Open Access Article
Open Access ArticleSelective elimination of enterovirus genotypes by activated sludge and chlorination†
Odile
Larivé
a,
Shotaro
Torii
 a,
Nicolas
Derlon
a,
Nicolas
Derlon
 b and
Tamar
Kohn
b and
Tamar
Kohn
 *a
*a
aLaboratory of Environmental Chemistry, School of Architecture, Civil and Environmental Engineering, École Polytechnique Fédérale de Lausanne (EPFL), CH-1015 Lausanne, Switzerland. E-mail: tamar.kohn@epfl.ch; Tel: +41 21 69 30891
bEawag, Swiss Federal Institute of Aquatic Science and Technology, Überlandstrasse 133, CH-8600, Dübendorf, Switzerland
First published on 11th May 2023
Abstract
Enteroviruses, which are commonly circulating viruses shed in the stool, are released into the sewage system and only partially removed or inactivated, resulting in the discharge of infectious enteroviruses into the environment. Activated sludge and chlorination remove or inactivate enterovirus genotypes to different extents, and thus have the potential to shape the population that will be discharged. The goal of this study was to evaluate how activated sludge and chlorination treatment shape an enterovirus population at the genotype level, using a population of eight genotypes commonly found in sewage: CVA9, CVB1, CVB2, CVB3, CVB4, CVB5, E25, E30. Our results show that the extent of inactivation varied among genotypes, but also across sludge samples. We find that the effluent of activated sludge systems will be depleted in CVA9, CVB1 and CVB2 while E25 together with CVB3, CVB4 and CVB5 will be prevalent. Furthermore, we found that microbial inactivation was the main mechanism of infectivity loss in the activated sludge, while adsorption to the sludge flocs was not significant. During effluent chlorination, we also observed that CVB5, CVB3 and to a lesser extent E25 were less susceptible to chlorination while E30 was readily inactivated, and activated sludge-derived EPS provided further protection against chlorination. This study contributes to a better understanding of the variability of sewage treatment efficacy against different enteroviruses.
Water impactEnteroviruses (EV) are important contaminants of water and wastewater. Their removal during wastewater treatment is typically measured as a bulk parameter. In this work we show that different EV genotypes exhibit different susceptibilities to wastewater treatment processes. This implies that the most persistent genotypes rather than the totality of EV should be used as indicators for treatment success. |
1. Introduction
Enteroviruses are commonly circulating viruses worldwide, that comprise 106 human genotypes. Many enteroviruses reproduce in the gastrointestinal tract and are therefore shed into the sewage, where they are subsequently subjected to various sewage treatment processes. Sewage treatment lowers the viral load, but it is well known that only partial removal of enteroviruses is achieved, implying that infectious viruses are regularly discharged into the environment.1,2Activated sludge and effluent chlorination are commonly used unit processes in sewage treatment that contribute to reductions in infectious virus loads. While the primary purpose of activated sludge treatment is the removal of nutrients and biological oxygen demand, activated sludge has also been found to reduce pathogen concentrations by 1 to 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10.1–4 The reduction in viral load as a result of chlorination in full scale treatment plants is more difficult to measure, because virus concentrations in chlorinated effluent are often very low.3 Costán-Longares et al.2 reported a reduction in the enterovirus load of 2.3 to 4.2
log10.1–4 The reduction in viral load as a result of chlorination in full scale treatment plants is more difficult to measure, because virus concentrations in chlorinated effluent are often very low.3 Costán-Longares et al.2 reported a reduction in the enterovirus load of 2.3 to 4.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 for tertiary treatment trains that include chlorination. In water reuse treatment trains which recycle sewage for potable or non-potable purposes, a 4
log10 for tertiary treatment trains that include chlorination. In water reuse treatment trains which recycle sewage for potable or non-potable purposes, a 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 virus removal credit is typically attributed to high dose chlorination.5
log10 virus removal credit is typically attributed to high dose chlorination.5
The reduction of the enterovirus load during sewage treatment is typically estimated for the total population of enteroviruses,1,3,4,6 though many different enterovirus genotypes can be found in sewage.7–9 This assumed homogeneity is due to the difficulty in measuring infectious enterovirus genotypes individually. However, exposure to activated sludge and chlorine remove or inactivate different enterovirus genotypes to differing extents.10,11 During activated sludge treatment, enteroviruses are removed by both adsorption to the sludge11,12 as well as by microbially mediated inactivation.12–14 Gerba et al.11 and Tao et al.15 found that the adsorption of enteroviruses onto sludge was genotype-specific. Similarly, inactivation in activated sludge can be expected to be genotype-specific, for several reasons. First, not all genotypes are equally susceptible to antiviral activity by microorganisms or their metabolites.16–18 Second, activated sludge operation temperatures between 4 °C and 32 °C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19 can result in differentiated inactivation for different enterovirus genotypes.20 And third, polysaccharides and peptidoglycans, two types of molecules present in extracellular polymeric substances (EPS) produced in activated sludge, have been shown to interact with the enterovirus capsid and influence its thermal and environmental stability in a genotype-specific fashion.21,22 Similar to activated sludge, the susceptibility of enteroviruses to disinfection treatment has also been shown to vary among genotypes when in the form of free chlorine,10,23–25 monochloramine26 or chlorine dioxide.27 Activated sludge and chlorination thus both have the capacity to shape the enterovirus population prior to discharge into the environment, yet the selectivity is not known.
19 can result in differentiated inactivation for different enterovirus genotypes.20 And third, polysaccharides and peptidoglycans, two types of molecules present in extracellular polymeric substances (EPS) produced in activated sludge, have been shown to interact with the enterovirus capsid and influence its thermal and environmental stability in a genotype-specific fashion.21,22 Similar to activated sludge, the susceptibility of enteroviruses to disinfection treatment has also been shown to vary among genotypes when in the form of free chlorine,10,23–25 monochloramine26 or chlorine dioxide.27 Activated sludge and chlorination thus both have the capacity to shape the enterovirus population prior to discharge into the environment, yet the selectivity is not known.
The goal of this study was to evaluate how activated sludge and chlorination treatment shape an enterovirus population at the genotype level and to identify the mechanisms involved in virus decay. To this end, a population of eight genotypes commonly found in sewage was exposed to activated sludge and free chlorine. Removal kinetics were monitored for each genotype, using ICC-RTqPCR as this method allows genotype-specific quantification of infectious enteroviruses.
2. Materials and methods
2.1 Preparation of viral stock solutions
Our starting enterovirus population was comprised of eight genotypes typically found in sewage.28 One environmental isolate was obtained for each genotype: coxsackieviruses B4 and B5 (CVB4 and CVB5) were previously isolated from Lausanne sewage (Meister et al.23 accession numbers MG845888 and MG845891) and coxsackieviruses A9, B1, B2 and B3 (CVA9, CVB1, CVB2, CVB3) and echoviruses 25 and 30 (E25 and E30) isolated from sewage were kindly provided by Soile Blomqvist and Carita Savolainen-Kopra (Finnish National Institute for Health and Welfare).Buffalo Green Monkey Kidney cells (BGMK; kindly provided by Spiez Laboratory, Switzerland) were used for the propagation of coxsackieviruses while Rhabdomyosarcoma cells (RD; ATCC CCL-136) were used to propagate echoviruses. BGMK and RD cells were grown at 37 °C in 5% CO2, on minimum essential medium (MEM, Life Technologies) and Dulbecco's modified Eagle medium (DMEM, Life Technologies) respectively, supplemented with 10% fetal bovine serum (FBS, Life Technologies) and 1% penicillin/streptomycin (Life Technologies). The maintenance media was prepared analogously to the growth media except that the FBS content was lowered to 2%.
To propagate viral stock solutions, confluent T-150 flasks (TPP™, 90150) were inoculated with one genotype in maintenance media and were incubated at 37 °C in 5% CO2 until full cytopathic effect (CPE) was observed. The flask underwent three cycles of freeze-thawing, the supernatant was collected, centrifuged 10 min at 300 × g and filtered at 0.2 μm (Sarstedt 83.1826.001). The stocks were aliquoted and stored at −20 °C. Concentrated stocks were prepared by concentrating the filtered supernatant 40- to 270-fold with Amicon centrifugal filters with a cutoff of 100 kDa (Millipore, UFC9100). The concentrated stocks were aliquoted and stored at −80 °C.
Titers of the stocks and concentrated stocks were determined by end-point dilution on their respective cell line. Specifically, the stocks were serially diluted in ten-fold series and 100 μL of each dilution was inoculated in five replicates on confluent 96 well plates (CELLSTAR® Greiner Bio-One, 7.655 180). The cytopathic effect in the wells was observed after five or six days of incubation at 37 °C and 5% CO2, and the Most Probable Number of infectious virus was calculated using the EPA MPN calculator.29
2.2 Virus enumeration
Two different methods were applied to enumerate viruses in experimental samples containing multiple enterovirus genotypes: reverse transcription RT-qPCR was used to quantify the total (infectious and inactivated) concentration of each genotype in a sample, and integrated cell culture reverse transcriptase quantitative PCR (ICC-RTqPCR) was used to enumerate the infectious concentration of each genotype.Cq values were converted to genome copy concentrations (N) by means of a standard curve. DNA standards were purchased for each genotype (GeneBlocks, Integrated DNA Technologies) and were diluted to create calibration curves ranging from 3.34 to 3.34 × 106 genome copies (gc) per μL. A calibration curve for each genotype of interest was included in each RT-qPCR run. Slopes and intercepts are given in Table S1.† The limit of quantification (LOQ) was determined for each primer pair in R using a curve-fitting method developed by Klymus et al.30 with a coefficient of variation (CV) threshold of 35%, applied to the measurement of ten replicates of the three lowest concentrations of the calibration curve. Concentrations of samples below the LOQ were set to the LOQ. RT-qPCR data were excluded if they exhibited interfering peaks in the melt analysis. Extraction blanks and no-template controls (molecular grade water) were analyzed in each run and yielded negative results. A checklist of experimental details as requested by the minimum information for publication of quantitative real-time PCR experiments (MIQE) guidelines31 is provided in Fig. S1.†
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 160) in duplicate plates. One plate was immediately processed (t0 sample), while the other plate was placed at 37 °C and 5% CO2 for 24 hours prior to processing (t24 sample). After 24 h, wells were checked to ensure that no cytopathic effect was visible. Each well was scraped and the entire content of the well was recovered and frozen at −20 °C. The frozen samples were stored for a maximum of 2.5 months and were then thawed, centrifuged for 10 min at 1000 × g and the supernatant was collected. RNA was extracted from 200 μL of the t0 and t24 supernatants, the genome copy number in each supernatant was quantified by RT-qPCR, and the increase in genome copies over 24 hours (deltagc (24 h)) was calculated. Deltagc (24 h) is proportional to the infectious concentration (C) of virus present in the sample, and calibration curves relating the deltagc (24 h) to the infectious concentration have been previously established for each genotype used in this study.28 The slope of these calibration curves was found to be reproducible, while the intercept varied depending on the age of the cells used. Here, we therefore used the calibration curve slopes determined previously (Table S2†), but determined the intercept for each ICC-RTqPCR run by analyzing a standard of known infectious concentration in parallel to the samples.
160) in duplicate plates. One plate was immediately processed (t0 sample), while the other plate was placed at 37 °C and 5% CO2 for 24 hours prior to processing (t24 sample). After 24 h, wells were checked to ensure that no cytopathic effect was visible. Each well was scraped and the entire content of the well was recovered and frozen at −20 °C. The frozen samples were stored for a maximum of 2.5 months and were then thawed, centrifuged for 10 min at 1000 × g and the supernatant was collected. RNA was extracted from 200 μL of the t0 and t24 supernatants, the genome copy number in each supernatant was quantified by RT-qPCR, and the increase in genome copies over 24 hours (deltagc (24 h)) was calculated. Deltagc (24 h) is proportional to the infectious concentration (C) of virus present in the sample, and calibration curves relating the deltagc (24 h) to the infectious concentration have been previously established for each genotype used in this study.28 The slope of these calibration curves was found to be reproducible, while the intercept varied depending on the age of the cells used. Here, we therefore used the calibration curve slopes determined previously (Table S2†), but determined the intercept for each ICC-RTqPCR run by analyzing a standard of known infectious concentration in parallel to the samples.
2.3 Activated sludge experiments
Viruses were spiked into the reactor as a mixture of all eight enteroviruses genotypes at approximately equal infectious concentration. Samples were taken at different time points to determine the physical and chemical parameters of the sludge (see section 2.3.2), as well as the virus concentrations in sludge and supernatant fractions (see section 2.3.3). At the end of each experiment, the residual sludge volume in the beaker was measured to determine the volume of evaporated liquid, and concentrations were adjusted correspondingly.
For chemical characterization, 5 to 30 mL of activated sludge were sampled, centrifuged for 10 min at 4000 × g, and the supernatant was filtered at 0.2 μm and exposed to UV for 20 min for sterilization. Samples were stored at 4 °C for a maximum of 7 days (with the exception of one instance where it was kept for 30 days) before being analyzed. Dissolved organic carbon (DOC), dissolved inorganic carbon (DIC) and total nitrogen (TN) were measured with a DOC/TOC Analyzer (Elementar Vario TOC Cube). DOC was in the range of 10–40, 10–15, 5–25 mg C L−1, in sludges 1, 2 and 3 respectively. Detailed measurements are shown in Fig. S3.† TN was in the range of 20–45, 30–45, 30–60 mgN L−1. Ammonium (NH4+), nitrate (NO3−), nitrite (NO2−), phosphate (PO43−), and chloride (Cl−) ions were measured by ion chromatography (Thermo Scientific Intergrion HPIC). Results are shown in Fig. S4.†
To exclude confounding effects from thermal inactivation, experiments were conducted in sterile phosphate-buffered saline (PBS; pH 7.4, Gibco™, 18912014, 10 mM Na3PO4, 2.68 mM KCl, 140 mM NaCl) and infectivity was monitored by ICC-RTqPCR over the course of five days.
Next, the experiment was repeated two more times using two different new samples of sludge (sludge 2 and sludge 3), to assess the variability in genotype inactivation among sludge samples. For this purpose, samples were only taken at times 30 min and 45 h. For sludge 2, this experiment simultaneously served as the control sample in experiments to determine the role of microbial activity (section 2.3.4).
The amount of polysaccharide in the EPS concentrate was determined using the phenol-sulfuric acid protocol based on Felz et al.34 and Dubois et al.35 Briefly, 400 μL of a sample were pipetted into a cuvette (Brand™, 759115), 10 μL of 80% phenol (Sigma Aldrich, 33517) were added, followed by 1 mL of concentrated sulfuric acid (95–98%, Sigma-Aldrich, 258105). The cuvette was left 10 min at room temperature, then 10 minutes in the water bath at 25 °C. The absorbance was read at 490 nm on a UV-vis spectrophotometer (UV-2550, Shimadzu) instrument. Glucose (Acros Organics, 410955000) diluted in MilliQ water to concentration between 0.1 and 100 mg L−1 were used as a calibration curve to allow an approximate quantification.
