Open Access Article
Open Access ArticleMethods to probe the formation of biofilms: applications in foods and related surfaces
Yating
Huang
 *ab,
Sayak
Chakraborty
b and
Hong
Liang
*ab,
Sayak
Chakraborty
b and
Hong
Liang
 b
b
aDepartment of Material Science and Mechanical Engineering, Beijing Technology and Business University, Gengyun Building, Beijing 100048, China. E-mail: huangyat@gmail.com
bJ. Mike Walker '66 Department of Mechanical Engineering, Texas A&M University, College Station, TX 77843-3123, USA. E-mail: hliang@tamu.edu
First published on 9th January 2020
Abstract
Biofilms of bacteria affect product quality and safety of food. Bacterial adhesion onto surfaces of processing equipment in contact with foods is indicated by the formation of biofilms. To date, little is known about the principles of nucleation and growth of such films. There are many factors promoting biofilms, such as food-surface contact, nature of the food product, types of bacteria, and parameters associated with food processing. The synergetic effects among them make it difficult to probe the formation of biofilms. In order to obtain a fundamental understanding of biofilms related to food and to identify effective methods to study the same, we reviewed the literature for roles of surface topography of bacteria and their attachment on a substrate. Specifically, we evaluated methods to characterize the morphology and detect chemicals of biofilms. Through this effort, we recommend three effective approaches to probe biofilms: (i) observation with various microscopic methods with different view fields at the same point; (ii) in-depth data analysis during microscopic image processing; (iii) combinative study using atomic force microscopy (AFM) and chemical analysis. This review intends to help researchers develop effective methods to provide insight into biofilm formation and to subsequently find ways to control it.
Introduction
Biofilms are one of the key problems in the food and beverage manufacturing industry and are formed by contaminations of bacterial cells and organisms. Biofilms cause not only the degradation of equipment performance, but also the failure of hygienic control.1 The subsequent cleaning processes will increase consumption of time, energy, and water, leading to higher production costs.2 In order to avoid the risk of cross-contamination3 and bio-corrosion,4,5 a better understanding of the mechanisms of biofilm-development is required.Biofilm formation is defined as the initial process whereby individual bacteria adhere to a surface and produce extracellular polymers facilitating the growth.6 When the single bacterial attachment to a surface switches from reversible adhesion to irreversible adhesion, the biofilm grows explosively until the bacteria reach a balance with the available resources. The initial rate of bacterial (Staphylococcus aureus ATCC 12600 or S. aureus) adhesion to a substratum surface is as low as 106 bacteria per cm2,7 and covers less than 1% of the surface. In contrast, a mature biofilm has a thickness of up to 200 μm in a dairy processing pipe8 which means that it contains 1010 bacteria per cm2.9 Due to the exponential nature of its growth, the prevention of initial biofilm formation is an effective option to control contamination in food processing.
To date, the formation of biofilm is still poorly understood.10 Biofilm formation is an integrated process regulated by the properties of substrate surfaces, microbiological factors and environmental conditions. These properties are interchangeable. Food products provide shelter and nutrients to bacteria. In return, a bacterial cell adheres to and subsequently colonizes surfaces, which in turn makes the surface conditions easier for food deposition.11 On the other hand, the surface conditioning of macromolecules (such as whey protein) modifies the surface topographical and physicochemical properties, leading to a physical barrier for bacterial growth.12 In order to pinpoint mechanisms of formation of biofilms, principles of characterization and properties of bacterial contaminated surfaces need to be understood.
In this paper, microscopic inspection techniques to study biofilm formation are analyzed. The methodology in morphological and chemical characterization of biofilms is studied. This study aims to provide a guideline for researchers to effectively probe and control the formation of biofilms.
Biofilm formation in food processing
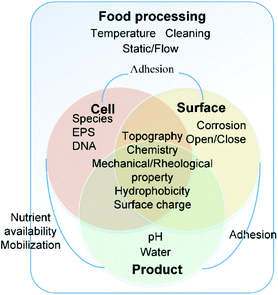
The interactions of bacteria and food processing surfaces during biofilm formation can be distinguished artificially into specific adhesion and non-specific adhesion13,14 to address different characteristics of interaction forces. Usually, specific adhesion and non-specific adhesion are studied by groups which focus on physical and biological properties, respectively.Non-specific adhesion is mostly studied by colloid science. It can be described typically by DLVO theory and its extensions. The dominant forces are Lifshitz–Van der Waals (LW), electrical double layer (EDL), and acid–base interaction (AB). It is influenced by the physical and physicochemical properties of bacteria, the surface and the near-surface environment, including chemistry, topography,15,16 mechanical properties, hydrophobicity17,18 and surface charge19 of both the bacterial cell and the food contact surface.20–24 These properties are not constant. The near-surface environment will change based on the food operating procedures, including the change of temperature,25 hydrodynamic conditions26 and equipment cleaning processes. Furthermore, the irreversible organic fouling will, in turn, alter surface properties11 (Fig. 1).
On the other hand, specific adhesion corresponds to the active attachment by the bacterial cell through sensing of the surface and communicating with other cells.27 In the presence of specific contributions, bacterial interaction forces are about 2 to 3 times stronger than those without specific contributions.13 Specific interactions will include specific information depending on the nature of the bacterial species. The bacteria sense the surface by chemical gradients, physical space and information from other bacteria's broadcast which is called “quorum sensing (QS)” or other signalling. Ihssen J and Ehli T28 stated that the growth rate of Escherichia coli is influenced via RpoS signalling. The bacteria detect the chemicals in the liquid and degradation of certain surface components. These chemical distributions have a great impact on physicochemical interaction29 and specific ligand–receptor interactions. The bacteria trap ions and small molecules to change pH in the microenvironment30 and to vary the DNA transformation rate in bacterial cells.27 Moreover, the bacteria feel and react to the constrained movement through appendages.31,32 The quorum sensing is a kind of communication between the individual cells by exchange of small extracellular molecules as messengers.33E. coli34 has been reported to communicate through controlling the surface charge through processing an ion-penetrable membrane and extracellular polymers. Exopolymeric substances (EPS) play an important role in this interaction.35
Despite the difference of focus, all the spatial morphology and substance chemical changes will leave clues on the surface and have direct or indirect impacts on the interaction. Different biofilm structures correspond to completely different growth mechanisms. “Thin” biofilms are formed in small scale flow cells at moderate to high hydrodynamic flow rates and will release a new cell progeny as soon as a cell attaches to the biofilm substratum. Then “thick” biofilms are formed in large-scale spaces with EPS. They are entirely different kinds of biofilms because the growth of the thin biofilm is limited by the supply rate of limiting-nutrient substrates, while the thick biofilm is limited because of its thickness and density due to diffusion-limitation of nutrients. The growth rate of both the attached and detached bacterial cells is a key factor to evaluate the spatial morphology of biofilms, which is important to understand the biofilm phenomena.
A detailed image of the surface will help to build the relationship between biofilm structures, interaction forces and other factors. Challenges are increasing due to the complicated interaction occurring at the length scale at a single cell level which is sometimes dramatically different from that at the macroscale. The bacteria only act in the microenvironment near themselves. The statistical pattern in the whole system is hard to apply to individual activities. Additionally, the biofilm formation involving live bacteria is time-dependent and environment-sensitive. The ideal observation of the biofilm formation process and evaluation of impact factors are supposed to be in vivo/in situ with chemical identification for uncultured microorganisms at high resolution in aqueous environments.
Characterization techniques
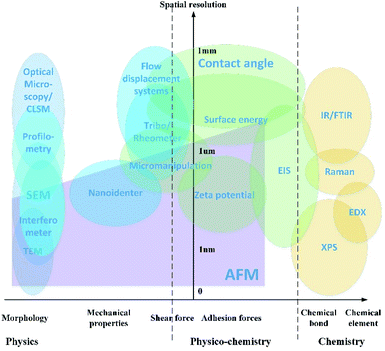
Detection strategies have been developed using the fundamentals of different subjects, including microscopic imaging of surface morphology, evaluation of interaction forces, and analysis of the chemical components. A map which presents the applications of the characterization approaches is shown in Fig. 2.The characterization method is evaluated by two aspects as shown in the map. The vertical y-axis is the approximate revolution of the instruments, varying from 0 at the base point to 1 millimeter. The horizontal x-axis indicates what kind of property is presented by each of the characterization technologies. The physical parameters include the surface morphology, mechanical properties for solids and rheology properties for liquids. The physicochemical properties include the adhesion forces and shear force in the length scale of nanometers. The chemical component analysis is the spectrum analysis of elements and chemical bonds.
In this paper, the application and development of the physical image and complementary technique for analysis of the surface chemicals are addressed based on the classification in the characterization map. Limitations and prospective evolution of the technology are stated.
Morphology of biofilms on food processing surfaces
The physical morphology of the surface shows the shape and texture information of surfaces. They provide direct evidence for the identification of the biofilm structures. The original surface topography of substrates and bacterial cells has a great impact on the first step of bacterial attachment. With the increase of the biofilm and the retention from the food products, the morphology of the substrate surface changes. The unique structures of the initial “thin” biofilm and further “thick” biofilm with EPS can be recognized directly by their physical morphology. Here we discuss several important microscopy approaches giving the morphology information of the biofilm from a picture (2D or 3D) of the sample surface.Lots of progress has been made in the visualization of the surface topography and biofilm structure from the nanometer scale to the micrometer scale. Both static and dynamic processes are observed. Confocal laser scanning microscopy (CLSM), scanning electron microscopy (SEM) and atomic force microscopy (AFM) are the three most important techniques. A brief comparison of the microscopic methods used for the biofilm study is summarized in Table 1.
| Microscopy technique | Application | Limitation | Sample preparation and environment |
|---|---|---|---|
| Confocal laser scanning microscopy (CLSM) | Reconstruction of the 3-D image, in situ visualization of the biofilm, mixed-species biofilm, quantitative assessment of bacteria, a combination with a flow chamber, high throughput detection | Low axial resolution. Slightly lower resolution. Limited laser penetration | Fluorescence label. No special requirement |
| Scanning electron microscopy (SEM) | Imaging sample structure details in high resolution, good at the observation of surface texture and membrane. Correlation study with other visualization methods for a qualitative study imaging functionalized AFM probe, combined with EDX | Complex sample preparation. Low axial resolution. Sample damage. Only solid samples allowed | Conductive or gold-coated samples. High-vacuum |
| Environmental-SEM | Imaging hydrate samples (cell, fouling), imaging functionalized AFM probe | Lower resolution compared with SEM. Sample damage | No special sample treatment. Low-vacuum |
| Focused ion beam-SEM (FIB) | Cross-sectional imaging, 3-D reconstruction by the “slice and view” process | Expensive and time-consuming. Sample damage | No special sample treatment. Vacuum |
| Atomic force microscopy (AFM) | 3-D reconstruction of the surface, roughness analysis, topography change with controlled environmental parameters | Small scan area (max: 150 × 150 μm). Sample damage. Fragile probe with wearable tip. Time-consuming | Cell immobilization and sample size limitation. No special requirement |
Confocal laser scanning microscopy-CLSM
Confocal laser scanning microscopy (CLSM) is an important tool for studying the structure of biofilms because of its strong capability of real-time visualization of fully hydrated, living samples. The limitation of the spatial resolution of light microscopy is improved by using a fluorescence technique which brings “optical microscopy into the nanodimension”. Both qualitative and quantitative information regarding the biofilm can be achieved with this “super-resolution optical microscopy”. From the references in this review, CLSM is the only method to observe in situ and even measure the growth rate of the biofilms and the cell behaviours of attachment, detachment or re-attachment and continue to accumulate producing high diffusion biofilms.Reconstruction of a 3-D image is the basic function of CLSM and is used for the analysis of the biofilm structure to reveal the diversity of biofilm architectures. Bridier et al.36 combined 96-well microtiter plates with CLSM to observe the three-dimensional structures of the biofilms of 60 pathogens. Images of some specific structures were captured in situ and with high resolution, such as the mushroom-like structures in Pseudomonas aeruginosa (P. aeruginosa) and hollow voids in Salmonella enterica (S. enterica) biofilms. They qualified the micrometer scale feature size of the biofilm structure from image stacks. The evidence from the CLSM images indicated the presence of bacteria-free exclusion zones (EZ) at the surface of a hydrophilic polymer material, such as Nafion.12 Some widespread bacteria such as Escherichia coli (E. coli) O157: H7, Staphylococcus aureus (S. aureus), and Listeria monocytogenes (L. monocytogenes) are included in this research. The thicknesses of EZ, measured from the CLSM images, ranged between 40 and 80 μm, for bacterial cells suspended in tryptic soy broth. The architecture of biofilms observed with CLSM shows their variation with strains, such as net-like adhesion cells,37 aerial filamentous structures,38 flat multilayers and honeycomb-like structures.39
CLSM and its associated fluorescence probe techniques are of great importance as they enable the in situ analysis of the kinetics of the biofilm formation. In situ detection by CLSM shows that the number of cells in the bottom layer is 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 higher as compared to that in the upper layer.40 The fact that the biofilm structure differed in its architecture proves the special growth behavior of Listeria. Olszewska41 recorded the three-dimensional development of the L. monocytogenes biofilm in five sanitizers. The live and dead cells are clearly visualized with fluorescent dye labels. Moreira42 marked the cellular extracts of E. coli with crystal violet. By doing so, they discovered that surface conditioning with cellular compartments will influence the initial bacterial adhesion. This work leads to a greater understanding of the factors that influence biofilm formation.
log10 higher as compared to that in the upper layer.40 The fact that the biofilm structure differed in its architecture proves the special growth behavior of Listeria. Olszewska41 recorded the three-dimensional development of the L. monocytogenes biofilm in five sanitizers. The live and dead cells are clearly visualized with fluorescent dye labels. Moreira42 marked the cellular extracts of E. coli with crystal violet. By doing so, they discovered that surface conditioning with cellular compartments will influence the initial bacterial adhesion. This work leads to a greater understanding of the factors that influence biofilm formation.
Furthermore, the fluorescence in situ hybridization (FISH) technique has been applied for the mixed-species biofilm study and also chemical composition identification. It is based on the CLSM observation of multiple fluorescent probe labels. FISH enables evaluation of competition and the symbiotic relationship between different microorganisms. For instance, the population proportions of Salmonella spp. and L. monocytogenes in milk samples are statistically analyzed with two fluorescent oligonucleotide probes.43 Garcia-Almendarez44 applied FISH to identify Lactococcus lactis (L. lactis) UQ2 on Listeria monocytogenes (L. monocytogenes). In addition, it can be used to identify microorganisms and organic material fouling. The protein from fish soil fouling and L. monocytogenes cells are marked in different colors.45 The heterogeneous distribution illustrates that organic material retained in the surface features increases the possibility of bacterial retention.
Combined with image processing software, CLSM provides a quantitative assessment of bacteria. Mathematical methods such as the multifractal analysis can not only quantitatively analyze the upper percentage coverage of the biofilms on the surfaces but also quantify the number of cell clusters.42 With a DNA-binding dye, a number of cell copies for a series of bacteria are presented, including Pseudomonas aeruginosa (P. aeruginosa), Burkholderia cenocepacia (B. cenocepacia), S. aureus, Propionibacterium acnes (P. acnes) and Candida albicans (C. albicans).46 For Salmonella spp. and L. monocytogenes, the probes targeting the rRNA (a conserved structure) are used.43 The surface-enhanced fluorescence (SEF) technique is introduced for distinguishing the bacterial cell surface deformation during adhesion. This technique offers access for the nanoscale monitoring of the influence of species and biomass during biofilm formation. The inability of most unmodified CLSM to detect fluorescence images is perhaps a disadvantage of such instruments. A special light source must be used in order to stimulate fluorescence.
In contrast to fluorescent proteins and dyes, bacterial bioluminescence is the production and emission of light by a living organism during the expression of the lux genes.47 No excitation light or the addition of exogenous substrates (such as the ATP bioluminescence48,49) is needed. It is the only way to measure real time metabolic rates and thus can be a particularly powerful technique for understanding biofilm processes. It should be noted that fluorescent proteins can produce a signal output that relates to the amount of biomass,50 whilst the output from lux-luminescence depends on the adenylate energy charge of the cell. If the emitted signals could be separated by peak positions, the two outputs together might be powerful tools for measuring instantaneous growth rates. Moreover, lux-luminescence is the only method that can be used to obtain accurate kill rates when studying actions of biocides or inhibitors on biofilms. Therefore, the development of more kinds of luciferase probes and persistence luminescence probes is needed.51
The flow chamber is powerful for evaluation of the influence of flow conditions when combined with CLSM. The hydrodynamic force plays a significant role in the micron-scale bacterial settlement. Under static fluid conditions, the surrounding liquid has hydrostatic pressure on the bacteria. Under dynamic flow conditions, the shear force and mass translation will influence the possibility of bacterial settlement on the surface. The control of the flow rate allows the estimation of the cell binding force through defined hydrodynamic shear forces to be applied (assuming laminar flow) and tapered flow cells used to produce defined shear force gradients. Boks et al.52 studied the forces involved in bacterial adhesion to hydrophilic and hydrophobic surfaces using a parallel-plate flow chamber. They concluded that the bond between a substratum surface and a bacterium becomes stronger after initial adhesion. Therefore, the first layer of attached living cells will remain in the steady state as a thin biofilm as the subsequent layers are easier to be removed by hydrodynamic forces. The microfluidic environment restricted by surface topology increases the complexity of fluid analysis.53 The so-called flow displacement systems are based on phase-contrast microscopy and a parallel plate flow chamber.7 Based on this technique, J. Li et al.54 reported an in situ method to observe adherent bacterial cells on non-transparent substrates and in a dynamic flow. Mathematical processing is necessary to enumerate the bacteria from the fluorescence image. Adhesions of S. aureus on polished and non-polished stainless steel, titanium alloy and polyvinyl chloride surfaces are observed within such a system. The results indicate that the number of bacteria adhering on the bottom increases linearly with distance from the inlet of the flow chamber. CLSM coupled with a controlled flow chamber is by far the most useful method to monitor experimental biofilms in real time from initiation to full maturity.
Multiple samples can be observed simultaneously in CLSM to ensure consistency of experimental conditions and to reduce experimental time. In Bridier's36 operation, CLSM combined with 96-well microtiter plates was employed to observe the three-dimensional structures of biofilms formed by as many as 60 pathogens simultaneously. Guilbaud39 takes images for 96 samples from varied environments. Therefore, CLSM has a strong ability for high throughput detection as compared with SEM and AFM, which are both time-consuming.
It is also possible to achieve readings of biofilms and the cell attachment using an ordinary light microscope mounted with a flow chamber, especially with a high magnificent (×100) lens. Only 2-D images can be recorded using a CCD, while CLSM provides the 3-D image of biofilms and thus the film thickness can be evaluated.
CLSM has become a standard technique for the study of in situ biofilm formation in an aqueous environment with appropriate fluorescent probes. Its significant application for studying biofilms exposed to a dynamic fluid flow is irreplaceable by SEM and AFM. So large-area imaging and deeper penetration of lasers are required for better description of the biofilm formation process. Moreover, new labelling techniques are required to improve the visibility of the fluorescent marks inside the biofilms and sub-liquid environments. Examples include quantum dots (Q-dots) and metallic nanoparticles. Fixation of the label remains a challenge. This means that new discoveries should confirm that “what you probe is what you see”.55 Zhang et al.56 employed AFM for the characterization of graphene Q-dots (a new kind of fluorescence label). The comparison of images in the bright field and the dark field shows that HeLa cells are successfully marked with the graphene Q-dots.
Scanning electron microscopy-SEM
A scanning electron microscope uses a beam of accelerated electrons as the source of illumination which scans across the sample surface, and the surface topography information is obtained from the signal of energy change. As the wavelength of an electron can be up to 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 times shorter than that of visible light photons, SEM can reveal the small structure of objects with resolution down to 0.5 nm using the most common signal of secondary electrons. It should be noted that SEM graphs have a large depth of field yielding a 3-D appearance, although lacking vertical resolution.57
000 times shorter than that of visible light photons, SEM can reveal the small structure of objects with resolution down to 0.5 nm using the most common signal of secondary electrons. It should be noted that SEM graphs have a large depth of field yielding a 3-D appearance, although lacking vertical resolution.57
Accordingly, SEM has been employed to observe the structure of biofilms. Its large depth of field is good for the visualization of the biofilm spatial structure.58,59 Borucki60 demonstrated a robust relationship between the strains and biofilm structure. A high-biofilm-forming strain produces a dense and three-dimensional biofilm structure. The biofilm structure is sensitive to the variation of pH values. The net-like biofilm structure is built at pH 7, while a monolayer biofilm is built at pH 6.61 Individual cells in a biofilm can also be visualized with SEM. Combined with dedicated imaging software, SEM can offer quantitative counting of bacteria.62
The SEM technique can be employed to access the biofilm structure during biofilm formation at different time periods. In Oliveira's63 model, the mature biofilm of L. monocytogenes adhered to a stainless-steel surface takes as long as 240 h to form. Samples of L. monocytogenes strains are collected for analysis of multi-layer structures.64 A mature biofilm is observed within 12 h and after 24 h, the cells are surrounded by a matrix.
Characterization of the surface with SEM is preferred in the study of nanocomposite coatings and surface texture. Santos65 shows the topography changes of modified stainless steel surfaces. The modifications include SiF3+ ion implantation, diamond-like carbon (DLC) sputtering, SiOx plasma enhanced chemical vapor deposition (PECVD), autocatalytic Ni–P-PTFE and silica coating. The TiN coatings with different silver contents are imaged by SEM combined with EDX (Energy-dispersive X-ray spectroscopy) for analysis of the film structure and chemical composition.66 SEM images provide visual evidence of how bacteria maximize the contact area and bind at different surface features with sizes lower than 1 micrometer. Examples are summarized in Table 2.
| Surface material | Surface topography | Bacteria | Conclusions | References |
|---|---|---|---|---|
| Stainless steel | 0.7 μm trenches | P. aeruginosa, P. putida, D. desulfuricans, Rhodococcus spp | Higher attachment; cells align with trenches | 71 |
| Polydimethylsiloxane (PDMS) | Post-array with a distance of 300 nm to 1 μm and space of 0.8–4 μm | P. aeruginosa, E. coli, B. subtilis | Adhesion changes significantly when the dimensions of confined spaces match those of bacterial cells | 72 and 73 |
| Silica/alumina | Silica: wells of 0.5 μm with 0.2 μm spacing; 1 × 1.5 μm rectangles with a spacing of 2 μm; 1 × 2 μm rectangles with a spacing of 0.5 μm; depth of all wells 27–32 nm; alumina: 20 or 200 nm pores | E. coli, L. innocua, P. fluorescens | Bacteria tend to maximize contact areas to bind to surface features | 74 |
| Polydimethylsiloxane (PDMS) | Hexagonal patterns of 2.7 μm in height, 3 μm in diameter, separated by 440 nm trenches | E. coli | Bacteria build net to cover unfavorable features with flagella to aid adhesion of additional cells | 32 |
| Polycarbonate | 120 nm pits with 300 nm center–center separation | S. aureus | Nanopatterning promotes the early-adhesion of S. aureus cells to the biomaterial surface | 75 |
Samples for traditional SEM observations should be conductive at high voltage. Insulated samples should be coated with gold or gold alloy films with a thickness of several nanometers.22,59,67 The sample preparation usually involves drying, fixing, dehydration and coating64 which are time-consuming and troublesome. Moreover, the fixation, dehydration, and coating with metal will kill the living bacteria which will result in the destruction of the biofilm structure and sample artifacts.68 An example is the shrinkage of biofilms due to the EPS collapse during drying.69 The limitation of SEM has resulted in an alternative application of environmental-SEM (ESEM), which retains the natural state of samples, despite sacrificing a little bit of the resolution.69,70
For most cases, ESEM investigates the sample without any pretreatment. It provides the opportunity of observation of hydrated biofilms76,77 or even fluids such as milk.78,79 Comparison studies with SEM show that ESEM will not lead to the dehydration and loss of mass.69,80 However, the damage caused by the energy of focused electron beams will increase at more than 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×.69,81 Therefore, the balance of resolution and influence on the sample should be considered.
000×.69,81 Therefore, the balance of resolution and influence on the sample should be considered.
SEM images, especially ESEM, provided the ex situ evidence of successful bacterial mobility on the AFM probe. Functionalized AFM tips with adhesion bacteria are widely used in the studies of interactions between the bacteria and the substrate surface. Such kinds of functionalized AFM tips can be shown by SEM pictures. Two decades ago, a modified AFM tip covered with many irregular E. coli cells was shown in SEM photos.82 Then the cells on the AFM cantilever or tips became smaller and smaller. Bowen21 observed the AFM colloid probes with active S. cerevisiae bacterial cells and Aspergillus niger spores on a cantilever. Furthermore, a single cell was attached onto the tip of the AFM probe.23
SEM can only be used for studying surface morphology. But the understanding of coating film adhesion and the biofilm structure offers useful information for biofilm formation. The SEM image of gradient Ni–P-PTFE coating in cross-sections helps improve the coating adhesion.83 When the cross-section is studied, the samples are usually cut off with a diamond saw blade and the cross-section should face the electron beam. On the other hand, the focused ion beam (FIB)-SEM technique should be considered. When a standard SEM with a backscattered electron image is coupled with a focused ion beam (FIB) milling tool, the “slice and view” processes provide multiple images to reconstruct the 3D model of the sample. The thickness of the FIB milling section is only 10 nm,57,69 much thinner compared to the 30–150 nm slices cut using a commercial diamond knife. FIB-SEM has been employed to study the connections of cell-to-cell and cell-to-EPS within the sessile communities.84 Limitations of FIB-SEM are similar to those of SEM, the high vacuum environment and possible damage of samples. Besides, the milling process is time-consuming and the sample will change during this long time processing.
The use of an electron beam as the source of illumination effectively reduces the diffraction effect, so SEM and ESEM techniques give an ultra-high resolution image in the xy dimension. But the 3-D appearance image cannot provide quantitative information in the axial direction. Complex sample preparation and sample damage will limit SEM from in situ observation, and only solid samples are allowed. In most study cases, SEM is one of the characterization methods, and complementary studies are required.
Atomic force microscopy-AFM
AFM is a powerful tool for obtaining true 3-D surface topography because of its featured functions. This section will discuss the imaging of biofilm formation with AFM, together with its challenges.AFM has been employed for visualizing surface morphology, along with examining microbiological and organic fouling, for nearly 30 years.98 One of the earliest AFM observations of biofilms was performed by Bremer et al.99 Hydrated freshwater bacterial biofilms on copper surfaces were imaged to show a heterogeneous biofilm, with depth and distribution varied with surface texture. Then biofilms grown on different types of steel surfaces were analyzed with AFM.76,100 Although the bacterial sample was still not in situ or in vivo, the researchers paved the way for getting qualitative and quantitative information on the biofilm structure in high resolution. Therefore, unlike SEM, the height evaluation of biomass and corrosion of surfaces can be investigated. AFM has rapidly evolved into a common tool for biofilm studies, with varied substratum surfaces and bacterial species. Examples can be found in a series of reviews.101–106 Topography examination with AFM has been widely used for visualization of single cells and biofilms on food contact surfaces with high resolution and sensitivity. Table 3 summarizes some of the studies in food processing.
| Bacteria | Substratum | Operation mode | Observations | Reference |
|---|---|---|---|---|
| S. aureus | Glass | Tapping | Surface grooves and perforation; cell debris | 85 |
| E. coli | Stainless steel | Tapping | Cell dimension | 86 |
| P. pastoris | Stainless steel | Tapping | Protective barrier failure; intracellular content leakage | 87 |
| S. typhimurium, S. aureus, L. monocytogenes | Mica sheet | Dynamic force | Morphology and cell count | 88 |
| S. xylosus, Z. bailii | Cell-tak-coated glass | Intermittent contact | Surface indentation | 89 |
| E. coli | Glass | Contact | Roughness change. Morphology | 90 |
| P. aeruginosa | Mica | Tapping | Matrix-like EPS materials among the cells | 91 |
| Sulfate-reducing bacteria (SRB) | Mica | Contact | Cell morphology | 92 |
| P. fluorescens | Gold | Contact | Cell and surface morphology | 93 |
| Salmonella | Mica | Contact | Cell and surface morphology | 75 and 94 |
| S. aureus | Nanopatterned polycarbonate | Intermittent contact mode | Cell and surface morphology | 75 |
| E. coli | Mica | Non-contact | Stain dimension | 95 |
| P. aeruginosa, S. aureus | Titanium oxide coating with 0.5 μm featured pattern | Contact | Cell morphology and dimension | 15 and 96 |
| C. botulinum | Mica | Tapping | Exosporium fragment morphology and dimension | 97 |
Single cell imaging has been used to investigate morphological variation due to the influence of sanitizers and environmental conditions. The bacteria on the substrate surface are qualitatively and quantitatively analyzed. Cui et al.85 observed the surface morphological alterations of Gram-positive and Gram-negative bacterial cells induced by epigallocatechin-3-gallate (EGCG) and H2O2. The aggregates, nanoscale perforations or microscale grooves in the cell envelopes were captured by AFM as the evidence for explaining cell lysis. The quantitative morphological parameters include height, lateral dimension and surface roughness (Ra and RMS), which can be obtained from the 3-D surface topography. The surface roughnesses of cells usually increase due to treatment with sanitizers.89,90 Kuda88 compared the amount of cells before and after drying on the stainless steel surface through AFM images. The results confirm that small food sediments provide sufficient protein and carbohydrates, which protect the adhesion pathogens on the surface. Only 0.05 mg dried nori, 5 mg fresh carrot, and 100 nL milk or soy milk on a surface of 10 mm in diameter were good enough for bacterial survival. Therefore, AFM is a promising approach to evaluate the dynamic changes of the biofilms.
Information regarding biofilm topography and cell-extracellular material on the food contact surface can be obtained by AFM. The sample preparation for AFM imaging developed from simple air drying to keeping live microbes in buffer solutions. High-resolution AFM images of dry samples can show surface topography details of the substrate, including the biofilm structure and EPS materials. The air drying process will immobilize the bacterial cell and surrounding materials. With this process, Yang et al.91 distinguished the matrix-like material in different P. aeruginosa biofilms with different EPS materials, Pel and Psl polysaccharides. The matrix-like material is seen in PAO1 and the PAO1DpilA mutant. On the other hand, they are absent in the case of the PAO1 ΔpilA ΔpelAΔpslBCD mutant. From these results, it is concluded that different EPS materials have different impacts during biofilm formation. Fang92 presented high-resolution topographical images of a single bacterium (sulfate-reducing bacteria, SRB) and biofilm on a mica surface. They proved the capability of AFM for observation of a single cell. Diaz93 studied the role of different structural factors of metal surfaces at the early stages of biofilms.
Differences between bacteria attached on the ordered or disordered nano/micro structure surfaces are observed and evaluated from the aspects of cell morphology, length, orientation, and flagellation. S. Aguayo75 imaged the nano-topography of S. aureus and the surrounding capsule to observe the cell attachment to the surface. In the AFM image, the capsules surround and partially cover the S. aureus cell on the flat surface, but they are unable to cover the cell on the surface with nanopatterns. Such kinds of capsules are destroyed during sample preparation in FIB-SEM. These research studies have important implications on the design of modified surfaces to prevent biofilm formation and to enhance biofouling removal processes. The roles of cellulose, curli, and surface proteins during bacterial growth in different cultivation environments are compared by AFM.94 Samples were obtained after different growth times in colonies on agar plates and those in a liquid environment. AFM images illustrate both curli and cellulose. Curli has an important impact on the formation and the morphology of a biofilm. But the effects of BapA are not seen. These studies demonstrate that AFM can efficiently monitor the morphology and surroundings of bacteria during biofilm formation.
Examination of biofilms in an aqueous environment using AFM is required for in situ observation because sometimes important structural changes would happen during dehydration. In Kailas's107 experiments, a comparison of AFM images of Clostridium botulinum (C. botulinum) exosporium fragments shows the variation of fibrils in air and water. The beads are found in dehydrated samples but not in samples underwater. The researchers attribute this to the dehydration process. However, AFM imaging of bio-cells in such an environment is not easy. First, the length scales of biofilms are beyond the measurement range of many commercial AFMs. AFM was originally designed for measurement of length scales from nanometers to micrometers. Sometimes, the biofilm thickness may exceed 200 μm (ref. 8) which would mean that only selected parts of the biofilm could be monitored with AFM. In addition, the initial attachment of bacterial cells onto the surface is via weak Lifshitz–Van der Waals forces. As a result, the adhesion cells are probably disturbed by the probe of the AFM. Thus although immobilization of cells is necessary, it sometimes impacts the original adhesion and makes the in situ process not as natural as expected. Kailas et al.107 monitored S. aureus in situ in culture media by AFM. They trapped cells mechanically on a lithographically patterned silicon wafer with a square lattice of holes. This physical immobilization method kept the bacteria alive. However, because the bacterial morphology is varied, the surface pattern for immobilization of different cells is still a big challenge. Furthermore, soft and gelatinous food material in a liquid environment increases the risk of fouling material attaching to the probe, especially in contact mode. So liquid cells for in situ AFM studies are a better choice for interaction-force studies and the situation that focuses on the change during dehydration, without restricting the substrate topography.
Another important function of AFM is surface roughness evaluation. Early research usually described surface roughness as quantitative statistical parameters such as Ra (arithmetical mean deviation) and RMS (root mean square). Ra is easy and sometimes effective for evaluation of surface cleanness.3,71,98 Experimental results show the close relationship between Ra and bacterial attachments to the surface.11 On the other hand, some research also provides conflicting results108 that indicate that there is no correlation between surface roughness and bacterial attachment when the surface roughness Ra of stainless steel changes from 0.01 μm to 0.9 μm. Ra is obviously not enough to describe the surface topography. Different surface profiles may present the same Ra value, which means the loss of some important surface topographical information. Special parameters are developed for various applications to capture the necessary surface topographical information.109 Nowadays, the theoretical model shows that surface topography changes the surface force distribution because every atom on the surface has a contribution based on its position. AFM provides a true 3-D reconstruction of the surface with high resolution which preserves the topographical information better than other microscopic methods and helps to draw useful information. In Aguayo's75 study, depth of nano-pits of about 70 nm could be measured quantitatively from the cross section by AFM. Moreover, topographical effects may compound with other factors such as surface energy, and adhesion mechanisms might ‘switch on’ at different topographical scales.110 No parameter can be defined to present these complex effects. Ra is not perfect but still useful and widely applied for the evaluation of surface roughness. In further studies, AFM surface topography imaging combined with interaction evaluation methods is expected to discover the mechanism of biofilm formation on various surface morphologies. More information will be revealed with image processing methods.
AFM has a high vertical resolution which is an important complementarity of other techniques. Besides, the potential for detection in a natural environment is exploited. However, the challenges increase with sample mobilization, wearable tip, and limitation of scan area. Moreover, the chemical information is hard to extract from the laser reflection signal. The high-resolution scanning probe microscopy (HR-SPM) technique for molecular structure identification is a way to ‘see’ the chemical components. However, the scan area is limited in several molecules and it needs special tip modification and the testing environment and sample preparation conditions are far from the comfort zone of the biofilm. Therefore, combination and comparison study with other techniques will provide complementarity of the potentialities.
Complementary study with multiple techniques
More evidence is needed for further understanding of the structure of biofilms and function of bacterial communities. Two strategies of complementary studies with multiple techniques are discussed in this section. The comparison study of microscopic methods at the same point will achieve a full picture of the biofilm from the nanoscale to mesoscale. The combination of microscopy methods and chemical analysis methods will allow the exploration of active attachment by the bacterial cell.Comparison study of microscopic methods
The relationship between the biofilm structure on the microscale and details of the biofilm topography on the nanoscale are needed to discover the mechanism of the biofilm formation process. The intracellular and extracellular chemicals will also leave traces in biofilms and the substrate surface. Observing the same position in different instruments is the most direct method. This will require transferability of samples between different instruments and micro-/nanometer relocating. The methods comprise image identification and mechanical positioning.SEM is an extremely convenient tool for surface feature identification because of its wide range of magnification (25× to 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×). The comparative analysis with SEM will provide qualitative support to other quantification methods. For example, the results of SEM compared with CLSM with fluorescence-labeled samples show clearly the position and posture of adhesion bacteria, with a wide range of magnifications.32,37,73,74,80 SEM also shows high correlation with chemical and microbiological methods for quantitative analysis.61,111–115 The correct interpretation of these results provides information on the three-dimensional development of the biofilm. During the SEM characterization operation of AFM probes, the magnification of SEM is continuously adjusted to focus on the position of the AFM tip and the adhesion bacteria. By doing so, it is easy to focus on a selected nanoscale feature.
000×). The comparative analysis with SEM will provide qualitative support to other quantification methods. For example, the results of SEM compared with CLSM with fluorescence-labeled samples show clearly the position and posture of adhesion bacteria, with a wide range of magnifications.32,37,73,74,80 SEM also shows high correlation with chemical and microbiological methods for quantitative analysis.61,111–115 The correct interpretation of these results provides information on the three-dimensional development of the biofilm. During the SEM characterization operation of AFM probes, the magnification of SEM is continuously adjusted to focus on the position of the AFM tip and the adhesion bacteria. By doing so, it is easy to focus on a selected nanoscale feature.
Advanced in-depth data mining techniques are supposed to improve the efficiency of comparative analysis. Image processing software and data statistical solutions are needed to obtain quantitative information from the microscopic images. Through the bacterial morphology recognition, the quality of bacteria is analyzed using a computer.116,117 Moreover, sometimes scientists will process large amounts of image data. Plenty of images can be saved using a high speed and high resolution camera during the observation of biofilm dynamic processes.118 Dozens of samples are pictured during the high throughput observation in CLSM.119 The edge detection and cell contours are still a challenge in the situation of touching cell recognition. Machine leaning has been used for object recognition in many areas, such as face recognition in a crowd. So new bacterial recognition strategies are expected to be built on a bacterial topography database.
CLSM and AFM are routinely used in the morphology imaging of neuron cells.120 The AFM results give the topography of ‘holes' in the cell surface varying from 0.01 to 3.5 μm2 with depth varying from 2 to 178 nm. Fluorescence images characterize the biological structure with a fluorescent label. The same physical structures in both techniques are used for the identification of corresponding positions. The advantages of CLSM in biofilm structure observation in the liquid are combined with cell surface topography evaluation by AFM to cover different length scales of each piece of equipment.121
Combined CLSM/AFM equipment can perform simultaneous topographic measurement and chemical identification with single molecule sensitivity. The system consists of an AFM scan head mounted on the inverted CLSM. The synchronization of image acquisition should be considered in the analysis. Fluorescence labels are employed to mark important chemicals, such as DNA, proteins, and lipids.122–125 Combined AFM and fluorescence spectral imaging of Rhodobacter sphaeroides (R. sphaeroides) helps to distinguish membrane fragments from other surface features.126 The membrane structural features and evolution associated with cellular signaling pathways are revealed.123,127 The relationship between bacterial viability and cell surface topography is assessed in special solvent.128 However, the fluorescence probe might be mobile which leads to untrusted results.129 Moreover, transparent substrates must be used for fluorescence light going through the observation system, which will limit the choice of substrates. Anyway, the integration of CLSM/FM (Fluorescence Microscopy) and AFM is a robust, widely used approach to study the state of bacteria and offers more chemical specificity than AFM alone.
Combination of microscopy and spectroscopy methods
The organic chemicals around the bacteria and inside the bacterial cells have an important impact on biofilm formation, especially to the specific adhesion. These chemicals are needed to be studied together with the surface morphology to evaluate the correlation and causal relationship between chemicals and biofilm formation. In CLSM microscopy, the chemicals are identified using a fluorescent probe. As discussed in Section 2.1, it is commonly used as a tracer, and this is not enough to qualify and evaluate the chemical effects. In the SEM technique, the chemical element information is obtained from the EDX spectrum. This is useless for the recognition of organic molecules because they are all made up of basic elements, carbon, hydrogen, oxygen etc. Therefore, other chemical analysis methods should be combined. As shown in Fig. 2, IR spectroscopy and Raman spectroscopy are widely used as chemical analysis methods. The combination with high-resolution microscopy methods will improve the spatial resolutions which are limited by the optical diffraction.Spectroscopy methods have been combined with CLSM and SEM approaches. FTIR (Fourier-transform infrared spectroscopy) and Raman analysis have been used for EPS detection during Salmonella biofilm formation in meat processing environments.35 The results reveal that the special chemical composition, EPS, consisting of polysaccharides and proteins, only shows up in the biofilm. On the other hand, these EPS chemicals are absent in the cultivation of corresponding planktonic cells. Therefore, EPS is proved to be an important marker in biofilm formation. The combination of these non-destructive and label-free chemical analysis techniques provides new insight into specific adhesion study. However, the combining CLSM or SEM with spectroscopy study is operated separately and can just provide the overall information of existence of EPS. But the distribution of EPS is spatially and temporally non-uniform.130 Therefore, point-to-point topology-chemical maps are required for a better understanding of EPS and biofilm formation.
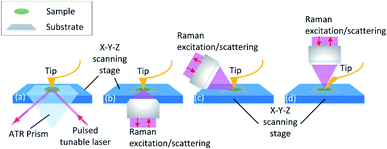
The equipment combing AFM with spectroscopies such as infrared spectroscopy (IR) and Raman spectroscopy is investigated. Through the adjustment of the tip position and the excitation spot, the topography and chemical information map can be collected point-to-point in nano-scale spatial resolution. The principle of the equipment set is illustrated in Fig. 3. Detailed explanation will be discussed in the following sections. The instrumental combination of AFM and vibrational spectroscopy provides the opportunity for a comprehensive understanding of the influence of surface topographical and chemical properties on biofilm formation.
