performed in 40 min by capillary forces and provides an LOD of 3 × 10−4 hemagglutinating units per reaction, which is sensitive enough for clinical applications. A custom-made smartphone-controlled, automated, and portable system was also developed which could serve as a platform for POC pathogen detection, particularly in resource-limited settings. Dou et al.201,202 have demonstrated a low-cost paper/polymer hybrid microfluidic POC device for rapid and accurate diagnosis of pertussis—a highly infectious respiratory disease caused by the bacterium Bordetella pertussis.203 Capillary flow thereby mediates the supply of sample and reagents for on-chip LAMP reactions in designated wells. Upon completion of the amplification process, results were visualized using a portable UV light pen illuminating the LAMP reaction products on the biochip. The generated fluorescence was observed by the naked eye or captured through a cellular phone camera where images were then processed using image analysis software. The system allows for detecting B. pertussis from NPS and nasal aspirate with high sensitivity and an LOD of 5 DNA copies per well. Sun et al. developed a microfluidic chip for LAMP-based detection of five bacterial and viral pathogens using a smartphone-based instrument for readout (Fig. 6B).204 The authors found that their system can reliably detect the specific target NA sequences without signal crosstalk. Their work represents the successful utilization of smartphone-based LAMP detection of pathogens for POC application. They also claimed that the portable microfluidic smartphone-based device can be used for detection of human pathogens such as SARS-CoV-2.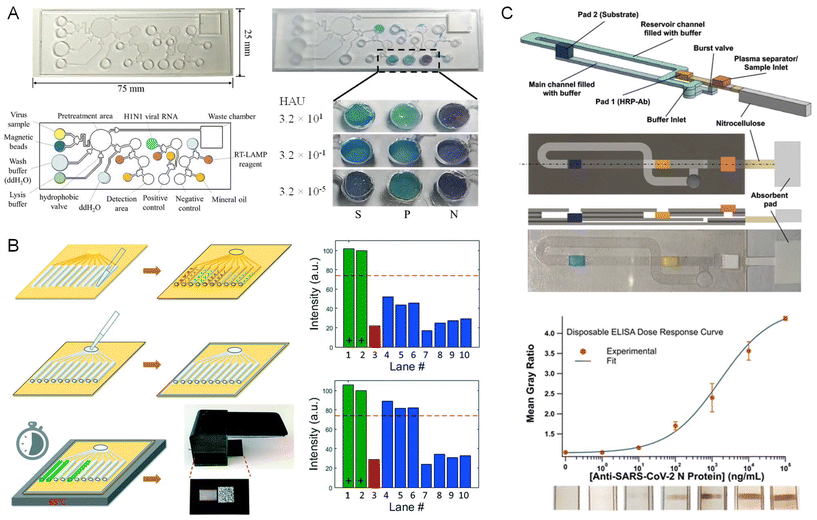 | ||
| Fig. 6 Detection of respiratory pathogens using capillary systems. (A) Multilayer microfluidic device for rapid detection of IAV H1N1 using RT-LAMP. Self-driven microfluidic capillary flow, a hydrophilic film and a vibration-generating motor enable the mixing and colorimetric detection of reagents and target samples. The images depict the LOD for IAV. S = test sample; P = positive control; and N = negative control. Reprinted with permission from ref. 200. Copyright © 2019 Elsevier. (B) LAMP assay on a microfluidic chip for fluorescence-based detection of bacterial and viral pathogens. Primers and LAMP reaction mixture are introduced using capillary flow. The microfluidic device is heated at 65 °C and then inserted into the fluorescence imaging module. Results are typically obtained after 30 min. Lanes in the intensity plots represent different samples with negative (upper panel) and some positive test results (lower panel). Reprinted with permission from ref. 204. Copyright © 2020 The Royal Society of Chemistry. (C) Schematics and photograph of a capillary-driven microfluidic device for Ab-based detection of SARS-CoV-2. The graph shows the dose–response curve of anti-N protein spiked into whole human blood obtained using microfluidic ELISA. Reprinted with permission from ref. 206. Copyright © 2023 The Royal Society of Chemistry. | ||
Clark et al.205 presented an electrochemical capillary-driven immunoassay made from polyethylene terephthalate and adhesive films for the detection of SARS-CoV-2. Upon sample loading, reagents and wash solutions are sequentially delivered to an integrated sensing zone where the carbon electrode pads are positioned. Subsequently, automated ELISA was conducted for detecting SARS-CoV-2 N protein using chronoamperometry measurements. The authors reported an estimated LOD of 68 equivalent pfu per mL. Building on these developments, they conceived a capillary-driven microfluidic device for serological testing at the point-of-need using enzymatic conversion of a chromogenic substrate (Fig. 6C).206 This device can quantify anti-SARS-CoV-2 N protein with an LOD of 2.8 ng mL−1, which is comparable to that of traditional well plate ELISA (1.2 ng mL−1). Furthermore, they quantify clinically relevant levels of SARS-CoV-2 N protein from NS in 20 min using smartphone images, achieving an LOD of 100 pfu mL−1.207 This capillary-driven approach also been demonstrated in conjunction with multiplexed immunoassays for the detection of H1N1 hemagglutinin and SARS-CoV-2 N protein.208
Robin et al. have developed a microfluidic DNA biosensor for the detection of non-amplified SARS-CoV-2 RNA.209 A specifically designed SARS-CoV-2 probe was covalently bound to the surface of a glass slide which was then integrated into the microfluidic system where it was hybridized with viral RNA extracted from human saliva. The DNA/RNA duplex was detected in the presence of SYBR Green I using an opto-electronic system, based on a high-power LED and a photo multiplier tube, converting the emitted fluorescence into an electrical signal that can be processed in less than 10 min. The LOD of the sensor reached 6 viral RNA copies per μL (corresponding to 10 aM). The work is distinguished from other approaches by the fact that it makes use of a relatively simple detection scheme without the need for intermediate transcription and amplification steps.
6.5 LF and paper-based devices
Wu et al.210 developed an automated, portable, and integrated paper-based microfluidic system for influenza A detection with a smartphone platform. The paper-based microfluidic chip consists of a reagent storage and a reaction module. The storage module, which comprises reagent chambers with dispensation channels, is designed for reagent storage and release. The reaction module consists of an absorbent pad and a nitrocellulose membrane which is functionalized with specific monoclonal Abs for immunoassay detection. Microfluidic Dot-ELISA was performed when the dispensed reagents flow through the membrane at a controllable speed and reach the absorbent pad. The preliminary results indicated that their device was more convenient and efficient in influenza A detection than traditional methods. Murdock et al.211 developed a POC μPAD for detecting influenza and determine antiviral susceptibility of the strain. The assay exploits the enzymatic activity of surface proteins present on all influenza strains, and potential false positive responses can be mitigated. A sample can be added to the device, distributed to 4 different reagent zones, and development of the enzymatic substrate under different buffer conditions takes place on the bottom of the device. Analysis can be performed by eye or through colorimetric image analysis. Garneret et al. developed a portable, paper-based microfluidic device for diagnosis of COVID-19.212 SARS-CoV-2 RNA is extracted, amplified isothermally by RT-LAMP, and detected using intercalating dyes or fluorescent probes. Depending on the viral load in samples, the detection takes between 20 min and 1 h. Using a set of 16 pools of NPS eluates, the authors found that LOD of their device was comparable to RT-PCR (i.e., 1 genome copy per μL of clinical sample) without cross-reaction among the eight major respiratory viruses currently circulating in Europe. Therefore, they proclaimed that they designed and fabricated an easy-to-use portable device (termed COVIDISC) to carry out the test at the point of need.Liu et al. described an LF recombinase polymerase amplification assay for rapid and sensitive detection of SARS-CoV-2 implemented into a single microfluidic chip.213 Analyte sequences are labelled with biotin and FAM during amplification, mixed with running buffer, and then delivered to the LF strip to provide easily interpreted positive or negative results. Testing requires only a simple NA extraction process along with subsequent loading and incubation to obtain results in approximately 30 min. SARS-CoV-2 armored RNA particles were used to validate the system, which showed an LOD of 1 copy per μL (or 30 copies per sample). Chip performance was further evaluated using clinically diagnosed cases of COVID-19 and revealed a sensitivity of 97% and specificity of 100%, highly comparable to current RT-PCR-based diagnostic assays. This assay is portable and comprises affordable materials, enabling mass production and decreased risk of contamination. The authors suggest that their assay can be used as a complement to RT-PCR for low-cost COVID-19 screening in resource-limited areas. Zhang et al. reported a one-pot, LF-based RT-LAMP assay for SARS-CoV-2 detection in clinical samples.214 In their study, they employed RT-LAMP to amplify virus RNA efficiently and rapidly in the sample without the need for specialized equipment. In addition, they used an LF assay that allows for interpretation of NA amplification results by direct visualization. When these technologies are integrated into one system, the analysis of a clinical sample can be achieved in less than 40 min from a swab sample to a diagnostic result. This sample-to-answer workflow exhibits the advantages of being simple, less costly, and portable for diagnosis. The authors claim that this technology is one step closer to achieve on-site rapid screening of pathogenic microorganisms, which is critical for infectious disease control and prevention.
Wang et al. developed a dual-mode LFIA that uses S protein-conjugated QDs for rapid detection of SARS-CoV-2-specific IgM and IgG in human serum.215 Colorimetric testing mode thereby allows for rapid screening of the patients with suspected SARS-CoV-2 infection without the need for specialized equipment while fluorescence testing mode simultaneously enables sensitive and quantitative analysis of IgM/IgG in the sample. According to the authors, the assay is 100 times more sensitive than a standard LFIA based on colloidal Au particles. Chen et al. have shown a rapid and sensitive LFIA that uses lanthanide-doped polystyrene NPs (LNPs) to detect anti-SARS-CoV-2 IgG in human serum.216 In their device, recombinant N protein of SARS-CoV-2 was dispensed onto a nitrocellulose membrane to capture specific IgG. Mouse anti-human IgG Ab labeled with self-assembled LNPs served as a fluorescent reporter. The authors suggest that this assay can achieve rapid (10 min) and sensitive detection of anti-SARS-CoV-2 IgG (in 100 μL aliquots of diluted human serum), and enables positive identification in suspicious cases; it can also be useful for monitoring the COVID-19 disease progression and evaluating patients’ response to treatment. Wang et al. developed an amplification-free NA immunoassay for fluorescence-based detection of SARS-CoV-2 RNA in less than 1 h.217 The assay uses monoclonal Ab-labelled europium-chelate-based fluorescent NPs to capture hybridized double strands formed by specially designed DNA probes and SARS-CoV-2 RNA on an LF strip (Fig. 7A). In a multi-hospital randomized double-blind trial involving 734 samples (593 TS and 141 sputum) provided by 670 individuals, the assay achieved 100% sensitivity and 99% specificity for both sample types.
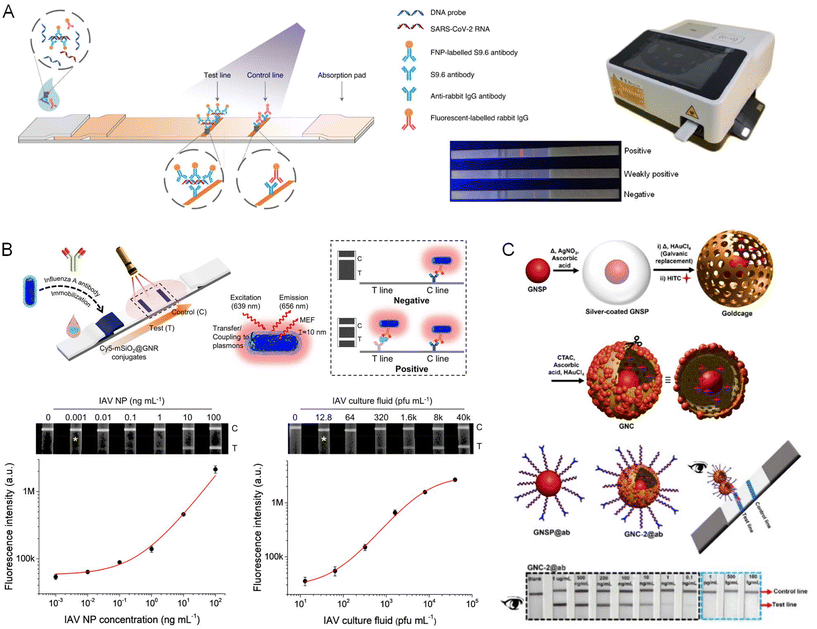 | ||
Fig. 7 Examples of LFIA implementations. (A) Principle of fluorescence-based SARS-CoV-2 detection along with representative results on an LF strip as detected by the fluorescence analysis device. Reprinted with permission from ref. 217. Copyright © 2020 Springer Nature. (B) Schematic representation of the sandwich immunoassay for detecting IAV using plasmonic fluorescence enhancement. Fluorescence images show the detection of serially diluted IAV N protein and cultured IAV H1N1. The respective plots reveal test line intensities for Ag concentrations of 0.001–100 ng mL−1 as well as culture fluid concentrations of 12.8–40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 pfu mL−1. Reprinted with permission from ref. 225. Copyright © 2023 American Chemical Society. (C) Schematic illustration of the synthesis of GNC used for colorimetric and SERS-based detection of SARS-CoV-2 S1 protein along with images of LFIA strips exposed to different analyte concentrations. Reprinted with permission from ref. 227. Copyright © 2024 American Chemical Society. 000 pfu mL−1. Reprinted with permission from ref. 225. Copyright © 2023 American Chemical Society. (C) Schematic illustration of the synthesis of GNC used for colorimetric and SERS-based detection of SARS-CoV-2 S1 protein along with images of LFIA strips exposed to different analyte concentrations. Reprinted with permission from ref. 227. Copyright © 2024 American Chemical Society. | ||
CRISPR in conjunction with CRISPR-associated (Cas) proteins constitutes a revolutionary gene-editing technology218,219 that is being integrated into LF devices for microbial and disease POC detection.220 For instance, Broughton et al.221 have demonstrated feasibility and validity of a CRISPR–Cas12-based LF device for detection of SARS-CoV-2 from extracted patient sample RNA. The assay implements standard tube-based RT-LAMP for RNA extracted from swab samples, followed by Cas12 detection of predefined coronavirus sequences, after which cleavage of a reporter molecule confirms detection of the virus. Similarly, Joung et al.222 have combined isothermal amplification and CRISPR-mediated detection for the diagnosis of SARS-CoV-2 infections. The methodology provides an LOD of 100 copies of viral genome input in saliva or NPS per reaction. Using LF readout, the entire process can be completed within 70 min, making this approach suitable for POC applications. Zhu et al. have described a CRISPR–Cas9 assay for the detection of M. pneumoniae.223 High amplification efficiency along with low temperatures for recombinase polymerase amplification enables detection of three copies at a preamplification temperature of 25 °C. The practical application of their device has been demonstrated with 100% accuracy by testing with 43 M. pneumoniae-infected specimens and 80 uninfected specimens. The entire detection process, including pretreatment, preamplification, CRISPR–Cas9 recognition, and visual analysis, can be completed in 30 min.
