Open Access Article
Open Access ArticleIn vivo delivery of a fluorescent FPR2/ALX-targeted probe using focused ultrasound and microbubbles to image activated microglia†
Sophie V.
Morse‡
 a,
Tamara
Boltersdorf‡
b,
Tiffany G.
Chan
a,
Tamara
Boltersdorf‡
b,
Tiffany G.
Chan
 ab,
Felicity N. E.
Gavins
*c,
James J.
Choi
ab,
Felicity N. E.
Gavins
*c,
James J.
Choi
 *a and
Nicholas J.
Long
*a and
Nicholas J.
Long
 *b
*b
aDepartment of Bioengineering, Imperial College London, London, SW7 2BP, UK. E-mail: j.choi@imperial.ac.uk
bDepartment of Chemistry, Imperial College London, Molecular Sciences Research Hub, London, W12 0BZ, UK. E-mail: n.long@imperial.ac.uk
cDepartment of Life Sciences, Brunel University London, Uxbridge, Middlesex UB8 3PH, UK. E-mail: felicity.gavins@brunel.ac.uk
First published on 23rd September 2020
Abstract
To image activated microglia, a small-molecule FPR2/ALX-targeted fluorescent probe was locally delivered into the brain using focused ultrasound and microbubbles. The probe did not co-localise with neurons or astrocytes but accumulated in activated microglia, making this a potential imaging tool for future drug discovery programs focused on neurological disorders.
Microglia are the resident immune cells of the brain and play a central role in maintaining homeostasis. They act as a host defense system by sensing and responding to changes in their environment. In brain injury and multiple disease states, there is often inflammation which is mediated by activated microglia.1–4 For instance, the progression of Alzheimer's disease, multiple sclerosis, Huntington's disease and Parkinson's disease have been linked to microglial activity, making these cells a potential marker of the onset and progression of neurological disease.5,6 As a result, there is a need for tools that enable the reliable imaging of activated microglia.
Microglia, in their resting state, have a ramified phenotype with a high number of processes that are constantly moving and interacting with neighbouring blood vessels, neurons and astrocytes.7 Microglial activation, however, is accompanied by several characteristic morphological changes within the cell, such as the retraction of the processes, which results in the microglia having an ameboid-shaped cell body and short or non-existent processes.7,8 Previous efforts to image activated microglia have focused around the development of radioligands that target the translocator protein (TSPO) receptor, which is upregulated in activated microglia and found on the outer mitochondrial membrane.9,10 Using this receptor as a microglial marker, however, can be problematic as TSPO is also abundantly expressed in a variety of healthy brain tissues. More recently, probes that allow the visualisation of cyclooxygenase (COX) enzymes, which are highly expressed key players in inflammation, have also been synthesised to image activated microglia. However, issues arising with these reported probes include low in vivo sensitivity and non-specific binding of these radioligands.11,12
Another possible target to visualise microglia is FPR2/ALX (formyl peptide receptor 2 or the lipoxin A4 receptor, previously known as formyl peptide receptor-like 1 [FPRL1]), a G-protein-coupled receptor which is expressed on the cell surface of immune cells.13 This receptor is known to produce ligand-dependent responses that can be both pro- or anti-inflammatory.14,15 In resting state microglia, FPR2/ALX is expressed at low levels, however it is upregulated in inflammatory conditions.16,17
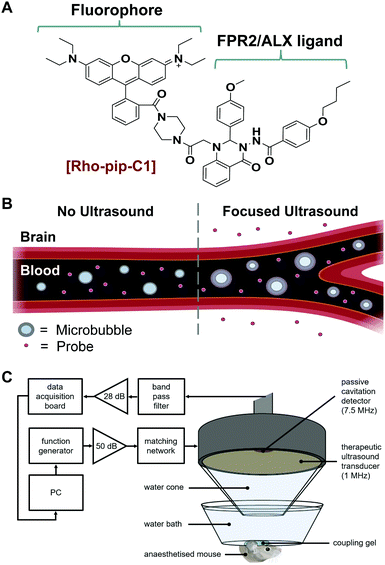
We have recently reported the design and evaluation of a targeted, small-molecule, fluorescent compound (Rho-pip-C1; Fig. 1A) that binds to neutrophils via FPR2/ALX.18 We hypothesised that Rho-pip-C1 could allow the selective visualisation of activated microglia and be used as a tool to study the role of activated microglia and this specific receptor in neuroinflammation.
Rho-pip-C1 was prepared by derivatizing Rhodamine B at the carboxylic acid position to form an amide bond with a piperazine amine.18 This modification prevents rhodamine spirocyclisation and as a result precludes formation of the non-fluorescent, cyclic rhodamine form.19 The strongly fluorescent rhodamine derivative was then combined with Quin C1,20 a FPR2/ALX-targeting ligand.21 The photophysical properties of the compound were found to be compatible with biological imaging (excitation wavelength: 560 nm, emission wavelength: 582 nm). We also previously demonstrated that Rho-pip-C1 is non-toxic to neutrophils in vitro at relevant concentrations (10−6–10−9 M) and is suitable for imaging neutrophils in vascular inflammation.18 Here, we sought to explore the possibility of this probe to act as a marker for activated microglia and the associated cerebral neuroinflammation.
Typically, small molecule probes, such as Rho-pip-C1 do not cross the blood–brain barrier (BBB) due to their size and the number of hydrogen bonds that they form.22 To circumvent this issue, a non-invasive and localised way to get such probes across the BBB is to use focused ultrasound and microbubbles.23 This method involves injecting the probe and clinically approved microbubbles into the bloodstream. Ultrasound is then focused onto the brain region of interest where the ultrasound pulses will drive the microbubbles to oscillate within the blood vessels (Fig. 1B). The oscillations exert mechanical stresses on the vessel walls within the region of interest, leading to an increased permeability of the BBB.
One of the major hurdles in the development of imaging agents for brain targets is the BBB. Traditionally, the way to achieve efficient passage across the BBB is to prepare lipophilic molecules under 400 Da in size with a lower propensity to form hydrogen bonds, which severely limits the scope of potential candidates.24 Furthermore, although these properties have been found to aid in crossing the BBB, they also lead to enhanced accumulation in peripheral tissue, higher toxicity and suitable concentrations within the brain are difficult to achieve.25–28
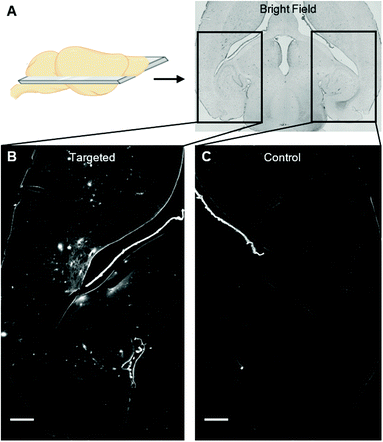
We instead delivered Rho-pip-C1 to a specific region of the brain in vivo using a focused ultrasound setup combined with systemically administered microbubbles (Fig. 1C). In this method, the left hippocampus of C57BL/6 mice was treated with ultrasound pulses (frequency: 1 MHz; peak-negative pressure: 530 kPa; pulse length: 10 ms; pulse repetition frequency: 0.5 Hz; sonication duration: 250 s), while the right hippocampus was treated as a control (no ultrasound treatment). During ultrasound exposure, SonoVue microbubbles were injected intravenously through the tail vein followed by Rho-pip-C1 (1.4 mg ml−1) and their presence was verified by listening to the microbubble's acoustic emissions with a passive cavitation detector. The ultrasound treatment in these experiments was not only used to get the probe into the brain but also to create an environment in which microglia would be activated. This was facilitated by choosing a high ultrasound pressure (530 kPa) which would lead to microglial activation,29 due to the microbubbles inducing higher stresses on the vessel walls. Following the ultrasound treatment, transcardial perfusion was performed and the brains were extracted, sectioned into 30 μm horizontal brain slices and imaged using bright-field and fluorescence microscopy (Fig. 2). Successful delivery to the left hippocampus (Fig. 2B), which had been treated using focused ultrasound to deliver the probe, was quantified via the normalised optical density (average fluorescence intensity in the targeted region subtracted by that of the control region).30,31 In the right hemisphere (Fig. 2C), where no ultrasound treatment had occurred, no fluorescent signal was observed. This indicates that the compound does not cross the BBB by itself, but that we can achieve localised delivery with focused ultrasound and microbubbles.
The probe was delivered in all brains (see normalised optical density quantification, Fig. S1, n = 3, ESI†) with a heterogeneous distribution pattern, resulting in areas with high accumulation of the probe within the same treated region as areas with little or no probe. This uneven distribution is likely to be a result of the ms-long ultrasound pulses and the high pressure used,30–32 parameters which were chosen to enable the probe to be delivered into the brain while inducing microglial activation.29,33 In future work, low pressure short pulses of ultrasound will be used to deliver this probe homogenously,31 as experiments will be performed on preclinical inflammatory models instead of (as performed here in this proof of principle study) using the ultrasound technique to induce the inflammation, i.e. microglial activation.31 To the best of our knowledge, this is the first time that a probe that targets activated microglia has been delivered to the brain in vivo using focused ultrasound.
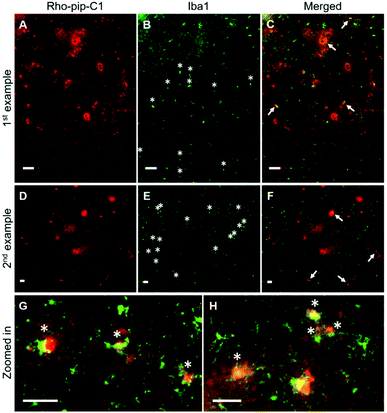
To assess whether the probe was able to target microglia once in the brain, brain sections were immunohistochemically stained for Iba1 (ionized calcium binding adaptor molecule 1). The Iba1 protein is specifically localised in microglia and is upregulated in activated microglia.34 Iba1 staining showed that microglia that displayed typical morphological changes for activation, including an ameboid-shaped cell body and short or non-existent processes,8 were present within the ultrasound targeted regions (Fig. 3B, E, G, H and Fig. S2, ESI†). Rho-pip-C1 was found to colocalize with activated microglia, which had a more ameboid-like shape (Fig. 3C, F–H), consistent with reports that activated microglia overexpress FPR2/ALX.16,17 We cannot rule out the possibility that the probe could be targeting some resting-state microglia and non-parenchymal macrophages; however, this will be quantified in future work with specific staining tools e.g. DAB staining and anti-TMEM119 antibodies.35,36 Colocalisation could also be due to the microglia phagocytosing Rho-pip-C1 to remove it from the brain.
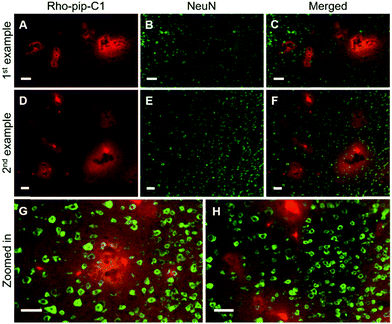
Immunohistochemical staining was also performed to check whether the probe colocalized with other cerebral specific cell types e.g. neurons (which are involved in signal transmission) and astrocytes (which have a number of functions including regulating the BBB permeability and maintaining the neuronal microenvironment).37,38
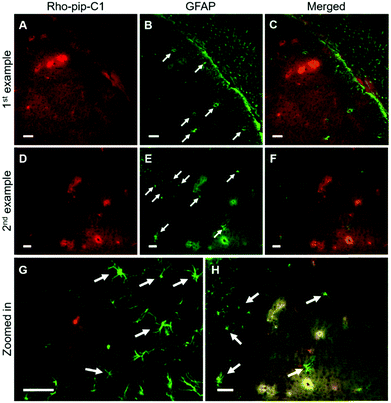
NeuN (neuronal nuclear protein) and GFAP (glial fibrillary acidic protein) staining were used to image neurons and astrocytes respectively. No colocalization between the probe and neurons (Fig. 4) was observed as expected, given that neurons do not express FPR2/ALX.39 Astrocytes, on the other hand, have been shown to express FPR2/ALX;39,40 however, no colocalization was detected between the probe and astrocytes using GFAP staining (Fig. 5). It should be noted that whilst GFAP is upregulated in reactive astrocytes, it is not present in all non-reactive astrocytes. Thus, in future work, additional astrocyte markers will be used to confirm that the probe does not colocalize with both reactive and all non-reactive astrocytes. Later time-points will also be explored to assess whether Rho-pip-C1 binds to astrocytes subsequently in time.
In this study, the delivery of Rho-pip-C1 was assessed ex vivo, in order to establish whether focused ultrasound would allow probe delivery to the brain. Future work will be performed to detect this probe in vivo by attaching MRI or PET imaging motifs, allowing for preclinical longitudinal studies.
Our study shows that Rho-pip-C1, an FPR2/ALX-targeted fluorescent probe, recently reported as a useful marker of vascular inflammation can also bind to activated microglia, which can be useful for biomedical research and preclinical drug discovery programs focusing on neuroinflammation. We initially demonstrated that whilst Rho-pip-C1 is unable to cross the BBB by itself, it can be delivered locally across the BBB using focused ultrasound and microbubbles. Using immunohistochemical staining, we established that Rho-pip-C1 does not bind to neurons or astrocytes but accumulates in activated microglial cells. These results suggest that Rho-pip-C1 could be an interesting future target to investigate the role of FPR2/ALX in activated microglia in neurological disorders.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The PhD studentships of S. V. M., T. B. and T. G. C. were funded by EPSRC Centre for Doctoral Training in Medical Imaging (EP/L015226/1) and the Centre for Neurotechnology (EP/L016737/1). We thank Javier Cudeiro Blanco for his support and the Facility for Imaging by Light Microscopy (FILM) at Imperial College London funded by the Wellcome Trust (grant 104931/ZS/14/Z) and BBSRC (grant BB/L015129/1).Notes and references
- S. A. Wolf, H. W. G. M. Boddeke and H. Kettenmann, Annu. Rev. Physiol., 2017, 79, 619–643 Search PubMed.
- M. W. Salter and B. Stevens, Nat. Med., 2017, 23, 1018–1027 Search PubMed.
- M. È. Tremblay, B. Stevens, A. Sierra, H. Wake, A. Bessis and A. Nimmerjahn, J. Neurosci., 2011, 31, 16064–16069 Search PubMed.
- F. Aloisi, Glia, 2001, 36, 165–179 Search PubMed.
- S. Hickman, S. Izzy, P. Sen, L. Morsett and J. El Khoury, Nat. Neurosci., 2018, 21, 1359–1369 Search PubMed.
- M. M. Bianchin, K. C. Martin, A. C. De Souza, M. A. De Oliveira and C. R. De Mello Rieder, Nat. Rev. Neurol., 2010, 6, 1–2 Search PubMed.
- M. T. Heneka, M. P. Kummer and E. Latz, Nat. Rev. Immunol., 2014, 14, 463–477 Search PubMed.
- M. E. Lull and M. L. Block, Neurotherapeutics, 2010, 7, 354–365 Search PubMed.
- L. Airas, M. Nylund and E. Rissanen, Front. Neurol., 2018, 9 Search PubMed.
- L. Beckers, D. Ory, I. Geric, L. Declercq, M. Koole, M. Kassiou, G. Bormans and M. Baes, Mol. Imaging Biol., 2018, 20, 94–102 Search PubMed.
- M. Laube, T. Kniess and J. Pietzsch, Molecules, 2013, 18, 6311–6355 Search PubMed.
- E. F. J. de Vries, J. Doorduin, R. A. Dierckx and A. van Waarde, Nucl. Med. Biol., 2008, 35, 35–42 Search PubMed.
- R. D. Ye, F. Boulay, M. W. Ji, C. Dahlgren, C. Gerard, M. Parmentier, C. N. Serhan and P. M. Murphy, Pharmacol. Rev., 2009, 61, 119–161 Search PubMed.
- D. A. Dorward, C. D. Lucas, G. B. Chapman, C. Haslett, K. Dhaliwal and A. G. Rossi, Am. J. Pathol., 2015, 185, 1172–1184 Search PubMed.
- J. G. Filep, M. Sekheri and D. El Kebir, Eur. J. Pharmacol., 2018, 833, 339–348 Search PubMed.
- Y. H. Cui, Y. Le, X. Zhang, W. Gong, K. Abe, R. Sun, J. Van Damme, P. Proost and J. M. Wang, Neurobiol. Dis., 2002, 10, 366–377 Search PubMed.
- Y.-H. Cui, Y. Le, W. Gong, P. Proost, J. Van Damme, W. J. Murphy and J. M. Wang, J. Immunol., 2002, 168, 434–442 Search PubMed.
- T. Boltersdorf, J. Ansari, E. Y. Senchenkova, J. Groeper, D. Pajonczyk, S. A. Vital, G. Kaur, J. Steve Alexander, T. Vogl, U. Rescher, N. J. Long and F. N. E. Gavins, Theranostics, 2020, 10, 6599–6614 Search PubMed.
- M. Beija, C. A. M. Afonso and J. M. G. Martinho, Chem. Soc. Rev., 2009, 38, 2410–2433 Search PubMed.
- M. Nanamori, X. Cheng, J. Mei, H. Sang, Y. Xuan, C. Zhou, M. W. Wang and R. D. Ye, Mol. Pharmacol., 2004, 66, 1213–1222 Search PubMed.
- T. Boltersdorf, J. Ansari, E. Y. Senchenkova, L. Jiang, A. J. P. White, M. Coogan, F. N. E. Gavins and N. J. Long, Dalton Trans., 2019, 48, 16764–16775 Search PubMed.
- W. M. Pardridge, J. Cereb. Blood Flow Metab., 2012, 32, 1959–1972 Search PubMed.
- K. Hynynen, N. McDannold, N. Vykhodtseva and F. A. Jolesz, Radiology, 2001, 220, 640–646 Search PubMed.
- W. M. Pardridge, NeuroRx, 2005, 2, 3–14 Search PubMed.
- P. J. Gaillard, C. C. Visser, C. C. M. Appeldoorn and J. Rip, Drug Discovery Today Technol., 2012, 9, e155–160 Search PubMed.
- V. A. Levin, J. Med. Chem., 1980, 23, 682–684 Search PubMed.
- E. G. Chikhale, K. Y. Ng, P. S. Burton and R. T. Borchardt, Pharm. Res. An Off. J. Am. Assoc. Pharm. Sci., 1994, 11, 412–419 Search PubMed.
- M. F. Salvatore, Y. Ai, B. Fischer, A. M. Zhang, R. C. Grondin, Z. Zhang, G. A. Gerhardt and D. M. Gash, Exp. Neurol., 2006, 202, 497–505 Search PubMed.
- Z. I. Kovacsa, S. Kima, N. Jikariaa, F. Qureshia, B. Miloa, B. K. Lewisa, M. Breslera, S. R. Burksa and J. A. Franka, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E75–E84 Search PubMed.
- J. J. Choi, K. Selert, F. Vlachos, A. Wong and E. E. Konofagou, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 16539–16544 Search PubMed.
- S. V. Morse, A. N. Pouliopoulos, T. G. Chan, M. J. Copping, J. Lin, N. J. Long and J. J. Choi, Radiology, 2019, 291, 459–466 Search PubMed.
- J. J. Choi, K. Selert, Z. Gao, G. Samiotaki, B. Baseri and E. E. Konofagou, J. Cereb. Blood Flow Metab., 2011, 31, 725–737 Search PubMed.
- Z. I. Kovacs, T. W. Tu, M. Sundby, F. Qureshi, B. K. Lewis, N. Jikaria, S. R. Burks and J. A. Frank, Theranostics, 2018, 8, 4837–4855 Search PubMed.
- D. Ito, Y. Imai, K. Ohsawa, K. Nakajima, Y. Fukuuchi and S. Kohsaka, Mol. Brain Res., 1998, 57, 1–9 Search PubMed.
- M. L. Bennett, F. C. Bennett, S. A. Liddelow, B. Ajami, J. L. Zamanian, N. B. Fernhoff, S. B. Mulinyawe, C. J. Bohlen, A. Adil, A. Tucker, I. L. Weissman, E. F. Chang, G. Li, G. A. Grant, M. G. Hayden Gephart and B. A. Barres, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E1738–E1746 Search PubMed.
- Q. Li and B. A. Barres, Nat. Rev. Immunol., 2018, 18, 225–242 Search PubMed.
- M. V. Sofroniew and H. V. Vinters, Acta Neuropathol., 2010, 119, 7–35 Search PubMed.
- K. Li, J. Li, J. Zheng and S. Qin, Aging Dis., 2019, 10, 664 Search PubMed.
- Z. Guo, Q. Hu, L. Xu, Z. N. Guo, Y. Ou, Y. He, C. Yin, X. Sun, J. Tang and J. H. Zhang, Stroke, 2016, 47, 490–497 Search PubMed.
- W. Liu, J. Huang, D. Doycheva, M. Gamdzyk, J. Tang and J. H. Zhang, Exp. Neurol., 2019, 320, 112982 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0cb00140f |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |