Emerging investigator series: interactions of engineered nanomaterials with the cell plasma membrane; what have we learned from membrane models?
Amir M.
Farnoud
 *ab and
Saeed
Nazemidashtarjandi
a
*ab and
Saeed
Nazemidashtarjandi
a
aDepartment of Chemical and Biomolecular Engineering, Chemical and Biomolecular Engineering Department, Ohio University, 168 Stocker Centre, Athens, Ohio 45701, USA. E-mail: farnoud@ohio.edu; Fax: +1 740 593 0873; Tel: +1 740 593 1426
bBiomedical Engineering Program, Russ College of Engineering and Technology, Ohio University, Athens, Ohio 45701, USA
First published on 16th October 2018
Abstract
With the increasing industrial and biomedical applications of engineered nanomaterials (ENMs), concerns have been raised regarding the increased risk of exposure. Exposure to ENMs can potentially lead to adverse health effects including cell toxicity. The plasma membrane, a lipid bilayer surrounding all cells, is the first cellular entity that comes into contact with foreign particles, and membrane damage by ENMs is one of the potential mechanisms through which ENMs induce cytotoxicity. In recent years, significant effort has been focused on elucidating the complex interactions at the particle–plasma membrane interface. Such studies have primarily relied on membrane models to tease out what particle physicochemical properties might perturb the structure and function of the cell plasma membrane. However, the diversity of membrane models has made it difficult to translate the results obtained from studies with one model to another and ultimately to live cells. This review summarizes the current knowledge on ENM–plasma membrane interactions based on studies in membrane models including lipid monolayers, supported lipid bilayers, and lipid vesicles. The mechanisms of membrane disruption by the ENMs in each model have been discussed in detail and the role of the membrane model itself in modulating the results has been described. In addition, results in membrane models have been compared with the current knowledge about cells. Finally, some of the challenges toward improving the current membrane models and enhancing the environmental relevance of studies are discussed.
Environmental significanceDisruption of the cell plasma membrane is one of the mechanisms through which nanomaterials induce cytotoxicity. Due to the complexity of the plasma membrane, various membrane models have been used to provide information on molecular interactions at the particle–membrane interface. This article provides an overview of nanoparticle–membrane interactions based on the information gathered from membrane models. The mechanisms of interaction are compared between various membrane models and the role of the membrane model in regulating the outcome of particle–membrane interactions is discussed. This information can be used for judicious selection of membrane models for mechanistic studies of nanoparticle-induced cell membrane damage, which can be valuable in evaluating the potential health hazards of nanomaterials. |
Introduction
The 21st century has seen an exponential rise in the use of engineered nanomaterials (ENMs). Electronics, cosmetics, coating, and even food industries are now actively using ENMs in their products1 while the use of ENMs for biomedical applications such as drug delivery, biomedical imaging, and biosensing continues to rise. The increasing use of ENMs enhances the possibility of unwanted exposure. Several cases of occupational exposure to ENMs have already been reported.2–4 Based on current estimates, the nanotechnology workforce is expected to surpass 6 million individuals by the year 2020;5 thus, the risk of occupational exposure to ENMs is expected to enhance. ENMs can also be released into the environment during their life cycle and contaminate soil,6,7 water resources,8,9 and air,10 thus furthering the risk of exposure and raising concerns regarding their ability to cause toxic and unwanted health effects in humans and other life forms.One important aspect of the toxicity of ENMs is their ability to kill or disrupt the function of cells. ENM-induced cytotoxicity has been reported with a wide variety of nanomaterials and in a large number of cells.11–14 The mechanisms of ENM-induced cytotoxicity are diverse (see e.g. Nel et al.15) and depend on ENM physicochemical properties and the type of cell. However, the most commonly reported mechanism is ENM-induced generation of reactive species, which can induce damage in cellular lipids, proteins, and nucleic acids or lead to a significant inflammatory response.16–19
While mechanisms related to oxidative damage require that cells internalize the ENMs, it is also possible for ENMs to cause damage to the cell plasma membrane without necessarily being internalized by the cell. The plasma membrane is a lipid bilayer, which separates the cell cytoplasm from the extracellular environment. Being at the interface, the plasma membrane comes into direct contact with all exogenous particles that reach the cell. Interactions between ENMs and the plasma membrane are known to directly contribute to ENM-induced cytotoxicity. ENMs have been shown to cause transient holes in the plasma membrane of live mammalian cells,20,21 increase membrane permeability,22 or change membrane potential with downstream effects on cell function.23,24 In agreement, ENM attachment to the plasma membrane has been reported to correlate with particle cytotoxicity25 and plasma membrane damage has been suggested as a primary mechanism of cytotoxicity in both bacterial26–28 and mammalian cells.29
Despite the evidence on the importance of ENM-induced plasma membrane damage in cytotoxicity, the mechanisms of ENM interactions with the cell plasma membrane remain largely obscure. This is in part due to the complexity of the cell plasma membrane. The plasma membrane is a dynamic and complex structure, which changes from one cell to another and can also change depending on diet30,31 and in disease.32,33 The complexity of the cell plasma membrane has led researchers to develop membrane models to study the mechanisms that regulate the complex interactions at the ENM–plasma membrane interface. However, given the wide variety of ENMs and membrane models used, there is still no consensus on what ENM physicochemical properties might be most disruptive to the plasma membrane and the mechanisms by which ENMs alter the structure or disrupt the integrity of the plasma membrane.
This manuscript aims to provide an overview of studies on ENM–membrane interactions using membrane models. The focus here is on the mechanisms of interaction in each model, how such mechanisms differ between different models, and the correlation between findings in membrane models and those observed in live cells. In addition, steps toward improving membrane models and enhancing the environmental relevance of studies on ENM–membrane interactions will be discussed. This manuscript will not focus on methods to study ENM–membrane interactions; multiple excellent reviews exist in that area and interested readers are referred to those reports.34–37
Common membrane models
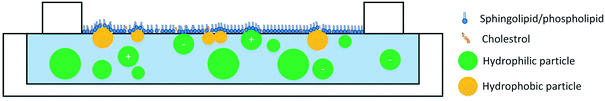
Three membrane models are predominantly used to investigate ENM interactions with the plasma membrane: lipid monolayers, supported lipid bilayers (SLBs), and lipid vesicles. Two other membrane models, namely, tethered bilayer lipid membranes (tBLMs) and suspended bilayers membranes are also starting to gain more attention in the context of ENM–membrane studies and have been briefly described below.Lipid monolayers consist of lipids spread on an air–water interface, generally in a Langmuir trough. Due to the amphiphilic nature of lipids, the hydrophilic headgroup will remain in water while the hydrophobic tail will face toward the air. This orientation has made lipid monolayers a popular model to study the interfacial properties of pulmonary surfactant,38–42 and more recently of the lipid tear film.43–45 However, lipid monolayers are also commonly used to study the exofacial leaflet (i.e. the leaflet facing outward) of the cell membrane. The use of monolayers as a model for the cell membrane provides multiple advantages. Lipids in monolayers have significant lateral mobility, as is expected for lipid molecules in the cell plasma membrane.46,47 In addition, the use of a Langmuir trough allows for compressing the lipids to a desirable mean molecular area to study the role of lipid packing in ENM–lipid interactions. This model allows for accurate and rapid measurement of monolayer surface tension and in conjunction with techniques such as fluorescence microscopy, Brewster angle microscopy, surface potentiometry, and atomic force microscopy (AFM) can also be used to study ENM effects on lipid phase separation, lipid headgroup orientation, and ENM localization in the monolayer. Naturally, due to their geometry, lipid monolayers have a few disadvantages: 1. only one lipid layer is used to model the plasma membrane, 2. lipids are at an air–water interface, and not entirely submerged in water, which is likely to affect ENM–lipid interactions, and 3. studies with techniques such as AFM require that lipids be deposited on solid substrates and cannot be performed in situ.40,41,48,49
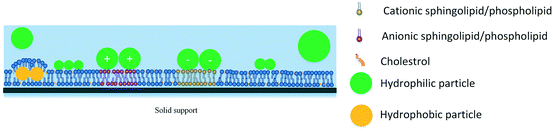
SLBs are lipid bilayers deposited on a solid substrate. SLB formation on a solid substrate is generally achieved by exposure of the substrate to lipid vesicles, which after reaching a critical concentration will fuse and form a bilayer on the substrate.50–52 SLBs have the advantage of being fixed on a solid support, which allows for evaluation of ENM–lipid interactions with techniques that are generally not possible to use with monolayers. Techniques such as AFM and the quartz crystal microbalance (QCM) method can be easily coupled with SLBs to study particle adsorption onto lipids and changes in lipid organization.53–55 However, several drawbacks are associated with the use of SLBs due to the fact that SLBs are attached to a solid support. First, SLBs are planar bilayers and do not mimic the curvature of the cell plasma membrane. Secondly, due to being fixed on a solid support, the mobility of lipids in SLBs is limited and the diffusion coefficient of SLB lipids is approximately two-fold less than that observed in vesicles.56,57 Finally, incorporation of transmembrane proteins into SLBs is a challenge. The water layer between the lower leaflet lipids and the solid substrate is only in the order of a few angstroms58 and is not thick enough to prevent direct interaction between the proteins and the substrate. This significantly limits the diffusion of proteins in the bilayer, leading to protein patches that appear to be fixed to the surface59 and/or protein denaturation. The use of “spacer” molecules between the lipids and the substrate was proposed as a method to alleviate some of the problems associated with SLBs and led to the development of tethered bilayer lipid membranes.60
Tethered bilayer lipid membranes (tBLMs) and suspended bilayer membranes are two planar membrane models that are starting to gain popularity in the context of ENM–membrane interactions.61,62 Tethered bilayer lipid membranes (tBLMs) are lipid bilayers connected to a solid support, via a spacer molecule, to prevent direct interactions between lipids/proteins in the bilayer with the substrate.63 Various tethering molecules, such as polymers,64 peptides,65 and proteins,66 have been used to generate tBLMs. Suspended bilayer lipid membranes, also known as black bilayer lipid membranes (BLM), are a freestanding lipid bilayer spread across the aperture of a chamber.67 The suspended bilayer separates the chamber into two aqueous phases in which electrodes are connected to a current measurement device. Both tBLMs and BLMs can be coupled with a variety of techniques and have been particularly useful to measure the electrical properties of the bilayer to investigate ion channel activity, membrane proteins, and other membrane properties.68,69 For example, hole formation in the suspended bilayer can be examined with high sensitivity by monitoring ionic currents. This method has been effectively used to study nanoparticle interactions with BLMs in a variety of nanomaterials including CdSe and CdSe/ZnS quantum dots,70 gold,71 and graphene oxide72 among others.
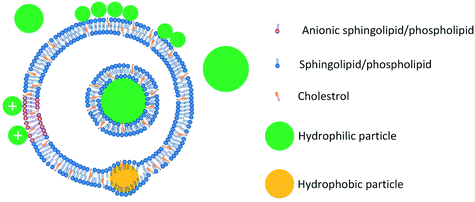
Lipid vesicles submerged in water are the most commonly used models to study ENM–membrane interactions. Vesicles are lipid bilayers in which lipids are laterally mobile. While lipid monolayers and SLBs are both flat surfaces, vesicles are curved and are closer to the morphology of the cell membrane. Lipid vesicles can be used to study ENM effects on membrane permeability, using the well-known vesicle leakage assay.73 Vesicles can also be used to study transmembrane potential,23,74 lipid phase segregation,75,76 and lipid packing.77,78 In addition, lipid vesicles can be deposited on a solid support and be studied using some of the experimental techniques commonly used for evaluation of SLBs such as QCM.79 Vesicles can be produced in different dimensions and lamellarities. Small unilamellar vesicles (SUVs) are generally less than 100 nm in diameter,80 large unilamellar vesicles (LUVs) can be in the order of 200–500 nm,81,82 and giant unilamellar vesicles (GUVs) are generally in the order of several microns.83,84 Multilamellar vesicles (MLVs) also exist and are in the same size range as LUVs, but with multiple lamellae.85 One potential disadvantage of using vesicles to study ENM–membrane interactions is that vesicles are generally one to several hundred nanometers in diameter and thus are much smaller than cells and in the same size order as ENMs. This is an important drawback when considering its implications for membrane curvature, which is inversely related to diameter. Mammalian cells vary widely in size, but most cells have a diameter of at least several microns (note that mammalian cells are not completely spherical and diameters are used simply as estimates). Thus, for example, a red blood cell, with a size of 6–8 μm, will have a much lower surface curvature compared to a spherical SUV of 100 nm in diameter. While the membrane of a mammalian cell is much less curved in comparison with a nanoparticle, the surface of a SUV might have a similar curvature compared to a nanoparticle. Not mimicking the curvature of the cell membrane is a shortcoming of most membrane models. While the use of vesicles, other than GUVs, over-represents the curvature of the membrane, the use of monolayers, BLMs, and SLBs underrepresents membrane curvature with potential implications for nanoparticle membrane interactions.
Compared to other vesicle models, GUVs are in the same size order as cells, but experimental studies on the interactions of GUVs with ENMs are more complicated. The most commonly used method for GUV synthesis, electroformation, is generally performed using custom-made set-ups, or recently developed commercial set-ups, that allow for the use of capacitors to provide an AC electric field to dried lipids while they are being rehydrated in a sealed chamber.76,86,87 The need to keep the vesicle suspension sealed makes it difficult to introduce ENMs to the system. While it is possible to introduce particles inside the sealed assembly, as shown by us88 and others,89 retaining vesicle stability while introducing the particles requires an immaculate experimental technique. In addition, achieving a good mixing of particles and vesicles is not easily achievable. The other commonly used method to synthesize GUVs, the gentle hydration method, requires the addition of 10–20% charged lipid, if performed in a high ionic strength medium, which limits the choice of lipid mixtures for studies of ENM–vesicle interactions.90
It is important to note that the ease with which the particle dose (i.e. the number of particles interacting with the membrane model) is evaluated depends on the membrane model. The ENM concentration is generally reported as the mass of particles suspended in a known volume of the buffer that is in contact with the membrane model. However, the actual number of particles that interact with the membrane model is different from the concentration of particles in the buffer. In the Langmuir trough, the particles are generally injected in the subphase, underneath the membrane model. Therefore, the number of particles that directly interact with the membrane model is difficult to estimate, particularly if particles are hydrophilic. In contrast, the particle dose is easier to quantify in SLBs and tBLMs. This is because these models are can be studied in conjunction with a QCM, in which frequency shifts are used to monitor changes in mass, thereby providing an accurate measurement of particle binding to the membrane. Similarly, BLMs also allow for accurate quantification of particles interacting with the membrane through measurement of alterations in the capacitance exhibited by the bilayer.62 In studies with vesicles, particles and vesicles are generally suspended in the same chamber, e.g. a cuvette, which can also be stirred (although stirring is not possible when working with GUVs). While stirring increases the possibility of continuous particle–vesicle interactions, it is difficult to quantify the number of particles that directly interact with vesicles.
Nanoparticle interactions with membrane models
Prior to discussing the studies on ENM–membrane interactions, clarifications should be made regarding the term “membrane disruption” in each membrane model. In a biological setting, ENM-induced disruption means that a membrane is not functioning as it was prior to the exposure. Membrane function, however, is difficult to measure in a non-living system and thus the definition of membrane disruption is somewhat subjective. In the following section, membrane disruption for lipid monolayers is defined as ENMs penetrating in between the lipids and changing the mean molecular area of lipid molecules. In SLBs, disruption is defined as ENMs depositing in the bilayer and removing lipids from the SLBs. Finally, in vesicles, disruption is defined as loss of membrane integrity as evidenced by leakage of the intravesicular content. It should be borne in mind that these definitions depend on the technique used to measure membrane properties. For example, in a vesicle leakage assay, the presence of ENMs might lead to complete leakage of the intravesicular content, which is sometimes accompanied with bilayer formation on the surface of the ENMs. In this case, even though the vesicle has been disrupted, the lipid bilayer still exists. In the following sections, the definitions noted above have been used for membrane disruption. However, the reader is cautioned that these definitions are somewhat subjective, and slightly different definitions of membrane disruption might be found in the literature.ENM–lipid monolayer interactions
Over the past decade, there have been several studies on the interactions of ENMs with lipid monolayers. In reviewing these reports, one should be careful to separate the studies that use lipid monolayers as a model for the cell plasma membrane from those that use them as a model to mimic the monolayer of lipids (and proteins) that cover the alveolar region of the lungs.48,91–93 There have also been studies that might be of interest for both systems given that the lining layer of the alveoli and the cell plasma membrane share a number of common lipids.42,94 Studies of ENM–monolayer interactions are generally performed in a Langmuir trough in which the lipids are compressed on an aqueous subphase and the changes in surface tension are recorded (commonly reported as a surface pressure vs. surface area isotherm, in which surface pressure is the surface tension of the pure subphase minus the surface tension in the presence of lipids). The interactions of ENMs with the monolayer can be studied by monitoring perturbations in the surface pressure isotherm.Table 1 summarizes the studies on ENM–lipid monolayer interactions focusing only on studies in which the monolayer has been used as a surrogate for the cell plasma membrane. It should be noted that while the majority of studies on ENM–membrane interactions focus on nanoparticles of interest for drug delivery (i.e. primarily polymeric particles), the mechanisms of interaction can be generalized to inform studies with nanoparticles of environmental interest. In Table 1, and other tables throughout this report, particle size refers to particle diameter, unless otherwise noted. The particle charge and diameter in the medium in which the experiment was performed (e.g. the hydrodynamic size of particle aggregates in the medium) has also been reported whenever such information was available in the original publication.
| Lipid model (molar ratios) | Nanoparticle propertiesb | Particle–lipid interactions | Reference |
|---|---|---|---|
| a Abbreviations: lipids: PC = phosphatidylcholine, PE = phosphatidylethanolamine, PS = phosphatidylserine, DOPC: dioleyl phosphatidylcholine, DPPC: dipalmitoyl phosphatidylcholine, DPPS: dipalmitoyl phosphatidylserine, DPPG: dipalmitoyl phosphatidylglycerol, DOPG: dioleyl phosphatidylglycerol, PI: phosphatidylinositol, DPPI: dipalmitoyl phosphatidylinositol, CL: cardiolipin, BMP: bis(monoacylglycero)phosphate, Chol: cholesterol. Surfactants: DMAB: didodecyldimethylammonium bromide, CTAB: cetyltrimethylammonium bromide, PVA: poly(vinyl alcohol), PVP: polyvinylpolypyrrolidone. b Nanoparticle size refers to nanoparticle diameter. Nanoparticle properties have been reported in the medium in which the experiment was performed whenever such information was available in the original publication. | |||
| Native artery's endothelial cell membrane composition: DPPC (56%), DPPE (24%), PI (8.5%), DPPS (4.3%), SM (6.5%), and CL (1.7%) | Polystyrene (20 and 60 nm): plain (−40.12 mV), amine (charge not reported), carboxyl-modified (−40.46 mV) | Amine-modified and plain particles, but not carboxyl-modified, particles penetrate the monolayer | Peetla and Labhasetwar (2008)95 |
| DPPC (56%), DPPE (24%), PI (8.5%), DPPS (4.3%), SM (6.5%), and CL (1.7%) | Polystyrene (130 to 170 nm) plain (7.84 mV) modified with surfactant: DMAB (57.74 mV), DTAB (39.25 mV), CTAB (32.86 mV), PVA (−14.35 mV) | Particles penetrate the monolayer depending on the hydrophobicity of the surfactant molecule | Peetla and Labhasetwar (2009)99 |
| DPPC | Endohedral metallofullerene (Dy@C82) (size and charge not reported) | Particles penetrate the monolayer due to their surface activity | Wang et al. (2009)100 |
| Plasma membrane model: DPPC (56%), DPPE (24%), DPPI (8%), DPPS (4.3%), SM (6%), CL (1.7%) | Poly(lactic-co-glycolic acid) (200–300 nm), plain (−19.31 mV), CTAB (14.31 mV), and DMAB (39.91 mV) modified | Particles penetrate the monolayer depending on the hydrophobicity of the surfactant molecule | Peetla et al. (2014)101 |
| Endosomal membrane model: DPPC (53%), DPPE (19%), DPPI (7%), DPPS (4%), SM (3%), BMP (14%) | |||
DPPC/DPPG (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and DOPC/DOPG (3 1) and DOPC/DOPG (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
Polymer-coated anionic (−63 mV) and cationic (46 mV) silver nanoparticles (14–20 nm) | Anionic particles condense the monolayer. Cationic monolayers condense unsaturated lipids, but penetrate in saturated lipids | Bothun et al. (2016)104 |
| DMPC | Citrate- (−36 mV) and 4-mercaptobenzoic acid (MBA)- (−33 mV) coated silver (∼8 nm) | Both particles penetrate the monolayer, citrate gets squeezed out, while MBA remains in the monolayer | Girón et al. (2016)256 |
DMPG/DPPG (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
Polyalkylcyanoacrylate (103–172 nm) with various PEG groups (−1 to −5 mV) | PEGylated particles penetrate the monolayer, PEG type plays a major role in penetration | Baghirov et al. (2017)102 |
| DPPC | Silver nanoparticles (5–10 nm) coated with a polyether block amide (particles were not charged) | Particles penetrate the monolayer due to their surface activity | Soriano et al. (2017)103 |
| DPPC, DPPG, DOPC, and Chol | PVP-coated silver nanoparticles (16–30 nm) | Particles condense the monolayer, leading to a reduction in surface pressure | Da Silva et al. (2018)257 |
SM/DOPC/Chol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
100 nm silica plain (−47.0 mV) or coated with amine (18.3 mV) and PEG 2 K (−2.9 mV), 5 K (−1.1 mV), and 20 K (8.0 mV) | PEGylated particles penetrate the monolayer due to their surface activity, amine-modified and plain particles do not penetrate the monolayer | Asgahri Adib et al. (2018)88 |
A close look at the studies in Table 1 reveals that a variety of lipid models have been used in the studies of ENM–monolayer interactions. While some studies have focused on mimicking the lipid composition of the cell plasma membrane, other studies have focused on singular, binary, or ternary lipid mixtures to garner mechanistic information. This variety in the composition of lipids has made it difficult to draw general conclusions regarding ENM–membrane interactions; however, several overarching trends can be deduced. The size of ENMs has an effect on their interactions with lipid monolayers with smaller particles disrupting the monolayer more than larger particles. This is evident both from studies in which monolayers are used as cell membrane models95 and those in which monolayers have been used as a model for pulmonary surfactant.96–98 A general trend also exists regarding particle hydrophobicity. Hydrophobic and surface-active particles penetrate the lipid monolayer at the air–water interface while hydrophilic particles are less likely to do so.88,99–103 The degree of particle hydrophobicity appears to directly correlate with their penetration to the monolayer.99,101
The role of particle surface charge in particle interactions with lipid monolayers has been investigated by a number of studies. A few studies report that positively-charged ENMs penetrate the monolayer95,104 while negatively charged ENMs are less likely to reach the air–water interface.49,95,104,105 However, the effects of particle charge depend on the lipid model. Phosphatidylserines (PS) and phosphatidylglycerols (PG) are negatively charged, and models that incorporate these lipids are more likely to see emphasized interactions between the lipid model and positively charged particles. In addition, even the interactions of oppositely charged particles and lipids appear to be somewhat dependent on the lipid structure and saturation level. For example, it has been reported that cationic particles condense unsaturated lipids, but penetrate in saturated lipids.104 In summary, three general trends can be observed: 1) smaller particles penetrate the monolayer more efficiently, 2) hydrophobic/surface-active particles penetrate the monolayer more than hydrophilic particles, and 3) positively charged particles tend to penetrate the monolayer more than negatively charged particles although this effect depends on the lipid model (Fig. 1).
ENM–SLB interactions
A summary of studies on ENM interactions with SLBs has been provided in Table 2. The overwhelming majority of these studies point to electrostatic interactions as the primary mechanism by which ENMs adsorb onto the bilayer. The importance of electrostatic forces in ENM–SLB interactions was first revealed by Hong and colleagues for poly(amidoamine) (PAMAM) dendrimers106 and was then confirmed by studies with other polymers and dendrimers,55,107,108 C60 fullerene nanoparticles,109,110 quantum dots,54 carbon nanotubes,111 graphene oxide,79 and gold nanoparticles.112–114 In all these cases, oppositely charged nanoparticles and lipid bilayers were shown to have a higher degree of interaction compared to lipids and particles of the same charge. The importance of electrostatic interactions in particle association with lipid bilayers has also been proven through a number of computational studies.115–117| Lipid model (molar ratios) | Nanoparticle propertiesb | Particle–lipid interactions | Reference |
|---|---|---|---|
| a Abbreviations: lipids: PC: phosphatidylcholine DMPC: dimyristoyl phosphatidylcholine, DMTAP: dimyristoyl-trimethylammonium-propane, DOTAP: dioleoyltrimethylammonium propane; particles: PAMAM: poly(amidoamine), PLL: poly-L-lysine, PEI: polyethylenimine, DEAE-DEX: diethylaminoethyl-dextran, MWCNT: multi-walled carbon nanotubes; chemicals: MUS: mercapto-undecanesulfonate, OT: octanethiol, PAA: poly(acrylic acid), PAH: poly(allylamine) hydrochloride, MPA: mercaptopropanoic acid, MPANH2: mercaptopropylamine, EG6 mercaptoundecanethiol ethyleneglycol hexamer, LPS: lipopolysaccharides. b Nanoparticle size refers to nanoparticle diameter. Nanoparticle properties been reported in the medium in which the experiment was performed whenever such information was available in the original publication. | |||
| DMPC | PAMAM dendrimers (∼8 nm) terminated with amine and acetamide groups | Cationic dendrimers form holes in the SLB, neutral dendrimers do not disrupt the SLB | Hong et al. (2004)106 |
| DMPC | G7 dendrimers (∼8 nm) | Particles disrupt the SLB by attaching to the liquid-condensed phase | Mecke et al. (2005)158 |
| DMPC | C60 fullerenes (∼0.7 nm, negatively charged), aggregates up to 15 nm in size (mean size of 4.7 nm) were observed | Particles adhere to the SLB without causing significant disruption, binding is charge-dependent | Spurlin and Gewirth (2006)109 |
| DMTAP | |||
| DMPC | PLL (17.59 mV), PEI (45.18 mV), DEAE-DEX (30.44 mV), and PAMAM (19.24 mV) | Cationic polymers, but not neutral polymers, disrupt the SLB | Hong et al. (2006)55 |
| DMPC | Gold (5–6 nm), silica (50 nm), PAMAM dendrimers: G3 (∼4 nm), G5 (∼5 nm), and G7 (∼8 nm); charge not reported, but all particles were cationic | Cationic particles disrupt the SLB, the extent of disruption is dependent on charge density of the particles | Leroueil et al. (2008)107 |
| POPC | Bare silica (50 nm) incubated with serum | Adherence of plasma proteins reduces the binding of particles to the SLB | Lesniak et al. (2008)25 |
| DMPC | PAMAM dendrimers: G2 (2.9 nm), G4 (4.2 nm), and G6 (6.7 nm) | Dendrimers disrupt the SLB by removing lipids. Particles with higher positive charge are more disruptive | Parimi et al. (2008)108 |
| Egg PC | Polystyrene (28, 62, 140 nm) all carboxyl-modified; zeta potentials changed between −54.94 and −29.48 mV depending on the medium | Particles remove lipids from the SLB. Lipid removal depends on particle hydrophobicity | Jing and Zhu (2011)118 |
| DOPC, | Amine-modified (15.99 mV) and carboxyl-modified (−22.86 mV) CdSe/ZnS (2–3 nm) | Cationic, but not anionic, particles bind to the SLB. Increasing ionic strength reduces the binding of cationic and increases the binding of anionic particles | Zhang and Yang (2011)54 |
DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOTAP (90 DOTAP (90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) 10) |
|||
DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOTAP (50 DOTAP (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) 50) |
|||
| DOPC and Asolectin | Silica (50 and 500 nm) plain and amine-modified | Particles disrupt the SLBs. Cationic particles are more disruptive at 500 nm, plain particles are more disruptive at 50 nm | de Planque et al. (2011)258 |
| Egg PC | Fullerene (139 nm) and fullerol (149 nm); zeta potentials were dependent on the pH, but ranged between −65 and −30 mV | Both particles adsorb onto the SLB. Adsorption increases when electrostatic repulsive forces are reduced | Hou et al. (2011)110 |
| Egg PC | Gold particles (5–100 nm) coated with tannic acid (charge not reported) or PVP (≃20 mV) | Smaller particles bind to SLBs more efficiently. PVP-coated particles bind less efficiently | Hou et al. (2012)113 |
POPC/POPS (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and POPC/POEPC (3 1) and POPC/POEPC (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
Graphene oxide 0.5–5 μm (−56 mV at pH = 4) | Anionic particles bind to cationic, but not anionic, SLBs | Frost et al. (2012)259 |
| DOPC | Carboxyl-modified MWCNT (112 nm, −16 mV) and carboxyl-modified CdSe/ZnS (12.7 nm, −9.8 mV) | Both particles disrupt the SLBs. Nanotubes traverse the bilayer resulting in leakage of ions and are more disruptive than CdSe/ZnS particles | Corredor et al. (2013)260 |
| DOPC | MWCNT (104–119 nm) | Particles bind to SLBs due to electrostatic interactions. Binding can be modulated by altering the pH | Yi and Chen (2013)111 |
| Egg PC | Carboxyl-modified polystyrene (130 nm) | Particle adsorption onto the SLB and disruption of the bilayer depends on the ions present in the medium | Jing et al. (2014)53 |
DOPC–DOTAP (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
Gold nanoparticles functionalized with MPA (9 nm, −29 to −27 mV) or PAH (6 nm, 32 to 38 mV) | Cationic particles bind more than anionic particles to anionic SLB | Troiano et al. (2015)114 |
| DOPC | Citrate-modified silver particles (49–65 nm, negatively-charged) with aggregates of ∼175 nm in 150 mM NaCl (pH = 7). Incubation with albumin resulted in aggregates of ∼100 nm. Aggregates of similar size but a positive charge were observed after incubation at pH = 2 | The albumin protein corona reduced particle adsorption onto the SLBs at neutral pH likely due to a reduction in particle surface energy or due to steric repulsion. Reducing the pH to 2 increased the adsorption of albumin-coated particles on the SLBs likely due to a reduction in steric repulsion caused by the albumin corona | Wang et al. (2016)248 |
SM/DOPC/Chol (60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20) and DOPC 20) and DOPC |
Gold nanoparticles (4 nm) functionalized with MPA (−28 to −21 mV) or MPNH2 (26 mV) | Cationic particles bind to anionic SLBs more than anionic particles. Particles bind more to phase-segregated lipids | Melby et al. (2016)112 |
POPE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) POPG (1 POPG (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
Carboxyl (negatively-charged) and amine-modified (positively-charged) polystyrene (20 nm) | Cationic particles disrupt the anionic SLBs more than anionic particles. Particle binding to and disruption of the bilayer is regulated by how strongly the bilayer is bound to the underlying substrate | Yousefi et al. (2016)123 |
| DOTAP, DOPC, and PG | Fullerene particles (170 nm, −19 mV) before and after incubation with humic acid (120 nm, −34 mV) or serum (∼200 nm and reduced zeta potential) | Humic acid corona reduced particle deposition in DOPC and PG, but not in DOTAP, due to electrostatic interactions. Serum corona reduced particle deposition in DOPC and DOTAP, but not in PG | Ha et al. (2016)255 |
| DOPC | Carboxyl-modified polystyrene of 20 nm (−42 mV) and 100 nm (−34 mV). Incubation with serum resulted in aggregates of 69.2 nm (−9 mV) and 173 nm (−10 mV) | 100 nm particles, unlike 20 nm particles, showed significant binding to and disruption of the SLBs. The presence of the serum biomolecular corona reduced particle binding to and disruption of the SLBs | Di Silvio et al. (2017)249 |
DOPC, DOPC/PI (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
Gold particles modified with MPA (20 nm, −38 mV), MPANH2 (660 nm, 27 mV), PAA (51 nm, −50 mV), PAH (52 nm, 40 mV), and EG6 (6 nm, −11 mV). Particle properties changed after incubation with serum: MPA (63 nm, −23 mV), MPANH2 (32 nm, −21 mV), EG6 (18 nm, −14 mV), PAA and PAH showed significant aggregation | Cationic particles, but not anionic particles, adsorbed onto SLBs. Adsorption of MPA-coated particles onto the SLBs increased after incubation with serum, while adsorption of MPANH2 decreased. Adsorption of EG6-coated particles onto DOPC/PI increased after incubation with serum | Melby et al. (2017)250 |
| POPC, POPC/smooth LPS (0.46 mol%), POPC/rough LPS (0.46 mol%), POPC/rough LPS (6.4 mol%) | PAH-modified nanodiamonds (17 nm, 21 mV). Particle properties changed after incubation with natural organic matter (NOM), aggregates of 34 nm and −33 mV were observed at high NOM to particle ratios | Particles adsorbed onto POPC; adsorption was slightly reduced at high amounts of rough LPS. The NOM corona significantly reduced particle adsorption onto SLBs at high NOM to particle ratios | Mensch et al. (2017)194 |
While evidence suggests that particle binding to the surface of SLBs is dependent on electrostatic interactions, ENM-induced pore formation in SLBs is likely controlled by factors other than surface charge. This is supported by the fact that many reports exist on binding of charged nanoparticles to SLBs without causing pore formation in the SLB.54,109–111,113,114 Studies of Jing and Zhu118 revealed that lipid adsorption on particle surfaces is primarily dependent on particle hydrophobicity. Experiments with carboxyl-modified polystyrene nanoparticles, which have a hydrophobic core but charged groups on the surface, showed that increasing the ionic strength of the solution, and thus screening the surface charge of the particles, enhances pore formation in SLBs, likely as a result of increased lipid adsorption on particle surfaces.118 Thus, particle-induced disruption of SLBs is a function of both the particle charge and particle hydrophobicity. Electrostatic attractions bring the ENMs in close proximity to SLBs while more hydrophobic particles are capable of removing lipids from the SLB (Fig. 2). There is a trade-off between these properties as increasing the particle charge reduces particle hydrophobicity.
Other investigated factors affecting ENM–SLB interactions include the particle size, the ionic composition of the medium, the interactions between the lipids and the underlying substrate, and the packing of lipids in the bilayer. Studies by Hou and colleagues113 have shown that adsorption of gold nanoparticles on SLBs is size-dependent, with smaller particles showing increased binding. However, caution should be taken in interpreting such studies, as changes in the size of particles might be accompanied by changes in the number of charged groups on the surface of the particles, as has been reported for gold nanoparticles119 and dendrimers.107,108 Changes in particle diameter also affect the ligand packing density and charge per unit nanoparticle surface area, and in some cases even the apparent pKa of charged ligands,120 thereby changing the electrostatic forces between the particles and the membrane. The ionic composition of the medium also plays an important role in ENM–SLB interactions.53 Pore formation by carboxyl-modified polystyrene particles in SLBs has been shown to follow the Hofmeister anion order (CH3COO− > Cl− > NO3− > SCN−).121 This effect was attributed to the differences in the ability of anions to bind to the hydrophobic core of the polystyrene. Ion binding reduces particle hydrophobicity and the ability of the particles to adsorb lipids from the bilayer.53 While these studies are performed in SLBs, it should be noted that the ionic strength and ionic composition are expected to affect ENM–membrane interactions in all membrane models. Increasing the ionic strength results in charge screening of both ENMs and lipid headgroups in all models and diminishes the importance of electrostatic interactions. Similarly, changes in ionic composition are expected to affect the behavior of ENMs and lipids in all models. For example, changing the composition of the subphase underneath lipid monolayers according to the Hofmeister ion series has been shown to affect the packing of phospholipid monolayers at air–water interfaces.122
The substrate underlying the SLBs can also play a role in modulating the physical properties of SLBs and their interactions with nanoparticles. To study how the adherence of the SLB to the substrate affects nanoparticle–SLB interactions, Yousefi and colleagues developed a SLB model the adherence of which to the underlying substrate could be modulated by changing the pH.123 A phospholipid mixture of POPE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) POPG (1
POPG (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), which has an overall negative charge, was used for the formation of SLBs, while aluminum, which is amphoteric, was used as the substrate. Changing the pH from 7.0 to 8.5 makes the surface of aluminum less positively charged, with little effect on the charge of the SLBs. Consequently, the SLB shows increased adherence to the substrate at a pH value of 7.0, but it becomes more “free-floating” as the pH is increased to 8.5. Using this model, it was shown that positively-charged, amine-modified polystyrene nanoparticles, with a nominal diameter of 20 nm, deposited more effectively on the free-floating bilayer compared to the highly bound bilayer. Similarly, bilayer damage by nanoparticles occurred earlier in the free-floating bilayer compared to the highly bound bilayer, pointing to the importance of the SLB–substrate interactions in modulating nanoparticle-induced membrane damage. The weakly adhered SLB model developed in this study could prove to be a useful model for free-floating cell plasma membrane.
2), which has an overall negative charge, was used for the formation of SLBs, while aluminum, which is amphoteric, was used as the substrate. Changing the pH from 7.0 to 8.5 makes the surface of aluminum less positively charged, with little effect on the charge of the SLBs. Consequently, the SLB shows increased adherence to the substrate at a pH value of 7.0, but it becomes more “free-floating” as the pH is increased to 8.5. Using this model, it was shown that positively-charged, amine-modified polystyrene nanoparticles, with a nominal diameter of 20 nm, deposited more effectively on the free-floating bilayer compared to the highly bound bilayer. Similarly, bilayer damage by nanoparticles occurred earlier in the free-floating bilayer compared to the highly bound bilayer, pointing to the importance of the SLB–substrate interactions in modulating nanoparticle-induced membrane damage. The weakly adhered SLB model developed in this study could prove to be a useful model for free-floating cell plasma membrane.
Finally, the role of lipid packing in ENM–lipid interactions is a less investigated but important area. Saturated sphingolipids and phospholipids are known to aggregate with cholesterol and result in the formation of ordered lipid phases, sometimes referred to as lipid domains, or rafts.75,124 A study by Melby and colleagues revealed that positively-charged gold nanoparticles bind significantly more to SLBs that contain lipid domains compared to SLBs with a similar surface charge that do not form ordered phases.112 Since ordered lipid phases have a height difference compared to disordered phases, the increased exposure of negatively-charged phosphate groups in lipids to ENMs is a likely explanation for this phenomenon.112 In contrast, one computational study has revealed that fullerene nanoparticles bind to the fluid phases of the bilayer and not the ordered domains.15 Further investigation is needed to elucidate the role of membrane phase segregation in particle–membrane interactions.
