Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A naphthoimidazolium-cholesterol derivative as a ratiometric fluorescence based chemosensor for the chiral recognition of carboxylates†
Xin
Zhang‡
 a,
Gyeongju
Ko‡
b,
Joonyoung F.
Joung‡
c,
Meng
Li
de,
Yerin
Jeong
b,
K. M. K.
Swamy
b,
Dayoung
Lee
b,
Yifan
Liu
b,
Songyi
Lee
a,
Gyeongju
Ko‡
b,
Joonyoung F.
Joung‡
c,
Meng
Li
de,
Yerin
Jeong
b,
K. M. K.
Swamy
b,
Dayoung
Lee
b,
Yifan
Liu
b,
Songyi
Lee
 *f,
Sungnam
Park
*f,
Sungnam
Park
 *c,
Tony D.
James
*c,
Tony D.
James
 *d and
Juyoung
Yoon
*d and
Juyoung
Yoon
 *b
*b
aNational Demonstration Center for Experimental Chemistry Education, College of Chemistry and Materials Science, Hebei Normal University, Shijiazhuang, 050024, China
bDepartment of Chemistry and Nano Science, Ewha Womans University, Seoul, 03760, Korea. E-mail: jyoon@ewha.ac.kr
cDepartment of Chemistry, Korea University, Seoul, 02841, Korea. E-mail: spark8@korea.ac.kr
dDepartment of Chemistry, University of Bath, Bath, BA2 7AY, UK. E-mail: t.d.james@bath.ac.uk
eDepartment of Environmental Science and Technology, North China Electric Power University, 689 Huadian Road, 071003, Baoding, China
fDepartment of Chemistry, Pukyong National University, Busan, 48513, Korea. E-mail: slee@pknu.ac.kr
First published on 2nd November 2018
Abstract
Fluorescent chemosensors for sensing chiral molecules have been actively studied in recent years. In the current study, we report a naphthoimidazolium-cholesterol derivative (NI-chol 1) as a fluorescence based chemosensor for chiral recognition, in which the naphthoimidazolium serves not only as a fluorophore but also as a recognition moiety for anions via imidazolium (C–H)+–anion binding and the cholesterol unit acts as a chiral barrier. In particular, NI-chol 1 displayed unique and distinct ratiometric changes with Boc-D-Phe, on the other hand, Boc-L-Phe induced a negligible change. Furthermore, a distinct downfield shift (from 9.64 ppm to 9.96 ppm) of the imidazolium C–H peak was observed for Boc-D-Phe (5 eq.) with severe broadening, which indicates strong ionic hydrogen bonding between the C–H proton and the carboxylate.
Since amino acids, the basic fundamental unit for peptides and proteins, are chiral, important processes in biology involve chiral interactions. Chiral fluorescence based chemosensors can convert enantioselective recognition into fluorescence changes,1,2 which results in simple and easy detection methods for chiral recognition compared to conventional methods, such as high-performance liquid chromatography (HPLC), capillary electrophoresis (CE), gas chromatography (GC) or NMR. Accordingly, fluorescence-based recognition for chiral anions has been extensively studied. Host–guest chemistry and molecular recognition have greatly affected the development of chiral fluorescence based chemosensors.3 The main requirement for the fluorescence based approaches to evaluate enantiomeric compositions is chiral sensors with the ability to differentially interact with opposite enantiomers of a chiral target in a manner that gives rise to different optical signal outputs.