2.4 Chlorination experiments
20 mL beakers were soaked overnight in a 200 mg L−1 sodium hypochlorite solution (Reactolab SA, Vaud, Switzerland) to remove chlorine demand. A working solution of sodium hypochlorite at a concentration between 2 and 4 mg L−1 free chlorine was prepared in 1 mM phosphate buffer. Before the experiment, the beakers were rinsed twice with MilliQ water and once with the working solution. They were then filled with 12.5 mL of the working solution and spiked with an enterovirus solution containing all eight genotypes to a concentration of approximately 1.1 × 108 MPN mL−1 per genotype. Two different enterovirus solutions were used for spiking: one solution consisted only of viruses in PBS, and the second solution contained viruses in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 PBS
1 PBS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EPS concentrate (pH adjusted to 7.4), for a final polysaccharide concentration of about 625 mg L−1. This latter matrix served to simulate the effect of activated sludge-derived EPS on the downstream chlorination process. Both PBS-free and PBS-containing enterovirus solutions were prepared in in triplicate, were incubated for 2 hours at room temperature, and were then spiked into separate reactors containing the chlorine working solution. Aliquots of 500 μL were taken after 10, 30, 45, 70 and 120 seconds and collected in a tube containing 5 μL of 5000 mg L−1 sodium thiosulfate (Sigma-Aldrich, Germany) to quench residual chlorine. The free chlorine in the reactor was measured prior and at the end of the experiment to estimate the exposure, using DPD Free Chlorine Reagent, Swiftest™ and DR300 pocket colorimeter (Hach Company, Loveland, CO). The chlorine dose (CT; mg min L−1) was calculated as the integral of free chlorine concentration over time of exposure, assuming a first-order decay of free chlorine concentration between the start and the end of the run. The initial concentration of chlorine had to be higher when the virus was in the EPS matrix, to compensate for the decrease in chlorine caused by the EPS matrix. Concentrations during each run are presented in Table S4.† We cannot exclude the formation of combined chlorine resulting from the reaction of free chlorine with ammonia residues in EPS. However, because combined chlorine it is much less efficient at inactivating enteroviruses than free chlorine,26 we here neglected its contribution to inactivation.
EPS concentrate (pH adjusted to 7.4), for a final polysaccharide concentration of about 625 mg L−1. This latter matrix served to simulate the effect of activated sludge-derived EPS on the downstream chlorination process. Both PBS-free and PBS-containing enterovirus solutions were prepared in in triplicate, were incubated for 2 hours at room temperature, and were then spiked into separate reactors containing the chlorine working solution. Aliquots of 500 μL were taken after 10, 30, 45, 70 and 120 seconds and collected in a tube containing 5 μL of 5000 mg L−1 sodium thiosulfate (Sigma-Aldrich, Germany) to quench residual chlorine. The free chlorine in the reactor was measured prior and at the end of the experiment to estimate the exposure, using DPD Free Chlorine Reagent, Swiftest™ and DR300 pocket colorimeter (Hach Company, Loveland, CO). The chlorine dose (CT; mg min L−1) was calculated as the integral of free chlorine concentration over time of exposure, assuming a first-order decay of free chlorine concentration between the start and the end of the run. The initial concentration of chlorine had to be higher when the virus was in the EPS matrix, to compensate for the decrease in chlorine caused by the EPS matrix. Concentrations during each run are presented in Table S4.† We cannot exclude the formation of combined chlorine resulting from the reaction of free chlorine with ammonia residues in EPS. However, because combined chlorine it is much less efficient at inactivating enteroviruses than free chlorine,26 we here neglected its contribution to inactivation.
2.5 Data analysis
Statistical analyses were performed in R using the packages stats36 and the segmented package.37 To determine the enterovirus decay rate constants in activated sludge, a segmented linear regression was fitted to ln-transformed decay curves of both solids and sludge using the segmented function from the segmented package, in order to determine the bending point of the curve. A linear regression was then fitted to the first segment, and the negative of the slope corresponded to the decay rate constant k(solids) (h−1) or k(supernatant) (h−1) (eqn (1) and (2)).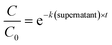 | (1) |
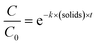 | (2) |
To determine if the inactivation by free chlorine differed depending on the genotypes, a one-way ANOVA (α = 0.05) was run, on each disinfection dose separately, using the aov function in R. It was then followed by a genotypes comparison using the TukeyHSD (α = 0.05) function. To determine if the addition of EPS had a significant effect on the decay rates, we first fitted the ln-transformed decay curves to a Chick–Watson model (eqn (3)).
 | (3) |
2.6 Data availability
Data will be made available on Zenodo upon acceptance of the manuscript.3. Results
3.1 Inactivation kinetics of different enterovirus genotypes in activated sludge reactors
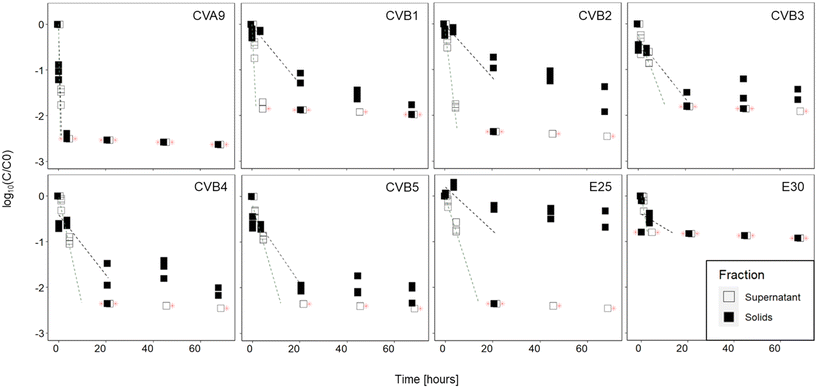
The infectious concentration of each genotype was monitored in the supernatant and in the solid fractions of activated sludge (sludge 1) over 68 hours (Fig. 2). Our results indicate that the inactivation was faster in the supernatant than in the solids of this sludge. In the supernatant, all genotypes decayed below the LOQ within 21 hours. In the solids, infectious viruses were still detected for all genotypes except CVA9 and E30 after 68 hours. The observed decay could not be attributed to thermal inactivation, because all genotypes remained stable in sterile PBS (Fig. S5†).In the supernatant, statistical analysis determined that the decay kinetics of the different genotypes varied (p < 0.001) and post-hoc analysis determined that the decays of CVA9 and CVB2 were significantly different from all other genotypes (Table S2†). CVA9 was reduced the fastest, with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 reduction in 30 min or less and CVB2 was among the fastest reduced, with a 1
log10 reduction in 30 min or less and CVB2 was among the fastest reduced, with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 reduction in less than 2.5 hours. Although CVB1 was reduced faster than CVB2, its decay rate was only statistically different from CVA9 and E25, probably due to the fact that only data up to 30 min were used in the linear regression of CVB1 (Table S5†). In the solids, infectivity loss followed two phase kinetics, with a rapid initial decay, followed by a later, slower one. A comparison of the initial decay phase (k values reported in Table 1) revealed that the decay kinetics were not significantly different across genotypes (p = 0.38), despite an important difference in k value between CVA9 and the rest of genotypes. Again the lack of statistical significance likely results from the limited number of measurable data points for CVA9 in solids (Table S5†).
