 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Body temperature protein X-ray crystallography at 37 °C: a rhenium protein complex seeking a physiological condition structure†
Francois J. F. Jacobs a,
John R. Helliwell
a,
John R. Helliwell *b and
Alice Brink
*b and
Alice Brink a
a
aDepartment of Chemistry, University of the Free State, Nelson Mandela Drive, Bloemfontein 9301, South Africa
bDepartment of Chemistry, University of Manchester, Oxford Road, Manchester M13 9PL, UK. E-mail: john.helliwell@manchester.ac.uk
First published on 9th October 2024
Abstract
The retention of the covalent binding of an organometalllic rhenium complex as a model for a technetium-99m imaging agent, to a protein at physiological body temperature 37 °C is described. Detailed structure comparisons are made to the related 100 K crystal structure. The generality of the need for this sort of analytical procedure for guiding ligand lead compound discovery is emphasised.
A general question about the strict relevance to biology of crystallography results, now predominantly based on X-ray diffraction data measured at cryo temperature (typically 100 K) has been raised by Halle, 2004: ‘In biomolecular cryocrystallography: structural changes occur during flash-cooling: conformational switching of solvent-exposed side chains and weak ligand binding are likely artefacts… also, the sites for molecular recognition, ligand binding or chemical catalysis, usually involve the solvent interfacial region where consequent cryo-artefacts are expected to be most pronounced.’2 My own laboratory's 1997 study3 was cited by Halle (2004) where we had noted in our Section 3.9 that several amino-acid side chains (of the concanavalin A protein) had adopted largely different conformations; most of the changes were in poorly determined residues in the room temperature structure, especially in the flexible loop regions; the number of detected solvent sites had more than doubled, to 319 at 110 K as compared with 149 at 293 K; and in particular there were 20 non-matching waters in the room temperature structure which were connected to the movement of side chains by freezing out of the low-energy conformations of Asp82, Ser117, His121 (in a loop region), Lys135 and Thr196. But there were no major differences in either the protein or solvent structure around the saccharide-binding. Vigilance is needed for such artefacts. A distinction should of course be made between structural details that change (at the two temperatures) and things that become visible at the cryo temperature which were not visible before at room temperature.
In a recent review by Fischer (2021)4 practical aspects were discussed for preparing, acquiring and analysing X-ray crystallography data at temperatures between 273–350 K and mentions the limited number of structures reported, constrained by radiation damage. Cryo-temperatures yield reduced radiation damage and allow for other advantages such as reducing dehydration and easy handling when remote data collection is required.5–7 A perspective by Hough et al.,8 too describes the recent developments on these topics. The pioneering study of multiple temperature crystal structures of ribonuclease was at nine temperatures9 (PDB codes 1RAT through to 9RAT) from 98 K right up to 320 K. These were undertaken to scrutinize protein structure differences with temperature. Unfortunately, the bound waters are not in these coordinates’ files (PDB codes 1RAT through to 9RAT) and the structure factors likewise are unavailable. To extend temperature considerations further for living organisms, can thermophilic proteins and extremophilic proteins be studied at these organisms’ temperatures, i.e. respectively, between 60 and 80 °C and >80 °C? Radiation damage by X-rays will presumably become the limiting factor, provided those protein crystals to be studied would be stable, but which could be circumvented by using neutrons for crystallography or by NMR. As to feasibility of ‘body temperature protein crystallography’, our interest in this paper, we mention that within an undergraduate physics project at York University in the mid-1980s, we showed that X-ray diffraction data could be recorded from a crystal of phenol insulin10 up to 50 °C. Also, that at about 55 °C the diffraction pattern disappeared but returned on cooling. Conducting crystallography checks at physiologically relevant temperatures, certainly 37 °C, is feasible and should be a routine objective where physiologically relevant structural results are claimed.
Our interest in radiopharmaceutical drug development utilising the chemical congener pair of rhenium (for therapeutic application) and its radioactive counterpart technetium-99m (for diagnostic application11,12) is structurally appropriate at primarily two temperatures: 100 K to obtain best resolution data with minimal radiation damage, including due to the presence of excess metals,13 and the body temperature (37 °C/310 K) appropriate for physiological conditions. Multiple temperature structural studies of proteins have previously elucidated aspects of protein folding and unfolding14–16 and of a glass transition at 200 K17,18 i.e. totally different themes. Recently we published a time-based series of freeze trap lysozyme rhenium-imidazole (HEWL-Re-Imi) crystal structures over a span of 38 weeks1 thereby of relevance to a patient's possible treatment period. The main rhenium covalent coordination was observed at His15, Asp101 and Asp119. Weak (i.e. noncovalent) interactions are observed at other aspartic, asparagine, proline, tyrosine and tryptophan side chains. We remarked that “the covalent bond stability at the three amino acid binding sites, their proximity to the solvent channel and the movement of residues to accommodate the metal were important, and (these results) may prove useful for future radiopharmaceutical development including target modification.” In this paper we investigate the same set of crystals for the stability of the covalent and non-covalent rhenium-imidazole binding to their amino acids in this protein when at body temperature i.e. 37 °C to test whether these structure based drug discovery fragments, identified at a very remote physiological temperature of 100 K, are still bound at a patient body temperature of 37 °C. Of course, these data are now evaluated at a later time date for the crystal in mother liquor time on the shelf (weeks 112 for 37 °C versus 38 weeks respectively). The X-ray diffractometer data collection required specific adaptations for the crystal mounting to prevent dehydration and reactivity to the cryoprotectant at the elevated temperature as well as observing the sensitivity to X-ray irradiation which became pronounced. Nevertheless, we achieved a diffraction data resolution of 2.2 Å on our home laboratory X-ray diffractometer for 37 °C (Fig. 1a and b).
Notably, the covalent binding at the three amino acid sites is retained (Fig. 2) at the elevated body temperature with minor variations in the function group orientations of the Re-complex. Overlays of the coordinated Re complex at Asp 119 shows they are nearly identical. However, the imidazole functional group has a weak interaction (2.7(1) Å) with a free Re214 atom in the 100 K data (8QCU) identified by the optimised anomalous density map. While Fo − Fc (4.0σ) density is present at 37 °C, it is insufficient to place a free Re atom with appropriate occupancy. What is noticeable is that the bound water interacting with the position of this free Re atom is prominent in both temperature studies (ca. 2.3(2) Å from Re214 to HOH) (Fig. 3).
 | ||
| Fig. 2 (a) A ribbon diagram overlay of the 37 °C and 100 K protein structures. The three main metal sites emphasised by blue circles. The red model is the 37 °C data set (this study) and the blue model the 100 K data set (PDB code 8QCU).1 (b) Detailed view of the three covalent binding sites at His15, Asp101 and Asp119. (Blue model = 100 K; red model = 37 °C.) Electron density maps of 37 °C in blue 2Fo − Fc contoured at 1.3 rms, in green Fo − Fc contoured at 3σ, in orange the anomalous difference Fourier maps contoured at 2.5σ. Here and in subsequent figures we colour code the protein for temperature, as stated, but not the rheniums or waters which are labeled with text. | ||
At Asp 101, movement at the aspartic acid side chain oxygen caused lateral shifting by 0.7(2) Å of the Re complex (Fig. 4), whereas at His15 there is little to no lateral shifting observed by the Re complex.
 | ||
| Fig. 4 Overlay at Asp101 indicating displacement of 37 °C versus 100 K. Viewing the 37 °C protein crystal structure and comparison overlay with the cryo crystal structure (PDB code 8QCU).1 The standard uncertainty on the rhenium atom shift was estimated using the Online_DPI webserver.19 | ||
The group of non-covalently interacting rhenium complex positions identified previously (100 K data versus 37 °C) had lower occupancies than the covalently bound ones. The non-covalent rhenium binding sites (i.e. free Re's) occur for two of the six sites identified at the 100 K and are in the vicinity of Arg14/His15 and Leu129 (Fig. 5). A possible third site at Tyr23 (indicated by Fo − Fc and 2Fo − Fc density) is conceivable however refined Re occupancy is low <7% and without clear anomalous density it is questionable if Re or rather Cl is a more suitable choice at this resolution. Non-observed Re sites in the 37 °C in comparison to that seen at 100 K are at Asp18, Pro70, Gln121.
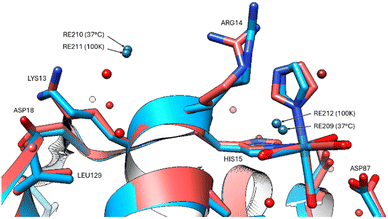 | ||
| Fig. 5 The two non-covalent Re binding sites indicated for both 37 °C (red model) and 100 K (blue model) in the vicinity of Arg14, His15 and Leu129. | ||
The protein chain appears mostly unaffected by the increase of temperature to 37 °C with only minor positional changes at Arg61, Ile78, Lys97, Asp101, Val109 in the 37 °C versus 100 K. A comparison of the bound waters at 100 K (PDB code 8QCU) with the electron density evidence available at 37 °C is of interest regarding the apparent greater mobility of some versus others (Table S2, ESI†). Obviously, the diffraction resolution is superior for the 100 K structure, and thereby more bound waters can be expected at the better resolution, but there seems to be a differential effect where some waters have remained more bound than others. Strongly bound waters occurring at both temperatures occur in regions of stability supported by at least two or with multiple HOH-chain interactions, such as sites, Gly102, Arg114, Asp66, Arg112, Arg128, Gly 121 (Fig. 6).
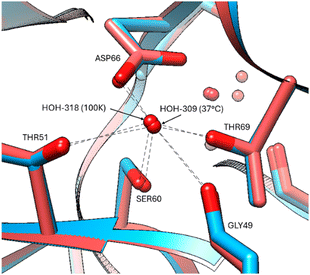 | ||
| Fig. 6 Stabilization of bound waters by multiple residues at all temperatures, indicated for the Asp66 residue at 37 °C (red model) and 100 K (blue model). | ||
A neutron protein crystallography structure20 of our protein rhenium bound crystal determined at both 37 °C and 100 K would allow us to compare the hydrogen bonding of the bound waters and comment directly on this apparent differential effect.
