Open Access Article
Open Access ArticleRational design of a stapled JAZ9 peptide inhibiting protein–protein interaction of a plant transcription factor†
Kaho
Suzuki
a,
Yousuke
Takaoka
 *a and
Minoru
Ueda
*a and
Minoru
Ueda
 *ab
*ab
aGraduate School of Science, Tohoku University, 6-3, Aramaki-Aza-Aoba, Aoba-ku, Sendai, 980-8578, Japan. E-mail: ytakaoka@tohoku.ac.jp
bGraduate School of Life Science, Tohoku University, 6-3, Aramaki-Aza-Aoba, Aoba-ku, Sendai, 980-8578, Japan
First published on 5th January 2021
Abstract
We herein describe the development of a stapled peptide inhibitor for a jasmonate-related transcription factor. The designed peptide selectively inhibited MYCs, master-regulators of jasmonate signaling, and selectively suppressed MYC-mediated gene expression in Arabidopsis thaliana. It is proposed as a novel chemical tool for the analysis of MYC related jasmoante signaling.
Plants are sessile and subjected to environmental changes, and have evolved unique signaling systems which include a myriad of transcription factors (TFs) under the control of small molecular plant hormones.1,2 One of these plant hormones, jasmonoyl-L-isoleucine (JA-Ile), contributes to the regulation of plant defense responses to environmental stresses.3,4 For example, JA-Ile causes protein–protein interactions (PPI) between F-box protein CORONATINE INSENSITIVE 1 (COI1) and jasmonate-ZIM-domain (JAZ) repressor protein,5–7 which triggers ubiquitination and subsequent degradation of the JAZ repressors through the 26S-proteasome pathway. In the absence of JA-Ile, JAZ repressor binds to and represses various JA-related transcriptional factors including MYC, the master regulator of JA-signaling.8,9 A variety of TFs are involved in this signaling process and responsible for a number of JA responses, including a defense response to pathogen and herbivore attack, secondary metabolite production, fertility, senescence, and growth inhibition.8,9 Therefore, to elucidate the function of each TF, chemical tools (agonists or antagonists) specific to each individual TF are needed.10–12
Herein, we focus on the JAZ repressor proteins which function as hubs in this complex signaling machinery. JAZ has dual functions as a component of COI1–JAZ co-receptor and repressor of many TFs. We developed TMR-JAZ9st3, a novel stapled peptide-based MYC inhibitor, the structure of which is based on the crystal structure of JAZ9 in complex with MYC3. TMR-JAZ9st3 selectively and strongly binds MYCs, but has almost no affinity with COI1 either in vitro or in planta, and therefore could serve as a competitive inhibitor of MYC-related JA-signaling in planta, little affecting MYC-independent JA-signaling. Thus, TMR-JAZ9st3 would be a powerful chemical tool for the MYC-related JA-signaling pathway.
The dual function of JAZ (as a component of the COI1–JAZ co-receptor and repressor of TFs) can be explained by reference to its structure – JAZ proteins incorporate in the C-terminal region the highly conserved Jas motif, which is an established binding domain for both COI1 and MYCs.9,13 In the reported 3D structures of the complexes of MYC3–JasJAZ9 and COI1–JA-Ile–JasJAZ1, the Jas motif had undergone a dynamic structural change: the JasJAZ9 peptide in complex with MYC3 forms an extended α-helix conformation, whereas the JasJAZ1 peptide in complex with COI1–JA-Ile formed a bipartite conformation (N-terminal loop and C-terminal extended α-helix) (Fig. 1a). Based on these findings, a covalently ‘stapled’ α-helical Jas peptide would be expected to function as a MYC-selective ligand, unable to effectively bind with COI1–JA-Ile. To test this hypothesis, we designed several types of helix-stapled JasJAZ9 peptides based on conventional hydrocarbon peptide stapling technology14,15 and guided by the reported crystal structure of MYC3-JasJAZ9. Initially, four amino acids (Q221, A225, A228 and E232) in JasJAZ9 were chosen as stapling sites based on their location not being in direct contact with MYC3 (Fig. 1b). Several groups recently reported bicyclic double-stapled peptides which demonstrated marked target affinity, efficient cellular uptake, and higher stability compared to the single-stapled peptides in mammalian cells.16,17 Therefore, we designed non-stapled peptide (JAZ9wt (1)), two single-stapled JAZ peptides (JAZ9st1 (2) and JAZ9st2 (3)), and a double-stapled JAZ9 peptide (JAZ9st3 (4)), as shown in Fig. 1c. All four were covalently conjugated with the reporter tetramethylrhodamine (TMR) and evaluated for their binding affinity with MYC3. Stapled and non-stapled peptides were synthesized using SPPS and purified by HPLC and characterized by MALDI-TOF MS analyses after cleavage from the resin (Scheme S1 and Fig. S1 and S2, ESI†).