Plasmonic approaches in LF devices have emerged as a promising technique for disease detection due to their high sensitivity, simplicity, and rapid response time. These approaches harness the unique properties of plasmonic NPs, which, when conjugated with capture probes, enable the detection of target analytes at very low concentrations by amplifying the signal generated upon analyte binding. Gupta et al. have shown an LFIA based on Ab-conjugated fluorescent gold nanorods that provides a 30-fold improvement in analytical performance (LOD and dynamic range) over standard ELISA and a 1000-fold improvement over conventional LFIAs using an inexpensive and portable fluorescence scanner for readout.224 Their plasmonic assay achieved 95% clinical sensitivity and 100% specificity for Ab in plasma and Ag in NPS from individuals with SARS-CoV-2 infection. Hong et al. have designed metal-enhanced fluorescence (MEF) probes based on a gold nanorod core, mesoporous silica shell, and Cy5 fluorophore (Fig. 7B).225 The authors investigated both theoretically and experimentally the distance dependence of plasmonic coupling between Cy5 and gold nanorods by adjusting the shell thickness to optimize the efficiency of MEF probes incorporated on an LFIA platform. Their assay enabled highly sensitive detection of IAV N protein with an LOD of 0.52 pg mL−1 within 20 min and showed high specificity and accuracy for determining IAV in clinical samples.
Liu et al.226 have demonstrated SERS for the simultaneous detection of IAV H1N1, SARS-CoV-2 and RSV in biological samples using an LFIA and high-performance magnetic SERS tags. The SERS tags can directly enrich and capture target viruses without pretreatment of samples, avoiding the interference of impurities in the samples as well as improving the sensitivity. The LOD of the SERS-based LFIA reached 85 copies per mL for H1N1, and 8 pg mL−1 for both SARS-CoV-2 and RSV. Atta et al.227 have integrated particles with a specially designed core–gap–shell morphology consisting of a gold shell decorated with external nanospheres, a structure referred to as gold nanocrown (GNC), labeled with a Raman reporter molecule 1,3,3,1′,3′,3′-hexamethyl-2,2′-indotricarbocyanine iodide to produce colorimetric and SERS signals, into an LFIA for the detection of SARS-CoV-2 S protein 1 (Fig. 7C). LOD for the colorimetric LFIA was determined to be 91.24 pg mL−1, while that of the SERS LFIA method was more than three orders of magnitude lower at 57.21 fg mL−1. The ultrahigh detection sensitivity of the GNC nanosensor along with excellent sensitivity, reproducibility, and rapid detection demonstrates the potential of plasmonic approaches for the early detection of respiratory virus infections.
7 Conclusions and outlook
In this review, we examined emerging concepts for the detection of ARTIs—an area where microfluidics has long been expected to shine. The developments presented here suggest that the technology—in large part—makes good on its promises and continues to take center stage when novel or well-established analytical methods are to be integrated into a compact, miniature format. Microfluidic devices can perform multiplexed assays, simultaneously testing for several target pathogens in a single sample, thereby expediting the diagnostic process. A number of examples highlight the possibility to enable rapid analysis of biological samples, such as blood, urine or saliva, allowing for diagnosis and treatment decisions to be made on-the-spot.For any microfluidic system to approach practical relevance, it must adhere to the ASSURED (affordable, sensitive, specific, user-friendly, rapid and robust, equipment-free and deliverable to end-users) criteria set forth by the WHO, which informs the development of novel diagnostic tests.228 Many of the emerging devices presented herein meet these conditions only partially. Some of the technical challenges that have hampered the transition of microfluidic systems into the clinical realm from the onset75,229 are still prevalent today. For example, sample preparation as well as signal detection and readout often depend on auxiliary equipment that is inadequate for deployment in resource-poor settings. Adsorption of assay components on device surfaces, degradation of sensitive reagents over time and fouling can drastically diminish reliability of the measurements and, ultimately, can cause the device to fail. Appropriate encapsulation of liquids, the use of dry reagents as well as adequate passivation of the inner surfaces are needed to provide acceptable delays in terms of shelf life and device performance. Also, many published demonstrations remain at the proof-of-concept level or were performed with spiked solutions instead of clinical samples; blood, saliva and urine are complex matrices that can be challenging to process and analyze. Ongoing research is continuously improving microfluidic technology by addressing these limitations. Commercial systems such as the Revogene® instrument59,165 and others highlighted in this review are testimony to these efforts. A striking example is also the Simple Plex™ system,230,231 an automated, microfluidic-based immunoassay platform developed by ProteinSimple, which provides significant improvement over traditional approaches in terms of sample requirements, test sensitivity, dynamic range, coefficient of variation, and reproducibility.
Clinical trials required to validate the practical utility of biomarkers, assays and devices have to comply with regulatory, legal, and ethical guidelines, and can pose a considerable obstacle in the development process of a novel diagnostic test.232 Regulatory approval is another administrative challenge facing the adoption of microfluidic-based diagnostics.232 Regulatory agencies therefore need to establish clear guidelines for the validation process of microfluidic devices, ensuring their safety, efficacy, and reliability. Additionally, reimbursement policies must be established to incentivize healthcare providers to adopt microfluidic technologies, particularly in resource-limited settings. Addressing these administrative hurdles requires close collaboration between regulatory agencies, healthcare providers, and industry stakeholders to streamline the approval process and facilitate market access for microfluidic-based diagnostic devices. According to some observers, a certain degree of reluctance persists within the medical community when it comes to the adoption of new diagnostic methods, which is an additional impediment for microfluidic systems to make it into the big picture.233,234 Established habits inherited through professional training and practice can indeed guide decision-making both at the individual and collective level.
To gain widespread acceptance by healthcare professionals, reliability and testing performance have to coincide with significant improvements in the way these novel systems are operated and maintained. They must also demonstrate substantial benefits over existing instrumentation in terms of data analysis, reporting and management. The integration of microfluidic devices with other technologies, such as biosensors and smartphone apps, further enhances their diagnostic capabilities. Biosensors can detect specific biological markers associated with ARTI, while smartphone apps can interpret the sensor data, providing real-time results and even guiding patients through the testing process. Analysis software that makes use of artificial intelligence (AI) can thereby help digest complex data output, identify patterns, and make accurate diagnostic predictions in real-time.235 Machine learning algorithms can leverage large data sets to improve the accuracy and reliability of disease diagnostics, enabling early detection of outbreaks and personalized treatment strategies.235,236 Furthermore, AI-driven decision support systems can assist healthcare providers in interpreting diagnostic results and guiding patient management, ultimately improving clinical outcomes and reducing healthcare costs. Adaptation of blockchain technology, on the other hand, can enhance data security, integrity, and traceability, enabling transparent and tamper-proof record-keeping throughout the diagnostic process.237,238 By securely storing patient information and test results on a decentralized ledger, blockchain technology can facilitate interoperability between healthcare providers and ensure patient privacy and confidentiality.
Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Conflicts of interest
The authors have declared that no competing interests exist.Acknowledgements
This article is dedicated to Prof. Michel G. Bergeron in retroactive recognition of his 80th birthday and on the occasion of the 50th anniversary of the research center he founded, le Centre de recherche en infectiologie (CRI) de l'Université Laval. We thank our colleagues Javier A. Hernández-Castro and Daniel Brassard for useful discussion, and Maurice Boissinot (CRI) for insightful feedback on the manuscript. We are grateful to Amaya Arcelus for her support.References
- C. Troeger, B. F. Blacker and I. A. Khalil, et al. , Lancet Infect. Dis., 2018, 18, 1191–1210 CrossRef PubMed.
- K. Mulholland, Pediatr. Pulmonol., 2003, 36, 469–474 CrossRef PubMed.
- B. G. Williams, E. Gouws and C. Boschi-Pinto, et al. , Lancet Infect. Dis., 2002, 2, 25–32 CrossRef PubMed.
- COVID-19 Coronavirus Pandemic, Worldometer, https://www.worldometers.info/coronavirus/ (accessed February 11, 2024).
- Economic Impact of the COVID-19 Pandemic, Wikipedia, https://en.wikipedia.org/wiki/Economic_impact_of_the_COVID-19_pandemic (accessed February 14, 2024).
- A. Zumla, J. A. Al-Tawfiq and V. I. Enne, et al. , Lancet Infect. Dis., 2014, 14, 1123–1135 CrossRef PubMed.
- C. Prendergast and J. Papenburg, Future Microbiol., 2013, 8, 435–444 CrossRef.
- L. Nshimyumukiza, X. Douville and D. Fournier, et al. , Influenza Other Respir. Viruses, 2016, 10, 113–121 CrossRef PubMed.
- Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report: 2022, World Health Organization, Geneva, Switzerland, https://www.who.int/publications/i/item/9789240062702 (accessed February 17, 2024).
- E. A. Tarim, B. Karakuzu and C. Oksuz, et al. , Emergent Mater., 2021, 4, 143–168 CrossRef PubMed.
- C. Wang, M. Liu and Z. Wang, et al. , Nano Today, 2021, 37, 101092 CrossRef.
- J. Shi, Y. Zhang and M. Yang, Biomicrofluidics, 2023, 17, 024104 CrossRef PubMed.
- K. Z. Mousaabadi, Z. T. Vandishi and M. Kermani, et al. , Trends Anal. Chem., 2023, 169, 117361 CrossRef.
- Z. Lin, Z. Zou and Z. Pu, et al. , Acta Pharm. Sin. B, 2023, 13, 2877–2896 CrossRef CAS PubMed.
- T. Lehnert and M. A. M. Gijs, Lab Chip, 2024, 24, 1441–1493 RSC.
- N. K. Brar and M. S. Niederman, Ther. Adv. Respir. Dis., 2011, 5, 61–78 CrossRef CAS PubMed.
- O. Ruuskanen, E. Lahti, L. C. Jennings and D. R. Murdoch, Lancet, 2011, 377, 1264–1275 CrossRef.
- T. van der Poll and S. M. Opal, Lancet, 2009, 374, 1543–1556 CrossRef CAS.
- G. Mackenzie, Pneumonia, 2016, 8, 14 CrossRef PubMed.
- J. I. F. Salluh, V. C. Souza-Dantas and P. Póvoa, Curr. Opin. Crit. Care, 2017, 23, 391–397 CrossRef PubMed.
- D. M. Musher and A. R. Thorner, N. Engl. J. Med., 2014, 371, 1619–1628 CrossRef PubMed.
- C. M. Luna, A. Famiglietti and R. Absi, et al. , Chest, 2000, 118, 1344–1354 CrossRef PubMed.
- M. Korppi, T. Heiskanen-Kosma and M. Kleemola, Respirology, 2004, 9, 109–114 CrossRef.
- C. Paules and K. Subbarao, Lancet, 2017, 390, 697–708 CrossRef.
- J. E. Bennett, R. Dolin and M. J. Blaser, Mandell, Douglas, and Bennett's Principles and Practices of Infectious Diseases, Elsevier/Saunders, Philadelphia, PA, 8th edn, 2015 Search PubMed.
- D. M. Morens and A. S. Fauci, J. Infect. Dis., 2007, 195, 1018–1028 CrossRef PubMed.
- A. Aassve, G. Alfani, F. Gandolfi and M. Le Moglie, Health Econ., 2021, 30, 840–857 CrossRef.
- A. R. Fehr and S. Perlman, Coronaviruses: An Overview of Their Replication and Pathogenesis, in Coronaviruses. Methods in Molecular Biology, ed. H. J. Maier, E. Bickerton and P. Britton, Humana Press, New York, NY, 2015, vol. 1282, pp. 1–23 Search PubMed.
- B. J. Bosch, R. van der Zee and C. A. M. de Haan, et al. , J. Virol., 2003, 77, 8801–8811 CrossRef.
- D. Schoeman and B. C. Fielding, Virol. J., 2019, 16, 69 CrossRef PubMed.
- Y. Guo, C. Korteweg, M. A. McNutt and J. Gu, Virus Res., 2008, 133, 4–12 CrossRef.
- J. W. LeDuc and M. A. Barry, Emerg. Infect. Dis., 2004, 10, e26 CrossRef.
- K. Stadler, V. Masignani and M. Eickmann, et al. , Nat. Rev. Microbiol., 2003, 1, 209–218 CrossRef.
- Y. Ding, L. He and Q. Zhang, et al. , J. Pathol., 2004, 203, 622–630 CrossRef CAS PubMed.
- B. Hu, H. Guo, P. Zhou and Z.-L. Shi, Nat. Rev. Microbiol., 2021, 19, 141–154 CrossRef CAS.
- D. Wang, B. Hu and C. Hu, et al. , J. Am. Med. Assoc., 2020, 323, 1061–1069 CrossRef CAS PubMed.
- N. Petrosillo, G. Viceconte and O. Ergonul, et al. , Clin. Microbiol. Infect., 2020, 26, 729–734 CrossRef CAS PubMed.
- Y. Wan, J. Shang and R. Graham, et al. , J. Virol., 2020, 94, e00127-20 CrossRef PubMed.
- A. Billah, M. Miah and N. Khan, PLoS One, 2020, 15, e0242128 CrossRef.
- E. Petersen, M. Koopmans and U. Go, et al. , Lancet Infect. Dis., 2020, 20, e238–e244 CrossRef.
- A. J. Kucharski and C. L. Althaus, Eurosurveillance, 2015, 20, 21167 CrossRef.
- Q. Li, X. Guan and P. Wu, et al. , N. Engl. J. Med., 2020, 382, 1199–1207 CrossRef PubMed.
- D. E. Dwyer, D. W. Smith, M. G. Catton and I. G. Barr, Med. J. Aust., 2006, 185, S48–S53 Search PubMed.
