Open Access Article
Open Access ArticleForced intercalation-induced light-up peptides as fluorogenic indicators for the HIV-1 TAR RNA-ligand assay†
En Ting Tabitha Lee,
Yusuke Sato *,
Akunna F. Ujuagu and
Seiichi Nishizawa
*,
Akunna F. Ujuagu and
Seiichi Nishizawa *
*
Department of Chemistry, Graduate School of Science, Tohoku University, Sendai 980-8578, Japan. E-mail: yusuke.sato.a7@tohoku.ac.jp; seiichi.nishizawa.c8@tohoku.ac.jp
First published on 29th May 2024
Abstract
Fluorescence indicators capable of binding to human immunodeficiency virus-1 (HIV-1) trans-activation responsive (TAR) RNA are powerful tools for the exploratory studies of the identification of anti-HIV drug candidates. This work presents a new design strategy for fluorogenic indicators with a transactivator of transcription (Tat)-derived peptide based on the forced intercalation of thiazole orange (TO) dyes (FIT). The developed 9-mer FIT peptide (RKKRR-TO-RRR: named FiLuP) features the TO unit integrated onto a Dap (2,3-diaminopropionic acid) residue in the middle of the Tat peptide sequence; the Q (glutamic acid) residue in the Tat peptide (RKKRR-Q-RRR) is replaced with TO as if it were an amino acid surrogate. This facilitates a significant light-up response (450-fold at λem = 541 nm, Φfree = 0.0057, and Φbound = 0.61) upon binding to TAR RNA. The response of FiLuP is highly selective to TAR RNA over other non-cognate RNAs, and FiLuP maintains strong binding affinity (Kd = 1.0 ± 0.6 nM). Significantly, in contrast to previously developed Tat peptide-based FRET probes, FiLuP is able to discriminate between “competitive” and “noncompetitive” inhibitors when used in the fluorescence indicator displacement (FID) assay. The FID assay under stringent screening conditions is also possible, enabling super-strong competitive binders toward TAR RNA to be sieved out.
Introduction
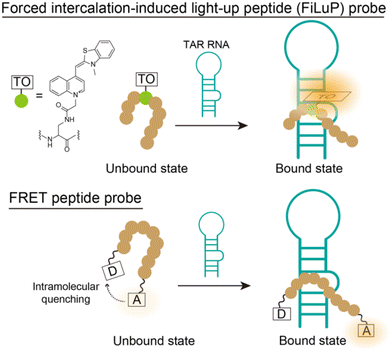
With increasing knowledge about the diverse roles of RNAs within the cells, RNA-binding ligands have received considerable attention in drug discovery.1,2 Human immunodeficiency virus-1 (HIV-1) trans-activation responsive (TAR) RNA plays a pivotal role in the replication of HIV-1 through binding to the transactivator of transcription (Tat) protein, making it an important therapeutic target for anti-HIV strategies.3 TAR RNA consists of 59 nucleotides located at the 5′ end of viral mRNAs, forming a stem-loop structure with a three-nucleotide (UCU) bulge (cf. Fig. 2A). It has been well known that Tat protein binds specifically to the three-nucleotide bulge region in TAR RNA, and the binding is mediated by a single arginine (R) within a nine residue stretch of basic amino acids (residues 49 to 57: R49KKRRQRRR57).4 The identification of TAR RNA-binding ligands capable of interfering with the formation of a Tat protein complex represents a promising avenue for potential anti-HIV drugs.5In this context, fluorescence methods are commonly employed for the exploratory studies of RNA-targeted ligands as they have several advantages such as sensitivity, speed and the convenient availability of equipment. In particular, the fluorescence indicator displacement (FID) assay is a simple and high-throughput method without fluorophore labeling of both target RNAs and test compounds used in screening experiments.6 While several fluorescent small molecules have been proposed for the FID assay,7 the most widely employed indicator for TAR RNA is based on Förster resonance energy transfer (FRET) with a truncated peptide comprising the basic domain of Tat protein (Tat49–57: R49K50K51R52R53Q54R55R56R57).8 A Tat49–57-containing 16-mer peptide (AAA-R49KKRRQRRR57-AAAC) labelled with 5-carboxyfluorescein and tetramethylrhodamine at the N-terminus and C-terminus, respectively, showed an increase in the FRET efficiency upon binding to TAR RNA, whereas the binding of the test compounds decreased the FRET efficiency (Fig. 1). To date, this FRET peptide probe has been widely used for functional evaluation of TAR RNA-binding ligands (Tat antagonists) discovered by in silico screening or structure-based design,9 and it has also been used for high-throughput screening (HTS) with ∼100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 compounds.10
000 compounds.10
Nevertheless, its fluorescence response for TAR RNA was quite modest, typically less than 3-fold,8,11 which poses a disadvantage in terms of the detection sensitivity of the displacement events. Recently, an aggregation induced emission (AIE) fluorogen (tetraphenylethylene: TPE) was newly used for the Tat peptide-based indicator design (TPE-G-R49KKRRQRRR57-PPQG);12 however, it did not result in an improvement in the fluorescence response (3-fold). Considering how the low fluorescence response of the indicator hampers the performance of the FID assay, including its reproducibility, robustness and reliability,6 there is a significant need to design Tat peptide-based indicators with large fluorescence signaling properties.
This paper presents a new design strategy for Tat peptide probes based on the forced intercalation of thiazole orange (TO) dyes (FIT).13 The Seitz group showed that a TO unit connected to a DNA backbone as a base surrogate by a short spacer could forcibly intercalate into predetermined base pairs of the duplex structure formed through hybridization with a complementary sequence.14
These FIT oligonucleotide probes (named FIT probes) display significant light-up properties, enabling the sensitive detection of target DNAs/RNAs. FIT was recently used for the design of DNA aptamers for the sensing of metal ions and proteins.15 FIT sensing was also achieved in peptide nucleic acid (PNA) probes for single-stranded RNAs,16 followed by in triplex-forming PNA probes17,18 for double-stranded RNAs by our group.19
Taking inspiration from these successes in oligonucleotide probes, we integrated FIT-sensing into the design of Tat peptide probes as novel fluorescence indicators for TAR RNA, where one amino acid in the Tat peptide (the basic domain of Tat protein: R49K50K51R52R53Q54R55R56R57) was replaced with TO as if it were an amino acid surrogate (Fig. 1 and 2A). Designing FIT peptides named FIT light-up peptide (FiLuP) probes, however, presents a challenge because of the inherent complexity of RNA–protein complexes compared to previous oligonucleotide scaffolds. We hypothesized that it could be achieved by strategically positioning the TO unit within the peptide sequence of the basic domain. Specifically, we aimed to place the TO unit in close proximity to the double-stranded RNA region for complex formation with TAR RNA while preserving the amino acid residues crucial for the binding of the Tat peptide. The designed 9-mer FiLuP probe (Fig. 2A, R49KKRR-TO-RRR57) exhibited a significant light-up response (450-fold) upon strong and selective binding to TAR RNA (Kd = 1.0 nM), which is clearly superior to the previously-developed 16-mer Tat peptide-based FRET probes (Kd = 23.6 to 286 nM).8,11 In comparison with a conventional peptide probe appended with the TO unit through a flexible spacer at the N-terminal (TO-R49KKRRQRRR57), we found that FiLuP had not only a larger response but also higher selectivity, showing the usefulness of FIT sensing for the fluorogenic probe design toward TAR RNA. Significantly, when used in the FID assay, FiLuP is able to discriminate between “competitive” and “noncompetitive” inhibitors, which has never been demonstrated with all previous indicators for TAR RNA.7,8,11,12 In addition, thanks to its strong binding affinity and significant light-up properties, the FID assay under stringent screening conditions can be performed, which sieves out the super-strong competitive inhibitor whose binding affinity is comparable to that of the wild Tat protein. These features of FiLuP are discussed as a basis for the advanced design of peptide-based fluorogenic probes for RNA sensing.
Experimental
The FRET probe (5-FAM-AAA-R49KKRRQRRR57-AAAK-TAMRA)11 was purchased from Hipep Laboratories (Kyoto, Japan). FiLuP (R49KKRR-TO-RRR57) and its variant probes were synthesized by Fmoc solid phase synthesis using a microwave-assisted peptide synthesizer. The crude probes were purified by reverse-phase HPLC (Fig. S1†), followed by verification by MALDI-TOF-MS (Table S1†). A conventional peptide probe appended with the TO unit through a lysine spacer at the N-terminal (TO-R49KKRRQRRR57) was also prepared (Table S1†). For experimental details and characterization, see the ESI.†Unless otherwise mentioned, all measurements were performed at 25 °C in 1 × PBS buffer (pH 7.4). Before measurements, annealing of RNA-containing samples was conducted as follows: heated at 95 °C for 10 min and gradually cooled to 25 °C (1 °C min−1). Errors denote the standard deviations obtained from at least three independent experiments.
The dissociation constant (Kd) for the binding of the probes to target RNA was determined by fluorescence titration experiments. The resulting titration curve was analysed based on a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding fitting model. Further details are provided in the ESI.†
1 binding fitting model. Further details are provided in the ESI.†
The FID assay was carried out with FiLuP as a fluorescence indicator. After the FiLuP/TAR RNA complex was formed by the annealing procedure as shown above, the test compounds were added and incubated for 10 min. The fluorescence response of the FiLuP/TAR RNA complex to the test compounds was then measured. The details are also shown in the ESI.†
Results and discussion
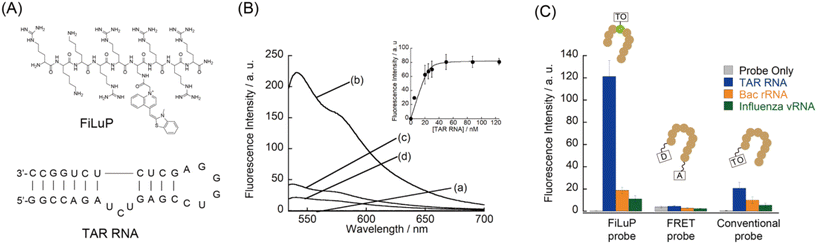
The FiLuP probe for TAR RNA was designed by integrating the TO unit into the Tat49–57 sequence (Fig. 2A). We decided to strategically replace the Q54 (glutamic acid) residue with a Dap (2,3-diaminopropionic acid) residue to which the TO unit was connected, based on the reports that the mutation of this residue minimally affects the RNA binding of the Tat peptide.20 Furthermore, the Q54 residue was shown to be located close to the double-stranded region of TAR RNA in the relevant Tat/TAR RNA complex structure.21 This observation suggests that the incorporation of the TO unit at this position would facilitate efficient FIT upon probe binding. The FiLuP probe (R49KKRR-TO-RRR57) was synthesized by Fmoc solid phase synthesis, where a TO derivative with a methylcarboxyl group on its quinoline ring was coupled with a Dap residue after the peptide assembly (see the ESI†). Fig. 2B shows the fluorescence response of the probe (50 nM) to 31-mer model TAR RNA at 25 °C in PBS buffer. The probe shows negligible emission of the TO unit (Φfree = 0.0057) in the absence of RNAs due to the free rotation between two heterorings.13 The addition of equimolar target RNA (50 nM) caused a significant light-up response (450-fold increase) of the TO unit (λem = 541 nm). This indicates that the TO unit is forced to intercalate into the base pairs of the double-stranded region of RNA upon probe binding.19The fluorescence quantum yield of the probe in the bound state with target RNA (Φbound) reaches as much as 0.61. Actually, the observed light-up response of FiLuP is superior to those of FIT oligonucleotide probes with the TO base surrogate,14,15,19 showing the usefulness of RNA-binding peptides as molecular scaffolds for FIT sensing. It is noteworthy that the light-up response of FiLuP is highly superior to that of the previously-developed Tat peptide-based FRET probe11 that shows a fluorescence increase of only 1.2-fold under identical conditions (Fig. 2C and S2A†). Also, the light-up response of FiLuP is highly superior to that of the Tat peptide-based probe with AIE fluorogens, where the fluorescence intensity was reported to increase by 3-fold in the presence of 600 nM TAR RNA.12 Moreover, our probe has excellent binding properties toward TAR RNA. The dissociation constant (Kd) of FiLuP based on a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 fitting model was determined to be 1.0 ± 0.6 nM by fluorescence titration experiments (inset of Fig. 2B). This indicates very strong binding to TAR RNA, which is comparable to even the wild Tat protein (Kd = 2–8 nM).22 Significantly, the affinity of FiLuP is found to be two orders of magnitude higher than that of the FRET probe examined under identical conditions (Kd = 360 ± 78 nM, Fig. S2B†). Hence, the forced intercalation of the TO unit confers advantage with regard to the binding affinity toward TAR RNA. The examination of various RNAs reveals that FiLuP is highly selective toward TAR RNA (Fig. 2C). The response for TAR RNA was more than 5-fold greater than that for non-cognate RNAs (the bacterial ribosomal RNA A-site and the influenza A virus RNA promoter region) (Fig. S3†).19e,f On the other hand, the FRET probe only exhibits moderate selectivity for TAR RNA (Fig. 2C and S3†), as reported previously.6,11 We reason that the observed difference in selectivity between the two probes stems from their working principles. In the case of the FRET probe, even non-specific binding would induce changes in FRET efficiency. In contrast, the FiLuP probe relies on the peptide's binding and the intercalation of the TO unit for the fluorescence response to occur. Non-specific binding events would not elicit a response unless the TO unit efficiently intercalates into the base pairs of target RNAs. This feature of the FIT system significantly improves the response selectivity for TAR RNA. It should be noted here that, prior to this work, our group reported that TO itself functioned as a light-up probe for TAR RNA and proposed its use for the FID assay.7d We believe TO is a useful indicator for the FID assay, but its binding affinity and signaling ability were only moderate (Kd = 60 nM, Φfree < 0.01, and Φbound = 0.198) compared to those of the FiLuP probe (Kd = 1.0 nM, Φfree = 0.0057, and Φbound = 0.61). Our group also reported that TO worked as a fluorogenic dye for nucleolar RNA imaging in living cells (Φbound = 0.16).23 The examination with synthetic RNA and DNA revealed that TO prefers RNA over DNA, with a significant response to double-stranded RNA (rG/rC and rA/rU) and single-stranded rG.23 In addition, TO and its derivative (TO-PRO-1) were reported as indicators in the FID assay for targeting various RNA structures including TAR RNA and the bacterial ribosomal RNA A-site,6,7a being apparently characterized as non-selective for TAR RNA. Apparently, our FiLuP probe does function distinctly from the previous Tat peptide-based probes8,11,12 as well as TO itself, having prominent binding and fluorescence properties toward TAR RNA.
1 fitting model was determined to be 1.0 ± 0.6 nM by fluorescence titration experiments (inset of Fig. 2B). This indicates very strong binding to TAR RNA, which is comparable to even the wild Tat protein (Kd = 2–8 nM).22 Significantly, the affinity of FiLuP is found to be two orders of magnitude higher than that of the FRET probe examined under identical conditions (Kd = 360 ± 78 nM, Fig. S2B†). Hence, the forced intercalation of the TO unit confers advantage with regard to the binding affinity toward TAR RNA. The examination of various RNAs reveals that FiLuP is highly selective toward TAR RNA (Fig. 2C). The response for TAR RNA was more than 5-fold greater than that for non-cognate RNAs (the bacterial ribosomal RNA A-site and the influenza A virus RNA promoter region) (Fig. S3†).19e,f On the other hand, the FRET probe only exhibits moderate selectivity for TAR RNA (Fig. 2C and S3†), as reported previously.6,11 We reason that the observed difference in selectivity between the two probes stems from their working principles. In the case of the FRET probe, even non-specific binding would induce changes in FRET efficiency. In contrast, the FiLuP probe relies on the peptide's binding and the intercalation of the TO unit for the fluorescence response to occur. Non-specific binding events would not elicit a response unless the TO unit efficiently intercalates into the base pairs of target RNAs. This feature of the FIT system significantly improves the response selectivity for TAR RNA. It should be noted here that, prior to this work, our group reported that TO itself functioned as a light-up probe for TAR RNA and proposed its use for the FID assay.7d We believe TO is a useful indicator for the FID assay, but its binding affinity and signaling ability were only moderate (Kd = 60 nM, Φfree < 0.01, and Φbound = 0.198) compared to those of the FiLuP probe (Kd = 1.0 nM, Φfree = 0.0057, and Φbound = 0.61). Our group also reported that TO worked as a fluorogenic dye for nucleolar RNA imaging in living cells (Φbound = 0.16).23 The examination with synthetic RNA and DNA revealed that TO prefers RNA over DNA, with a significant response to double-stranded RNA (rG/rC and rA/rU) and single-stranded rG.23 In addition, TO and its derivative (TO-PRO-1) were reported as indicators in the FID assay for targeting various RNA structures including TAR RNA and the bacterial ribosomal RNA A-site,6,7a being apparently characterized as non-selective for TAR RNA. Apparently, our FiLuP probe does function distinctly from the previous Tat peptide-based probes8,11,12 as well as TO itself, having prominent binding and fluorescence properties toward TAR RNA.
