Open Access Article
Open Access ArticleAmino acid ester-coupled caffeoylquinic acid derivatives as potential hypolipidemic agents: synthesis and biological evaluation†
Xi Zhang‡
,
Dong-yun Liu‡,
Hai Shang,
Yi Jia,
Xu-Dong Xu,
Yu Tian * and
Peng Guo*
* and
Peng Guo*
Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences, Peking Union Medical College, No. 151, Malianwa North Road, Haidian District, Beijing 100193, P. R. China. E-mail: ytian@implad.ac.cn; 18010100068@163.com
First published on 5th January 2021
Abstract
Pandanus tectorius (L.) Parkins. (PTPs) is rich in caffeoylquinic acids and amino acids, especially some essential amino acids, such as valine, phenylalanine, and so forth. A series of novel amino acid ester-coupled caffeoylquinic acid derivatives have been designed and synthesized. Biological evaluation suggested that some amino acid ester-coupled derivatives exhibited varying degrees of lipid-lowering effects on oleic acid-elicited lipid accumulation in HepG2 liver cells. Particularly, derivatives 6c, 6d, 6e and 6f exhibited comparable potential lipid-lowering effect with the positive control simvastatin and chlorogenic acid. Further studies on the mechanism of 6c, 6d, 6e and 6f revealed that the lipid-lowering effects were related to their regulation of TG levels and mRNA levels of lipometabolic-modulating genes, and merit further investigation.
Introduction
Hyperlipidemia, one of the most common metabolic and endocrine disorder diseases, is characterized by the decreasing concentration of high density lipoprotein cholesterol (HDL-C) and increasing concentration of triglycerides (TG), total cholesterol (TC), and low density lipoprotein cholesterol (LDL-C).1–4 It is well known that hyperlipidemia is one of the key risk factors for cerebral and cardiovascular diseases, and the increasing levels of TG show a significant influence on coronary heart disease (CHD) and atherosclerosis (AS).5–7 Several studies have provided evidence that sterol-regulatory element binding protein 1 (SREBP1) is involved in low-density lipoprotein gene transcription and sterol synthesis, and acetyl-CoA carboxylase (ACC) and fatty acid synthase (FASN) are responsible for fatty acid synthesis; these three genes are all associated with lipid synthesis.8–11 Therefore, it is quite beneficial to research and explore anti-hypolipidemic agents that could modulate the dysregulation of lipid metabolism, and decrease the elevated levels of serum TG, the gene SREBP1, ACC, and FASN.Pandanus tectorius (L.) Parkins. (PTPs, Fig. 1), shaped like a pineapple, is rich in phenolic acids and amino acids, and often used as a traditional medicine in the folk for treating leprosy, bronchitis, measles, dermatitis and diabetes diseases.12–14 The content of amino acids in PTPs fruit is as high as 10.5 μg g−1, including seven high content of essential amino acids, such as leucine (1.0 μg g−1), valine (0.8 μg g−1), phenylalanine (0.5 μg g−1) and so on. These essential amino acids, which must be supplied by food, participate in many physiological functions of the human body.14 As the ester of caffeic acid and quinic acid, chlorogenic acid (3-O-caffeoylquinic acid, CA, 1, Fig. 1) was also found abundantly in PTPs. CA was reported to have multiple potential activities, including antioxidant, anti-inflammatory, antimicrobial, and anti-hyperglycemic effects.15–18 Our previous researches revealed that the caffeoylquinic acid analogues extracted from PTPs were confirmed to possess obvious anti-hyperlipidemia activity, and could significantly inhibit lipid accumulation and TG levels in the liver.13,19 In addition, we also found that CA derivatives (chlorogenic acid diketal, 2, Fig. 1) could decrease lipid accumulation in HepG2 cells, with the ketal substituent on the 1,4,5-position hydroxyl and carboxyl group.20 Recently, our preliminary SARs of chlorogenic acid suggested that the phenolic hydroxyl, not the carboxyl group, on the caffeic acid moiety is vital for the lipid-lowering activity of CA, and CA amide derivatives (3 and 4, Fig. 1) exhibited more potential lipid-lowering effect than the positive control simvastatin and chlorogenic acid.20,21 However, despite the anticipated pharmacological properties of caffeoylquinic acid amide analogues, synthesis and investigation of the novel amino acid ester-coupled caffeoylquinic acid derivatives lipid-decreasing effects has never been researched yet.
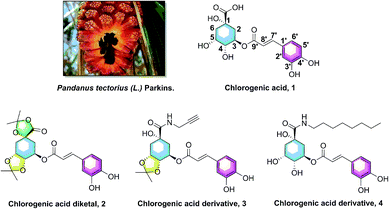 | ||
| Fig. 1 The Pandanus tectorius (L.) Parkins. fruit and the structures of chlorogenic acid (1), and its active analogues chlorogenic acid diketal (2), derivatives 3 and 4. | ||
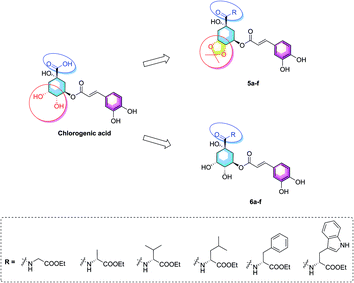
Considering of the high content of essential amino acids in PTPs fruit and the difference structures of amino acids, herein, we choose six kind of ethyl amino acid esters as the coupling groups (glycine, alanine, valine, leucine, phenylalanine, and tryptophan ethyl esters) which were coupled to the carboxyl group on 1-position of chlorogenic acid, and report the synthesis of twelve novel amino acid ester-coupled caffeoylquinic acid derivatives with various amino acid esters. Meanwhile, in consideration of diketal analogues of chlorogenic acid attenuated lipid accumulation, we design the derivatives with 4,5-position hydroxyl groups substituted by ketal (isopropylidene). Therefore, this research was aim to synthesize novel amino acid ester-coupled caffeoylquinic acid derivatives (5a–f, 6a–f, Fig. 2) and investigate the effects of derivatives on oleic acid-elicited lipid accumulation in HepG2 cells. The underlying lipid-lowering effect mechanisms of the amino acid ester-coupled derivatives were also elucidated by intracellular TG quantification and quantitative real-time PCR.