On the other hand, imidazolium based fluorescent chemosensors have been actively studied since they can show strong ionic hydrogen bonding interactions such as imidazolium (C–H)+–anions.4 Over the last decade, fluorescence based imidazolium derivatives have found application in the area of sensing simple anions,5 nucleotides,6 IP3 and IP6,7 RNA or DNA,8 anionic surfactants,9 bacteria10 and CO2 gas,11etc.12 Our group has actively investigated imidazolium based fluorescent chemosensors for various targets.8 However, there are a paucity of papers for chiral recognition using fluorescence changes using imidazolium groups.13,14 Yu and coworkers introduced an imidazolium moiety into BINOL (1,1′-bi-2-naphthol), which displayed red shifts with fluoride and acetate in acetonitrile and large fluorescence quenching effects with t-Boc Ala anions producing a KL/KD value of 4.5 in acetonitrile.13a
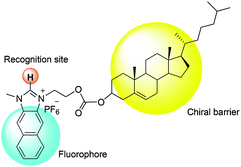
Compared to binaphthyl based chemosensors, there are relatively few examples in which the cholesterol moiety has been used as a chiral barrier.14 In the current study, we report the example of a naphthoimidazolium-cholesterol derivative as a chiral fluorescence based chemosensor. Naphthoimidazoliums are inherently fluorescent, so an extra fluorophore is not required as in the case with imidazolium based fluorescence based chemosensors. NI-chol 1 displayed a unique ratiometric change with Boc-D-Phe, while Boc-L-Phe induced very little change compared to that of the D-isomer. Similar but less distinct changes were also observed for Boc-D-Val over Boc-L-Val. In addition, a distinct downfield shift (from 9.64 ppm to 9.96 ppm) of the imidazolium C–H peak of NI-chol 1 was observed for Boc-D-Phe (5 eq.) with severe broadening, which can be attributed to the strong ionic hydrogen bonding interaction between the C–H proton and the carboxylate.
Compound 2 was synthesized according to literature procedures.5c Compound 3 was prepared from cholesteryl chloroformate and 2-iodoethanol in the presence of pyridine in a yield of 80.8% after column chromatography using hexane/dichloromethane as eluents (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). Then, 2 and 3 were refluxed in acetonitrile to afford 4 as shown in Scheme 1, which was then treated with aqueous saturated KFP6 to afford 1 (NI-chol 1) as a white solid. Detailed synthetic procedures are given in the Experimental section. All new compounds were fully characterized by 1H and 13C NMR as well as high-resolution mass spectroscopy (Fig. S1–S9, ESI†).
1, v/v). Then, 2 and 3 were refluxed in acetonitrile to afford 4 as shown in Scheme 1, which was then treated with aqueous saturated KFP6 to afford 1 (NI-chol 1) as a white solid. Detailed synthetic procedures are given in the Experimental section. All new compounds were fully characterized by 1H and 13C NMR as well as high-resolution mass spectroscopy (Fig. S1–S9, ESI†).
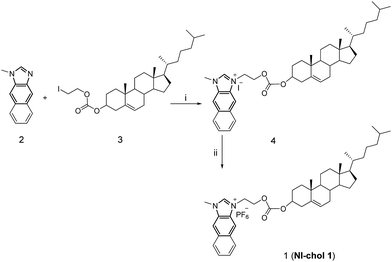 | ||
| Scheme 1 The synthetic procedure of NI-chol 1. (i) Acetonitrile, N2, reflux, 24 h, 81.1%; (ii) DMF, KPF6, 98.6%. | ||
Fig. 1 illustrates the design strategy towards chiral probe 1, in which naphthoimidazolium serves not only as a fluorophore but also as a recognition moiety for anions via imidazolium (C–H)+–anion binding and the cholesterol unit acts as a chiral barrier.
The enantioselectivity of NI-chol 1 for Boc-D-Phe detection was investigated through fluorescence emission spectroscopy by adding various amino acids to a solution of compound NI-chol 1 in CH3CN–DMSO (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, v/v). The chiral recognition was examined with tetrabutylammonium salts of Boc-D- and Boc-L-amino acid anions. In the absence of amino acids, NI-chol 1 displayed an emission at 452 nm and the fluorescence quantum yield (Φf) of NI-chol 1 was 0.27, using 9,10-diphenylanthracene standard with a Φf of 0.97 in cyclohexane (Fig. S10, ESI†). Among phenylalanine (Phe), valine (Val), serine (Ser) glutamine (Glu), leucine (Leu) and alanine (Ala), as shown in Fig. 2, NI-chol 1 displayed unique ratiometric changes with Boc-D-Phe with a Φf of 0.30. The blue shifted emission peak at 370 nm increases while the original emission at 452 nm decreases. A clear iso-emissive point appears at 401 nm. Even though a relatively smaller effect was observed for D-Val, a ratiometric change was clear enough to be observed. The blue shift to 370 nm from 452 nm can be attributed to the imidazolium C–H interaction with the carboxylate group of the amino acid. Electron withdrawing effects of the imidazolium moiety decrease when strong ionic hydrogen bonding occurs. Naphthoimidazolium has a donor–acceptor system and can undergo internal charge transfer (ICT) from naphthalene to the imidazolium upon excitation by light, with the imidazolium acting as an acceptor. The electron withdrawing effect of the imidazolium moiety decreases when strong ionic hydrogen bonding occurs.5a Hence, NI-chol 1 behaves as a ratiometric fluorescence based probe for Boc-D-Phe against Boc-L-Phe.