Overall, we commend that especially in lead compound drug discovery any promising bound ligand is checked for its presence using ‘body temperature (37 °C) protein crystallography’ and the network of bound waters be considered from the aspect of kinetic stability occurring within the structure. Accordingly, it would be advantageous if synchrotron radiation and XFEL (X-ray free electron laser) beamlines made such a utility of body temperature single crystal data collection readily available for users. The same experimental capability of body temperature measurements should be provided at the neutron macromolecular crystallography instruments. Their use provides a near complete hydrogenated structure and hydrogen bonding details, which X-rays and electrons as probes cannot, and be radiation damage free even at 37 °C.
We thank the South African National Research Foundation for funding of the University of the Free State X-ray diffractometer. This research is supported in part by the National Research Foundation of South Africa (Grant Numbers: 137759; 116180), the SA-DHET Future Professors Program and the University of Manchester. We thank Diamond Light Source for access to the I04 beamline through the South African Structural Biology Consortium.
Data availability
Protein crystallographic data is found on RCSB PDB (https://www.rcsb.org/; PDB code: 9GHX) and raw diffraction images are available for download at Zenodo (DOI: https://doi.org/10.5281/zenodo.13331546). Additional data information found in the ESI.†Conflicts of interest
There are no conflicts to declare.Notes and references
- F. J. F. Jacobs, J. R. Helliwell and A. Brink, IUCrJ, 2024, 11, 359–373 CrossRef CAS PubMed.
- B. Halle, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 4793–4798 CrossRef CAS PubMed.
- A. Deacon, T. Gleichmann, A. J. Kalb (Gilboa), H. Price, J. Raftery, G. Bradbrook, J. Yariv and J. R. Helliwell, Faraday Trans., 1997, 93, 4305–4312 RSC.
- M. Fischer, Q. Rev. Biophys., 2021, 54, e1 CrossRef PubMed.
- E. Garman, Curr. Opin. Struct. Biol., 2003, 13, 545–551 CrossRef CAS PubMed.
- E. Garman and M. Weik, Curr. Opin. Struct. Biol., 2023, 82, 102662 CrossRef CAS PubMed.
- M. Weik and J.-P. Colletier, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, D66, 437–446 CrossRef PubMed.
- M. A. Hough, F. Prischi and J. A. R. Worrall, Front. Mol. Biosci., 2023, 10, 1113762 CrossRef CAS PubMed.
- R. F. Tilton, J. C. Dewan and G. A. Petsko, Biochemistry, 1992, 31, 2469–2481 CrossRef CAS PubMed.
- U. Derewenda, Z. Derewenda, E. J. Dodson, G. G. Dodson, C. D. Reynolds, G. D. Smith, C. Sparks and D. Swenson, Nature, 1989, 338, 594–596 CrossRef CAS PubMed.
- R. Alberto, Inorg. Chem., 2023, 62, 20539–20548 CrossRef CAS PubMed.
- S. Jürgens, W. A. Herrman and F. E. Kühn, J. Organomet. Chem., 2014, 751, 83–89 CrossRef.
- K. L. Shelley and E. Garman, Acta Crystallogr., Sect. D: Struct. Biol., 2024, 80, 314–327 CrossRef CAS PubMed.
- M. C. Thompson, Chapter Nine – Combining temperature perturbations with X-ray crystallography to study dynamic macromolecules: A thorough discussion of experimental methods, in Methods in Enzymology, ed. N. Ando, Academic Press, 2023, vol. 688, pp. 255–305 Search PubMed.
- M. L. Scalley and D. Baker, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 10636–10640 CrossRef CAS PubMed.
- G. Feller, Semin. Cell Dev. Biol., 2018, 84, 129–137 CrossRef CAS PubMed.
- D. Vitkup, D. Ringe and G. Petsko, et al., Nat. Struct. Mol. Biol., 2000, 7, 34–38 CrossRef CAS PubMed.
- D. Ringe and G. A. Petsko, Biophys. Chem., 2003, 105, 667–680 CrossRef CAS PubMed.
- K. S. D. Kumar, M. Gurusaran, S. N. Satheesh, P. Radha, S. Pavithra, K. P. S. Thulaa Tharshan, J. R. Helliwell and K. Sekar, J. Appl. Crystallogr., 2015, 48, 939–942 CrossRef CAS.
- M. P. Blakeley, M. Cianci, J. R. Helliwell and P. J. Rizkallah, Chem. Soc. Rev., 2004, 33, 548–557 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedure. Table S1. X-Ray crystallographic data and model-refinement statistics for the 37 °C crystal structure. Statistics for the highest-resolution shell are shown in parentheses. Table S2. Comparison of all bound ligands (Cl, Na, Re, etc.) and bound waters at 100 K and at 37 °C specifying B-factors and occupancies where appropriate. Table S3. Rhenium occupancy values, anomalous difference map peak heights and residual Fo − Fc densities as found at 100 K and at 37 °C. The number of weeks soaked in mother liquor is indicated.1 See DOI: https://doi.org/10.1039/d4cc04245j |
| This journal is © The Royal Society of Chemistry 2024 |