Related to this work, DNA-binding peptides conjugated with TO were developed for DNA analysis.24 In these works, the TO unit was appended to the terminal of the peptide through a flexible spacer that enabled the TO unit to fold back and to intercalate into the DNA duplex. We thus synthesized a Tat47–57 peptide probe appended with the TO unit at the N-terminal via a lysine linkage (named a conventional probe: TO-R49KKRRQRRR57). In comparison with FiLuP, the conventional probe exhibited a modest light-up response (35-fold) upon binding to TAR RNA (Fig. 2C). This mainly results from inefficient intercalation of the TO unit. In addition, we noticed that the background emission of the conventional probe is more than 2-fold higher than that of FiLuP, leading to a lower light-up response. This would be because of the emission originating from the back-folding of the TO unit into the peptide itself, as reported in other TO-appended PNA probes.25 We also found that the selectivity of the conventional probe for TAR RNA was inferior (Fig. 2C and S3†), presumably due to the non-selective intercalation of the TO unit.26 Hence, the connection of the TO unit with a short spacer at the middle of the Tat47–57 peptide as if it were a substitute for the amino acids that make up the Tat peptide, a key design characteristic of FiLuP, is crucial for significant light-up response and selectivity for TAR RNA.
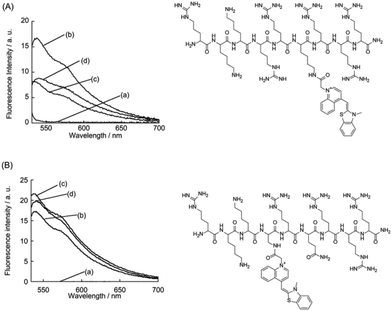
Furthermore, two other variant probes were examined to validate the FiLuP design (Fig. 3). The first variant had TO attached to a longer lysine residue instead of Dap (R49KKRR-(Lys-TO)-RRR57). This resulted in a significant decrease in fluorescence response, although selectivity towards TAR RNA remained intact. We reason that the longer linker length results in higher flexibility that impedes the intercalation of TO into the RNA. The second variant altered the position of TO in the probe, substituting it with R52 (arginine) instead (R49KK-TO-RQRRR57). This variant lacked selectivity for TAR RNA, matching previous literature that reported how R52 is crucial for TAR RNA recognition.4a–c These experiments verified the significance of the TO unit's environment within the RNA-binding peptide on the probe's functions.
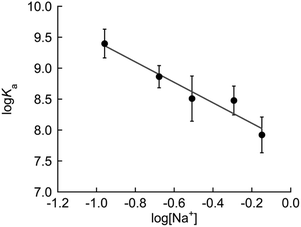
We then examined the thermodynamics of the binding of FiLuP to TAR RNA according to the polyelectrolyte theory proposed by Record et al. (ESI†).27 We here determined the binding constants (Ka = 1/Kd) from the fluorescence titration experiments in phosphate buffer (pH 7.4) containing NaCl concentrations (100–700 mM) (Fig. S4†). From the examination of the salt dependence of the binding affinity (Fig. 4), the observed binding free energy (ΔGobs) was divided into polyelectrolyte (ΔGpe) and non-polyelectrolyte (ΔGt) contributions. The ΔGpe contribution arises from the release of counterions from RNA upon binding, while the ΔGt contribution arises from other molecular interactions such as hydrogen bonds and van der Waals interactions. Interestingly, the previous study of positively charged 15-mer Tat peptide (SYG-RKKRRQRRR-PPQ) binding to TAR RNA revealed that electrostatic interaction contributes only to a limited extent to the free energy change, with the major role being played by non-polyelectrolyte interactions (ΔGobs = −9.9 kcal mol−1, ΔGpe = −1.9 kcal mol−1 and ΔGt = −8.0 kcal mol−1 at 80 mM Na+ and pH 7.5).28 In their analysis, the value of the slope of the linear plot of salt dependence (cf. Fig. 4) was obtained as −1.14, indicating the release of one counterion upon binding. The authors suggested that out of the six arginine (R) molecules present in the peptide, only one arginine is involved in electrostatic interaction with TAR RNA. This is consistent with the previous studies showing that a single arginine residue in the peptide is responsible for specific binding to TAR RNA.4 In the case of the binding of FiLuP to TAR RNA, we found a linear relationship between the double logarithmic plot of the binding affinity and the salt concentrations ranging from 100 mM NaCl to 700 mM NaCl. The slope was determined to be −1.65 (Fig. 4). ΔGt was found to contribute more greatly to the binding events (ΔGobs = −12.0 kcal mol−1, ΔGpe = −2.1 kcal mol−1 and ΔGt = −9.9 kcal mol−1 at 110 mM Na+ and pH 7.4). It is highly likely that FiLuP can bind to TAR RNA in a similar manner to the parent Tat peptide.
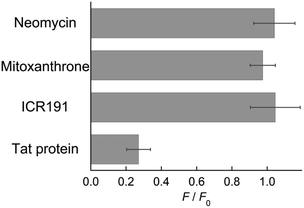
Finally, a mock FID assay revealed the potential of FiLuP as the fluorescence indicator for TAR RNA (Fig. 5A). The FID assay is based on a model in which the indicator bound to target RNA is displaced by a test compound in a competitive manner. Thus, the binding capability of the test compounds can be evaluated by monitoring the change in the indicator's fluorescence during the displacement event. Three well-known small molecules capable of binding to TAR RNA with distinct modes (mitoxanthrone,9b neomycin29 and ICR 1917c) were explored as test compounds. Mitoxanthrone was first identified through a virtual screening and then experimentally validated to bind to TAR RNA (Kd = 55 nM) and inhibit its interaction with Tat.9b Neomycin is also a strong binder for TAR RNA (Kd = 160 nM),29a but it is a noncompetitive inhibitor with respect to Tat that can form a ternary complex with Tat-TAR RNA; the three-nucleotide bulge (UCU, cf. Fig. 2A) is not the primary contact site for neomycin.29b ICR 191 was reported as a fluorescent indicator for TAR RNA with Kd = ∼100 nM. The intercalative binding of ICR 191 at the bulge site was revealed by a docking experiment, while the multi-binding to TAR RNA (TAR/ICR, TAR/ICR2, and TAR/ICR3) was revealed by fluorescence and MS experiments.7c Chloramphenicol was also used as a negative control as it specifically binds to 50S rRNA, not TAR RNA.30 We examined the fluorescence changes in the 500 nM TAR RNA/FiLuP complex upon addition of 60 equivalents of the test compounds (30 μM). In the absence of the test compounds, strong emission of the complex was observed due to the light-up response of FiLuP upon binding to TAR RNA (Fig. 5B). The addition of chloramphenicol resulted in a negligible change in the emission of the complex. This indicates that competitive binding does not occur because of the inability of chloramphenicol to bind to TAR RNA.30 In sharp contrast, the addition of mitoxanthrone led to a drastic decrease in FiLuP's emission. Given that mitoxanthrone was reported to bind to the Tat-binding site in TAR RNA,9b the observed fluorescence decrease can be explained by the displacement event with the FiLuP probe. Here, importantly, almost no response was obtained for neomycin, a noncompetitive inhibitor of the Tat-TAR RNA complex. Considering its binding affinity (Kd = 160 nM) and the current experimental conditions ([FiLuP] = [TAR RNA] = 500 nM, [neomycin] = 30 μM), it is likely that neomycin forms a ternary complex with FiLuP-TAR RNA, similar to the binding to Tat-TAR RNA. In the case of ICR 191, only a small decrease in FiLuP's emission was observed, indicating its relatively weak competitive ability. It is likely that the majority of CR191 binds to TAR RNA in a noncompetitive manner with FiLuP, forming ternary and/or higher order complexes. These results verify the unique feature of the FID assay with the FiLuP indicator for the identification of TAR RNA bulge-binding molecules, being able to discriminate between “competitive” and “noncompetitive” inhibitors. Such discrimination ability has never been achieved for all the previous indicators for TAR RNA including Tat peptide-based probes8,11,12 and small molecule-based indicators.7 All the previous FID assays showed considerable responses to neomycin. For example, in the FID assay with TO as the indicator, its fluorescence was totally quenched in the presence of 5 equiv. of neomycin ([TO] = [TAR RNA] = 500 nM, [neomycin] = 2.5 μM).7d In the case of the FID assay with the FRET probe (Kd = 23.6 nM),11a over 25% displacement was obtained for neomycin, and it was almost comparable to that for mitoxanthrone ([probe] = 50 nM, [TAR RNA] = 40 nM, [test compound] = 10 μM). While the exact reason is not clear for now, the binding of FiLuP is likely to take place almost in the same manner as the binding of the parent Tat peptide to TAR RNA, which may also be supported by its high affinity and selectivity for TAR RNA (cf. Fig. 2) and its binding thermodynamics (cf. Fig. 4).
FiLuP has another promising feature as an indicator in the FID assay. As described above, FiLuP features strong binding (Kd = 1.0 nM) and a large light-up response (450-fold increase) for TAR RNA compared to the FRET probe (Kd = 360 nM, 1.2-fold). This facilitates the FID assay to take place under stringent screening conditions. We carried out the mock FID assay using a 2 nM TAR RNA/FiLuP complex, where the wild Tat protein (9.8 kDa) was newly examined as a test compound in addition to TAR RNA-binding small molecules examined above (Fig. 5). We observed the emission enhancement of FiLuP upon addition of TAR RNA (Fig. S5†). This shows that FiLuP binding occurs even at such a low RNA concentration, in sharp contrast to the case of the previous FRET probe (Fig. S6†). The addition of Tat protein to the FiLuP/TAR RNA complex caused a large decrease in FiLuP's emission, whereas negligible response was seen for small molecules (Fig. 6). The response to Tat protein was concentration-dependent (Fig. S7†). This shows that the binding of Tat protein with TAR RNA resulted in the dissociation of FiLuP, which is reasonable considering that the Tat protein can compete with FiLuP to bind to TAR RNA due to its comparable affinity. It should be noted that mitoxanthrone with reasonably strong affinity (Kd = 55 nM)9b was unable to displace FiLuP bound to TAR RNA under the present stringent conditions, implying that the present FID assay can sieve out the super-strong competitive inhibitor whose binding affinity is comparable to that of the wild Tat protein (Kd = 2–8 nM).22 Given the crucial role of ultra-high affinity for TAR RNA in the development of anti-HIV drugs,31 we expect FiLuP to be a useful fluorescence indicator for the exploratory studies of TAR RNA-targeting drug candidates, ultimately contributing to the development of novel anti-HIV therapies.
Conclusions
In conclusion, FiLuP has been developed as a useful fluorogenic probe for TAR RNA by replacing one amino acid in the Tat peptide with TO as if it were an amino acid surrogate. We here designed FiLuP as an advanced version of oligonucleotide-based FIT probes with TO as a base surrogate. To the best of our knowledge, this is the first report on FIT-based fluorescence sensing on peptide scaffolds. In contrast to previous FRET peptide probes, FiLuP does exhibit a significant light-up response for TAR RNA (450-fold increase at λem = 541 nm, Φfree = 0.0057, and Φbound = 0.61), and responses are highly selective to TAR RNA over other non-cognate RNAs. Also, FiLuP does maintain a strong binding affinity (Kd = 1.0 ± 0.6) and is likely to bind to TAR RNA in a similar manner to the parent Tat peptide. The unique feature of FiLuP as an indicator in the FID assay is its ability to distinguish between competitive and noncompetitive inhibitors, which has never been demonstrated with all previous indicators for TAR RNA. Furthermore, thanks to its strong binding affinity and significant light-up properties, the FID assay under stringent screening conditions is possible. These features would facilitate a rigorous screening system as well as streamlining the subsequent screening procedures.19eIt should be noted here that, as demonstrated for oligonucleotide-based FIT probes,14,16,19 the TO unit would be replaced by other cyanine dyes including deep-red emissive BIQ (benzo[c,d]indole-quinoline cyanine)19d for an improved FID assay that does not suffer from compound optical interference.32 We also envision that the present design strategy is applicable to various peptide-based fluorogenic indicators targeting other druggable RNAs. In fact, preliminary experiments have shown that another FIT peptide (FiLuP), designed based on a λN protein-derived peptide,33 exhibits useful binding and fluorescence properties for its target boxB RNA (data not shown). Further studies are underway in these directions.
Author contributions
Y. S. conceived the study; E. T. T. L. and A. F. U. synthesized the probes and characterized their functions. All authors analyzed the data. E. T. T. L., Y. S. and S. N. wrote the paper.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by JSPS KAKENHI Grant Number 22H02099 and 23H00297.References
- J. R. Thomas and P. J. Hergenrother, Chem. Rev., 2008, 108, 1171–1224 CrossRef CAS PubMed.
- (a) B. S. Morgan, J. E. Forte and A. E. Hargrove, Nucleic Acids Res., 2018, 46, 8025–8037 CrossRef CAS PubMed; (b) A. U. Juru and A. E. Hargrove, J. Biol. Chem., 2021, 296, 100191 CrossRef PubMed; (c) A. E. Hargrove, Chem. Commun., 2020, 56, 14744–14756 RSC; (d) J. P. Falese, A. Donlic and A. E. Hargrove, Chem. Soc. Rev., 2021, 50, 2224–2243 RSC.
- (a) C. Dingwall, I. Ernberg, M. J. Gait, S. M. Green, S. Heaphy, J. Karn, A. D. Lowe, M. Singh and M. A. Skinner, EMBO J., 1990, 9, 4145–4153 CrossRef CAS PubMed; (b) D. Harrich, C. Ulich, L. F. Garcia-Martinez and R. B. Gaynor, EMBO J., 1997, 16, 1224–1235 CrossRef CAS PubMed.
- (a) B. J. Calnan, B. Tidor, S. Biancalana, D. Hudson and A. D. Frankel, Science, 1991, 252, 1167–1171 CrossRef CAS PubMed; (b) J. Tao and A. D. Frankel, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 2723–2726 CrossRef CAS PubMed; (c) J. Tao and A. D. Frankel, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 1571–1575 CrossRef CAS PubMed; (d) J. D. Puglisi, R. Tan, B. J. Calnan, A. D. Frankel and J. R. Williamson, Science, 1992, 257, 76–80 CrossRef CAS PubMed.
- R. Paul, D. Dutta, R. Paul and J. Dash, Angew. Chem., Int. Ed., 2020, 59, 12407–12411 CrossRef CAS PubMed.
- S. L. Wicks and A. E. Hargrove, Methods, 2019, 167, 3–14 CrossRef CAS PubMed.
- (a) P. N. Asare-Okai and C. S. Chow, Anal. Biochem., 2011, 408, 269–276 CrossRef CAS PubMed; (b) S. Kumar, P. Kellish, W. E. Robinson Jr., D. Wang, D. H. Appella and D. P. Arya, Biochemistry, 2011, 51, 2331–2347 CrossRef PubMed; (c) L. Qi, J. Zhang, T. He, Y. Huo and Z.-Q. Zhang, Spectrochim. Acta, Part A, 2017, 173, 93–98 CrossRef CAS PubMed; (d) Y. Ito, Y. Sato, N. Teramae and S. Nishizawa, Bunseki Kagaku, 2021, 70, 149–157 CrossRef CAS.
- C. Matsumoto, K. Hamasaki, H. Mihara and A. Ueno, Bioorg. Med. Chem. Lett., 2000, 10, 1857–1861 CrossRef CAS PubMed.
- (a) S. Kumar, P. Kellish, W. E. Robinson, D. Wang, D. H. Appella and D. P. Arya, Biochemistry, 2012, 51, 2331–2347 CrossRef CAS PubMed; (b) A. C. Stelzer, A. T. Frank, J. D. Kratz, M. D. Swanson, M. J. Gonzalez-Hernandez, J. Lee, I. Andricioaei, D. M. Markovitz and H. M. Al-Hashimi, Nat. Chem. Biol., 2011, 7, 553–559 CrossRef CAS PubMed; (c) N. N. Patwardhan, L. R. Ganser, G. J. Kapral, C. S. Eubanks, J. Lee, B. Sathyamoorthy, H. M. Al-Hashimi and A. E. Hargrove, MedChemComm, 2017, 8, 1022–1036 RSC.
- L. R. Ganser, J. Lee, A. Rangadurai, D. K. Merriman, M. L. Kelly, A. D. Kansal, B. Sathyamooorthy and H. M. Al-Hashimi, Nat. Struct. Mol. Biol., 2018, 25, 425–434 CrossRef CAS PubMed.
- (a) N. N. Patwardhan, Z. Cai, C. N. Newson and A. E. Hargrove, Org. Biomol. Chem., 2019, 17, 1778–1786 RSC; (b) N. N. Patwardhan, Z. Cai, A. U. Juru and A. E. Hargrove, Org. Biomol. Chem., 2019, 17, 9313–9320 RSC.
- J. Li, Y.-Y. Fan, J. Wen, J. Zhang and Z.-Q. Zhang, Anal. Chem., 2022, 94, 4695–4702 CrossRef CAS PubMed.
- (a) B. A. Armitage, Top. Curr. Chem., 2005, 253, 55–76 CrossRef CAS; (b) B. A. Armitage, Top. Heterocycl. Chem., 2008, 14, 11–29 CAS; (c) O. Suss, L. Motiei and D. Margulies, Molecules, 2021, 206, 2828 CrossRef PubMed.
- (a) O. Köhler, D. V. Jarikote and O. Seitz, ChemBioChem, 2005, 6, 69–77 CrossRef PubMed; (b) F. Hövelmann, I. Gaspar, A. Ephrussi and O. Seitz, J. Am. Chem. Soc., 2013, 135, 19025–19032 CrossRef PubMed; (c) F. Hövelmann, I. Gaspar, J. Chemiolo, M. Kasper, J. Steffen, A. Ephrussi and O. Seitz, Chem. Sci., 2016, 7, 128–135 RSC.
- S. B. Ebrahimi, D. Samata, H. F. Cheng, L. I. Nathan and C. A. Mirkin, J. Am. Chem. Soc., 2019, 141, 13744–13748 CrossRef CAS PubMed.
- (a) N. Kolevzon, D. Hashoul, S. Naik, A. Rubinstein and E. Yavin, Chem. Commun., 2016, 52, 2405–2407 RSC; (b) I. Peled and E. Yavin, ACS Omega, 2018, 3, 3813–3818 CrossRef CAS PubMed; (c) D. Hashoul, R. Shapira, M. Falchenko, O. Tepper, V. Paviov, A. Nissan and E. Yavin, Biosens. Bioelectron., 2019, 137, 271–278 CrossRef CAS PubMed; (d) O. Tepper, H. Zheng, D. H. Appella and E. Yavin, Chem. Commun., 2021, 57, 540–543 RSC; (e) O. Tepper, I. Peled, Y. Fastman, A. Heinberg, V. Mitesser, R. Dzikowski and E. Yavin, ACS Sens., 2022, 7, 50–59 CrossRef CAS PubMed; (f) O. Tepper, D. H. Appella, H. Zheng, R. Dzikowski and E. Yavin, ACS Sens., 2024, 9, 1458–1464 CrossRef CAS PubMed.
- (a) M. Li, T. Zengeya and E. Rozners, J. Am. Chem. Soc., 2010, 132, 8676–8681 CrossRef CAS PubMed; (b) P. Gupta, T. Zengeya and E. Rozners, Chem. Commun., 2011, 47, 11125–11127 RSC; (c) T. Zengeya, M. Li and E. Rozners, Bioorg. Med. Chem. Lett., 2011, 21, 2121–2124 CrossRef CAS PubMed; (d) P. Gupta, O. Muse and E. Rozners, Biochemistry, 2012, 51, 11125–11127 CrossRef PubMed; (e) T. Zengeya, P. Gupta and E. Rozners, Angew. Chem., Int. Ed., 2012, 51, 12593–12596 CrossRef CAS PubMed; (f) O. Muse, T. Zengeya, J. Mwaura, D. Hnedzko, D. W. McGee, C. T. Grewer and E. Rozners, ACS Chem. Biol., 2013, 8, 1683–1686 CrossRef CAS PubMed; (g) T. Endoh, D. Hnedzko, E. Rozners and N. Sugimoto, Angew. Chem., Int. Ed., 2016, 55, 899–903 CrossRef CAS PubMed; (h) T. Endoh, C. Annoni, D. Hnedzko, E. Rozners and N. Sugimoto, Phys. Chem. Chem. Phys., 2016, 18, 32002–32006 RSC; (i) I. Kumpina, N. Brodyagin, J. A. MacKay, S. D. Kennedy, M. Katkevics and E. Rozners, J. Org. Chem., 2019, 84, 13276–13298 CrossRef CAS PubMed; (j) C. A. Ryan, V. Baskevics, M. Katkevics and E. Rozners, Chem. Commun., 2022, 58, 7148–7151 RSC; (k) C. A. Ryan, M. M. Rahman, V. Kumar and E. Rozners, J. Am. Chem. Soc., 2023, 145, 10497–10504 CrossRef CAS PubMed.
- (a) G. Devi, Z. Yuan, Y. Lu, Y. Zhao and G. Chen, Nucleic Acids Res., 2014, 42, 4008–4018 CrossRef CAS PubMed; (b) D.-F. K. Toh, G. Devi, K. M. Patil, Q. Qu, M. Maraswami, Y. Xiao, T. P. Loh, Y. Zhao and G. Chen, Nucleic Acids Res., 2016, 44, 9071–9082 CAS; (c) A. A. L. Ong, D.-F. K. Toh, K. M. Patil, Z. Meng, Z. Yuan, M. S. Krishna, G. Devi, P. Haruehanroengra, Y. Lu, K. Xia, K. Okamura, J. Sheng and G. Chen, Biochemistry, 2019, 58, 1319–1331 CrossRef CAS PubMed; (d) J. Kesy, K. M. Patil, S. R. Kumar, Z. Shu, H. Y. Yong, L. Zimmermann, A. A. L. Ong, D.-D. K. Toh, M. S. Krishna, L. Yang, J.-L. Decout, D. Luo, M. Prabakaran, G. Chen and E. Kierzek, Bioconjugate Chem., 2019, 30, 931–943 CrossRef CAS PubMed; (e) M. S. Krishna, D.-F. Toh, Z. Meng, A. A. L. Ong, Z. Wang, Y. Lu, K. Xia, M. Prabakaran and G. Chen, Anal. Chem., 2019, 91, 5331–5338 CrossRef CAS PubMed.
- (a) T. Sato, Y. Sato and S. Nishizawa, J. Am. Chem. Soc., 2016, 138, 9397–9400 CrossRef CAS PubMed; (b) T. Sato, Y. Sato and S. Nishizawa, Chem. – Eur. J., 2017, 23, 4079–4088 CrossRef CAS PubMed; (c) T. Chiba, T. Sato, Y. Sato and S. Nishizawa, Org. Biomol. Chem., 2017, 15, 7765–7769 RSC; (d) Y. Yoshino, Y. Sato and S. Nishizawa, Anal. Chem., 2019, 91, 14254–14260 CrossRef CAS PubMed; (e) E. T. T. Lee, Y. Sato and S. Nishizawa, Chem. Commun., 2020, 56, 14976–14979 RSC; (f) Y. Sato, H. Miura, T. Tanabe, C. U. Okeke, A. Kikuchi and S. Nishizawa, Anal. Chem., 2022, 94, 7814–7822 CrossRef CAS PubMed; (g) C. U. Okeke, H. Miura, Y. Sato and S. Nishizawa, Org. Biomol. Chem., 2023, 21, 3402–3410 RSC.
- B. J. Calnan, S. Biancalan, D. Hudson and A. D. Frankel, Genes Dev., 1991, 5, 201–210 CrossRef CAS PubMed.
- K. Anand, A. Schulte, K. Vogel-Bachmayr, K. Scheffzek and M. Geyer, Nat. Struct. Mol. Biol., 2008, 15, 1287–1292 CrossRef CAS PubMed.
- O. Chaloin, J.-C. Peter, J.-P. Briand, B. Masquida, C. Desgranges, S. Muller and J. Hoebeke, Cell. Mol. Life Sci., 2005, 62, 355–361 CrossRef CAS PubMed.
- M. He, Y. Sato and S. Nishizawa, Analyst, 2023, 148, 636–642 RSC.
- (a) J. R. Carreon, K. P. Mahon and S. O. Kelley, Org. Lett., 2004, 6, 517–519 CrossRef CAS PubMed; (b) M. Thompson and N. W. Woodbury, Biochemistry, 2000, 39, 4327–4338 CrossRef CAS PubMed; (c) M. Thompson, Biomacromolecules, 2007, 8, 3628–3633 CrossRef CAS PubMed.
- N. Svanvik, J. Nygren, G. Westman and M. Kubista, J. Am. Chem. Soc., 2001, 123, 803–809 CrossRef CAS PubMed.
- T. Sato, Y. Sato, K. Iwai, S. Kuge, S. Nishizawa and N. Teramae, Chem. Commun., 2015, 51, 1421–1423 RSC.
- M. T. Record Jr., C. F. Anderson and T. M. Lohman, Q. Rev. Biophys., 1978, 11, 103–178 CrossRef CAS PubMed.
- H. Suryawanshi, H. Sabharwal and S. Maiti, J. Phys. Chem. B, 2010, 114, 11155–11163 CrossRef CAS PubMed.
- (a) T. D. Bradrick and J. P. Marino, RNA, 2004, 10, 1459–1468 CrossRef CAS PubMed; (b) S. Wang, P. W. Huber, M. Cui, A. W. Czarnik and H.-Y. Mei, Biochemistry, 1998, 37, 5549–5557 CrossRef CAS PubMed.
- (a) K. S. Long and B. T. Porse, Nucleic Acids Res., 2003, 31, 7208–7215 CrossRef CAS PubMed; (b) D. Moazed and H. F. Noller, Biochemie, 1987, 69, 879–884 CrossRef CAS PubMed.
- M. D. Shortridge, P. T. Wille, A. N. Jones, A. Davidson, J. Bogdanovic, E. Arts, J. Karn, J. A. Robinson and G. Varani, Nucleic Acids Res., 2019, 47, 1523–1531 CrossRef CAS PubMed.
- (a) Y. Sato, S. Yajima, A. Taguchi, K. Baba, M. Nakagomi, Y. Aiba and S. Nishizawa, Chem. Commun., 2019, 55, 3183–3186 RSC; (b) Y. Sato, Y. Aiba, S. Yajima, T. Tanabe, K. Higuchi and S. Nishizawa, ChemBioChem, 2019, 20, 2752–2756 CrossRef CAS PubMed.
- H. S. Jeoing, S. Choi, H. Kim, J. W. Park, H. N. Park, S. M. Park, S. K. Jang, Y. M. Rhee and B. H. Kim, Mol. Biosyst., 2013, 9, 948–951 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, probe synthesis, fluorescence response, salt dependence of the binding affinity, and FID results. See DOI: https://doi.org/10.1039/d4an00530a |
| This journal is © The Royal Society of Chemistry 2024 |