log10 reduction in less than 2.5 hours. Although CVB1 was reduced faster than CVB2, its decay rate was only statistically different from CVA9 and E25, probably due to the fact that only data up to 30 min were used in the linear regression of CVB1 (Table S5†). In the solids, infectivity loss followed two phase kinetics, with a rapid initial decay, followed by a later, slower one. A comparison of the initial decay phase (k values reported in Table 1) revealed that the decay kinetics were not significantly different across genotypes (p = 0.38), despite an important difference in k value between CVA9 and the rest of genotypes. Again the lack of statistical significance likely results from the limited number of measurable data points for CVA9 in solids (Table S5†).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 reduction in infectious virus titer
log10 reduction in infectious virus titer
| Genotype | k(supernatant) (h−1) | ±95% CI | Hours to reach 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 reduction log10 reduction |
k(solids) (h−1) | ±95% CI | Hours to reach 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 reduction log10 reduction |
|---|---|---|---|---|---|---|
| CVA9 | 7.1 | 4.2 | 0.3 | 4.8 | 3.7 | 0.5 |
| CVB1 | 2.4 | 4.3 | 0.9 | 0.1 | 0.0 | 10 |
| CVB2 | 1.0 | 0.1 | 2.5 | 0.1 | 0.1 | 10 |
| CVB3 | 0.3 | 0.3 | 10 | 0.1 | 0.0 | 10 |
| CVB4 | 0.5 | 0.1 | 5 | 0.2 | 0.1 | 10 |
| CVB5 | 0.4 | 0.2 | 5 | 0.2 | 0.0 | 10 |
| E25 | 0.4 | 0.1 | 5 | 0.1 | 0.1 | 10 |
| E30 | 0.7 | 3.7 | 3.3 | 0.1 | 0.4 | 33.3 |
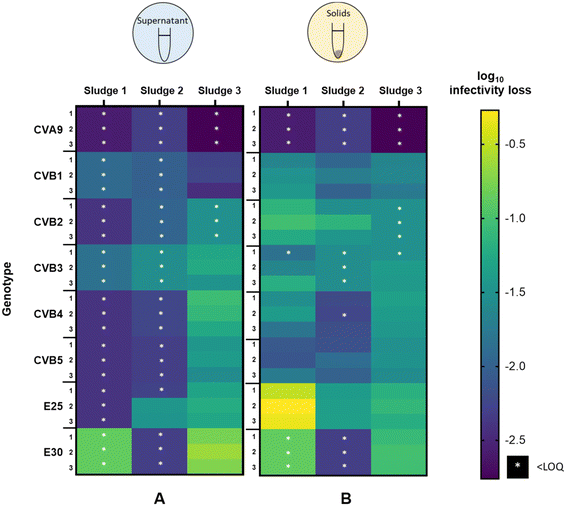
3.2 Variability in virus decay among different sludge samples
To determine if the extent of infectivity loss and the observed differences among genotypes were generalizable across sludge samples, we analyzed virus removal in two additional batches of sludge. The inactivation of each genotype after 45 hours in the supernatant and solid fractions is shown in Fig. 3A and B respectively, for all three activated sludge samples tested (sludge 1, sludge 2, sludge 3). In the supernatant (Fig. 3A), the three sludges exhibited variability in the magnitude of infectivity loss across sludges, with most genotypes in sludges 1 and 2 decaying below the LOQ within 45 h. In contrast, most genotypes retained quantifiable infectious concentrations in the supernatant of sludge 3 and displayed a wide range of infectivity loss. CVA9 was readily removed from the supernatant (>2.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10), while E30 was the most stable (<0.8
log10), while E30 was the most stable (<0.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10). In the supernatant of sludge 2, E25 was the least removed (<1.5
log10). In the supernatant of sludge 2, E25 was the least removed (<1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10) and was the only genotype that remained quantifiable after 45 h.
log10) and was the only genotype that remained quantifiable after 45 h.
In the solid fraction (Fig. 3B), infectious concentrations of most genotypes were still measurable after 45 hours. Virus decay in the solid fraction ranged from 0.3 to >2.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 in sludge 1, from 1.2 to >2.3
log10 in sludge 1, from 1.2 to >2.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 in sludge 2 and from 0.6
log10 in sludge 2 and from 0.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 to >2.8
log10 to >2.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 in sludge 3. CVA9 was readily removed in the solids of all sludges, while E25 was among the least removed. CVB5 was consistently well removed compared to the other genotypes (except CVA9), whereas the other genotypes exhibited variable relative removal across the three sludge samples. For example, E30 was the least removed in sludge 3, but was among the most removed in sludge 2.
log10 in sludge 3. CVA9 was readily removed in the solids of all sludges, while E25 was among the least removed. CVB5 was consistently well removed compared to the other genotypes (except CVA9), whereas the other genotypes exhibited variable relative removal across the three sludge samples. For example, E30 was the least removed in sludge 3, but was among the most removed in sludge 2.
3.3 Mechanism of virus removal in sludge: adsorption to solids versus microbial activity
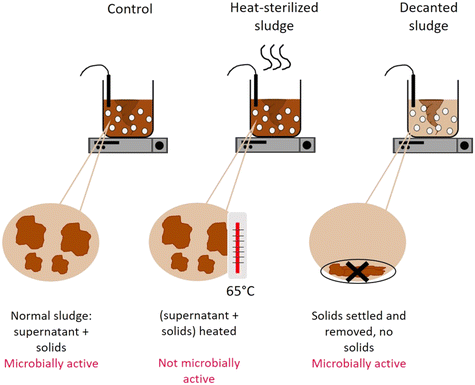
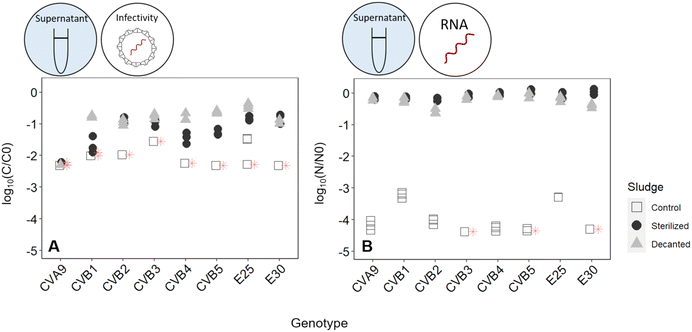
To determine the mechanism by which activated sludge leads to virus decay, we next investigated the contributions of adsorption to solids and microbial activity. To this end, virus removal was measured in the supernatant of a regular activated sludge reactor (control), in the supernatant phase after decanting and removal of the solids (decanted) and in the supernatant of heat-sterilized activated sludge (sterilized) (Fig. 1). In the decanted reactor, some microbial activity was likely maintained, while the solid fraction and hence the possibility for adsorption to particles was largely eliminated. In the sterilized reactor, biological activity was likely reduced, but the solid fraction was maintained as a site for virus adsorption. Note that heat sterilization did not alter the adsorption capacity of the solid fraction compared to the regular activated sludge after 30 minutes: in a time span that allows sufficient time for partitioning between sludge and supernatant while also limiting the extent of inactivation (30 min), the fraction of spiked virus found on the solids was similar in both the control and the heat sterilized sludge (Fig. S6A†), indicating that the amount of virus adsorbed was similar in both reactors.
Fig. 4 illustrates the infectivity loss and reduction in genome copies of each genotype in the supernatant fraction after 45 hours in the different reactors. In the control reactor, the infectious concentration of most genotypes decayed below the LOQ, corresponding to at least 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 loss in infectious titer (Fig. 4A). Simultaneously, a loss in genome copies of up to >4
log10 loss in infectious titer (Fig. 4A). Simultaneously, a loss in genome copies of up to >4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 was observed for most genotypes (Fig. 4B).
log10 was observed for most genotypes (Fig. 4B).
When the solids were removed from the reactor, inactivation was still observed, which we attribute to either microbial activity or chemical inactivation. However, it was greatly reduced compared the control reactor, with most genotypes except CVA9 experiencing around 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 loss in infectivity (Fig. 4A). The reduction in genome copies was even smaller, with a maximum loss of 0.6
log10 loss in infectivity (Fig. 4A). The reduction in genome copies was even smaller, with a maximum loss of 0.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log and 0.5
log and 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log for CVB2 and E30 respectively (Fig. 4B). Decanting the sludge thus leads to a partial suppression of virus decay, confirming that solids play an important role in virus removal. This role could either be to serve as an adsorption site for physical removal from the supernatant, or to serve as a source of microorganisms that facilitate virus inactivation.
log for CVB2 and E30 respectively (Fig. 4B). Decanting the sludge thus leads to a partial suppression of virus decay, confirming that solids play an important role in virus removal. This role could either be to serve as an adsorption site for physical removal from the supernatant, or to serve as a source of microorganisms that facilitate virus inactivation.