- A. H. L. Bruning, M. M. G. Leeflang and J. M. B. W. Vos, et al. , Clin. Infect. Dis., 2017, 65, 1026–1032 CrossRef.
- T. Lynch, L. Bialy and J. D. Kellner, et al. , PLoS One, 2010, 5, e11989 CrossRef.
- E. Prina, O. T. Ranzani and A. Torres, Lancet, 2015, 386, 1097–1108 CrossRef.
- J. J. Petrozzino, C. Smith and M. J. Atkinson, J. Emerg. Med., 2010, 39, 476–490 CrossRef.
- D. R. Peaper and M. L. Landry, Clin. Lab. Med., 2014, 34, 365–385 CrossRef.
- F. Chua, D. Armstrong-James and S. R. Desai, et al. , Lancet Respir. Med., 2020, 8, 438–440 CrossRef PubMed.
- A. M. Caliendo, D. N. Gilbert and C. C. Ginocchio, et al. , Clin. Infect. Dis., 2013, 57, S139–S170 CrossRef PubMed.
- A. Hosseini, R. Pandey and E. Osman, et al. , ACS Sens., 2020, 5, 3328–3345 CrossRef.
- K. E. Hanson, M. M. Azar and R. Banerjee, et al. , Clin. Infect. Dis., 2020, 71, 2744–2751 CrossRef.
- M. Ieven, J. Clin. Virol., 2007, 40, 259–276 CrossRef PubMed.
- S. J. Courtney, Z. R. Stromberg and J. Z. Kubicek-Sutherland, Biosensors, 2021, 11, 47 CrossRef PubMed.
- Nucleic Acid Based Tests, U.S. Food & Drug Administration, https://www.fda.gov/medical-devices/in-vitro-diagnostics/nucleic-acid-based-tests#microbial (accessed March 3, 2024).
- S. Das, S. Dunbar and Y.-W. Tang, Front. Microbiol., 2018, 9, 2478 CrossRef PubMed.
- P. Pokhrel, C. Hu and H. Mao, ACS Sens., 2020, 5, 2283–2296 CrossRef PubMed.
- COVID-19 Tests Granted Traditional Marketing Authorization by the FDA, U.S. Food & Drug Administration, https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/covid-19-tests-granted-traditional-marketing-authorization-fda (accessed March 17, 2024).
- R. F. Omar, M. Boissinot, A. Huletsky and M. G. Bergeron, Infect. Dis. Rep., 2024, 16, 216–227 CrossRef PubMed.
- B. Jia, X. Li and W. Liu, et al. , Front. Microbiol., 2019, 10, 2860 CrossRef PubMed.
- L. Becherer, N. Borst and M. Bakheit, et al. , Anal. Methods, 2020, 12, 717–746 RSC.
- D. R. Murdoch, L. C. Jennings, N. Bhat and T. P. Anderson, Infect. Dis. Clin. North Am., 2010, 24, 791–807 CrossRef PubMed.
- A. M. Werno and D. R. Murdoch, Clin. Infect. Dis., 2008, 46, 926–932 CrossRef PubMed.
- W. Chen, Z. Xu and J. Mu, et al. , J. Med. Microbiol., 2004, 53, 435–438 CrossRef PubMed.
- F. Amanat, D. Stadlbauer and S. Strohmeier, et al. , Nat. Med., 2020, 26, 1033–1036 CrossRef PubMed.
- A. Petherick, Lancet, 2020, 395, 1101–1102 CrossRef PubMed.
- G. Bauer, J. Med. Virol., 2021, 93, 311–322 CrossRef PubMed.
- T. Nguyen, D. D. Bang and A. Wolff, Micromachines, 2020, 11, 306 CrossRef PubMed.
- R. W. Y. Chan, M. C. W. Chan, J. M. Nicholls and J. S. M. Peiris, Virus Res., 2013, 178, 133–145 CrossRef PubMed.
- T. Dilnessa and H. Zeleke, J. Microbiol. Mod. Tech., 2017, 2, 102 Search PubMed.
- D. S. Leland and C. C. Ginocchio, Clin. Microbiol. Rev., 2007, 20, 49–78 CrossRef PubMed.
- F. C. Tenover, Clin. Infect. Dis., 2011, 52, S338–S345 CrossRef PubMed.
- A. Hematian, N. Sadeghifard and R. Mohebi, et al. , Osong Public Health Res. Perspect., 2016, 7, 77–82 CrossRef PubMed.
- P. Yager, T. Edwards and E. Fu, et al. , Nature, 2006, 442, 412–418 CrossRef PubMed.
- L. Gervais, N. de Rooij and E. Delamarche, Adv. Mater., 2011, 23, H151–H176 Search PubMed.
- A. M. Foudeh, T. F. Didar, T. Veres and M. Tabrizian, Lab Chip, 2012, 12, 3249–3266 RSC.
- Y. Yang, Y. Chen and H. Tang, et al. , Small Methods, 2020, 4, 1900451 CrossRef.
- K.-Z. Liu, G. Tian and A. C.-T. Ko, et al. , Biomed. Microdevices, 2020, 22, 29 CrossRef PubMed.
- G. M. Whitesides, Nature, 2006, 442, 368–373 CrossRef PubMed.
- T. M. Squires and S. R. Quake, Rev. Mod. Phys., 2005, 77, 977–1026 CrossRef.
- D. Mark, S. Haeberle and G. Roth, et al. , Chem. Soc. Rev., 2010, 39, 1153–1182 RSC.
- J. B. Nielsen, R. L. Hanson and H. M. Almughamsi, et al. , Anal. Chem., 2020, 92, 150–168 CrossRef PubMed.
- S. F. Berlanda, M. Breitfeld, C. L. Dietsche and P. S. Dittrich, Anal. Chem., 2021, 93, 311–331 CrossRef PubMed.
- S.-Y. Teh, R. Lin, L.-H. Hung and A. P. Lee, Lab Chip, 2008, 8, 198–220 RSC.
- L. Shang, Y. Cheng and Y. Zhao, Chem. Rev., 2017, 117, 7964–8040 CrossRef PubMed.
- Y. Ding, P. D. Howes and A. J. DeMello, Anal. Chem., 2020, 92, 132–149 CrossRef CAS PubMed.
- P. Zhu and L. Wang, Lab Chip, 2017, 17, 34–75 RSC.
- B. J. Hindson, K. D. Ness and D. A. Masquelier, et al. , Anal. Chem., 2011, 83, 8604–8610 CrossRef PubMed.
- R. Nyaruaba, C. Mwaliko and D. Dobnik, et al. , Clin. Microbiol. Rev., 2022, 35, e00168-21 CrossRef PubMed.
- M. Madou, J. Zoval and G. Jia, et al. , Annu. Rev. Biomed. Eng., 2006, 8, 601–628 CrossRef PubMed.
- M. Focke, F. Stumpf and G. Roth, et al. , Lab Chip, 2010, 10, 3210–3212 RSC.
- O. Strohmeier, M. Keller and F. Schwemmer, et al. , Chem. Soc. Rev., 2015, 44, 6187–6229 RSC.
- A. Glière and C. Delattre, Sens. Actuators, A, 2006, 130/131, 601–608 CrossRef.
- J. Siegrist, R. Gorkin and L. Clime, et al. , Microfluid. Nanofluid., 2010, 9, 55–63 CrossRef.
