Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Live chicken egg embryos as an alternative in vivo tumour model for deep surface enhanced Raman spectroscopy†
Samantha M.
McCabe
 abc,
Hannah
Gardiner
b,
Calum
Mullen
abc,
Hannah
Gardiner
b,
Calum
Mullen
 b,
Gregory Q.
Wallace
b,
Gregory Q.
Wallace
 a,
Neil C.
Shand
c,
Alexander B.
Mullen
b,
Linda
Horan
b,
Duncan
Graham
a,
Neil C.
Shand
c,
Alexander B.
Mullen
b,
Linda
Horan
b,
Duncan
Graham
 a,
Karen
Faulds
a,
Karen
Faulds
 *a and
Marie
Boyd
*b
*a and
Marie
Boyd
*b
aDepartment of Pure and Applied Chemistry, Technology and Innovation Centre, University of Strathclyde, 99 George Street, Glasgow, G1 1RD, UK. E-mail: karen.faulds@strath.ac.uk
bStrathclyde Institute of Pharmacy and Biomedical Sciences, University of Strathclyde, 161 Cathedral Street, Glasgow, G4 0RE, UK
cThe Defence Science and Technology Laboratory (Dstl), Porton Down, Salisbury, SP4 0JQ, UK
First published on 4th June 2024
Abstract
Live chicken egg embryos offer new opportunities for evaluation and continuous monitoring of tumour growth for in vivo studies compared to traditional rodent models. Here, we report the first use of surface enhanced Raman scattering (SERS) mapping and surface enhanced spatially offset Raman scattering (SESORS) for the detection and localisation of targeted gold nanoparticles in live chicken egg embryos bearing a glioblastoma tumour.
The chicken egg embryo is an alternative in vivo platform which can be used for studying tumour biology,1 drug distribution,2 angiogensis3 and toxicology.4 The chorioallantoic membrane (CAM) is a highly vascularised membrane that attaches to the eggshell and forms around the chicken embryo.5 The CAM develops from day 3 until day 14 with embryonic development. The first use of the CAM model for assessment of tumour growth was demonstrated in 1911,6 and since then there has been a plethora of research which has advanced the use of the model for multidisciplinary purposes.7–9 The main advantage of using a chicken egg embryo in cancer research is that tumours in vivo are highly vascularised and heterogeneous, which is well reflected in the CAM model, allowing for research into angiogenesis, tumour formation and metastasis. Furthermore, the in vivo CAM model contributes towards a reduction and/or replacement model for rodents. The use of rodents for in vivo cancer research is expensive, requires an animal license and is time consuming as well as having ethical considerations. The chicken egg embryo model is considerably less expensive, has lower sentience than rodents, potentially higher throughput, does not require a home office license for use before day 14, and allows for easy visualisation of the tumour and tumour vasculature.
Glioblastoma (GBM) is a stage IV, lethal form of brain cancer, with survival rates at 25% for more than 1 year, and 5% for more than 5 years.10 Patients who are usually treated with radio-chemotherapy suffer side effects caused by the inability to specifically identify tumour cells or margins due to the difficulty in distinguishing between cancerous and non-cancerous tissues. Techniques such as surface enhanced Raman scattering (SERS) and surface enhanced spatially offset Raman scattering (SESORS) have previously been used for in vitro cancer detection and imaging.11,12 SERS for imaging applications generally uses metallic nanoparticles, most notably gold nanoparticles (AuNPs), with a Raman reporter adsorbed onto the surface, to achieve an enhancement in the Raman signal. Spatially offset Raman scattering (SORS) uses a spatial offset between the excitation and collection optics to detect Raman scattered photons at depth, through a barrier.13 SESORS couples the two techniques to create the non-invasive detection of Raman scattered photons at depth through turbid media, such as bone and tissue.14–16 AuNPs can be functionalised with targeting antibodies specific towards a tumour of interest. Tenascin-C is a glycoprotein that is expressed in the extracellular matrix of U87-MG glioblastoma cancer cells.17 Therefore, attaching a tenascin-C antibody to the surface of AuNPs could exploit tumour specific targeting opportunities.