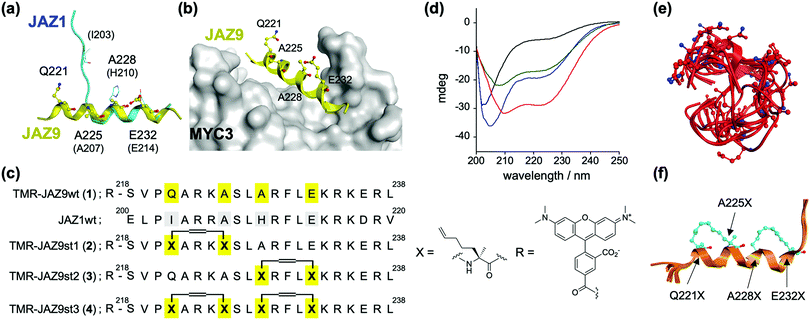 | ||
| Fig. 1 (a) Superimposed structures of JasJAZ1 (cyan) complexed with COI1–JA-Ile (PDB ID: 3OGL) or JasJAZ9 (yellow) complexed with MYC3 (PDB ID: 4RS9). (b) Crystal structure of MYC3–JAZ9 showing the stapling sites (PDB ID: 4RS9). (c) Molecular design of stapled JAZ9-peptides with amino acid sequences of JasJAZ1/JasJAZ9, and chemical structures of the building block for stapling (X) and tetramethylrhodamine (R). (d) CD spectra of TMR-JAZ9 peptides (each 10 μM) (1; black, 2; blue, 3; green, 4; red) in PBS buffer (pH 7.4). (e and f) Superposition of the most stable top 5 model structures obtained by conformational searching with MD simulation; (e) JAZ9wt, (f) JAZ9st3. The amino acids at the stapling sites (Q221X, A225X, A228X, and E232X) are shown as stick models. | ||
The helicities of the synthesized stapled peptides were examined by circular dichroism (CD) experiments. From these results (Fig. 1d), 1 was inferred to form a random loop structure in the buffer solution (the helix content was calculated to be 6%, and random structure to be 81%, respectively).18 The 3D structure of this peptide was predicted to be mainly disordered and random, by conformational searching with an MD simulation (Fig. 1e). In contrast, the ellipticity values of the stapled peptides at both 208 nm and 220 nm grow negatively, and the helix contents of these peptides were calculated to be 25, 25, and 45%, for 2, 3, and 4, respectively (Fig. 1d). The predicted 3D structures by the in silico MD simulation also supported these results (Fig. 1e, f and Fig. S3, ESI†). The helicities of the top five predicted structures of these peptides were calculated to be 8%, 31%, 32%, or 68% for JAZ9wt, JAZ9st1, JAZ9st2, or JAZ9st3, respectively, consistent with the analyses by CD. These results demonstrated that the 3D structure of JasJAZ9 peptide is intrinsically disordered, and the helicities of our stapled peptides increased with increasing stapling number.
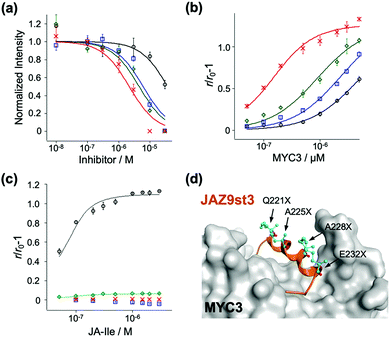
To examine the affinity of these stapled peptides for MYCs and COI1–JA-Ile, we prepared the JAZ-binding domains of MYC and COI1 (His6-SUMO-tagged MYC2(55-259), MYC3(44-238), MYC4(55-253), and GST-tagged COI1(full-length)) according to the previous reports.7,10,19 The interaction between MYC3 and non-stapled or stapled peptides (1–4) was confirmed using the AlphaScreen inhibitory assays between His6-SUMO-MYC3 and biotin-conjugated JasJAZ9.19 As shown in Fig. 2a, all four peptides competitively inhibited the MYC-JasJAZ9 interaction; the IC50 value of 4 was determined to be 2.7 μM, 10-fold lower than that of 1, and clearly demonstrating that our stapled peptides are more potent inhibitors of MYC3 compared with non-stapled JAZ9 peptide (Table 1).
| MYC3 | MYC2 | MYC4 | COI1–JA-Ile | ||
|---|---|---|---|---|---|
| IC50a/μM | K d /μM | K d /μM | K d /μM | K d /nM | |
| a The values were obtained by AlphaScreen, shown in Fig. 2a. b The values were obtained by FA experiments, shown in Fig. 2b, c and Fig. S4 (ESI). c The value could not be determined due to the low signal change. | |||||
| TMR-JAZ9wt (1) | 37 ± 10 | 5.0 ± 1.1 | 3.6 ± 0.8 | 11 ± 3.7 | 15 ± 4.3 |
| TMR-JAZ9st1 (2) | 6.0 ± 1.6 | 2.4 ± 0.5 | 1.5 ± 0.2 | 2.6 ± 0.3 | n.d.c |
| TMR-JAZ9st2 (3) | 4.5 ± 1.4 | 0.88 ± 0.2 | 0.73 ± 0.1 | 1.5 ± 0.2 | 29 ± 11 |
| TMR-JAZ9st3 (4) | 2.2 ± 0.5 | 0.10 ± 0.02 | 0.16 ± 0.06 | 0.88 ± 0.2 | n.d.c |
Quantitative evaluation of the binding affinity was accomplished in a fluorescence anisotropy (FA) assay; the FA values of these peptides were significantly increased upon addition of His6-SUMO-MYC3 in a dose dependent manner (Fig. 2b). The apparent Kd value of 1 with His6-SUMO-MYC3 was calculated to be 5.0 μM (Table 1). In contrast, the Kd value of 4 was 0.10 μM, which is 50-fold, 24-fold, or 8.8-fold lower than that of 1, 2 or 3, respectively (Table 1). The significant enhancement in the affinity of the double-stapled peptide was also observed in the cases of MYC3 isoforms, MYC2 and MYC4 (Fig. S4, ESI† and Table 1). Moreover, the stapled peptide 4 showed similar affinity to all MYC isoforms (0.10, 0.16, or 0.88 μM for MYC3, MYC2, or MYC4, respectively), which is consistent with the sequence homology analysis of the binding sites of these three MYCs for a JAZ (Fig. S5, ESI†). These results suggest that the secondary structure of the Jas motif comprises an extended helix when interacting with MYC2 or MYC4, as is the case with MYC3.
To verify the target selectivity of our stapled peptides, we undertook a binding assay with COI1–JA-Ile, by FA-based methods.20 Upon addition of JA-Ile to a solution of GST-COI1 and 1, the FA values were significantly increased (Fig. 2c). The apparent Kd value with 1 and GST-COI1 was 13 nM, consistent with previous results (Table 1) (Kd was found to be 10 nM, by using fluorescein-conjugated JAZ9wt).20 A similar value was obtained with 3, a single stapled peptide in the C-terminal region of the Jas motif which will have the same N-terminal random loop conformation when it binds with COI1 (Fig. 2c and Table 1). In contrast, no significant change in FA value was observed for 2 and 4, which are stapled in the N-terminal region of the Jas motif (Fig. 2c). These results clearly demonstrate that the double-stapling strategy markedly enhances the binding affinity for MYCs and selectivity for MYCs over COI1.
To better understand the binding between the designed peptides with MYC3, we undertook an in silico molecular docking simulation. As shown in Fig. 2d and Fig. S6 (ESI†), the double stapled JAZ9 peptide (JAZ9st3) was found to tightly bind to MYC3, without perturbation of this interaction by the stapling sites of JAZ9st3. These results suggest that our design strategy was largely successful both thermodynamically and structurally.
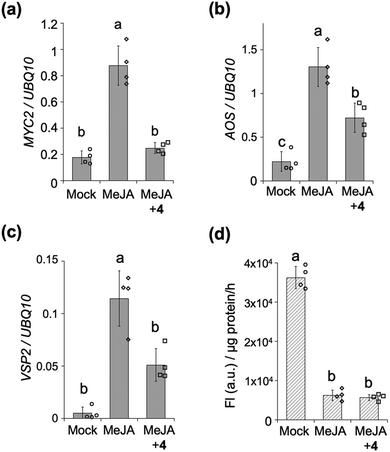
We next assessed the inhibitory effect of the stapled peptide 4 on MYC-mediated gene expression in Arabidopsis seedlings – treatment of which with methyl jasmonate (MeJA, a precursor of JA-Ile) was previously reported to strongly upregulate the expression of MYC-regulated marker genes such as MYC2, JAZ1, LOX2, AOS (JA-Ile biosynthesis) or VSP2 (defense responses) (Fig. 3a–c and Fig. S7, ESI†).21,22 Upon co-treatment of the seedlings with MeJA and 4, the expression of these MYC-mediated marker genes was significantly suppressed (Fig. 3a–c and Fig. S7, ESI†). The extent of inhibition of the expression of AOS and VSP2 was found to depend on the dose of 4, up to 20 μM (Fig. S8, ESI†), whereas the expression of AOS was upregulated in the presence of 50 μM of 4 (data not shown). Very little inhibition of the expression of AOS or VSP2 was observed with 1 (Fig. S9, ESI†), indicating that the inhibition of MeJA-mediated expressions of MYC-regulated genes in vivo by stapled 4 can be attributed to its much higher binding affinity for the MYCs compared to the non-stapled 1.
The MYC-selective affinity of the double-stapled peptide 4 suggested it to have a COI1-independent function in planta. COI1-dependent signaling in planta can be quantified using transgenic β-glucuronidase (GUS)-reporter line (35S: JAZ1-GUS): MeJA causes degradation of the JAZ1-GUS fusion protein through COI1, and the downstream signaling could be quantified as reduced GUS staining (Fig. 3d and Fig. S10, ESI†). This degradation was also observed even in the presence of 4, demonstrating that 4 does not affect the interaction between COI1 and JAZ in vivo as well as in vitro.
Conclusions
In conclusion, a double-stapled JAZ9 peptide selective inhibitor of MYC TFs, the master regulator of jasmonate signaling, has been designed based on the crystal structure of MYC3–JasJAZ9, and tested.19 The 3D structure of this peptide resembles a helix structure as MYC3-binding form, resulting in a 50-fold higher affinity for MYC3 compared to the non-stapled peptide – perhaps due to chemical stabilization of the 3D structure of the peptide entropically favoring binding with MYC3. Such affinity enhancement was also observed for MYC2 and 4, suggesting that the MYC family proteins bind to the Jas motif in a manner similar to that for MYC3; analysis of the 3D structures of MYC2 and 4 is pending. In contrast, no binding was observed between our double-stapling peptide and COI1–JA-Ile – presumably because this peptide cannot form a loop structure as COI1-binding form.7 Moreover, our double-stapled peptide 4 could suppress the jasmonate-induced MYC-related gene expression in Arabidopsis thaliana, demonstrating that our peptide would be a useful chemical tool for analyses of the MYC-related jasmonate signaling, as well as open the door to develop various peptide-based functional ligands in planta in the future.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
We thank Prof. S. Nishizawa and Dr. Y. Sato (Tohoku University) for the use of the circular dichroism spectrometer. We also thank Mr. D. Kanayama (Tohoku University) for his technical support on AlphaScreen assay, and Ms. Ika N. Azizah (Tohoku University) for her technical support on protein preparation and peptide synthesis. This work was financially supported by a Grant-in-Aid for Scientific Research from JSPS, Japan (no. 17H06407, 18KK0162, and 20H00402 for MU, and 19H05283 for YT), JSPS A3 Foresight Program (MU), JSPS Core-to-Core Program Asian Chemical Biology Initiative (MU), JST-PREST Grant JPMJPR16Q4 (YT), and Takeda Science Foundation (YT).Notes and references
- A. Santner and M. Estelle, Nature, 2009, 459, 1071–1078 CrossRef CAS.
- T. A. Meraj, J. Fu, M. A. Raza, C. Zhu, Q. Shen, D. Xu and Q. Wang, Genes, 2020, 11, 346 CrossRef CAS.
- C. Wasternack and B. Hause, Ann. Bot., 2013, 111, 1021–1058 CrossRef CAS.
- S. Fonseca, A. Chini, M. Hamberg, B. Adie, A. Porzel, R. Kramell, O. Miersch, C. Wasternack and R. Solano, Nat. Chem. Biol., 2009, 5, 344–350 CrossRef CAS.
- A. Chini, S. Fonseca, G. Fernandez, B. Adie, J. M. Chico, O. Lorenzo, G. Garcia-Casado, I. Lopez-Vidriero, F. M. Lozano, M. R. Ponce, J. L. Micol and R. Solano, Nature, 2007, 448, 666–671 CrossRef CAS.
- B. Thines, L. Katsir, M. Melotto, Y. Niu, M. Ajin, G. Liu, K. Nomura, S. Y. He, G. A. Howe and J. Browse, Nature, 2007, 448, 661–665 CrossRef CAS.
- L. B. Sheard, X. Tan, H. Mao, J. Withers, G. Ben-Nissan, T. R. Hinds, Y. Kobayashi, F. F. Hsu, M. Sharon, J. Browse, S. Y. He, J. Rizo, G. A. Howe and N. Zheng, Nature, 2010, 468, 400–405 CrossRef CAS.
- L. Pauwels and A. Goossens, Plant Cell, 2011, 23, 3089–3100 CrossRef CAS.
- A. Chini, S. Gimenez-Ibanez, A. Goossens and R. Solano, Curr. Opin. Plant Biol., 2016, 33, 147–156 CrossRef CAS.
- Y. Takaoka, M. Iwahashi, A. Chini, H. Saito, Y. Ishimaru, S. Egoshi, N. Kato, M. Tanaka, K. Bashir, M. Seki, R. Solano and M. Ueda, Nat. Commun., 2018, 9, 3654 CrossRef.
- C. Meesters, T. Monig, J. Oeljeklaus, D. Krahn, C. S. Westfall, B. Hause, J. M. Jez, M. Kaiser and E. Kombrink, Nat. Chem. Biol., 2014, 10, 830–836 CrossRef CAS.
- I. Monte, M. Hamberg, A. Chini, S. Gimenez-Ibanez, G. Garcia-Casado, A. Porzel, F. Pazos, M. Boter and R. Solano, Nat. Chem. Biol., 2014, 10, 671–676 CrossRef CAS.
- A. Wager and J. Browse, Front. Plant Sci., 2012, 3, 41 CAS.
- C. E. Schafmeister, J. Po and G. L. Verdine, J. Am. Chem. Soc., 2000, 122, 5891–5892 CrossRef CAS.
- P. M. Cromm, J. Spiegel and T. N. Grossmann, ACS Chem. Biol., 2015, 10, 1362–1375 CrossRef CAS.
- G. H. Bird, N. Madani, A. F. Perry, A. M. Princiotto, J. G. Supko, X. He, E. Gavathiotis, J. G. Sodroski and L. D. Walensky, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 14093–14098 CrossRef CAS.
- G. J. Hilinski, Y.-W. Kim, J. Hong, P. S. Kutchukian, C. M. Crenshaw, S. S. Berkovitch, A. Chang, S. Ham and G. L. Verdine, J. Am. Chem. Soc., 2014, 136, 12314–12322 CrossRef CAS.
- A. Micsonai, F. Wien, L. Kernya, Y.-H. Lee, Y. Goto, M. Réfrégiers and J. Kardos, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E3095–E3103 CrossRef CAS.
- F. Zhang, J. Yao, J. Ke, L. Zhang, V. Q. Lam, X. F. Xin, X. E. Zhou, J. Chen, J. Brunzelle, P. R. Griffin, M. Zhou, H. E. Xu, K. Melcher and S. Y. He, Nature, 2015, 525, 269–273 CrossRef CAS.
- Y. Takaoka, K. Nagumo, I. N. Azizah, S. Oura, M. Iwahashi, N. Kato and M. Ueda, J. Biol. Chem., 2019, 294, 5074–5081 CrossRef CAS.
- O. Lorenzo, J. M. Chico, J. J. Sanchez-Serrano and R. Solano, Plant Cell, 2004, 16, 1938–1950 CrossRef CAS.
- F. Schweizer, P. Fernandez-Calvo, M. Zander, M. Diez-Diaz, S. Fonseca, G. Glauser, M. G. Lewsey, J. R. Ecker, R. Solano and P. Reymond, Plant Cell, 2013, 25, 3117–3132 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0cb00204f |
| This journal is © The Royal Society of Chemistry 2021 |