- G. Czilwik, T. Messinger and O. Strohmeier, et al. , Lab Chip, 2015, 15, 3749–3759 RSC.
- H. van Heeren, Lab Chip, 2012, 12, 1022–1025 RSC.
- Y. Temiz, R. D. Lovchik, G. V. Kaigala and E. Delamarche, Microelectron. Eng., 2015, 132, 156–175 CrossRef CAS.
- M. Geissler, L. Clime and X. D. Hoa, et al. , Anal. Chem., 2015, 87, 10565–10572 CrossRef CAS PubMed.
- J.-M. Park, Y.-K. Cho and B.-S. Lee, et al. , Lab Chip, 2007, 7, 557–564 RSC.
- K. Abi-Samra, L. Clime and L. Kong, et al. , Microfluid. Nanofluid., 2011, 11, 643–652 CrossRef CAS.
- L. Clime, D. Brassard, M. Geissler and T. Veres, Lab Chip, 2015, 15, 2400–2411 RSC.
- D. Brassard, M. Geissler and M. Descarreaux, et al. , Lab Chip, 2019, 19, 1941–1952 RSC.
- L. Clime, J. Daoud and D. Brassard, et al. , Microfluid. Nanofluid., 2019, 23, 29 CrossRef.
- M. Geissler, D. Brassard and L. Clime, et al. , Analyst, 2020, 145, 6831–6845 RSC.
- M. Geissler, D. Brassard and N. Adam, et al. , Lab Chip, 2024, 24, 668–679 RSC.
- B.-U. Moon, M. G. de Vries and C. A. Cordeiro, et al. , Anal. Chem., 2013, 85, 10949–10955 CrossRef CAS PubMed.
- L. Clime, X. D. Hoa and N. Corneau, et al. , Biomed. Microdevices, 2015, 17, 17 CrossRef PubMed.
- V. Gnyawali, B.-U. Moon and J. Kieda, et al. , Soft Matter, 2017, 13, 4011–4016 RSC.
- L. Malic, J. Daoud and M. Geissler, et al. , Analyst, 2019, 144, 6541–6553 RSC.
- V. Gnyawali, M. Saremi, M. C. Kolios and S. S. H. Tsai, Biomicrofluidics, 2017, 11, 034104 CrossRef PubMed.
- G. A. Cooksey, C. G. Sip and A. Folch, Lab Chip, 2009, 9, 417–426 RSC.
- T. Satoh, G. Narazaki and R. Sugita, et al. , Lab Chip, 2016, 16, 2339–2348 RSC.
- S. Yasotharan, S. Pinto and J. G. Sled, et al. , Lab Chip, 2015, 15, 2660–2669 RSC.
- K. Boissinot, R. Peytavi and S. Chapdelaine, et al. , Analyst, 2021, 146, 4226–4234 RSC.
- B.-U. Moon, D. K. Hwang and S. S. H. Tsai, Lab Chip, 2016, 16, 2601–2608 RSC.
- Y.-T. Kao, T. S. Kaminski and W. Postek, et al. , Lab Chip, 2020, 20, 54–63 RSC.
- B.-U. Moon, N. Abbasi and S. G. Jones, et al. , Anal. Chem., 2016, 88, 3982–3989 CrossRef PubMed.
- B. F. L. Lai, R. X. Z. Lu and L. Davenport Huyer, et al. , Nat. Protoc., 2021, 16, 2158–2189 CrossRef PubMed.
- K. Choi, A. H. C. Ng, R. Fobel and A. R. Wheeler, Annu. Rev. Anal. Chem., 2012, 5, 413–440 CrossRef PubMed.
- R. B. Fair, Microfluid. Nanofluid., 2007, 3, 245–281 CrossRef.
- X. Ding, P. Li and S.-C. S. Lin, et al. , Lab Chip, 2013, 13, 3626–3649 RSC.
- T. P. Hunt, D. Issadore and R. M. Westervelt, Lab Chip, 2008, 8, 81–87 RSC.
- J. Z. Chen, S. M. Troiana, A. A. Darhuber and S. Wagner, J. Appl. Phys., 2005, 97, 014906 CrossRef.
- U. Lehmann, S. Hadjidj and V. K. Parashar, et al. , Sens. Actuators, B, 2006, 117, 457–463 CrossRef CAS.
- Y. Zhang and N.-T. Nguyen, Lab Chip, 2017, 17, 994–1008 RSC.
- P. Y. Chiou, H. Moon and H. Toshiyoshi, et al. , Sens. Actuators, A, 2003, 104, 222–228 CrossRef CAS.
- J. Lamanna, E. Y. Scott and H. S. Edwards, et al. , Nat. Commun., 2020, 11, 5632 CrossRef CAS PubMed.
- J. Peng, C. Chan and S. Zhang, et al. , Chem. Sci., 2023, 14, 2887–2900 RSC.
- L. Malic, D. Brassard, T. Veres and M. Tabrizian, Lab Chip, 2010, 10, 418–431 RSC.
- M. Abdelgawad and A. R. Wheeler, Microfluid. Nanofluid., 2008, 4, 349–355 CrossRef CAS.
- V. Jain, T. P. Raj, R. Deshmukh and R. Patrikar, Microsyst. Technol., 2017, 23, 389–397 CrossRef CAS.
- J. Li and C.-J. Kim, Lab Chip, 2020, 20, 1705–1712 RSC.
- M. J. Jebrail, M. S. Bartsch and K. D. Patel, Lab Chip, 2012, 12, 2452–2463 RSC.
- E. Samiei, M. Tabrizian and M. Hoorfar, Lab Chip, 2016, 16, 2376–2396 RSC.
- X. Rui, S. Song, W. Wang and J. Zhou, Biomicrofluidics, 2020, 14, 061503 CrossRef CAS PubMed.
- L. Gervais and E. Delamarche, Lab Chip, 2009, 9, 3330–3337 RSC.
- M. Geissler, E. Roy and G. A. Diaz-Quijada, et al. , ACS Appl. Mater. Interfaces, 2009, 1, 1387–1395 CrossRef CAS PubMed.
- L. Gervais, M. Hitzbleck and E. Delamarche, Biosens. Bioelectron., 2011, 27, 64–70 CrossRef CAS PubMed.
- D. Brassard, L. Clime and K. Li, et al. , Lab Chip, 2011, 11, 4099–4107 RSC.
- R. Safavieh and D. Juncker, Lab Chip, 2013, 13, 4180–4189 RSC.
- A. Olanrewaju, M. Beaugrand, M. Yafia and D. Juncker, Lab Chip, 2018, 18, 2323–2347 RSC.
- Y. Zhu and K. Petkovic-Duran, Microfluid. Nanofluid., 2010, 8, 275–282 CrossRef.
- M. Zimmermann, H. Schmid, P. Hunziker and E. Delamarche, Lab Chip, 2007, 7, 119–125 RSC.
- M. L. Salva, Y. Temiz and M. Rocca, et al. , Sci. Rep., 2019, 9, 17242 CrossRef PubMed.
- M. Yafia, O. Ymbern and A. O. Olanrewaju, et al. , Nature, 2022, 605, 464–469 CrossRef CAS PubMed.