5, v/v). The chiral recognition was examined with tetrabutylammonium salts of Boc-D- and Boc-L-amino acid anions. In the absence of amino acids, NI-chol 1 displayed an emission at 452 nm and the fluorescence quantum yield (Φf) of NI-chol 1 was 0.27, using 9,10-diphenylanthracene standard with a Φf of 0.97 in cyclohexane (Fig. S10, ESI†). Among phenylalanine (Phe), valine (Val), serine (Ser) glutamine (Glu), leucine (Leu) and alanine (Ala), as shown in Fig. 2, NI-chol 1 displayed unique ratiometric changes with Boc-D-Phe with a Φf of 0.30. The blue shifted emission peak at 370 nm increases while the original emission at 452 nm decreases. A clear iso-emissive point appears at 401 nm. Even though a relatively smaller effect was observed for D-Val, a ratiometric change was clear enough to be observed. The blue shift to 370 nm from 452 nm can be attributed to the imidazolium C–H interaction with the carboxylate group of the amino acid. Electron withdrawing effects of the imidazolium moiety decrease when strong ionic hydrogen bonding occurs. Naphthoimidazolium has a donor–acceptor system and can undergo internal charge transfer (ICT) from naphthalene to the imidazolium upon excitation by light, with the imidazolium acting as an acceptor. The electron withdrawing effect of the imidazolium moiety decreases when strong ionic hydrogen bonding occurs.5a Hence, NI-chol 1 behaves as a ratiometric fluorescence based probe for Boc-D-Phe against Boc-L-Phe.
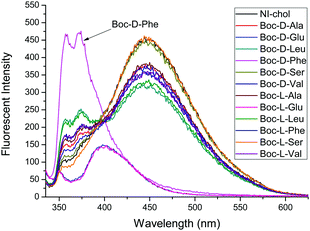 | ||
Fig. 2 Fluorescence changes of NI-chol 1 (10 μM) in CH3CN–DMSO (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, v/v) with D and L isomers of Ala, Glu, Leu, Phe, Ser and Val (500 μM). (λex = 325 nm, slit 3 × 5). 5, v/v) with D and L isomers of Ala, Glu, Leu, Phe, Ser and Val (500 μM). (λex = 325 nm, slit 3 × 5). | ||
Fig. 3 and 4 illustrate the fluorescence titrations of Boc-D-Phe and Boc-L-Phe. The blue shift to 370 nm from 452 nm can be attributed to the imidazolium C–H interaction with the carboxylate group of the amino acid. The association constant (KD) was calculated using the software (SigmaPlot 2000) to be 2.13 × 103 M−1 based on the ratio of I370nm/I452nm for Boc-D-Phe.
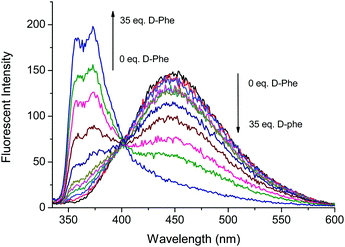 | ||
Fig. 3 Fluorescence based titrations of NI-chol 1 (10 μM) in CH3CN–DMSO (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, v/v) with various equivalents of Boc-D-Phe. 5, v/v) with various equivalents of Boc-D-Phe. | ||
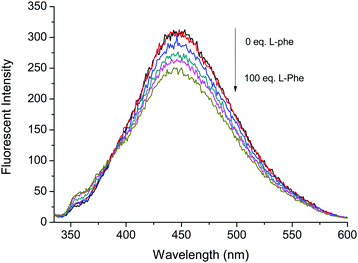 | ||
Fig. 4 Fluorescence based titrations of NI-chol 1 (10 μM) in CH3CN–DMSO (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, v/v) with various equivalents of Boc-L-Phe. 5, v/v) with various equivalents of Boc-L-Phe. | ||
However, the addition of Boc-L-Phe induced only a small change as illustrated in Fig. 4. The association constant (KL) for Boc-L-Phe was 1.5 × 102 M−1. Thus, the enantioselectivity could be obtained via KD/KL as ca. 14, which is higher than those for most of the reported imidazolium-based chiral sensors. In addition, the fluorescence titrations with D-Val are illustrated in the ESI† (Fig. S11 and S12). Even though the changes were smaller than those with Boc-D-Phe, similar ratiometric changes were observed at 370 nm and 452 nm.
To understand the interaction of NI-chol 1 with Boc-D-Phe, 1H NMR titrations were performed in CD3CN-DMSO-d6 (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, v/v) (Fig. 5 and Fig. S13, ESI†). When 1.0 eq. of Boc-D-Phe was added to the solution of NI-chol 1, the imidazolium C–H peak at 9.64 ppm downfield shifted to 9.96 ppm, which indicates a hydrogen bonding interaction between the C–H proton and the carboxylate. Furthermore, severe broadening of this peak was observed, which also supports the presence of strong ionic hydrogen bonding. With >5.0 eq. of Boc-D-Phe, the peak of imidazole C–H of NI-chol 1 disappeared due to partial deprotonation. However, a smaller shift to 9.87 ppm was observed upon the addition of Boc-L-Phe (5 eq.) and the sharp singlet of Ha was preserved (Fig. S13, ESI†). In addition, there were slight downfield shifts for ethylene linkers between the naphthoimidazolium and the cholesteryl moiety. Since there are no significant changes in the 1H NMR spectra for other peaks, it is not easy to predict the exact binding mode. Hydrogen bonding interactions between carboxylates and imidazolium C–H protons could be clearly deduced based on NMR data and the cholesteryl unit successfully served as a chiral barrier based on the selectivity for D-isomers of Phe and Val.
5, v/v) (Fig. 5 and Fig. S13, ESI†). When 1.0 eq. of Boc-D-Phe was added to the solution of NI-chol 1, the imidazolium C–H peak at 9.64 ppm downfield shifted to 9.96 ppm, which indicates a hydrogen bonding interaction between the C–H proton and the carboxylate. Furthermore, severe broadening of this peak was observed, which also supports the presence of strong ionic hydrogen bonding. With >5.0 eq. of Boc-D-Phe, the peak of imidazole C–H of NI-chol 1 disappeared due to partial deprotonation. However, a smaller shift to 9.87 ppm was observed upon the addition of Boc-L-Phe (5 eq.) and the sharp singlet of Ha was preserved (Fig. S13, ESI†). In addition, there were slight downfield shifts for ethylene linkers between the naphthoimidazolium and the cholesteryl moiety. Since there are no significant changes in the 1H NMR spectra for other peaks, it is not easy to predict the exact binding mode. Hydrogen bonding interactions between carboxylates and imidazolium C–H protons could be clearly deduced based on NMR data and the cholesteryl unit successfully served as a chiral barrier based on the selectivity for D-isomers of Phe and Val.
 | ||
Fig. 5 Partial 1H NMR spectra (300 MHz) of NI-chol 1 (5 mM) upon the addition of Boc-D-Phe (tetrabutylammonium salt) in CD3CN-DMSO-d6 (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, v/v). 5, v/v). | ||
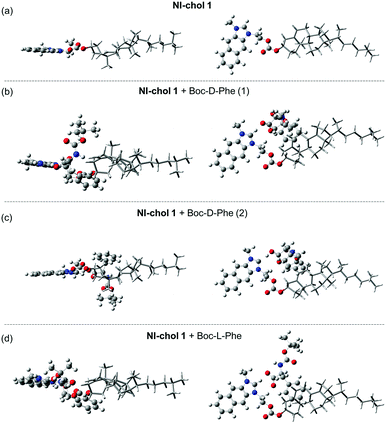
Furthermore, DFT calculations were used to understand the enantioselectivity of NI-chol 1 for Boc-D-Phe and the ratiometric fluorescence change. First, the optimized structures of NI-chol 1 with Boc-D-Phe or Boc-L-Phe were obtained as shown in Fig. 6. In the optimized structures, the oxygen atom in the carboxyl group of Phe is found to form a hydrogen-bond to Ha. This result agrees well with the NMR spectra in which the Ha proton is significantly influenced when NI-chol 1 binds to Boc-D-Phe in Fig. 5. As shown in Fig. 6 and movie clips (ESI†), both phenyl and Boc groups of Boc-D-Phe interact with the cholesterol moiety of NI-chol 1. However, only the phenyl group of Boc-L-Phe interacts with the cholesterol moiety of NI-chol 1. The energy of NI-chol 1 with Boc-D-Phe (Fig. 6(b)) is found to be 98.2 kcal mol−1 lower than that of NI-chol 1 with Boc-L-Phe (Fig. 6(d)). This DFT calculation result indicates that the interaction between Boc-D-Phe and NI-chol 1 is much stronger than the interaction between Boc-L-Phe and NI-chol 1. Such a significant difference in the binding energy between NI-chol 1 and Boc-D-Phe or Boc-L-Phe is responsible for the enantioselectivity of NI-chol 1 for Boc-D-Phe.
Fig. 6(b and c) display the two optimized structures of NI-chol 1 with Boc-D-Phe and their calculated absorption spectra are shown in Fig. S14 (ESI†). Note that the Ha proton of the NI unit is transferred to the carboxyl group of Boc-D-Phe, shown in Fig. 6(c). This result indicates that the absorption spectrum of NI is blue-shifted when the Ha proton moves significantly away from the NI unit. Similar results are expected from the emission spectra of NI units. To confirm this, the emission spectra of protonated and deprotonated NI units were calculated in Fig. S15(a) (ESI†). The calculated emission spectra of NI units agree well with the fluorescence spectra of NI-chol 1 with and without Boc-D-Phe as shown in Fig. S15 (ESI†). This result indicates that after Boc-D-Phe binds to NI-chol 1, the NI unit is deprotonated by the carbonyl group of Boc-D-Phe and as a result, its fluorescence spectrum is blue-shifted.
In summary, we have developed a new naphthoimidazolium-cholesteryl derivative NI-chol 1 for chiral anion recognition. The naphthoimidazolium moiety serves not only as a hydrogen bonding donor but also as a fluorescence unit. On the other hand, the cholesteryl moiety acts as a chiral barrier to induce enantiomer selectivity. Chiral fluorescence based receptor 1 displayed a unique ratiometric change in its emission with Boc-D-Phe. When Boc-D-Phe is added, the blue shifted emission at 370 nm increases with a concurrent decrease of the original emission at 452 nm, which results in a clear ratiometric fluorescence change. On the other hand, Boc-L-Phe did not induce any significant change. D-Val also induces similar but smaller ratiometric changes. The ionic hydrogen bonding interactions between the imidazolium C–H proton and carboxylate anions were confirmed by NMR analysis. Finally, the DFT calculations explain (1) the enantioselectivity of NI-chol 1 for Boc-D-Phe, (2) emission spectra of NI-chol 1 with and without Boc-D-Phe, and (3) the mechanism for ratiometric fluorescence detection of Boc-D-Phe by NI-chol 1.
This work was supported by the National Research Foundation of Korea (NRF), which is funded by the Korea government (MSIP) (No. 2012R1A3A2048814 for J. Y.; No. 2017R1A6A3A04004954 for S. L.; No. 2016R1A2B4011522 for S. P.). X. Z. thanks the National Natural Science Foundation of China (21502040) and the Youth Top-notch Talent Foundation of the Education Department of Hebei Province (No. BJ2014039) for support. TDJ thanks the Royal Society for a Wolfson Research Merit Award. The mass spectral data were obtained at Korea Basic Science Institute (Seoul) using MS Q-TOF/G6550A (Agilent).
Conflicts of interest
There are no conflicts to declare.Notes and references
- (a) X. Zhang, J. Yin and J. Yoon, Chem. Rev., 2014, 114, 4918 CrossRef CAS PubMed; (b) L. Pu, Acc. Chem. Res., 2012, 45, 150 CrossRef CAS PubMed; (c) A. Accetta, R. Corradini and R. Marchelli, Top. Curr. Chem., 2011, 300, 175 CrossRef CAS PubMed.
