A heteromultivalent host–guest sensor array for cell recognition and discrimination†
Xin-Yue
Hu
 *,
Zong-Ying
Hu
,
Jia-Hong
Tian
,
Lin
Shi
,
Fei
Ding
,
Hua-Bin
Li
and
Dong-Sheng
Guo
*,
Zong-Ying
Hu
,
Jia-Hong
Tian
,
Lin
Shi
,
Fei
Ding
,
Hua-Bin
Li
and
Dong-Sheng
Guo
 *
*
College of Chemistry, Key Laboratory of Functional Polymer Materials (Ministry of Education), State Key Laboratory of Elemento-Organic Chemistry, National Demonstration Center for Experimental Chemistry Education, Nankai University, Tianjin, 300071, China. E-mail: huxinyue@nankai.edu.cn; dshguo@nankai.edu.cn
First published on 4th November 2022
Abstract
We present a supramolecular sensor array based on a series of heteromultivalent macrocyclic coassemblies using amphiphilic calixarenes and cyclodextrin as the building blocks for cell recognition. The corresponding cross-reactivity between the coassemblies and cells served as the unique fingerprint for cell classification, and successfully identified the normal cell lines, cancerous cell lines, and cross-contaminated cells.
Rapid characterization of different types of cells is crucial for prognosis,1 designing the therapeutic strategy for the development of precision medicine,2,3 tracking biological events,4 and controlling cell quality.5 The conventional technique used for cell characterization involves the use of highly specific biomarker probes, such as antibodies,6 aptamers,4 and small organic molecules.7 These approaches are effective because of the presence of strong interactions present between the biomarker and the cell surface that help in cell line recognition. However, there are several limitations. For example, biomarkers that are specific to a given cell line are difficult to identify.8 The analytical methods, such as flow cytometery9 or immunohistochemistry,6 used for cell line identification are cost-inefficient, complex, and time-consuming. Researchers have reported the use of materials that exhibit non-specific interactions to develop sensor arrays (“chemical nose or tongue”) to address the problems.10–16 The sensor array works on the principle of non-specific binding (unlike specific sensing) between analytes and an array of cross-reactive receptors.17 Several groups have used fluorescent polymers,18,19 fluorescent proteins,20 gold nanoparticles-DNA21 to construct sensor arrays and successfully identify different cell lines. The multivalent interactions between these materials and cells help in enhancing the selectivity of the array. However, the complex cell surface bears multiple and diverse binding sites.22 A heteromultivalent recognition system23–25 (and not a monovalent, heterotopic, or homomultivalent system) is in high demand, as it can be effectively used to differentiate between various cell types.
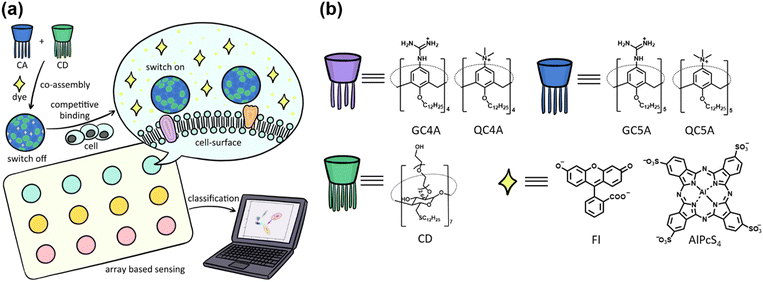
Herein, we propose a supramolecular strategy to achieve heteromultivalent recognition by coassembling amphiphilic cyclodextrin (CD) and calixarene (CA) for cell recognition and differentiation. These heteromultivalent coassemblies showed great binding affinities and selectivity to peptides and proteins,23,24 which showed their potential to identify a larger scaled system. The properties of the coassembly could be effectively tuned, as the constituents of the coassembly could be varied and the ratio of the components could be efficiently tuned.26 We prepared a series of sensor units and constructed a sensor array to optically differentiate cell lines (Scheme 1a) by tuning the CA components and constituent ratios.
Guanidinium and quaternary ammonium-modified calixarenes (GC4A, GC5A, QC4A, and QC5A), and CD were chosen to construct these macrocyclic amphiphile coassemblies (Scheme 1b). The synergistic effect of non-covalent interactions, such as electrostatic interactions, hydrogen bonds, and hydrophobic effects,27 help the four positively charged CAs to bind to anionic guests, which helped to interact with the negatively charged cell membrane. The different cavity sizes and the decoration groups of CAs, and cavities of CD may fit for different binding sites on the cell surface. These factors help achieve the distinct interactions between the coassemblies and cell surface and generate the responses that are unique for each cell type (responses are generated when cells interact with the array). An efficient sensor array should possess effective recognition ability and produce easily observable signals to reflect the recognition events between the sensors and analytes. In the present system, the output signals were generated by conducting the indicator displacement assay (IDA):14 fluorescent dyes were bound to a host that result in fluorescence quenching, and after the addition of the analytes, the competitive binding to hosts caused the release of the dyes and the fluorescence signals were obtained. We chose fluorescein (Fl) and sulfonated aluminum phthalocyanine (AlPcS4) as the reporter dyes (based on the structural characteristics of the CAs) to generate the fluorescent signals.
The hydrated diameters of the four coassemblies were in the range of 45–65 nm and the surface potential was approximately 45 mV (Fig. S1a and b, ESI†). Significant fluorescence quenching could be achieved by binding the two dyes with the co-assemblies (Fig. S1c and d, ESI†), which helped promote the fabrication of switch-on sensors.
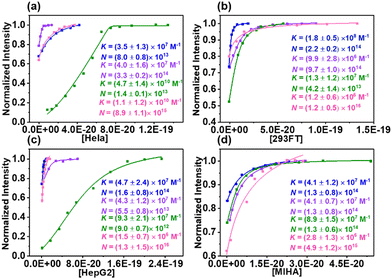
Before constructing sensor array, it is necessary to validate whether the CA–CD coassemblies can exhibit variable fluorescence responses while interacting with multiple cell lines. The cross-reactivity between the sensors and analytes forms the basis of differential sensing. Four cell lines, human cervical cancer cells (HeLa), human embryonic kidney cells (293FT), human liver cancer cells (HepG2), and human normal liver cells (MIHA), were chosen for the experiments. These four cell lines have different tissue origins and states, which can reflect whether the CA–CD coassemblies exhibit different binding abilities toward different cell lines. We used Fl as the reporter dye to investigate the binding constants between the CA–CD coassemblies and cells (Fig. S2–S5, ESI†). IDA was carried out during which the fluorescence intensities increased gradually post the addition of the cells. The increase in the fluorescent intensities can be attributed to the displacement of Fl from the CA cavities by cell surfaces (Fig. 1). We fit these data to an N:1 competitive binding model (which assumes different binding sites to be identical and independent to simplify the fitting procedure) to obtain the quantified results.28 The fitted binding constants (K values that reflect the binding strength of a single unit of CA–CD coassembly bearing a single site on the cell surface) and N values (which reflect the number of binding sites) were determined. The properties of the cell surface are complex. Analysis of the K and N values can help quantify the differences in the recognition properties and abilities of the coassemblies when they interact with various types of cells. The same coassembly also exhibits different K and N values for different cell lines. These orthogonal recognition data form the chemical basis for the construction of a host–guest chemical nose for the efficient differentiation of cells.
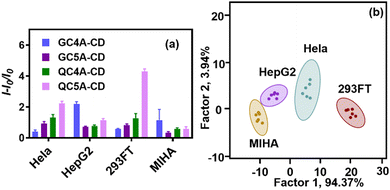
It is important to identify the species and nature (cancerous or non-cancerous) of cells in real conditions. These four cell lines were used to conduct a preliminary test to determine whether the sensor array consisting of the coassemblies (AlPcS4 as the reporter dyes) could identify the different cell lines. The four sensor units exhibited changes in the fluorescence intensities post the addition of the cells, and each cell line exhibited a unique response pattern (Fig. 2a). The linear discriminant analysis (LDA) method was used to statistically characterize the changes in the fluorescence intensities. The two canonical factors contained 94.37% and 3.94% of the variation (accuracy of cell type identification: 100%; Fig. 2b). The normal cell lines (MIHA and 293FT) and the cancerous cell lines (HeLa and HepG2) could be identified.
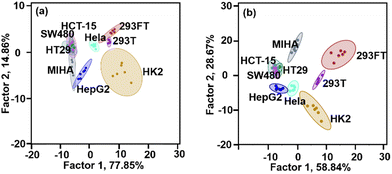
We further tested if the heteromultivalent sensor array could be used to identify more cell lines. Human kidney cells (HK-2), human embryonic kidney cells (293T), human colon cancer cells (SW480), human colon cancer cells (HCT-15), and human colon cancer cells (HT29) were added to test the discrimination ability of the sensor array. Fig. 3a shows the results of the LDA analysis conducted using the sensor array constructed using the four CA–CD coassemblies (CA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CD = 1
CD = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The confidence circles of MIHA, HepG2, HT29, SW480, and HCT-12 could not be separated, indicating that the four-channel sensor array could not identify these cell lines. By adding more sensor units, more information between sensor units and analytes will be provided, which enrich the “fingerprint” of each cell.
1). The confidence circles of MIHA, HepG2, HT29, SW480, and HCT-12 could not be separated, indicating that the four-channel sensor array could not identify these cell lines. By adding more sensor units, more information between sensor units and analytes will be provided, which enrich the “fingerprint” of each cell.
Therefore, it is reasonable to expect that different coassemblies exhibit different interaction properties when they interact with the cells. Because of the dynamic nature of assembly, we could add more heteromultivalent sensor units by changing their coassembly ratio instead of synthesizing new amphiphilic macrocyclic compounds.26 We introduced two sensor units (GC4A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CD = 2
CD = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and GC5A
1 and GC5A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CD = 2
CD = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to construct a six-channel sensor array, which could be used to identify seven cell types with 83% accuracy (Fig. 3b). The four normal cell lines were discriminated from the cancerous cell line, and the cells from different cancer were also been discriminated. The three colon cancer cells could not be separately identified. This could be attributed to the fact that the three cancer cell lines had similar origins (all were colon cancer cells). The homologous similarities exhibited by these cells were higher than the homologous similarities exhibited by other cancer and normal cells.
1) to construct a six-channel sensor array, which could be used to identify seven cell types with 83% accuracy (Fig. 3b). The four normal cell lines were discriminated from the cancerous cell line, and the cells from different cancer were also been discriminated. The three colon cancer cells could not be separately identified. This could be attributed to the fact that the three cancer cell lines had similar origins (all were colon cancer cells). The homologous similarities exhibited by these cells were higher than the homologous similarities exhibited by other cancer and normal cells.
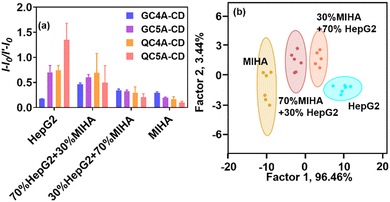
Besides the cell line identification, the discrimination of cancer cells from cancer/normal cell mixtures is also important to monitor the infestation process. We chose HepG2 and MIHA as representative examples of mixed cells because these two cells have their origin in the human liver cell lines. A sensor array constructed from four CA–CD coassemblies (ratio: 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was used to conduct the experiments. The two pure cells and two mixed cells were classified into four distinct clusters (90% identification accuracy, Fig. 4). The cancerous HepG2 cells could be identified in the presence of the normal MIHA cells. This result also showed that the sensor array could discriminate against cancer cells in a semi-quantitative analysis.
1) was used to conduct the experiments. The two pure cells and two mixed cells were classified into four distinct clusters (90% identification accuracy, Fig. 4). The cancerous HepG2 cells could be identified in the presence of the normal MIHA cells. This result also showed that the sensor array could discriminate against cancer cells in a semi-quantitative analysis.
In summary, we developed a heteromultivalent host–guest sensor array for cell identification. The heteromultivalent recognition of cells was achieved by coassembling two types of macrocyclic amphiphiles. The coassembling components can be varied, and the coassembling ratio can be fine-tuned. We studied the recognition abilities of the different coassemblies using different types of cells. The cross-reactivity of the fluorescence output helps in the sensor array-based discrimination of the cell lines (discrimination between normal cell lines and different cancer cell lines). The sensor array could be efficiently used to study the cross-contaminated cells and discriminate the cancer cells following a semiquantitative analysis approach. Numerous sensor units can be fabricated by varying the coassembling components and the reporter dyes. This makes the heteromultivalent coassembly a desirable material for constructing sensor arrays to distinguish complex systems, especially for larger-scale species with multiple and diverse functional sites.
This work was supported by NSFC (22101142 and U20A20259), the Fundamental Research Funds for the Central Universities, and NCC Fund (grant no. NCC2020FH04). The authors also thank Prof. Bart Jan Ravoo at Westfälische Wilhelms-Universität Münster for supplying the amphiphilic cyclodextrin used in this work.
Conflicts of interest
There are no conflicts to declare.Notes and references
- L. A. M. Gravendeel, M. C. M. Kouwenhoven, O. Gevaert, J. J. de Rooi, A. P. Stubbs, J. E. Duijm, A. Daemen, F. E. Bleeker, L. B. C. Bralten, N. K. Kloosterhof, B. De Moor, P. H. C. Eilers, P. J. van der Spek, J. M. Kros, P. A. E. Sillevis Smitt, M. J. van den Bent and P. J. French, Cancer Res., 2009, 69, 9065–9072 CrossRef.
- J. Lamb, E. D. Crawford, D. Peck, J. W. Modell, I. C. Blat, M. J. Wrobel, J. Lerner, J.-P. Brunet, A. Subramanian, K. N. Ross, M. Reich, H. Hieronymus, G. Wei, S. A. Armstrong, S. J. Haggarty, P. A. Clemons, R. Wei, S. A. Carr, E. S. Lander and T. R. Golub, Science, 2006, 313, 1929 CrossRef PubMed.
- A. B. Parsons, A. Lopez, I. E. Givoni, D. E. Williams, C. A. Gray, J. Porter, G. Chua, R. Sopko, R. L. Brost, C.-H. Ho, J. Wang, T. Ketela, C. Brenner, J. A. Brill, G. E. Fernandez, T. C. Lorenz, G. S. Payne, S. Ishihara, Y. Ohya, B. Andrews, T. R. Hughes, B. J. Frey, T. R. Graham, R. J. Andersen and C. Boone, Cell, 2006, 126, 611–625 CrossRef.
- M. L. Whitfield, G. Sherlock, A. J. Saldanha, J. I. Murray, C. A. Ball, K. E. Alexander, J. C. Matese, C. M. Perou, M. M. Hurt, P. O. Brown and D. Botstein, Mol. Biol. Cell, 2002, 13, 1977–2000 CrossRef CAS.
- J. L. Page, M. C. Johnson, K. M. Olsavsky, S. C. Strom, H. Zarbl and C. J. Omiecinski, Toxicol. Sci., 2007, 97, 384–397 CrossRef CAS.
- C. Solier and H. Langen, Proteomics, 2014, 14, 774–783 CrossRef CAS.
- R. Kubota and I. Hamachi, Chem. Soc. Rev., 2015, 44, 4454–4471 RSC.
- A. Vidyasagar, N. A. Wilson and A. Djamali, Fibrog. Tissue Repair, 2012, 5, 7 CrossRef CAS.
- A. Adan, G. Alizada, Y. Kiraz, Y. Baran and A. Nalbant, Crit. Rev. Biotechnol., 2017, 37, 163–176 CrossRef CAS PubMed.
- J. Freudenberg, F. Hinkel, D. Jänsch and U. H. F. Bunz, Top. Curr. Chem., 2017, 375, 67 CrossRef.
- Z. Jiang, N. D. B. Le, A. Gupta and V. M. Rotello, Chem. Soc. Rev., 2015, 44, 4264–4274 RSC.
- L. You, D. Zha and E. V. Anslyn, Chem. Rev., 2015, 115, 7840–7892 CrossRef CAS.
- H. Bai, Z. Liu, T. Zhang, J. Du, C. Zhou, W. He, J. H. C. Chau, R. T. K. Kwok, J. W. Y. Lam and B. Z. Tang, ACS Nano, 2020, 14, 7552–7563 CrossRef CAS.
- J. Han, H. Cheng, B. Wang, M. S. Braun, X. Fan, M. Bender, W. Huang, C. Domhan, W. Mier, T. Lindner, K. Seehafer, M. Wink and U. H. F. Bunz, Angew. Chem., Int. Ed., 2017, 56, 15246–15251 CrossRef CAS.
- J. Chen, B. L. Hickey, L. Wang, J. Lee, A. D. Gill, A. Favero, R. Pinalli, E. Dalcanale, R. J. Hooley and W. Zhong, Nat. Chem., 2021, 13, 488–495 CrossRef CAS PubMed.
- C. Jirayupat, K. Nagashima, T. Hosomi, T. Takahashi, B. Samransuksamer, Y. Hanai, A. Nakao, M. Nakatani, J. Liu, G. Zhang, W. Tanaka, M. Kanai, T. Yasui, Y. Baba and T. Yanagida, Chem. Commun., 2022, 58, 6377–6380 RSC.
- Y. Geng, W. J. Peveler and V. M. Rotello, Angew. Chem., Int. Ed., 2019, 58, 5190–5200 CrossRef.
- A. Bajaj, O. R. Miranda, I.-B. Kim, R. L. Phillips, D. J. Jerry, U. H. F. Bunz and V. M. Rotello, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 10912–10916 CrossRef.
- S. Tomita, S. Ishihara and R. Kurita, ACS Appl. Mater. Interfaces, 2019, 11, 6751–6758 CrossRef PubMed.
- A. Bajaj, S. Rana, O. R. Miranda, J. C. Yawe, D. J. Jerry, U. H. F. Bunz and V. M. Rotello, Chem. Sci., 2010, 1, 134–138 RSC.
- X. Yang, J. Li, H. Pei, Y. Zhao, X. Zuo, C. Fan and Q. Huang, Anal. Chem., 2014, 86, 3227–3231 CrossRef CAS.
- J. Huskens, L. Prins, R. Haag and B. J. Ravoo, Multivalency concepts, research & applications, Wiley-VCH, New Jersey, 2017 Search PubMed.
- Y.-C. Pan, H. Wang, X. Xu, H.-W. Tian, H. Zhao, X.-Y. Hu, Y. Zhao, Y. Liu, G. Ding, Q. Meng, B. J. Ravoo, T. Zhang and D.-S. Guo, CCS Chem., 2020, 2, 2485–2497 Search PubMed.
- Z. Xu, S. Jia, W. Wang, Z. Yuan, B. Jan Ravoo and D. S. Guo, Nat. Chem., 2019, 11, 86–93 CrossRef CAS PubMed.
- H. Wang, X. Xu, Y.-C. Pan, Y. Yan, X.-Y. Hu, R. Chen, B. J. Ravoo, D.-S. Guo and T. Zhang, Adv. Mater., 2021, 33, 2006483 CrossRef CAS.
- J.-H. Tian, X.-Y. Hu, Z.-Y. Hu, H.-W. Tian, J.-J. Li, Y.-C. Pan, H.-B. Li and D.-S. Guo, Nat. Commun., 2022, 13, 4293 CrossRef.
- Z. Zheng, W.-C. Geng, J. Gao, Y.-Y. Wang, H. Sun and D.-S. Guo, Chem. Sci., 2018, 9, 2087–2091 RSC.
- G. Ghale, A. G. Lanctôt, H. T. Kreissl, M. H. Jacob, H. Weingart, M. Winterhalter and W. M. Nau, Angew. Chem., Int. Ed., 2014, 53, 2762–2765 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2cc04963e |
| This journal is © The Royal Society of Chemistry 2022 |