We have previously applied shell isolated AuNPs functionalised with a tenascin-C antibody to in vitro 3D tumour mimics, multicellular tumour spheroids (MTS).18 Although MTS provide a bridge between 2-dimensional cell culture and in vivo models for understanding NP uptake, they still do not fully represent a vascularised tumour. In this regard, an in vivo animal model is the next progression in evaluating the potential of these NPs, for both uptake and tumour targeting. Whilst rodent models are most commonly used for exploring in vivo detection of AuNPs,16,19 there are many requirements and ethical considerations for their use in laboratory experiments. Therefore, we applied antibody functionalised AuNPs to the chicken embryo tumour model as it is a suitable alternative in vivo cancer model to rodent tumour models providing promising opportunities for assessing the use of AuNP detection in cancer nanomedicine. This model also has the potential to allow for monitoring tumour/NP interactions over longer time periods than would be possible with rodent models which are subject to time limitations or culled before measurements. In this work, shell isolated AuNPs (AuNP-SHINs) were for the first-time detected by SESORS and SERS in U87-MG glioblastomas grown on the CAM of live chicken egg embryos.
Shell isolated AuNPs (AuNP-SHINs) were formed using a 50 nm AuNP seed, synthesised using a modified Turkevich citrate reducing method.20 A non-resonant Raman reporter, (1H-pyrazol-4-yl) pyridine (PPY) (Fig. S1†) was added to the surface of the gold. The shell was formed around the AuNPs using (3-aminopropyl)trimethoxysilane (APTMS) and sodium silicate,21 which stabilises the AuNPs for the addition of AuNP-SHINs into cells and cell media.22 Additionally, controlled aggregation of the AuNPs prior to shell formation allowed for the AuNP-SHIN to have an increased SERS signal, desirable for SERS and SESORS. A tenascin-C antibody was immobilised onto the AuNP-SHIN nanotags to create Ab nanotags, synthesised using the same procedure as reported in our previous work.18 U87-MG cells are glioblastoma (GBM) brain cancer cells that overexpress the protein tenascin-C.23,24 Tenascin-C was chosen as a biomarker of interest because it is only overexpressed in adult tissues that are diseased or injured,17 making it a potentially strong candidate for targeting. The PPY-AuNP-SHIN nanotags and antibody functionalised PPY-AuNP-SHIN nanotags (Ab nanotags) had an average diameter of 66 nm and 82 nm respectively. The synthesis and characterisation of these can be found in the ESI (Fig. S1–S3 and Table S1†).
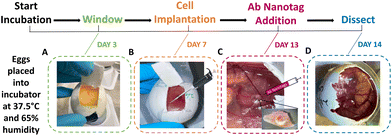
Fertilised white leghorn chicken eggs were placed into a 45° rotating incubator set to 37.5 °C with 65% humidity. This was determined as being day 0 of the incubation period. On day 3, the rotation was stopped, and the eggs were removed. A small puncture was created in the bottom of the egg to allow the CAM to be released from the eggshell, ∼3 mL of albumin was then removed and electrical tape was applied to the puncture. A small window was created in the eggshell to check viability (Fig. 1A). If viable, magic tape was placed over the window to keep it closed to prevent infection or drying out, and the eggs were placed back into the incubator. On day 7, the viable embryos had 2 × 106 U87-MG glioblastoma cells added to the surface of the CAM (Fig. 1B). Cell preparation involved resuspending the cells in a mixture of phosphate buffered saline (PBS) and Matrigel (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 v/v). Trauma was induced to the main blood vessel using sterile tissue paper and 0.05% of trypsin-EDTA was added to aid in cell invasion immediately before cell implantation. The U87-MG glioblastoma cells were then added to the CAM and incubated for 6 days. On day 13, the embryos were checked for viability and tumour growth. Where tumours had grown, the Ab nanotags (0.67 pM) were added to the surface of the tumour and CAM area (Fig. 1C). The nanotags were left for 24 hours to allow for internalisation into the tumour and CAM and were found to be non-toxic to the embryos as they were still viable after 24-hour incubation. On day 14, after 24 hours of nanotag incubation, SESORS measurements were taken, followed by dissection of the tumour (Fig. 1D). All embryos were culled by decapitation on day 14 as this was the quickest and most humane method.
50 v/v). Trauma was induced to the main blood vessel using sterile tissue paper and 0.05% of trypsin-EDTA was added to aid in cell invasion immediately before cell implantation. The U87-MG glioblastoma cells were then added to the CAM and incubated for 6 days. On day 13, the embryos were checked for viability and tumour growth. Where tumours had grown, the Ab nanotags (0.67 pM) were added to the surface of the tumour and CAM area (Fig. 1C). The nanotags were left for 24 hours to allow for internalisation into the tumour and CAM and were found to be non-toxic to the embryos as they were still viable after 24-hour incubation. On day 14, after 24 hours of nanotag incubation, SESORS measurements were taken, followed by dissection of the tumour (Fig. 1D). All embryos were culled by decapitation on day 14 as this was the quickest and most humane method.
To propose the chicken embryo tumour model as a potential in vivo model for investigation of AuNP localisation and uptake, SESORS and SERS measurements were performed. Previously,18 we reported the use of SERS mapping in MTS to demonstrate the successful uptake and localisation of the same type of Ab nanotags. Although MTS models provide a more accurate insight into how the nanotags may distribute in a tumour due to their 3D microenvironment compared to 2D cell cultures, they still lack the overall complexity of an in vivo tumour.
Therefore, we wanted to progress from the 3D in vitro investigation to an in vivo model to observe whether the uptake and distribution of the Ab nanotags remained similar. This was carried out using the chicken embryo tumour model due to the various advantages over traditional rodent models. For example, the simplicity, lower cost, minimal ethical restrictions, and the potential ability to monitor the tumour and tumour vasculature continuously and easily over time.
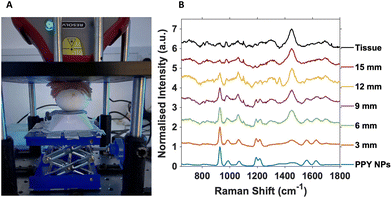
For SESORS depth prediction, the top of the eggshell was removed to allow the full tumour and embryo to be accessible, with care not to puncture or damage the CAM, and to allow alignment of the spectrometer to the tumour. Once aligned, subsequent layers of porcine tissue with a thickness of ∼3 mm were added to the top of the open eggshell (Fig. 2A). This was carried out to better reflect an in vivo tumour located underneath mammalian tissue that would be present in an animal model, such as rodents. A handheld SORS instrument with an 830 nm excitation wavelength and 8 mm spatial offset was used with a total exposure time of 10 seconds (2 s integration time with 5 accumulations).
All SORS measurements were carried out with the nose cone on to allow contact with the porcine tissue and ensure no air gap was located between the spectrometer and the sample. Porcine tissue layers were successively added until the 928 cm−1 PPY peak of the nanotags was no longer observed and the 1440 cm−1 tissue spectrum completely dominated the overall spectrum (Fig. 2B).
Fig. 2B is the resulting stacked plot from the depth study and shows the PPY signal from the Raman reporter on the Ab nanotags, that can be observed through up to 12 mm of tissue. With increasing tissue layer additions, the tissue peak at 1440 cm−1 becomes visible and increases with thickness as the main PPY peak at 928 cm−1, from the nanotags decreases. At 15 mm, the tissue spectrum dominates and the nanotags can no longer be observed. The tissue spectrum arises from the CH2 lipid stretch present in mammalian tissue.25 Importantly, in tumours that did not have any nanotags added, and used as controls, the characteristic peaks of the PPY were not present (Fig. S5†), indicating that the peaks from the SESORS measurements were in fact those from the PPY, and hence nanotags. Live chicken egg embryos with nanotag incubated tumours were investigated in triplicate and showed consistent results (Fig. S6†). It is also important to note that the embryos were still alive after the experiment and therefore the experimental conditions used did not cause embryonic death. Therefore, live chicken egg embryos harbouring a human tumour impregnated with the Ab nanotags, were for the first time, detected using a handheld SORS instrument. The fact that the embryos were still alive after measurements clearly demonstrates that this approach could be used to monitor tumours/NP interactions over time as repeat measurements could be carried out on the same embryo.
Although 12 mm is not as deep as observed previously for in vivo depth studies using SESORS,19 it is important to note that the 12 mm relates to the thickness of the porcine tissue on top of the egg. In the previous study, the authors also used a transmission geometry, which would not be optimal for this model due to the tumour being on the surface of the CAM, and the incoming photons would have to penetrate through the albumin and the embryo within the shell before reaching the tumour. In addition, here we used a handheld SORS instrument which is easier to use and better translates into a clinical setting. Not only is the chicken embryo model easy to control, inexpensive and shows rapid tumour growth, but it also allows for continuous monitoring and imaging through the window formed in the eggshell. The embryos can be removed from the incubators, analysed, and placed back without causing any detrimental effects to the embryo or experiment. Additionally, there is no need for anaesthetic and subsequent culling during experiments, which would be required for rodent studies for ethical reasons. Therefore, in terms of longitudinal monitoring, the chicken embryo tumour model is a promising potential alternative to in vivo rodent models allowing for monitoring of NP uptake into the same tumour over time and to investigate the effects of NP uptake on viable continuously growing tumours using SORS.
While SESORS imaging provides non-invasive insight into the detection of the nanotags through a barrier at depth, currently, it does not provide sufficient resolution to understand how NPs are distributed throughout the tumour. Therefore, as was previously done with the MTS models,18 SERS mapping was carried out to understand more fully the results from the SESORS maps and the location of the nanotags in the tumours.
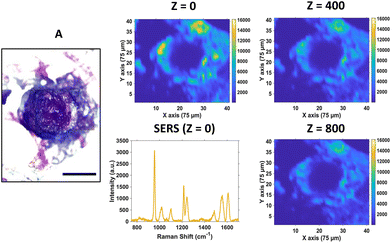
For SERS mapping, the tumours were dissected, subsequently fixed using paraformaldehyde and then placed onto a microscope slide. The white light image of the tumour (Fig. 3A) is provided to illustrate the margins of the tumour and where the CAM starts, since part of the CAM is also removed during dissection of the tumour. To determine the localisation of the nanotags throughout the tumour, a series of 2D XY maps covering the entire lateral dimensions of the tumour (3 mm × 3 mm) were acquired at numerous depths separated by 100 μm. Fig. 3 shows a selection of the resulting SERS intensity maps of the 956 cm−1 peak of PPY at various Z depths.
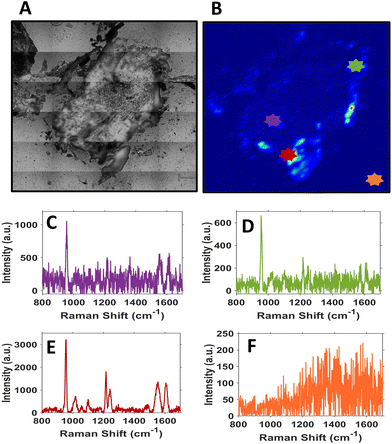
It is evident from the SERS intensity maps that the nanotags have accumulated within the CAM surrounding the tumour. This is not surprising because the CAM is a very thin membrane, therefore it is easy for the Ab nanotags to enter. Additionally, there are no Ab nanotags present within the main bulk of the tumour, demonstrated by the lack of nanotag signal. Nevertheless, there is still evidence that there are Ab nanotags within the outer rim of the tumour. Since it was not possible to determine whether these nanotags had not fully penetrated the tumour because of tumour biology or due to the penetration limitations of Raman through the tumour volume, further investigation was required. Therefore, the tumours were sectioned into 10 μm thick sections to ascertain the location of the Ab nanotags within the tumour (Fig. 4).
Sectioning of the tumour clearly identified that there were nanotags present throughout the tumour. Fig. 4 illustrates that the nanotags seem to be accumulating mostly around the outer edge of the tumour, where the highest intensity point chosen shows a strong PPY spectrum (Fig. 4E). This was carried out on n = 3 different sections and they all showed similar trends (Fig. S8†). Given that the sections correspond to different vertical positions of the tumour, through the various SERS maps of the section, it can be ascertained that the nanotags do distribute throughout the tumour.
Similarly to what we reported previously in an MTS model using the same cell type,18 there was successful uptake of the Ab nanotags and likewise they were localised only to a small area of the tumour, with very minimal uptake throughout the main bulk of the tumours. It is important to note that in both cases, the MTS were simply incubated with nanotags or the nanotags were merely dropped on top of the tumour. Further understanding and investigation into the introduction of the nanotags to the tumours is required, as this could provide a means of increasing their uptake into the bulk tumour, and hence advance the potential depth detection through SESORS.
Conclusions
The chicken egg embryo model is easy to control, inexpensive, supports rapid tumour growth, allows for continuous monitoring of the tumour, and has limited ethical restrictions and requirements compared to rodent models. This preliminary, proof of concept work demonstrates the potential chicken embryo tumour models could provide in the future for in vivo investigation of AuNPs. In this work, SESORS and SERS mapping were performed on chicken egg embryo models harbouring a tumour formed from U87-MG cells, impregnated with Ab nanotags. Using a clinically relevant handheld SORS instrument, SESORS was used to detect the Ab nanotags through a tissue barrier in ovo, without the need to remove the tumour from the embryo, or the embryo from the eggshell. Subsequent SERS mapping of the dissected whole and sectioned tumours provided crucial insight into how the nanotags were distributed throughout the tumour and surrounding CAM. Specifically, it was found that the nanotags were preferentially found around the margins of the tumour and in the immediate surrounding CAM.Since this work is, to the best of our knowledge, the first use of the chicken egg embryo model for investigation, analysis and detection using Raman spectroscopies, there is a plethora of potential avenues that need to be explored before the model can be widely adopted. The means of introducing the nanotags into the tumour and the concentration of the nanotags used, are of immediate interest. In this work, the nanotags were simply added on top of the tumour. Future work should investigate the injection of the nanotags into the main blood vessels of the CAM, which could better reflect injection into the vasculature of other animal models. Improved targeting is consistently an area of interest as maximising nanotag uptake and having a more controlled localisation of the nanotags both remain important steps towards clinical relevance. This model offers excellent potential to understand more about NP targeting, uptake, and localisation within a vascularised live tumour model to allow optimisation of targeted nanotag design before moving to live rodent models. Given the number of conclusions that need to be fully optimised, and therefore the necessary number of models needed, then it is evident that having an effective, low-cost steppingstone towards in vivo settings is required which could be fulfilled by using the chicken egg embryo models.
Author contributions
S. M. M., H. G., and C. M. developed the chicken embryo tumour model to be an appropriate model for tumour growth. G. Q. W conducted the data processing, helped practically with SERS mapping and editing of the manuscript. L. H. provided initial training on the model. A. B. M. and M. B. provided the initial idea of the use of the model in the department and provided academic input throughout. N. C. S. provided the funding towards this project. M. B. obtained the NC3Rs grant to allow work to be conducted on the model. D. G., K. F., and M. B., collectively provided supervision throughout the work and assisted in the writing and editing of the manuscript. S. M. M. wrote the manuscript, created the figures, deduced the data provided by G. Q. W. into graphs, conducted the synthesis, characterisation, and conjugation of the nanotags. As well as the growth of the U87 tumours on the egg models and subsequent SESORS and SERS mapping on the intact and dissected tumours.Data availability
The research data associated with this paper is available at the following link: https://doi.org/10.15129/bd4589d4-3273-4baf-95ac-343693e19252.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the University of Strathclyde and DSTL for the financial support. S. M. M., D. G., and K. F. thank DSTL for the funding that supported this work. D. G., K. F., and G. Q. W. acknowledge support from the Leverhulme Trust, RPG-2020-400. M. B. acknowledges the support from the NC3Rs, NCW001055/1.References
- D. Ribatti, Exp. Cell Res., 2014, 328, 314–324 CrossRef CAS PubMed.
- D. Zosen, M. G. Hadera, J. S. Lumor, J. M. Andersen and R. E. Paulsen, J. Pharmacol. Toxicol. Methods, 2021, 112, 107105 CrossRef CAS PubMed.
- D. Ribatti, T. Annese and R. Tamma, Microvasc. Res., 2020, 131, 104026 CrossRef CAS PubMed.
- S. Ghimire, X. Zhang, J. Zhang and C. Wu, Chem. Res. Toxicol., 2022, 35, 550–568 Search PubMed.
- P. Nowak-Sliwinska, T. Segura and M. L. Iruela-Arispe, Angiogenesis, 2014, 17, 779–804 CrossRef CAS PubMed.
- P. Rous and J. B. Murphy, J. Am. Med. Assoc., 1911, 56, 741–742 Search PubMed.
- K. H. Kain, J. W. I. Miller, C. R. Jones-Paris, R. T. Thomason, J. D. Lewis, D. M. Bader, J. V. Barnett and A. Zijlstra, Dev. Dyn., 2014, 243, 216–228 CrossRef PubMed.
- N. A. Lokman, A. S. F. Elder, C. Ricciardelli and M. K. Oehler, Int. J. Mol. Sci., 2012, 13, 9959–9970 CrossRef CAS PubMed.
- R. A. Ranjan, J. K. Muenzner, P. Kunze, C. I. Geppert, M. Ruebner, H. Huebner, P. A. Fasching, M. W. Beckmann, T. Bäuerle, A. Hartmann, W. Walther, M. Eckstein, R. Erber and R. Schneider-Stock, Cancers, 2023, 15, 1704 CrossRef CAS PubMed.
- Brain Tumour Research, https://www.braintumourresearch.org/glioblastoma-multiforme, (accessed 30 May 2023).
- A. Kapara, V. Brunton, D. Graham and K. Faulds, Chem. Sci., 2020, 11, 5819–5829 RSC.
- F. Nicolson, L. E. Jamieson, S. Mabbott, K. Plakas, N. C. Shand, M. R. Detty, D. Graham and K. Faulds, Chem. Commun., 2018, 54, 8530–8533 RSC.
- F. Nicolson, M. F. Kircher, N. Stone and P. Matousek, Chem. Soc. Rev., 2021, 50, 566–568 RSC.
- F. Nicolson, L. E. Jamieson, S. Mabbott, K. Plakas, N. C. Shand, M. R. Detty, D. Graham and K. Faulds, Chem. Sci., 2018, 9, 3788–3792 RSC.
- M. E. Berry, S. M. McCabe, S. Sloan-Dennison, S. Laing, N. C. Shand, D. Graham and K. Faulds, ACS Appl. Mater. Interfaces, 2022, 14, 31613–31624 CrossRef CAS PubMed.
- F. Nicolson, B. Andreiuk, C. Andreou, H.-T. Hsu, S. Rudder and M. F. Kircher, Theranostics, 2019, 9, 5899–5913 CrossRef CAS PubMed.
- M. Tucić, V. Stamenković and P. Andjus, Front. Cell Dev. Biol., 2021, 9, 674199 CrossRef PubMed.
- S. M. McCabe, G. Q. Wallace, S. Sloan-Dennison, W. J. Tipping, N. C. Shand, D. Graham, M. Boyd and K. Faulds, Analyst, 2023, 148, 3247–3256 RSC.
- P. Dey, A. Vaideanu, S. Mosca, M. Salimi, B. Gardner, F. Palombo, I. Uchegbu, J. Baumberg, A. Schatzlein, P. Matousek and N. Stone, Nanotheranostics, 2022, 6, 337–349 CrossRef PubMed.
- J. Turkevich, P. C. Stevenson and J. Hillier, Discuss. Faraday Soc., 1951, 11, 55–75 RSC.
- J. F. Li, X. D. Tian, S. B. Li, J. R. Anema, Z. L. Yang, Y. Ding, Y. F. Wu, Y. M. Zeng, Q. Z. Chen, B. Ren, Z. L. Wang and Z. Q. Tian, Nat. Protoc., 2013, 8, 52 CrossRef CAS PubMed.
- J. F. Li, Y. F. Huang, Y. Ding, Z. L. Yang, S. B. Li, X. S. Zhou, F. R. Fan, W. Zhang, Z. Y. Zhou, D. Y. Wu, B. Ren, Z. L. Wang and Z. Q. Tian, Nature, 2010, 464, 392–395 CrossRef CAS PubMed.
- G. Orend and R. Chiquet-Ehrismann, Cancer Lett., 2006, 244, 143–163 CrossRef CAS PubMed.
- Q. Zhang, B. Xu, F. Hu, X. Chen, X. Liu, Q. Zhang and Y. Zuo, J. Mol. Neurosci., 2021, 71, 1636–1647 CrossRef CAS PubMed.
- H. Li, T. Ning, F. Yu, Y. Chen, B. Zhang and S. Wang, Molecules, 2021, 26, 921 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4an00617h |
| This journal is © The Royal Society of Chemistry 2024 |