- (a) X. Chen, Z. Huang, S.-Y. Chen, K. Li, X. Q. Yu and L. Pu, J. Am. Chem. Soc., 2010, 132, 7297 CrossRef CAS PubMed; (b) H. Liu, Q. Peng, Y. Wu, D. Chen, X. Hou, M. Sabat and L. Pu, Angew. Chem., Int. Ed., 2010, 49, 602 CrossRef CAS PubMed; (c) X. Zhang, L. Chi, S. Ji, Y. Wu, P. Song, K. Han, H. Guo, T. D. James and J. Zhao, J. Am. Chem. Soc., 2009, 131, 17452 CrossRef CAS PubMed; (d) J. Zhao, M. G. Davidson, M. F. Mahon, G. Kociok-Kohn and T. D. James, J. Am. Chem. Soc., 2004, 126, 16179 CrossRef CAS PubMed; (e) L. Zhu and E. V. Anslyn, J. Am. Chem. Soc., 2004, 126, 3676 CrossRef CAS PubMed; (f) D. Ryu, E. Park, D. S. Kim, S. Yan, J. Y. Lee, B. Y. Chang and K. H. Ahn, J. Am. Chem. Soc., 2008, 130, 2394 CrossRef CAS PubMed; (g) H. Miyaji, S. Hong, S. Jeong, D. Yoon, H. Na, J. Hong, S. Ham, J. L. Sessler and C. Lee, Angew. Chem., Int. Ed., 2007, 46, 2508 CrossRef CAS PubMed.
- J. Yoon and D. J. Cram, J. Am. Chem. Soc., 1997, 119, 11796 CrossRef CAS.
- (a) Z. Xu, S. K. Kim and J. Yoon, Chem. Soc. Rev., 2010, 39, 1457 RSC; (b) J. Shang, B. M. Rambo, X. Hao, J.-F. Xiang, H.-Y. Gong and J. L. Sessler, Chem. Sci., 2016, 7, 4148–4157 RSC; (c) P. Sabater, F. Zapata, A. Caballero, I. Fernandez, C. Ramirez de Arellano and P. Molina, J. Org. Chem., 2016, 81, 3790 CrossRef CAS PubMed; (d) A. Caballero, N. G. White and P. D. Beer, Angew. Chem., Int. Ed. Engl., 2011, 50, 1845 CrossRef CAS PubMed; (e) D. Zhang, H. Yang, A. Martinez, K. Jamieson, J. P. Dutasta and G. Gao, Chem. – Eur. J., 2014, 20, 17161 CrossRef CAS PubMed; (f) M. W. Ahmad, S. H. Kim and H.-S. Kim, Tetrahedron Lett., 2011, 52, 6743 CrossRef CAS.
- (a) Z. Xu, N. J. Singh, S. K. Kim, D. R. Spring, K. S. Kim and J. Yoon, Chem. – Eur. J., 2011, 17, 1163 CrossRef CAS PubMed; (b) N. R. Song, J. H. Moon, E. J. Jun, J. Choi, Y. Kim, S.-J. Kim, J. Y. Lee and J. Yoon, Chem. Sci., 2013, 4, 1765 RSC; (c) Z. Xu, S. K. Kim, S. J. Han, C. Lee, G. Kociok-Kohn, T. D. James and J. Yoon, Eur. J. Org. Chem., 2009, 3058 CrossRef CAS; (d) Y. Chun, N. J. Singh, I. C. Hwang, J. W. Lee, S. U. Yu and K. S. Kim, Nat. Commun., 2013, 4, 1797 CrossRef PubMed; (e) H. Zhou, Y. Zhao, G. Gao, S. Li, J. Lan and J. You, J. Am. Chem. Soc., 2013, 135, 14908 CrossRef CAS PubMed; (f) V. Amendola, M. Boiocchi, L. Fabbrizzi and N. Fusco, Eur. J. Org. Chem., 2011, 6434 CrossRef CAS.
- J. Y. Kwon, N. J. Singh, H. Kim, S. K. Kim, K. S. Kim and J. Yoon, J. Am. Chem. Soc., 2004, 126, 8892 CrossRef CAS PubMed.