To further explore the role of solids, we compared experiments in the control reactor to those in heat-sterilized sludge. In the sterilized sludge, infectivity loss was again greatly reduced compared to the control reactor (Fig. 4A). Nevertheless, a residual decay in infectious viruses was observed, ranging from 0.7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 for E30 to >2.2
log10 for E30 to >2.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 for CVA9 (Fig. 4A). Furthermore, the genome copies of all genotypes were almost fully recovered in the supernatant after 45 h (Fig. 4B). Only approximately 10% of the spiked virus were adsorbed to the solids, and this fraction remained stable over the course of the experiment (Fig. S6†). This latter finding indicates that irreversible adsorption of viral particles to the solids is not an important mechanism contributing to the observed virus removal in our experimental system. Instead, as only microbially active solids cause substantial inactivation, the role of solids is likely to serve as a source of microorganisms. The slight inactivation observed in sterilized sludge may then stem from residual microbial or enzymatic activity retained in the solid after heat treatment, or from chemical inactivation by solution components of activated sludge supernatant.
log10 for CVA9 (Fig. 4A). Furthermore, the genome copies of all genotypes were almost fully recovered in the supernatant after 45 h (Fig. 4B). Only approximately 10% of the spiked virus were adsorbed to the solids, and this fraction remained stable over the course of the experiment (Fig. S6†). This latter finding indicates that irreversible adsorption of viral particles to the solids is not an important mechanism contributing to the observed virus removal in our experimental system. Instead, as only microbially active solids cause substantial inactivation, the role of solids is likely to serve as a source of microorganisms. The slight inactivation observed in sterilized sludge may then stem from residual microbial or enzymatic activity retained in the solid after heat treatment, or from chemical inactivation by solution components of activated sludge supernatant.
3.4 Inactivation of different enterovirus genotypes by chlorination
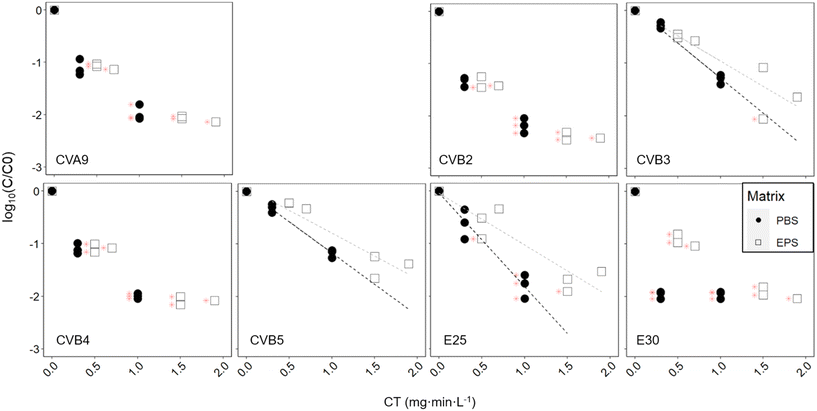
Finally, we assessed how chlorination affects the fate of different enterovirus genotypes. Exposure of the enterovirus population to chlorine resulted in a broad range of responses from the different genotypes (Fig. 5, black dots, Table 2). At a CT of 0.3 mg min L−1, E30 was inactivated below the LOQ with around 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 inactivation, while CVB3 and CVB5 were inactivated by less than 0.5
log10 inactivation, while CVB3 and CVB5 were inactivated by less than 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10. Statistical analysis determined that there were three groups of genotypes that had significantly different degrees of inactivation across groups but not within the group (Table S6†). CVB3, CVB5 and E25 were reduced the slowest; CVA9, CVB2 and CVB4 exhibited intermediate inactivation; and inactivation of E30 was most significantly faster than all the other genotypes. At a CT of 1 mg min L−1, only CVB3 and CVB5 could still be quantified by ICC-RTqPCR with between 1.1 and 1.4
log10. Statistical analysis determined that there were three groups of genotypes that had significantly different degrees of inactivation across groups but not within the group (Table S6†). CVB3, CVB5 and E25 were reduced the slowest; CVA9, CVB2 and CVB4 exhibited intermediate inactivation; and inactivation of E30 was most significantly faster than all the other genotypes. At a CT of 1 mg min L−1, only CVB3 and CVB5 could still be quantified by ICC-RTqPCR with between 1.1 and 1.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 inactivation, while all other genotypes were below the LOQ. CVB3 and CVB5 were both found to have significantly different inactivation than all other genotypes, except each other. It should be noted that the CVB3 primers are not fully selective and also measure CVB5, albeit at a much lower efficiency.28 We can therefore not exclude that the inactivation results for CVB3 were influenced by the presence of CVB5, in particular if CVB5 was present at a much higher infectious concentration.
log10 inactivation, while all other genotypes were below the LOQ. CVB3 and CVB5 were both found to have significantly different inactivation than all other genotypes, except each other. It should be noted that the CVB3 primers are not fully selective and also measure CVB5, albeit at a much lower efficiency.28 We can therefore not exclude that the inactivation results for CVB3 were influenced by the presence of CVB5, in particular if CVB5 was present at a much higher infectious concentration.
| Genotype | CT = 0.3 mg min L−1 | CT = 1 mg min L−1 | ||||
|---|---|---|---|---|---|---|
| Log10 inactivation (number of observations) | Lower 95% CI | Upper 95% CI | Log10 inactivation (number of observations) | Lower 95% CI | Upper 95% CI | |
| CVA9 | 1.1 (n = 3) | 0.9 | 1.3 | >2 (n = 3) | 1.8 | 2.2 |
| CVB2 | 1.3 (n = 3) | 1.2 | 1.4 | >2.2 (n = 3) | 2 | 2.4 |
| CVB3 | 0.3 (n = 3) | 0.2 | 0.4 | 1.3 (n = 3) | 1.2 | 1.4 |
| CVB4 | 1.1 (n = 3) | 1.0 | 1.2 | >2 (n = 3) | 1.9 | 2.1 |
| CVB5 | 0.3 (n = 3) | 0.2 | 0.4 | 1.2 (n = 3) | 1.1 | 1.3 |
| E25 | 0.6 (n = 3) | 0.3 | 1.1 | >1.8 (n = 3) | 1.6 | 2.1 |
| E30 | >2 (n = 3) | 1.9 | 2.0 | >2 (n = 3) | 1.9 | 2.0 |
| Genotype | PBS | EPS | P value ANCOVA | ||
|---|---|---|---|---|---|
| k (number of observations) | 95% CI | k (number of observations) | 95% CI | ||
| CVB3 | 3.1 (n = 9) | 0.1 | 2.2 (n = 9) | 0.3 | 0.08 |
| CVB5 | 2.7 (n = 9) | 0.1 | 2.0 (n = 9) | 0.2 | 0.05 |
| E25 | 4.1 (n = 9) | 0.4 | 2.3 (n = 9) | 0.3 | <0.01 |
4. Discussion
In this work, we investigated the effect of activated sludge treatment and chlorination on eight commonly encountered enterovirus genotypes. The variability of viral decay among genotypes and across sludge samples, and the mechanisms of decay in activated sludge are discussed here, as well as the variability of responses to chlorination.Genotype-dependence of virus decay during activated sludge treatment
Inactivation occurred both in the supernatant and in the solid fraction, and the relative degree of inactivation in each fraction was dependent on the activated sludge sample (Fig. 2, 3 and S7†). In all sludge samples tested, most enteroviruses retained infectivity for 45 hours (Fig. 3), suggesting that solids may be a source of infectious virus, either by resuspension into the supernatant or when used in land application. The overall extent of infectivity loss after 45 hours is sludge dependent (Fig. 3), as is the difference in decay observed among genotypes. Nevertheless, some consistent patterns can be identified. Sludge treatment will likely lead to a depletion of the enterovirus population in CVA9, CVB1, and CVB2 which were removed more readily than the other genotypes in the supernatant of both sludge 1 and 3, and were also removed below the LOQ in sludge 2 (Fig. 3 and Table 1). Furthermore, the enterovirus population will be enriched in E25, since it was among the least removed in sludges 1 and 2, and relatively stable in sludge 3. CVB3, CVB4 and CVB5 were also among the most stable in sludge 1 (Table 1) and sludge 3 (Fig. 3), though their relative stability could not be assessed in sludge 2.Potential drivers of variability in virus decay among sludge samples
The variability in virus decay among the three sludge samples is likely linked to differences in sludge characteristics, though it is not clear which sludge parameter drives the observed differences. A potential contributor of virus decay is TSS, because sludge with higher TSS offers more surface area for virus removal by adsorption. In this study, the TSS in all sludge samples used was within the lower range of typical activated sludge reactors (Table S3†). Counter-intuitively, the highest TSS concentration (sludge sample 3) was associated with a low overall virus decay in the supernatant (Fig. 3), suggesting that TSS is not an important driver virus decay.Instead, microbial processes exerted by the sludge may exhibit a greater influence on virus stability. In this work, sludges 1 and 2 appeared to display nitrification (Fig. S4†), while sludge sample 3 did not. This difference, however, is also unlikely to be linked to the observed virus removal, because previous literature suggests a minor effect of nitrification on virus stability.1,38 Alternatively, the bacterial production of flocs may dictate virus decay. Specifically, Kim and Unno12 reported inactivation of poliovirus only occurred in presence of floc-forming bacteria. Floc-forming ability is linked to secretion of EPS, which has been found to be a place of high concentration of hydrolytic enzymes,39–41 and may thus enhance virus inactivation. Floc production was not addressed in this study but may be an interesting parameter to investigate in future work.