Results and discussion
Chemistry
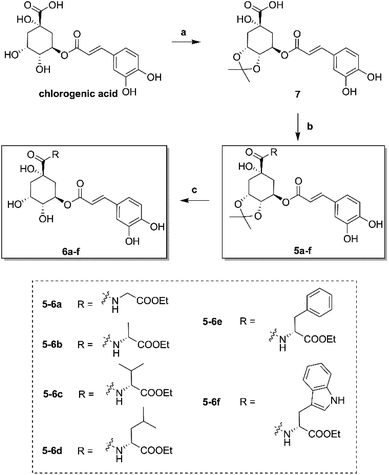
The synthesis of amino acid ester-coupled caffeoylquinic acids derivatives 5a–f and 6a–f were outlined in Scheme 1. Chlorogenic acid (1) was treated with dimethyoxypropane (DMP) and p-toluenesulfonic acid (TsOH) in dry acetone to gain chlorogenic acid ketal the intermediate 7. Then the target products 5a–f were attained via amidation with various amino acid ethyl esters hydrochloride (glycine ethyl ester hydrochloride, L-alanine ethyl ester hydrochloride, L-valine ethyl ester hydrochloride, L-leucine ethyl ester hydrochloride, L-phenylalanine ethyl ester hydrochloride, L-tryptophan ethyl ester hydrochloride) of carboxyl group on 1-position of intermediate 7 in BOP (benzotriazol-1-yl-oxytris(dimethylamino)phosphoniumhexa-fluorophosphate) and DIEA (dimethyltriethylamine) conditions, and then followed by deprotection of ketal with TFA (trifluoroacetic acid) to obtain another series of target products 6a–f.Biological results and discussion
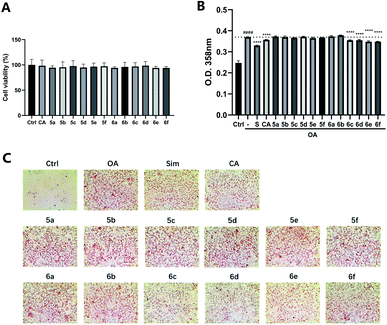
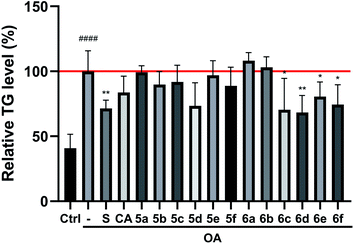
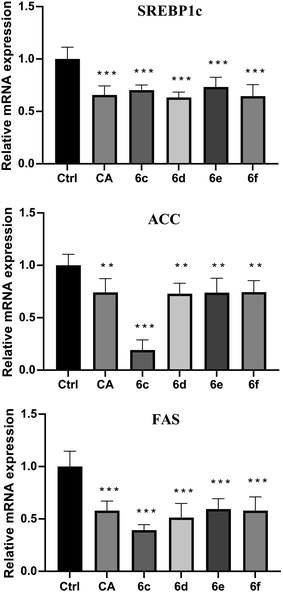
The preliminary structure–activity relationships (SARs) suggested that, compared to the positive group Sim and CA, the amino acid ester-coupled caffeoylquinic acids derivatives with 4,5-position hydroxyl groups substituted by ketal were weaker potent than those with 4,5-position hydroxyl groups exposed. Beyond that, the derivatives 6c, 6d, 6e and 6f with valine, leucine, phenylalanine, and tryptophan esters groups could better regulate lipid accumulation after oleic acid treatment, indicating that the introduction of valine, leucine, phenylalanine, and tryptophan esters groups could ameliorate lipid accumulation. Among them, derivative 6c and 6d exhibited more potent lipid-lowering effects, compared to 6e and 6f, which means that aliphatic group with on the amino acid is sort of favourable substituent for regulation lipid accumulation than aromatic group in HepG2 liver cells. However, derivatives 6a and 6b, including glycine and alanine esters groups, were inert, suggesting that such unbranched aliphatic glycine and alanine esters groups was negative factor for the derivatives. In addition, the above derivatives showed lipid-lowering effects in Fig. 3 and 4, illustrating that blockage of 4,5-position hydroxyl groups on the chlorogenic acid with isopropylidene may deteriorate regulating lipid effects, such as compound 5c vs. 6c and 5e vs. 6e, and different amino acid esters groups on 1-position carboxyl group influenced lipid-lowering activities obviously.
Experimental
General information
All the reagents were used without further purification unless otherwise specified. Solvents were dried and redistilled before used in the reaction. Analytical TLC was performed using silica gel HF254. Preparative column chromatography was performed with silica gel H. 1H and 13C NMR spectra were recorded on a Bruker Advance III 600 MHz spectrometer. HRMS were obtained on a Thermofisher LTQ-Obitrap XL. Chlorogenic acid, DMP (dimethyoxypropane), TsOH (p-toluenesulfonic acid), BOP (benzotriazol-1-yl-oxytris (dimethylamino)phosphonium-hexa-fluorophosphate), DIEA (dimethyltriethylamine), glycine ethyl ester hydrochloride, L-alanine ethyl ester hydrochloride, L-valine ethyl ester hydrochloride, L-leucine ethyl ester hydrochloride, L-phenylalanine ethyl ester hydrochloride, L-tryptophan ethyl ester hydrochloride, TFA (trifluoroacetic acid), DCM (dichloromethane) were purchased from the InnoChem Company. Dulbecco's Modified Eagle Medium (DMEM), fetal bovine serum were purchased from Corning Inc (CA, USA). Penicillin and streptomycin were procured from Hyclone (Logan, Utah, USA). Simvastatin, oil-red O, oleic acid (OA) and dimethyl sulphoxide DMSO were purchased from Sigma-Aldrich (St. Louis, MO, USA). The kits for triglyceride (TG) were purchased from Jian Cheng Biotechnology Company (Nanjing, China). Total RNA extraction reagent Trizol (Ambion, USA), PrimeScript RT reagent kit and SYBR-Green PCR kit were purchased from Transgene Biotech, Inc. (Beijing, China).Chemistry
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to offer pure light yellow solid compound 7 (9.6 g, 82% yield).
1) to offer pure light yellow solid compound 7 (9.6 g, 82% yield).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to obtain pure compounds 5a–f as white solid.
1) to obtain pure compounds 5a–f as white solid.![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2CH3), 3.95 (s, 2H, NH–C
2CH3), 3.95 (s, 2H, NH–C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 2.38–2.35 (m, 1H, H-2), 2.19–2.16 (m, 1H, H-2), 2.02–1.99 (m, 1H, H-6), 1.94–1.89 (m, 1H, H-6), 1.53 (s, 3H, CH3–C–O), 1.35 (s, 3H, C
2), 2.38–2.35 (m, 1H, H-2), 2.19–2.16 (m, 1H, H-2), 2.02–1.99 (m, 1H, H-6), 1.94–1.89 (m, 1H, H-6), 1.53 (s, 3H, CH3–C–O), 1.35 (s, 3H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3–C–O), 1.26 (t, J = 7.1 Hz, 3H, COOCH2C
3–C–O), 1.26 (t, J = 7.1 Hz, 3H, COOCH2C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3); 13C-NMR (150 MHz, MeOD) δ: 178.5 (
3); 13C-NMR (150 MHz, MeOD) δ: 178.5 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ONH), 171.2 (
ONH), 171.2 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) OOEt), 168.6 (C-9′), 149.6 (C-4′), 147.2 (C-7′), 146.8(C-3′), 127.7 (C-1′), 123.1 (C-6′), 116.5 (C-5′), 115.1 (C-2′), 115.0 (C-8′), 110.4 (O–C–O), 78.6 (C-3), 76.1 (C-1), 75.5 (C-4), 72.5 (C-5), 62.4 (COO
OOEt), 168.6 (C-9′), 149.6 (C-4′), 147.2 (C-7′), 146.8(C-3′), 127.7 (C-1′), 123.1 (C-6′), 116.5 (C-5′), 115.1 (C-2′), 115.0 (C-8′), 110.4 (O–C–O), 78.6 (C-3), 76.1 (C-1), 75.5 (C-4), 72.5 (C-5), 62.4 (COO![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2CH3), 42.0 (NH
H2CH3), 42.0 (NH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 37.7 (C-2), 35.3 (C-6), 28.4 (
H2), 37.7 (C-2), 35.3 (C-6), 28.4 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3–C–O), 26.2 (
H3–C–O), 26.2 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3–C–O), 14.4 (COOCH2
H3–C–O), 14.4 (COOCH2![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3); HRMS (ESI): calcd for [M + Na]+ C23H29NNaO10: 502.1689, found 502.1691.
H3); HRMS (ESI): calcd for [M + Na]+ C23H29NNaO10: 502.1689, found 502.1691.![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) CH3), 4.21–4.16 (m, 3H, H-5, COOC
CH3), 4.21–4.16 (m, 3H, H-5, COOC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2CH3), 2.35–2.31 (m, 1H, H-2), 2.18–2.15 (m, 1H, H-2), 1.97–1.90 (m, 2H, H-6), 1.53 (s, 3H, CH3–C–O), 1.42 (d, J = 7.3 Hz, 3H, NHCHC
2CH3), 2.35–2.31 (m, 1H, H-2), 2.18–2.15 (m, 1H, H-2), 1.97–1.90 (m, 2H, H-6), 1.53 (s, 3H, CH3–C–O), 1.42 (d, J = 7.3 Hz, 3H, NHCHC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3), 1.35 (s, 3H, C
3), 1.35 (s, 3H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3–C–O), 1.27 (t, J = 7.2 Hz, COOCH2C
3–C–O), 1.27 (t, J = 7.2 Hz, COOCH2C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3); 13C-NMR (150 MHz, MeOD) δ: 177.6 (
3); 13C-NMR (150 MHz, MeOD) δ: 177.6 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ONH), 173.9 (
ONH), 173.9 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) OOEt), 168.6 (C-9′), 149.7 (C-4′), 147.2 (C-7′), 146.8 (C-3′), 127.7 (C-1′), 123.0 (C-6′), 116.5 (C-5′), 115.1 (C-2′), 115.0 (C-8′), 110.4 (O–C–O), 78.6 (C-3), 76.0 (C-1), 75.4 (C-4), 72.5 (C-5), 62.5 (COO
OOEt), 168.6 (C-9′), 149.7 (C-4′), 147.2 (C-7′), 146.8 (C-3′), 127.7 (C-1′), 123.0 (C-6′), 116.5 (C-5′), 115.1 (C-2′), 115.0 (C-8′), 110.4 (O–C–O), 78.6 (C-3), 76.0 (C-1), 75.4 (C-4), 72.5 (C-5), 62.5 (COO![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2CH3), 49.5 (NH
H2CH3), 49.5 (NH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) HCH3), 37.6 (C-2), 35.3 (C-6), 28.4 (
HCH3), 37.6 (C-2), 35.3 (C-6), 28.4 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3–C–O), 26.2 (
H3–C–O), 26.2 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3–C–O), 17.5 (NHCH
H3–C–O), 17.5 (NHCH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3), 14.4 (COOCH2
H3), 14.4 (COOCH2![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3); HRMS (ESI): calcd for [M + Na]+ C24H31NNaO10: 516.1846, found 516.1846.
H3); HRMS (ESI): calcd for [M + Na]+ C24H31NNaO10: 516.1846, found 516.1846.![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ), 4.25–4.16 (m, 3H, H-5, COOC
), 4.25–4.16 (m, 3H, H-5, COOC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2CH3), 2.36–2.33 (m, 1H, H-2), 2.22–2.14 (m, 2H, H-2, NHCHC
2CH3), 2.36–2.33 (m, 1H, H-2), 2.22–2.14 (m, 2H, H-2, NHCHC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ), 1.96–1.91 (m, 2H, H-6), 1.53 (s, 3H, CH3–C–O), 1.34 (s, 3H, C
), 1.96–1.91 (m, 2H, H-6), 1.53 (s, 3H, CH3–C–O), 1.34 (s, 3H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3–C–O), 1.28 (t, J = 7.1 Hz, COOCH2C
3–C–O), 1.28 (t, J = 7.1 Hz, COOCH2C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3), 0.97–0.92 (m, 6H, 2 × CH3); 13C-NMR (150 MHz, MeOD) δ: 177.7 (
3), 0.97–0.92 (m, 6H, 2 × CH3); 13C-NMR (150 MHz, MeOD) δ: 177.7 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ONH), 172.7 (
ONH), 172.7 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) OOEt), 168.6 (C-9′), 149.7 (C-4′), 147.3 (C-7′), 146.9 (C-3′), 127.6 (C-1′), 123.1 (C-6′), 116.5 (C-5′), 115.1 (C-2′), 115.0 (C-8′), 110.4 (O–C–O), 78.6 (C-3), 76.2 (C-1), 75.4 (C-4), 72.4 (C-5), 62.4 (COO
OOEt), 168.6 (C-9′), 149.7 (C-4′), 147.3 (C-7′), 146.9 (C-3′), 127.6 (C-1′), 123.1 (C-6′), 116.5 (C-5′), 115.1 (C-2′), 115.0 (C-8′), 110.4 (O–C–O), 78.6 (C-3), 76.2 (C-1), 75.4 (C-4), 72.4 (C-5), 62.4 (COO![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2CH3), 58.8 (NH
H2CH3), 58.8 (NH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H), 37.9 (C-2), 35.1 (C-6), 32.1 (NHCH
H), 37.9 (C-2), 35.1 (C-6), 32.1 (NHCH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H), 28.4 (
H), 28.4 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3–C–O), 26.2 (
H3–C–O), 26.2 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3–C–O), 19.4 (CH
H3–C–O), 19.4 (CH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3), 18.2 (CH
H3), 18.2 (CH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3), 14.5 (COOCH2
H3), 14.5 (COOCH2![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3); HRMS (ESI): calcd for [M + Na]+ C26H35NNaO10: 544.2159, found 544.2161.
H3); HRMS (ESI): calcd for [M + Na]+ C26H35NNaO10: 544.2159, found 544.2161.![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ), 4.21–4.14 (m, 3H, H-5, COOC
), 4.21–4.14 (m, 3H, H-5, COOC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2CH3), 2.35–2.31 (m, 1H, H-2), 2.18–2.15 (m, 1H, H-2), 1.94–1.92 (m, 2H, H-6), 1.71–1.64 (m, 3H, NHCHC
2CH3), 2.35–2.31 (m, 1H, H-2), 2.18–2.15 (m, 1H, H-2), 1.94–1.92 (m, 2H, H-6), 1.71–1.64 (m, 3H, NHCHC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2C
2C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ), 1.53 (s, 3H, CH3–C–O), 1.34 (s, 3H, C
), 1.53 (s, 3H, CH3–C–O), 1.34 (s, 3H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3–C–O), 1.26 (t, J = 7.1 Hz, COOCH2C
3–C–O), 1.26 (t, J = 7.1 Hz, COOCH2C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3), 0.95–0.94 (m, 3H, CH3), 0.92–0.91 (s, 3H, CH3); 13C-NMR (150 MHz, MeOD) δ: 177.8 (CONH), 173.9 (
3), 0.95–0.94 (m, 3H, CH3), 0.92–0.91 (s, 3H, CH3); 13C-NMR (150 MHz, MeOD) δ: 177.8 (CONH), 173.9 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) OOEt), 168.6 (C-9′), 149.7 (C-4′), 147.3 (C-7′), 146.8 (C-3′), 127.7 (C-1′), 123.1 (C-6′), 116.5 (C-5′), 115.1 (C-2′), 115.0 (C-8′), 110.4 (O–C–O), 78.6 (C-3), 76.1 (C-1), 75.4 (C-4), 72.5 (C-5), 62.4 (COO
OOEt), 168.6 (C-9′), 149.7 (C-4′), 147.3 (C-7′), 146.8 (C-3′), 127.7 (C-1′), 123.1 (C-6′), 116.5 (C-5′), 115.1 (C-2′), 115.0 (C-8′), 110.4 (O–C–O), 78.6 (C-3), 76.1 (C-1), 75.4 (C-4), 72.5 (C-5), 62.4 (COO![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2CH3), 52.1 (NH
H2CH3), 52.1 (NH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H), 41.4 (NHCH
H), 41.4 (NHCH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 37.7 (C-2), 35.3 (C-6), 28.4 (
H2), 37.7 (C-2), 35.3 (C-6), 28.4 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3–C–O), 26.2 (
H3–C–O), 26.2 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3–C–O), 26.0 (
H3–C–O), 26.0 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H(CH3)2), 23.3 (CH
H(CH3)2), 23.3 (CH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3), 21.9 (CH
H3), 21.9 (CH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3), 14.5 (COOCH2
H3), 14.5 (COOCH2![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3); HRMS (ESI): calcd for [M + Na]+ C27H37NNaO10: 558.2315, found 558.2319.
H3); HRMS (ESI): calcd for [M + Na]+ C27H37NNaO10: 558.2315, found 558.2319.![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ), 4.52–4.50 (m, 1H, H-4), 4.19–4.14 (m, 3H, H-5, COOC
), 4.52–4.50 (m, 1H, H-4), 4.19–4.14 (m, 3H, H-5, COOC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2CH3), 3.19 (dd, J = 13.8 Hz, 5.6 Hz, 1H, ph′′C
2CH3), 3.19 (dd, J = 13.8 Hz, 5.6 Hz, 1H, ph′′C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 3.06 (dd, J = 13.8 Hz, 8.3 Hz, 1H, ph′′C
2), 3.06 (dd, J = 13.8 Hz, 8.3 Hz, 1H, ph′′C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 2.32–2.29 (m, 1H, H-2), 2.11–2.08 (m, 1H, H-2), 1.76–1.68 (m, 2H, H-6), 1.51 (s, 3H, CH3–C–O), 1.34 (s, 3H, CH3–C–O), 1.24 (t, J = 7.2 Hz, COOCH2C
2), 2.32–2.29 (m, 1H, H-2), 2.11–2.08 (m, 1H, H-2), 1.76–1.68 (m, 2H, H-6), 1.51 (s, 3H, CH3–C–O), 1.34 (s, 3H, CH3–C–O), 1.24 (t, J = 7.2 Hz, COOCH2C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3); 13C-NMR (150 MHz, MeOD) δ: 177.4 (
3); 13C-NMR (150 MHz, MeOD) δ: 177.4 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ONH), 172.7 (
ONH), 172.7 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) OOEt), 168.6 (C-9′), 149.7 (C-4′), 147.3 (C-7′), 146.8 (C-3′), 137.7 (C–Ph′′), 130.4 (2C–Ph′′), 129.6 (2C–Ph′′), 128.0 (C–Ph′′), 127.7 (C-1′), 123.1 (C-6′), 116.5 (C-5′), 115.2 (C-2′), 115.1 (C-8′), 110.4 (O–C–O), 78.5 (C-3), 76.0 (C-1), 75.4 (C-4), 72.5 (C-5), 62.6 (COO
OOEt), 168.6 (C-9′), 149.7 (C-4′), 147.3 (C-7′), 146.8 (C-3′), 137.7 (C–Ph′′), 130.4 (2C–Ph′′), 129.6 (2C–Ph′′), 128.0 (C–Ph′′), 127.7 (C-1′), 123.1 (C-6′), 116.5 (C-5′), 115.2 (C-2′), 115.1 (C-8′), 110.4 (O–C–O), 78.5 (C-3), 76.0 (C-1), 75.4 (C-4), 72.5 (C-5), 62.6 (COO![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2CH3), 54.8 (NH
H2CH3), 54.8 (NH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) HCH2), 38.4 (NHCH
HCH2), 38.4 (NHCH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 37.7 (C-2), 35.0 (C-6), 28.4 (
H2), 37.7 (C-2), 35.0 (C-6), 28.4 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3–C–O), 26.2 (
H3–C–O), 26.2 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3–C–O), 14.4 (COOCH2
H3–C–O), 14.4 (COOCH2![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3); HRMS (ESI): calcd for [M + Na]+ C30H35NNaO10: 592.2159, found 592.2163.
H3); HRMS (ESI): calcd for [M + Na]+ C30H35NNaO10: 592.2159, found 592.2163.![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ), 4.50–4.48 (m, 1H, H-4), 4.15–4.09 (m, 3H, H-5, COOC
), 4.50–4.48 (m, 1H, H-4), 4.15–4.09 (m, 3H, H-5, COOC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2CH3), 2.30–2.29 (m, 2H, indole–C
2CH3), 2.30–2.29 (m, 2H, indole–C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 2.30–2.27 (m, 1H, H-2), 2.10–2.07 (m, 1H, H-2), 1.74–1.73 (m, 2H, H-6), 1.50 (s, 3H, CH3–C–O), 1.31 (s, 3H, CH3–C–O), 1.18 (t, J = 7.3 Hz, COOCH2C
2), 2.30–2.27 (m, 1H, H-2), 2.10–2.07 (m, 1H, H-2), 1.74–1.73 (m, 2H, H-6), 1.50 (s, 3H, CH3–C–O), 1.31 (s, 3H, CH3–C–O), 1.18 (t, J = 7.3 Hz, COOCH2C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3); 13C-NMR (150 MHz, MeOD) δ: 177.5 (CONH), 173.2 (
3); 13C-NMR (150 MHz, MeOD) δ: 177.5 (CONH), 173.2 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) OOEt), 168.6 (C-9′), 149.7 (C-4′), 147.2 (C-7′), 146.9 (C-3′), 138.0 (Cindole′′), 128.7 (C–indole′′), 127.7 (C-1′), 124.6 (C–indole′′), 123.1 (C-6′), 122.5 (C–indole′′), 119.9 (C–indole′′), 119.3 (C–indole′′), 116.5 (C-5′), 115.1 (C-2′), 115.0 (C-8′), 112.3 (C–indole′′), 110.4 (O–C–O), 110.2 (C–indole′′), 78.6 (C-3), 76.0 (C-1), 75.4 (C-4), 72.4 (C-5), 62.6 (COO
OOEt), 168.6 (C-9′), 149.7 (C-4′), 147.2 (C-7′), 146.9 (C-3′), 138.0 (Cindole′′), 128.7 (C–indole′′), 127.7 (C-1′), 124.6 (C–indole′′), 123.1 (C-6′), 122.5 (C–indole′′), 119.9 (C–indole′′), 119.3 (C–indole′′), 116.5 (C-5′), 115.1 (C-2′), 115.0 (C-8′), 112.3 (C–indole′′), 110.4 (O–C–O), 110.2 (C–indole′′), 78.6 (C-3), 76.0 (C-1), 75.4 (C-4), 72.4 (C-5), 62.6 (COO![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2CH3), 54.5 (NH
H2CH3), 54.5 (NH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) HCH2), 37.5 (C-2), 35.1 (C-6), 28.4 (
HCH2), 37.5 (C-2), 35.1 (C-6), 28.4 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3–C–O), 28.3 (NHCH
H3–C–O), 28.3 (NHCH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 26.2 (
H2), 26.2 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3–C–O), 14.3 (COOCH2
H3–C–O), 14.3 (COOCH2![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3); HRMS (ESI): calcd for [M + Na]+ C32H36N2NaO10: 631.2268, found 631.2269.
H3); HRMS (ESI): calcd for [M + Na]+ C32H36N2NaO10: 631.2268, found 631.2269.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to gain pure compound 6a–f as white solid.
1) to gain pure compound 6a–f as white solid.![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2CH3), 3.97–3.90 (m, 2H, NH–C
2CH3), 3.97–3.90 (m, 2H, NH–C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 3.72–3.70 (m, 1H, H-5), 2.19–2.17 (m, 1H, H-2), 2.09–2.06 (m, 1H, H-2), 2.02–1.98 (m, 2H, H-6), 1.27–1.24 (m, 3H, COOCH2C
2), 3.72–3.70 (m, 1H, H-5), 2.19–2.17 (m, 1H, H-2), 2.09–2.06 (m, 1H, H-2), 2.02–1.98 (m, 2H, H-6), 1.27–1.24 (m, 3H, COOCH2C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3); 13C-NMR (150 MHz, MeOD) δ: 177.5 (
3); 13C-NMR (150 MHz, MeOD) δ: 177.5 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ONH), 171.2 (
ONH), 171.2 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) OOEt), 169.1 (C-9′), 149.6 (C-4′), 147.0 (C-7′), 146.8 (C-3′), 127.8 (C-1′), 123.0 (C-6′), 116.5 (C-5′), 115.4 (C-2′), 115.1 (C-8′), 77.8 (C-3), 74.4 (C-1), 72.6 (C-4), 71.9 (C-5), 62.4 (COO
OOEt), 169.1 (C-9′), 149.6 (C-4′), 147.0 (C-7′), 146.8 (C-3′), 127.8 (C-1′), 123.0 (C-6′), 116.5 (C-5′), 115.4 (C-2′), 115.1 (C-8′), 77.8 (C-3), 74.4 (C-1), 72.6 (C-4), 71.9 (C-5), 62.4 (COO![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2CH3), 42.0 (NH
H2CH3), 42.0 (NH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 39.9 (C-2), 38.7 (C-6), 14.4 (COOCH2
H2), 39.9 (C-2), 38.7 (C-6), 14.4 (COOCH2![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3); HRMS (ESI): calcd for [M + Na]+ C20H25NNaO10: 462.1376, found 462.1378.
H3); HRMS (ESI): calcd for [M + Na]+ C20H25NNaO10: 462.1376, found 462.1378.![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ), 4.25–4.24 (m, 1H, H-4), 4.21–4.13 (m, 2H, COOC
), 4.25–4.24 (m, 1H, H-4), 4.21–4.13 (m, 2H, COOC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2CH3), 3.72–3.70 (m, 1H, H-5), 2.15–2.12 (m, 1H, H-2), 2.06–1.99 (m, 3H, H-2, H-6), 1.41 (d, J = 7.3 Hz, 1H, NHCHC
2CH3), 3.72–3.70 (m, 1H, H-5), 2.15–2.12 (m, 1H, H-2), 2.06–1.99 (m, 3H, H-2, H-6), 1.41 (d, J = 7.3 Hz, 1H, NHCHC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ), 1.26 (m, J = 7.3 Hz, 3H, COOCH2C
), 1.26 (m, J = 7.3 Hz, 3H, COOCH2C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3); 13C-NMR (150 MHz, MeOD) δ: 176.6 (
3); 13C-NMR (150 MHz, MeOD) δ: 176.6 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ONH), 174.0 (
ONH), 174.0 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) OOEt), 169.1 (C-9′), 149.6 (C-4′), 147.0 (C-7′), 146.8 (C-3′), 127.8 (C-1′), 123.0 (C-6′), 116.5 (C-5′), 115.3 (C-2′), 115.1 (C-8′), 77.7 (C-3), 74.3 (C-1), 72.6 (C-4), 71.9 (C-5), 62.5 (COO
OOEt), 169.1 (C-9′), 149.6 (C-4′), 147.0 (C-7′), 146.8 (C-3′), 127.8 (C-1′), 123.0 (C-6′), 116.5 (C-5′), 115.3 (C-2′), 115.1 (C-8′), 77.7 (C-3), 74.3 (C-1), 72.6 (C-4), 71.9 (C-5), 62.5 (COO![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2CH3), 49.5 (NH
H2CH3), 49.5 (NH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) HCH3), 39.8 (C-2), 38.6 (C-6), 17.5 (NHCH
HCH3), 39.8 (C-2), 38.6 (C-6), 17.5 (NHCH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3), 14.4 (COOCH2
H3), 14.4 (COOCH2![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3); HRMS (ESI): calcd for [M + Na]+ C21H27NNaO10: 476.1533, found 476.1537.
H3); HRMS (ESI): calcd for [M + Na]+ C21H27NNaO10: 476.1533, found 476.1537.![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ), 4.25–4.23 (m, 1H, H-4), 4.22–4.17 (m, 2H, COOC
), 4.25–4.23 (m, 1H, H-4), 4.22–4.17 (m, 2H, COOC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2CH3), 3.72–3.70 (m, 1H, H-5), 2.22–1.96 (m, 5H, H-2, H-6, NHCHC
2CH3), 3.72–3.70 (m, 1H, H-5), 2.22–1.96 (m, 5H, H-2, H-6, NHCHC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ), 1.28 (t, J = 7.1 Hz, COOCH2C
), 1.28 (t, J = 7.1 Hz, COOCH2C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3), 0.95–0.93 (m, 6H, 2×CH3); 13C-NMR (150 MHz, MeOD) δ: 176.7 (
3), 0.95–0.93 (m, 6H, 2×CH3); 13C-NMR (150 MHz, MeOD) δ: 176.7 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ONH), 172.7 (
ONH), 172.7 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) OOEt), 169.0 (C-9′), 149.6 (C-4′), 147.1 (C-7′), 146.8 (C-3′), 127.7 (C-1′), 123.0 (C-6′), 116.5 (C-5′), 115.3 (C-2′), 115.1 (C-8′), 77.9 (C-3), 74.3 (C-1), 72.6 (C-4), 71.8 (C-5), 62.4 (COO
OOEt), 169.0 (C-9′), 149.6 (C-4′), 147.1 (C-7′), 146.8 (C-3′), 127.7 (C-1′), 123.0 (C-6′), 116.5 (C-5′), 115.3 (C-2′), 115.1 (C-8′), 77.9 (C-3), 74.3 (C-1), 72.6 (C-4), 71.8 (C-5), 62.4 (COO![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2CH3), 58.7 (NH
H2CH3), 58.7 (NH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H), 40.2 (C-2), 38.5 (C-6), 32.1 (NHCH
H), 40.2 (C-2), 38.5 (C-6), 32.1 (NHCH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H), 19.4 (CH
H), 19.4 (CH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3), 18.2 (CH
H3), 18.2 (CH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3), 14.5 (COOCH2
H3), 14.5 (COOCH2![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3); HRMS (ESI): calcd for [M + Na]+ C23H31NNaO10: 504.1846, found 504.1849.
H3); HRMS (ESI): calcd for [M + Na]+ C23H31NNaO10: 504.1846, found 504.1849.![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ), 4.25–4.23 (m, 1H, H-4), 4.20–4.14 (m, 2H, COOC
), 4.25–4.23 (m, 1H, H-4), 4.20–4.14 (m, 2H, COOC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2CH3), 3.72–3.70 (m, 1H, H-5), 2.13–2.00 (m, 4H, H-2, H-6), 1.71–1.63 (m, 3H, NHCHC
2CH3), 3.72–3.70 (m, 1H, H-5), 2.13–2.00 (m, 4H, H-2, H-6), 1.71–1.63 (m, 3H, NHCHC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2C
2C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ), 0.95–0.94 (m, 3H, CH3), 0.92–0.91 (s, 3H, CH3); 13C-NMR (150 MHz, MeOD) δ: 176.8 (CONH), 173.9 (
), 0.95–0.94 (m, 3H, CH3), 0.92–0.91 (s, 3H, CH3); 13C-NMR (150 MHz, MeOD) δ: 176.8 (CONH), 173.9 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) OOEt), 169.1 (C-9′), 149.6 (C-4′), 147.1 (C-7′), 146.8 (C-3′), 127.8 (C-1′), 123.0 (C-6′), 116.5 (C-5′), 115.3 (C-2′), 115.2 (C-8′), 77.8 (C-3), 74.3 (C-1), 72.6 (C-4), 71.9 (C-5), 62.4 (COO
OOEt), 169.1 (C-9′), 149.6 (C-4′), 147.1 (C-7′), 146.8 (C-3′), 127.8 (C-1′), 123.0 (C-6′), 116.5 (C-5′), 115.3 (C-2′), 115.2 (C-8′), 77.8 (C-3), 74.3 (C-1), 72.6 (C-4), 71.9 (C-5), 62.4 (COO![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2CH3), 52.1 (NH
H2CH3), 52.1 (NH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H), 41.4 (NHCH
H), 41.4 (NHCH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 39.9 (C-2), 38.6 (C-6), 26.1 (
H2), 39.9 (C-2), 38.6 (C-6), 26.1 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3–C–O), 23.3 (CH
H3–C–O), 23.3 (CH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3), 21.9 (CH
H3), 21.9 (CH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3), 14.5 (COOCH2
H3), 14.5 (COOCH2![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3); HRMS (ESI): calcd for [M + Na]+ C23H31NNaO8: 518.2002, found 518.2009.
H3); HRMS (ESI): calcd for [M + Na]+ C23H31NNaO8: 518.2002, found 518.2009.![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ), 4.21–4.20 (m, 1H, H-4), 4.18–4.14 (m, 2H, COOC
), 4.21–4.20 (m, 1H, H-4), 4.18–4.14 (m, 2H, COOC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2CH3), 3.67–3.65 (m, 1H, H-5), 3.19–3.16 (m, 2H, NHCHC
2CH3), 3.67–3.65 (m, 1H, H-5), 3.19–3.16 (m, 2H, NHCHC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 3.07–3.04 (m, 1H, NHCHC
2), 3.07–3.04 (m, 1H, NHCHC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 2.02–1.79 (m, 4H, H-2, H-6), 1.23 (m, J = 7.3 Hz, 3H, COOCH2C
2), 2.02–1.79 (m, 4H, H-2, H-6), 1.23 (m, J = 7.3 Hz, 3H, COOCH2C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3); 13C-NMR (150 MHz, MeOD) δ: 176.5 (
3); 13C-NMR (150 MHz, MeOD) δ: 176.5 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ONH), 172.6 (
ONH), 172.6 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) OOEt), 169.0 (C-9′), 149.6 (C-4′), 147.0 (C-7′), 146.8 (C-3′), 137.8 (C–Ph′′), 130.4 (2C–Ph′′), 129.5 (2C–Ph′′), 128.0 (C–Ph′′), 127.8 (C-1′), 123.0 (C-6′), 116.5 (C-5′), 115.3 (C-2′), 115.1 (C-8′), 77.7 (C-3), 74.3 (C-1), 72.5 (C-4), 71.8 (C-5), 62.5 (COO
OOEt), 169.0 (C-9′), 149.6 (C-4′), 147.0 (C-7′), 146.8 (C-3′), 137.8 (C–Ph′′), 130.4 (2C–Ph′′), 129.5 (2C–Ph′′), 128.0 (C–Ph′′), 127.8 (C-1′), 123.0 (C-6′), 116.5 (C-5′), 115.3 (C-2′), 115.1 (C-8′), 77.7 (C-3), 74.3 (C-1), 72.5 (C-4), 71.8 (C-5), 62.5 (COO![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2CH3), 54.7 (NH
H2CH3), 54.7 (NH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) HCH2), 39.9 (NHCH
HCH2), 39.9 (NHCH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 38.5 (C-2), 38.3 (C-6), 14.4 (COOCH2
H2), 38.5 (C-2), 38.3 (C-6), 14.4 (COOCH2![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3); HRMS (ESI): calcd for [M + Na]+ C27H31NNaO10: 552.1846, found 552.1850.
H3); HRMS (ESI): calcd for [M + Na]+ C27H31NNaO10: 552.1846, found 552.1850.![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ), 4.20–4.19 (m, 1H, H-4), 4.13–4.08 (m, 2H, COOC
), 4.20–4.19 (m, 1H, H-4), 4.13–4.08 (m, 2H, COOC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2CH3), 3.67–3.65 (m, 1H, H-5), 3.30–3.29 (m, 2H, NHCHC
2CH3), 3.67–3.65 (m, 1H, H-5), 3.30–3.29 (m, 2H, NHCHC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 2.01–1.85 (m, 4H, H-2, H-6), 1.17 (m, J = 7.1 Hz, 3H, COOCH2C
2), 2.01–1.85 (m, 4H, H-2, H-6), 1.17 (m, J = 7.1 Hz, 3H, COOCH2C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3); 13C-NMR (150 MHz, MeOD) δ: 175.1 (CONH), 171.8 (
3); 13C-NMR (150 MHz, MeOD) δ: 175.1 (CONH), 171.8 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) OOEt), 167.6 (C-9′), 148.2 (C-4′), 145.6 (C-7′), 145.4 (C-3′), 136.6 (C–indole′′), 127.3 (C–indole′′), 126.4 (C-1′), 123.2 (C–indole′′), 121.6 (C-6′), 121.1 (C–indole′′), 118.5 (C–indole′′), 117.9 (C–indole′′), 115.1 (C-5′), 114.0 (C-2′), 113.7 (C-8′), 110.9 (C–indole′′), 108.8 (C–indole′′), 76.3 (C-3), 72.9 (C-1), 71.1 (C-4), 70.4 (C-5), 61.1 (COO
OOEt), 167.6 (C-9′), 148.2 (C-4′), 145.6 (C-7′), 145.4 (C-3′), 136.6 (C–indole′′), 127.3 (C–indole′′), 126.4 (C-1′), 123.2 (C–indole′′), 121.6 (C-6′), 121.1 (C–indole′′), 118.5 (C–indole′′), 117.9 (C–indole′′), 115.1 (C-5′), 114.0 (C-2′), 113.7 (C-8′), 110.9 (C–indole′′), 108.8 (C–indole′′), 76.3 (C-3), 72.9 (C-1), 71.1 (C-4), 70.4 (C-5), 61.1 (COO![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2CH3), 53.1 (NH
H2CH3), 53.1 (NH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) HCH2), 38.4 (C-2), 37.1 (C-6), 26.9 (NHCH
HCH2), 38.4 (C-2), 37.1 (C-6), 26.9 (NHCH![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 12.9 (COOCH2
H2), 12.9 (COOCH2![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3); HRMS (ESI): calcd for [M + Na]+ C29H32N2NaO10: 591.1955, found 591.1960.
H3); HRMS (ESI): calcd for [M + Na]+ C29H32N2NaO10: 591.1955, found 591.1960.The spectrograms of the derivatives 5a–f and 6a–f were shown in ESI.†
Evaluation of the biological activity
The primers used are shown in the ESI Table 1.†
Conclusions
The results of the present research indicated that amino acid ester-coupled caffeoylquinic acids derivatives 6c, 6d, 6e and 6f exerts a profound lipid-lowering activity against oleic acid-elicited lipid accumulation for HepG2 liver cells through regulation of TG levels and mRNA levels of lipometabolic-modulating genes. Preliminary SAR analysis has shown that the aliphatic group on the amino acid ester-coupled derivatives had a good impact on the protective effect. Further studies are in progress as the promising results demonstrate that these amino acid ester-coupled caffeoylquinic acids derivatives merit further investigation as potential hypolipidemic agents.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Beijing Natural Science Foundation (grant no. 7192129), the National Natural Sciences Foundation of China (grant no. 81302656), and the CAMS Innovation Fund for Medical Science (CIFMS) (grant no. 2016-I2M-1-012).Notes and references
- T. J. Lan, Q. F. Li, M. Chang, C. L. Yin, D. Zhu, Z. Wu, X. L. Li, W. Q. Zhang, B. W. Yue, J. L. Shi, H. B. Yuan, Z. H. Su and H. W. Guo, J. Ethnopharmacol., 2020, 260, 112989 CrossRef CAS.
- H. L. Gao, Y. Z. Long, X. Q. Jiang, Z. C. Liu, D. C. Wang, Y. Zhang, D. W. Li and B. L. Sun, Exp. Gerontol., 2013, 48, 572–578 CrossRef CAS.
- S. Gao, G. S. Hu, D. Li, M. Z. Sun and D. H. Mou, J. Funct. Foods, 2020, 66, 103837 CrossRef CAS.
- M. J. Hao, Z. J. Guan, Y. Gao, J. L. Xing, X. X. Zhou, C. Y. Wang, J. Xu and W. M. Li, Phytomedicine, 2020, 78, 153292 CrossRef CAS.
- T. T. Zeng, D. J. Tang, Y. X. Yuan, J. Su and H. Jiang, Gene, 2017, 626, 319–325 CrossRef CAS.
- V. Bittner, L. Q. Deng, R. S. Rosenson, B. Taylor, S. P. Glasser, S. T. Kent, M. E. Farkouh and P. Maul, J. Am. Coll. Cardiol., 2015, 66, 1864–1872 CrossRef.
- Y. X. Zhang, Y. Y. Gu, Y. B. Chen, Z. Y. Huang, M. Li, W. H. Jiang, J. H. Chen, W. T. Rao, S. F. Luo, Y. Y. Chen, J. Q. Chen, L. J. Li, Y. H. Jia, M. H. Liu and F. H. Zhou, J. Ethnopharmacol., 2021, 266, 113436 CrossRef CAS.
- J. Bordoloia, D. Ozahc, T. Borac, J. Kalitaa and P. Mannaa, J. Nutr. Biochem., 2019, 70, 174–184 CrossRef.
- Z. S. Xie, X. M. Wan, L. J. Zhong, H. Yang, P. Li and X. J. Xu, J. Funct. Foods, 2017, 31, 217–228 CrossRef CAS.
- M. W. Martin, L. D. R. Jr, H. B. Li, S. E. R. Schiller, A. V. Toms, Z. G. Wang, K. W. Bair, J. Castro, S. Fessler, D. Gotur, S. E. Hubbs, G. S. Kauffman, M. Kershaw, G. P. Luke, C. McKinnon, L. Yao, W. Lu and D. S. Millan, Bioorg. Med. Chem. Lett., 2019, 29, 1001–1006 CrossRef CAS.
- L. Q. Han, T. Y. Gao, G. Y. Yang and J. J. Loor, J. Dairy Sci., 2018, 101, 6523–6531 CrossRef CAS.
- Y. Andriani, N. M. Ramli, D. F. Syamsumir, M. N. I. Kassim, J. Jaafar, N. A. Aziz, L. Marlina, N. S. Musa and H. Mohamad, Arabian J. Chem., 2019, 12, 3555–3564 CrossRef CAS.
- C. M. Wu, X. P. Zhang, X. Zhang, H. Luan, G. B. Sun, X. B. Sun, X. L. Wang, P. Guo and X. D. Xu, J. Nutr. Biochem., 2014, 25, 412–419 CrossRef CAS.
- L. H. Peng, J. W. Liu, J. K. Zheng, J. L. Cheng and M. Y. Xian, Chin. J. Ethnomed. Ethnopharm., 2012, 5, 24–25 Search PubMed.
- I. Tomaca, M. Šerugaa and J. Labudab, Food Chem., 2020, 325, 126787 CrossRef.
- V. Francisco, G. Costa, A. Figueirinha, C. Marques, P. Pereira, B. M. Neves, M. C. Lopes, M. T. Cruz and M. T. Batista, J. Ethnopharmacol., 2013, 148, 126–134 CrossRef CAS.
- D. Liu, S. Meng, Z. H. Xiang, N. J. He and G. W. Yang, Phytochemistry, 2019, 163, 1–10 CrossRef CAS.
- D. Y. Wang, X. M. Zhao and Y. L. Liu, Int. J. Biol. Macromol., 2017, 102, 396–404 CrossRef CAS.
- X. P. Zhang, C. M. Wu, H. F. Wu, L. H. Sheng, Y. Su, X. Zhang, H. Luan, G. B. Sun, X. B. Sun, Y. Tian, Y. B. Ji, P. Guo and X. D. Xu, PLoS One, 2013, 8, 61922 CrossRef.
- X. X. Cao, C. M. Wu, Y. Tian and P. Guo, RSC Adv., 2019, 9, 12247–12254 RSC.
- Y. Tian, X. X. Cao, H. Shang, C. M. Wu, X. Zhang, P. Guo, X. P. Zhang and X. D. Xu, Molecules, 2019, 24, 964 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra09621k |
| ‡ These two authors contributed equally to this paper. |
| This journal is © The Royal Society of Chemistry 2021 |