- J. Y. Jung, E. J. Jun, Y.-U. Kwon and J. Yoon, Chem. Commun., 2012, 48, 7928 RSC.
- (a) H. N. Kim, E.-H. Lee, Z. Xu, H.-E. Kim, H.-S. Lee, J.-H. Lee and J. Yoon, Biomaterials, 2012, 33, 2282 CrossRef CAS PubMed; (b) M. Yousuf, I. S. Youn, J. Yun, L. Rasheed, R. Valero, G. Shi and K. S. Kim, Chem. Sci., 2016, 7, 3581 RSC; (c) W. Zhang, F. Li, Y. Hu, S. Gan, D. Han, Q. Zhang and L. Niu, J. Mater. Chem. B, 2014, 2, 3142 RSC; (d) N. Ahmed, B. Shirinfar, V. M. Miriyala, S.-K. Choi, K.-M. Lee, W. B. Jeon, Y. S. Park and H. G. Nam, Supramol. Chem., 2014, 27, 478 CrossRef; (e) G. Li, X. Zhou, P. Yang, Y. Jian, T. Deng, H. Shen and Y. Bao, Tetrahedron Lett., 2014, 55, 7054 CrossRef CAS.
- (a) X. Chen, S. Kang, M. J. Kim, J. Kim, Y. S. Kim, H. Kim, B. Chi, S.-J. Kim, J. Y. Lee and J. Yoon, Angew. Chem., Int. Ed., 2010, 49, 1422 CrossRef CAS PubMed; (b) S. Hussain, A. H. Malik and P. K. Iyer, ACS Appl. Mater. Interfaces, 2015, 7, 3189 CrossRef CAS PubMed.
- S. Lee, H. Cheng, M. Chi, Q. Xu, X. Chen, C.-Y. Eom, T. D. James, S. Park and J. Yoon, Biosens. Bioelectron., 2016, 77, 1016 CrossRef CAS PubMed.
- (a) Z. Guo, N. R. Song, J. H. Moon, M. Kim, E. J. Jun, J. Choi, J. Y. Lee, C. W. Bielawski, J. L. Sessler and J. Yoon, J. Am. Chem. Soc., 2012, 134, 17846 CrossRef CAS PubMed; (b) Q. Xu, S. Lee, Y. Cho, M. H. Kim, J. Bouffard and J. Yoon, J. Am. Chem. Soc., 2013, 135, 17751 CrossRef CAS PubMed.
- (a) Y. Yang, X. Wang, Q. Cui, Q. Cao and L. Li, ACS Appl. Mater. Interfaces, 2016, 8, 7440 CrossRef CAS PubMed; (b) S. Kumar, P. Singh, G. Hundal, M. S. Hundal and S. Kumar, Chem. Commun., 2013, 49, 2667 RSC; (c) Q. Cui, X. Wang, Y. Yang, S. Li, L. Li and S. Wang, Chem. Mater., 2016, 28, 4661 CrossRef CAS.
- (a) Q. Lu, L. Dong, J. Zhang, J. Li, L. Jiang, Y. Huang, S. Qin, C. Hu and X. Yu, Org. Lett., 2009, 11, 669 CrossRef CAS PubMed; (b) L. Yang, S. Qin, X. Su, F. Yang, J. You, C. Hu, R. Xie and J. Lan, Org. Biomol. Chem., 2010, 8, 339 RSC; (c) X. Su, K. Luo, Q. Xiang, J. Lan and R. Xie, Chirality, 2009, 21, 539 CrossRef CAS PubMed.
- (a) S. Liu, K. Y. Law, Y. He and W. H. Chan, Tetrahedron Lett., 2006, 47, 7857 CrossRef CAS; (b) H. Wang, W. H. Chan and A. W. M. Lee, Org. Biomol. Chem., 2008, 6, 929 RSC; (c) S. Shinoda, T. Okazaki, T. N. Player, H. Misaki, K. Hori and H. Tsukube, J. Org. Chem., 2005, 70, 1835 CrossRef CAS PubMed; (d) M. W. Ahmad, S. H. Kim and H.-S. Kim, Tetrahedron Lett., 2011, 52, 6743 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details, 1H NMR spectra of compound 1. See DOI: 10.1039/c8cc06262e |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |