Influence of ring size in conformationally restricted ring I analogs of paromomycin on antiribosomal and antibacterial activity†
Michael G.
Pirrone
 abc,
Sven N.
Hobbie
abc,
Sven N.
Hobbie
 d,
Andrea
Vasella
e,
Erik C.
Böttger
d and
David
Crich
d,
Andrea
Vasella
e,
Erik C.
Böttger
d and
David
Crich
 *abf
*abf
aDepartment of Pharmaceutical and Biomedical Sciences, University of Georgia, 250 West Green Street, Athens, GA 30602, USA. E-mail: David.crich@uga.edu
bComplex Carbohydrate Research Center, University of Georgia, 315 Riverbend Road, Athens, GA 30602, USA
cDepartment of Chemistry, Wayne State University, 5101 Cass Avenue, Detroit, MI 48202, USA
dInstitute of Medical Microbiology, University of Zurich, Gloriastrasse 28, 8006 Zürich, Switzerland
eOrganic Chemistry Laboratory, ETH Zürich, Vladimir-Prelog-Weg 1-5/10, 8093 Zürich, Switzerland
fDepartment of Chemistry, University of Georgia, 140 Cedar Street, Athens, GA 30602, USA
First published on 5th August 2021
Abstract
In order to further investigate the importance of the conformation of the ring I side chain in aminoglycoside antibiotic binding to the ribosomal target several derivatives of paromomycin were designed with conformationally locked side chains. By changing the size of the appended ring between O-4′ and C-6′ used to restrict the motion of the side chain, the position of the C-6′ hydroxy group was fine tuned to probe for the optimal conformation for inhibition of the ribosome. While the changes in orientation of the 6′-hydroxy group cannot be completely dissociated from the size and hydrophobicity of the conformation-restricting ring, overall, it is apparent that the preferred conformation of the ring I side chain for interaction with A1408 in the decoding A site of the bacterial ribosome is an ideal gt conformation, which results in the highest antimicrobial activity as well as increased selectivity for bacterial over eukaryotic ribosomes.
Introduction
The 2-deoxystreptamine (DOS) aminoglycoside antibiotics (AGAs) form a class of polyamino pseudosaccharides, whose antibacterial activity derives from their ability to bind to the decoding A site of the bacterial ribosome and so to interfere with bacterial protein synthesis. These antibiotics are currently undergoing a renaissance in the face of the ever-growing threat of multidrug-resistant infections.1–5 AGAs derive a substantial part of their affinity for the decoding A site from the electrostatic interaction of the negatively charged backbone of the ribosome with the multiple amino groups of AGAs, which are protonated at physiological pH and positively charged.6,7 More subtly directed hydrogen bonding and other non-covalent specific drug–ribosome interactions make additional contributions and provide the basis for the discrimination between prokaryotic and eukaryotic ribosomes.8–16 In our laboratories we have focused on the 4,5-disubstituted and apramycin classes of DOS-AGAs, so as to avoid the increasingly prevalent ribosomal methyltransferase (RMTase) mechanism of AGA resistance that plagues the 4,6-DOS AGAs in wide clinical usage,17–19 with the emphasis on modifications designed to improve antibacterial activity, while increasing selectivity at the target level, and reducing susceptibility to the main mechanisms of AGA-resistance – derivatization by the aminoglycoside modifying enzymes (AMEs).20–23A key AGA-ribosome interaction is the pseudo base pair interaction between the AGA ring I and A1408 in the decoding A site in which ring I both donates to and accepts a hydrogen bond from A1408 with either the 6′-hydroxy group or the protonated 6′-amino group, depending on the series, serving as donor and the pyranoside ring oxygen (O5′) as acceptor (Fig. 1). In multiple AGA-ribosome crystal structures24–26 the side chain of ring I takes up the gauche,trans (gt) conformation in the pseudo base pair with A1408, whereas in free solution the side chain of typical glucopyranosides and their 2-deoxy-2-amino analogs is an approximately 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50
50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 mixture of the gt, gauche,gauche (gg), and trans,gauche (tg) staggered conformers,27–32 suggesting that AGA analogs with the ring I side chain artificially restricted to the gt conformation would show increased activity. This concept was reduced to practice with the paromomycin analog 1 with the trans-decalinoid ring I, whose equatorial 6′-hydroxy group mimics the ideal gt side chain conformation. A second diastereoisomeric analog 2, with the side chain artificially held in the gg conformation was substantially less active than 1 and the parent paromomycin 3, providing strong support for this hypothesis.33 Further support was derived from the relative activities of the 6′R and 6′S-methyl paromomycin derivatives 4 and 5 in which the former, with the predominant gt conformation outperforms the latter with the predominant gg conformation.34