Further work is needed to capture the full extent and drivers of variability in virus decay among sludges. In this respect, the current work has two important limitations. First, we only investigated three sludges, whereas a greater number of samples is needed to fully characterize inter-sludge variability in virus decay. Replicate experiments with the same sludge would furthermore allow to estimate the intra-sludge variability in viral inactivation. And second, we used a hydraulic retention time that was higher than in a regular activated sludge reactor, where it is around 4–24 hours,42 with continuous feeding of the sludge. Haun et al.43 found that hydraulic retention time and sludge retention time had an effect on virus removal. Therefore, the extent of, and variability in, inactivation in a real activated sludge reactor may differ from that observed in this study.
Mechanisms of virus decay during activated sludge treatment
Our data suggests that inactivation mediated by microorganisms in the sludge is an important cause of viral infectivity loss in the supernatant. Virus stability was enhanced if microbially active solids were decanted, indicating their importance in the viral decay. Similarly, virus stability was enhanced if solids were heat-sterilized, confirming that only microbially active solids promote inactivation. In contrast, adsorption to solids was a negligible source of virus removal, since genome copies of spiked virus could be almost fully measured in supernatant of heat-sterilized sludge. We cannot exclude additional protective mechanisms exerted by sludge heating, such as the lysis of cells and resulting increase in DOC (Fig. S3†), which may include protective EPS. Nevertheless, reducing microbial activity, either by removing the solids or heat-sterilizing them, coincides with higher virus stability, which suggests an important role of microbial activity in virus removal and inactivation by sludge. The measurement of microbial activity under the different conditions studied herein was beyond the scope of this study but will contribute to ascertain these hypotheses in future experiments.Viruses were still inactivated (though not physically removed) in the supernatant when the sludge was heat sterilized, though to a lesser extent than in the control. A similar extent of inactivation was also observed after decanting microbially active solids. This small extent of inactivation may result from residual microbial activity in the decanted and sterilized sludge. In addition, inactivation may be mediated by chemical processes induced by molecules in the supernatant. A contribution of chemical inactivation is supported by a shift in inactivation mechanism: in microbially active sludge, virus loss is accompanied by extensive loss in viral RNA (Fig. 4), indicating that both the viral capsid and genome were readily degraded. In contrast, in the absence of microbially active solids, the number of genome copies remained constant, consistent with an inactivation mechanism that mainly targets the viral capsid while protecting the RNA from complete degradation. These mechanisms remain to be confirmed in future work with additional sludges.
Adsorption onto the solids has previously been described as one of the main sources of virus removal in activated sludge.11,43 In contrast, here we show that compared to microbial activity, adsorption is a negligible process in virus decay in our experimental system. While adsorption onto the solid fraction of the sludge did occur, and while the concentration of enteroviruses on the solid fraction can be important, adsorption did not explain the extent of infectivity loss in the supernatant. Our findings are consistent with other studies which have found that adsorption was not the main virus removal mechanism during activated sludge treatment.13,44 However, in contrast to our findings, Kelly et al.,13 Knowlton and Ward14 and Kim and Unno12 found no inactivation in the supernatant after removal of solids, indicating an absence of chemical inactivation in their experimental system. Nevertheless, these authors reach the same conclusion as this work that virus infectivity loss mainly occurs in the presence of microbially active solids. Specifically, they conclude that inactivation is due to microbial activity, and that antiviral compounds are either short-lived or active only when associated with microorganisms.14 Knowlton and Ward14 furthermore found that untreated mixed liquor suspended solids caused inactivation of poliovirus 1 and release of its RNA, which is consistent with our observations that enterovirus inactivation resulted in RNA degradation (Fig. 4).
Variability of responses to chlorination
Similar to activated sludge treatment, we found that enterovirus genotypes exhibited a wide range of susceptibilities to free chlorine disinfection. Among the genotypes studied, E30 was the most chlorine susceptible, and CVB5 and CVB3 the most stable. Torii et al.,24 who compared the chlorine sensitivities of CVB3, CVB4 and CVB5 also found CVB5 to be the most chlorine resistant, and Meister et al.23 found that among environmental strains of CVB, CVB5 and CVB1 were more resistant to free chlorine than CVB4.We furthermore found that the presence of EPS was protective against chlorine treatment for CVB3, CVB5, and E25, although not significantly so for CVB3. This protection was provided by polysaccharide extracted from wet sludge at a concentration of 625 mg L−1 glucose equivalent. For the other genotypes tested, inactivation in the presence of EPS remained sufficiently rapid that they readily decayed below the LOQ, and we could therefore not determine if there was a protective effect. Waldman et al.21 found substantial protection of E30 from chlorination provided by commercial lipopolysaccharides and peptidoglycans at a concentration of 1000 mg L−1. Our experiment was in the same concentration range, but with unknown composition of the polysaccharides, and possible uncertainty in the polysaccharide concentration estimation.34 The protection was less pronounced than that observed by Waldman et al.,21 though the EPS used herein is likely more representative of the composition and concentration of EPS in sewage. Some protection to chlorination may be provided by attachment of enteroviruses to polysaccharides from the activated sludge, and consequently higher chlorine doses may be required to achieve the targeted extent of inactivation. Finally, the extent of protection will likely differ between, as shown by Waldman et al.,21 and as we see here with E25 that seems to get more protection than CVB5 and CVB3, though it would require further investigation to confirm it.
Shifts in enterovirus population composition by sequential treatment by activated sludge and chlorine
Overall, our results demonstrate that not only the enterovirus load, but also the population composition will change in a sewage treatment train that includes activated sludge and chlorination. This shaping of the population is illustrated in Fig. 6, which visualizes how these treatment processes affect the relative genotype abundance in a hypothetical enterovirus population with equal starting concentrations. Hereby, we assume a hydraulic residence time of 6 hours in the sludge with the decay rates observed in sludge 1 supernatant (k(supernatant)), and a chlorine residual of 0.3 mg min L−1. It is evident that CVA9 and CVB1 were readily depleted in activated sludge, while E25 is enriched. CVB3, CVB4 and CVB5, which have medium sensitivity to activated sludge treatment, also remain prevalent in the effluent. The prevalence of CVB3, CVB5 and E25 in the effluent was then further enhanced during chemical disinfection, due to their relative resistance to chlorine treatment, in particular in the presence of protective EPS. We emphasize that the population shifts shown in Fig. 6 may differ between sludges and treatment systems.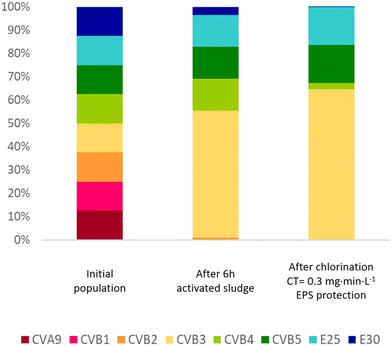 | ||
| Fig. 6 Simulation of the change in composition of an enterovirus population subjected to activated sludge treatment followed by chlorination. The initial population is assumed to consist of all eight genotypes studied at equal concentrations. Inactivation rate constants from the supernatant of sludge 1 (Table 1) were used to estimate the decay in infectious virus concentrations after 6 hours exposure to the sludge. Inactivation values listed in Table 2 were used to estimate titer reductions during chlorination. Since no data was collected for inactivation of CVB1 by free chlorine, the same inactivation as CVB5 was used, as Meister et al.23 and Liu and McGowan10 found similar inactivation by free chlorine for both genotypes. To evaluate the protection provided by EPS, inactivation at 0.3 and 1 mg min L−1 were estimated for CVB3, CVB5 and E25 based on the k values in EPS matrix (Table 3). | ||
A shift in composition may affect the susceptibility of the overall enterovirus population to both wastewater treatment and natural virus decay. While the load of enteroviruses in wastewater continues to decrease during treatment, the different unit processes continuously select for the most resistant phenotypes (here CVB3, CVB5 and E25; Fig. 6), which may lead to decreasing success in virus removal over the course of the treatment. The final enterovirus population in the wastewater effluent is then enriched in resistant genotypes, which ultimately become released into the environment. Treatment resistance has previously found to be associated with environmental persistence,45 such that genotypes released from wastewater treatment plants can be expected to also better withstand natural stressors than treatment-susceptible genotypes. Knowledge of such population shifts are important to identify the most critical serotypes to focus on with respect to assessment of public health risks arising from wastewater discharge.
While our findings for enteroviruses cannot be directly generalized to other virus families, similar population shifts may occur in other virus families of common enteric viruses containing multiple genotypes or genogroups, such as rotavirus, norovirus, adenovirus. Future studies should address the population dynamics of these other virus populations, to identify resistant genotypes or genogroups to focus on during wastewater treatment and environmental risk assessment.
5. Conclusion
This study investigated the decay of eight ubiquitous enterovirus genotypes during exposure to three different activated sludge samples, and found that microbial inactivation is the main driver of infectivity loss. To a lesser degree, viruses also underwent chemical inactivation, while adsorption to the solids was found to be a negligible contributor to virus removal. The extent of inactivation varied among genotypes and activated sludge samples. Overall, our results suggest that effluent of activated sludge will be depleted in CVA9 and CVB1, while E25 will be prevalent, along with CVB3, CVB4 and CVB5. CVB5 and CVB3, and to a lesser extent E25, were also less susceptible to chlorination compared to the other genotypes, such that they are further enriched in the final effluent. Activated sludge-derived EPS provided them further protection against chlorination, suggesting that activated sludge treatment prior to chlorination reduces the disinfection outcome for these viruses. Taken across the whole sewage treatment process, CVB3, CVB5 and E25 emerged as suitable indicators to monitor virus control.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This work was supported by the Swiss National Science Foundation (grant no. 182468). The authors thank Soile Blomqvist and Carita Savolainen-Kopra (Finnish National Institute for Health and Welfare) for providing environmental enterovirus isolates.References
- J. Hewitt, M. Leonard, G. E. Greening and G. D. Lewis, Influence of wastewater treatment process and the population size on human virus profiles in wastewater, Water Res., 2011, 45, 6267–6276 CrossRef CAS PubMed.
- A. Costán-Longares, L. Mocé-Llivina, A. Avellón, J. Jofre and F. Lucena, Occurrence and distribution of culturable enteroviruses in wastewater and surface waters of north-eastern Spain, J. Appl. Microbiol., 2008, 105, 1945–1955 CrossRef PubMed.
- Y. Qiu, B. E. Lee, N. Neumann, N. Ashbolt, S. Craik, R. Maal-Bared and X. L. Pang, Assessment of human virus removal during municipal wastewater treatment in Edmonton, Canada, J. Appl. Microbiol., 2015, 119, 1729–1739 CrossRef CAS PubMed.
- H. Katayama, E. Haramoto, K. Oguma, H. Yamashita, A. Tajima, H. Nakajima and S. Ohgaki, One-year monthly quantitative survey of noroviruses, enteroviruses, and adenoviruses in wastewater collected from six plants in Japan, Water Res., 2008, 42, 1441–1448 CrossRef CAS PubMed.
- J. A. Soller, S. E. Eftim and S. P. Nappier, Direct Potable Reuse Microbial Risk Assessment Methodology: Sensitivity Analysis and Application to State
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log Credit Allocations, Water Res., 2018, 128, 286–292 CrossRef CAS PubMed.
log Credit Allocations, Water Res., 2018, 128, 286–292 CrossRef CAS PubMed. - G. La Rosa, M. Pourshaban, M. Iaconelli and M. Muscillo, Quantitative real-time PCR of enteric viruses in influent and effluent samples from wastewater treatment plants in Italy, Ann. Ist. Super. Sanita, 2010, 46, 266–273 CAS.
- K. S. M. Benschop, J. C. Rahamat-Langendoen, H. G. A. M. van der Avoort, E. C. J. Claas, S. D. Pas, R. Schuurman, J. J. Verweij, K. C. Wolthers, H. G. M. Niesters and M. P. G. Koopmans, VIRO-TypeNed, systematic molecular surveillance of enteroviruses in the Netherlands between 2010 and 2014, Eurosurveillance, 2016, 21, 1–9 Search PubMed.
- R. Delogu, A. Battistone, G. Buttinelli, S. Fiore, S. Fontana, C. Amato, K. Cristiano, S. Gamper, J. Simeoni, R. Frate, L. Pellegrinelli, F. Pennino, C. Germinario, V. Balena, A. Cicala, P. Mercurio and L. Fiore, Poliovirus and Other Enteroviruses from Environmental Surveillance in Italy, 2009 – 2015, Food Environ. Virol., 2018, 10, 333–342 CrossRef PubMed.
- T. Hovi, M. Stenvik and M. Rosenlew, Relative abundance of enterovirus serotypes in sewage differs from that in patients: Clinical and epidemiological implications, Epidemiol. Infect., 1996, 116, 91–97 CrossRef CAS PubMed.
- O. C. Liu and F. McGowan, Effect of chlorination on human enteric viruses in partially treated water from the Potomac river estuary, Environ. Prot. Agency Water supply programs Div. Northeast. Water Supply Lab, 1971 Search PubMed.
- C. Gerba, S. Goyal, C. Hurst and R. Labelle, Type and strain dependence of enterovirus adsorption to activated sludge, soils and estuarine sediments, Water Res., 1980, 14, 1197–1198 CrossRef.
- T.-D. Kim and H. Unno, The roles of microbes in the removal and inactivation of viruses in a biological wastewater treatment system, Water Sci. Technol., 1996, 33, 243–250 CrossRef CAS.
- S. Kelly, W. W. Sanderson and C. Neidl, Removal of Enteroviruses from Sewage by Activated Sludge, J. - Water Pollut. Control Fed., 1961, 33, 1056–1062 Search PubMed.
- D. R. Knowlton and R. L. Ward, Characterization of virucidal agents in activated sludge, Appl. Environ. Microbiol., 1987, 53, 621–626 CrossRef CAS PubMed.
- Z. Tao, Z. Wang, X. Lin, S. Wang, H. Wang, H. Yoshida, A. Xu and Y. Song, One-year Survey of human enteroviruses from sewage and the factors affecting virus adsorption to the suspended solids, Sci. Rep., 2016, 6, 1–9 CrossRef PubMed.
- M. Olive, F. Moerman, X. Fernandez-Cassi, F. Altermatt and T. Kohn, Removal of Waterborne Viruses by Tetrahymena pyriformis Is Virus-Specific and Coincides with Changes in Protist Swimming Speed, Environ. Sci. Technol., 2022, 56, 4062–4070 CrossRef CAS PubMed.
- M. H. Corre, V. Bachmann and T. Kohn, Bacterial matrix metalloproteases and serine proteases contribute to the extra-host inactivation of enteroviruses in lake water, ISME J., 2022, 16, 1970–1979 CrossRef CAS PubMed.
- D. O. Cliver and J. E. Hermann, Proteolytic and Microbial inactivation of Enteroviruses, Water Res., 1972, 6, 797–805 CrossRef CAS.
- M. R. Beychok, Performance of surface-aerated basins, Chem. Eng. Prog., Symp. Ser., 1971, 67, 322–339 CAS.
- S. Lo, J. Gilbert and F. Hetrick, Stability of Human Enteroviruses in Estuarine and Marine Waters, Appl. Environ. Microbiol., 1976, 32, 245–249 CrossRef CAS PubMed.
- P. Waldman, A. Meseguer, F. Lucas, L. Moulin and S. Wurtzer, Interaction of Human Enteric Viruses with Microbial Compounds: Implication for Virus Persistence and Disinfection Treatments, Environ. Sci. Technol., 2017, 51, 13633–13640 CrossRef CAS PubMed.
- C. M. Robinson, P. R. Jesudhasan and J. K. Pfeiffer, Bacterial lipopolysaccharide binding enhances virion stability and promotes environmental fitness of an enteric virus, Cell Host Microbe, 2014, 15, 36–46 CrossRef CAS PubMed.
- S. Meister, M. E. Verbyla, M. Klinger and T. Kohn, Variability in Disinfection Resistance between Currently Circulating Enterovirus B Serotypes and Strains, Environ. Sci. Technol., 2018, 52, 3696–3705 CrossRef CAS PubMed.
- S. Torii, M.-H. Corre, F. Miura, M. Itamochi, K. Haga, K. Katayama, H. Katayama and T. Kohn, Genotype-dependent kinetics of enterovirus inactivation by free chlorine and ultraviolet (UV) irradiation, Water Res., 2022, 220, 118712 CrossRef CAS PubMed.
- P. Payment, M. Tremblay and M. Trudel, Relative resistance to chlorine of poliovirus and coxsackievirus isolates from environmental sources and drinking water, Appl. Environ. Microbiol., 1985, 49, 981–983 CrossRef CAS PubMed.
- T. L. Cromeans, A. M. Kahler and V. R. Hill, Inactivation of adenoviruses, enteroviruses, and murine norovirus in water by free chlorine and monochloramine, Appl. Environ. Microbiol., 2010, 76, 1028–1033 CrossRef CAS PubMed.
- S. Harakeh, The behavior of viruses on disinfection by chlorine dioxide and other disinfectants in effluent, FEMS Microbiol. Lett., 1987, 44, 335–341 CrossRef CAS.
- O. Larivé, J. Brandani, M. Dubey and T. Kohn, An integrated cell culture reverse transcriptase quantitative PCR (ICC-RTqPCR) method to simultaneously quantify the infectious concentrations of eight environmentally relevant enterovirus serotypes, J. Virol. Methods, 2021, 296, 114225 CrossRef PubMed.
- US Environmental Protection Agency, Most Probable Number (MPN) Calculator Version 2.0 - User and System Installation and Administration Manual, 2013, pp. 3–4 Search PubMed.
- K. E. Klymus, C. M. Merkes, M. J. Allison, C. A. Jackson, C. S. Goldberg, C. C. Helbing, M. E. Hunter, R. F. Lance, A. M. Mangan, E. M. Monroe, A. J. Piaggio, J. P. Stokdyk, C. C. Wilson and C. A. Richter, Reporting the limits of detection and quantification for environmental DNA assays, Environ. DNA, 2020, 2, 271–282 CrossRef.
- S. A. Bustin, V. Benes, J. A. Garson, J. Hellemans, J. Huggett, M. Kubista, R. Mueller, T. Nolan, M. W. Pfaffl and G. L. Shipley, The MIQE Guidelines : Minimum Information for Publication of Quantitative Real-Time PCR Experiments, Clin. Chem., 2009, 55, 611–622 CrossRef CAS PubMed.
- APHA, in Standard Methods for the Examination of Water and Wastewater, American Public Health Association, American Water Works Association, Water Environment Federation, Washington, DC, 21st edn, 2005, pp. 55–61 Search PubMed.
- S. Felz, S. Al-Zuhairy, O. A. Aarstad, M. C. M. van Loosdrecht and Y. M. Lin, Extraction of structural extracellular polymeric substances from aerobic granular sludge, J. Visualized Exp., 2016, 2016, 1–8 Search PubMed.
- S. Felz, P. Vermeulen, M. C. M. van Loosdrecht and Y. M. Lin, Chemical characterization methods for the analysis of structural extracellular polymeric substances (EPS), Water Res., 2019, 157, 201–208 CrossRef CAS PubMed.
- M. Dubois, K. A. Gilles, J. K. Hamilton, P. A. Rebers and F. Smith, Colorimetric Method for Determination of Sugars and Related Substances, Anal. Chem., 1956, 28, 350–356 CrossRef CAS.
- R Core Team, R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria, 2020 Search PubMed.
- V. M. Muggeo, Interval estimation for the breakpoint in segmented regression: a smoothed score-based approach, Aust. N. Z. J. Stat., 2017, 59, 311–322 CrossRef.
- H. N. Bischel, A. Schertenleib, A. Fumasoli, K. M. Udert and T. Kohn, Inactivation kinetics and mechanisms of viral and bacterial pathogen surrogates during urine nitrification, Environ. Sci.: Water Res. Technol., 2015, 1, 65–76 RSC.
- A. Guellil, M. Boualam, H. Quiquampoix, P. Ginestet, J. Audic and J. Block, Hydrolysis of wastewater colloidal organic matter by extracellular enzymes extracted from activated sludge flocs, Water Sci. Technol., 2001, 43, 33–40 CrossRef CAS PubMed.
- B. Frolund, T. Griebe and P. H. Nielsen, Enzymatic activity in the activated-sludge floc matrix, Appl. Microbiol. Biotechnol., 1995, 43, 755–761 CrossRef CAS.
- R. Goel, T. Mino, H. Satoh and T. Matsuo, Enzyme activities under anaerobic and aerobic conditions in activated sludge sequencing batch reactor, Water Res., 1998, 32, 2081–2088 CrossRef CAS.
- State of Maine Department of Environmental Protection, Notes on Activated Sludge Process Control, 2009 Search PubMed.
- E. Haun, K. Ulbricht, R. Nogueira and K.-H. Rosenwinkel, Virus elimination in activated sludge systems: from batch tests to mathematical modeling, Water Sci. Technol., 2014, 70, 1115–1121 CrossRef CAS PubMed.
- R. M. Chaudhry, K. L. Nelson and J. E. Drewes, Mechanisms of Pathogenic Virus Removal in a Full-Scale Membrane Bioreactor, Environ. Sci. Technol., 2015, 49, 2815–2822 CrossRef CAS PubMed.
- A. Carratalà, V. Bachmann, T. R. Julian and T. Kohn, Adaptation of Human Enterovirus to Warm Environments Leads to Resistance against Chlorine Disinfection, Environ. Sci. Technol., 2020, 54, 11292–11300 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ew00050h |
| This journal is © The Royal Society of Chemistry 2023 |