ENM–vesicle interactions
A summary of studies on ENM–vesicle interactions is provided in Table 3. Since vesicle lamellarity and size are important properties in the context of ENM–vesicle interactions, the vesicle model and vesicle size have also been included in Table 3 whenever such information was available. When the vesicle size was not reported in the publication, but vesicles were reported to be extruded through a filter, the pore size of the filter has been reported as an approximate value for the vesicle size.| Lipid model (molar ratios) | Nanoparticle propertiesb | Particle–lipid interactions | Reference |
|---|---|---|---|
| a Abbreviations: DLPC: dilauroyl phospotidylcholine, DPTAP: dipalmitoyltrimethylammoniumpropane, DOEPC: dioleoyl ethylphosphotidylcholine, POPG: palmitoyloleyl phosphatidylglycerol, DOPA: dioleoylphosphatidic acid. Chemicals: DAD: poly(diallyldimethylammonium chloride), SOD: sodium polyacrylate. b Nanoparticle size refers to nanoparticle diameter. Nanoparticle properties been reported in the medium in which the experiment was performed whenever such information was available in the original publication. | |||
DOTAP and DOPC/DOPS (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 2 1, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 1 1, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (SUV) 1) (SUV) |
Silica (110 nm) | Vesicle disruption by particles is charge-dependent and occurs in cationic, neutral, and low negatively-charged vesicles, but not in vesicles that are highly anionic | Mornet et al. (2005)125 |
| DLPC (SUV) 200 nm | Carboxyl-modified polystyrene (20 nm) | Nanoparticles stabilize the liposomes without causing disruption | Zhang and Granick (2006)136 |
| DPPC (SUV) size was temperature dependent, but between 15 and 120 nm at 25 °C (sizes are for vesicle-particle assemblies) | Silver (5.7 nm) | Nanoparticles increase the vesicle fluidity without causing disruption | Bothun et al. (2008)146 |
| Egg PC and Egg PC/DOTAP (95/5) and various other lipids; both SUVs (46 nm) and GUVs (size not specified) | CdSe/ZnS (10–20 nm) conjugated to peptides, all particles were negatively-charged | Anionic particles bind to cationic vesicles, but not neutral vesicles. Particles disrupt SUVs, but not GUVs | Dif et al. (2008)126 |
| DOPC, DLPC, and DPPC (LUV) | Carboxyl-modified (0.91e− nm−2) and amine-modified (0.25 e+ nm−2) polystyrene (20 nm) | Anionic particles induce lipid gelation, cationic particles fluidize the lipids | Wang et al. (2008)131 |
| DOPC (LUV) 200 nm | Silica nanoparticles (15–190 nm) | Particles are internalized in vesicles. This process is size-dependent, particles <30 nm do not internalize in vesicles | Le Bihan et al. (2009)138 |
DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPG (1 DOPG (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (LUV) 200 nm 1) (LUV) 200 nm |
SWNT (negatively-charged) and SWNT with adsorbed lysozyme on the surface (positively-charged, size not reported) | SWNT-lysozyme causes vesicle disruption more than SWNT alone. This effect is charge-dependent and is abrogated once the vesicle charge is reduced or pH is increased to a point where lysozyme is not charged | Hirano et al. (2010)127 |
DOPC, DOPC/DOPG (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), and DOPC/DOTAP (9 1), and DOPC/DOTAP (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (GUV) 1) (GUV) |
Core–shell (iron oxide-silica) particles with positive (13 mV) and negative (−36 mV) surface charges, (35 nm) | Cationic particles adsorb identically on the surface of anionic, neutral, and cationic vesicles. Anionic nanoparticles do not adsorb onto the membrane regardless of the charge | Laurencin et al. (2010)132 |
| DMPC and DPPC (SUV and LUV) 52, 96, and 156 nm | Silica (5–100 nm), zeta potential between −23 mV and −65 mV depending on the size and medium | Particles do not disrupt the vesicles in low ionic strength media. Addition of salt results in vesicle disruption and formation of bilayers on particles | Savarala et al. (2011)137 |
DPPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPTAP (3 DPTAP (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (SUV), 68 nm 1) (SUV), 68 nm |
SPIONs 16 nm (−39.8 mV) and 30 nm (−26.5 mV) | Small particles bind to vesicles, large particles cause vesicle disruption | Chen and Bothun (2011)140 |
POPC (MLV) and POPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cholesterol (80 cholesterol (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20) (GUV) 20) (GUV) |
C60 fullerenes (2 nm), aggregates (15–50 nm) were observed (−26 mV) | Particles disrupt GUVs but not MLVs, unless MLVs are subject to freeze-thawing | Zupanc et al. (2012)141 |
| DOPC (LUV), 100 nm | Au-DAD (16.2 nm, 22.0 mV), Au-tannic acid (18.2 nm, −36.8 mV), and Au-PVP (21.9 nm, −36.8 mV), TiO2-DAD (10.9 nm, 31.2 mV), TiO2-SOD (7.7 nm, −45.8 mV) | Cationic particles disrupt anionic vesicles more than anionic particles | Moghadam et al. (2012)133 |
DOPC and DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPG (SUV), 45, 70, and 125 nm DOPG (SUV), 45, 70, and 125 nm |
Silica (8 and 33 nm) | Particles disrupt vesicles dependent on the particle and vesicle charge. Larger particles are more disruptive than smaller particles | Pera et al. (2012)128 |
POPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE DOPE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPS DOPS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cholesterol at 9 cholesterol at 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 weight ratio, (LUV) 400 nm 4 weight ratio, (LUV) 400 nm |
Silica (14 nm), agglomerates of ∼500 nm observed in cell culture medium | Particles disrupt vesicles by causing transient pores. Effects are dependent on the lipid and buffer composition | Mu et al. (2012)151 |
| DOPC (GUV), 4–20 μm | Silica (18, 78, 182 nm) | Small particles, but not large particles, induce pores in vesicles | Zhang et al. (2012)139 |
DOPC, DOPC/DOEPC (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), and DOPC/DOPA (3 1), and DOPC/DOPA (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), (SUV) 100 nm 1), (SUV) 100 nm |
Carboxyl- (16.8 nm, −26.5 mV) and amine-modified (20.5 nm, 32.2 mV) polystyrene | Particles induce pores in all vesicles. Cationic particles are more disruptive to DOPC than anionic particles | Negoda et al. (2013)261 |
Egg PC–cholesterol (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, weight ratio), 400 nm 1, weight ratio), 400 nm |
Single-walled carbon nanotubes (diameter: 0.8 nm, L: 1.5 μm) covered by negatively charged surfactant | Particles do not cause vesicle disruption due to both particles and vesicles being anionic | Shi et al. (2013)262 |
DPPC–DOPC (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), DOPG, DOPE, DOPS, and DPPC–DOPC–Chol (1 1), DOPG, DOPE, DOPS, and DPPC–DOPC–Chol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (GUV) 1) (GUV) |
Amine- and carboxyl-modified polystyrene (20 nm), amine-modified polystyrene (120 nm) | Cationic particles, but not anionic particles, form pores in vesicles. Smaller particles are more disruptive than larger particles | Li and Malmstadt (2013)89 |
| DOPC (SUV) 91–94 nm | Graphene oxide (120–130 nm), aggregates of up to 600 nm observed at high ionic strength | Particles adsorb onto vesicles due to electrostatic interactions and cause vesicle leakage | Liu and Chen (2015)79 |
DOPC and DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) POPG (2 POPG (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (LUV), 400 nm 1) (LUV), 400 nm |
Plain, amine-, and carboxyl-modified silica (50 to 500 nm), plain and carboxyl-modified polystyrene (50–200 nm), zeta potentials between −22 mV and −50 mV | Large particles cause more disruption than small particles. Particle-induced disruption is more emphasized in zwitterionic vesicles compared to anionic vesicles | Alkhammash et al. (2015)129 |
DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPPG (9 DPPG (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 weight ratio) and DOPC 1 weight ratio) and DOPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DPTAP (9 DPTAP (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (weight ratio), (GUV) 1) (weight ratio), (GUV) |
SiO2 (∼50 nm and ∼200 nm), positively charged (6.19 mV), negatively charged (−5.48 mV), and highly negatively charged (−25.59 mV to −32.64 mV) | Particles cause significant disruption to vesicles of opposite charges | Wei et al. (2015)130 |
SM/DOPC/Chol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) LUV (149.1 nm) 1) LUV (149.1 nm) |
100 nm silica plain (−47.0 mV) or coated with amine (18.3 mV) and PEG 2K (−2.9 mV), 5K (−1.1 mV), and 20K (8.0 mV) | Plain and amine-modified particles caused significant vesicle leakage. Only PEG20K-modified particles caused leakage in vesicles | Asghari Adib et al. (2018)88 |
| DOPC, DOPC/DPPC, and DOPC/DOPG, DPPC, SUV (∼80 nm) and giant multilamellar vesicles | Gold (1.7, 2.4, and 4.2 nm) coated with MUS and MUS/OT | Particles in the 2.5–3 nm range cause significant vesicle fusion. Particles bind exclusively to liquid phases | Atukorale et al. (2018)149 |
Studies on ENM–vesicle interactions, similar to the studies on ENM interaction with SLBs, point to the importance of electrostatic forces on vesicle disruption by ENMs. The role of electrostatic forces in regulating ENM–vesicle interactions was first demonstrated by Mornet and colleagues.125 In this study, it was reported that vesicles disrupt and form bilayers on the surface of negatively-charged silica nanoparticles in a charge-dependent manner. Vesicle disruption was observed in vesicles that were positively-charged, neutral, and even vesicles with a low negative charge, but not in vesicles with a high negative charge due to the repulsive forces between the particles and vesicles.125 Similar effects were reported by other studies for similar or different nanoparticle and vesicle systems. Dif et al.126 reported that negatively-charged quantum dots bind to positively charged vesicles, but not neutral vesicles. Hirano et al.127 reported that single wall carbon nanotubes cause little leakage in negatively-charged vesicles, but once positively-charged proteins are adsorbed on the particle's surface, significant leakage is observed. Negatively-charged silica nanoparticles were shown to disrupt zwitterionic vesicles more than negatively-charged vesicles.128,129 Similarly, Wei and colleagues demonstrated that silica nanoparticles disrupt vesicles of opposite charges more than similarly charged vesicles.130
While electrostatic interactions between nanoparticles and vesicles are expected, they are somewhat complicated by the fact that even neutral phospholipids are zwitterionic and include both positive and negative charges in their lipid headgroup. The importance of the zwitterionic headgroup in ENM–vesicle interactions was shown in a pioneering study by Wang and colleagues, who demonstrated that negatively-charged particles interact preferentially with the N+ headgroup of phospholipids and are able to alter the tilting angle of lipids and make the vesicles more ordered, while positively-charged nanoparticles increase the fluidity of ordered bilayers and are more likely to disrupt them.131 In agreement, a number of studies have reported that positively-charged nanoparticles adsorb to the surface of zwitterionic vesicles89,132 more than negatively-charged particles and show more vesicle disruption.35,133 These experimental findings have been confirmed by several computational studies using planar lipid bilayers.117,134,135 Consistent with these findings, addition of negatively-charged nanoparticles has been shown to stabilize vesicles in multiple studies.136,137 It should be noted that the importance of zwitterionic lipid headgroups is not limited to ENM interactions with vesicles and should be considered in electrostatic forces between ENMs and lipids in all membrane models.
The effect of nanoparticle size on vesicles requires careful consideration of the relative size of particles and vesicles. Alkhammash and colleagues129 reported that 200 nm and 500 nm silica particles, but not 50 nm particles, cause significant leakage in 400 nm DOPC vesicles. It was proposed that when particles and vesicles are similar in size, their interaction results in vesicle disruption and formation of lipid bilayers on ENMs. In contrast, smaller particles can only cause a local perturbation in the vesicle structure as only a small surface area of the vesicle is associated with the particles.129 This observation suggests that not the absolute size but the relative size of the ENMs and vesicles is the primary factor when considering the effect of nanoparticle size. Two microscopy studies confirm this proposed mechanism. The study of Le Bihan and colleagues138 revealed that particles larger than 30 nm can be engulfed inside ∼300 nm vesicles, while particles smaller than 30 nm adsorb onto the membrane without internalization. Particles were engulfed through an invagination process and were covered by lipid bilayers once inside the vesicles. Thus, it is possible that if the particle size is close to the vesicle size, the amount of lipids required to cover the particles will result in complete disruption of the vesicles. Similarly, the study of Zhang and colleagues139 reported that small (18 nm) silica nanoparticles are trapped on the membrane of the vesicle while the membrane wraps around larger (78 nm and 182 nm) particles and eventually collapses.
The proposed mechanism on the importance of the relative size of particles and vesicles explains a number of phenomena observed in previous studies. For example, CdSe/ZnS particles (10–12 nm) were shown to disrupt SUVs, which are less than 100 nm in diameter, but not GUVs that are micron-sized.126 Similarly, a study on the interactions of 16 and 30 nm super paramagnetic iron oxide nanoparticles (SPIONs) with 75 nm vesicles reported that while smaller particles are capable of adsorbing onto the vesicles, only the larger particles cause complete vesicle disruption.140 A similar trend has been observed in another study, in which 33 nm silica particles have been shown to cause greater vesicle leakage compared to 8 nm particles.128 But, there has been at least one study in disagreement with this mechanism. The study of Zupanc and colleagues reported that C60 fullerenes (with individual radii of ∼2 nm) disrupt GUVs, but not the smaller MLVs.141 However, fullerenes in that study included some large aggregates with diameters >1 μm, which were thus in the same size range as GUVs. In addition, the differences in the lamellarity of MLVs (multilamellar) and GUVs (unilamellar) might have contributed to differences in their disruption by the fullerenes. Thus, based on current evidence, it appears that the relative size of ENMs and vesicles, and not the absolute size of particles, is a primary factor in ENM-induced vesicle disruption.
Hydrophobic nanoparticles are more disruptive to lipid vesicles than hydrophilic particles. Particles with hydrophobic cores have been shown to disrupt vesicles142 and surface functionalization with charged groups has been reported to reduce the disruption caused by hydrophobic particles.129 The disruptive effects of hydrophobic particles on vesicles stem from the fact that such particles are more likely to localize at the core of the bilayer and thus have higher potential to disrupt the bilayer. The ability of hydrophobic particles to localize at the core of the bilayer has been shown in a series of studies by Bothun and colleagues.143–146 In these studies, incorporation of hydrophobic particles was shown to affect vesicle lipid packing143,144,146 and release properties.143 The mechanisms of ENM interactions with vesicles are summarized in Fig. 3.
Other less studied factors also play a role in the outcome of ENM–vesicle interactions. While several studies have focused on how ENMs alter membrane fluidity,131,140,147,148 a recent study by Atukorale et al. reported that membrane fluidity and phase segregation play an important role in ENM–membrane interactions.149 Using phase segregated vesicles, it was shown that surface-modified gold nanoparticles only bind to fluid phases in vesicles. Interestingly, these findings seem to be in disagreement with the study of Melby et al., which showed increased binding of gold nanoparticles to phase segregated SLBs.112 The differences in the membrane model (vesicles vs. SLBs) as well as the differences in nanoparticle properties might have contributed to the differences in the two studies. The particles used by Melby and colleagues112 were mercaptopropylamine (MPNH2)-coated gold nanoparticles (core diameter of 4 nm). The particles used by Atukorale and colleagues149 were also gold nanoparticles; however, these particles which were coated with mercaptoundecanesulfonate (MUS)/octanethiol (OT) in a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio had a slightly different core diameter (2.5 nm) and were negatively charged in aqueous medium.150 While the particles used by the two studies have different physicochemical properties, thes differences in results point to the need for further studies on the role of membrane phase segregation in its interaction with ENMs. Finally, the effect of the aqueous medium in which ENMs and vesicles interact cannot be ignored. The pH of the buffer can modulate the charges on particles and vesicles;128 increasing the ionic strength of the buffer can result in screening of electrostatic charges on the particles and vesicles,137 while the macromolecules and ions in the medium can induce particle aggregation, which in turn will affect particle–vesicle interactions.151
1 ratio had a slightly different core diameter (2.5 nm) and were negatively charged in aqueous medium.150 While the particles used by the two studies have different physicochemical properties, thes differences in results point to the need for further studies on the role of membrane phase segregation in its interaction with ENMs. Finally, the effect of the aqueous medium in which ENMs and vesicles interact cannot be ignored. The pH of the buffer can modulate the charges on particles and vesicles;128 increasing the ionic strength of the buffer can result in screening of electrostatic charges on the particles and vesicles,137 while the macromolecules and ions in the medium can induce particle aggregation, which in turn will affect particle–vesicle interactions.151
Experimental evidence on the role of particle shape in ENM–membrane interactions is scarce, primarily due to the difficulties of synthesizing small particles with an accurate shape. However, this area has benefitted from computational approaches, which have reported on the importance of shape anisotropy in particle–membrane interactions. For instance, it has been shown that a disk-shaped particle with a large diameter, thus having a large contact area with the membrane, induces pore formation in planar lipid bilayers, while particles with a small diameter translocate the bilayer without significant disruption.152 Anisotropic particles have also been shown to undergo reorientation, driven by electrostatic forces, which will maximize their contact area with the membrane, and significantly disrupt the bilayer in the process of translocation.153 Particle reorientation has also been reported for hydrophobic particles; however, for these particles, reorientation occurs after the particles have penetrated the bilayer. A study by Baoukina and colleagues154 on the interactions between carbon nanotubes and DOPC bilayers revealed that non-functionalized, hydrophobic carbon nanotubes with a diameter of 1.23 nm and three different lengths (4.1, 6.5, and 9.7 nm) all reorient horizontally inside the core of the bilayer with the reorientation time depending on the particle length. In summary, evidence from computational studies suggests that particle translocation into the bilayer depends on the particle–membrane contact area and can be affected by particle reorientation before and after entrance into the bilayer.
How does the membrane model affect ENM–membrane interactions?
While a considerable body of literature is available on the interactions of lipid monolayers, SLBs, and lipid vesicles with ENMs, little information is available on how different membrane models compare in their interactions with ENMs. Given the widespread use of all three models, it is useful to consider how changing the membrane model affects ENM–lipid interactions, so that caution can be applied in comparing the results of one model to another and ultimately to the findings in cells.The effect of changing the membrane model on the outcome of ENM–membrane interactions becomes clear once the overall trends on particle–membrane interactions for each model are considered. A comparison of studies in lipid monolayers vs. those in SLBs and vesicles reveals a discrepancy on the role of particle charge. In studies with lipid monolayers, charged particles are generally reported to not significantly disrupt lipid films.49,155,156 This is because charged particles are more likely to remain in the aqueous subphase and not interact with lipids. In contrast, in studies of SLBs and vesicles, electrostatic interactions are the main driving force for particle-binding to the membrane and charged particles are generally more disruptive. This discrepancy roots in one of the disadvantages noted earlier for lipid monolayers: the fact that lipids are at an air–water interface and not completely submerged in water. As a result, highly charged ENMs are likely to remain in the aqueous subphase and do not come into direct contact with lipids at the air–water interface. By extension, the presence of lipids at the air–water interface also results in surface-active particles being more disruptive to lipids. This was recently demonstrated by comparing the effects of charged and PEGylated silica nanoparticles on lipid monolayers vs. lipid vesicles.88 Charged particles disrupted the vesicles, as evidenced by significant leakage, while PEGylated particles were not disruptive to the vesicles (except for the cases in which the particles were only partially covered by PEG). In contrast, PEGylated particles, unlike charged particles, showed significant penetration in the monolayers.88 The effects of PEGylated particles on the monolayer are due to the fact that PEGylation makes the particles more surface-active, thus making them more likely to penetrate the air–water interface.157 This example clearly shows that changing the membrane model affects ENM–lipid interactions. While the above-mentioned study focuses on lipid monolayers and vesicles, the role of the substrate in modulating the interactions of ENMs with SLBs should also be considered. Mica is a commonly used substrate for SLB formation.55,106,107,109 However, the substrate might affect electrostatic interactions between ENMs and lipids. In fact, while studies have shown that positively-charged ENMs can disrupt SLBs,107,158 the use of mica, a negatively-charged surface in water,159 as the substrate has been suggested to have played a role in such interactions.138 Thus, electrostatic interactions in SLBs should be interpreted following careful consideration of the role of the substrate.
Another inconsistency between the membrane models is found when the role of particle size in ENM–lipid interactions is compared between lipid monolayers, SLBs, and vesicles. In studies with lipid monolayers and SLBs, it is generally reported that smaller particles are more disruptive to the membrane.95–98,113 However, it is not the absolute value of the particle size but the ratio of particles to vesicles that plays an important role. In fact, in most cases, larger particles have been reported to be more disruptive to vesicles than smaller particles. This discrepancy between the models is the collective result of two phenomena. First, there is a difference in the interactions of ENMs with monolayers and SLBs compared to vesicles. In lipid monolayers and in SLBs, particle interaction with lipids is caused by particle diffusion to the air–water interface or to the surface of the SLB. However, in vesicles, it is not particle diffusion alone but wrapping of the membrane around the particle and particle internalization inside the vesicles that play an important role in the outcome of the interaction.138,139 Secondly, as discussed in the drawbacks of vesicles in the previous section, vesicles, except for GUVs, are in the same size order as ENMs. Therefore, their wrapping around ENMs is size dependent. Wrapping around a larger ENM will result in the loss of more lipids and eventually bilayer formation on the surface of the particle,138 and will thus be more disruptive to vesicle integrity. On a similar note, vesicle lamellarity is also likely to play a role in ENM–vesicle interactions. While the majority of studies on the interactions of ENMs with vesicles have focused on unilamellar vesicles, studies with multilamellar vesicles also exist.141,149 Since lipid bilayer formation on the surface of the particles is known to reduce their ability to disrupt vesicles,160 multilamellar vesicles are likely to be more resistant by ENMs. Studies on the role of vesicle lamellarity on their interactions with ENMs do not exist and will help address some of the experimental uncertainties.
The role of membrane curvature should also be taken into account when comparing different membrane models. As noted in the discussion of their shortcomings in the previous section, monolayers and SLBs are both planar surfaces and do not mimic the curved surface of the cell plasma membrane. Membrane curvature is known to play an important role in ENM–membrane interactions. As shown by both simulations and experiments, amphiphilic gold nanoparticles penetrate curved lipid surfaces significantly more than planar surfaces.71 This is due to the fact that the incorporation of these particles into the membrane requires hydrophobic defects in the bilayer, which allows for the contact of hydrophobic groups on the particles to interact with acyl chains in lipids.71 This finding suggests that particles are more likely to bind to the curved surface of vesicles than the planar surfaces of SLBs and monolayers. As noted earlier, while monolayers and SLBs do not mimic membrane curvature, the highly curved surface of SUVs and smaller LUVs (with diameters of one to several hundreds of nanometers) exaggerates the plasma membrane curvature (with a diameter of ∼6–50 μm in mammalian cells). GUVs are likely the best representatives of the cell plasma membrane curvature among the currently used models.
The three different membrane models are also likely to present different intermolecular distances between the lipids. Studies with lipid monolayers are generally performed using a monolayer that is initially fully expanded but is compressed in the presence of ENMs, resulting in a surface pressure isotherm that spans a large range of surface pressure values (in the range of 0 to 70 mN m−1) in a relatively short amount of time. In some cases, the particles disrupt the monolayer at low surface pressure values (i.e. high intermolecular distance), but their effects are diminished once the film is compressed.88,99 However, SLBs formed via the common vesicle fusion method have an intermolecular distance that is similar to that of monolayers at a surface pressure of 38 mN m−1.161 The heterogeneity of the plasma membrane and the presence of membrane proteins make it difficult to estimate the intermolecular distance for membrane lipids. However, it is useful to compare the above-mentioned surface pressure values with those reported for the plasma membrane. A series of elegant studies by Van Deenen's group,162–164 using phospholipases that are dependent on surface pressure for their enzymatic activity, revealed a surface pressure range of 31 to 34.8 mN m−1 for erythrocyte plasma membrane and a surface pressure of 34 mN m−1 for platelet plasma membrane. Thus, SLBs have a surface pressure close to that observed in the plasma membrane while only a small surface pressure range in the Langmuir trough mimics the surface pressure observed in the plasma membrane. In vesicles, the intermolecular distance and surface pressure depend on the vesicle curvature, regulated by the vesicle size and the type of vesicle (e.g. SUV vs. GUV), and also by the ionic strength of the medium, which reduces the distance between lipid headgroups by screening electrostatic charges.
In addition to the above, the lateral mobility of lipids between various models is also different. The lateral mobility of lipids in the plasma membrane ensures proper protein dynamics and has been suggested to play a role in protein localization,165 folding,166 and transport of molecules across the membrane.167 Lateral diffusion of lipids is generally evaluated by measuring the diffusion coefficient of lipids, which is approximately 0.1–0.2 μm2 s−1 in the cell plasma membrane.46 The diffusion coefficient of lipids in membrane models highly depends on the model used. For example, the lateral diffusion of lipids in a DOPC SLB has been reported to be 3.1 ± 0.3 μm2 s−1, which is lower than the diffusion coefficient of the same lipid in the same medium in vesicles (7.8 ± 0.8 μm2 s−1), as measured by fluorescence correlation spectroscopy.168 Similar results have been observed for other lipids in SLBs and in vesicles measured using fluorescence recovery after photobleaching.57 This difference is due to the fact that in SLBs the lipids are fixed in a solid support and are thus not capable of moving as freely as they are in a vesicle. The lateral diffusion of lipids in monolayers has also been measured and is a function of surface pressure, which itself is a function of the surface area available to the lipids. For instance, the lateral diffusion of lipids in a DMPC monolayer at a surface pressure of 40 mN m−1 is 6.5 ± 0.8 μm2 s−1,169 which is similar to that of vesicles, but this number can be adjusted by altering the surface pressure. It should be emphasized that the diffusion coefficient values reported above are for single phospholipids and the presence of other lipids, in particular sterols, and the lipid chemical structure (i.e. acyl chain length, degree of saturation, and headgroup structure) will highly affect lateral diffusivity. However, as can be deduced from the numbers reported above, the lateral diffusion of lipids in membrane models is generally higher than that in a native membrane. This phenomenon has also been reported for membrane proteins170 and is due to the crowded membrane environment in live cells, which contains a large number of proteins and is likely also affected by the attachment of the membrane to the underlying cell cytoskeleton.
The differences in the diffusion coefficient of lipids in different models also have implications for membrane fluidity. As noted above, the role of membrane fluidity in ENM–membrane interactions still needs more investigation, but at least one study has reported that the presence of lipid domains in SLBs increases the binding of gold nanoparticles to the lipids.112 Given the differences in the diffusion coefficient of lipids between vesicles and SLBs,56,57 it is likely that the same lipids in a vesicle, or a monolayer, will be more fluid compared to SLBs. Thus, the membrane model should be considered when examining the role of membrane fluidity in ENM–membrane interactions.
Another difference between the membrane models is their response to differences in particle hydrophobicity. In studies with lipid monolayers, hydrophobic particles are generally considered to be damaging toward the membrane due to the fact that they penetrate the air–water interface and can localize in between the lipid molecules and/or adsorb lipid molecules on their surface.99–101 Similarly, in SLBs, hydrophobic ENMs are considered to be more disruptive than hydrophilic particles given their ability to localize in and remove lipids from the bilayer.118 In contrast, hydrophobic ENMs are not always disruptive to vesicles. If hydrophobic ENMs are small enough, in the order of the thickness of the bilayer, they could localize at the hydrophobic core of the bilayer without disrupting the vesicles. This has been demonstrated with various hydrophobic ENMs such as 5.7 nm thiol-coated silver nanoparticles,146 ∼2 nm thiol-coated gold nanoparticles,171,172 5 nm oleic acid-capped maghemite nanoparticles,145 and ∼3 nm trioctylphosphine oxide-coated CdSe quantum dots.173 Similar observations have also been made computationally with small (<4 nm) hydrophobic particles shown to localize in the hydrophobic core of the bilayer.174
Taken together, it is clear that changing the membrane model affects ENM interactions with the membrane. The choice of the membrane model to a large extent depends on the property that the researchers are interested in studying and the instruments that are available. Planar models do not mimic membrane curvature, but, when coupled with other instruments, can provide information on surface properties such as surface tension and surface potential.97,105,175 Planar models are also useful in providing a quantitative understanding of particle binding to the membrane and ion transport through ENM-induced pores in the membrane. Vesicles, on the other hand, mimic membrane curvature and allow for studies on membrane integrity, lipid packing, and lipid phase segregation. In the case of GUVs, vesicles can also be used for in situ imaging of ENM–lipid interactions. While all models provide important information, in their current form, no model is a perfect mimic of the plasma membrane. All models can be modified to better represent the complexity of the cell plasma membrane as discussed below. Comprehensive information on ENM–membrane interactions will likely require a combination of models and a close examination of the correlations between observations in the model and in live cells.
How do the findings in membrane models correlate to cells?
The diversity of the structure and composition of the plasma membrane in different cells has made it difficult to develop general principles of ENM-induced cytotoxicity. However, more than two decades of cytotoxicity studies with ENMs has led to some general principles on particle cytotoxicity, some of which have been developed based on information from studies in cell membrane models.It is commonly accepted that cationic particles are more disruptive to cells than anionic particles.20,176,177 This is in agreement with several studies with cell membrane models demonstrating that positively-charged particles are more disruptive to the cell membrane than negatively-charged particles.35,89,132,133 In addition, a few recent studies have shown that the presence of a biomolecular corona reduces the cytotoxicity of ENMs.160,178 This is also in agreement with studies in membrane models where the presence of a lipid bilayer around the particles or adsorption of proteins on the particles has been shown to reduce their disruptive effects on membrane models.129,138 Nanoparticle PEGylation is also known to reduce particle toxicity to the cells.179,180 This agrees with studies on the interactions of PEGylated particles with vesicles88 but disagrees with studies in monolayers.88,102 This discrepancy is due to the differences between the two membrane models and how membrane disruption is defined in each model. PEG surface-activity drives particle interaction with lipids. In the case of vesicles, this might lead to particle localization at the membrane, without significant leakage, or invagination of PEGylated particles by the vesicles. However, in the case of monolayers, the ability to invaginate does not exist, and particle co-localization with lipids leads to changes in surface activity, which is defined as membrane disruption.
Smaller particles are known to be more toxic to cells than larger particles.181–184 However, studies with membrane models, in particular those with vesicles, frequently report that larger particles are disruptive to membrane integrity.89,138 For example, using gold nanoparticles with sizes ranging from 1.4 nm to 15 nm, Broda and colleagues185 reported that larger gold particles are more disruptive to the integrity of suspended lipid bilayers compared to smaller particles. However, when the same particles were tested against cells, the smaller particles caused greater toxicity than larger particles.185 This could mean that the cytotoxicity of such particles has a completely separate mechanism from membrane damage, even if damage is observed in studies with membrane models. However, these contradictory findings also point to an important limitation of studies with membrane models: the inability to account for lipid biogenesis. Larger particles can be internalized in vesicles and in the process get covered by lipid bilayers.138 This loss of membrane lipids is the likely reason behind vesicle disruption by such particles. However, it is unlikely that such a process would lead to toxicity in live cells, at least in short time scales, due to the ability of cells to replenish lost membrane material. In fact, large, micron-sized, particles can be engulfed by cells without leading to immediate cell death.186,187 It is also known that hydrophobic nanoparticles are more toxic than hydrophilic ones. For example, modifying the surface of hydrophobic C60 fullerenes with hydrophilic moieties has been shown to significantly reduce their cytotoxicity.188 Interestingly, the hydrophobic particles were shown to cause significant plasma membrane damage in cells. This is in agreement with studies in all three membrane models in which hydrophobic nanoparticles have been shown to disrupt lipid monolayers,88,99–103 remove lipids from SLBs118 and disrupt the integrity of vesicles.129,142
Parallel studies in model membranes and live cells have also been performed to uncover the mechanisms by which ENMs induce cytotoxicity. For example, Goodman and colleagues189 examined the interactions of mixed monolayer protected gold clusters (core size of 2 nm, but aggregates of up to 9.9 nm in suspension190), with positive and negative charges, with erythrocytes, COS1 cells, and Escherichia coli. Positively-charged particles resulted in significantly higher cytotoxicity compared to negatively-charged particles. Positively-charged particles were also more disruptive to negatively-charged vesicles, as evidenced by a vesicle leakage assay, suggesting that their cytotoxicity might have been caused by damage in the cell plasma membrane, which has an overall negative charge.189 Other examples of simultaneous use of membrane models and live cells exist in the literature. In one such study, penicillamine-coated CdSe/ZnS nanoparticles (4 nm) were shown to passively internalize in erythrocytes without disrupting the erythrocyte cell membrane. QCM experiments with SLBs mimicking the composition of the erythrocyte plasma membrane showed that particles do not cause pores in the bilayer but lead to a more flexible membrane, which is likely to increase their traversal into the cells without membrane disruption.191 Similar studies have also been used to examine the mechanisms of cytotoxicity of fullerene nanoparticles. Aqueous suspensions of fullerene C60 (sizes between 0.7–2.8 nm) were able to disrupt BLMs of phosphatidylcholine, as evidenced by an increase in specific conductivity and capacity of the membrane, suggesting that the presence of the particles disrupts the bilayer integrity. The same particles were also shown to cause cell swelling and increased the exposure of phosphatidylserine in the outer leaflet of the membrane, which serves as a measure of cytotoxicity.192 In addition to these studies, there have also been efforts to examine the effects of ENMs in membrane models mimicking the outer membrane of Gram-negative bacteria as well as live bacterial cells.193,194 These studies have been described in more detail in the next section.
It is also important to point out how studies in membrane models have been used to elucidate some of the mechanisms of ENM internalization in cells. A decade ago, it was shown that amphiphilic gold nanoparticles (4 nm) coated with alternating hydrophobic (octanethiol, OT) and hydrophilic (mercaptoundecane sulfonate, MUS) groups are able to passively translocate the cell membrane and enter the cytosol, even at 4 °C, a temperature at which endocytosis is not active.150,195 Follow-up studies in membrane models demonstrated that amphiphilic nanoparticles, unlike hydrophobic nanoparticles of the same size, are able to penetrate to the core of the bilayer, without causing vesicle disruption.62,71 Studies using SLBs revealed that internalization of amphiphilic particles requires contact between the hydrophobic groups on the particle surface and acyl chains in the membrane and preferentially occurs in areas of membrane defects.196 In summary, while internalization of amphiphilic gold nanoparticles was first observed in cells, information on the mechanisms of internalization was garnered using membrane models.
Future challenges and opportunities
The primary motivation for studies of ENM–membrane interactions is to understand whether exposure to ENMs might result in cytotoxicity. However, while studies in simple membrane models have generated a significant body of knowledge, many of the important structural features of the cell plasma membrane are not captured in currently used models and their role in ENM–membrane interactions remains obscure. Naturally, models cannot replicate all the structural complexities of the cell membrane; however, simple steps can be taken to build up on currently available models and increase the biological relevance of ENM–membrane studies. In addition to advancing membrane models, increasing the biological relevance of ENM–membrane studies also requires a consideration of the particle properties by the time they reach the cell membrane. Particles will always be exposed to biological fluids prior to reaching the cells. While the concept of a “biomolecular corona” is well established, the presence of the corona has not been considered in the majority of ENM–membrane interaction studies. Considering the lipid/protein corona around the particles will significantly enhance the environmental relevance of studies on particle-membrane interactions. Thus, two primary future challenges in studies of ENM–membrane interactions include 1) increasing the biological relevance of membrane models and 2) considering the role of the biomolecular corona in ENM–plasma membrane interactions. Each of these areas has been discussed in more detail below.Increasing the biological relevance of membrane models
A first step in increasing the biological relevance of membrane models is to consider what types of cells are being mimicked and what lipids constitute the membrane of that cell. This is important particularly given the structural diversity of cells in eukaryotes vs. prokaryotes. Eukaryotes have only one membrane surrounding the cell, the plasma membrane. Gram-positive prokaryotic cells are also covered by only one membrane, which itself is covered by a peptidoglycan layer. Gram-negative prokaryotes, however, have two membranes: a cytoplasmic membrane, also covered by a peptidoglycan layer, as well as an outer membrane. Studies on membrane models should consider the fact that membrane lipids are very different between eukaryotes and prokaryotes and even between Gram-positive and Gram-negative prokaryotes.197 A famous example of such differences between the membranes of different cells is the amount and types of sterols in mammalian, fungal, and bacterial cells. Sterols have received significant attention due to their role in increasing the lipid order, leading to the formation of lipid domains. The primary sterol in mammalian is cholesterol, while the primary sterol in fungal cells is ergosterol. In contrast, only a few bacteria, e.g. Helicobacter,198,199 contain sterols. Some bacteria recruit sterols from host cells,200 while some bacteria synthesize hopanoids, sterol-like molecules, which have been suggested to promote lipid domain formation.201 Studies in which the lipid composition of the membrane model is matched with cells are surprisingly rare. Close attention to the plasma membrane lipid composition of cells that are to be mimicked and using relevant lipids in models is an important first step in ensuring the biological relevance of membrane models.Another important consideration to increase the biological relevance of cells is the presence of membrane glycans. Both Gram-positive and Gram-negative bacteria contain glycans in their membrane although their structures differ. The membrane of Gram-negative bacteria contains lipopolysaccharide (LPS) while the membrane of Gram-positive bacteria contains a significant amount of lipoteichoic acid (LPA).202 Membrane carbohydrates also exist in some mammalian cells in the form of the glycocalyx, although in much lower amounts compared to bacteria. Studies in live bacterial cells have shown that membrane glycans in Gram-negative bacteria can protect bacteria against ENMs.203 Studies in membrane models have started to incorporate LPS into the membrane, in an effort to increase the biological relevance of membrane models. Studies using a POPC SLB showed that addition of LPS, which is negatively-charged, to the SLB increases the binding of positively-charged gold ENMs to the bilayer.193 Binding was dependent on the LPS structure and nanoparticles adhered more to the SLB including smooth LPS compared to the SLB including rough LPS. Rough LPS is a truncated form of smooth LPS, which has less negatively-charged phosphate groups and acidic sugars.204 Thus, increased binding of ENMs to SLBs containing smooth LPS suggests that ENM binding to the membrane is controlled by electrostatic attractive forces. The presence of LPS might also hinder the access of ENMs to the membrane due to steric hindrance. As shown by Mensch and colleagues,194 addition of 6.4 mol% rough LPS to POPC SLBs results in a significant negative charge but hinders the attachment of positively-charged nanodiamonds to the bilayer. LPS has also been incorporated into synthetic vesicles;205 however, LPS-containing vesicles have not been used in the context of ENM–membrane interactions. It should also be noted that significant advances have been made in computational studies of the outer membrane of Gram-negative bacteria206 with focus on the role of LPS structural diversity,207 LPS interactions with functional groups on nanoparticle surfaces,208 and protein self-assembly in the outer membrane.209 To the best of our knowledge, these advances in computational modeling of the bacterial outer membrane have not yet been used in the context of ENM–membrane interactions, but they provide a venue for a molecular level understanding of the interactions between ENMs and the bacterial membrane.
Membrane proteins add another layer of complexity to the physical interactions between ENMs and the cell membrane, which is often ignored. Current membrane models are overwhelmingly focused on lipids; as a result, there is currently very little information available on how ENMs might affect the function of membrane proteins. Such studies are particularly important given the fact that ENMs are known to affect membrane polarization.23,210,211 On the other hand, such studies are also needed to examine how membrane proteins might modulate ENM effects on membrane integrity. Proteovesicles, lipid vesicles that contain proteins, are a commonly used tool in membrane biology;212–214 however, they have not been used in the context of ENM–membrane interactions. Proteovesicles made with proteins whose function can be easily measured, e.g. bacteriorhodopsin, a protein that converts light into an electrochemical proton gradient,215 can be used as a starting point to investigate the effects of ENMs on protein function in membranes.
Current membrane models have also ignored an important structural complexity of the cell membrane, namely, membrane asymmetry. The cell membrane is asymmetric with respect to the phospholipid composition. Phosphatidylserine and phosphatidylethanolamine are predominantly located on the cytofacial leaflet, which faces the cytoplasm, while the majority of phosphatidylcholines, sphingomyelins, and sphingolipids are located in the exofacial leaflet, which faces the extracellular environment.216,217 Membrane asymmetry is known to play an important role in a number of important biological processes such as cell division, programmed cell death, and blood coagulation.218,219 However, its role in modulating ENM–membrane interactions has been so far overlooked. Our preliminary efforts have shown that membrane asymmetry is indeed important in the context of ENM-induced cell membrane damage.220 However, there is clearly more work to be done in this area given that membrane asymmetry can affect the behavior of membrane proteins,221 and alter lipid domain formation in vesicles,222,223 the latter of which is known to affect the interactions of vesicles with ENMs.112,149
Another area, which has not been explored in significant detail, is the role of structural support for the membrane in the form of cytoskeleton in eukaryotic cells and the peptidoglycan layer in Gram-negative bacteria. In addition to playing a crucial role in membrane dynamics (e.g. during cell migration and endocytosis),224 the cytoskeleton provides structural support to the plasma membrane. While cell-based studies have started to examine the role of the cytoskeleton in ENM penetration into the cell,225 its role in providing structural support to the plasma membrane upon interaction with ENMs has not been examined in detail. In a similar vein, the role of the peptidoglycan layer in providing support to the outer membrane in Gram-negative bacteria upon interaction with ENMs is not well understood. Tethered bilayers, in which the components of the cytoskeleton and peptidoglycan layer are used as tethering molecules, can provide a suitable model to examine the role of these structures in the interactions of the plasma membrane with ENMs.
In summary, a number of steps can be taken to improve the biological relevance of membrane models. These include 1) directly correlating the lipid composition of the membrane to the cell it is intended to mimic, 2) considering the types of cells the model is intended to mimic and accordingly incorporating membrane glycans, 3) incorporating membrane proteins and using the membrane model setting to examine the role of ENMs in protein function, and considering the role of 4) membrane asymmetry and 5) cytoskeleton in mammalian cells and peptidoglycan in Gram-negative bacteria. While attempts to address each of these complexities have been started, the majority of ENM–membrane studies have been focused on simple lipid membranes, and examining the role of each of these factors is an area for future investigation that could significantly improve the current understanding of ENM–membrane interactions.
Considering the biomolecular corona following environmental exposure
In humans, there are three major routes of environmental exposure to ENMs: skin exposure, ingestion, and inhalation;1 ocular exposure also presents a minor route.226,227 Other routes might also exist in animals, for example, the gill surface presents a major route of exposure to ENMs in fish.228,229 Of the primary routes of exposure in humans, skin exposure is less significant in the context of cytotoxicity because in in vivo settings, ENMs do not penetrate the skin deep enough to come into contact with keratinocytes.230 However, ENMs can come into contact with cells following ingestion or inhalation. In either route, ENMs will first be exposed to biological fluids and will develop a biomolecular corona prior to reaching the cells. To increase the environmental relevance of ENM–membrane interaction studies, attention should be given to the fact that each route of exposure is associated with its own biomolecular corona and that the presence of the corona will affect the ENMs' physicochemical properties and their interactions with the cell and its plasma membrane.Upon ingestion, ENMs get exposed to fluids with different pH values, enzymes, and ionic strengths. Given the complexity of these fluids, a number of artificial media (e.g. artificial saliva231 and gastric fluid232,233) have been developed. Studies with these artificial fluids have shown that particle properties change following exposure to biological fluids; however, changes in particle properties are not universal and depend on the physicochemical properties of pristine particles. For example, there have been reports on significant particle agglomeration after incubation in artificial saliva234,235 but also reports of only minor particle agglomeration231,236,237 depending on the properties of pristine particles. Changes in particle properties after exposure to artificial gastric juice are more significant; the high acidity of this environment can disrupt the binding of stabilizing agents231 and is reported to result in significant particle agglomeration.231,236,238,239 Gastric fluids can also entirely or partially dissolve ENMs, affecting the particle size and leading to the release of ions.238–240
Following inhalation, particles come into contact with the pulmonary surfactant, a mixture of lipids and proteins that lines the deep lungs,42 prior to interacting with cells. Experiments and simulations using carbon nanotubes,241 silver,242 polystyrene,242 and magnetite nanoparticles243 have shown that ENMs with different surface properties have similar lipids but different protein profiles in their corona after interaction with the pulmonary surfactant. These studies and others244,245 suggest that protein adsorption, but not lipid adsorption, on ENM surfaces depends on the physicochemical properties of pristine particles. Particles can also enter the circulation following both ingestion246 and inhalation247 and the presence of the primary corona will affect the formation of the secondary corona in the bloodstream.241
Understanding of the evolution of the biomolecular corona after exposure to biological fluids is currently very limited. However, a few studies have started to investigate the role of the corona on nanoparticle interactions with the cell plasma membrane. Early studies by Lesniak and colleagues25 showed that adherence of plasma proteins on the surface of silica nanoparticles (50 nm) significantly reduces their attachment to POPC SLBs compared to bare nanoparticles. Similarly, coating citrate-coated silver nanoparticles (49–65 nm) with human serum albumin was shown to reduce their adhesion to DOPC SLBs.248 In agreement, coating carboxyl-modified polystyrene nanoparticles (20 and 100 nm) with serum biomolecular corona reduced the disruptive effects on DOPC SLBs compared to bare particles.249 The studies noted above suggest that the presence of a corona is associated with a reduction in particle binding to SLBs. However, this is not always the case. A study by Melby and colleagues,250 using gold nanoparticles (4–5 nm), showed that the biomolecular corona can increase the binding of particles to SLBs. Further, it was shown that binding of corona-coated particles with SLBs is not purely driven by electrostatic interactions as particles with similar zeta potentials but different profiles of proteins in their corona showed very different binding behaviors to SLBs. It should also be noted that while the majority of studies on ENM–membrane interactions in mammalian cells have focused on the plasma membrane, ENM interactions with intracellular membranes is also of importance in the context of ENM-induced cytotoxicity.251–254 The biomolecular corona in this context is understudied but is expected to primarily consist of cytoplasmic biomolecules.
A particle corona composed of natural organic matter (NOM) is also starting to gain attention. A coating of humic acid on fullerene particles (∼120 nm) was shown to reduce their binding to solid supported lipid membranes and reduce their toxicity to epithelial cells.255 In agreement, Mensch and colleagues showed that the formation of a NOM corona around PAH-modified nanodiamonds reduces their binding to DOPC bilayers as well as their membrane toxicity to bacterial cells.194 Further studies on the NOM corona are needed to gain information on the ecotoxicity of ENMs.
Prediction of the biomolecular corona and its role in modulating ENM interactions has been a central challenge in nanotechnology for more than a decade. Many challenges remain in the field which are of significant interest to studies of ENM–membrane interactions. One central issue is understanding how the presence of a corona affects ENM binding to lipids and subsequently ENM-induced membrane damage. A few studies, summarized above, have already started to address this question. However, a general understanding of the role of the corona in ENM–membrane interactions is currently lacking and is hampered by the inability to definitively predict the corona, and this remains a challenge for the future. Another future challenge is understanding the role of the corona developed following various exposure routes in ENM binding to lipids. The majority of current studies have focused on a corona made up of plasma proteins. However, the corona following particle inhalation or ingestion will be different from the serum corona and the effects of these coronas on ENM binding to lipid membranes remain understudied. The role of the NOM corona in particle interactions with the cellular membranes has also received little attention and is a topic for future research. While studies have started to emerge on the role of the NOM corona in ENM–membrane interactions, many more studies are needed to elucidate how the NOM corona depends on particle properties and how it might affect particle interactions with biological membranes. Elucidating the role of the biomolecular corona in regulating ENM–membrane interactions will likely remain a challenge well into the future, but addressing some of the challenges noted above might provide a starting point for future studies in this area.
Conclusions
With the increasing concern regarding the toxicity of nanomaterials, membrane models have provided a simple working model to tease out the mechanisms of plasma membrane damage by ENMs. Valuable information has been gained using lipid monolayers, SLBs, and lipid vesicles. However, care should be taken when using such membrane models, as the findings from one model might not translate to another model and ultimately to the cell membrane. This is partially illustrated by the fact that the mechanisms by which ENMs cause membrane damage are different depending on the model used. The use of membrane models that capture some of the structural complexities of the cell membrane and considering the particle corona following an exposure could increase the biological and environmental relevance of studies on nanoparticle interactions with the cell plasma membrane.Conflicts of interest
There are no conflicts to declare.Acknowledgements
AF acknowledges financial support from the Russ College of Engineering and Technology and the Department of Chemical and Biomolecular Engineering at Ohio University.References
- M. E. Vance, T. Kuiken, E. P. Vejerano, S. P. McGinnis, M. F. Hochella Jr, D. Rejeski and M. S. Hull, Nanotechnology in the real world: Redeveloping the nanomaterial consumer products inventory, Beilstein J. Nanotechnol., 2015, 6, 1769–1780 CrossRef CAS PubMed.
- A. Pietroiusti and A. Magrini, Engineered nanoparticles at the workplace: current knowledge about workers' risk, Occup. Med., 2014, 64, 319–330 CrossRef CAS PubMed.
- J. I. Phillips, F. Y. Green, J. C. Davies and F. Jill Murray Mbch, Pulmonary and systemic toxicity following exposure to nickel nanoparticles, Am. J. Ind. Med., 2010, 53, 763–767 Search PubMed.
- Y. Song, X. Li and X. Du, Exposure to nanoparticles is related to pleural effusion, pulmonary fibrosis and granuloma, Eur. Respir. J., 2009, 34, 559–567 CrossRef CAS PubMed.
- M. C. Roco, The long view of nanotechnology development: the National Nanotechnology Initiative at 10 years, J. Nanopart. Res., 2011, 13(2), 427–445 CrossRef.
- A. A. Keller, S. McFerran, A. Lazareva and S. Suh, Global life cycle releases of engineered nanomaterials, J. Nanopart. Res., 2013, 15, 1692 CrossRef.
- F. Gottschalk, T. Sonderer, R. W. Scholz and B. Nowack, Modeled environmental concentrations of engineered nanomaterials (TiO2, ZnO, Ag, CNT, fullerenes) for different regions, Environ. Sci. Technol., 2009, 43, 9216–9222 CrossRef CAS PubMed.
- C. M. Park, K. H. Chu, N. Her, M. Jang, M. Baalousha, J. Heo and Y. Yoon, Occurrence and Removal of Engineered Nanoparticles in Drinking Water Treatment and Wastewater Treatment Processes, Sep. Purif. Rev., 2017, 46, 255–272 CrossRef CAS.
- K. Tiede, S. F. Hanssen, P. Westerhoff, G. J. Fern, S. M. Hankin, R. J. Aitken, Q. Chaudhry and A. B. Boxall, How important is drinking water exposure for the risks of engineered nanoparticles to consumers?, Nanotoxicology, 2016, 10, 102–110 CAS.
- M. Baalousha, Y. Yang, M. E. Vance, B. P. Colman, S. McNeal, J. Xu, J. Blaszczak, M. Steele, E. Bernhardt and M. F. Hochella, Outdoor urban nanomaterials: the emergence of a new, integrated, and critical field of study, Sci. Total Environ., 2016, 557, 740–753 CrossRef PubMed.
- J. Lovrić, H. S. Bazzi, Y. Cuie, G. R. Fortin, F. M. Winnik and D. Maysinger, Differences in subcellular distribution and toxicity of green and red emitting CdTe quantum dots, J. Mol. Med., 2005, 83, 377–385 CrossRef PubMed.
- P. Ariano, P. Zamburlin, A. Gilardino, R. Mortera, B. Onida, M. Tomatis, M. Ghiazza, B. Fubini and D. Lovisolo, Interaction of spherical silica nanoparticles with neuronal cells: size-dependent toxicity and perturbation of calcium homeostasis, Small, 2011, 7, 766–774 CrossRef CAS PubMed.
- P. Vedantam, G. Huang and T. J. Tzeng, Size-dependent cellular toxicity and uptake of commercial colloidal gold nanoparticles in DU-145 cells, Cancer Nanotechnol., 2013, 4, 13–20 CrossRef CAS PubMed.
- R. Coradeghini, S. Gioria, C. P. García, P. Nativo, F. Franchini, D. Gilliland, J. Ponti and F. Rossi, Size-dependent toxicity and cell interaction mechanisms of gold nanoparticles on mouse fibroblasts, Toxicol. Lett., 2013, 217, 205–216 CrossRef CAS PubMed.
- A. E. Nel, L. Mädler, D. Velegol, T. Xia, E. M. Hoek, P. Somasundaran, F. Klaessig, V. Castranova and M. Thompson, Understanding biophysicochemical interactions at the nano–bio interface, Nat. Mater., 2009, 8, 543–557 CrossRef CAS PubMed.
- A. Manke, L. Wang and Y. Rojanasakul, Mechanisms of nanoparticle-induced oxidative stress and toxicity, Biomed. Res. Int., 2013, 2013, 942916 Search PubMed.
- C. T. Ng, L. Q. Yong, M. P. Hande, C. N. Ong, L. E. Yu, B. H. Bay and G. H. Baeg, Zinc oxide nanoparticles exhibit cytotoxicity and genotoxicity through oxidative stress responses in human lung fibroblasts and Drosophila melanogaster, Int. J. Nanomed., 2017, 12, 1621 CrossRef CAS PubMed.
- T. Xia, M. Kovochich, M. Liong, L. Mädler, B. Gilbert, H. Shi, J. I. Yeh, J. I. Zink and A. E. Nel, Comparison of the mechanism of toxicity of zinc oxide and cerium oxide nanoparticles based on dissolution and oxidative stress properties, ACS Nano, 2008, 2, 2121–2134 CrossRef CAS PubMed.
- P. Møller, N. R. Jacobsen, J. K. Folkmann, P. H. Danielsen, L. Mikkelsen, J. G. Hemmingsen, L. K. Vesterdal, L. Forchhammer, H. Wallin and S. Loft, Role of oxidative damage in toxicity of particulates, Free Radical Res., 2010, 44, 1–46 CrossRef PubMed.
- J. Chen, J. A. Hessler, K. Putchakayala, B. K. Panama, D. P. Khan, S. Hong, D. G. Mullen, S. C. DiMaggio, A. Som and G. N. Tew, Cationic nanoparticles induce nanoscale disruption in living cell plasma membranes, J. Phys. Chem. B, 2009, 113, 11179–11185 CrossRef CAS PubMed.
- P. Ruenraroengsak, P. Novak, D. Berhanu, A. J. Thorley, E. Valsami-Jones, J. Gorelik, Y. E. Korchev and T. D. Tetley, Respiratory epithelial cytotoxicity and membrane damage (holes) caused by amine-modified nanoparticles, Nanotoxicology, 2012, 6, 94–108 CrossRef CAS PubMed.
- L. Wang, X. Jiang, Y. Ji, R. Bai, Y. Zhao, X. Wu and C. Chen, Surface chemistry of gold nanorods: origin of cell membrane damage and cytotoxicity, Nanoscale, 2013, 5, 8384–8391 RSC.
- E. A. Warren and C. K. Payne, Cellular binding of nanoparticles disrupts the membrane potential, RSC Adv., 2015, 5, 13660–13666 RSC.
- R. R. Arvizo, O. R. Miranda, M. A. Thompson, C. M. Pabelick, R. Bhattacharya, J. D. Robertson, V. M. Rotello, Y. Prakash and P. Mukherjee, Effect of nanoparticle surface charge at the plasma membrane and beyond, Nano Lett., 2010, 10, 2543–2548 CrossRef CAS PubMed.
- A. Lesniak, A. Salvati, M. J. Santos-Martinez, M. W. Radomski, K. A. Dawson and C. Åberg, Nanoparticle adhesion to the cell membrane and its effect on nanoparticle uptake efficiency, J. Am. Chem. Soc., 2013, 135, 1438–1444 CrossRef CAS PubMed.
- S. Kang, M. Herzberg, D. F. Rodrigues and M. Elimelech, Antibacterial effects of carbon nanotubes: size does matter!, Langmuir, 2008, 24, 6409–6413 CrossRef CAS PubMed.
- S. Kang, M. Pinault, L. D. Pfefferle and M. Elimelech, Single-walled carbon nanotubes exhibit strong antimicrobial activity, Langmuir, 2007, 23, 8670–8673 CrossRef CAS PubMed.
- L. Lai, S.-J. Li, J. Feng, P. Mei, Z.-H. Ren, Y.-L. Chang and Y. Liu, Effects of Surface Charges on the Bactericide Activity of CdTe/ZnS Quantum Dots: A Cell Membrane Disruption Perspective, Langmuir, 2017, 33, 2378–2386 CrossRef CAS PubMed.
- T. Yu, A. Malugin and H. Ghandehari, Impact of silica nanoparticle design on cellular toxicity and hemolytic activity, ACS Nano, 2011, 5, 5717–5728 CrossRef CAS PubMed.
- A. Ibrahim and S. Natarajan, Dietary trans–fatty acids alter adipocyte plasma membrane fatty acid composition and insulin sensitivity in rats, Metabolism, 2005, 54, 240–246 CrossRef CAS PubMed.
- W. Molee, M. Bouillier-Oudot, A. Auvergne and R. Babilé, Changes in lipid composition of hepatocyte plasma membrane induced by overfeeding in duck, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 2005, 141, 437–444 CrossRef CAS PubMed.
- L. Hsiao, R. Howard, M. Aikawa and T. Taraschi, Modification of host cell membrane lipid composition by the intra-erythrocytic human malaria parasite Plasmodium falciparum, Biochem. J., 1991, 274, 121–132 CrossRef CAS PubMed.
- P. N. Tran, S. H. Brown, M. Rug, M. C. Ridgway, T. W. Mitchell and A. G. Maier, Changes in lipid composition during sexual development of the malaria parasite Plasmodium falciparum, Malar. J., 2016, 15, 73 CrossRef PubMed.
- K. L. Chen and G. D. Bothun, Nanoparticles meet cell membranes: probing nonspecific interactions using model membranes, Environ. Sci. Technol., 2013, 48, 873–880 CrossRef PubMed.
- A. Negoda, Y. Liu, W.-C. Hou, C. Corredor, B. Y. Moghadam, C. Musolff, L. Li, W. Walker, P. Westerhoff and A. J. Mason, Engineered nanomaterial interactions with bilayer lipid membranes: screening platforms to assess nanoparticle toxicity, Int. J. Biomed. Nanosci. Nanotechnol., 2013, 3, 52–83 CrossRef CAS.
- E. Rascol, J.-M. Devoisselle and J. Chopineau, The relevance of membrane models to understand nanoparticles–cell membrane interactions, Nanoscale, 2016, 8, 4780–4798 RSC.
- L. Wu and X. Jiang, Recent developments in methodology employed to study the interactions between nanomaterials and model lipid membranes, Anal. Bioanal. Chem., 2016, 408, 2743–2758 CrossRef CAS PubMed.
- R. H. Notter, S. A. Tabak and R. D. Mavis, Surface properties of binary mixtures of some pulmonary surfactant components, J. Lipid Res., 1980, 21, 10–22 CAS.
- A. von Nahmen, A. Post, H. J. Galla and M. Sieber, The phase behavior of lipid monolayers containing pulmonary surfactant protein C studied by fluorescence light microscopy, Eur. Biophys. J., 1997, 26, 359–369 CrossRef CAS PubMed.
- H. Zhang, Q. Fan, Y. E. Wang, C. R. Neal and Y. Y. Zuo, Comparative study of clinical pulmonary surfactants using atomic force microscopy, Biochim. Biophys. Acta, Biomembr., 2011, 1808, 1832–1842 CrossRef CAS PubMed.
- H. Zhang, Y. E. Wang, Q. Fan and Y. Y. Zuo, On the low surface tension of lung surfactant, Langmuir, 2011, 27, 8351–8358 CrossRef CAS PubMed.
- Y. Y. Zuo, R. A. W. Veldhuizen, A. W. Neumann, N. O. Petersen and F. Possmayer, Current perspectives in pulmonary surfactant--inhibition, enhancement and evaluation, Biochim. Biophys. Acta, Biomembr., 2008, 1778, 1947–1977 CrossRef CAS PubMed.
- M. S. Bhamla, C. Chai, N. I. Rabiah, J. M. Frostad and G. G. Fuller, Instability and breakup of model tear films, Invest. Ophthalmol. Visual Sci., 2016, 57, 949–958 CrossRef CAS PubMed.
- M. S. Bhamla, W. L. Nash, S. Elliott and G. G. Fuller, Influence of lipid coatings on surface wettability characteristics of silicone hydrogels, Langmuir, 2014, 31, 3820–3828 CrossRef PubMed.
- P. Mudgil and T. J. Millar, Surfactant properties of human meibomian lipids, Invest. Ophthalmol. Visual Sci., 2011, 52, 1661–1670 CrossRef CAS PubMed.
- M. de Brabander, R. Nuydens, A. Ishihara, B. Holifield, K. Jacobson and H. Geerts, Lateral diffusion and retrograde movements of individual cell surface components on single motile cells observed with Nanovid microscopy, Int. J. Biochem. Cell Biol., 1991, 112, 111–124 CAS.
- R. Peters, Lateral mobility of proteins and lipids in the red cell membrane and the activation of adenylate cyclase by β-adrenergic receptors, FEBS Lett., 1988, 234, 1–7 CrossRef CAS PubMed.
- Q. Fan, Y. E. Wang, X. Zhao, J. S. C. Loo and Y. Y. Zuo, Adverse biophysical effects of hydroxyapatite nanoparticles on natural pulmonary surfactant, ACS Nano, 2011, 5, 6410–6416 CrossRef CAS PubMed.
- A. M. Farnoud and J. Fiegel, Low concentrations of negatively charged sub-micron particles alter the microstructure of DPPC at the air-water interface, Colloids Surf., A, 2012, 415, 320–327 CrossRef CAS.
- N.-J. Cho, C. W. Frank, B. Kasemo and F. Höök, Quartz crystal microbalance with dissipation monitoring of supported lipid bilayers on various substrates, Nat. Protoc., 2010, 5, 1096 CrossRef CAS PubMed.
- T. H. Anderson, Y. Min, K. L. Weirich, H. Zeng, D. Fygenson and J. N. Israelachvili, Formation of supported bilayers on silica substrates, Langmuir, 2009, 25, 6997–7005 CrossRef CAS PubMed.
- R. P. Richter, J. L. K. Him and A. Brisson, Supported lipid membranes, Mater. Today, 2003, 6, 32–37 CrossRef CAS.
- B. Jing, R. C. Abot and Y. Zhu, Semihydrophobic nanoparticle-induced disruption of supported lipid bilayers: specific ion effect, J. Phys. Chem. B, 2014, 118, 13175–13182 CrossRef CAS PubMed.
- X. Zhang and S. Yang, Nonspecific adsorption of charged quantum dots on supported zwitterionic lipid bilayers: real-time monitoring by quartz crystal microbalance with dissipation, Langmuir, 2011, 27, 2528–2535 CrossRef CAS PubMed.
- S. Hong, P. R. Leroueil, E. K. Janus, J. L. Peters, M.-M. Kober, M. T. Islam, B. G. Orr, J. R. Baker Jr and M. M. Banaszak Holl, Interaction of polycationic polymers with supported lipid bilayers and cells: nanoscale hole formation and enhanced membrane permeability, Bioconjugate Chem., 2006, 17, 728–734 CrossRef CAS PubMed.
- R. Macháň and M. Hof, Recent developments in fluorescence correlation spectroscopy for diffusion measurements in planar lipid membranes, Int. J. Mol. Sci., 2010, 11, 427–457 CrossRef PubMed.
- L. Guo, J. Y. Har, J. Sankaran, Y. Hong, B. Kannan and T. Wohland, Molecular diffusion measurement in lipid bilayers over wide concentration ranges: a comparative study, ChemPhysChem, 2008, 9, 721–728 CrossRef CAS PubMed.
- S. J. Johnson, T. M. Bayerl, D. C. McDermott, G. W. Adam, A. R. Rennie, R. K. Thomas and E. Sackmann, Structure of an adsorbed dimyristoylphosphatidylcholine bilayer measured with specular reflection of neutrons, Biophys. J., 1991, 59, 289–294 CrossRef CAS PubMed.
- S. Goennenwein, M. Tanaka, B. Hu, L. Moroder and E. Sackmann, Functional incorporation of integrins into solid supported membranes on ultrathin films of cellulose: impact on adhesion, Biophys. J., 2003, 85, 646–655 CrossRef CAS PubMed.
- M. Tanaka, F. F. Rossetti and S. Kaufmann, Native supported membranes: creation of two-dimensional cell membranes on polymer supports, Biointerphases, 2008, 3, FA12–FA16 CrossRef CAS PubMed.
- R. V. Goreham, V. C. Thompson, Y. Samura, C. T. Gibson, J. G. Shapter and I. Köper, Interaction of silver nanoparticles with tethered bilayer lipid membranes, Langmuir, 2015, 31, 5868–5874 CrossRef CAS PubMed.
- R. P. Carney, Y. Astier, T. M. Carney, K. Voïtchovsky, P. H. Jacob Silva and F. Stellacci, Electrical method to quantify nanoparticle interaction with lipid bilayers, ACS Nano, 2013, 7, 932–942 CrossRef CAS PubMed.
- A. l. Coutable, C. Thibault, J. r. m. Chalmeau, J. M. François, C. Vieu, V. Noireaux and E. Trévisiol, Preparation of tethered-lipid bilayers on gold surfaces for the incorporation of integral membrane proteins synthesized by cell-free expression, Langmuir, 2014, 30, 3132–3141 CrossRef CAS PubMed.
- J. C. Munro and C. W. Frank, Adsorption of lipid-functionalized poly (ethylene glycol) to gold surfaces as a cushion for polymer-supported lipid bilayers, Langmuir, 2004, 20, 3339–3349 CrossRef CAS PubMed.
- C. Peggion, F. Formaggio, C. Toniolo, L. Becucci, M. R. Moncelli and R. Guidelli, A peptide-tethered lipid bilayer on mercury as a biomimetic system, Langmuir, 2001, 17, 6585–6592 CrossRef CAS.
- F. Giess, M. G. Friedrich, J. Heberle, R. L. Naumann and W. Knoll, The protein-tethered lipid bilayer: A novel mimic of the biological membrane, Biophys. J., 2004, 87, 3213–3220 CrossRef CAS PubMed.
- I. Köper, Insulating tethered bilayer lipid membranes to study membrane proteins, Mol. BioSyst., 2007, 3, 651–657 RSC.
- H. T. Tien, Cyclic voltammetry of bilayer lipid membranes, J. Phys. Chem., 1984, 88, 3172–3174 CrossRef CAS.
- M. Winterhalter, Black lipid membranes, Curr. Opin. Colloid Interface Sci., 2000, 5, 250–255 CrossRef CAS.
- S. Ramachandran, N. E. Merrill, R. H. Blick and D. W. van der Weide, Colloidal quantum dots initiating current bursts in lipid bilayers, Biosens. Bioelectron., 2005, 20, 2173–2176 CrossRef CAS PubMed.
- R. C. Van Lehn, P. U. Atukorale, R. P. Carney, Y.-S. Yang, F. Stellacci, D. J. Irvine and A. Alexander-Katz, Effect of particle diameter and surface composition on the spontaneous fusion of monolayer-protected gold nanoparticles with lipid bilayers, Nano Lett., 2013, 13, 4060–4067 CrossRef CAS PubMed.
- B. Luan, S. Zhou, D. Wang and R. Zhou, Detecting interactions between nanomaterials and cell membranes by synthetic nanopores, ACS Nano, 2017, 11, 12615–12623 CrossRef CAS PubMed.
- T. Benachir and M. Lafleur, Study of vesicle leakage induced by melittin, Biochim. Biophys. Acta, Biomembr., 1995, 1235, 452–460 CrossRef.
- E. H. Shin, Y. Li, U. Kumar, H. V. Sureka, X. Zhang and C. K. Payne, Membrane potential mediates the cellular binding of nanoparticles, Nanoscale, 2013, 5, 5879–5886 RSC.
- S. N. Ahmed, D. A. Brown and E. London, On the origin of sphingolipid/cholesterol-rich detergent-insoluble cell membranes: physiological concentrations of cholesterol and sphingolipid induce formation of a detergent-insoluble, liquid-ordered lipid phase in model membranes, Biochemistry, 1997, 36, 10944–10953 CrossRef CAS PubMed.
- S. L. Veatch and S. L. Keller, Separation of liquid phases in giant vesicles of ternary mixtures of phospholipids and cholesterol, Biophys. J., 2003, 85, 3074–3083 CrossRef CAS PubMed.
- P. Pathak and E. London, Measurement of lipid nanodomain (raft) formation and size in sphingomyelin/POPC/cholesterol vesicles shows TX-100 and transmembrane helices increase domain size by coalescing preexisting nanodomains but do not induce domain formation, Biophys. J., 2011, 101, 2417–2425 CrossRef CAS PubMed.
- J. Wang, Megha and E. London, Relationship between sterol/steroid structure and participation in ordered lipid domains (lipid rafts): implications for lipid raft structure and function, Biochemistry, 2004, 43, 1010–1018 CrossRef CAS PubMed.
- X. Liu and K. L. Chen, Interactions of graphene oxide with model cell membranes: Probing nanoparticle attachment and lipid bilayer disruption, Langmuir, 2015, 31, 12076–12086 CrossRef CAS PubMed.
- H. Komatsu, P. T. Guy and E. S. Rowe, Effect of unilamellar vesicle size on ethanol-induced interdigitation in dipalmitoylphosphatidylcholine, Chem. Phys. Lipids, 1993, 65, 11–21 CrossRef CAS PubMed.
- M. Hope, M. Bally, G. Webb and P. Cullis, Production of large unilamellar vesicles by a rapid extrusion procedure. Characterization of size distribution, trapped volume and ability to maintain a membrane potential, Biochim. Biophys. Acta, Biomembr., 1985, 812, 55–65 CrossRef CAS.
- L. Mayer, M. Hope and P. Cullis, Vesicles of variable sizes produced by a rapid extrusion procedure, Biochim. Biophys. Acta, Biomembr., 1986, 858, 161–168 CrossRef CAS.
- A. Moscho, O. Orwar, D. T. Chiu, B. P. Modi and R. N. Zare, Rapid preparation of giant unilamellar vesicles, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 11443–11447 CrossRef CAS.
- T. Pott, H. Bouvrais and P. Méléard, Giant unilamellar vesicle formation under physiologically relevant conditions, Chem. Phys. Lipids, 2008, 154, 115–119 CrossRef CAS PubMed.
- M. Hope, M. Bally, L. Mayer, A. Janoff and P. Cullis, Generation of multilamellar and unilamellar phospholipid vesicles, Chem. Phys. Lipids, 1986, 40, 89–107 CrossRef.
- M. Angelova, S. Soléau, P. Méléard, F. Faucon and P. Bothorel, in Trends in Colloid and Interface Science VI, Springer, 1992, pp. 127–131 Search PubMed.
- M. D. Collins and S. E. Gordon, Giant liposome preparation for imaging and patch-clamp electrophysiology, J. Visualized Exp., 2013, 76, 50227 Search PubMed.
- A. Asghari Adib, S. Nazemidashtarjandi, A. Kelly, A. Kruse, K. Cimatu, A. E. David and A. M. Farnoud, Engineered silica nanoparticles interact differently with lipid monolayers compared to lipid bilayers, Environ. Sci.: Nano, 2018, 5(2), 289–303 RSC.
- S. Li and N. Malmstadt, Deformation and poration of lipid bilayer membranes by cationic nanoparticles, Soft Matter, 2013, 9, 4969–4976 RSC.
- K.-i. Akashi, H. Miyata, H. Itoh and K. Kinosita Jr, Preparation of giant liposomes in physiological conditions and their characterization under an optical microscope, Biophys. J., 1996, 71, 3242–3250 CrossRef CAS PubMed.
- A. M. Farnoud and J. Fiegel, Calf lung surfactant recovers surface functionality after exposure to aerosols containing polymeric particles, J. Aerosol Med. Pulm. Drug Delivery, 2016, 29(1), 10–23 CrossRef CAS PubMed.
- M. S. Bakshi, L. Zhao, R. Smith, F. Possmayer and N. O. Petersen, Metal nanoparticle pollutants interfere with pulmonary surfactant function in vitro, Biophys. J., 2008, 94, 855–868 CrossRef CAS PubMed.
- T. Kanishtha, R. Banerjee and C. Venkataraman, Effect of particle emissions from biofuel combustion on surface activity of model and therapeutic pulmonary surfactants, Environ. Toxicol. Pharmacol., 2006, 22, 325–333 CrossRef CAS PubMed.
- J. A. Virtanen, K. H. Cheng and P. Somerharju, Phospholipid composition of the mammalian red cell membrane can be rationalized by a superlattice model, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 4964–4969 CrossRef CAS.
- C. Peetla and V. Labhasetwar, Biophysical characterization of nanoparticle-endothelial model cell membrane interactions, Mol. Pharmaceutics, 2008, 5, 418–429 CrossRef CAS PubMed.
- D. Stuart, R. Lobenberg, T. Ku, S. Azarmi, L. Ely, W. Roa and E. J. Prenner, Biophysical investigation of nanoparticle interactions with lung surfactant model systems, J. Biomed. Nanotechnol., 2006, 2, 245–252 CrossRef CAS.
- T. Ku, S. Gill, R. Lobenberg, S. Azarmi, W. Roa and E. J. Prenner, Size dependent interactions of nanoparticles with lung surfactant model systems and the significant impact on surface potential, J. Nanosci. Nanotechnol., 2008, 8, 2971–2978 CrossRef CAS PubMed.
- C. Schleh, C. Muhlfeld, K. Pulskamp, A. Schmiedl, M. Nassimi, H. D. Lauenstein, A. Braun, N. Krug, V. J. Erpenbeck and J. M. Hohlfeld, The effect of titanium dioxide nanoparticles on pulmonary surfactant function and ultrastructure, Respir. Res., 2009, 10, 90–101 CrossRef PubMed.
- C. Peetla and V. Labhasetwar, Effect of molecular structure of cationic surfactants on biophysical interactions of surfactant-modified nanoparticles with a model membrane and cellular uptake, Langmuir, 2009, 25, 2369–2377 CrossRef CAS PubMed.
- Z. Wang, X. Li and S. Yang, Studies of dipalmitoylphosphatidylcholine (DPPC) monolayers embedded with endohedral metallofullerene (Dy@ C82), Langmuir, 2009, 25, 12968–12973 CrossRef CAS PubMed.
- C. Peetla, S. Jin, J. Weimer, A. Elegbede and V. Labhasetwar, Biomechanics and thermodynamics of nanoparticle interactions with plasma and endosomal membrane lipids in cellular uptake and endosomal escape, Langmuir, 2014, 30, 7522–7532 CrossRef CAS PubMed.
- H. Baghirov, S. Melikishvili, Y. Mørch, E. Sulheim, A. K. Åslund, T. Hianik and C. de Lange Davies, The effect of poly (ethylene glycol) coating and monomer type on poly (alkyl cyanoacrylate) nanoparticle interactions with lipid monolayers and cells, Colloids Surf., B, 2017, 150, 373–383 CrossRef CAS PubMed.
- G. B. Soriano, R. da Silva Oliveira, F. F. Camilo and L. Caseli, Interaction of non-aqueous dispersions of silver nanoparticles with cellular membrane models, J. Colloid Interface Sci., 2017, 496, 111–117 CrossRef CAS PubMed.
- G. Bothun, N. Ganji, I. Khan, A. Xi and C. Bobba, Anionic and Cationic Silver Nanoparticle Binding Restructures Net-Anionic PC/PG Monolayers with Saturated or Unsaturated Lipids, Langmuir, 2016, 33, 353–360 CrossRef PubMed.
- A. M. Farnoud and J. Fiegel, Interaction of dipalmitoyl phosphatidylcholine monolayers with a particle-laden subphase, J. Phys. Chem. B, 2013, 117, 12124–12134 CrossRef CAS PubMed.
- S. Hong, A. U. Bielinska, A. Mecke, B. Keszler, J. L. Beals, X. Shi, L. Balogh, B. G. Orr, J. R. Baker Jr and M. M. Banaszak Holl, Interaction of poly (amidoamine) dendrimers with supported lipid bilayers and cells: hole formation and the relation to transport, Bioconjugate Chem., 2004, 15, 774–782 CrossRef CAS PubMed.
- P. R. Leroueil, S. A. Berry, K. Duthie, G. Han, V. M. Rotello, D. Q. McNerny, J. R. Baker, B. G. Orr and M. M. Banaszak Holl, Wide varieties of cationic nanoparticles induce defects in supported lipid bilayers, Nano Lett., 2008, 8, 420–424 CrossRef CAS PubMed.
- S. Parimi, T. J. Barnes and C. A. Prestidge, PAMAM dendrimer interactions with supported lipid bilayers: a kinetic and mechanistic investigation, Langmuir, 2008, 24, 13532–13539 CrossRef CAS PubMed.
- T. A. Spurlin and A. A. Gewirth, Effect of C60 on solid supported lipid bilayers, Nano Lett., 2007, 7, 531–535 CrossRef CAS PubMed.
- W.-C. Hou, B. Y. Moghadam, P. Westerhoff and J. D. Posner, Distribution of fullerene nanomaterials between water and model biological membranes, Langmuir, 2011, 27, 11899–11905 CrossRef CAS PubMed.
- P. Yi and K. L. Chen, Interaction of multiwalled carbon nanotubes with supported lipid bilayers and vesicles as model biological membranes, Environ. Sci. Technol., 2013, 47, 5711–5719 CrossRef CAS PubMed.
- E. S. Melby, A. C. Mensch, S. E. Lohse, D. Hu, G. Orr, C. J. Murphy, R. J. Hamers and J. A. Pedersen, Formation of supported lipid bilayers containing phase-segregated domains and their interaction with gold nanoparticles, Environ. Sci.: Nano, 2016, 3, 45–55 RSC.
- W.-C. Hou, B. Y. Moghadam, C. Corredor, P. Westerhoff and J. D. Posner, Distribution of functionalized gold nanoparticles between water and lipid bilayers as model cell membranes, Environ. Sci. Technol., 2012, 46, 1869–1876 CrossRef CAS PubMed.
- J. M. Troiano, L. L. Olenick, T. R. Kuech, E. S. Melby, D. Hu, S. E. Lohse, A. C. Mensch, M. Dogangun, A. M. Vartanian and M. D. Torelli, Direct probes of 4 nm diameter gold nanoparticles interacting with supported lipid bilayers, J. Phys. Chem. C, 2014, 119, 534–546 CrossRef.
- R. Gupta and B. Rai, Effect of size and surface charge of gold nanoparticles on their skin permeability: a molecular dynamics study, Sci. Rep., 2017, 7, 45292 CrossRef CAS PubMed.
- J. Lin, H. Zhang, Z. Chen and Y. Zheng, Penetration of lipid membranes by gold nanoparticles: insights into cellular uptake, cytotoxicity, and their relationship, ACS Nano, 2010, 4, 5421–5429 CrossRef CAS PubMed.
- Y. Li and N. Gu, Thermodynamics of charged nanoparticle adsorption on charge-neutral membranes: a simulation study, J. Phys. Chem. B, 2010, 114, 2749–2754 CrossRef CAS PubMed.
- B. Jing and Y. Zhu, Disruption of supported lipid bilayers by semihydrophobic nanoparticles, J. Am. Chem. Soc., 2011, 133, 10983–10989 CrossRef CAS PubMed.
- J. M. Troiano, T. R. Kuech, A. M. Vartanian, M. D. Torelli, A. Sen, L. M. Jacob, R. J. Hamers, C. J. Murphy, J. A. Pedersen and F. M. Geiger, On electronic and charge interference in second harmonic generation responses from gold metal nanoparticles at supported lipid bilayers, J. Phys. Chem. C, 2016, 120, 20659–20667 CrossRef CAS.
- D. Wang, R. J. Nap, I. Lagzi, B. Kowalczyk, S. Han, B. A. Grzybowski and I. Szleifer, How and why nanoparticle's curvature regulates the apparent p K a of the coating ligands, J. Am. Chem. Soc., 2011, 133, 2192–2197 CrossRef CAS PubMed.
- W. Kunz, J. Henle and B. W. Ninham, ‘Zur Lehre von der Wirkung der Salze’(about the science of the effect of salts): Franz Hofmeister's historical papers, Curr. Opin. Colloid Interface Sci., 2004, 9, 19–37 CrossRef CAS.
- A. Aroti, E. Leontidis, E. Maltseva and G. Brezesinski, Effects of Hofmeister anions on DPPC Langmuir monolayers at the air-water interface, J. Phys. Chem. B, 2004, 108, 15238–15245 CrossRef CAS.
- N. Yousefi, A. Wargenau and N. Tufenkji, Toward more free-floating model cell membranes: method development and application to their interaction with nanoparticles, ACS Appl. Mater. Interfaces, 2016, 8, 14339–14348 CrossRef CAS PubMed.
- K. Simons and E. Ikonen, Functional rafts in cell membranes, Nature, 1997, 387, 569–572 CrossRef CAS PubMed.
- S. Mornet, O. Lambert, E. Duguet and A. Brisson, The formation of supported lipid bilayers on silica nanoparticles revealed by cryoelectron microscopy, Nano Lett., 2005, 5, 281–285 CrossRef CAS PubMed.
- A. Dif, E. Henry, F. Artzner, M. Baudy-Floc'h, M. Schmutz, M. Dahan and V. Marchi-Artzner, Interaction between water-soluble peptidic CdSe/ZnS nanocrystals and membranes: formation of hybrid vesicles and condensed lamellar phases, J. Am. Chem. Soc., 2008, 130, 8289–8296 CrossRef CAS PubMed.
- A. Hirano, K. Uda, Y. Maeda, T. Akasaka and K. Shiraki, One-dimensional protein-based nanoparticles induce lipid bilayer disruption: carbon nanotube conjugates and amyloid fibrils, Langmuir, 2010, 26, 17256–17259 CrossRef CAS PubMed.
- H. Pera, J. M. Kleijn and F. A. Leermakers, Interaction of silica nanoparticles with phospholipid membranes, Chem. Lett., 2012, 41, 1322–1324 CrossRef CAS.
- H. I. Alkhammash, N. Li, R. Berthier and M. R. de Planque, Native silica nanoparticles are powerful membrane disruptors, Phys. Chem. Chem. Phys., 2015, 17, 15547–15560 RSC.
- X. Wei, W. Jiang, J. Yu, L. Ding, J. Hu and G. Jiang, Effects of SiO 2 nanoparticles on phospholipid membrane integrity and fluidity, J. Hazard. Mater., 2015, 287, 217–224 CrossRef CAS PubMed.
- B. Wang, L. Zhang, S. C. Bae and S. Granick, Nanoparticle-induced surface reconstruction of phospholipid membranes, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 18171–18175 CrossRef CAS PubMed.
- M. Laurencin, T. Georgelin, B. Malezieux, J.-M. Siaugue and C. Ménager, Interactions between giant unilamellar vesicles and charged core−shell magnetic nanoparticles, Langmuir, 2010, 26, 16025–16030 CrossRef CAS PubMed.
- B. Y. Moghadam, W.-C. Hou, C. Corredor, P. Westerhoff and J. D. Posner, Role of nanoparticle surface functionality in the disruption of model cell membranes, Langmuir, 2012, 28, 16318–16326 CrossRef CAS PubMed.
- P. Chen, Z. Huang, J. Liang, T. Cui, X. Zhang, B. Miao and L.-T. Yan, Diffusion and directionality of charged nanoparticles on lipid bilayer membrane, ACS Nano, 2016, 10, 11541–11547 CrossRef CAS PubMed.
- X. Lin, C. Wang, M. Wang, K. Fang and N. Gu, Computer simulation of the effects of nanoparticles' adsorption on the properties of supported lipid bilayer, J. Phys. Chem. C, 2012, 116, 17960–17968 CrossRef CAS.
- L. Zhang and S. Granick, How to stabilize phospholipid liposomes (using nanoparticles), Nano Lett., 2006, 6, 694–698 CrossRef CAS PubMed.
- S. Savarala, S. Ahmed, M. A. Ilies and S. L. Wunder, Stabilization of soft lipid colloids: competing effects of nanoparticle decoration and supported lipid bilayer formation, ACS Nano, 2011, 5, 2619–2628 CrossRef CAS PubMed.
- O. Le Bihan, P. Bonnafous, L. Marak, T. Bickel, S. Trépout, S. Mornet, F. De Haas, H. Talbot, J.-C. Taveau and O. Lambert, Cryo-electron tomography of nanoparticle transmigration into liposome, J. Struct. Biol., 2009, 168, 419–425 CrossRef CAS PubMed.
- S. Zhang, A. Nelson and P. A. Beales, Freezing or wrapping: the role of particle size in the mechanism of nanoparticle–biomembrane interaction, Langmuir, 2012, 28, 12831–12837 CrossRef CAS PubMed.
- Y. Chen and G. D. Bothun, Cationic Gel-phase liposomes with “decorated” anionic SPIO nanoparticles: Morphology, colloidal, and bilayer properties, Langmuir, 2011, 27, 8645–8652 CrossRef CAS PubMed.
- J. Zupanc, D. Drobne, B. Drasler, J. Valant, A. Iglic, V. Kralj-Iglic, D. Makovec, M. Rappolt, B. Sartori and K. Kogej, Experimental evidence for the interaction of C-60 fullerene with lipid vesicle membranes, Carbon, 2012, 50, 1170–1178 CrossRef CAS.
- G. Rusciano, A. De Luca, G. Pesce and A. Sasso, On the interaction of nano-sized organic carbon particles with model lipid membranes, Carbon, 2009, 47, 2950–2957 CrossRef CAS.
- M. R. Preiss, A. Hart, C. Kitchens and G. D. Bothun, Hydrophobic nanoparticles modify the thermal release behavior of liposomes, J. Phys. Chem. B, 2017, 121, 5040–5047 CrossRef CAS PubMed.
- G. Von White, Y. Chen, J. Roder-Hanna, G. D. Bothun and C. L. Kitchens, Structural and thermal analysis of lipid vesicles encapsulating hydrophobic gold nanoparticles, ACS Nano, 2012, 6, 4678–4685 CrossRef CAS PubMed.
- Y. Chen, A. Bose and G. D. Bothun, Controlled release from bilayer-decorated magnetoliposomes via electromagnetic heating, ACS Nano, 2010, 4, 3215–3221 CrossRef CAS PubMed.
- G. D. Bothun, Hydrophobic silver nanoparticles trapped in lipid bilayers: Size distribution, bilayer phase behavior, and optical properties, J. Nanobiotechnol., 2008, 6, 13 CrossRef PubMed.
- S.-H. Park, S.-G. Oh, J.-Y. Mun and S.-S. Han, Effects of silver nanoparticles on the fluidity of bilayer in phospholipid liposome, Colloids Surf., B, 2005, 44, 117–122 CrossRef CAS PubMed.
- S.-H. Park, S.-G. Oh, J.-Y. Mun and S.-S. Han, Loading of gold nanoparticles inside the DPPC bilayers of liposome and their effects on membrane fluidities, Colloids Surf., B, 2006, 48, 112–118 CrossRef CAS PubMed.
- P. U. Atukorale, Z. P. Guven, A. Bekdemir, R. P. Carney, R. C. Van Lehn, D. S. Yun, P. H. Jacob Silva, D. Demurtas, Y.-S. Yang and A. Alexander-Katz, Structure–property relationships of amphiphilic nanoparticles that penetrate or fuse lipid membranes, Bioconjugate Chem., 2018, 29(4), 1131–1140 CrossRef CAS PubMed.
- A. Verma, O. Uzun, Y. Hu, Y. Hu, H.-S. Han, N. Watson, S. Chen, D. J. Irvine and F. Stellacci, Surface-structure-regulated cell-membrane penetration by monolayer-protected nanoparticles, Nat. Mater., 2008, 7, 588 CrossRef CAS PubMed.
- Q. Mu, N. S. Hondow, Ł. Krzemiński, A. P. Brown, L. J. Jeuken and M. N. Routledge, Mechanism of cellular uptake of genotoxic silica nanoparticles, Part. Fibre Toxicol., 2012, 9, 29 CrossRef CAS PubMed.
- K. Yang and Y.-Q. Ma, Computer simulation of the translocation of nanoparticles with different shapes across a lipid bilayer, Nat. Nanotechnol., 2010, 5, 579–583 CrossRef CAS PubMed.
- S. Nangia and R. Sureshkumar, Effects of nanoparticle charge and shape anisotropy on translocation through cell membranes, Langmuir, 2012, 28, 17666–17671 CrossRef CAS PubMed.
- S. Baoukina, L. Monticelli and D. P. Tieleman, Interaction of pristine and functionalized carbon nanotubes with lipid membranes, J. Phys. Chem. B, 2013, 117, 12113–12123 CrossRef CAS PubMed.
- E. Guzmán, L. Liggieri, E. Santini, M. Ferrari and F. Ravera, Influence of silica nanoparticles on phase behavior and structural properties of DPPC–palmitic acid Langmuir monolayers, Colloids Surf., A, 2011, 413, 280–287 CrossRef.
- E. Guzman, L. Liggieri, E. Santini, M. Ferrari and F. Ravera, DPPC-DOPC Langmuir monolayers modified by hydrophilic silica nanoparticles: phase behaviour, structure and rheology, Colloids Surf., A, 2012, 417, 174–183 CrossRef.
- M. Winterhalter, H. Bürner, S. Marzinka, R. Benz and J. Kasianowicz, Interaction of poly (ethylene-glycols) with air-water interfaces and lipid monolayers: investigations on surface pressure and surface potential, Biophys. J., 1995, 69, 1372–1381 CrossRef CAS PubMed.
- A. Mecke, D.-K. Lee, A. Ramamoorthy, B. G. Orr and M. M. Banaszak Holl, Synthetic and natural polycationic polymer nanoparticles interact selectively with fluid-phase domains of DMPC lipid bilayers, Langmuir, 2005, 21, 8588–8590 CrossRef CAS PubMed.
- Y. Roiter and S. Minko, Adsorption of polyelectrolyte versus surface charge: in situ single-molecule atomic force microscopy experiments on similarly, oppositely, and heterogeneously charged surfaces, J. Phys. Chem. B, 2007, 111, 8597–8604 CrossRef CAS PubMed.
- R. A. Roggers, M. Joglekar, J. S. Valenstein and B. G. Trewyn, Mimicking red blood cell lipid membrane to enhance the hemocompatibility of large-pore mesoporous silica, ACS Appl. Mater. Interfaces, 2014, 6, 1675–1681 CrossRef CAS PubMed.
- E. B. Watkins, C. E. Miller, W.-P. Liao and T. L. Kuhl, Equilibrium or quenched: fundamental differences between lipid monolayers, supported bilayers, and membranes, ACS Nano, 2014, 8, 3181–3191 CrossRef CAS PubMed.
- R. Zwaal, B. Roelofsen, P. Comfurius and L. Van Deenen, Organization of phospholipids in human red cell membranes as detected by the action of various purified phospholipases, Biochim. Biophys. Acta, Biomembr., 1975, 406, 83–96 CrossRef CAS.
- R. Demel, W. G. Van Kessel, R. Zwaal, B. Roelofsen and L. Van Deenen, Relation between various phospholipase actions on human red cell membranes and the interfacial phospholipid pressure in monolayers, Biochim. Biophys. Acta, Biomembr., 1975, 406, 97–107 CrossRef CAS.
- H. Chap, R. Zwaal and L. Van Deenen, Action of highly purified phospholipases on blood platelets. Evidence for an asymmetric distribution of phospholipids in the surface membrane, Biochim. Biophys. Acta, Biomembr., 1977, 467, 146–164 CrossRef CAS.
- A. R. Siafakas, L. C. Wright, T. C. Sorrell and J. T. Djordjevic, Lipid rafts in Cryptococcus neoformans concentrate the virulence determinants phospholipase B1 and Cu/Zn superoxide dismutase, Eukaryotic Cell, 2006, 5, 488–498 CrossRef CAS PubMed.
- S. R. Maurya, D. Chaturvedi and R. Mahalakshmi, Modulating lipid dynamics and membrane fluidity to drive rapid folding of a transmembrane barrel, Sci. Rep., 2013, 3, 1989 CrossRef PubMed.
- C. Le Grimellec, G. Friedlander, E. El Yandouzi, P. Zlatkine and M.-C. Giocondi, Membrane fluidity and transport properties in epithelia, Kidney Int., 1992, 42, 825–836 CrossRef CAS PubMed.
- M. Przybylo, J. Sýkora, J. Humpolíčková, A. Benda, A. Zan and M. Hof, Lipid diffusion in giant unilamellar vesicles is more than 2 times faster than in supported phospholipid bilayers under identical conditions, Langmuir, 2006, 22, 9096–9099 CrossRef CAS PubMed.
- M. Gudmand, M. Fidorra, T. Bjørnholm and T. Heimburg, Diffusion and partitioning of fluorescent lipid probes in phospholipid monolayers, Biophys. J., 2009, 96, 4598–4609 CrossRef CAS PubMed.
- K. Jacobson, A. Ishihara and R. Inman, Lateral diffusion of proteins in membranes, Annu. Rev. Physiol., 1987, 49, 163–175 CrossRef CAS PubMed.
- W. H. Binder, R. Sachsenhofer, D. Farnik and D. Blaas, Guiding the location of nanoparticles into vesicular structures: a morphological study, Phys. Chem. Chem. Phys., 2007, 9, 6435–6441 RSC.
- M. R. Rasch, E. Rossinyol, J. L. Hueso, B. W. Goodfellow, J. Arbiol and B. A. Korgel, Hydrophobic gold nanoparticle self-assembly with phosphatidylcholine lipid: membrane-loaded and janus vesicles, Nano Lett., 2010, 10, 3733–3739 CrossRef CAS PubMed.
- G. Gopalakrishnan, C. Danelon, P. Izewska, M. Prummer, P. Y. Bolinger, I. Geissbühler, D. Demurtas, J. Dubochet and H. Vogel, Multifunctional lipid/quantum dot hybrid nanocontainers for controlled targeting of live cells, Angew. Chem., Int. Ed., 2006, 45, 5478–5483 CrossRef CAS PubMed.
- V. V. Ginzburg and S. Balijepalli, Modeling the thermodynamics of the interaction of nanoparticles with cell membranes, Nano Lett., 2007, 7, 3716–3722 CrossRef CAS PubMed.
- M. Diociaiuti, I. Ruspantini, C. Giordani, F. Bordi and P. Chistolini, Distribution of GD3 in DPPC monolayers: a thermodynamic and atomic force microscopy combined study, Biophys. J., 2004, 86, 321–328 CrossRef CAS PubMed.
- P. Ruenraroengsak and T. D. Tetley, Differential bioreactivity of neutral, cationic and anionic polystyrene nanoparticles with cells from the human alveolar compartment: robust response of alveolar type 1 epithelial cells, Part. Fibre Toxicol., 2015, 12, 19 CrossRef PubMed.
- C. Hoskins, A. Cuschieri and L. Wang, The cytotoxicity of polycationic iron oxide nanoparticles: common endpoint assays and alternative approaches for improved understanding of cellular response mechanism, J. Nanobiotechnol., 2012, 10, 15 CrossRef CAS PubMed.
- D. Docter, C. Bantz, D. Westmeier, H. J. Galla, Q. Wang, J. C. Kirkpatrick, P. Nielsen, M. Maskos and R. H. Stauber, The protein corona protects against size-and dose-dependent toxicity of amorphous silica nanoparticles, Beilstein J. Nanotechnol., 2014, 5, 1380 CrossRef PubMed.
- M. Peracchia, E. Fattal, D. Desmaele, M. Besnard, J. Noel, J. Gomis, M. Appel, J. d'Angelo and P. Couvreur, Stealth® PEGylated polycyanoacrylate nanoparticles for intravenous administration and splenic targeting, J. Controlled Release, 1999, 60, 121–128 CrossRef CAS PubMed.
- N. A. Stasko, C. B. Johnson, M. H. Schoenfisch, T. A. Johnson and E. L. Holmuhamedov, Cytotoxicity of polypropylenimine dendrimer conjugates on cultured endothelial cells, Biomacromolecules, 2007, 8, 3853–3859 CrossRef CAS PubMed.
- M. Baek, M. Kim, H. Cho, J. Lee, J. Yu, H. Chung and S. Choi, Factors influencing the cytotoxicity of zinc oxide nanoparticles: particle size and surface charge, J. Phys.: Conf. Ser., 2011, 304, 012044 CrossRef.
- M. V. Park, A. M. Neigh, J. P. Vermeulen, L. J. de la Fonteyne, H. W. Verharen, J. J. Briedé, H. van Loveren and W. H. de Jong, The effect of particle size on the cytotoxicity, inflammation, developmental toxicity and genotoxicity of silver nanoparticles, Biomaterials, 2011, 32, 9810–9817 CrossRef CAS PubMed.
- M. Ekkapongpisit, A. Giovia, C. Follo, G. Caputo and C. Isidoro, Biocompatibility, endocytosis, and intracellular trafficking of mesoporous silica and polystyrene nanoparticles in ovarian cancer cells: effects of size and surface charge groups, Int. J. Nanomed., 2012, 7, 4147 CAS.
- R. Y. Prasad, J. K. McGee, M. G. Killius, D. A. Suarez, C. F. Blackman, D. M. DeMarini and S. O. Simmons, Investigating oxidative stress and inflammatory responses elicited by silver nanoparticles using high-throughput reporter genes in HepG2 cells: effect of size, surface coating, and intracellular uptake, Toxicol. In Vitro, 2013, 27, 2013–2021 CrossRef CAS PubMed.
- J. Broda, J. Setzler, A. Leifert, J. Steitz, R. Benz, U. Simon and W. Wenzel, Ligand-lipid and ligand-core affinity control the interaction of gold nanoparticles with artificial lipid bilayers and cell membranes, Nanomedicine: Nanotechnology, Biol. Med., 2016, 12, 1409–1419 CAS.
- S. Chono, T. Tanino, T. Seki and K. Morimoto, Efficient drug targeting to rat alveolar macrophages by pulmonary administration of ciprofloxacin incorporated into mannosylated liposomes for treatment of respiratory intracellular parasitic infections, J. Controlled Release, 2008, 127, 50–58 CrossRef CAS PubMed.
- K. Hirota, T. Hasegawa, H. Hinata, F. Ito, H. Inagawa, C. Kochi, G.-I. Soma, K. Makino and H. Terada, Optimum conditions for efficient phagocytosis of rifampicin-loaded PLGA microspheres by alveolar macrophages, J. Controlled Release, 2007, 119, 69–76 CrossRef CAS PubMed.
- C. M. Sayes, J. D. Fortner, W. Guo, D. Lyon, A. M. Boyd, K. D. Ausman, Y. J. Tao, B. Sitharaman, L. J. Wilson and J. B. Hughes, The differential cytotoxicity of water-soluble fullerenes, Nano Lett., 2004, 4, 1881–1887 CrossRef CAS.
- C. M. Goodman, C. D. McCusker, T. Yilmaz and V. M. Rotello, Toxicity of gold nanoparticles functionalized with cationic and anionic side chains, Bioconjugate Chem., 2004, 15, 897–900 CrossRef CAS PubMed.
- C. M. McIntosh, E. A. Esposito, A. K. Boal, J. M. Simard, C. T. Martin and V. M. Rotello, Inhibition of DNA transcription using cationic mixed monolayer protected gold clusters, J. Am. Chem. Soc., 2001, 123, 7626–7629 CrossRef CAS PubMed.
- T. Wang, J. Bai, X. Jiang and G. U. Nienhaus, Cellular uptake of nanoparticles by membrane penetration: a study combining confocal microscopy with FTIR spectroelectrochemistry, ACS Nano, 2012, 6, 1251–1259 CrossRef CAS PubMed.
- S. Prylutska, R. Bilyy, M. Overchuk, A. Bychko, K. Andreichenko, R. Stoika, V. Rybalchenko, Y. Prylutskyy, N. Tsierkezos and U. Ritter, Water-soluble pristine fullerenes C60 increase the specific conductivity and capacity of lipid model membrane and form the channels in cellular plasma membrane, J. Biomed. Nanotechnol., 2012, 8, 522–527 CrossRef CAS PubMed.
- K. H. Jacobson, I. L. Gunsolus, T. R. Kuech, J. M. Troiano, E. S. Melby, S. E. Lohse, D. Hu, W. B. Chrisler, C. J. Murphy and G. Orr, Lipopolysaccharide density and structure govern the extent and distance of nanoparticle interaction with actual and model bacterial outer membranes, Environ. Sci. Technol., 2015, 49, 10642–10650 CrossRef CAS PubMed.
- A. C. Mensch, R. T. Hernandez, J. E. Kuether, M. D. Torelli, Z. V. Feng, R. J. Hamers and J. A. Pedersen, Natural organic matter concentration impacts the interaction of functionalized diamond nanoparticles with model and actual bacterial membranes, Environ. Sci. Technol., 2017, 51, 11075–11084 CrossRef CAS PubMed.
- R. P. Carney, T. M. Carney, M. Mueller and F. Stellacci, Dynamic cellular uptake of mixed-monolayer protected nanoparticles, Biointerphases, 2012, 7, 1–9 CrossRef PubMed.
- R. C. Van Lehn, M. Ricci, P. H. Silva, P. Andreozzi, J. Reguera, K. Voïtchovsky, F. Stellacci and A. Alexander-Katz, Lipid tail protrusions mediate the insertion of nanoparticles into model cell membranes, Nat. Commun., 2014, 5, 4482 CrossRef CAS PubMed.
- R. M. Epand and R. F. Epand, Bacterial membrane lipids in the action of antimicrobial agents, J. Pept. Sci., 2011, 17, 298–305 CrossRef CAS PubMed.
- M. Haque, Y. Hirai, K. Yokota and K. Oguma, Steryl glycosides: a characteristic feature of the Helicobacter spp.?, J. Bacteriol., 1995, 177, 5334–5337 CrossRef CAS PubMed.
- Y. Hirai, M. Haque, T. Yoshida, K. Yokota, T. Yasuda and K. Oguma, Unique cholesteryl glucosides in Helicobacter pylori: composition and structural analysis, J. Bacteriol., 1995, 177, 5327–5333 CrossRef CAS PubMed.
- J. T. Crowley, A. M. Toledo, T. J. LaRocca, J. L. Coleman, E. London and J. L. Benach, Lipid exchange between Borrelia burgdorferi and host cells, PLoS Pathog., 2013, 9, e1003109 CrossRef CAS PubMed.
- J. P. Sáenz, E. Sezgin, P. Schwille and K. Simons, Functional convergence of hopanoids and sterols in membrane ordering, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 14236–14240 CrossRef PubMed.
- R. M. Epand and R. F. Epand, Lipid domains in bacterial membranes and the action of antimicrobial agents, Biochim. Biophys. Acta, Biomembr., 2009, 1788, 289–294 CrossRef CAS PubMed.
- A. Ivask, E. Suarez, T. Patel, D. Boren, Z. Ji, P. Holden, D. Telesca, R. Damoiseaux, K. A. Bradley and H. Godwin, Genome-wide bacterial toxicity screening uncovers the mechanisms of toxicity of a cationic polystyrene nanomaterial, Environ. Sci. Technol., 2012, 46, 2398–2405 CrossRef CAS PubMed.
- M. Triantafilou, K. Triantafilou and N. Fernandez, Rough and smooth forms of fluorescein-labelled bacterial endotoxin exhibit CD14/LBP dependent and independent binding that is influencedby endotoxin concentration, FEBS J., 2000, 267, 2218–2226 CAS.
- H. Nikaido and T. Nakae, Permeability of model membranes containing phospholipids and lipopolysaccharides: some preliminary results, J. Infect. Dis., 1973, 128, S30–S34 CrossRef.
- H. Ma, F. J. Irudayanathan, W. Jiang and S. Nangia, Simulating Gram-negative bacterial outer membrane: a coarse grain model, J. Phys. Chem. B, 2015, 119, 14668–14682 CrossRef CAS PubMed.
- H. Ma, D. D. Cummins, N. B. Edelstein, J. Gomez, A. Khan, M. D. Llewellyn, T. Picudella, S. R. Willsey and S. Nangia, Modeling diversity in structures of bacterial outer membrane lipids, J. Chem. Theory Comput., 2017, 13, 811–824 CrossRef CAS PubMed.
- J. T. Buchman, A. Rahnamoun, K. M. Landy, X. Zhang, A. M. Vartanian, L. M. Jacob, C. J. Murphy, R. Hernandez and C. L. Haynes, Using an environmentally-relevant panel of Gram-negative bacteria to assess the toxicity of polyallylamine hydrochloride-wrapped gold nanoparticles, Environ. Sci.: Nano, 2018, 5, 279–288 RSC.
- H. Ma, A. Khan and S. Nangia, Dynamics of OmpF trimer formation in the bacterial outer membrane of Escherichia coli, Langmuir, 2017, 19, 5623–5634 Search PubMed.
- M. De Nicola, S. Bellucci, E. Traversa, G. De Bellis, F. Micciulla and L. Ghibelli, Carbon nanotubes on Jurkat cells: effects on cell viability and plasma membrane potential, J. Phys.: Condens. Matter, 2008, 20, 474204 CrossRef.
- W. Lin, I. Stayton, Y.-w. Huang, X.-D. Zhou and Y. Ma, Cytotoxicity and cell membrane depolarization induced by aluminum oxide nanoparticles in human lung epithelial cells A549, Toxicol. Environ. Chem., 2008, 90, 983–996 CrossRef CAS.
- N. Kahya, D. A. Brown and P. Schwille, Raft partitioning and dynamic behavior of human placental alkaline phosphatase in giant unilamellar vesicles, Biochemistry, 2005, 44, 7479–7489 CrossRef CAS PubMed.
- L. Kalvodova, N. Kahya, P. Schwille, R. Ehehalt, P. Verkade, D. Drechsel and K. Simons, Lipids as modulators of proteolytic activity of BACE involvement of cholesterol, glycosphingolipids, and anionic phospholipids in vitro, J. Biol. Chem., 2005, 280, 36815–36823 CrossRef CAS PubMed.
- H. Heise, T. Bayerl, G. Isenberg and E. Sackmann, Human platelet P-235, a talin-like actin binding protein, binds selectively to mixed lipid bilayers, Biochim. Biophys. Acta, Biomembr., 1991, 1061, 121–131 CrossRef CAS.
- N. Kahya, D. A. Wiersma, B. Poolman and D. Hoekstra, Spatial organization of bacteriorhodopsin in model membranes light-induced mobility changes, J. Biol. Chem., 2002, 277, 39304–39311 CrossRef CAS PubMed.
- M. S. Bretscher, Asymmetrical lipid bilayer structure for biological membranes, Nature, 1972, 236, 11–12 CrossRef CAS.
- P. F. Devaux, Static and dynamic lipid asymmetry in cell membranes, Biochemistry, 1991, 30, 1163–1173 CrossRef CAS PubMed.
- B. Fadeel and D. Xue, The ins and outs of phospholipid asymmetry in the plasma membrane: roles in health and disease, Crit. Rev. Biochem. Mol. Biol., 2009, 44, 264–277 CrossRef CAS PubMed.
- B. R. Lentz, Exposure of platelet membrane phosphatidylserine regulates blood coagulation, Prog. Lipid Res., 2003, 42, 423–438 CrossRef CAS PubMed.
- S. Nazemidashtarjandi, A. Kelly, A. David and A. Farnoud, The role of membrane asymmetry in nanoparticle-induced plasma membrane damage, Biophys. J., 2018, 114, 176a–177a CrossRef.
- Q. Lin and E. London, The influence of natural lipid asymmetry upon the conformation of a membrane-inserted protein (Perfringolysin O), J. Biol. Chem., 2014, 289, 5467–5478 CrossRef CAS PubMed.
- F. A. Heberle, D. Marquardt, M. Doktorova, B. Geier, R. F. Standaert, P. Heftberger, B. Kollmitzer, J. D. Nickels, R. A. Dick and G. W. Feigenson, Subnanometer structure of an asymmetric model membrane: interleaflet coupling influences domain properties, Langmuir, 2016, 32, 5195–5200 CrossRef CAS PubMed.
- Q. Lin and E. London, Ordered raft domains induced by outer leaflet sphingomyelin in cholesterol-rich asymmetric vesicles, Biophys. J., 2015, 108, 2212–2222 CrossRef CAS PubMed.
- J. Saarikangas, H. Zhao and P. Lappalainen, Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides, Physiol. Rev., 2010, 90, 259–289 CrossRef CAS PubMed.
- A. Aalipour, A. M. Xu, S. Leal-Ortiz, C. C. Garner and N. A. Melosh, Plasma membrane and actin cytoskeleton as synergistic barriers to nanowire cell penetration, Langmuir, 2014, 30, 12362–12367 CrossRef CAS PubMed.
- W. Wu, L. Yan, Q. Wu, Y. Li, Q. Li, S. Chen, Y. Yang, Z. Gu, H. Xu and Z. Q. Yin, Evaluation of the toxicity of graphene oxide exposure to the eye, Nanotoxicology, 2016, 10, 1329–1340 CrossRef CAS PubMed.
- W. An, Y. Zhang, X. Zhang, K. Li, Y. Kang, S. Akhtar, X. Sha and L. Gao, Ocular toxicity of reduced graphene oxide or graphene oxide exposure in mouse eyes, Exp. Eye Res., 2018, 174, 59–69 CrossRef CAS PubMed.
- T. M. Scown, E. M. Santos, B. D. Johnston, B. Gaiser, M. Baalousha, S. Mitov, J. R. Lead, V. Stone, T. F. Fernandes and M. Jepson, Effects of aqueous exposure to silver nanoparticles of different sizes in rainbow trout, Toxicol. Sci., 2010, 115, 521–534 CrossRef CAS PubMed.
- A. Ale, C. Bacchetta, A. S. Rossi, J. Galdopórpora, M. F. Desimone, R. Fernando, S. Gervasio and J. Cazenave, Nanosilver toxicity in gills of a neotropical fish: Metal accumulation, oxidative stress, histopathology and other physiological effects, Ecotoxicol. Environ. Saf., 2018, 148, 976–984 CrossRef CAS.
- C. S. Campbell, L. R. Contreras-Rojas, M. B. Delgado-Charro and R. H. Guy, Objective assessment of nanoparticle disposition in mammalian skin after topical exposure, J. Controlled Release, 2012, 162, 201–207 CrossRef CAS PubMed.
- L. Böhmert, M. Girod, U. Hansen, R. Maul, P. Knappe, B. Niemann, S. M. Weidner, A. F. Thünemann and A. Lampen, Analytically monitored digestion of silver nanoparticles and their toxicity on human intestinal cells, Nanotoxicology, 2014, 8, 631–642 CrossRef PubMed.
- M. Vertzoni, J. Dressman, J. Butler, J. Hempenstall and C. Reppas, Simulation of fasting gastric conditions and its importance for the in vivo dissolution of lipophilic compounds, Eur. J. Pharm. Biopharm., 2005, 60, 413–417 CrossRef CAS PubMed.
- M. Vertzoni, A. Diakidou, M. Chatzilias, E. Söderlind, B. Abrahamsson, J. B. Dressman and C. Reppas, Biorelevant media to simulate fluids in the ascending colon of humans and their usefulness in predicting intracolonic drug solubility, Pharm. Res., 2010, 27, 2187–2196 CrossRef CAS PubMed.
- E. Roblegg, E. Froehlich, C. Meindl, B. Teubl, M. Zaversky and A. Zimmer, Evaluation of a physiological in vitro system to study the transport of nanoparticles through the buccal mucosa, Nanotoxicology, 2012, 6, 399–413 CrossRef CAS PubMed.
- B. J. Teubl, C. Schimpel, G. Leitinger, B. Bauer, E. Fröhlich, A. Zimmer and E. Roblegg, Interactions between nano-TiO2 and the oral cavity: impact of nanomaterial surface hydrophilicity/hydrophobicity, J. Hazard. Mater., 2015, 286, 298–305 CrossRef CAS PubMed.
- L. Pinďáková, V. Kašpárková, K. Kejlová, M. Dvořáková, D. Krsek, D. Jírová and L. Kašparová, Behaviour of silver nanoparticles in simulated saliva and gastrointestinal fluids, Int. J. Pharm., 2017, 527, 12–20 CrossRef PubMed.
- B. J. Teubl, M. Absenger, E. Fröhlich, G. Leitinger, A. Zimmer and E. Roblegg, The oral cavity as a biological barrier system: design of an advanced buccal in vitro permeability model, Eur. J. Pharm. Biopharm., 2013, 84, 386–393 CrossRef CAS PubMed.
- S. K. Mwilu, A. M. El Badawy, K. Bradham, C. Nelson, D. Thomas, K. G. Scheckel, T. Tolaymat, L. Ma and K. R. Rogers, Changes in silver nanoparticles exposed to human synthetic stomach fluid: effects of particle size and surface chemistry, Sci. Total Environ., 2013, 447, 90–98 CrossRef CAS PubMed.
- A. P. Walczak, R. Fokkink, R. Peters, P. Tromp, Z. E. Herrera Rivera, I. M. Rietjens, P. J. Hendriksen and H. Bouwmeester, Behaviour of silver nanoparticles and silver ions in an in vitro human gastrointestinal digestion model, Nanotoxicology, 2012, 7, 1198–1210 CrossRef PubMed.
- S.-W. Bian, I. A. Mudunkotuwa, T. Rupasinghe and V. H. Grassian, Aggregation and dissolution of 4 nm ZnO nanoparticles in aqueous environments: influence of pH, ionic strength, size, and adsorption of humic acid, Langmuir, 2011, 27, 6059–6068 CrossRef CAS PubMed.
- M. Gasser, B. Rothen-Rutishauser, H. F. Krug, P. Gehr, M. Nelle, B. Yan and P. Wick, The adsorption of biomolecules to multi-walled carbon nanotubes is influenced by both pulmonary surfactant lipids and surface chemistry, J. Nanobiotechnol., 2010, 8, 31 CrossRef CAS PubMed.
- Q. Hu, X. Bai, G. Hu and Y. Y. Zuo, Unveiling the molecular structure of pulmonary surfactant corona on nanoparticles, ACS Nano, 2017, 11, 6832–6842 CrossRef CAS PubMed.
- S. S. Raesch, S. Tenzer, W. Storck, A. Rurainski, D. Selzer, C. A. Ruge, J. Perez-Gil, U. F. Schaefer and C.-M. Lehr, Proteomic and lipidomic analysis of nanoparticle corona upon contact with lung surfactant reveals differences in protein, but not lipid composition, ACS Nano, 2015, 9, 11872–11885 CrossRef CAS PubMed.
- C. A. Shaw, G. M. Mortimer, Z. J. Deng, E. S. Carter, S. P. Connell, M. R. Miller, R. Duffin, D. E. Newby, P. W. Hadoke and R. F. Minchin, Protein corona formation in bronchoalveolar fluid enhances diesel exhaust nanoparticle uptake and pro-inflammatory responses in macrophages, Nanotoxicology, 2016, 10, 981–991 CrossRef CAS PubMed.
- N. V. Konduru, R. M. Molina, A. Swami, F. Damiani, G. Pyrgiotakis, P. Lin, P. Andreozzi, T. C. Donaghey, P. Demokritou and S. Krol, Protein corona: implications for nanoparticle interactions with pulmonary cells, Part. Fibre Toxicol., 2017, 14, 42 CrossRef PubMed.
- A. Zane, C. McCracken, D. A. Knight, T. Young, A. D. Lutton, J. W. Olesik, W. J. Waldman and P. K. Dutta, Uptake of bright fluorophore core-silica shell nanoparticles by biological systems, Int. J. Nanomed., 2015, 10, 1547 CrossRef CAS PubMed.
- A. Nemmar, P. M. Hoet, B. Vanquickenborne, D. Dinsdale, M. Thomeer, M. Hoylaerts, H. Vanbilloen, L. Mortelmans and B. Nemery, Passage of inhaled particles into the blood circulation in humans, Circulation, 2002, 105, 411–414 CrossRef CAS PubMed.
- Q. Wang, M. Lim, X. Liu, Z. Wang and K. L. Chen, Influence of solution chemistry and soft protein coronas on the interactions of silver nanoparticles with model biological membranes, Environ. Sci. Technol., 2016, 50, 2301–2309 CrossRef CAS PubMed.
- D. Di Silvio, M. Maccarini, R. Parker, A. Mackie, G. Fragneto and F. B. Bombelli, The effect of the protein corona on the interaction between nanoparticles and lipid bilayers, J. Colloid Interface Sci., 2017, 504, 741–750 CrossRef CAS PubMed.
- E. S. Melby, S. E. Lohse, J. E. Park, A. M. Vartanian, R. A. Putans, H. B. Abbott, R. J. Hamers, C. J. Murphy and J. A. Pedersen, Cascading effects of nanoparticle coatings: Surface functionalization dictates the assemblage of complexed proteins and subsequent interaction with model cell membranes, ACS Nano, 2017, 11, 5489–5499 CrossRef CAS PubMed.
- K. C. Nguyen, P. Rippstein, A. F. Tayabali and W. G. Willmore, Mitochondrial toxicity of cadmium telluride quantum dot nanoparticles in mammalian hepatocytes, Toxicol. Sci., 2015, 146, 31–42 CrossRef CAS PubMed.
- L. Sun, Y. Li, X. Liu, M. Jin, L. Zhang, Z. Du, C. Guo, P. Huang and Z. Sun, Cytotoxicity and mitochondrial damage caused by silica nanoparticles, Toxicol. In Vitro, 2011, 25, 1619–1629 CrossRef CAS PubMed.
- J. S. Teodoro, A. M. Simões, F. V. Duarte, A. P. Rolo, R. C. Murdoch, S. M. Hussain and C. M. Palmeira, Assessment of the toxicity of silver nanoparticles in vitro: a mitochondrial perspective, Toxicol. In Vitro, 2011, 25, 664–670 CrossRef CAS PubMed.
- S. M. Santos, A. M. Dinis, F. Peixoto, L. Ferreira, A. S. Jurado and R. A. Videira, Interaction of fullerene nanoparticles with biomembranes: from the partition in lipid membranes to effects on mitochondrial bioenergetics, Toxicol. Sci., 2013, 138, 117–129 CrossRef PubMed.
- Y. Ha, X. Wang, H. M. Liljestrand, J. A. Maynard and L. E. Katz, Bioavailability of fullerene under environmentally relevant conditions: Effects of humic acid and fetal bovine serum on accumulation in lipid bilayers and cellular uptake, Environ. Sci. Technol., 2016, 50, 6717–6727 CrossRef CAS PubMed.
- J. V. M. Girón, R. V. Vico, B. Maggio, E. Zelaya, A. Rubert, G. Benítez, P. Carro, R. C. Salvarezza and M. E. Vela, Role of the capping agent in the interaction of hydrophilic Ag nanoparticles with DMPC as a model biomembrane, Environ. Sci.: Nano, 2016, 3, 462–472 RSC.
- R. L. C. G. da Silva, H. F. O. da Silva, L. H. da Silva Gasparotto and L. Caseli, How the interaction of PVP-stabilized Ag nanoparticles with models of cellular membranes at the air-water interface is modulated by the monolayer composition, J. Colloid Interface Sci., 2018, 512, 792–800 CrossRef PubMed.
- M. R. de Planque, S. Aghdaei, T. Roose and H. Morgan, Electrophysiological characterization of membrane disruption by nanoparticles, ACS Nano, 2011, 5, 3599–3606 CrossRef CAS PubMed.
- R. Frost, G. E. Jönsson, D. Chakarov, S. Svedhem and B. Kasemo, Graphene oxide and lipid membranes: interactions and nanocomposite structures, Nano Lett., 2012, 12, 3356–3362 CrossRef CAS PubMed.
- C. Corredor, W.-C. Hou, S. A. Klein, B. Y. Moghadam, M. Goryll, K. Doudrick, P. Westerhoff and J. D. Posner, Disruption of model cell membranes by carbon nanotubes, Carbon, 2013, 60, 67–75 CrossRef CAS.
- A. Negoda, K.-J. Kim, E. D. Crandall and R. M. Worden, Polystyrene nanoparticle exposure induces ion-selective pores in lipid bilayers, Biochim. Biophys. Acta, Biomembr., 2013, 1828, 2215–2222 CrossRef CAS PubMed.
- L. Shi, D. Shi, M. U. Nollert, D. E. Resasco and A. Striolo, Single-walled carbon nanotubes do not pierce aqueous phospholipid bilayers at low salt concentration, J. Phys. Chem. B, 2013, 117, 6749–6758 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |