Open Access Article
Open Access ArticleIn situ monitoring of the shikimate pathway: a combinatorial approach of Raman reverse stable isotope probing and hyperspectral imaging†
Jiro
Karlo
,
Aryan
Gupta
and
Surya Pratap
Singh
 *
*
Department of Biosciences and Bioengineering, Indian Institute of Technology Dharwad, Dharwad, Karnataka 580011, India. E-mail: ssingh@iitdh.ac.in
First published on 25th March 2024
Abstract
Sensing and visualization of metabolites and metabolic pathways in situ are significant requirements for tracking their spatiotemporal dynamics in a non-destructive manner. The shikimate pathway is an important cellular mechanism that leads to the de novo synthesis of many compounds containing aromatic rings of high importance such as phenylalanine, tyrosine, and tryptophan. In this work, we present a cost-effective and extraction-free method based on the principles of stable isotope-coupled Raman spectroscopy and hyperspectral Raman imaging to monitor and visualize the activity of the shikimate pathway. We also demonstrated the applicability of this approach for nascent aromatic amino acid localization and tracking turnover dynamics in both prokaryotic and eukaryotic model systems. This method can emerge as a promising tool for both qualitative and semi-quantitative in situ metabolomics, contributing to a better understanding of aromatic ring-containing metabolite dynamics across various organisms.
Introduction
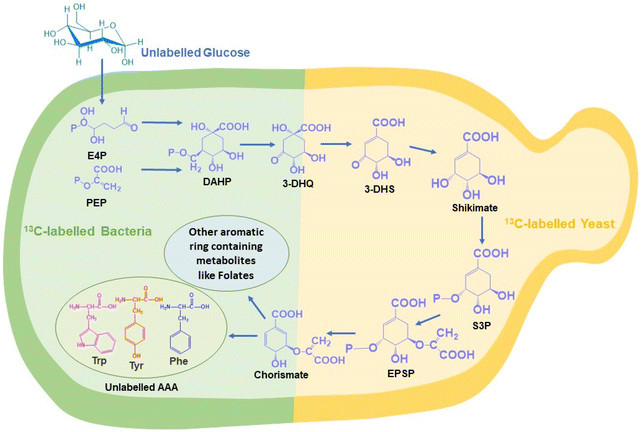
The shikimate pathway is the bridge between carbon source metabolism and biosynthesis of many aromatic ring-containing metabolites. The major products of this pathway are three aromatic amino acids, i.e., phenylalanine, tyrosine and tryptophan. This pathway is present only in plants and microbes, like bacteria and yeast, but never in animals.1,2 It not only produces aromatic amino acids but also many other important aromatic intermediates and aromatic biomolecules such as chorismate, dehydroshikimate, folates, salicylic acid, resveratrol, flavonoids, dopamine, and alkaloids. These molecules have a wide range of applications in developing value-added aromatic compounds for the food, agrochemical, fuel additive, cosmetic, dye, pharmaceutical and other industries (Fig. 1).2,3 Over 20% of all fixed carbon flows through this pathway.4 The shikimate pathway also plays an important role mediating the production of antiviral drugs such as oseltamivir.5 The quinoid core found in benzoquinone and ubiquinone is a derivative of the shikimate pathway in bacteria and yeast.6 Novel approaches that can help one to understand or visualize metabolites and metabolic pathways in situ (in their native position) or in vivo (inside the living system) are still at the inception stage. This is primarily attributed to the metabolome complexity and dynamic nature of biomolecules.The common conventional approaches for profiling metabolites and monitoring metabolic pathways are gas chromatography-mass spectrometry and liquid chromatography-mass spectrometry (GC-MS and LC-MS).4–8 These are highly efficient and sensitive methods for both qualitative and quantitative analyses of the entire pathway at each step until the final product is delivered. However, these approaches require the extraction of metabolites and do not provide information related to the dynamic nature of metabolic pathways.7,8 Fluorescence spectroscopy-based methods are an alternative but it involves the introduction of foreign molecules, which can perturb the natural metabolic processes.7 Raman spectroscopy is an efficient label-free, reagent-free, and non-destructive analytical tool for analysing biomolecules in situ. Stable isotope probes, such as 2H, 13C, 15N, and 18O, can be used as tags for analysing the de novo biosynthesis of different biomolecules simultaneously using Raman spectroscopy. When the parent atom is replaced by its heavier isotope, the reduced mass increases. As the relationship between the wavenumber and the square root of reduced mass is not directly proportional, due to the isotopic effect, the spectral signal shifts to a lesser wavenumber known as the red shift of the peak. However, this approach is limited by the fact that stable isotope sources are not abundantly available and their relative cost is higher when compared to that of their unlabelled counterparts. To overcome this, we have used the strategy of Raman reverse stable isotope probing (RrSIP). A reverse isotopic effect is observed when the heavier isotope is replaced by the lighter isotope resulting in the reduction of reduced mass and a shift towards a higher wavenumber (blue shift).9,10
In this study, for the first time, we report in situ monitoring of the activity of the shikimate pathway at a community level in Escherichia coli (a prokaryotic microbe) and at a single-cell level in Saccharomyces cerevisiae (a eukaryotic microbe). The primary reason for using these two model systems is their well-known utility in industrial fermentation and a wide range of biotechnological applications. We here report the visualization of shikimate pathway activity via sensing the spatial distribution of nascent phenylalanine peaks through hyperspectral Raman imaging combined with RrSIP with time. We also performed multivariate curve resolution (MCR) to extract each component with interpretable spectra from a complex mixture of spectral data. This approach of Raman spectroscopy and imaging combined with RrSIP can be a promising analytical tool to explore different metabolic pathways involving aromatic rings.
Experimental methods
Microbial rSIP culture conditions and sample preparation
In our study, the bacterial strain Escherichia coli K12 and yeast strain Saccharomyces cerevisiae were used. For the primary culture, a single colony was inoculated in a carbon-free growth medium with 5 g L−1 of uniformly labelled 13C glucose (Cambridge Isotope Laboratories) as the sole carbon source. This secondary broth culture was incubated overnight according to standard microbial culture protocols. An aliquot of the 13C-labelled microbial cells was taken and centrifuged at 7000g. Subsequently, the cells were washed twice with phosphate-buffered saline (PBS, Sisco Research Laboratories) to eliminate any residual traces of the 13C-labelled growth medium. Immediately after the washing steps, the cell pellets were reintroduced into the growth medium, this time with 5 g L−1 of unlabelled (12C) glucose (Sigma-Aldrich) as the exclusive carbon source. This culture was then subjected to standard incubation protocols. An inhibition assay was conducted using glyphosate, a well-known shikimate pathway inhibitor (Sigma-Aldrich). A working concentration of 700 μg mL−1 glyphosate was introduced into M9 minimal media containing 5 g L−1 unlabelled carbon before inoculating the 13C-labelled cells.11 Following a similar procedure to that with the mentioned glyphosate treatment, a supplement of three aromatic amino acids of concentration 30 μg mL−1 was added to the growth medium. Subsequently, growth-monitoring experiments were performed for all cultures using the optical density method at 600 nm (OD600). The values were recorded at various time points starting from 0 hour to 24 hours as shown in Fig. S1 and S2.†Raman spectral measurement and analysis
Aliquots of microbial samples were collected from both control and treated groups at various time intervals, i.e. 0, 1, 2, 4, 8, 16 and 24 hours. Appropriate dilutions were performed to maintain approximately equal numbers of cells. The sample was immediately centrifuged at 7000 rpm for 5 min, followed by washing twice with PBS. A similar procedure was performed with yeast samples. The cells were placed on a clean ethanol-washed slide and Raman spectra were acquired using a WITec confocal micro-Raman spectrometer, equipped with a 532 nm laser source and 100× objective lens with a numerical aperture of 0.8. A single spectrum was averaged over 5 accumulations with 10-second exposure time in the spectral range of 200 to 3200 cm−1 with 600 gr mm−1 diffraction grating. The Raman spectral data were pre-processed using MATLAB R2021B. Smoothening was applied using the Savitzky Golay filter; a 5th order polynomial was used for baseline correction and unit normalization was performed. All spectral plots and figures including two dimensional correlation maps were generated using Origin Pro 2023b software.Single-cell Raman imaging and MCR analysis
For Raman imaging, yeast cells were collected using the procedure mentioned earlier. The cells were diluted to an optimal concentration to make sure single cells can be obtained. After dilution, 10 μL was spread on an ethanol-washed calcium fluoride substrate and was subjected to Raman imaging using the above mentioned WITec confocal micro-Raman spectrometer. The step size of the scan was maintained at 0.33 microns with 5 s laser exposure time. The Raman hyperspectral data were pre-processed and the Raman image was generated using in-house scripts in MATLAB 2021B software. Multivariate curve resolution analysis (MCR) was performed using RamApp.12 MCR analysis was initiated to resolve the complex spectral matrix into two components while these two MCR components retain their interpretable spectral information.Results and discussion
In situ sensing of shikimate pathway activity and monitoring the de novo synthesis of phenylalanine using Raman spectroscopy
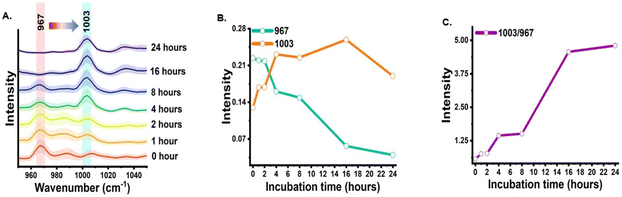
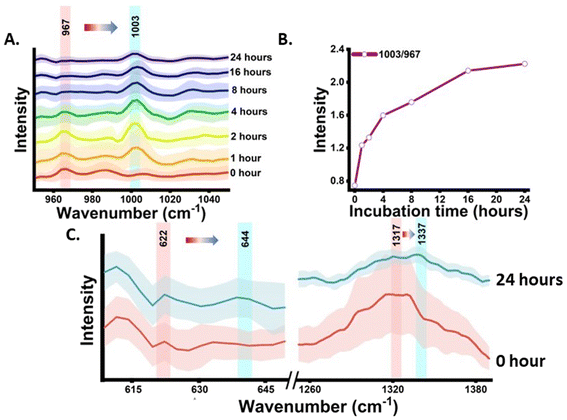
The shikimate pathway is directly related to the biosynthesis of aromatic amino acids. Therefore, the signal for phenylalanine was taken as the representative signature peak. The signal shows a strong unique peak in the Raman spectra of cells at 1003 cm−1 for unlabelled phenylalanine and at 967 cm−1 for 13C-labelled phenylalanine, as detailed in Table 1. This peak arises due to the ring-breathing vibration. In the Raman reverse stable isotope probing approach, the 13C-labelled E. coli cells were allowed to grow in unlabelled media and their Raman spectra at various time points of 0, 1, 2, 4, 8, 16 and 24 h post-incubation were acquired, as shown in Fig. 2(A). Initially, at 0 h, the peak intensity at 967 cm−1 is high and very low at 1003 cm−1. This indicates that nearly the entire aromatic amino acid pool inside cells is 13C-labelled. However, with the increase in the incubation time, we observed dynamic changes in the peak intensity. The peak intensity at 967 cm−1 gradually decreases while simultaneously there is a gradual increase in peak intensity at 1003 cm−1. At 1 and 2 h, we observed a small signal at 1003 cm−1; this blue shift in the phenylalanine peak indicates the incorporation of 12C into the newly synthesized phenylalanine from the supplemented unlabelled glucose, which suggests positive shikimate pathway activity in the E. coli cells. At 4 h post-incubation, we observed that the intensity of both bands is nearly equal. At the later time points of 8, 16 and 24 h, a significant decrease in the peak intensity at 967 cm−1 and an exponential increase in the peak intensity at 1003 cm−1 can be observed. This blue shift of the peak with time can be interpreted as the old pool of aromatic amino acids becoming moderately substituted by the newly synthesised aromatic amino acids via the active shikimate pathway in the cells. This can be better visualized in the line plots shown in Fig. 2(B) and (C), which represent dynamic intensity vs. time relationships showing the incorporation of 12C into the phenylalanine pool of the cells. This enables us to track the carbon flow from the supplement carbon source to the aromatic amino acid through the shikimate pathway. The time-dependent change in the ratiometric intensity of 1003/967 cm−1 is plotted in Fig. 2(C), which shows the turnover of the nascent unlabelled phenylalanine in the cells.| 13C-labelled peaks (cm−1) | Unlabelled peaks (cm−1) | Assignment | Significance | Ref. |
|---|---|---|---|---|
| 622 | 644 | Tyrosine | 622 cm−1 is overlapped by unlabelled phenylalanine and 13C-labelled tyrosine. Possible qualitative marker | 15–18 |
| 967 | 1003 | Phenylalanine | Evident qualitative and quasi-quantitative marker | 10, 15–17, 19–21 |
| 1317 | 1337 | Tryptophan | Band overlap by adenine and guanine. Possible qualitative marker | 15–17 |
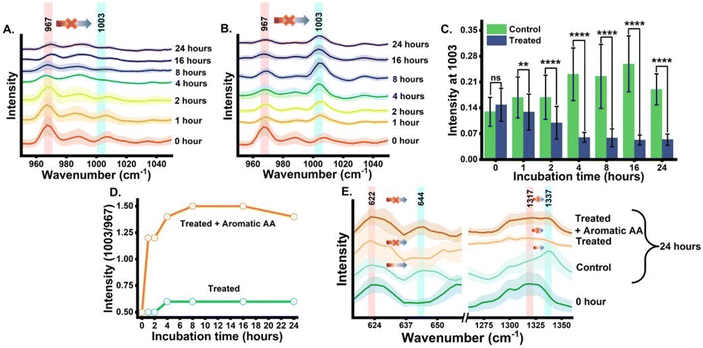
The next analysis was completed to establish and validate that the origin of the blue-shifted phenylalanine reference peak is via the shikimate pathway only. To achieve this, we monitored the growth of cells with and without glyphosate treatment. Glyphosate (N-[phosphonomethyl]-glycine) is a well-known inhibitor of the shikimate pathway in E. coli.13,14 To check the inhibitory action of glyphosate, OD600 values from the treated culture were plotted against the incubation time, as shown in Fig. S1.† Post-treatment, we can see that the OD600 values do not increase with the incubation time, confirming the inhibitory effect of glyphosate on the phenylalanine synthesis of E. coli. Post-treatment Raman spectra from the cells were recorded at time points of 0, 1, 2, 4, 8, 16 and 24 h post-incubation and are shown in Fig. 3(A). As can be seen, the blue shift of the phenylalanine peak stopped due to the disruption in shikimate pathway activity. There are negligible changes in peak intensity at 1003 cm−1 with the increase in incubation time, which is expected to derive from the nascent phenylalanine when compared with the peak intensity of the untreated group, as shown in Fig. 2(A). This observation suggests that the assigned peak at 1003 cm−1 indeed has its origin from shikimate pathway activity.
Furthermore, we also performed a complementary analysis by reversing the inhibitory effect of glyphosate on the shikimate pathway of the cells. The glyphosate-treated growth medium was exogenously supplemented with the three unlabelled aromatic amino acids that are essential for cell survivability. The Raman spectra of all three supplemented unlabelled aromatic amino acids were recorded (Fig. S3†) to ensure the purity and peaks of interest. The Raman spectra from the cells were recorded at various time points of 0, 1, 2, 4, 8, 16 and 24 h, as shown in Fig. 3(B). We successfully monitored the incorporation of exogenous aromatic amino acids from the growth media into the glyphosate-treated cells by Raman spectroscopy in an extraction-free manner. At 1 h, we noticed a rapid increment of intensity at 1003 cm−1 when compared to that of the untreated cells. However, with the incubation time, the intensity increases gradually. It was seen that this increase in the peak at 1003 cm−1 was not accompanied by the peak signal diminishing at 967 cm−1 after 1 h and so on. This indicates that there is no blue shift in the phenylalanine band confirming the inhibited shikimate pathway, despite the signal at 1003 cm−1 being from supplemented phenylalanine. This validates the fact that the peak at 1003 cm−1 from the cells, either the phenylalanine is de novo synthesized or exogenously supplied is indeed derived from the unlabelled phenylalanine only. Furthermore, to verify the statistical significance of the Raman peak intensity difference between treated and untreated cell groups for nascent phenylalanine at 1003 cm−1, we performed a Student's t-test (Welch corrected). In Fig. 3(C), the significance (p-value ≤0.05) of the intensity difference can be observed from the 1 h incubation time. Likewise, the dynamic ratiometric intensity difference was calculated as seen in the line plot of Fig. 3(D) for the treated and treated and supplemented with aromatic amino acid groups.
Tracking shikimate pathway dynamics using tyrosine and tryptophan Raman spectral markers and 2D correlation spectroscopy
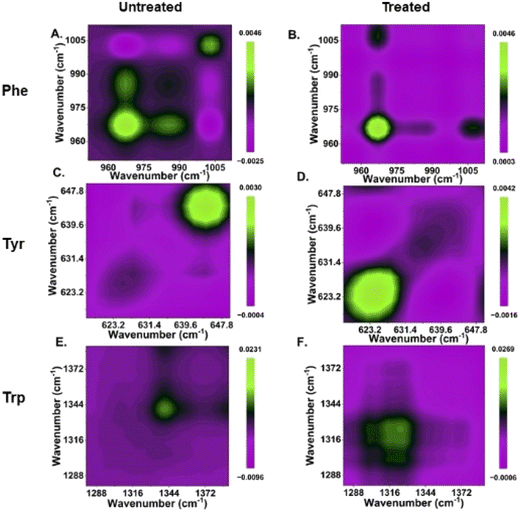
The phenylalanine position is known to be represented by a sharp single peak.15 Therefore, phenylalanine peaks at 967 and 1003 cm−1 can act as both qualitative and quasi-quantitative markers for studying shikimate pathway dynamics in a non-destructive manner. In addition to this, peaks assigned to other major products of the shikimate pathway, such as tyrosine and tryptophan, can also act as qualitative markers as shown in Fig. 3(E). These peaks cannot be preferred for semi-quantitative analysis as they are influenced by other biomolecular Raman bands, as shown in Table 1. In previous studies, the peaks at around 622 and 1317 cm−1 have been assigned to 13C-labelled tyrosine and tryptophan, respectively, whereas the peaks at around 644 and 1337 cm−1 have been assigned to unlabelled tyrosine and tryptophan, respectively, as shown in Table 1.15,17 At 0 h, we observed the intensified signals at 622 and 1317 cm−1 and negligible peak intensity at 644 and 1337 cm−1. However, after 24 h of incubation, we observed intensified signals from the new peaks at 644 and 1337 cm−1, giving us insight into the active shikimate pathway qualitatively. Unlike 13C-labelled phenylalanine peaks, 13C-labelled tyrosine and tryptophan peaks do not change much. This is because during the reverse labelling process, the band at 622 cm−1 is also overlapped by the band for the newly synthesized unlabelled phenylalanine. Likewise, the band at 1317 cm−1 is contributed to by other biomolecular bands such as those for adenine and guanine (Table 1). Furthermore, the inhibition of the shikimate pathway by glyphosate treatment also shows a negative effect on the blue shift of these bands, and the supplement of aromatic amino acids exogenously shows the reversal effect. This further confirms that tyrosine and tryptophan can also act as possible qualitative Raman spectral markers for monitoring the shikimate pathway in situ.Furthermore, to resolve and validate the time-dependent overlapping Raman peak dynamics, we performed a two-dimensional correlation analysis. A synchronous correlation map was generated from the mean Raman spectra at various time points, as shown in Fig. 4. The auto peak along the diagonal corresponds to the dynamics of that peak with time. We observed a strong auto peak in the correlation map of the untreated group at the unlabelled position of nascent phenylalanine, tyrosine, and tryptophan. This corresponds to the fact that these peaks have changed over time due to the active shikimate pathway resulting in the turnover of nascent aromatic acids. In contrast, we observed strong auto peaks at 967, 1317 and 622 cm−1 in the treated group, which supports the observation that the 13C-labelled peak has undergone significant changes with time. Furthermore, we observed a negative cross peak for 13C-labelled and an unlabelled aromatic amino acid peak position in the untreated group. This shows the negative correlation between two peak positions, that is, the dynamics is in opposite directions, which supports the fact that shikimate pathway activity takes place in cells. Additionally, to verify if the outcomes of the 2D correlation study are specific for the spectral range, we also performed the same analysis in the 600–1400 cm−1 range, which includes all the three bands. The results are shown in the ESI, Fig. S4.† Due to the presence of multiple variables (spectral bands) in this range, the analysis did not provide any interpretable results. Therefore, the outcome of the 2D correlation analysis in the present study is specific for bands involved in the dynamics interpretation of the shikimate pathway.
In situ monitoring and visualization of de novo phenylalanine synthesis at the single-cell level in the eukaryotic system
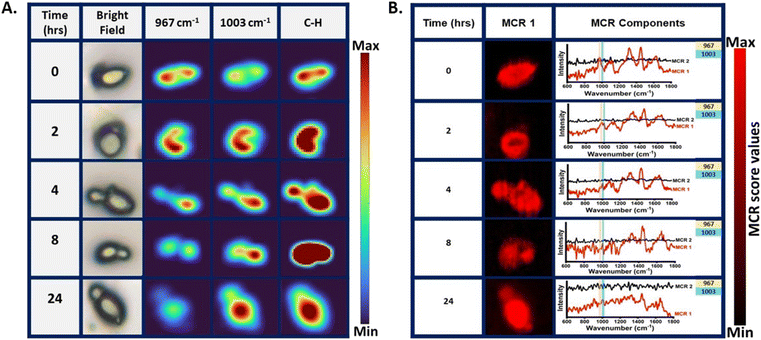
After demonstrating the potential of Raman spectroscopy in monitoring shikimate pathway activity using nascent phenylalanine peak dynamics at 1003 cm−1 at the community level in microbes, we monitored the shikimate pathway at the single-cell level using Saccharomyces cerevisiae by employing the same reverse stable isotope probing strategy. Raman spectra were recorded from a single yeast cell at various incubation time points of 0, 1, 2, 4, 8, 16 and 24 h as shown in Fig. 5(A). At the initial time point, the peak intensity at 967 cm−1 was high while that at 1003 cm−1 was low, indicating a 13C-labelled aromatic amino acid pool inside the cell. Over time, we observed that the peak at 1003 cm−1 intensifies through a blue shift of the peak at 967 cm−1. This temporal shift implies that Raman spectroscopy can sense positive shikimate pathway activity at the single-cell level and the gradual turnover of nascent phenylalanine replacing the older pool of phenylalanine can be objectively monitored. The time-dependent increase in the ratiometric intensity plot depicted in Fig. 5(B) illustrates the incorporation of 12C into the nascent phenylalanine and the relative turnover process over time at the single-cell level. Furthermore, we monitored the qualitative spectral marker of shikimate pathway dynamics, which is the peak assigned to tyrosine and tryptophan, as shown in Fig. 5(C). At 0 h, we observed negligible peak intensity at 644 cm−1 and 1337 cm−1 a low intensity peak and its changes with time suggesting the turnover of the newly synthesized tyrosine and tryptophan from the shikimate pathway.Raman hyperspectral imaging combines spectroscopy with imaging, which provides information not only about the biomolecular components but also their spatial distribution within the subcellular resolution at different pixel positions of the Raman image. We mapped the peak intensity of phenylalanine at 967 and 1003 cm−1 from the single S. cerevisiae cell at various time points of 0, 2, 4, 8 and 24 h, as shown in Fig. 6(A). The time points for Raman imaging were decided by analysing the notable changes in the mean Raman spectra of the single cell, as shown in Fig. 5(A). The bright field and the corresponding Raman image of the C–H band show the cellular boundary. The Raman image of the cell at 0 h shows a highly intensified image at 967 cm−1 with respect to that at 1003 cm−1, indicating the initial phenylalanine pool to be 13C-labelled. This distribution of phenylalanine comes from the free phenylalanine of the cytoplasm as well as the protein structures. The Raman image of the cell at 2 h post-inoculation shows the nearly equivalent spatial intensity distribution of phenylalanine at 967 and 1003 cm−1. This confirms positive shikimate pathway activity and nearly equivalent distribution of the 13C-labelled and unlabelled phenylalanine in the cellular phenylalanine pool, as shown in the mean Raman spectra from a single cell in Fig. 5(A). Likewise, with the increase in the incubation time, we see that the spatial distribution of the 13C-labelled phenylalanine diminishes while that of the unlabelled nascent phenylalanine increases confirming the visualization of the phenylalanine turnover dynamics. We also noticed that a newly formed bud of the budding yeast showed an intensified signal at 1003 cm−1, giving insight into the carbon flow and newly synthesized aromatic amino acids in the bud connected to the mother cell. The Raman image was constructed targeting the distribution of the single peak intensity in each pixel. Multivariate curve resolution (MCR) was performed to resolve the complex mixture of spectral data of the Raman image into various components by conserving the chemical meaning in each component. We observed that the two components had most of the information of interest for understanding the biomolecular spectral pattern. For quenching out the biomolecular Raman spectral pattern from the mixed complex hyperspectral Raman data, MCR analysis with two components was performed. Among the two MCR components under study, one component had phenylalanine and other biomolecular information of the cell of interest and the other one had a negligibly interpretable biomolecular spectral pattern that consists of background information, as shown in Fig. 6(B). An MCR 1 component map at various time points showed a similar cell boundary and pattern when compared to the bright field images shown in Fig. 6(A). The corresponding MCR1 component loading plot can be seen with the phenylalanine peak intensity marked in Fig. 6(B). We can see the dynamics in the phenylalanine bands at 967 cm−1 and 1003 cm−1 with time at the single-cell level.
Overall findings suggest that Raman spectroscopy and hyperspectral imaging combined with reverse stable isotope probing could serve as an extraction-free and non-destructive approach for metabolic pathway monitoring. Previously, Weng J. et al. have reported high single-cell phenotypic heterogeneity of carbon uptake and hydrogen uptake in carotenoids using Raman stable isotope probing (RSIP).22 Li J. et al. using Raman active cell sorting SIP identified, sorted and isolated the active toluene degrader single cell Pigmentiphaga.23 Li Menqui et al. have shown the potential of RSIP using phenylalanine and thymine as Raman spectral biomarkers to track the carbon flow in the predator–prey model at the single-cell level.20 Although these studies explored the possibilities of RSIP, we considered RrSIP a cost-effective approach with respect to RSIP, as preparing the labelled cell for inoculation, and growing in unlabelled media will require an extremely low amount of stable isotope. Additionally, employing RrSIP reveals a blue shift in the Raman spectra, signifying the migration of labelled metabolite peaks toward the position of unlabelled metabolite peaks over time. This shift can be readily confirmed, as the positions of unlabelled bands in the cell spectra have been thoroughly investigated in previous studies.10,21 In one of our recent studies, we reported the utility of this approach for global proteome monitoring.10 Due to the easy availability and low cost of unlabelled supplements, it is an efficient approach for requirements to working with large growth medium volumes. This offers an attractive opportunity in the future for sensing and visualizing metabolites and metabolic pathways inside cells in situ or in vivo. As cells have carbon everywhere at the genome, proteome, and metabolome levels, carbon-based RrSIP can give insight into sensing the de novo synthesis and turnover of different metabolites without any requirement for extraction. In our work, we have described its potential in prokaryotic and eukaryotic model systems. A remarkable finding in this work is the observation that the phenylalanine peaks at 967 and 1003 cm−1 are established as Raman metabolic spectral markers for studying shikimate pathway activity, de novo phenylalanine synthesis, and the turnover dynamics of nascent phenylalanine over time. We have verified that the signal arising from newly synthesized unlabelled phenylalanine originates solely from the shikimate pathway via an inhibition assay and bypasses the effect of inhibition with exogenous supplements.
One of the multiple beneficial attributes of this approach is that along with the 1D temporal spectral information, spatial information can also be derived using Raman hyperspectral imaging. This provides an image representation of the sample with different pseudo-colours that correspond to the intensity of the biomolecular peak for visualization and quantification. Along with the identification, it also gives information about the heterogeneity of the scanned area without the requirement of endogenous and exogenous labels or markers, unlike conventional fluorescence-based methods. The endogenous labels like fluorescent proteins and exogenous labels like dyes (Nile red and BIODPY) or quantum dots can interfere with the metabolic activity or can cause photobleaching or phototoxicity.7,24–29 Raman hyperspectral imaging combined with multivariate curve resolution validates the sensitivity and specificity of our approach. Visualizing the spatial distribution and extracting the spectral pattern of phenylalanine from intricate spectral data not only contributes to the validation of our study but also enhances our ability to explore shikimate pathway activity at various incubation times in situ. Furthermore, we have identified peak positions linked to tyrosine and tryptophan, which can serve as potential qualitative markers. This identification has been further corroborated through the application of two-dimensional correlation spectroscopy, adding validation to our findings.
Conclusions
Overall, the findings of our study demonstrate the immense potential of this combinatorial approach in objectively unravelling the dynamics and complexities of metabolic pathways. The presented approach is cost-effective and has potential to be translated into a preferred method for monitoring the production of commercially or medically relevant metabolites and byproducts.Author contributions
Jiro Karlo – conceptualization, investigation, methodology, formal analysis, software, visualization, and writing – original draft. Aryan Gupta – investigation, methodology, and formal analysis. Surya Pratap Singh – resources, funding acquisition, supervision, and writing – review & editing.Conflicts of interest
The authors have no conflicts of interest to declare.Acknowledgements
This work was carried out under research grant project no (37/1739/23/EMR-II) supported by the Council of Scientific and Industrial Research (CSIR), Government of India and project no. IIRP-2023-1734 from the Indian Council of Medical Research (ICMR), Government of India.References
- K. M. Herrmann and L. M. Weaver, Annu. Rev. Plant Physiol. Plant Mol. Biol., 1999, 50, 473–503 CrossRef CAS PubMed.
- H. Liu, Q. Xiao, X. Wu, H. Ma, J. Li, X. Guo, Z. Liu, Y. Zhang and Y. Luo, Commun. Chem., 2023, 6, 152 CrossRef CAS PubMed.
- J.-H. Lee and V. F. Wendisch, J. Biotechnol., 2017, 257, 211–221 CrossRef CAS PubMed.
- K. M. Herrmann, Plant Cell, 1995, 7, 907–919 CrossRef CAS.
- H.-N. Lee, S.-Y. Seo, H.-J. Kim, J.-H. Park, E. Park, S.-S. Choi, S. J. Lee and E.-S. Kim, J. Ind. Microbiol. Biotechnol., 2021, 48(9–10), kuab043 CrossRef CAS PubMed.
- R. Meganathan, FEMS Microbiol. Lett., 2001, 203, 131–139 CrossRef CAS PubMed.
- C. Lima, H. Muhamadali and R. Goodacre, Annu. Rev. Anal. Chem., 2021, 14, 323–345 CrossRef CAS PubMed.
- J. Karlo, R. Prasad and S. P. Singh, J. Agric. Food Res., 2023, 11, 100482 CAS.
- Y. Wang, W. E. Huang, L. Cui and M. Wagner, Curr. Opin. Biotechnol, 2016, 41, 34–42 CrossRef CAS PubMed.
- J. Karlo, A. K. Dhillon, S. Siddhanta and S. P. Singh, J. Biophotonics, 2024, 17(2), e202300341 CrossRef CAS PubMed.
- V. Ramachandran, V. Ramachandran, R. Singh, S. Yang, T. Ragadeepthi, S. Mohapatra, S. Khandelwal and S. Patel, Advances and Applications in Bioinformatics and Chemistry, 2013, pp. 1 Search PubMed.
- R. Vanna, G. De Poli, A. Masella, E. Broggio, D. Polli and M. Bregonzio, RamApp Search PubMed.
- D. Patriarcheas, T. Momtareen and J. E. G. Gallagher, Curr. Genet., 2023, 69, 203–212 CrossRef CAS PubMed.
- B. Ely and J. Pittard, J. Bacteriol., 1979, 138, 933–943 CrossRef CAS PubMed.
- F. Weber, T. Zaliznyak, V. P. Edgcomb and G. T. Taylor, Appl. Environ. Microbiol., 2021, 87(22), e0146021 CrossRef PubMed.
- F. S. de Siqueira e Oliveira, H. E. Giana and L. Silveira, J. Biomed. Opt., 2012, 17, 107004 CrossRef PubMed.
- S. Sil, R. Mukherjee, N. S. Kumar, S. Aravind and U. K. Singh, Def. Life Sci. J., 2017, 2, 435 CrossRef.
- C. Fan, Z. Hu, A. Mustapha and M. Lin, Appl. Microbiol. Biotechnol., 2011, 92, 1053–1061 CrossRef CAS PubMed.
- J. Karlo, A. K. Dhillon, S. Siddhanta and S. P. Singh, J. Biophotonics, 2023, 16(4), e202200341 CrossRef CAS PubMed.
- M. Li, W. E. Huang, C. M. Gibson, P. W. Fowler and A. Jousset, Anal. Chem., 2013, 85, 1642–1649 CrossRef CAS PubMed.
- Y. Wang, Y. Song, Y. Tao, H. Muhamadali, R. Goodacre, N.-Y. Zhou, G. M. Preston, J. Xu and W. E. Huang, Anal. Chem., 2016, 88, 9443–9450 CrossRef CAS PubMed.
- J. Weng, K. Müller, O. Morgaienko, M. Elsner and N. P. Ivleva, Analyst, 2023, 148, 128–136 RSC.
- J. Li, D. Zhang, C. Luo, B. Li and G. Zhang, Environ. Sci. Technol., 2023, 57, 17087–17098 CrossRef CAS PubMed.
- D. Fu, Curr. Opin. Chem. Biol., 2017, 39, 24–31 CrossRef CAS PubMed.
- G. Barzan, A. Sacco, L. Mandrile, A. M. Giovannozzi, C. Portesi and A. M. Rossi, Appl. Sci., 2021, 11, 3409 CrossRef CAS.
- M. Hosokawa, M. Ando, S. Mukai, K. Osada, T. Yoshino, H. Hamaguchi and T. Tanaka, Anal. Chem., 2014, 86, 8224–8230 CrossRef CAS PubMed.
- C. Kallepitis, M. S. Bergholt, M. M. Mazo, V. Leonardo, S. C. Skaalure, S. A. Maynard and M. M. Stevens, Nat. Commun., 2017, 8, 14843 CrossRef CAS PubMed.
- H. Noothalapati, T. Sasaki, T. Kaino, M. Kawamukai, M. Ando, H. Hamaguchi and T. Yamamoto, Sci. Rep., 2016, 6, 27789 CrossRef CAS PubMed.
- H. Noothalapati, K. Iwasaki and T. Yamamoto, Anal. Sci., 2017, 33, 15–22 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4an00203b |
| This journal is © The Royal Society of Chemistry 2024 |