Prospects and applications of synergistic noble metal nanoparticle-bacterial hybrid systems
Alba
Vázquez-Arias
 ab,
Jorge
Pérez-Juste
ab,
Jorge
Pérez-Juste
 ab,
Isabel
Pastoriza-Santos
ab,
Isabel
Pastoriza-Santos
 *ab and
Gustavo
Bodelon
*ab and
Gustavo
Bodelon
 *ab
*ab
aCINBIO, Universidade de Vigo, Departamento de Química Física, Campus Universitario Lagoas, Marcosende, 36310 Vigo, Spain. E-mail: gbodelon@uvigo.es; pastoriza@uvigo.es
bGalicia Sur Health Research Institute (IIS Galicia Sur), SERGAS-UVIGO, 36312 Vigo, Spain
First published on 4th October 2021
Abstract
Hybrid systems composed of living cells and nanomaterials have been attracting great interest in various fields of research ranging from materials science to biomedicine. In particular, the interfacing of noble metal nanoparticles and bacterial cells in a single architecture aims to generate hybrid systems that combine the unique physicochemical properties of the metals and biological attributes of the microbial cells. While the bacterial cells provide effector and scaffolding functions, the metallic component endows the hybrid system with multifunctional capabilities. This synergistic effort seeks to fabricate living materials with improved functions and new properties that surpass their individual components. Herein, we provide an overview of this research field and the strategies for obtaining hybrid systems, and we summarize recent biological applications, challenges and current prospects in this exciting new arena.
1. Introduction
Living organisms naturally produce composite functional materials based on biomolecules and minerals. Inspired by such abiotic–biotic interfaces, a new paradigm has emerged at the intersection of materials science, nanotechnology, chemistry and biology that seeks to combine synthetic materials with living cells to develop novel biological applications. This research avenue aims to create synthetic biological pathways, boost natural ones, and fabricate hybrid systems with unprecedented functions.1 In this context, research efforts have led to new concepts such as cyborg cells,2 living functional materials,1,3–6 materials synthetic biology,7 nano(bio)hybrids,8–10 and nanobionics.11,12 In general, these approaches involve the rational combination of nanomaterials with cells in innovative applications like the manipulation of cell behavior,13,14 optogenetics,15,16 photosynthesis and energy conversion,11,12,17–22 sensing,23–25,26 bioelectronics,27–29 fabrication of artificial organelles,30 nanoreactors,31–33 as well as microrobots for biomedicine.9,34–37 The aforementioned hybrid systems combine the advantages of both the biotic and abiotic worlds; while the living organism provides metabolic pathways, self-organization, autonomous motion, sensing, actuation, and self-replication, the material provides optical, electronic, magnetic and catalytic features.A wide range of synthetic elements have been explored in the abovementioned hybrid systems, ranging from inorganic (e.g., minerals, clays, metals, semiconductors, carbons, ceramics) to organic materials (e.g., small molecules, polymers, lipids) and composites (e.g., metal–organic frameworks and metal-phenolic networks).8 For instance, inspired by biochemical reactions taking place in natural organelles, Lee and collaborators fabricated a silica-confined magnetothermia-induced nanoreactor for catalyzing palladium-driven carbocyclization reactions.32 The multifunctional composite consists of an iron oxide core inside a hollow silica nanoshell bearing palladium nanocrystals. While the Fe3O4 magnetic core serves as a magnetic field-induced nanoheater to trigger catalytic reactions, the porous silica enclosure acts as a molecular sieving element that prevents the entry of large biomolecules that could poison the Pd surface. In this study, the intracellular catalytic transformation of non-fluorescent procoumarin to a highly fluorescent coumarin derivative was demonstrated without compromising the cell viability. Semiconductor and metal nanoparticles bearing efficient light-absorption capacity and tunable charge separation have been used to produce higher order chemicals in living microbes by artificial photosynthesis.38 In such hybrid systems, the combination of synthetic and biological components can produce solar-to-fuel and solar-to-chemical conversion pathways that are not feasible by natural or artificial systems alone, which has found vast potential.39 In one study, Ding and collaborators fabricated different core–shell quantum dots, with controlled bandgap energies ranging from ultraviolet to near-infrared (NIR), for the renewable production of various biofuels and chemicals from carbon dioxide, water, and nitrogen in non-photosynthetic microbial species.40 In another study, Sakimoto and collaborators used cadmium sulphide nanoparticles as light collectors on the surface of a non-photosynthetic bacterium with the aim of producing acetic acid from carbon dioxide and thus sustaining the cellular metabolism.17 Carbon-based nanomaterials such as carbon nanotubes, graphene oxide, fullerenes and nanodiamonds have excellent mechanical, electrical and biomimetic properties for biomedical applications. Early studies showed that carbon nanotubes possess the ability to interact intimately with the cellular membrane promoting neuronal electrical activity, synapse formation, axon growth, and excitability in cultured cells.41–43 Strikingly, these studies show that instructive cues promoted by the interfacing nanomaterial may be translated into cellular signals for driving synaptic network development. Therefore, it is speculated that similar approaches employing biohybrid systems may help to develop unconventional therapeutic applications in neuromedicine. In this framework, Usmani and collaborators showed that carbon nanotube scaffolds can promote axon regeneration in spinal cord injury rats, thereby improving motor function recovery and neuronal regrowth in vivo.44 Even though this field of research has recently emerged, a number of comprehensive reviews already exist describing the integration of a wide range of nanomaterials and cellular systems, ranging from virus to multicellular organisms.1,3–5,7–10
In general, nanomaterials can be either incorporated into living organisms by in situ biogenic synthesis, harnessing the natural biosynthetic capabilities of the cell,45 or by means of physicochemical and biomolecular interactions employing preformed nanoparticles.46 The manufacturing of complex, hierarchically ordered, biohybrid composites can take advantage of biofabrication techniques like micropatterning, microfluidics, and three-dimensional (3D) printing at multiple scales and customized geometries.6 However, despite significant progress, the seamless integration of synthetic nanomaterials with living biological systems is challenging due to the intrinsic differences between both abiotic and biotic worlds. Toxicity and uncontrolled reactivity with the cellular environment are significant issues. Also, dilution of the synthetic material in the cellular population after prolonged cell divisions can be a concern for certain applications.
Owing to their ease of culture, genetic manipulation, and short generation time for many species, bacterial cells have been intensely used as a model organism in an ample variety of research disciplines. Pioneering work on hybrid materials powered by synthetic biology initially focused on Escherichia coli, primarily due to the extensive collection of genetic parts and tools available.1 The evolution of genomics and synthetic biology has facilitated to expand the field to other many bacterial species aiming to take advantage of their biological properties such as photosynthesis, energy conversion, extremophilic abilities, tolerance to desiccation, and mineralization.4,9 Researchers have also learned to manipulate microbial consortia with programmed behaviors for enhanced bioproduction, substrate usage, as well as the production of functional materials.47 The aforementioned advancements have made possible a number of biotechnological and biomedical applications based on the combination of inorganic nanomaterials and bacterial cells.4,9
Noble metal nanoparticles are characterized by their excellent physicochemical properties, controlled chemical synthesis, tuneable surface functionalization, high stability and biocompatibility. These nanomaterials exhibit resonance electron oscillation known as localized surface plasmon resonance.48 The unique plasmonic properties of noble metal nanoparticles, such as the large electromagnetic field enhancements, rich spectral responses and high photothermal conversion efficiency, endow them with high potential in a variety of sensing applications.49 In particular, the biological applications of noble metal nanoparticles has sparked the interest from a large community of researchers with different backgrounds, ranging from materials science, chemistry, and optics to biomedicine and biodiagnostics.50 In this context, the capability to rationally combine noble metal nanoparticles and biological systems with nontoxicity holds great potential to develop new research avenues. In-depth reviews regarding the toxicity of noble metal nanoparticles to bacterial cells have been published elsewhere.51,52
In this minireview, we will focus on noble metal nanoparticles and their integration with bacteria to achieve functional hybrid systems with synergistic properties. We define synergistic hybrid systems as rationally designed conjugates of nanoparticles and microbes, in which both components interact and co-operate towards a gain of function or activity. Therefore, the resultant hybrids are endowed with improved functions that recapitulate, or even surpass, their individual components. From a fundamental scientific viewpoint, the interfaced abiotic–biotic hybrids have provided new insights into the molecular mechanisms underlying cell–material interactions. Most importantly, this multidisciplinary field of research paves the way for potential applications in the fields of energy generation and storage, electronics, catalysis, sensing, and biomedicine. Therefore, herein, we will discuss the strategies for interfacing bacterial cells with noble metal nanoparticles and summarize the most relevant applications. Finally, we will provide the current challenges and prospects for the field.
2. Strategies for the functionalization of bacteria with noble metal nanoparticles
Interfacing noble metal nanoparticles and bacterial cells can take place either by biogenic and non-biogenic strategies. The first approach is based on the biosynthesis of the nanomaterial through endogenous (i.e., cellular) enzymatic and non-enzymatic reactions. The second one relies on the use of pre-made nanoparticles that are implemented in the hybrids through approaches for promoting their interaction with the bacterial cell. These interactions are generally classified into physisorption and chemisorption. The microbes serve as templates enabling the organization of nanoparticles into well-defined structures at the nano and micro-scales.2.1. Biogenic approaches
Metal ions are essential for many cellular processes, however in excess they can be toxic. Thus, bacteria have evolved detoxification mechanisms to counter their potential toxic effects, including their reduction to nanoparticles. For instance, Pseudomonas stutzeri can withstand high concentrations of silver ions by the production of crystalline silver nanoparticles (AgNPs) in vacuole-like granules inside the bacterial cell (Fig. 1a).53 This natural ability of bacteria has been harnessed for the bioremediation of heavy metals, and the green-production of metallic nanoparticles. In a seminal study, Beveridge and Murray reported the extracellular deposition of gold nanoparticles (AuNPs) on the cell wall of Bacillus subtilis in the presence of a gold chloride solution.54 The biogenic synthesis of metal nanoparticles can take place both intracellularly and extracellularly, and it has been recently reviewed in several studies (Fig. 1b).45,55–58 Although the precise biochemical and physiological pathways for the individual metals remain for the most part unknown, it is widely accepted that enzymes with NADH-dependent reductase activity, proteins as capping agents, and other biomolecular cofactors are involved.55,56,58 Even though the biogenic procedure is cost-effective, its efficiency is rather low, and it is often difficult to obtain metal nanoparticles with a well-defined size, shape and monodispersity,59,60 which may be detrimental for certain downstream applications in hybrid systems. However, through appropriate strain selection, the development of genetically engineered microbes, synthetic gene circuits, and optimization of culture conditions it could be possible to overcome the aforementioned limitations.45 For instance, the recombinant expression of metal-binding proteins has proven useful for enhancing the biogenic synthesis of metal nanoparticles. In one study, Choi and collaborators co-expressed metallothionein (mt) and phytochelatin synthase (pcs) in E. coli,61 which bind heavy metal ions through their cysteine residues facilitating their reduction, and produced approximately 60 different types of crystalline and amorphous nanomaterials, including AuNPs and AgNPs, covering 34 elements from the periodic table.61 As in any colloidal synthesis of nanoparticles, precursor concentration, temperature, pH, pressure and reaction time are key parameters known to affect the size and shape of the metal nanoparticles. Interestingly, Choi et al., showed that the bioproduction and crystallinity of the nanomaterial could be predicted by Pourbaix diagram analyses, which define what chemical species predominate for a given reduction potential and pH (Fig. 1c). Based on the Pourbaix diagram analyses, the initial pH of the reaction was changed from 6.5 to 7.5, leading to the biosynthesis of various crystalline nanomaterials.61 Curli fibres are a class of extracellular proteinaceous amyloid fibers that consist of self-assembled CsgA protein monomers. The production of curli fibres can be genetically controlled by the expression of CsgA under the control of chemically inducible promoters. This has facilitated the application of this synthetic biology tool in various hybrid systems.28,62 In one study, Nguyen and collaborators engineered E. coli to secrete curli fibers composed of CsgA genetically fused to a peptide programmed to carry out biotemplating of AgNPs in one-dimensional nanowires from a solution of AgNO3.62 Seker and coworkers explored a similar approach to biosynthesize and assemble AuNPs, as well as gold nanowires, in curli fibers in a controllable way in the presence of sodium citrate and gold(III) chloride.28 In this study, both unmodified CsgA and a genetically modified version of the protein linked to different nucleating peptides served as templates for nanoparticle synthesis. The size of the nanoparticles could be tuned as a function of the metal-binding affinity of the curli fibers. The interfaced hybrid systems were used to mediate tunable electrical conductivity.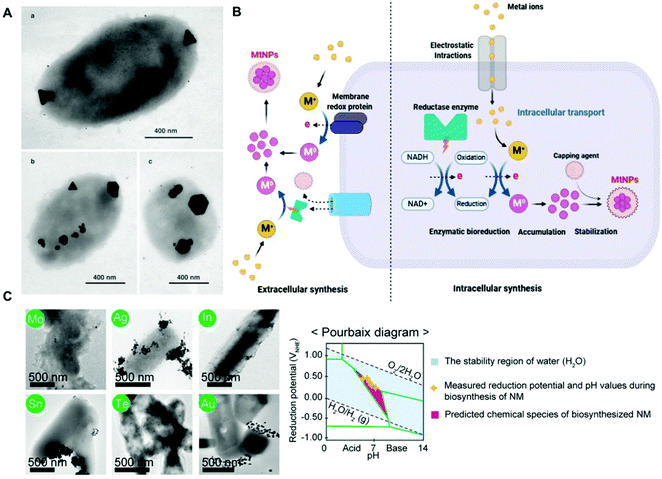 | ||
| Fig. 1 (A) TEM images of silver nanoparticles of different sizes and morphologies biosynthesized inside P. stutzeri bacteria. The scale bar indicates 400 nm. Reproduced from ref. 53 with permission from PNAS. Copyright 1999 National Academy of Sciences. (B) Schematic representation of potential mechanisms for the extracellular and intracellular biogenic synthesis of metal nanoparticles in bacteria. Reproduced from ref. 56 with permission from Creative Commons CC BY. (C) TEM images of crystalline nanoparticles synthesized in vivo by E. coli co-expressing metallothionein and phytochelatin synthase. The corresponding elements are labeled in green circles, and the scale bar indicates 500 nm. Pourbaix diagram and various inorganic nanomaterials biosynthesized in vivo at the initial pH of 7.5. Reproduced from ref. 61 with permission from PNAS. Copyright 2018 National Academy of Sciences. | ||
Jian and collaborators biofabricated AuNPs (3–4 nm) inside bacterial cells for electron microscopy imaging at the single-molecule level.63 This approach consisted of the intracellular expression of cysteine-rich polypeptides genetically fused to the protein targets of interest. The cysteine-rich peptides reacted with thiol ligands (RSH) (i.e. 2-mercaptoethanol and D-penicillamine), which subsequently reduced HAuCl4 to form thiolate-Au(I) complexes, which are finally reduced by NaBH4 to form AuNPs. The size of the AuNPs could be tuned by varying the RSH-to-Au ratio and the pH.63 As mentioned earlier, the extracellular biogenic synthesis of metal nanoparticles takes advantage of reductive enzymes present in the bacterial cell wall or secreted to the extracellular medium. In one study, Qi and collaborators biosynthesized palladium nanoparticles (PdNPs) on the surface of E. coli by the action of molybdoenzymes, which reduced Pd2+ in the presence of sodium formate.64 Interestingly, PdNPs subsequently promoted the in situ polymerization of photoactive polyphenyleneethynylene with light-harvesting capabilities. In another study, a cell surface-associated NADPH-dependent reductase activity was employed to reduce Au3+ to Au0 leading to the deposition of AuNPs on the bacterial surface.65 An additional method, called bacterial surface display,66 is based on the expression of membrane proteins genetically linked to peptides with the capacity to bind nanoparticles or their surface ligands by covalent, and/or electrostatic bonds between charged residue and surface groups of the substrate. Tsai and coworkers evaluated the efficacy of various bacterial display systems to translocate metal-binding peptides to the cell surface of E. coli and Ralstonia eutropha aiming to promote the green synthesis of gold and platinum nanoparticles. The whole-cell biocatalysts were tested for the reduction of 4-nitroaniline.67
2.2. Non-biogenic approaches
Non-biogenic approaches for the fabrication of noble metal nanoparticle-bacterial hybrids rely on the synthesis of metal nanoparticles with desired size, shape and composition and their further integration, intracellular or extracellular, in the bacterial cell.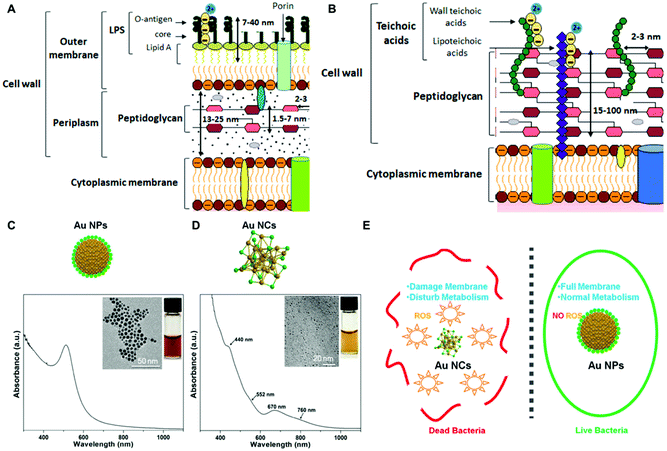 | ||
| Fig. 2 (A and B) Differences between Gram-positive and Gram-negative bacterial cell walls, respectively. Reproduced from ref. 78 with permission from Journal of Nanoparticle Research. Copyright 2019 http://creativecommons.org/licenses/by/4.0/. (C and D) UV–vis absorption and TEM characterization of AuNPs and AuNCs. Reproduced from ref. 71 with permission from ACS nano. Copyright 2017 American Chemical Society. (E) Illustration of the antibacterial effect elicited by AuNCs as compared to AuNPs larger in size due to ROS production and membrane destabilization. Reproduced from ref. 71 with permission from ACS nano. Copyright 2017 American Chemical Society. | ||
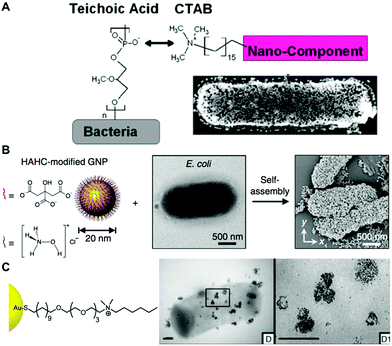 | ||
| Fig. 3 (A) Electrostatic functionalization of CTAB-stabilized gold nanospheres on B. cereus. Reproduced from ref. 80 with permission from Journal of the American Chemical Society. Copyright 2015 American Chemical Society. (B) Self-assembly of hydroxylamine hydrochloride (HAHC)-modified gold nanoparticles on the surface of E. coli. Reproduced from ref. 86 with permission from Nano-Micro letters. Copyright 2018. (C) Interaction of cationic NP1 AuNPs functionalized with hexyl-substituted ammonium-functionalized thiol on the B. subtilis surface. Reproduced from ref. 87 with permission from Journal of the American Chemical Society. Copyright 2012 American Chemical Society. | ||
In the case of electrostatic interactions between bacteria and noble metal nanoparticles, the zeta potential of both systems is a key parameter that can determine the strength of the interaction. For instance, Pajerski and collaborators demonstrated the direct correlation between the number of citrate-stabilized AuNPs assembled on the bacterial surface and the zeta potential of the different bacterial species (Fig. 4a and b).78 Tadesse and collaborators investigated electrostatic interactions between CTAB-stabilized gold nanorods and different Gram-positive and negative bacterial species for label-free surface-enhanced Raman scattering (SERS) applications.89 In this study, it was shown that surface charges indeed play a significant role, as Gram-positive Staphylococcus epidermidis and Staphylococcus aureus bacteria bearing higher negative surface charge density than Gram-negative E. coli and Serratia marcescens led to stronger electrostatic interactions with the positively charged nanorods, thereby yielding significantly higher SERS signals. Cryo-EM analysis showed higher nanoparticle coverage in the Gram-positive S. epidermidis than in the Gram-negative E. coli (Fig. 4c). A drawback of this functionalization strategy is related with the potential toxic activities of the assembled nanoparticles when conjugated to cationic ligands such as CTAB.81 Interestingly, Tadesse and collaborators demonstrated that the removal of CTAB from gold nanorods to a minimal surfactant coverage did not significantly affect the bacterial viability.89 Also, in order to reduce the potential toxicity of CTAB-functionalized nanoparticles, this molecule could be replaced with biocompatible molecules by ligand exchange.90
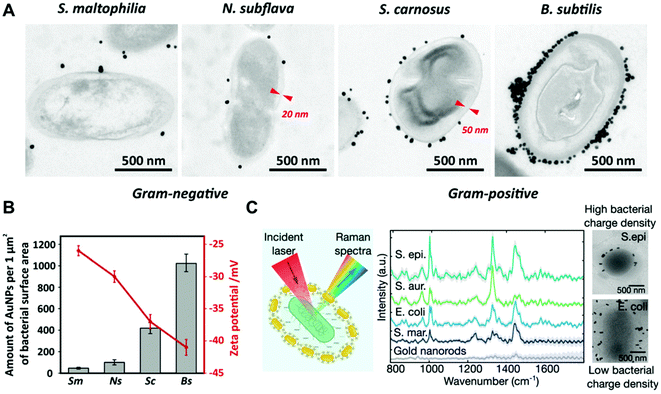 | ||
| Fig. 4 (A) TEM images of the interaction of citrate-stabilized AuNPs with different bacterial species. Red markers indicate the thickness of cell walls. Reproduced from ref. 78 with permission from Journal of Nanoparticle Research. Copyright 2019. (B) Relationship between the zeta potential of bacteria (red plot) and the amount of adhered citrate-stabilized AuNPs (grey bars). Sm, S. maltophilia; Ns, N. subflava; Sc, S. carnosus; Bs, B. Subtilis. Reproduced from ref. 78 with permission from Journal of Nanoparticle Research. Copyright 2019 (C) SERS spectra and cryo-EM images of S. epidermidis (S. epi) and E. coli conjugated with CTAB-stabilized gold nanorods. Reproduced from ref. 89 with permission from Nano Letters. Copyright 2020 American Chemical Society. | ||
The mechanism of antibacterial activity of large metallic nanoparticles, in the range of 80–100 nm, unable to freely translocate across the bacterial cell membrane has been associated with the alteration of the cell membrane.68,69 Recently, Linklater et al., investigated the antibacterial mechanism action of 100 nm AuNPs in Pseudomonas aeruginosa and S. aureus.91 To this aim, they explored nanoparticles bearing different surface areas (quasi-spherical vs. nanostars) functionalized with either mercaptohexanoic acid (MHA) to yield hydrophilic nanoparticles or with a hydrophobic zwitterionic sulfobetaine ligand (ZwBuEt) (Fig. 5a and b). The quasi-spherical nanoparticles (AuNSPs) coated with MHA exhibited a greater bactericidal action than the hydrophobic nanoparticles due to a lower dispersity and higher interactive affinity, resulting in greater cell death. A biophysical model enabled the authors to propose that the adsorption of the nanoparticles to the bacterial surface triggers an increase in membrane tension and mechanical deformation, thereby leading to cell death.91 The surface charge of bacteria may be also modified by functionalization with polyelectrolytes employing the layer-by-layer method.2,92,93 Feng and collaborators investigated the self-assembly of AuNPs on the surface of Campylobacter jejuni for obtaining bacterial cells displaying chiroptical activity.94 Initially, a direct adsorption approach was performed with positively charged Au nanocolloids, leading to non-uniform deposition. This was attributed to the uneven negative charge distribution on the bacterial surfaces. Next, the authors employed polydiallyldimethylammonium (PDDA), a positively charged polyelectrolyte, to functionalize the surface of C. jejuni (Fig. 5c). This strategy promoted an homogeneous adsorption of negatively charged AuNPs with much higher uniformity as compared with the alternative approach that employed positively charged AuNPs.94 Park and coworkers covered Clostridium novyi-NT spores with positively charged branched polyethylenimine (bPEI, 600 Da), which facilitated the assembly of 4-mercaptobenzoic acid stabilized branched gold nanoparticles (BGNPs) through electrostatic interactions.85 Another popular strategy to obtain noble metal nanoparticle-bacterial hybrids consists of the conjugation of the nanoparticle with functional groups that bind specifically to bacterial cell wall components, such as vancomycin which targets Gram-positive peptidoglycans, or polymyxin which targets lipopolysaccharides of Gram-negative bacteria. For instance, Wang and coworkers reported an orthogonal approach based on a trans-cyclooctene derivative of vancomycin (Van-TCO) that specifically binds to the cell envelope of Gram-positive bacteria by forming hydrogen bonds with the C-terminal D-Ala-D-Ala motif of a peptidoglycan precursor. In a second step, Van-TCO can orthogonally conjugate with tetrazine-functionalized AuNPs via instantaneous cycloaddition leading to the deposition of the nanoparticles on the cellular surface.95
 | ||
| Fig. 5 (A and B) Schematic depiction of gold nanoparticles coated with zwitterionic ligands MHA and ZwBuEt, and potential mechanism of the AuNP–membrane interaction of large nanoparticles with a diameter greater than the thickness of the cell membrane (>10 nm). Reproduced from ref. 91 with permission from Advanced Materials. Copyright 2020. (C) Schematic illustration for the preparation of BGNP-coated C. novyi-NT spores through electrostatic deposition employing the cationic polymer bPEI. Reproduced from ref. 85 with permission from Small. Copyright 2017 PMC. (D and F) TEM images of bacteria without (D) and with (F) induction of the displayed AuNP-binding peptide. Scale bar indicates 200 nm. (E and G) Schematic illustration of the repulsive interaction between the negatively charged cell membrane and negatively charged AuNPs in the absence of binding peptides (E). This repulsive force is overcome by means of the surface-displayed peptide (G). Reproduced from ref. 106 with permission from Langmuir. Copyright 2018 American Chemical Society. | ||
In general, the non-covalent approaches (physisorption) are straightforward means for the functionalization of bacteria. However, these interactions are labile and may be easily disrupted by ionic strength and pH, thereby affecting the robustness of the hybrid system. Covalent conjugation provides a stable adhesion between the microbe and the nanomaterial, and therefore they can be implemented to overcome the limitations of the aforementioned strategy. In this context, the surface chemistry of metal nanoparticles can be easily engineered with multiple ligands and functional groups to promote their covalent binding to biomolecules present on the cell envelope through carbodiimide and click chemistry.96,97 The accessible amines on the cell surface can react with N-hydroxysuccinimide esters, cyanuric chloride-activated molecules, and unsaturated ester aldehyde moieties through azaelectrocyclization.98 Thiols can also react with maleimide-functionalized molecules to form a stable thioether bond. Also, bioorthogonal reaction pairs such as azides and terminal alkynes, azides and phosphines, or aldehydes/ketones can be employed.99 However, since this approach is not specific, unintended cell surface biomolecules may be also chemically modified in an uncontrolled way, which in turn may affect their biological function affecting the viability of the hybrid system.
The specificity of the nanoparticle assembly on the bacterial surface can be achieved using ligand–receptor and other biomolecular interactions such as biotin–streptavidin,100 antigen–antibody,101 as well as carbohydrate–protein interactions.102,103 Another approach takes advantage of the coordinative binding between electron donor groups on the imidazole ring of histidine and divalent cations. This strategy was employed to assemble nickel nitrilotriacetic acid-conjugated gold nanoparticles in polyhistidine-tagged CsgA subunits27,104 and TasA amyloid fibrils.105 Dong and coworkers employed a bacterial surface display system based on the enhanced circularly permuted outer membrane protein X (eCPX) scaffold to translocate metal-binding peptides on the surface of E. coli, which in turn enabled the deposition of citrate-stabilized AuNPs (Fig. 5d–g).106 The authors speculated that the binding of the peptide to the Au surface originated the displacement of citrate anions, which in turn allows to overcome the repulsive electrostatic forces between the bacterial membrane and the citrate-stabilized AuNPs, both negatively charged. As the viability of this hybrid system was not compromised, the authors suggested that those approaches having the capability of distancing the inorganic moiety from the cell wall, may avoid potential toxic interactions.106
3. Biological applications of hybrid systems
The interfacing of noble metal nanoparticles with living bacterial cells and biofilms has yielded an ample variety of hybrid systems with enhanced properties and functionalities not reachable by each component separately. In the next section, we highlight recent investigations and applications of this synergistic combination focused on the fabrication of living electronics, solar fuel production, and bacterial microswimmers.3.1 Living electronics
Noble metals are known for their high electrical conductivity, which makes possible their implementation in conductive hybrid systems and electrical biodevices. Moreover, the discovery of electron transport through bacterial biofilms offers new possibilities in bacteria-electrode interactions and bioelectronics. In this context, metallic-like conductivity in the biofilms of Geobacter sulfurreducens has been reported, showing an electronic conductivity of ∼5 mS cm.107 Interestingly, the electron transfer capabilities of bacterial biofilms could be improved by interfacing them with metal nanoparticles. Saraf's group demonstrated the potential of hybrid composite devices of bacteria and noble metal nanoparticles toward the development of highly conductive systems.80,82,83 In one study, they combined the conductive properties of AuNPs and the biological response of B. cereus to fabricate a humidity-based electronic system.83 The device consisted of bacterial cells coated with poly-lysine-functionalized AuNPs (30 nm) and two bridging electrodes. Variations in humidity modulate the inter-particle distance and thereby the tunneling current of the system. The authors demonstrated that the cell viability was maintained during the manufacture of the device.83 He and coworkers developed a hybrid system consisting of Bacillus subtilis ribbons coated by AuNPs (20 nm). The ribbons with a width of ca. 1 μm and several millimeters in length were conductive and displayed Ohmic behavior.108 However, the preparation process required aging for 4 weeks, which was toxic to bacteria inducing massive cellular death. Dissimilatory metal-reducing bacteria, including Shewanella and Geobacter species, can inherently carry out extracellular electron transfer (EET) to drive extracellular reduction reactions. This feature has been harnessed for the green synthesis of inorganic nanomaterials.109 For instance, Chen and collaborators investigated the capability of G. sulfurreducens biofilms to biosynthesize AuNPs, and the resulting hybrid system displayed increased conductivity and enhanced EET.110 The biogenic fabrication of the nanoparticles was performed in situ by dropping NaAuCl4 into the bacterial biofilm, in a process that did not impair cellular viability. In a different study, Seker and coworkers employed genetically controlled CsgA curli fibers aiming to fabricate gold nanowires in E. coli with tunable electrical conductance.28 Furthermore, the authors showed that the expression of various types of gold-binding peptides genetically fused to CsgA promoted their mineralization with different size distributions. Taking advantage of this phenomenon, the choice of a particular nucleating peptide enabled them to tune the electrical conductivity of the bacterial biofilms. In one approach, nickel-nitrilotriacetic acid (Ni-NTA)-coated AuNPs were bound to the fibers based on interactions between Ni-NTA and the histidine residues of CsgA-P1. Next, gold enhancement was used to fabricate metallic gold nanowires adhered to the curli fibers on the bacterial surface (Fig. 6a and b). Conductance measurements on the cellular populations revealed a significant increase in conductance when curli-fiber-organized gold nanowires were chemically induced versus the uninduced state (Fig. 6c). The authors also synthesized curli fibers with gold nucleation followed by gold enhancement, albeit the generated gold nanowires displayed lower conductivity levels.28 In a similar approach, Chen and coworkers employed an inducible genetic system to express amyloid fibrils in E. coli composed of polyhistidine-tagged CsgA (CsgA-His) for further conjugation with Ni-NTA AuNPs. The AuNPs endowed the resulting biofilms with conductive properties that were externally controlled as in an electronic switch.27 In another study, an engineered curli system was exploited to produce and assemble extracellular curli fibrils in bacterial colonies arranged into 3D patterns.104 The assembly of Ni-NTA AuNPs on the curli fibrils gave rise to domelike structures with electrical properties. When opposed to one another, such hybrid materials formed a miniature electronic device capable of sensing external pressure.104 As living entities, bacterial biofilms can self-replicate and self-repair and their conductivity can be tuned by regulating gene expression. With further developments, this new class of engineered living conductive materials could be exploited in a number of applications, including improved catalytic coatings for microbial fuel-cell electrodes, and next-generation living components of bioelectronic devices.7,111 For instance, Huang and collaborators engineered and fabricated viscoelastic biofilms of B. subtilis in microstructures with diverse 3D shapes employing 3D printing and microencapsulation techniques.105 Microbial fuel cells (MFCs) can directly convert the chemical energy stored in organic matter to electricity by harvesting the energy generated through the bacterial metabolism with electrodes. Since MFCs can use a wide range of organic fuels to create electricity, the technology is of considerable interest for renewable power generation from biomass and wastewater treatment.112 Among the bacteria used to power these systems, Shewanella is particularly well-suited for this purpose. However, despite considerable efforts to improve these systems, current MFCs often suffer from low current and power densities largely limited by the inefficient electron-transfer processes between the microbes and the anode. In order to enhance the charge-extraction efficiency, Cao et al., developed a novel strategy for boosting the performance of Shewanella MFCs that consists of coating the bacteria with biosynthesized AgNPs.113 This study showed that when bacterial biofilms are developed on a reduced graphene oxide/silver nanoparticle (rGO/Ag) anode, silver nanoparticles become associated with the cellular membranes, greatly enhancing their electron-transfer efficiency. Interestingly, the STEM image and EDX elemental mapping studies on ultrathin sections of the hybrids showed the presence of abundant nanoparticles inside, and across the bacterial membrane. Cao et al., speculated that the rGO/Ag electrode releases Ag ions, which diffuse towards Shewanella and are reduced in situ to form AgNPs by the electrons generated as a result of the cellular metabolism. The cell-associated nanoparticles can potentially act as metallic shortcuts making direct contact with external electrodes for more-efficient charge extraction. Importantly, the authors showed that silver did not seem to compromise the viability of the bacteria in the hybrid system. In another study, Zhu and coworker's implemented a layer-by-layer strategy to alternatively coat Au and CdS NPs onto the cell surface of E. coli for the fabrication of photo-bioanodes in an MFC.114 Interestingly, the CdS layer was shown to protect the bacterial viability from light-induced inactivation.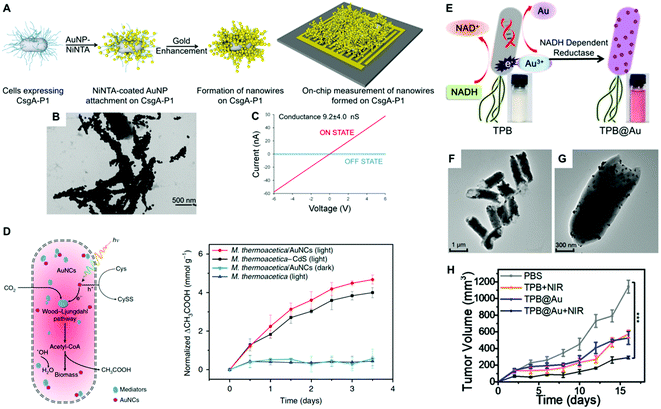 | ||
| Fig. 6 (A) Conductive nanowire formation on cell-synthesized CsgA curli fibers with preformed gold nanoparticles followed by gold enhancement. (B) TEM image of gold nanowires in bacterial curli. (C) Conductance measurements on the cellular populations induced by aTc (ON STATE) versus the uninduced state (OFF STATE). Reproduced from ref. 28 with permission from ACS Synthetic Biology. Copyright 2017 American Chemical Society. (D) Schematic diagram of the photosynthetic biohybrid system composed of M. thermoacetica/AuNC that enables the photosynthesis of acetic acid from CO2 and normalized photosynthetic production of acetic acid by M. thermoacetica, M. thermoacetica/AuNCs PBSs under continuous low-intensity illumination and dark conditions. Reproduced from ref. 19 with permission from Nature Nanotechnology. Copyright 2018 Springer Nature. (E) Illustration of biosynthesis mechanism of AuNPs by TPB through enzymatic reduction. (F and G) TEM images of TPB@Au. (H) Tumor volume after the indicated treatments. Reproduced from ref. 65 with permission from Nano Letters. Copyright 2018 American Chemical Society. | ||
3.2 Solar fuel production
An emerging attractive technology that aims to convert light energy into chemical bonds is based on semi-artificial photosynthesis, which combines the power of synthetic catalysts, for harvesting light energy, with the unmatched efficiency and specificity of biological catalysts, for chemical reactions. This emerging technology involves the fabrication of artificial photosynthetic hybrid systems, where synthetic light-harvesting materials are coupled with microbial cells. Typically, this approach entails the use of non-photosynthetic microorganisms as they usually harbor biochemical pathways for more elaborate products than their photosynthetic partners. In such hybrid systems, light-harvesting nanoparticles are selected due to their superior optical and electronic properties, high surface area and nanoscale dimensions. The field of photosensitizing microorganisms is still in its infancy, and most studies have been focused on semiconducting materials such as CdS nanoparticles.20 Therefore, the number of reports regarding the use of metal nanoparticles is still very scarce. An in depth review covering hybrid systems based on other inorganic nanomaterials for bioenergy conversion from sunlight can be found elsewhere.115 The study by Zhang and coworkers may pave the way for the implementation of noble metal nanoparticles and their composites for solar fuel production.19 In their study, biocompatible AuNCs, as the light absorber, were interfaced with non-photosynthetic M. thermoacetica bacteria to fabricate an artificial photosynthetic hybrid system (Fig. 6b). The AuNCs enabled the intracellular photosynthesis of acetic acid from CO2via the acetyl coenzyme A or the Wood–Ljungdahl pathway,116 thereby bypassing the slow mass transport and energy consumption across the cell membrane. Moreover, the AuNCs can scavenge ROS, resulting in high cell survival. Taking advantage of its light absorption capabilities and biocompatibility, this hybrid system can efficiently harvest sunlight and transfer photogenerated electrons to the cellular metabolism, thereby realizing CO2 fixation continuously over several days.19 The aforementioned hybrid brings together the high light-harvesting efficiency of solid-state semiconductors and the superior catalytic performance of living bacterial cells, thus realizing renewable and sustainable fuel production by artificial photosynthesis. In another study, Wang and co-workers fabricated bacterial flagella-templated 3D Au nanochains with broad optical absorption for solar-thermal energy conversion and electricity generation.117 To this aim, purified flagella served as templates for the assembly of pre-synthesized AuNPs (∼3 nm) along the cylindrical outer wall of the templates. By a seed-mediated growth process, the individual AuNPs on the templates became larger (up to 50 nm) and simultaneously the inter-particle distance was gradually reduced, to finally form 3D nanochains allowing the strong coupling of surface plasmons. Upon assembly into porous films, the 3D Au nanochains could effectively convert nearly the full spectrum of solar energy into heat, which was further efficiently converted into electricity through a commercial thermoelectric generation unit. Interestingly, the porous film structure formed by the 3D nanochains can effectively enhance the absorption of sunlight and reduce the loss of light through surface reflection. The reported findings may represent a potential avenue for optical property manipulation and solar energy related applications based on composites of living biofilms and noble metal nanoparticles.3.3 Bacterial microswimmers
Bacterial cells have evolved an impressive diversity of motility mechanisms, which can involve surface appendages, such as flagella, and internal structures such as gas vesicles.118 This property has been exploited for the fabrication of hybrid microswimmers composed of at least one living bacteria and one inanimate object.34 The combination of bacterial cells with abiotic systems, such as metal nanoparticles, leads to advanced levels of functionalization not reachable by each component separately. For example, the capability of bacteria to sense, move and respond to environmental stimuli, combined with the inherent physicochemical properties of the nanoparticles, can be harnessed to develop future therapeutic tools and motile sensors.23 This is a fast-growing research field that is attracting enormous attention because of its many potential applications. Huo and collaborators recently reported a comprehensive review of the principles of bacterial–nanoparticle hybrids for tumor therapy.9 For cancer treatment, the concentration of the therapeutic agents in tumor tissue is the key factor that determines treatment efficiency. Although a vast array of nanomaterials have been developed for tumor therapy, the lack of self-replicating and motility capabilities limits their clinical efficacy. Conversely, therapeutic bacteria can preferentially grow and actively spread in tumor-specific microenvironments.119 Therefore, the selective colonization of tumors by bacterial cells as carriers can make possible the delivery of therapeutics that can be toxic systemically, and thereby improve efficacy and safety profiles. Such bacterial vectors can be genetically programmed to produce anti-tumoral proteins, toxins, pro-drugs, and immunostimulants as therapeutic agents.120,121 The on-site controlled delivery of such therapeutics by engineered bacteria will increase antitumor efficacy, eliciting less toxic effects than current systemic anticancer treatment regimens.121 In a seminal study, Akin et al., coined the term “microbot” to describe a hybrid system consisting of attenuated Listeria Monocytogenes and streptavidin-coated polystyrene nanoparticles joined together through biotinylated antibodies.122 Bacteria decorated with nanoparticles and biotinylated plasmids were internalized by mammalian cells in vitro leading to the expression of the green fluorescent protein encoded in the plasmid vector. Mice injected with the microbots successfully expressed the genes in vivo, thereby demonstrating that the potential of the reported approach for gene delivery in an animal model. Metal nanoparticles could serve as potent radiosensitizers and transducers, as well as a functional carrier for the delivery of cytotoxic drugs for cancer treatment. Therefore, the rational combination of noble metal nanoparticles and bacteria may lead to hybrids with enhanced therapeutic capabilities. Park and collaborators labeled C. novyi-NT spores with branched AuNPs for intra-procedural X-ray CT monitoring of spore accumulation in tumors.85 This approach takes advantage of the tropism of certain bacterial species towards hypoxic regions of solid tumors, which are often refractory to therapeutic treatment. Importantly, this work assessed that nanoparticle coating had no impact on spore germination and proliferation rates, which is fundamental for the expected functionality of the hybrid system. Fan and co-workers developed a hybrid system for photothermal tumor therapy, namely thermally sensitive programmable bacteria (TPB), consisting of E. coli bacteria functionalized with biomineralized AuNPs bearing a heat-triggered TNF-α-producing genetic circuit.65 The photothermic gold nanoparticles were biosynthesized by TPB via the spontaneous redox reaction of bacteria to form TPB@Au (Fig. 6e). A specific NADPH-dependent reductase (e.g., nitrate reductase) could promote Au3+ to Au0 conversion through electron shuttle enzymatic metal reduction, leading to the deposition of AuNPs on the bacteria surface (Fig. 6f and g). The bacterial expression of therapeutic TNF-α was controlled by NIR irradiation, which raised the temperature at the tumor site due to the photothermal conversion of AuNPs, significantly reducing tumor volumes in mouse tumor models. In another study, gold nanorods were conjugated on the surface of Bifidobacterium breve for the photothermal ablation of tumors upon NIR light excitation.101 Unfortunately, it was found that the B. breve–AuNP hybrid system did not lead to a significant therapeutic effect. Most recently, attenuated Salmonella typhi Ty21a was investigated for the delivery of AuNPs to the tumor's hypoxic region and eventually enhanced the efficacy of radiation therapy.123In Table 1 we have summarized selected examples of hybrid systems indicating the bacterial species, nanoparticle size, whether they were biosynthesized or not, their surface ligand, and their toxicity.
| Bacterial species | NP | Size | Biogenic synthesis | Surface ligand | Application | Toxicitya | Ref. |
|---|---|---|---|---|---|---|---|
| a Toxicity of the NPs towards the inhibition of bacterial growth. N.D. Not determined | |||||||
| B. cereus | AuNP | 30 nm | No | Poly(L-lysine) | Living electronics | N.D. | 83 |
| E. coli | AuNP | 5 nm | No | Ni-NTA | Living electronics | N.D. | 28 |
| B. subtilis | AuNP | 20 nm | No | Citrate | Living electronics | Yes | 108 |
| G. sulfurreducens | AuNP | 5–50 nm | Yes | N.D. | Living electronics | No | 110 |
| S. oneidensis | AgNP | N.D. | Yes | N.D. | Living electronics | No | 113 |
| E. coli | AuNP | 10 nm | No | Ni-NTA | Living electronics | N.D. | 104 |
| E. coli | AuNP | 18 nm | No | PDDA | Solar fuel production | No | 124 |
| M. thermoacetica | AuNC | N.D. | Yes | N.D. | Solar fuel production | No | 19 |
| E. coli | AuNP | 60 nm | Yes | N.D. | Bacterial microswimmers | No | 101 |
| S. typhi | AuNP | N.D. | No | Folic acid | Bacterial microswimmers | No | 123 |
| E. coli | AuNP | <14 nm | Yes | N.D. | Biocatalysis | N.D. | 67 |
| E. coli | PdNP | N.D. | Yes | N.D. | Biocatalysis | Yes | 64 |
4. Perspectives and conclusions
This present mini-review explores current synergistic hybrid systems of noble metal nanoparticles and bacteria. Although researchers have long started to investigate the interactions and potential uses of nanoparticle–bacteria hybrid systems, recent advancements of nanotechnology and synthetic biology have opened new avenues to fully exploit their capabilities. The physicochemical properties and sensing capabilities of noble metal nanoparticles, together with the biological properties of the microbial cells such as self-replication, movement, chemotaxis, communication, metals homeostasis and resistance to metals, as well as programmable gene expression, have been harnessed in hybrid systems towards the development of future applications. Additionally, the native immunogenicity of bacteria offers another valuable asset, enabling them to be leveraged as powerful adjuvants and vaccine vectors.Herein, we have summarized the design and fabrication strategies, as well as recent applications in various fields including electronics, energy production and therapy. Owing to the inherent physicochemical properties and biocompatibility of metallic nanoparticles, most studies have been focused on gold, as silver displays antibacterial activities. However, certain bacterial species that naturally tolerate high concentrations of metal ions such as those employed for biogenic synthesis could be investigated in future hybrid systems. Other noble metals such as platinum and palladium have been explored in this field, although to a much lesser extent. In particular, palladium nanostructures show great potential owing to their catalytic effect, high thermal stability, oxidation resistance, and tunable optical response.125
As discussed here, by genetic manipulation it is now possible to program and rationally tune the implementation of nanomaterials in hybrid systems. Moreover, biogenic approaches enable to synthesize the metal nanoparticles inside bacteria, which paves the way for exciting new applications such as the development of artificial organelles and energy generation. The possibility to fabricate hybrid cells composed of noble metal nanoparticles bound to extracellular proteins and fibers (e.g. curli) is a new means to the generation of living electronics and sensors. For instance, the metal nanoparticles in hybrids can be used as high-sensitivity plasmonic nanosensors to monitor environmental pollution and report changes in a timely way.56 Bioimaging and biodiagnostics are other potential applications in which the bacterial–nanoparticle hybrids could be explored. As mentioned, the plasmonic properties of the metallic nanoparticles could be harnessed in the hybrid systems to fabricate living sensors and optical bio-probes based on LSPR and SERS. Also, metal nanoparticles have been combined with bacterial cellulose to fabricate an ample variety of biosensors, as recently reviewed.126,127 Typically, these composite materials are obtained after the extraction of the native cellulose. However, their use in live bacteria could be a new viable route for the fabrication of living hybrids. In a different application, the development of programmable biofilms with nanoparticle biotemplating capabilities holds great potential to fabricate large-scale designable biomaterials.62
This field is still in its infancy and many avenues need to be further investigated, such as testing a wider range of metal nanoparticles, alloys and their composites, as well as the exploration of new bacterial species and pathways for nanoparticle biosynthesis. Biogenic nanoparticles pose a few challenges which need to be addressed including the lack of monodispersity, low production rates, and batch-to-batch variations, which hamper their use for large-scale applications.128 We envision that the development of new microscopy techniques with improved resolution such as structured illumination microscopy will allow us to directly visualize the implementation and monitoring of the metal nanoparticles in live bacteria. It is important to note that both the strengths and limitations of the generated hybrids should be properly addressed in order to meet the requirements of living organisms, as well as to enable their translation towards real-world applications. For instance, the viability of the bacterial cell should not be compromised in the hybrid composite, as it is highly relevant for many applications. Clearly, this is a key issue that should be further investigated in future studies. Therefore, the development of non-toxic and robust strategies to functionalize microbial cells is of outermost importance.
The synergistic assembly and integration of noble metal nanoparticles and bacteria in a single architecture have realized the generation of “cyborg” cells with enhanced and new properties. The yet untapped potential of this synergistic combination offers an excellent opportunity to reveal new discoveries, exciting applications and tools.
Conflicts of interest
The authors declare no conflict of interest.Funding
Acknowledgements
This work was supported by the Spanish Ministry of Science and Innovation (grant PID2019-109669RB-I00 and PID2019-108954RB-I00), FOODSENS cofunded by FEDER through the program Interreg V-A España-Portugal (POCTEP) 2014–2020 and Xunta de Galicia/FEDER (grant GRC ED431C 2016-048). G. B. acknowledges financial support from the Xunta de Galicia (Centro singular de investigación de Galicia accreditation 2019–2022) and the European Union (European Regional Development Fund – ERDF).References
- C. Gilbert and T. Ellis, ACS Synth. Biol., 2019, 8, 1–15 CrossRef CAS PubMed.
- R. F. Fakhrullin, A. I. Zamaleeva, R. T. Minullina, S. A. Konnova and V. N. Paunov, Chem. Soc. Rev., 2012, 41 RSC.
- A. Y. Chen, C. Zhong and T. K. Lu, ACS Synth. Biol., 2015, 4, 8–11 CrossRef CAS PubMed.
- P. Q. Nguyen, N. D. Courchesne, A. Duraj-Thatte, P. Praveschotinunt and N. S. Joshi, Adv. Mater., 2018, 30, e1704847 CrossRef PubMed.
- W. V. Srubar 3rd, Trends Biotechnol., 2021, 39, 574–583 CrossRef PubMed.
- L. Shang, C. Shao, J. Chi and Y. Zhao, Acc. Mater. Res., 2020, 2, 59–70 CrossRef.
- T.-C. Tang, B. An, Y. Huang, S. Vasikaran, Y. Wang, X. Jiang, T. K. Lu and C. Zhong, Nat. Rev. Mater., 2020, 6, 332–350 CrossRef.
- Z. Guo, J. J. Richardson, B. Kong and K. Liang, Sci. Adv., 2020, 6, eaaz0330 CrossRef CAS PubMed.
- M. Huo, L. Wang, Y. Chen and J. Shi, Nano Today, 2020, 32, 100854 CrossRef CAS.
- Y. Chen, M. Du, J. Yu, L. Rao, X. Chen and Z. Chen, BIO Integr., 2020, 1, 25–36 CrossRef.
- S.-Y. Kwak, J. P. Giraldo, M. H. Wong, V. B. Koman, T. T. S. Lew, J. Ell, M. C. Weidman, R. M. Sinclair, M. P. Landry, W. A. Tisdale and M. S. Strano, Nano Lett., 2017, 17, 7951–7961 CrossRef CAS PubMed.
- J. P. Giraldo, M. P. Landry, S. M. Faltermeier, T. P. McNicholas, N. M. Iverson, A. A. Boghossian, N. F. Reuel, A. J. Hilmer, F. Sen, J. A. Brew and M. S. Strano, Nat. Mater., 2014, 13, 400–408 CrossRef CAS PubMed.
- G. Bodelón, C. Costas, J. Pérez-Juste, I. Pastoriza-Santos and L. M. Liz-Marzán, Nano Today, 2017, 13, 40–60 CrossRef.
- J. Song, X. Jia and K. Ariga, Small Methods, 2020, 4, 2000500 CrossRef CAS.
- S. Chen, A. Z. Weitemier, X. Zeng, L. He, X. Wang, Y. Tao, A. J. Y. Huang, Y. Hashimotodani, M. Kano, H. Iwasaki, L. K. Parajuli, S. Okabe, D. B. L. Teh, A. H. All, I. Tsutsui-Kimura, K. F. Tanaka, X. Liu and T. J. McHugh, Science, 2018, 359, 679–684 CrossRef CAS PubMed.
- A. H. All, X. Zeng, D. B. L. Teh, Z. Yi, A. Prasad, T. Ishizuka, N. Thakor, Y. Hiromu and X. Liu, Adv. Mater., 2019, 31, 1803474 CrossRef CAS PubMed.
- K. K. Sakimoto, A. B. Wong and P. Yang, Science, 2015, 351, 74–77 CrossRef PubMed.
- J. Guo, M. Suástegui, K. K. Sakimoto, V. M. Moody, G. Xiao, D. G. Nocera and N. S. Joshi, Science, 2018, 362, 813–816 CrossRef CAS PubMed.
- H. Zhang, H. Liu, Z. Tian, D. Lu, Y. Yu, S. Cestellos-Blanco, K. K. Sakimoto and P. Yang, Nat. Nanotechnol., 2018, 13, 900–905 CrossRef CAS PubMed.
- M. Martins, C. Toste and I. A. C. Pereira, Angew. Chem., 2021, 60, 9055–9062 CrossRef CAS PubMed.
- I. Santana, H. Wu, P. Hu and J. P. Giraldo, Nat. Commun., 2020, 11, 2045 CrossRef CAS PubMed.
- H. Wu, N. Tito and J. P. Giraldo, ACS Nano, 2017, 11, 11283–11297 CrossRef CAS PubMed.
- Z. Sun, P. Popp, C. Loderer and A. Revilla-Guarinos, Sensors, 2019, 20, 180 CrossRef PubMed.
- S. Liu and W. Xu, Front. Sens., 2020, 1, 586300 CrossRef.
- T. T. S. Lew, M. Park, J. Cui and M. S. Strano, Adv. Mater., 2020, 33, 2005683 CrossRef PubMed.
- L. K. Rivera-Tarazona, Z. T. Campbell and T. H. Ware, Soft Matter, 2021, 17, 785–809 RSC.
- A. Y. Chen, Z. Deng, A. N. Billings, U. O. S. Seker, M. Y. Lu, R. J. Citorik, B. Zakeri and T. K. Lu, Nat. Mater., 2014, 13, 515–523 CrossRef CAS PubMed.
- U. O. S. Seker, A. Y. Chen, R. J. Citorik and T. K. Lu, ACS Synth. Biol., 2016, 6, 266–275 CrossRef PubMed.
- S. Joshi, E. Cook and M. S. Mannoor, Nano Lett., 2018, 18, 7448–7456 CrossRef CAS PubMed.
- R. Oerlemans, S. Timmermans and J. C. M. van Hest, ChemBioChem, 2021, 22, 2051–2078 CrossRef CAS PubMed.
- A. Kumar, S. Kumar, N. Kumari, S. H. Lee, J. Han, I. J. Michael, Y.-K. Cho and I. S. Lee, ACS Catal., 2018, 9, 977–990 CrossRef.
- J. Lee, S. Dubbu, N. Kumari, A. Kumar, J. Lim, S. Kim and I. S. Lee, Nano Lett., 2020, 20, 6981–6988 CrossRef CAS PubMed.
- A. Sousa-Castillo, J. R. Couceiro, M. Tomás-Gamasa, A. Mariño-López, F. López, W. Baaziz, O. Ersen, M. Comesaña-Hermo, J. L. Mascareñas and M. A. Correa-Duarte, Nano Lett., 2020, 20, 7068–7076 CrossRef CAS PubMed.
- J. Bastos-Arrieta, A. Revilla-Guarinos, W. E. Uspal and J. Simmchen, Front. Robot. AI, 2018, 5, 97 CrossRef PubMed.
- J. Shao, M. Xuan, H. Zhang, X. Lin, Z. Wu and Q. He, Angew. Chem., Int. Ed., 2017, 56, 12935–12939 CrossRef CAS PubMed.
- M. Hu, X. Ge, X. Chen, W. Mao, X. Qian and W.-E. Yuan, Pharmaceutics, 2020, 12, 665 CrossRef CAS PubMed.
- L. P. Jahromi, M. A. Shahbazi, A. Maleki, A. Azadi and H. A. Santos, Adv. Sci., 2021, 8, 2002499 CrossRef CAS PubMed.
- K. A. Brown and P. W. King, Photosynth. Res., 2019, 143, 193–203 CrossRef PubMed.
- X. Fang, S. Kalathil and E. Reisner, Chem. Soc. Rev., 2020, 49, 4926–4952 RSC.
- Y. Ding, J. R. Bertram, C. Eckert, R. R. Bommareddy, R. Patel, A. Conradie, S. Bryan and P. Nagpal, J. Am. Chem. Soc., 2019, 141, 10272–10282 CrossRef CAS PubMed.
- V. Lovat, D. Pantarotto, L. Lagostena, B. Cacciari, M. Grandolfo, M. Righi, G. Spalluto, M. Prato and L. Ballerini, Nano Lett., 2005, 5, 1107–1110 CrossRef CAS PubMed.
- G. Cellot, E. Cilia, S. Cipollone, V. Rancic, A. Sucapane, S. Giordani, L. Gambazzi, H. Markram, M. Grandolfo, D. Scaini, F. Gelain, L. Casalis, M. Prato, M. Giugliano and L. Ballerini, Nat. Nanotechnol., 2008, 4, 126–133 CrossRef PubMed.
- A. Fabbro, A. Villari, J. Laishram, D. Scaini, F. M. Toma, A. Turco, M. Prato and L. Ballerini, ACS Nano, 2012, 6, 2041–2055 CrossRef CAS PubMed.
- S. Usmani, A. F. Biagioni, M. Medelin, D. Scaini, R. Casani, E. R. Aurand, D. Padro, A. Egimendia, P. Ramos-Cabrer, M. Scarselli, M. De Crescenzi, M. Prato and L. Ballerini, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 25212–25218 CrossRef CAS PubMed.
- R. Dhanker, T. Hussain, P. Tyagi, K. J. Singh and S. S. Kamble, Front. Microbiol., 2021, 12, 638003 CrossRef PubMed.
- M. S. Saveleva, K. Eftekhari, A. Abalymov, T. E. L. Douglas, D. Volodkin, B. V. Parakhonskiy and A. G. Skirtach, Front. Chem., 2019, 7, 179 CrossRef CAS PubMed.
- N. S. McCarty and R. Ledesma-Amaro, Trends Biotechnol., 2019, 37, 181–197 CrossRef CAS PubMed.
- B. Sepulveda, P. C. Angelome, L. M. Lechuga and L. M. Liz-Marzan, Nano Today, 2009, 4, 244–251 CrossRef CAS.
- P. K. Jain, X. H. Huang, I. H. El-Sayed and M. A. El-Sayed, Acc. Chem. Res., 2008, 41, 1578–1586 CrossRef CAS PubMed.
- J. N. Anker, W. P. Hall, O. Lyandres, N. C. Shah, J. Zhao and R. P. Van Duyne, Nat. Mater., 2008, 7, 442–453 CrossRef CAS PubMed.
- Y. N. Slavin, J. Asnis, U. O. Hafeli and H. Bach, J. Nanobiotechnol., 2017, 15, 65 CrossRef PubMed.
- G. V. Vimbela, S. M. Ngo, C. Fraze, L. Yang and D. A. Stout, Int. J. Nanomed., 2017, 12, 3941–3965 CrossRef CAS PubMed.
- T. Klaus, R. Joerger, E. Olsson and C. G. Granqvist, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 13611–13614 CrossRef CAS PubMed.
- T. J. Beveridge and R. G. Murray, J. Bacteriol., 1980, 141, 876–887 CrossRef CAS PubMed.
- X. Fang, Y. Wang, Z. Wang, Z. Jiang and M. Dong, Energies, 2019, 12, 190 CrossRef CAS.
- H. Bahrulolum, S. Nooraei, N. Javanshir, H. Tarrahimofrad, V. S. Mirbagheri, A. J. Easton and G. Ahmadian, J. Nanobiotechnol., 2021, 19, 86 CrossRef PubMed.
- T. Klaus-Joerger, R. Joerger, E. Olsson and C.-G. Granqvist, Trends Biotechnol., 2001, 19, 15–20 CrossRef CAS PubMed.
- Y. Choi and S. Y. Lee, Nat. Rev. Chem., 2020, 4, 638–656 CrossRef CAS.
- S. Iravani, Int. Scholarly Res. Not., 2014, 2014, 1–18 CrossRef PubMed.
- D. Zhang, X.-l. Ma, Y. Gu, H. Huang and G.-W. Zhang, Front. Chem., 2020, 8, 799 CrossRef CAS PubMed.
- Y. Choi, T. J. Park, D. C. Lee and S. Y. Lee, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 5944–5949 CrossRef CAS PubMed.
- P. Q. Nguyen, Z. Botyanszki, P. K. R. Tay and N. S. Joshi, Nat. Commun., 2014, 5, 4945 CrossRef CAS PubMed.
- Z. Jiang, X. Jin, Y. Li, S. Liu, X.-M. Liu, Y.-Y. Wang, P. Zhao, X. Cai, Y. Liu, Y. Tang, X. Sun, Y. Liu, Y. Hu, M. Li, G. Cai, X. Qi, S. Chen, L.-L. Du and W. He, Nat. Methods, 2020, 17, 937–946 CrossRef CAS PubMed.
- R. Qi, H. Zhao, X. Zhou, J. Liu, N. Dai, Y. Zeng, E. Zhang, F. Lv, Y. Huang, L. Liu, Y. Wang and S. Wang, Angew. Chem., 2021, 60, 5759–5765 CrossRef CAS PubMed.
- J.-X. Fan, Z.-H. Li, X.-H. Liu, D.-W. Zheng, Y. Chen and X.-Z. Zhang, Nano Lett., 2018, 18, 2373–2380 CrossRef CAS PubMed.
- L. Han, Y. Zhao, S. Cui and B. Liang, Appl. Biochem. Biotechnol., 2017, 185, 396–418 CrossRef PubMed.
- D.-Y. Tsai, Y.-J. Tsai, C.-H. Yen, C.-Y. Ouyang and Y.-C. Yeh, RSC Adv., 2015, 5, 87998–88001 RSC.
- G. Bodelon, V. Montes-Garcia, J. Perez-Juste and I. Pastoriza-Santos, Front. Cell. Infect. Microbiol., 2018, 8, 143 CrossRef PubMed.
- J. Lin, H. Zhang, Z. Chen and Y. Zheng, ACS Nano, 2010, 4, 5421–5429 CrossRef CAS PubMed.
- K. Zheng, M. I. Setyawati, D. T. Leong and J. Xie, Bioact. Mater., 2021, 6, 941–950 CrossRef CAS PubMed.
- K. Zheng, M. I. Setyawati, D. T. Leong and J. Xie, ACS Nano, 2017, 11, 6904–6910 CrossRef CAS PubMed.
- K. Zheng, M. I. Setyawati, D. T. Leong and J. Xie, Chem. Mater., 2018, 30, 2800–2808 CrossRef CAS.
- J. D. Prajapati, U. Kleinekathöfer and M. Winterhalter, Chem. Rev., 2021, 121, 5158–5192 CrossRef CAS PubMed.
- X.-H. N. Xu, W. J. Brownlow, S. V. Kyriacou, Q. Wan and J. J. Viola, Biochemistry, 2004, 43, 10400–10413 CrossRef CAS PubMed.
- L. M. Browning, K. J. Lee, P. K. Cherukuri, P. D. Nallathamby, S. Warren, J.-M. Jault and X.-H. N. Xu, RSC Adv., 2016, 6, 36794–36802 RSC.
- L. M. Browning, K. J. Lee, P. K. Cherukuri, T. Huang, P. Songkiatisak, S. Warren and X.-H. N. Xu, Analyst, 2018, 143, 1599–1608 RSC.
- P. K. Cherukuri, P. Songkiatisak, F. Ding, J.-M. Jault and X.-H. N. Xu, ACS Omega, 2020, 5, 1625–1633 CrossRef CAS PubMed.
- W. Pajerski, D. Ochonska, M. Brzychczy-Wloch, P. Indyka, M. Jarosz, M. Golda-Cepa, Z. Sojka and A. Kotarba, J. Nanopart. Res., 2019, 21, 186 CrossRef.
- A. S. Joshi, P. Singh and I. Mijakovic, Int. J. Mol. Sci., 2020, 21, 7658 CrossRef CAS PubMed.
- V. Berry, A. Gole, S. Kundu, C. J. Murphy and R. F. Saraf, J. Am. Chem. Soc., 2005, 127, 17600–17601 CrossRef CAS PubMed.
- Z. V. Feng, I. L. Gunsolus, T. A. Qiu, K. R. Hurley, L. H. Nyberg, H. Frew, K. P. Johnson, A. M. Vartanian, L. M. Jacob, S. E. Lohse, M. D. Torelli, R. J. Hamers, C. J. Murphy and C. L. Haynes, Chem. Sci., 2015, 6, 5186–5196 RSC.
- V. Berry, S. Rangaswamy and R. F. Saraf, Nano Lett., 2004, 4, 939–942 CrossRef CAS.
- V. Berry and R. F. Saraf, Angew. Chem., Int. Ed., 2005, 44, 6668–6673 CrossRef CAS PubMed.
- Y. Zheng, W. Liu, Z. Qin, Y. Chen, H. Jiang and X. Wang, Bioconjugate Chem., 2018, 29, 3094–3103 CrossRef CAS PubMed.
- W. Park, S. Cho, X. Huang, A. C. Larson and D.-H. Kim, Small, 2017, 13, 1602722 CrossRef PubMed.
- K. Whang, J.-H. Lee, Y. Shin, W. Lee, Y. W. Kim, D. Kim, L. P. Lee and T. Kang, Light: Sci. Appl., 2018, 7, 68 CrossRef PubMed.
- S. C. Hayden, G. Zhao, K. Saha, R. L. Phillips, X. Li, O. R. Miranda, V. M. Rotello, M. A. El-Sayed, I. Schmidt-Krey and U. H. F. Bunz, J. Am. Chem. Soc., 2012, 134, 6920–6923 CrossRef CAS PubMed.
- J. T. Buchman, A. Rahnamoun, K. M. Landy, X. Zhang, A. M. Vartanian, L. M. Jacob, C. J. Murphy, R. Hernandez and C. L. Haynes, Environ. Sci. Nano, 2018, 5, 279–288 RSC.
- L. F. Tadesse, C.-S. Ho, D.-H. Chen, H. Arami, N. Banaei, S. S. Gambhir, S. S. Jeffrey, A. A. E. Saleh and J. Dionne, Nano Lett., 2020, 20, 7655–7661 CrossRef CAS PubMed.
- X. Shi, H. L. Perry and J. D. E. T. Wilton-Ely, Nanotheranostics, 2021, 5, 155–165 CrossRef PubMed.
- D. P. Linklater, V. A. Baulin, X. Le Guével, J. B. Fleury, E. Hanssen, T. H. P. Nguyen, S. Juodkazis, G. Bryant, R. J. Crawford, P. Stoodley and E. P. Ivanova, Adv. Mater., 2020, 32, 2005679 CrossRef PubMed.
- T. Liu, Y. Wang, W. Zhong, B. Li, K. Mequanint, G. Luo and M. Xing, Adv. Healthcare Mater., 2019, 8, 1800939 CrossRef PubMed.
- B. Dai, L. Wang, Y. Wang, G. Yu and X. Huang, ChemistrySelect, 2018, 3, 7208–7221 CrossRef CAS.
- W. Feng, U. Kadiyala, J. Yan, Y. Wang, V. J. DiRita, J. S. VanEpps and N. A. Kotov, Chirality, 2020, 32, 899–906 CrossRef CAS PubMed.
- H. Wang, W. Ouyang, X. Zhang, J. Xue, X. Lou, R. Fan, X. Zhao, L. Shan and T. Jiang, J. Mater. Chem. B, 2019, 7, 4630–4637 RSC.
- J. Conde, J. T. Dias, V. Grazú, M. Moros, P. V. Baptista and J. M. de la Fuente, Front. Chem., 2014, 2, 48 Search PubMed.
- A. Heuer-Jungemann, N. Feliu, I. Bakaimi, M. Hamaly, A. Alkilany, I. Chakraborty, A. Masood, M. F. Casula, A. Kostopoulou, E. Oh, K. Susumu, M. H. Stewart, I. L. Medintz, E. Stratakis, W. J. Parak and A. G. Kanaras, Chem. Rev., 2019, 119, 4819–4880 CrossRef CAS PubMed.
- X. Bi, J. Yin, A. C. Guanbang and C.-F. Liu, Chem. – Eur. J., 2018, 24, 8042–8050 CrossRef CAS PubMed.
- S. Gautam, T. J. Gniadek, T. Kim and D. A. Spiegel, Trends Biotechnol., 2013, 31, 258–267 CrossRef CAS PubMed.
- S. Lim, O. K. Koo, Y. S. You, Y. E. Lee, M.-S. Kim, P.-S. Chang, D. H. Kang, J.-H. Yu, Y. J. Choi and S. Gunasekaran, Sci. Rep., 2012, 2, 456 CrossRef PubMed.
- C.-H. Luo, C.-T. Huang, C.-H. Su and C.-S. Yeh, Nano Lett., 2016, 16, 3493–3499 CrossRef CAS PubMed.
- P. M. Chaudhary, S. Sangabathuni, R. V. Murthy, A. Paul, H. V. Thulasiram and R. Kikkeri, Chem. Commun., 2015, 51, 15669–15672 RSC.
- J. K. Ajish, A. B. Kanagare, K. S. A. Kumar, M. Subramanian, A. D. Ballal and M. Kumar, ACS Appl. Bio Mater., 2019, 3, 1307–1317 CrossRef.
- Y. Cao, Y. Feng, M. D. Ryser, K. Zhu, G. Herschlag, C. Cao, K. Marusak, S. Zauscher and L. You, Nat. Biotechnol., 2017, 35, 1087–1093 CrossRef CAS PubMed.
- J. Huang, S. Liu, C. Zhang, X. Wang, J. Pu, F. Ba, S. Xue, H. Ye, T. Zhao, K. Li, Y. Wang, J. Zhang, L. Wang, C. Fan, T. K. Lu and C. Zhong, Nat. Chem. Biol., 2018, 15, 34–41 CrossRef PubMed.
- H. Dong, D. A. Sarkes, J. J. Rice, M. M. Hurley, A. J. Fu and D. N. Stratis-Cullum, Langmuir, 2018, 34, 5837–5848 CrossRef CAS PubMed.
- N. S. Malvankar, M. Vargas, K. P. Nevin, A. E. Franks, C. Leang, B.-C. Kim, K. Inoue, T. Mester, S. F. Covalla, J. P. Johnson, V. M. Rotello, M. T. Tuominen and D. R. Lovley, Nat. Nanotechnol., 2011, 6, 573–579 CrossRef PubMed.
- Y. He, J. Yuan, F. Su, X. Xing and G. Shi, J. Phys. Chem. B, 2006, 110, 17813–17818 CrossRef PubMed.
- L. Zou, F. Zhu, Z.-e. Long and Y. Huang, J. Nanobiotechnol., 2021, 19, 120 CrossRef PubMed.
- M. Chen, X. Zhou, X. Liu, R. J. Zeng, F. Zhang, J. Ye and S. Zhou, Biosens. Bioelectron., 2018, 108, 20–26 CrossRef CAS PubMed.
- L. J. Bird, E. L. Onderko, D. A. Phillips, R. L. Mickol, A. P. Malanoski, M. D. Yates, B. J. Eddie and S. M. Glaven, MRS Commun., 2019, 9, 505–517 CrossRef CAS.
- C. Santoro, C. Arbizzani, B. Erable and I. Ieropoulos, J. Power Sources, 2017, 356, 225–244 CrossRef CAS PubMed.
- B. Cao, Z. Zhao, L. Peng, H.-Y. Shiu, M. Ding, F. Song, X. Guan, C. K. Lee, J. Huang, D. Zhu, X. Fu, G. C. L. Wong, C. Liu, K. Nealson, P. S. Weiss, X. Duan and Y. Huang, Science, 2021, 373, 1336–1340 CrossRef CAS PubMed.
- P. P. Li, Y. J. Jiang, R. B. Song, J. R. Zhang and J. J. Zhu, J. Mater. Chem. B, 2021, 9, 1638–1646 RSC.
- Q. Pan, X. Tian, J. Li, X. Wu and F. Zhao, Appl. Energy, 2021, 292, 116885 CrossRef CAS.
- V. Müller, Appl. Environ. Microbiol., 2003, 69, 6345–6353 CrossRef PubMed.
- L. Wang, P. H. Qiu, T. Yang, N. Y. Zhou, M. M. Zhai, Y. Li, Y. D. Zhou, S. L. Zou, M. Y. Yang and C. B. Mao, Mater. Horiz., 2021, 8, 2097–2105 RSC.
- K. F. Jarrell and M. J. McBride, Nat. Rev. Microbiol., 2008, 6, 466–476 CrossRef CAS PubMed.
- N. S. Forbes, Nat. Rev. Cancer, 2010, 10, 785–794 CrossRef CAS PubMed.
- L. Rong, Q. Lei and X.-Z. Zhang, ACS Appl. Bio Mater., 2020, 3, 8136–8145 CrossRef CAS.
- H. Bayley, T. Harimoto and T. Danino, Emerging Top. Life Sci., 2019, 3, 623–629 CrossRef PubMed.
- D. Akin, J. Sturgis, K. Ragheb, D. Sherman, K. Burkholder, J. P. Robinson, A. K. Bhunia, S. Mohammed and R. Bashir, Nat. Nanotechnol., 2007, 2, 441–449 CrossRef CAS PubMed.
- A. Kefayat, F. Ghahremani, H. Motaghi, S. Rostami and M. A. Mehrgardi, J. Drug Targeting, 2018, 27, 315–324 CrossRef PubMed.
- P. Li, Y. Jiang, R.-B. Song, J.-R. Zhang and J.-J. Zhu, J. Mater. Chem. B, 2021, 9, 1638–1646 RSC.
- S. De Marchi, S. Núñez-Sánchez, G. Bodelón, J. Pérez-Juste and I. Pastoriza-Santos, Nanoscale, 2020, 12, 23424–23443 RSC.
- J. Van Rie and W. Thielemans, Nanoscale, 2017, 9, 8525–8554 RSC.
- F. G. Torres, O. P. Troncoso, K. N. Gonzales, R. M. Sari and S. Gea, Med. Devices Sens., 2020, 3, e10102 CAS.
- S. Ghosh, R. Ahmad, M. Zeyaullah and S. K. Khare, Front. Chem., 2021, 9, 626834 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2021 |