- B. O'Farrell, Lateral Flow Immunoassay Systems: Evolution from the Current State of the Art to the Next Generation of Highly Sensitive, Quantitative Rapid Assays, in The Immunoassay Handbook: Theory and Applications of Ligand Binding, ELISA and Related Techniques, ed. D. Wild, Elsevier, Oxford, UK, 4th edn, 2013, pp. 89–108 Search PubMed.
- W. C. Mak, V. Beni and A. P. F. Turner, Trends Anal. Chem., 2016, 79, 297–305 CrossRef CAS.
- J. Budd, B. S. Miller and N. E. Weckman, et al. , Nat. Rev. Bioeng., 2023, 1, 13–31 CrossRef.
- X. Nan, X. Yao, L. Yang and Y. Cui, Analyst, 2023, 148, 4573–4590 RSC.
- C. Carrell, A. Kava and M. Nguyen, et al. , Microelectron. Eng., 2019, 206, 45–54 CrossRef CAS.
- E. Noviana, T. Ozer and C. S. Carrell, et al. , Chem. Rev., 2021, 121, 11835–11885 CrossRef CAS PubMed.
- M. B. Kulkarni, N. H. Ayachit, T. M. Aminabhavi and B. W. Pogue, Biochem. Eng. J., 2023, 198, 109027 CrossRef CAS.
- H. A. Silva-Neto, I. V. S. Arantes and A. L. Ferreira, et al. , Trends Anal. Chem., 2023, 158, 116893 CrossRef CAS.
- K. Sun, Y. Fan, M. Hebda and Y. Zhang, Biomed. Mater. Diagn. Devices, 2023, 1, 388–401 CrossRef.
- K. Alby, E. B. Popowitch and M. B. Miller, J. Clin. Microbiol., 2013, 51, 352–353 CrossRef CAS PubMed.
- S. Nie, R. B. Roth and J. Stiles, et al. , J. Clin. Microbiol., 2014, 52, 3339–3344 CrossRef CAS PubMed.
- M. E. Andersson, S. Olofsson and M. Lindh, Scand. J. Infect. Dis., 2014, 46, 897–901 CrossRef CAS PubMed.
- S. D. Goldenberg and J. D. Edgeworth, Expert Rev. Mol. Diagn., 2015, 15, 23–32 CrossRef CAS PubMed.
- W. S. Stevens, L. Scott and L. Noble, et al., Impact of the GeneXpert MTB/RIF Technology on Tuberculosis Control, in Tuberculosis and the Tubercle Bacillus, ed. W. R. Jacobs Jr., H. McShane, V. Mizrahi and I. M. Orme, ASM Press, Washington, DC, 2nd edn, 2017, pp. 389–410 Search PubMed.
- D. Banerjee, J. Michael and B. Schmitt, et al. , J. Clin. Microbiol., 2020, 58, e01775-19 CrossRef PubMed.
- R. Batra, L. G. Olivieri and D. Rubin, et al. , J. Clin. Virol., 2020, 132, 104645 CrossRef CAS PubMed.
- C. W. Chan, S. Shahul and C. Coleman, et al. , Am. J. Clin. Pathol., 2021, 155, 286–295 CrossRef CAS PubMed.
- J. Xu, T. Kirtek and Y. Xu, et al. , Am. J. Clin. Pathol., 2021, 155, 815–822 CrossRef CAS PubMed.
- S. Sachdeva, R. W. Davis and A. K. Saha, Front. Bioeng. Biotechnol., 2021, 8, 602659 CrossRef PubMed.
- L. Bissonnette and M. G. Bergeron, Micromachines, 2016, 7, 94 CrossRef PubMed.
- C. Sheridan, Nat. Biotechnol., 2020, 38, 515–518 CrossRef CAS PubMed.
- D. A. Mistry, J. Y. Wang and M.-E. Moeser, et al. , BMC Infect. Dis., 2021, 21, 828 CrossRef CAS PubMed.
- F. Stumpf, F. Schwemmer and T. Hutzenlaub, et al. , Lab Chip, 2016, 16, 199–207 RSC.
- M. Rombach, S. Hin and M. Specht, et al. , Analyst, 2020, 145, 7040–7047 RSC.
- G. Huang, Q. Huang and L. Xie, et al. , Sci. Rep., 2017, 7, 6441 CrossRef PubMed.
- F. Tian, C. Liu and J. Deng, et al. , Sci. China: Chem., 2020, 63, 1498–1506 CrossRef CAS PubMed.
- K. G. de Oliveira, P. F. N. Estrela and G. de Melo Mendes, et al. , Analyst, 2021, 146, 1178–1187 RSC.
- Y. Chen, N. Zong and F. Ye, et al. , Anal. Chem., 2022, 94, 9603–9609 CrossRef CAS PubMed.
- R. R. G. Soares, A. S. Akhtar and I. F. Pinto, et al. , Lab Chip, 2021, 21, 2932–2944 RSC.
- L. Malic, D. Brassard and D. Da Fonte, et al. , Lab Chip, 2022, 22, 3157–3171 RSC.
- L. Malic, L. Clime and B.-U. Moon, et al. , Lab Chip, 2024, 24, 4755–4765 RSC.
- H. Van Nguyen, V. M. Phan and T. S. Seo, Sens. Actuators, B, 2024, 399, 134771 CrossRef.
- Q. Lin, D. Wen and J. Wu, et al. , Anal. Chem., 2020, 92, 9454–9458 CrossRef CAS PubMed.
- S. Liu, Y. Hou and Z. Li, et al. , ACS Sens., 2023, 8, 3520–3529 CrossRef CAS PubMed.
- H. Wang, Z. Ma and J. Qin, et al. , Biosens. Bioelectron., 2019, 126, 373–380 CrossRef CAS PubMed.
- C. Zhu, A. Hu and J. Cui, et al. , Micromachines, 2019, 10, 537 CrossRef PubMed.
- R.-Q. Zhang, S.-L. Hong and C.-Y. Wen, et al. , Biosens. Bioelectron., 2018, 100, 348–354 CrossRef CAS PubMed.
- Y.-T. Yeh, K. Gulino and Y. Zhang, et al. , Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 895–901 CrossRef CAS PubMed.
- Y. Kim, A. T. Abafogi and B. M. Tran, et al. , Micromachines, 2020, 11, 203 CrossRef PubMed.
- A. Ganguli, A. Mostafa and J. Berger, et al. , Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 22727–22735 CrossRef CAS PubMed.
- D. Najjar, J. Rainbow and S. S. Timilsina, et al. , Nat. Biomed. Eng., 2022, 6, 968–978 CrossRef CAS PubMed.
- Y. Zai, C. Min and Z. Wang, et al. , Lab Chip, 2022, 22, 3436–3452 RSC.
- I. Rutten, D. Daems, K. Leirs and J. Lammertyn, Biosensors, 2023, 13, 100 CrossRef CAS PubMed.
- N. Yamaguchi, Y. Tokunaga and S. Goto, et al. , Sci. Rep., 2017, 7, 3092 CrossRef PubMed.
- R. Singh, S. Hong and J. Jang, Sci. Rep., 2017, 7, 42771 CrossRef CAS PubMed.
- J. Yang, V. M. Phan and C.-K. Heo, et al. , Sens. Actuators, B, 2023, 380, 133331 CrossRef CAS PubMed.
- E. Wulff-Burchfield, W. A. Schell and A. E. Eckhardt, et al. , Diagn. Microbiol. Infect. Dis., 2010, 67, 22–29 CrossRef CAS PubMed.
- R. Prakash, K. Pabbaraju and S. Wong, et al. , J. Electrochem. Soc., 2014, 161, B3083–B3093 CrossRef CAS PubMed.
- Y. Wang, Q. Ruan and Z.-C. Lei, et al. , Anal. Chem., 2018, 90, 5224–5231 CrossRef CAS PubMed.
- T. Zhang, W. Zhao and X. Chen, et al. , Anal. Chem., 2022, 94, 15472–15480 CrossRef CAS PubMed.
- S. Anderson, B. Hadwen and C. Brown, Lab Chip, 2021, 21, 962–975 RSC.
- M. Ho, N. Sathishkumar and A. A. Sklavounos, et al. , Lab Chip, 2024, 24, 63–73 RSC.
- C. Dong, F. Li and Y. Sun, et al. , Lab Chip, 2024, 24, 3850–3862 RSC.
- N. Ramalingam, T. C. San and T. J. Kai, et al. , Microfluid. Nanofluid., 2009, 7, 325–336 CrossRef CAS PubMed.
- Y.-D. Ma, Y.-S. Chen and G.-B. Lee, Sens. Actuators, B, 2019, 296, 126647 CrossRef CAS.
- M. Dou, J. Sanchez and H. Tavakoli, et al. , Anal. Chim. Acta, 2019, 1065, 71–78 CrossRef CAS PubMed.
- M. Dou, N. Macias and F. Shen, et al. , EClinicalMedicine, 2019, 8, 72–77 CrossRef PubMed.
- P. E. Kilgore, A. M. Salim, M. J. Zervos and H.-J. Schmitt, Clin. Microbiol. Rev., 2016, 29, 449–486 CrossRef CAS PubMed.
- F. Sun, A. Ganguli and J. Nguyen, et al. , Lab Chip, 2020, 20, 1621–1627 RSC.
- K. M. Clark, M. S. Schenkel and T. W. Pittman, et al. , ACS Meas. Sci. Au, 2022, 2, 584–594 CrossRef CAS PubMed.
- C. Carrell, I. Jang and J. Link, et al. , Anal. Methods, 2023, 15, 2721–2728 RSC.
- J. S. Link, C. S. Carrell and I. Jang, et al. , Anal. Chim. Acta, 2023, 1277, 341634 CrossRef CAS PubMed.
- J. S. Link, J. O'Donnell-Sloan and S. Curdts, et al. , Anal. Chem., 2024, 96, 4111–4119 CrossRef CAS PubMed.
- P. Robin, L. Barnabei and S. Marocco, et al. , Biosens. Bioelectron.: X, 2023, 13, 100302 CAS.
- D. Wu, J. Zhang and F. Xu, et al. , Microfluid. Nanofluid., 2017, 21, 43 CrossRef.
- R. C. Murdock, K. M. Gallegos and J. A. Hagen, et al. , Lab Chip, 2017, 17, 332–340 RSC.
- P. Garneret, E. Coz and E. Martin, et al. , PLoS One, 2021, 16, e0243712 CrossRef CAS PubMed.
- D. Liu, H. Shen and Y. Zhang, et al. , Lab Chip, 2021, 21, 2019–2026 RSC.
- C. Zhang, T. Zheng and H. Wang, et al. , Anal. Chem., 2021, 93, 3325–3330 CrossRef CAS PubMed.
- C. Wang, X. Yang and B. Gu, et al. , Anal. Chem., 2020, 92, 15542–15549 CrossRef CAS PubMed.
- Z. Chen, Z. Zhang and X. Zhai, et al. , Anal. Chem., 2020, 92, 7226–7231 CrossRef CAS PubMed.
- D. Wang, S. He and X. Wang, et al. , Nat. Biomed. Eng., 2020, 4, 1150–1158 CrossRef CAS PubMed.
- K. S. Makarova, D. H. Haft and R. Barrangou, et al. , Nat. Rev. Microbiol., 2011, 9, 467–477 CrossRef CAS PubMed.
- F. Hille, H. Richter and S. P. Wong, et al. , Cell, 2018, 172, 1239–1259 CrossRef CAS PubMed.
- F. Nafian, S. Nafian, B. Kamali Doust Azad and M. Hashemi, Mol. Biotechnol., 2023, 65, 497–508 CAS.
- J. P. Broughton, X. Deng and G. Yu, et al. , Nat. Biotechnol., 2020, 38, 870–874 CrossRef CAS PubMed.
- J. Joung, A. Ladha and M. Saito, et al. , N. Engl. J. Med., 2020, 383, 1492–1494 CrossRef CAS PubMed.
- R. Zhu, H. Jiang and C. Li, et al. , Anal. Chim. Acta, 2023, 1257, 341175 CrossRef CAS PubMed.
- R. Gupta, P. Gupta and S. Wang, et al. , Nat. Biomed. Eng., 2023, 7, 1556–1570 CrossRef CAS PubMed.
- D. Hong, E.-J. Jo and D. Bang, et al. , ACS Nano, 2023, 17, 16607–16619 CrossRef CAS PubMed.
- Z. Liu, C. Wang and S. Zheng, et al. , Nanomedicine, 2023, 47, 102624 CrossRef CAS PubMed.
- S. Atta, Y. Zhao, J. Q. Li and T. Vo-Dinh, Anal. Chem., 2024, 96, 4783–4790 CrossRef CAS PubMed.
- C. S. Kosack, A. L. Page and P. R. Klatser, Bull. W. H. O., 2017, 95, 639–645 CrossRef PubMed.
- S. Baratchi, K. Khoshmanesh and C. Sacristán, et al. , Biotechnol. Adv., 2014, 32, 333–346 CrossRef CAS PubMed.
- P. Aldo, G. Marusov and D. Svancara, et al. , Am. J. Reprod. Immunol., 2016, 75, 678–693 CrossRef CAS PubMed.
- M. Dysinger, G. Marusov and S. Fraser, J. Immunol. Methods, 2017, 451, 1–10 CrossRef CAS PubMed.
- V. Iyer, Z. Yang and J. Ko, et al. , Lab Chip, 2022, 22, 3110–3121 RSC.
- L. Kim, Overview of the Microfluidic Diagnostics Commercial Landscape, in Microfluidic Diagnostics – Methods and Protocols, ed. G. Jenkins and C. D. Mansfield, Humana Press, Totowa, NJ, 2013, vol. 949, pp. 65–83 Search PubMed.
- S. Nahavandi, S. Baratchi and R. Soffe, et al. , Lab Chip, 2014, 14, 1496–1514 RSC.
- A. Panesar, Machine Learning and AI for Healthcare: Big Data for Improved Health Outcomes, Apress, Berkeley, CA, 2nd edn, 2020 Search PubMed.
- S. M. D. A. C. Jayatilake and G. U. Ganegoda, J. Healthcare Eng., 2021, 2021, 6679512 Search PubMed.
- C. C. Agbo, Q. H. Mahmoud and J. M. Eklund, Healthcare, 2019, 7, 56 CrossRef PubMed.
- T. McGhin, K.-K. R. Choo, C. Z. Liu and D. He, J. Netw. Comput. Appl., 2019, 135, 62–75 CrossRef.
| This journal is © The Royal Society of Chemistry 2025 |