0 mixture of the gt, gauche,gauche (gg), and trans,gauche (tg) staggered conformers,27–32 suggesting that AGA analogs with the ring I side chain artificially restricted to the gt conformation would show increased activity. This concept was reduced to practice with the paromomycin analog 1 with the trans-decalinoid ring I, whose equatorial 6′-hydroxy group mimics the ideal gt side chain conformation. A second diastereoisomeric analog 2, with the side chain artificially held in the gg conformation was substantially less active than 1 and the parent paromomycin 3, providing strong support for this hypothesis.33 Further support was derived from the relative activities of the 6′R and 6′S-methyl paromomycin derivatives 4 and 5 in which the former, with the predominant gt conformation outperforms the latter with the predominant gg conformation.34
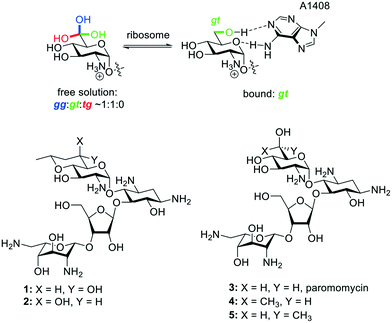 | ||
| Fig. 1 Unbound and bound aminoglycoside ring I side chain conformations, and existing compounds 1–5. | ||
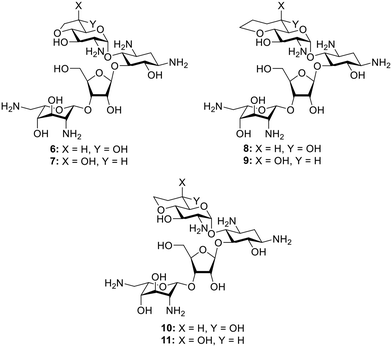
In spite of the increased activity of analog 1 with its gt side chain conformation, as compared to its gg isomer 2, the relatively low resolution (2.5–3 Å) of typical AGA-ribosome crystal structures coupled with the minor adjustments necessarily forced on the pseudo base pair interaction by motions in the ribosome during the course of translation35 left open the possibility that locking of the side chain into the perfect gt conformation may not provide the optimal compound. To probe this possibility we designed analogs 6 and 8 in which the side chain is held in an approximate but not ideal gt conformation and in which greater mobility is afforded by the fused five and seven-membered rings36 as compared to the rigid decalinoid system 1 embodying the ideal gt conformation. For completeness we also prepared the pseudoaxial diastereoisomers 7 and 9. Finally, although the methyl substituent in 1 and 2 was originally incorporated to match the optimum chain of 4′-susbtituted paromomycin derivatives,33,37,38 cognizant of the varied effects of methyl groups in medicinal chemistry,39,40 we prepared the desmethyl analogs 10 and 11 (Fig. 2). We describe the synthesis of 6–11, and their evaluation in cell-free translation assays with wild-type bacterial ribosomes and hybrid bacterial ribosomes carrying complete eukaryotic decoding A sites,41 and in antibacterial screens for activity against representative bacteria. We find that the trans-decalinoid analog 1, with its rigidly equatorial 6′-hydroxy group and pendant methyl group, is the optimal compound leading to the conclusion that the pseudo base pair interaction with A1408 is favored by the ideal gt conformation of the ring I side chain and that the methyl does confer an additional small advantage.
Results and discussion
Paromomycin derivatives with a fused five membered ring connecting O-4′ and C-6′ were synthesized starting with compound 12,34 which was oxidized to the aldehyde using Swern conditions. This was followed by alkylation with the vinyl Grignard reagent to give an inseparable 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
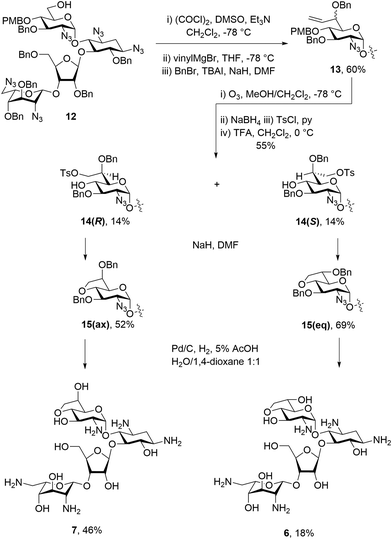
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of diastereomers. The mixture was then subjected to benzylation conditions using benzyl bromide and sodium hydride with TBAI to give an inseparable mixture of compounds 13 in 60% yield. Compounds 13 were then subjected to ozonolysis, followed by reduction and tosylation of the resulting 7′-hydroxyl group before removal of the PMB ether at the 4′-position using TFA to give diastereoisomers 14(R) and 14(S) each isolated in 14% yield after normal phase HPLC. Ring closing displacement of the 7′-O-tosylates using sodium hydride then gave compounds 15(ax) and 15(eq) in 52% and 69% yields, respectively. The configuration at C-6′ was assigned based on the 3JH5′,H6′ values of 4.3 Hz in 15(ax) and 7.4 Hz in 15(eq). Finally, both 15(ax) and 15(eq) were subjected to hydrogenolysis to give 7 and 6 in 46% and 18% yield, respectively (Scheme 1). The assignment of configuration at C-6′ in these compounds was verified during the conformational analysis of the deprotected compounds shown below.
1 mixture of diastereomers. The mixture was then subjected to benzylation conditions using benzyl bromide and sodium hydride with TBAI to give an inseparable mixture of compounds 13 in 60% yield. Compounds 13 were then subjected to ozonolysis, followed by reduction and tosylation of the resulting 7′-hydroxyl group before removal of the PMB ether at the 4′-position using TFA to give diastereoisomers 14(R) and 14(S) each isolated in 14% yield after normal phase HPLC. Ring closing displacement of the 7′-O-tosylates using sodium hydride then gave compounds 15(ax) and 15(eq) in 52% and 69% yields, respectively. The configuration at C-6′ was assigned based on the 3JH5′,H6′ values of 4.3 Hz in 15(ax) and 7.4 Hz in 15(eq). Finally, both 15(ax) and 15(eq) were subjected to hydrogenolysis to give 7 and 6 in 46% and 18% yield, respectively (Scheme 1). The assignment of configuration at C-6′ in these compounds was verified during the conformational analysis of the deprotected compounds shown below.
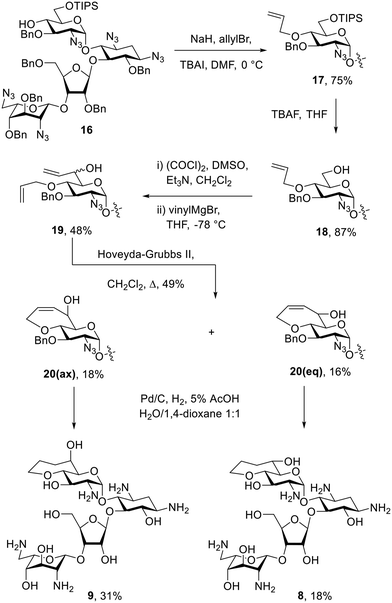
The bicyclic paromomycin derivatives with a fused 7-membered ring containing O-4′ and C-6′ were prepared starting with allylation of the 4′-hydroxyl group of 16 using allyl bromide, TBAI, and sodium hydride to give compound 17 in 76% yield. Compound 17 was subjected to TBAF in THF to remove of the silyl group and to give 18 in 89% yield. Alcohol 18 was then oxidized using Swern conditions, followed by alkylation using the vinyl Grignard reagent to give compounds 19 as an inseparable mixture of diastereoisomers in 47% yield. Mixture 19 was subjected to ring closing metathesis using the Hoveyda–Grubbs second generation catalyst,42 resulting in a mixture of diastereoisomers 20 in 49% yield, from which 20(ax) and 20(eq) were isolated in 18% and 16% yield, respectively. The configuration at C-6′ in these compounds was assigned based on the 3J5′,6′ values of 2.4 Hz for 20(ax) and 9.1 Hz for 20(eq). Compounds 20(ax) and 20(eq) were deprotected by hydrogenolysis to give 9 and 8 in 31% and 18% yield, respectively (Scheme 2). The configuration at C-6′ was further verified during conformational analysis of the deprotected compounds as described below.
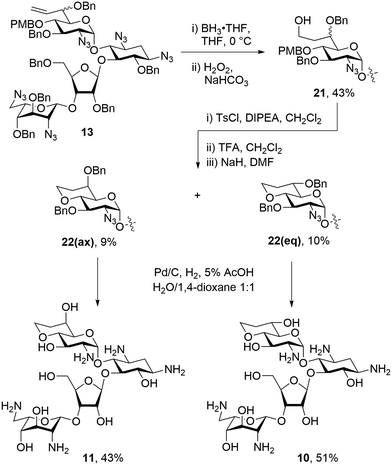
To make a more direct comparison between fused ring sizes, 6-membered bicyclic derivatives related to the previous alcohols 1 and 2 were synthesized that lack the 8′-methyl group. To this end mixture 13 was subjected to hydroboration with BH3·THF followed by workup with hydrogen peroxide to give the 8′-hydroxy compounds 21 as an inseparable mixture of diastereoisomers in 43% yield. Tosylation of the mixture of 21 with tosyl chloride and Hünig's base followed by PMB ether removal using TFA, and ring closing displacement of the tosylates gave 22(eq) and 22(ax) in 10% and 9% yield, respectively. The configuration at C-6′ of compounds 22(eq) and 22(ax) was assigned based on the 3JH5′,H6′ values of 9.4 Hz and 2.5 Hz, respectively. These assignments were further verified during the conformational analysis of the deprotected compounds below. Both isomers of 22 were subjected to hydrogenolysis to give 10 and 11 in 51% and 43%, respectively (Scheme 3).
Inspection of the 3JH,H coupling constants around ring I of the various deprotected derivatives reveals that the orientation of the 6′-hydroxy group varies with the size of the appended ring (Table 1), while the pyranoside ring itself retains the usual 4C1 chair conformation with little to no distortion.
| Cmpd | Fused ring size | Coupling constant (Hz) | ||||
|---|---|---|---|---|---|---|
| 3 J H1′,H2′ | 3 J H2′,H3′ | 3 J H3′,H4′ | 3 J H4′,H5′ | 3 J H5′,H6′ | ||
| 1 | 6 | 4.1 | 9.9 | 9.5 | 9.5 | 9.4 |
| 2 | 6 | 4.1 | — | — | 10.0 | 2.8 |
| 6 | 5 | 4.3 | 9.8 | 9.8 | 10.0 | 7.9 |
| 7 | 5 | 4.2 | 9.9 | 9.9 | 9.9 | 4.5 |
| 10 | 6 | 4.1 | 10.0 | 10.0 | 9.5 | 9.4 |
| 11 | 6 | 4.1 | 10.2 | 10.2 | 9.4 | 2.0 |
| 8 | 7 | 3.8 | 10.8 | 9.1 | 9.4 | 10.0 |
| 9 | 7 | 3.9 | 10.5 | 9.5 | 9.5 | 3.5 |
Cell-free translation assays
Cell free ribosomal translation assays were conducted for all compounds with both wild-type bacterial ribosomes and hybrid bacterial ribosomes carrying the complete decoding A sites of the human mitochondrial (Mit13), A1555G mutant mitochondrial (A1555G) and cytoplasmic (Cyt14) ribosomes (Table 2).41 The data for the bacterial ribosome shows a trend of activity increasing as the orientation of the C-6′ hydroxy group progresses towards that defined by the gt conformation of the freely rotating side chain in the parent. Ignoring compounds 1 and 2 with the additional pendant methyl group, the bacterial IC50 trend shows a progression from least to most active 11 < 9 < 7 < 6 < 10 < 8, in agreement with the progression from the imposed gg to the imposed gt conformation. Interestingly, compound 8 with the fused seven-membered ring and the ideal gt orientation of the 6′-hydroxy group is more active than its six-membered counterpart 10, and approaches the activity of the initial hit 1 with the fused six-membered ring and the additional methyl group. Overall, it is apparent that an imposed gt-like conformation provides a more optimal H-bonding interaction with A1408 in the decoding A site than a gg-like conformation (Fig. 3). As 8, 10, and 1 all have the near ideal gt orientation of their 6′-hydroxy groups, as judged from the magnitude of their 3JH5′,H6′ coupling constants (Table 1), it is apparent that the larger hydrophobic patch in 8 and 1 contributes to their enhanced activity over 10. A similar contribution of the additional methyl group to inhibition of the bacterial ribosome is seen from the comparison of 2 and 11 with their otherwise identical structures. The positive contribution of a hydrophobic patch in the locus of the 4′-position has been previously noted in propylamycin (4′-deoxy-4′-propyl)paromomycin.32,38| Cmpd | Fused ring size | Predominant conformation | IC50a (μM ± STD) | Selectivity | |||||
|---|---|---|---|---|---|---|---|---|---|
| Bacterial | Mit13 | A1555G | Cyt14 | Mit13 | A1555G | Cyt14 | |||
| a All values were determined in duplicate or more with the exception of MIT13, A1555G and Cyt14 assays for 7, 9 and 11 for which limited material was available. Selectivities are calculated by dividing IC50 for the mutant ribosome by that for the bacterial ribosome. | |||||||||
| Parom | — | — | 0.033 ± 0.006 | 131 ± 63 | 12 ± 3 | 37 ± 9 | 3970 | 364 | 1121 |
| 2 | 6 | gg | 0.89 ± 0.14 | 372 ± 125 | 327 ± 6 | 266 ± 12 | 418 | 367 | 299 |
| 9 | 6 | gg | 1.5 ± 0.4 | 514 | 545 | — | 342 | 363 | — |
| 11 | 7 | gg > gt | 3.3 ± 0.9 | 554 | 602 | 557 | 168 | 182 | 169 |
| 7 | 5 | gg > gt | 0.2 ± 0.005 | 131 | 117 | 276 | 655 | 585 | 1380 |
| 6 | 5 | gg < gt | 0.13 ± 0.05 | 151 ± 27 | — | 40 ± 2 | 943 | — | 250 |
| 10 | 7 | gt | 0.12 ± 0.01 | 231 ± 44 | 132 ± 51 | 120 ± 43 | 1925 | 1100 | 1000 |
| 8 | 6 | gt | 0.071 ± 0.020 | 243 ± 75 | 42 ± 2 | 56 ± 9 | 3423 | 592 | 789 |
| 1 | 6 | gt | 0.063 ± 0.017 | 250 ± 57 | 20 ± 6 | 34 ± 7 | 3968 | 317 | 540 |
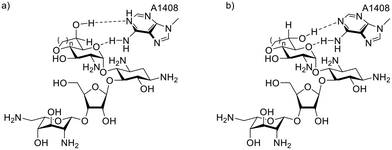 | ||
| Fig. 3 Axial (a) and equatorial (b) derivatives with their imposed gg- and gt-like side chain conformations have different H-bonding geometries with A1408. | ||
Aminoglycosides carrying hydrophobic chains have been demonstrated to have significant off-target non-ribosomal activity arising from their ability to induce plasma membrane permeabilization.43,44 However, as the contribution from the relatively minor hydrophobic patch in the present series of compounds is detected in cell-free translation, we see no reason consider membrane permeabilization as a significant contributor to the activity of this series of compounds. In passing, we note that the clinical aminoglycosides gentamicin, sisomicin, and plazomicin are all 3′,4′-dideoxy compounds and consequently have hydrophobic patches in the approximate vicinity of those described here, leading to the speculation that such a patch is generally beneficial.
In terms of selectivity, the imposition of the ideal gt orientation on the 6′-hydroxy group of compounds 1, 8, and 10 does not significantly affect selectivity for inhibition of the bacterial over the mitochondrial ribosome (Mit13). On the other hand, the decreased antibacterioribosomal activity of compounds 2, 9, 11, and 7 with the approximate gg orientation of the 6′-hydroxy group is accompanied by a reduction in selectivity. Selectivity for inhibition of the bacterial ribosome over either the A1555G mutant mitochondrial ribosome (A1555G) or the cytoplasmic ribosome (Cyt14) is not impacted in a major way by the change in size of the fused ring nor by the orientation, (pseudo)axial or (pseudo)equatorial of the 6′-hydroxy group.
The trend from the antibacterial assays with wild-type strains follows that from the cell-free ribosomal translation assays (Table 3). The 8′-methyl group on compounds 1 and 2 offers no significant improvement in activity over 10 and 11.
| Cmpd | Fused ring size | Predominant conformation | MIC (mg L−1) | |||||
|---|---|---|---|---|---|---|---|---|
| MRSAb | E. coli | A. Baum.b | K. Pneum.b | E. cloacae | P. aeruginosa | |||
| AG038 | AG001 | AG225 | AG215 | AG290 | AG031c | |||
| a All measurements were conducted in duplicate. b MRSA, methicillin-resistant Staphylococcus aureus; E. coli, Escherichia coli; A. baum, Acinetobacter baumanii; K. pneum, Klebsiella pneumoniae; E. cloacae, Enterobacter cloacae; P. aeruginosa, Pseudomonas aeruginosa. c This strain carries the gene for the APH(3′)-II AME. | ||||||||
| Parom | — | — | 2 | 2–4 | 2–4 | 1 | 2 | >128 |
| 2 | 6 | gg | 32 | 32 | 32 | 16–32 | 32–64 | >128 |
| 11 | 6 | gg | 32 | 128 | 64 | 64 | 64 | >128 |
| 9 | 7 | gg > gt | 32 | 64 | 32 | 32–64 | 32–64 | >128 |
| 7 | 5 | gg > gt | 8 | 16 | 8–16 | 4 | 8 | >64 |
| 6 | 5 | gg < gt | 16–32 | 32 | 32 | 16–32 | 32 | >128 |
| 8 | 7 | gt | 2–4 | 4–8 | 4 | 2–4 | 2 | 64 |
| 10 | 6 | gt | 2–4 | 8 | 8 | 4 | 4 | >128 |
| 1 | 6 | gt | 4–8 | 4–8 | 4–8 | 2 | 2 | 32 |
Compounds 1 and 10 also regained some activity against P. aeruginosa strain AG031 (Table 3), which carries the APH(3′)-II AME gene, likely due to the added bulk on O-4′ hindering the approach of the enzyme to the adjacent C-3′-hydroxy group. In contrast to paromomycin and as expected, all bicyclic derivatives retained activity in strains of MRSA containing the ANT(4′)-I AME, as well as in an E. coli strain containing the ANT(4′).
Conclusion
The antiribosomal and antibacterial activity of the bicyclic compounds is a function of the size of the fused ring and the orientation of the C-6′ hydroxy group; it is optimal for the equatorial orientation of the C-6′OH in the fused six- and seven-membered rings, that best approximate the gt conformation of the freely rotating side chain. The pendant methyl group in compound 1 enhances inhibition of the bacterial ribosome in comparison to the desmethyl analog 10 indicating the positive influence of a small hydrophobic patch, as supported by the superior performance of the seven-membered analog 8 in comparison to 10.Conflicts of interest
E. C. B., D. C., S. N. H. and A. V. are cofounders of and maintain an equity interest in Juvabis AG, a biotech startup working in the aminoglycoside field. M. G. P. declares no conflict of interest.Acknowledgements
We thank the NIH (AI123352) for financial support of this work.References
- E. C. Böttger and D. Crich, Aminoglycosides: Time for Resurrection of a Neglected Class of Antibacterials?, ACS Infect. Dis., 2020, 6, 168–172 CrossRef PubMed.
- T. Brennan-Krohn, R. Manetsch, G. A. O'Doherty and J. E. Kirby, New Strategies and Structural Considerations in Development of Therapeutics for Carbapenem-Resistant Enterobacteriaceae, Transl. Res., 2020, 2020, 14–32 CrossRef PubMed.
- M. Y. Fosso, Y. Li and S. Garneau-Tsodikova, New Trends in the Use of Aminoglycosides, Med. Chem. Commun., 2014, 5, 1075–1091 RSC.
- Y. Takahashi and M. Igarashi, Destination of Aminoglycoside Antibiotics in the ‘Post-Antibiotic Era’, J. Antibiot., 2018, 71, 4–14 CrossRef CAS PubMed.
- E. S. Armstrong, C. F. Kostrub, R. T. Cass, H. E. Moser, A. W. Serio and G. H. Miller, Aminoglycosides, in Antibiotic Discovery and Development, ed. T. J. Dougherty and M. J. Pucci, Springer Science+Business Media, New York, 2012, pp. 229–269 Search PubMed.
- M. Kaul, C. M. Barbieri, J. E. Kerrigan and D. S. Pilch, Coupling of Drug Protonation to the Specific Binding of Aminoglycosides to the A Site of 16 S rRNA: Elucidation of the Number of Drug Amino Groups Involved and their Identities, J. Mol. Biol., 2003, 326, 1373–1387 CrossRef CAS PubMed.
- M. Kaul and D. S. Pilch, Thermodynamics of Aminoglycoside-rRNA Recognition: The Binding of Neomycin-Class Aminoglycosides to the A Site of 16S rRNA, Biochemistry, 2002, 41, 7695–7706 CrossRef CAS PubMed.
- C. M. Barbieri, M. Kaul, M. Bozza-Hingos, F. Zhao, Y. Tor, T. Hermann and D. S. Pilch, Defining the Molecular Forces That Determine the Impact of Neomycin on Bacterial Protein Synthesis: Importance of the 2-Amino Functionality, Antimicrob. Agents Chemother., 2007, 51, 1760–1769 CrossRef CAS PubMed.
- J. Kondo, M. Hainrichson, I. Nudelman, D. Shallom-Shezifi, C. M. Barbieri, D. S. Pilch, E. Westhof and T. Baasov, Differential Selectivity of Natural and Synthetic Aminoglycosides towards the Eukaryotic and Prokaryotic Decoding A Sites, ChemBioChem, 2007, 8, 1700–1709 CrossRef CAS PubMed.
- F. Corzana, I. Cuesta, F. Freire, J. Revuelta, M. Torrado, A. Bastida, J. Jiménez-Barbero and J. L. Asensio, The Pattern of Distribution of Amino Groups Modulates the Structure and Dynamics of Natural Aminoglycosides: Implications for RNA Recognition, J. Am. Chem. Soc., 2007, 129, 2849–2865 CrossRef CAS PubMed.
- L. Chen, M. Hainrichson, D. Bourdetsky, A. Mor, S. Yaron and T. Baasov, Structure–Toxicity Relationship of Aminoglycosides: Correlation of 2′-Amine Basicity with Acute Toxicity in Pseudo-Disaccharide Scaffolds, Bioorg. Med. Chem., 2008, 16, 8940–8951 CrossRef CAS PubMed.
- A. Sonousi, J. C. K. Quirke, P. Waduge, T. Janusic, M. Gysin, K. Haldimann, S. Xu, S. N. Hobbie, S.-H. Sha, J. Schacht, C. S. Chow, A. Vasella, E. C. Böttger and D. Crich, An Advanced Apralog with Increased in-vitro and in-vivo Activity toward Gram-negative Pathogens and Reduced ex-vivo Cochleotoxicity, ChemMedChem, 2021, 16, 335–339 CrossRef CAS PubMed.
- T. Matt, C. L. Ng, K. Lang, S.-H. Sha, R. Akbergenov, D. Shcherbakov, M. Meyer, S. Duscha, J. Xie, S. R. Dubbaka, D. Perez-Fernandez, A. Vasella, V. Ramakrishnan, J. Schacht and E. C. Böttger, Dissociation of Antibacterial Activity and Aminoglycoside Ototoxicity in the 4-Monosubstituted 2-Deoxystreptamine Apramycin, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 10984–10989 CrossRef CAS PubMed.
- E. C. Böttger and J. Schacht, The Mitochondrion: A Perpetrator of Acquired Hearing Loss, Hear. Res., 2013, 303, 12–19 CrossRef PubMed.
- Y. Qian and M.-X. Guan, Interaction of Aminoglycosides with Human Mitochondrial 12S rRNA Carrying the Deafness-Associated Mutation, Antimicrob. Agents Chemother., 2009, 53, 4612–4618 CrossRef CAS PubMed.
- R. Akbergenov, D. Shcherbakov, T. Matt, S. Duscha, M. Meyer, D. Perez-Fernandez, R. Pathak, S. Harish, I. Kudyba, S. R. Dubbaka, S. Silva, M. Ruiz Ruiz, S. Salian, A. Vasella and E. C. Böttger, Decoding and Deafness: Two Sides of a Coin, in Ribosomes: Structure, Function, and Dynamics, ed. M. V. Rodnina, W. Wintermeyer and R. Green, Springer-Verlag, Vienna, 2011, pp. 249–261 Search PubMed.
- K. K. Kumarasamy, M. A. Toleman, T. R. Walsh, J. Bagaria, F. Butt, R. Balakrishnan, U. Chaudhary, M. Doumith, C. G. Giske, S. Irfan, P. Krishnan, A. V. Kumar, S. Maharjan, S. Mushtaq, T. Noorie, D. L. Paterson, A. Pearson, C. Perry, R. Pike, B. Rao, U. Ray, J. B. Sarma, M. Sharma, E. Sheridan, M. A. Thirunarayan, J. Turton, S. Upadhyay, M. Warner, W. Welfare, D. M. Livermore and N. Woodford, Emergence of a New Antibiotic Resistance Mechanism in India, Pakistan, and the UK: A Molecular, Biological, and Epidemiological Study, Lancet Infect. Dis., 2010, 10, 597–602 CrossRef CAS PubMed.
- Z. Liu, A. Yoshihara, C. Kelly, J. T. Heap, M. H. S. Marqvorsen, S. F. Jenkinson, M. R. Wormald, J. M. Otero, A. Estévez, A. Kato, G. W. J. Fleet, R. J. Estévez and K. Izumori, 6-Deoxyhexoses from L-Rhamnose in the Search for Inducers of the Rhamnose Operon: Synergy of Chemistry and Biotechnology, Chem. – Eur. J., 2016, 22, 12557–12565 CrossRef CAS PubMed.
- M. Plattner, M. Gysin, K. Haldimann, K. Becker and S. N. Hobbie, Epidemiologic, Phenotypic, and Structural Characterization of Aminoglycoside-Resistance Gene AAC(3)-IV, Int. J. Mol. Sci., 2020, 21, 6133 CrossRef CAS PubMed.
- R.-B. Yan, M. Yuan, Y. Wu, X. You and X.-S. Ye, Rational Design and Synthesis of Potent Aminoglycoside Antibiotics Against Resistant Bacterial Strains, Bioorg. Med. Chem., 2011, 19, 30–40 CrossRef CAS PubMed.
- V. R. Bacot-Davis, A. V. Bassenden and A. M. Berghuis, Drug-Target Networks in Aminoglycoside Resistance: Hierarchy of Priority in Structural Drug Design, Med. Chem. Commun., 2016, 7, 103–113 RSC.
- C. Hobson, A. N. Chan and G. D. Wright, The Antibiotic Resistome: A Guide for the Discovery of Natural Products as Antimicrobial Agents, Chem. Rev., 2021, 121, 3464–3494 CrossRef CAS PubMed.
- S. Garneau-Tsodikova and K. J. Labby, Mechanisms of Resistance to Aminoglycoside Antibiotics: Overview and Perspectives, Med. Chem. Commun., 2016, 7, 11–27 RSC.
- A. P. Carter, W. M. Clemons, D. E. Brodersen, R. J. Morgan-Warren, B. T. Wimberly and V. Ramakrishnan, Functional Insights from the Structure of the 30S Ribosomal Subunit and its Interactions with Antibiotics, Nature, 2000, 407, 340–348 CrossRef CAS PubMed.
- P. Pfister, S. Hobbie, Q. Vicens, E. C. Böttger and E. Westhof, The Molecular Basis for A-Site Mutations Conferring Aminoglycoside Resistance: Relationship between Ribosomal Susceptibility and X-ray Crystal Structures, ChemBioChem, 2002, 4, 1078–1088 CrossRef PubMed.
- D. Perez-Fernandez, D. Shcherbakov, T. Matt, N. C. Leong, I. Kudyba, S. Duscha, H. Boukari, R. Patak, S. R. Dubbaka, K. Lang, M. Meyer, R. Akbergenov, P. Freihofer, S. Vaddi, P. Thommes, V. Ramakrishnan, A. Vasella and E. C. Böttger, 4′-O-Substitutions Determine Aminoglycoside Selectivity at the Drug Target Level, Nat. Commun., 2014, 5, 3112, DOI:10.1038/ncomms4112.
- R. H. Marchessault and S. Perez, Conformations of the Hydroxymethyl Group in Crystalline Aldohexopyranoses, Biopolymers, 1979, 18, 2369–2374 CrossRef CAS.
- T. B. Grindley, Structure and Conformation of Carbohydrates, in Glycoscience: Chemistry and Chemical Biology, ed. B. Fraser-Reid, K. Tatsuta and J. Thiem, Springer, Berlin, 2001, vol. 1, pp. 3–51 Search PubMed.
- V. S. R. Rao, P. K. Qasba, P. V. Balaji and R. Chandrasekaran, Conformation of Carbohydrates, Harwood Academic Publishers, Amsterdam, 1998, p. 359 Search PubMed.
- K. Bock and J. O. Duus, A Conformational Study of Hydroxymethyl Groups in Carbohydrates Investigated by 1H NMR Spectroscopy, J. Carbohydr. Chem., 1994, 13, 513–543 CrossRef CAS.
- T. Kato, A. Vasella and D. Crich, Stereospecific Synthesis of Methyl 2-Amino-2-deoxy-(6S)-deuterio-α,β-D-glucopyranoside and Methyl 2,6-Diamino-2,6-dideoxy-(6R)-deuterio-α,β-D-glucopyranoside: Side Chain Conformations of the 2-Amino-2-deoxy and 2,6-Diamino-2,6-Dideoxyglucopyranosides, Carbohydr. Res., 2017, 448, 10–17 CrossRef CAS PubMed.
- M. G. Pirrone, T. Matsushita, A. Vasella and D. Crich, Stereospecific Synthesis of Methyl 2-Amino-2,4-dideoxy-6S-deuterio-α-D-xylopyranoside and Methyl 2-Amino-2,4-dideoxy-6S-deuterio-4-propyl-α-D-glucopyranoside: Side Chain Conformation of the Novel Aminoglycoside Antibiotic Propylamycin, Carbohydr. Res., 2020, 491, 107984 CrossRef CAS PubMed.
- A. R. Mandhapati, G. Yang, T. Kato, D. Shcherbakov, S. N. Hobbie, A. Vasella, E. C. Böttger and D. Crich, Structure-Based Design and Synthesis of Apramycin-Paromomycin Analogues. Importance of the Configuration at the 6′-Position and Differences Between the 6′-Amino and Hydroxy Series, J. Am. Chem. Soc., 2017, 139, 14611–14619 CrossRef CAS PubMed.
- M. G. Pirrone, M. Gysin, K. Haldimann, S. N. Hobbie, A. Vasella and D. Crich, Predictive Analysis of the Side Chain Conformation of the Higher Carbon Sugars: Application to the Preorganization of the Aminoglycoside Ring 1 Side Chain for Binding to the Bacterial Ribosomal Decoding A Site, J. Org. Chem., 2020, 85, 16043–16059 CrossRef CAS PubMed.
- M. R. Wasserman, A. Pulk, Z. Zhou, R. B. Altman, J. C. Zinder, K. D. Green, S. Garneau-Tsodikova, J. H. Doudna Cate and S. C. Blanchard, Chemically Related 4,5-Linked Aminoglycoside Antibiotics Drive Subunit Rotation in Opposite Directions, Nat. Commun., 2015, 6, 7896 CrossRef CAS PubMed.
- E. L. Eliel and S. H. Wilen, Stereochemistry of Organic Compounds, Wiley, New York, 1994 Search PubMed.
- S. Duscha, H. Boukari, D. Shcherbakov, S. Salian, S. Silva, A. Kendall, T. Kato, R. Akbergenov, D. Perez-Fernandez, B. Bernet, S. Vaddi, P. Thommes, J. Schacht, D. Crich, A. Vasella and E. C. Böttger, Identification and Evaluation of Improved 4′-O-(Alkyl) 4,5-Disubstituted 2-Deoxystreptamines as Next Generation Aminoglycoside Antibiotics, MBio, 2014, 5(5), e01827 CrossRef CAS PubMed.
- T. Matsushita, G. C. Sati, N. Kondasinghe, M. G. Pirrone, T. Kato, P. Waduge, H. S. Kumar, A. Cortes Sanchon, M. Dobosz-Bartoszek, D. Shcherbakov, M. Juhas, S. N. Hobbie, T. Schrepfer, C. S. Chow, Y. S. Polikanov, J. Schacht, A. Vasella, E. C. Böttger and D. Crich, Design, Multigram Synthesis, and in Vitro and in Vivo Evaluation of Propylamycin: A Semisynthetic 4,5-Deoxystreptamine Class Aminoglycoside for the Treatment of Drug-Resistant Enterobacteriaceae and Other Gram-Negative Pathogens, J. Am. Chem. Soc., 2019, 141, 5051–5061 CrossRef CAS PubMed.
- C. S. Leung, S. S. F. Leung, J. Tirado-Rives and W. L. Jorgensen, Methyl Effects on Protein−Ligand Binding, J. Med. Chem., 2012, 55, 4489–4500 CrossRef CAS PubMed.
- E. J. Barreiro, A. E. Kümmerle and C. A. M. Fraga, The Methylation Effect in Medicinal Chemistry, Chem. Rev., 2011, 111, 5215–5246 CrossRef CAS PubMed.
- S. N. Hobbie, S. K. Kalapala, S. Akshay, C. Bruell, S. Schmidt, S. Dabow, A. Vasella, P. Sander and E. C. Böttger, Engineering the rRNA Decoding Site of Eukaryotic Cytosolic Ribosomes in Bacteria, Nucleic Acids Res., 2007, 35, 6086–6093 CrossRef CAS PubMed.
- S. B. Garber, J. S. Kingsbury, B. L. Gray and A. H. Hoveyda, Efficient and Recyclable Monomeric and Dendritic Ru-Based Metathesis Catalysts, J. Am. Chem. Soc., 2000, 122, 8168–8179 CrossRef CAS.
- I. M. Herzog and M. Fridman, Design and Synthesis of Membrane-Targeting Antibiotics: From Peptides- to Aminosugar-Based Antimicrobial Cationic Amphiphiles, MedChemComm, 2014, 5, 1014–1026 RSC.
- G. Guchhait, A. Altieri, B. Gorityala, X. Yang, B. Findlay, G. G. Zhanel, N. Mookherjee and F. Schweizer, Amphiphilic Tobramycins with Immunomodulatory Properties, Angew. Chem., Int. Ed., 2015, 54, 6278–6282 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1md00214g |
| This journal is © The Royal Society of Chemistry 2021 |