AFM-IR detects the sample thermal expansion with radiation from the infrared-pulsed light with the AFM tip at the atomic scale. The sample is placed on an IR transparent prism made of ZnSe and irradiated with a tuned internal reflection laser (Fig. 3(a)). The AFM tip is sensitive to the thermal expansion due to the heat converted from the absorption light with the corresponding wavelength, and the induced ringing of the cantilever is recorded.131 Then the IR spectrum is obtained with FFT of the original cantilever ring-down signal in the time domain. Dazzi132 proved that the light absorption of a single E. coli cell response to different wavenumbers measured with AFM-IR is consistent with that in its biofilm measured with FT-IR. Barlow also showed good agreement between AFM-IR and FT-IR spectra of Pseudomonas protegens monolayer biofilms on a polyether–polyurethane (PU) film.133
AFM-IR performs well for the investigation of microorganism samples on micro- and nanoscales. The samples range from a single cell to biofilm. For single cell investigation,132 distributions of protein and phosphate groups (related to the DNA) inside E. coli are mapped through the amide I band (1660 cm−1) and amide III band (1240 cm−1). The amides are more concentrated in the center of a bacterial cell. This study shows potential of AFM-IR in the chemical analysis with the spatial resolution down to dozens of nanometers within the bacteria. AFM topography and the corresponding AFM-IR map at 1740 cm−1 IR absorption reveal the lipid production of Streptomyces bacteria in the first 12 hours of culture growth.134 This indicates that AFM-IR is sensitive to chemicals inside the bacteria cell and at the biofilm surface. It is possible to resolve some of the characterization problems, such as detection of untouchable bio-material, bioproduction and material inside the biofilm during the specific adhesion. AFM-IR is suitable for samples with low thermal conductivities and high thermal expansion. The spatial resolution is as low as 200 nm. However for non-isolated objects, thermal diffusion can limit the resolution. Samples are all dried in the presented studies to avoid the IR absorption by water. Some unexpected probe–sample interactions would result in the variation in local spectra, so this effect should be removed through the correction of mapping data with traditional FT-IR spectra. Moreover, an IR transparent prism underneath the sample limits the choice of substrate materials. Some of the multilayer polymer films for food packaging have been studied.131 Food contact materials such as stainless steel for industrial food processing cannot be easily operated in such a system.
Raman spectroscopy accesses the chemical content through the detection of Raman scattering. The biggest challenges of Raman spectroscopy are the poor Raman signal compared with strong background signals. The combination of AFM and Raman is more than the functional superimposing of chemical analysis on ultra-high resolution AFM. The Raman signals in local areas between the tip and surface can be enhanced up to 108 theoretically using a conductive metal such as gold or silver nanoparticle coated AFM tips.135 This tip-enhanced Raman spectroscopy (TERS) was invented by laboratories of Zenobi,136 Kawata137 and Anderson138 in the year 2000, and then it was developed by many researchers. Thus the correlated topography and chemical information on the interested molecules located under the AFM tip are collected simultaneously. Therefore, this technique combines the chemical fingerprint recognition provided by Raman spectroscopy with a nanoscale topographic resolution provided by AFM. Because the equipment for Raman spectral detection is stable and robust with limited numbers of standard and modular parts, the combination of Raman spectroscopy and AFM is easier and the TERS instruments are quickly commercialized. The applications of TERS in food processing related bacterial cell characterization are listed in Table 4.
| Bacteria | AFM operation mode | Tip modification | Morphology observations | Chemical observations | Reference |
|---|---|---|---|---|---|
| B. subtilis | Tapping | 14 nm Au-nanoparticle coating | Phase map of the selected region on the spore surface | External glycoprotein layer | 142 |
| S. epidermidis | Non-contact (intermittent contact) | Coated with 20 nm Ag film | 3-D topography of cells | Surface: peptide or a protein; lipids and saccharide/peptide mixtures | 141 |
| S. epidermidis | Non-contact | Coated with an Ag film | 3-D topographic of a single cell | DNA and RNA; surface: sugar and peptide | 139 and 140 |
| S. cerevisiae | Tapping for topography; contact for elastic properties | Electrochemical etching gold wire | 3-D topography of cells and surface profiles | Protein, lipids, and polysaccharide in the cell. Highlight the GDH protein spectrum analysis | 143 |
AFM and TERS approaches offer the opportunity for deeper insight into cell characterization, which can build the point-to-point variation between cell morphology and chemical information. N Neugebauer139,140 and Budich141 reported the 3-D topographic image of S. epidermidis cells and the corresponding TERS measurement. The chemical components of DNA or RNA inside the cell and sugar and peptide at the cell surface are distinguished from the Raman spectra and crucially related to the sample point on cell topography. G. Rusciano et al.142 correlated the AFM phase map and Raman spectra of B. subtilis with TERS. Their results demonstrate that there is a denser arrangement of both proteins and carbohydrates on specific spore surface regions. This AFM-TERS combination method also opens a window into the cell's life for on-line monitoring of special components. The single yeast cell is kept alive during the AFM imaging and TERS detection for the cell wall study.143 The acquired maps can be used to reveal the chemical distribution of a complex biological system in its natural environment.
To the knowledge of the authors, there is no report of TERS application in the spatial scale of a whole biofilm. But the association of AFM and TERS is supposed to have great potential in biofilm investigation in food processing. Many important species of food-borne bacteria, such as E. coli,144S. enterica,145 and S. xylosus146 have been detected by similar techniques such as surface-enhanced Raman spectroscopy (SERS). In SERS, the bacterial cell needs to be fabricated with metallic nanoparticles for enhancement of the Raman signal. The operation in TERS is based on AFM and has the advantages of easy and more nature sample preparation, which is important for maintaining clues to biofilm formation.
Furthermore, with AFM-IR, the choice of the substrate surface can be expanded to both opaque and transparent samples, such as metal, graphene and other opaque materials.147,148 This is due to the flexible illumination and detection solution. As shown in Fig. 3(b–d), the side-illumination (b) and top-illumination (c) have been developed from the traditional bottom-illumination. Side-illumination is widely used because most of the commercial probes can be used, but a special tip is needed for top-illumination. The disadvantage of the side-illumination is the signal loss; thus higher laser power is required to compensate for the insufficient signal acquisition. This is convenient for the selection of substrates to simulate the biofilm formation environment in food processing.
The limitation of AFM-TERS arises from both AFM and Raman technology. The TER probe is difficult to fabricate and has poor reliability and short lifetime, especially with Ag modification, as Ag nanoparticles are chemically active and easily oxidized by oxygen or sulfur in laboratory atmospheric or samples.149,150 This increases the time and cost during the experiments. In addition, the Raman spectra correspond to a functional group instead of biological molecules. Some big organic molecules have the same chemical groups but varied with a spatial structure or arrangement, such as proteins and lipids. This will result in the same vibration and make it hard to distinguish in the Raman spectra. Moreover, different groups may also have the same Raman band. Assignment analysis is a challenge. For example, both amino and imino groups have the 1144 cm−1 vibrational band, which might be allocated to many amino acids in the peptide chain, such as asparagine, glutamine, lysine, and arginine.135 Besides, the mechanism of the band shift is not clear yet. All these unambiguous band assignments are extremely challenging for complex chemical analysis in biological systems.
The combination of AFM and the mentioned label-free chemical methods (IR/Raman) retains the advantages of the high resolution and is less destructive to samples of AFM, making up for the lack of chemical analysis of AFM. The spatial resolution of the tip-enhanced Raman scattering imaging is down to 3–15 nanometers using commercial equipment. Thus the point-to-point chemical analysis can be processed simultaneously with topological structure measurement on the nanoscale. The technique is extremely valuable for the study of the formation and organization of a microfilm system whose properties strongly depend on the basic surface topography and chemical composition.
Conclusions
Reducing the formation of biofilms is crucial for microbial-contamination control in the food industry. Observing the micro- and nano-features and tracking the growth process of biofilms are essential approaches to understand biofilm formation. The biofilm micro-structure complexity and its changing nature with time increase the difficulty in understanding the mechanisms that govern biofilm formation.This review focused on biofilm formation from the fundamental aspects of adhesion of bacteria onto surfaces of products in food processing. The characterization of surface morphology and substance chemical compositions was discussed to probe biofilm formation. A map of characterization techniques is generated to show the spatial resolution and function of existing instruments. Confocal laser scanning microscopy (CLSM), scanning electron microscopy (SEM) and atomic force microscopy (AFM) are the three most important techniques for static and dynamic process observation with micro- or nano-scale spatial resolution. CLSM has been successfully used to reveal 3D structures and shows the expression of genes during biofilm formation in aqueous environments, but it is not adapted to the visualization of ultra-structures due to the intrinsic limitations of light microscopy. Finding a fluorescent label and coloring for specific components is not easy. SEM has a high lateral resolution but low vertical resolution. However, the testing environment is not natural and some fragile features of the sample may get affected during the electron scan. AFM is the only method which can provide nanoscale spatial resolution in three dimensions, and thus the surface topography and surface roughness can be measured quantitatively. It also has great potential for in situ studies with environmental control. Meanwhile, the scan area is limited and unexpected sample damages are revealed. There is also a need to coordinate the conflict between bio-sample immobilization and in situ observation of the samples. Complementary studies at the same position with multiple techniques are discussed from two aspects, comparative analysis at different length scales and the chemical identifications combined with microscopy methods. The application and limitation of these techniques are evaluated from the aspects of range of the view field, spatial and time resolution, working environment, sample preparation approach and sample state, as well as cost.
To effectively probe and study the formation of biofilms, strategies to combine selected techniques are recommended. The following suggestions are proposed:
(i) A nano-/micrometer repositioning protocol is required in correlative microscopic studies. Feasible methods comprise use of scanning electron microscopic (SEM) images taken at different magnifications to correlative confocal laser scanning microscopy (CLSM) and atomic force microscopy (AFM) and use of advanced object recognition techniques for image comparative analysis.
(ii) Obtaining quantitative assessment of the biofilm from microscopic image data mining. Object recognition and adaptive thresholding routines will be the most crucial aspects that can be improved by machine learning.
(iii) Integration of AFM and chemical analysis methods (IR/Raman/Fluorescence Microscopy) should be developed. This will lead to a clearer perception of the influence of chemical and surface mechanical properties on cell-to-cell communication in bacteria and biofilm formation.
These approaches will enhance the understanding of biofilm formation and advance the development of new technologies in an attempt to control microorganisms in biofilms.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 51675297 and 31901817). Yating Huang was financially supported by Beijing Technology and Business University. Thanks for the help of the surface science group in Texas A&M University. Assistance provided by the Texas A&M University Writing Center, Lian Ma, and Eugene Chen was acknowledged.References
- P. J. Fryer and K. Asteriadou, A prototype cleaning map: A classification of industrial cleaning processes, Trends Food Sci. Technol., 2009, 20(6–7), 255–262 CrossRef CAS.
- D. Lukenhaus, H. Cao, K. D. Dearn and S. Bakalis, Tribology of Cleaning Processes, 2016, 11(2), 298–307 Search PubMed.
- J. Verran and J. Redfern, Testing Surface Cleanability in Food Processing, in Handbook of Hygiene Control in the Food Industry, ed. H. Lelieveld, J. Holah and D. Gabrić, Woodhead Publishing, San Diego, 2nd edn, 2016, ch. 42, pp. 651–61 Search PubMed.
- J. Verran, D. L. Rowe, D. Cole and R. D. Boyd, The use of the atomic force microscope to visualise and measure wear of food contact surfaces, Int. Biodeterior. Biodegrad., 2000, 46(2), 99–105 CrossRef.
- W. B. Beech and J. Sunner, Biocorrosion: towards understanding interactions between biofilms and metals, Curr. Opin. Biotechnol., 2004, 15(3), 181–186 CrossRef PubMed.
- V. Carniello, B. W. Peterson, H. C. van der Mei and H. J. Busscher, Physico-chemistry from initial bacterial adhesion to surface-programmed biofilm growth, Adv. Colloid Interface Sci., 2018, 261, 1–14 CrossRef CAS PubMed.
- J. Y. Li, H. J. Busscher, W. Norde and J. Sjollema, Analysis of the contribution of sedimentation to bacterial mass transport in a parallel plate flow chamber, Colloids Surf., B, 2011, 84(1), 76–81 CrossRef CAS PubMed.
- A. Jang, J. Szabo, A. A. Hosni, M. Coughlin and P. L. Bishop, Measurement of chlorine dioxide penetration in dairy process pipe biofilms during disinfection, Appl. Microbiol. Biotechnol., 2006, 72(2), 368–376 CrossRef CAS PubMed.
- J. Sjollema, M. Rustema-Abbing, H. C. van der Mei and H. J. Busscher, Generalized Relationship between Numbers of Bacteria and Their Viability in Biofilms, Appl. Environ. Microbiol., 2011, 77(14), 5027–5029 CrossRef CAS PubMed.
- F. Cappitelli, A. Polo and F. Villa, Biofilm Formation in Food Processing Environments is Still Poorly Understood and Controlled, Food Eng. Rev., 2014, 6(1–2), 29–42 CrossRef.
- K. A. Whitehead and J. Verran, Formation, architecture and functionality of microbial biofilms in the food industry, Curr. Opin. Food Sci., 2015, 2, 84–91 CrossRef.
- Y. F. Cheng and C. I. Moraru, Long-range interactions keep bacterial cells from liquid-solid interfaces: Evidence of a bacteria exclusion zone near Nafion surfaces and possible implications for bacterial attachment, Colloids Surf., B, 2018, 162, 16–24 CrossRef CAS PubMed.
- H. J. Busscher, W. Norde and H. C. Van der Mei, Specific molecular recognition and nonspecific contributions to bacterial interaction forces, Appl. Environ. Microbiol., 2008, 74(9), 2559–2564 CrossRef CAS PubMed.
- L. Ploux, A. Ponche and K. Anselme, Bacteria/Material Interfaces: Role of the Material and Cell Wall Properties, J. Adhes. Sci. Technol., 2010, 24(13–14), 2165–2201 CrossRef CAS.
- K. A. Whitehead and J. Verran, The effect of surface properties and application method on the retention of Pseudomonas aeruginosa on uncoated and titanium-coated stainless steel, Int. Biodeterior. Biodegrad., 2007, 60(2), 74–80 CrossRef CAS.
- Y. Chaturongkasumrit, H. Takahashi, S. Keeratipibul, T. Kuda and B. Kimura, The effect of polyesterurethane belt surface roughness on Listeria monocytogenes biofilm formation and its cleaning efficiency, Food Control, 2011, 22(12), 1893–1899 CrossRef CAS.
- H. C. van der Mei, R. Bos and H. J. Busscher, A reference guide to microbial cell surface hydrophobicity based on contact angles, Colloids Surf., B, 1998, 11(4), 213–221 CrossRef CAS.
- R. L. Meyer, A. Arpanaei, S. Pillai, N. Bernbom, J. J. Enghild and Y. Y. Ng, et al., Physicochemical characterization of fish protein adlayers with bacteria repelling properties, Colloids Surf., B, 2013, 102, 504–510 CrossRef CAS PubMed.
- B. A. Jucker, H. Harms and A. J. B. Zehnder, Adhesion of the positively charged bacterium Stenotrophomonas (Xanthomonas) maltophilia 70401 to glass and teflon, J. Bacteriol., 1996, 178(18), 5472–5479 CrossRef CAS PubMed.
- R. Bos, H. C. van der Mei and H. J. Busscher, Physico-chemistry of initial microbial adhesive interactions - its mechanisms and methods for study, FEMS Microbiol. Rev., 1999, 23(2), 179–230 CrossRef CAS PubMed.
- W. R. Bowen, R. W. Lovitt and C. J. Wright, Atomic force microscopy study of the adhesion of Saccharomyces cerevisiae, J. Colloid Interface Sci., 2001, 237(1), 54–61 CrossRef CAS PubMed.
- K. A. Whitehead, P. Benson, L. A. Smith and J. Verran, The use of physicochemical methods to detect organic food soils on stainless steel surfaces, Biofouling, 2009, 25(8), 749–756 CrossRef CAS PubMed.
- A. Harimawan, A. Rajasekar and Y. P. Ting, Bacteria attachment to surfaces - AFM force spectroscopy and physicochemical analyses, J. Colloid Interface Sci., 2011, 364(1), 213–218 CrossRef CAS PubMed.
- J. Palmer, S. Flint and J. Brooks, Bacterial cell attachment, the beginning of a biofilm, J. Ind. Microbiol. Biotechnol., 2007, 34(9), 577–588 CrossRef CAS PubMed.
- E. C. R. Bonsaglia, N. C. C. Silva, A. Fernades Junior, J. P. Araujo Junior, M. H. Tsunemi and V. L. M. Rall, Production of biofilm by Listeria monocytogenes in different materials and temperatures, Food Control, 2014, 35(1), 386–391 CrossRef CAS.
- F. Gaboriaud, M. L. Gee, R. Strugnell and J. F. L. Duval, Coupled electrostatic, hydrodynamic, and mechanical properties of bacterial interfaces in aqueous media, Langmuir, 2008, 24(19), 10988–10995 CrossRef CAS PubMed.
- M. C. M. Vanloosdrecht, J. Lyklema, W. Norde and A. J. B. Zehnder, Influence of Interfaces on Microbial Activity, Microbiol. Rev., 1990, 54(1), 75–87 CrossRef CAS.
- J. Ihssen and T. Egli, Specific growth rate and not cell density controls the general stress response in Escherichia coli, Microbiology, 2004, 150(6), 1637–1648 CrossRef CAS PubMed.
- K. L. Meibom, X. B. B. Li, A. T. Nielsen, C. Y. Wu, S. Roseman and G. K. Schoolnik, The Vibrio cholerae chitin utilization program, Proc. Natl. Acad. Sci. U. S. A., 2004, 101(8), 2524–2529 CrossRef CAS PubMed.
- L. Ponsonnet, M. Boureanu, N. Jaffrezic, A. Othmane, C. Dorel and P. Lejeune, Local pH variation as an initial step in bacterial surface-sensing and biofilm formation, Mater. Sci. Eng., C, 2008, 28(5–6), 896–900 CrossRef CAS.
- H. H. Tuson and D. B. Weibel, Bacteria-surface interactions, Soft Matter, 2013, 9(17), 4368–4380 RSC.
- R. S. Friedlander, H. Vlamakis, P. Kim, M. Khan, R. Kolter and J. Aizenberg, Bacterial flagella explore microscale hummocks and hollows to increase adhesion, Proc. Natl. Acad. Sci. U. S. A., 2013, 110(14), 5624–5629 CrossRef CAS PubMed.
- W. L. Ng and B. L. Bassler, Bacterial Quorum-Sensing Network Architectures, Annual Review of Genetics. Annual Review of Genetics, Annual Reviews, Palo Alto, 2009, vol. 43, pp. 197–222 Search PubMed.
- K. E. Eboigbodin, J. R. A. Newton, A. F. Routh and C. A. Biggs, Bacterial quorum sensing and cell surface electrokinetic properties, Appl. Microbiol. Biotechnol., 2006, 73(3), 669–675 CrossRef CAS PubMed.
- H. Wang, S. Ding, G. Wang, X. Xu and G. Zhou, In situ characterization and analysis of Salmonella biofilm formation under meat processing environments using a combined microscopic and spectroscopic approach, Int. J. Food Microbiol., 2013, 167(3), 293–302 CrossRef CAS PubMed.
- A. Bridier, F. Dubois-Brissonnet, A. Boubetra, V. Thomas and R. Briandet, The biofilm architecture of sixty opportunistic pathogens deciphered using a high throughput CLSM method, J. Microbiol. Methods, 2010, 82(1), 64–70 CrossRef CAS PubMed.
- E. J. Marsh, H. L. Luo and H. Wang, A three-tiered approach to differentiate Listeria monocytogenes biofilm-forming abilities, FEMS Microbiol. Lett., 2003, 228(2), 203–210 CrossRef PubMed.
- S. Renier, C. Chagnot, J. Deschamps, N. Caccia, J. Szlavik and S. A. Joyce, et al., Inactivation of the SecA2 protein export pathway in Listeria monocytogenes promotes cell aggregation, impacts biofilm architecture and induces biofilm formation in environmental condition, Environ. Microbiol., 2014, 16(4), 1176–1192 CrossRef CAS PubMed.
- M. Guilbaud, P. Piveteau, M. Desvaux, S. Brisse and R. Briandet, Exploring the Diversity of Listeria monocytogenes Biofilm Architecture by High-Throughput Confocal Laser Scanning Microscopy and the Predominance of the Honeycomb-Like Morphotype, Appl. Environ. Microbiol., 2015, 81(5), 1804–1810 CrossRef PubMed.
- M. S. Chae and H. Schraft, Comparative evaluation of adhesion and biofilm formation of different Listeria monocytogenes strains, Int. J. Food Microbiol., 2000, 62(1–2), 103–111 CrossRef CAS PubMed.
- M. A. Olszewska, T. Zhao and M. P. Doyle, Inactivation and induction of sublethal injury of Listeria monocytogenes in biofilm treated with various sanitizers, Food Control, 2016, 70, 371–379 CrossRef CAS.
- J. M. R. Moreira, L. C. Gomes, K. A. Whitehead, S. Lynch, L. A. Tetlow and F. J. Mergulhao, Effect of surface conditioning with cellular extracts on Escherichia coli adhesion and initial biofilm formation, Food Bioprod. Process., 2017, 104, 1–12 CrossRef CAS.
- M. Oliveira, L. Blasco, S. Ferrer and F. Bernardo, Rapid and simultaneous detection of Salmonella spp. and Listeria monocytogenes in milk by fluorescent in situ hybridisation, Rev. Port. Cienc. Vet., 2004, 552, 215 Search PubMed.
- B. E. Garcia-Almendarez, I. K. O. Cann, S. E. Martin, I. Guerrero-Legarreta and C. Regalado, Effect of Lactococcus lactis UQ2 and its bacteriocin on Listeria monocytogenes biofilms, Food Control, 2008, 19(7), 670–680 CrossRef CAS.
- K. A. Whitehead, P. S. Benson and J. Verran, Developing application and detection methods for Listeria monocytogenes and fish extract on open surfaces in order to optimize cleaning protocols, Food Bioprod. Process., 2015, 93, 224–233 CrossRef.
- E. Peeters, H. J. Nelis and T. Coenye, Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates, J. Microbiol. Methods, 2008, 72(2), 157–165 CrossRef CAS PubMed.
- G. M. Robinson, K. M. Tonks, R. M. S. Thorn and D. M. Reynolds, Application of Bacterial Bioluminescence To Assess the Efficacy of Fast-Acting Biocides, Antimicrob. Agents Chemother., 2011, 55(11), 5214–5219 CrossRef CAS PubMed.
- M. W. Griffiths, Applications of bioluminescence in the dairy industry, J. Dairy Sci., 1993, 76(10), 3118–3125 CrossRef CAS PubMed.
- B. Bottari, M. Santarelli and E. Neviani, Determination of microbial load for different beverages and foodstuff by assessment of intracellular ATP, Trends Food Sci. Technol., 2015, 44(1), 36–48 CrossRef CAS.
- N. T. Liu, X. Nou, G. R. Bauchan, C. Murphy, A. M. Lefcourt and D. R. Shelton, et al., Effects of Environmental Parameters on the Dual-Species Biofilms Formed by Escherichia coli O157:H7 and Ralstonia insidiosa, a Strong Biofilm Producer Isolated from a Fresh-Cut Produce Processing Plant, J. Food Prot., 2015, 78(1), 121–127 CrossRef CAS PubMed.
- J.-M. Liu, Z.-H. Wang, H. Ma and S. Wang, Probing and Quantifying the Food-Borne Pathogens and Toxins: From In Vitro to In Vivo, J. Agric. Food Chem., 2018, 66(5), 1061–1066 CrossRef CAS PubMed.
- N. P. Boks, W. Norde, H. C. van der Mei and H. J. Busscher, Forces involved in bacterial adhesion to hydrophilic and hydrophobic surfaces, Microbiology, 2008, 154, 3122–3133 CrossRef CAS PubMed.
- J. M. R. Moreira, J. D. P. Araujo, J. M. Miranda, M. Simoes, L. F. Melo and F. J. Mergulhao, The effects of surface properties on Escherichia coli adhesion are modulated by shear stress, Colloids Surf., B, 2014, 123, 1–7 CrossRef CAS PubMed.
- J. Y. Li, H. J. Busscher, H. C. van der Mei and J. Sjollema, Surface enhanced bacterial fluorescence and enumeration of bacterial adhesion, Biofouling, 2013, 29(1), 11–19 CrossRef PubMed.
- T. R. Neu and J. R. Lawrence, Innovative techniques, sensors, and approaches for imaging biofilms at different scales, Trends Microbiol., 2015, 23(4), 233–242 CrossRef CAS PubMed.
- Q. F. Zhuang, Y. Wang and Y. N. Ni, Solid-phase synthesis of graphene quantum dots from the food additive citric acid under microwave irradiation and their use in live-cell imaging, Luminescence, 2016, 31(3), 746–753 CrossRef CAS PubMed.
- J. Azeredo, N. F. Azevedo, R. Briandet, N. Cerca, T. Coenye and A. R. Costa, et al., Critical review on biofilm methods, Crit. Rev. Microbiol., 2017, 43(3), 313–351 CrossRef CAS PubMed.
- C. Hung, Y. Z. Zhou, J. S. Pinkner, K. W. Dodson, J. R. Crowley and J. Heuser, et al., Escherichia coli Biofilms Have an Organized and Complex Extracellular Matrix Structure, Mbio, 2013, 4(5), 10 CrossRef PubMed.
- D. Rodrigues, M. Banobre-Lopez, B. Espina, J. Rivas and J. Azeredo, Effect of magnetic hyperthermia on the structure of biofilm and cellular viability of a food spoilage bacterium, Biofouling, 2013, 29(10), 1225–1232 CrossRef CAS PubMed.
- M. K. Borucki, J. D. Peppin, D. White, F. Loge and D. R. Call, Variation in biofilm formation among strains of Listeria monocytogenes, Appl. Environ. Microbiol., 2003, 69(12), 7336–7342 CrossRef CAS PubMed.
- H. Xu, H. Y. Lee and J. Ahn, Characteristics of biofilm formation by selected foodborne pathogens, J. Food Saf., 2011, 31(1), 91–97 CrossRef.
- C. Ceresa, F. Tessarolo, I. Caola, G. Nollo, M. Cavallo and M. Rinaldi, et al., Inhibition of Candida albicans adhesion on medical-grade silicone by a Lactobacillus-derived biosurfactant, Journal of Applied Microbiology, 2015, 118(5), 1116–1125 CrossRef CAS PubMed.
- M. M. M. de Oliveira, D. F. Brugnera, E. Alves and R. H. Piccoli, Biofilm formation by Listeria monocytogenes on stainless steel surface and biotransfer potential, Braz. J. Microbiol., 2010, 41(1), 97–106 CrossRef PubMed.
- S. P. Doijad, S. B. Barbuddhe, S. Garg, K. V. Poharkar, D. R. Kalorey and N. V. Kurkure, et al., Biofilm-Forming Abilities of Listeria monocytogenes Serotypes Isolated from Different Sources, PLoS One, 2015, 10(9), 14 CrossRef PubMed.
- O. Santos, T. Nylander, R. Rosmaninho, G. Rizzo, S. Yiantsios and N. Andritsos, et al., Modified stainless steel surfaces targeted to reduce fouling - surface characterization, J. Food Eng., 2004, 64(1), 63–79 CrossRef.
- K. Whitehead, P. Kelly, H. Q. Li and J. Verran, Surface topography and physicochemistry of silver containing titanium nitride nanocomposite coatings, J. Vac. Sci. Technol., B: Microelectron. Nanometer Struct.--Process., Meas., Phenom., 2010, 28(1), 180–187 CrossRef CAS.
- Y. Wyart, G. Georges, C. Demie, C. Amra and P. Moulin, Membrane characterization by microscopic methods: Multiscale structure, J. Membr. Sci., 2008, 315(1–2), 82–92 CrossRef CAS.
- C. Hannig, M. Follo, E. Hellwig and A. Al-Ahmad, Visualization of adherent micro-organisms using different techniques, J. Med. Microbiol., 2010, 59(1), 1–7 CrossRef CAS PubMed.
- M. Alhede, K. Qvortrup, R. Liebrechts, N. Hoiby, M. Givskov and T. Bjarnsholt, Combination of microscopic techniques reveals a comprehensive visual impression of biofilm structure and composition, FEMS Immunol. Med. Microbiol., 2012, 65(2), 335–342 CrossRef CAS PubMed.
- B. Little, P. Wagner, R. Ray, R. Pope and R. Scheetz, Biofilms - an esem evaluation of artifacts introduced during SEM preparation, J. Ind. Microbiol., 1991, 8(4), 213–221 CrossRef CAS.
- E. Medilanski, K. Kaufmann, L. Y. Wick, O. Wanner and H. Harms, Influence of the surface topography of stainless steel on bacterial adhesion, Biofouling, 2002, 18(3), 193–203 CrossRef.
- A. K. Epstein, A. I. Hochbaum, P. Kim and J. Aizenberg, Control of bacterial biofilm growth on surfaces by nanostructural mechanics and geometry, Nanotechnology, 2011, 22(49), 8 CrossRef PubMed.
- A. I. Hochbaum and J. Aizenberg, Bacteria Pattern Spontaneously on Periodic Nanostructure Arrays, Nano Lett., 2010, 10(9), 3717–3721 CrossRef CAS PubMed.
- L. C. Hsu, J. Fang, D. A. Borca-Tasciuc, R. W. Worobo and C. I. Moraru, Effect of Micro- and Nanoscale Topography on the Adhesion of Bacterial Cells to Solid Surfaces, Appl. Environ. Microbiol., 2013, 79(8), 2703–2712 CrossRef CAS PubMed.
- S. Aguayo, A. Strange, N. Gadegaard, M. J. Dalby and L. Bozec, Influence of biomaterial nanotopography on the adhesive and elastic properties of Staphylococcus aureus cells, RSC Adv., 2016, 6(92), 89347–89355 RSC.
- I. B. Beech, C. W. S. Cheung, D. B. Johnson and J. R. Smith, Comparative studies of bacterial biofilms on steel surfaces using atomic force microscopy and environmental scanning electron microscopy, Biofouling, 1996, 10(1–3), 65–& CrossRef CAS PubMed.
- A. Pompilio, S. De Nicola, V. Crocetta, S. Guarnieri, V. Savini and E. Carretto, et al., New insights in Staphylococcus pseudintermedius pathogenicity: antibiotic-resistant biofilm formation by a human wound-associated strain, BMC Microbiol., 2015, 15, 14 CrossRef PubMed.
- C. Cragnell, K. Hansson, T. Andersson, B. Jonsson and M. Skepo, Underlying mechanisms behind adhesion of fermented milk to packaging surfaces, J. Food Eng., 2014, 130, 52–59 CrossRef CAS.
- K. Huang and J. M. Goddard, Influence of fluid milk product composition on fouling and cleaning of Ni-PTFE modified stainless steel heat exchanger surfaces, J. Food Eng., 2015, 158, 22–29 CrossRef CAS.
- A. Bridier, T. Meylheuc and R. Briandet, Realistic representation of Bacillus subtilis biofilms architecture using combined microscopy (CLSM, ESEM and FESEM), Micron, 2013, 48, 65–69 CrossRef CAS PubMed.
- L. Muscariello, F. Rosso, G. Marino, A. Giordano, M. Barbarisi and G. Cafiero, et al., A critical overview of ESEM applications in the biological field, J. Cell. Physiol., 2005, 205(3), 328–334 CrossRef CAS PubMed.
- Y. L. Ong, A. Razatos, G. Georgiou and M. M. Sharma, Adhesion forces between E-coli bacteria and biomaterial surfaces, Langmuir, 1999, 15(8), 2719–2725 CrossRef CAS.
- Q. Zhao, Y. Liu, H. Muller-Steinhagen and G. Liu, Graded Ni-P-PTFE coatings and their potential applications, Surf. Coat. Technol., 2002, 155(2–3), 279–284 CrossRef CAS.
- P. K. Wallace, B. Arey and W. F. Mahaffee, Subsurface examination of a foliar biofilm using scanning electron- and focused-ion-beam microscopy, Micron, 2011, 42(6), 579–585 CrossRef CAS PubMed.
- Y. Cui, Y. J. Oh, J. Lim, M. Youn, I. Lee and H. K. Pak, et al., AFM study of the differential inhibitory effects of the green tea polyphenol (-)-epigallocatechin-3-gallate (EGCG) against Gram-positive and Gram-negative bacteria, Food Microbiol., 2012, 29(1), 80–87 CrossRef CAS PubMed.
- J. F. Zhang and H. S. Yang, Effects of potential organic compatible sanitisers on organic and conventional fresh-cut lettuce (Lactuca sativa Var. Crispa L), Food Control, 2017, 72, 20–26 CrossRef CAS.
- L. Zhao, Y. Zhang and H. S. Yang, Efficacy of low concentration neutralised electrolysed water and ultrasound combination for inactivating Escherichia coli ATCC 25922, Pichia pastoris GS115 and Aureobasidium pullulans 2012 on stainless steel coupons, Food Control, 2017, 73, 889–899 CrossRef.
- T. Kuda, G. Shibata, H. Takahashi and B. Kimura, Effect of quantity of food residues on resistance to desiccation of food-related pathogens adhered to a stainless steel surface, Food Microbiol., 2015, 46, 234–238 CrossRef PubMed.
- M. Hyldgaard, D. S. Sutherland, M. Sundh, T. Mygind and R. L. Meyer, Antimicrobial Mechanism of Monocaprylate, Appl. Environ. Microbiol., 2012, 78(8), 2957–2965 CrossRef CAS PubMed.
- S. V. Bhat, S. C. Booth, E. A. N. Vantomme, S. Afroj, C. K. Yost and T. E. Dahms, Oxidative stress and metabolic perturbations in Escherichia coli exposed to sublethal levels of 2,4-dichlorophenoxyacetic acid, Chemosphere, 2015, 135, 453–461 CrossRef CAS PubMed.
- L. Yang, Y. F. Hu, Y. Liu, J. D. Zhang, J. Ulstrup and S. Molin, Distinct roles of extracellular polymeric substances in Pseudomonas aeruginosa biofilm development, Environ. Microbiol., 2011, 13(7), 1705–1717 CrossRef CAS PubMed.
- H. H. P. Fang, K. Y. Chan and L. C. Xu, Quantification of bacterial adhesion forces using atomic force microscopy (AFM), J. Microbiol. Methods, 2000, 40(1), 89–97 CrossRef CAS PubMed.
- C. Diaz, P. L. Schilardi, R. C. Salvarezza and M. F. L. de Mele, Nano/Microscale order affects the early stages of Biofilm formation on metal surfaces, Langmuir, 2007, 23(22), 11206–11210 CrossRef CAS PubMed.
- K. Jonas, H. Tomenius, A. Kader, S. Normark, U. Romling and L. M. Belova, et al., Roles of curli, cellulose and BapA in Salmonella biofilm morphology studied by atomic force microscopy, BMC Microbiol., 2007, 7, 9 CrossRef PubMed.
- H. Yang and Y. Wang, Application of Atomic Force Microscopy on Rapid Determination of Microorganisms for Food Safety, J. Food Sci., 2008, 73(8), N44–N50 CrossRef CAS PubMed.
- K. A. Whitehead, D. Rogers, J. Colligon, C. Wright and J. Verran, Use of the atomic force microscope to determine the effect of substratum surface topography on the ease of bacterial removal, Colloids Surf., B, 2006, 51(1), 44–53 CrossRef CAS PubMed.
- T. K. Janganan, N. Mullin, S. B. Tzokov, S. Stringer, R. P. Fagan and J. K. Hobbs, et al., Characterization of the spore surface and exosporium proteins of Clostridium sporogenes; implications for Clostridium botulinum group I strains, Food Microbiol., 2016, 59, 205–212 CrossRef CAS PubMed.
- J. Verran and R. D. Boyd, The relationship between substratum surface roughness and microbiological and organic soiling: a review, Biofouling, 2001, 17(1), 59–+ CrossRef.
- P. J. Bremer, G. G. Geesey and B. Drake, Atomic force microscopy examination of the topography of a hydrated bacterial biofilm on a copper surface, Curr. Microbiol., 1992, 24(4), 223–230 CrossRef CAS.
- A. Steele, D. T. Goddard and I. B. Beech, An atomic-force microscopy study of the biodeterioration of stainless-steel in the presence of bacterial biofilms, Int. Biodeterior. Biodegrad., 1994, 34(1), 35–46 CrossRef CAS.
- S. Y. Liu and Y. F. Wang, Application of AFM in Microbiology: A Review, Scanning, 2010, 32(2), 61–73 CrossRef CAS PubMed.
- C. J. Wright, M. K. Shah, L. C. Powell and I. Armstrong, Application of AFM From Microbial Cell to Biofilm, Scanning, 2010, 32(3), 134–149 CrossRef CAS PubMed.
- L. S. Dorobantu and M. R. Gray, Application of Atomic Force Microscopy in Bacterial Research, Scanning, 2010, 32(2), 74–96 CrossRef CAS PubMed.
- S. A. James, L. C. Powell and C. J. Wright, Atomic Force Microscopy of Biofilms—Imaging, Interactions, and Mechanics. Microbial Biofilms-Importance and Applications, IntechOpen, 2016 Search PubMed.
- A. D. Ozkan, A. E. Topal, F. B. Dikecoglu, M. O. Guler, A. Dana and A. B. Tekinay, Probe microscopy methods and applications in imaging of biological materials, Semin. Cell Dev. Biol., 2018, 73, 153–164 CrossRef CAS PubMed.
- Q. Liu and H. Yang, Application of atomic force microscopy in food microorganisms, Trends Food Sci. Technol., 2019, 87, 73–83 CrossRef CAS.
- L. Kailas, E. C. Ratcliffe, E. J. Hayhurst, M. G. Walker, S. J. Foster and J. K. Hobbs, Immobilizing live bacteria for AFM imaging of cellular processes, Ultramicroscopy, 2009, 109(7), 775–780 CrossRef CAS PubMed.
- L. R. Hilbert, D. Bagge-Ravn, J. Kold and L. Gram, Influence of surface roughness of stainless steel on microbial adhesion and corrosion resistance, Int. Biodeterior. Biodegrad., 2003, 52(3), 175–185 CrossRef CAS.
- P. Champigneux, M. L. Delia and A. Bergel, Impact of electrode micro - and nano-scale topography on the formation and performance of microbial electrodes, Biosens. Bioelectron., 2018, 118, 231–246 CrossRef CAS PubMed.
- Y. F. Cheng, G. P. Feng and C. I. Moraru, Micro- and Nanotopography Sensitive Bacterial Attachment Mechanisms: A Review, Front. Microbiol., 2019, 10, 17 CrossRef PubMed.
- G. Di Bonaventura, A. Spedicato, D. D'Antonio, I. Robuffo and R. Piccolomini, Biofilm formation by Stenotrophomonas maltophilia: Modulation by quinolones, trimethoprim-sulfamethoxazole, and ceftazidime, Antimicrob. Agents Chemother., 2004, 48(1), 151–160 CrossRef CAS PubMed.
- G. Di Bonaventura, A. Pompilio, C. Picciani, M. Iezzi, D. D'Antonio and R. Piccolomini, Biofilm formation by the emerging fungal pathogen Trichosporon asahii: Development, architecture, and antifungal resistance, Antimicrob. Agents Chemother., 2006, 50(10), 3269–3276 CrossRef CAS PubMed.
- G. Di Bonaventura, R. Piccolomini, D. Paludi, V. D'Orio, A. Vergara and M. Conter, et al., Influence of temperature on biofilm formation by Listeria monocytogenes on various food-contact surfaces: relationship with motility and cell surface hydrophobicity, J. Appl. Microbiol., 2008, 104(6), 1552–1561 CrossRef CAS PubMed.
- S. Hasan, M. Danishuddin and A. U. Khan, Inhibitory effect of zingiber officinale towards Streptococcus mutans virulence and caries development: in vitro and in vivo studies, BMC Microbiol., 2015, 15, 14 CrossRef PubMed.
- T. A. Van Laar, T. Chen, T. You and K. P. Leung, Sublethal Concentrations of Carbapenems Alter Cell Morphology and Genomic Expression of Klebsiella pneumoniae Biofilms, Antimicrob. Agents Chemother., 2015, 59(3), 1712–1722 CrossRef PubMed.
- H. Daims, S. Lucker and M. Wagner, daime, a novel image analysis program for microbial ecology and biofilm research, Environ. Microbiol., 2006, 8(2), 200–213 CrossRef CAS PubMed.
- C. A. Gross, C. K. Reddy and F. B. Dazzo, CMEIAS Color Segmentation: An Improved Computing Technology to Process Color Images for Quantitative Microbial Ecology Studies at Single-Cell Resolution, Microb. Ecol., 2010, 59(2), 400–414 CrossRef PubMed.
- L. E. C. de Paz, Image Analysis Software Based on Color Segmentation for Characterization of Viability and Physiological Activity of Biofilms, Appl. Environ. Microbiol., 2009, 75(6), 1734–1739 CrossRef PubMed.
- O. Sliusarenko, J. Heinritz, T. Emonet and C. Jacobs-Wagner, High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics, Mol. Microbiol., 2011, 80(3), 612–627 CrossRef CAS PubMed.
- J. Laishram, S. Kondra, D. Avossa, E. Migliorini, M. Lazzarino and V. Torre, A morphological analysis of growth cones of DRG neurons combining Atomic Force and Confocal Microscopy, J. Struct. Biol., 2009, 168(3), 366–377 CrossRef PubMed.
- Z. Q. Qin, J. D. Zhang, Y. F. Hu, Q. J. Chi, N. P. Mortensen and D. Qu, et al., Organic compounds inhibiting S. epidermidis adhesion and biofilm formation, Ultramicroscopy, 2009, 109(8), 881–888 CrossRef CAS PubMed.
- A. R. Burns, Domain structure in model membrane bilayers investigated by simultaneous atomic force microscopy and fluorescence imaging, Langmuir, 2003, 19(20), 8358–8363 CrossRef CAS.
- Z. Deng, T. Zink, H. Y. Chen, D. Walters, F. T. Liu and G. Y. Liu, Impact of Actin Rearrangement and Degranulation on the Membrane Structure of Primary Mast Cells: A Combined Atomic Force and Laser Scanning Confocal Microscopy Investigation, Biophys. J., 2009, 96(4), 1629–1639 CrossRef CAS PubMed.
- M. Baclayon, W. H. Roos and G. J. L. Wuite, Sampling Protein Form and Function with the Atomic Force Microscope, Mol. Cell. Proteomics, 2010, 9(8), 1678–1688 CrossRef CAS PubMed.
- J. M. Kim, T. Ohtani, S. Sugiyama, T. Hirose and H. Muramatsu, Simultaneous topographic and fluorescence imaging of single DNA molecules for DNA analysis with a scanning near-field optical/atomic force microscope, Anal. Chem., 2001, 73(24), 5984–5991 CrossRef CAS PubMed.
- R. Kassies, K. O. Van der Werf, A. Lenferink, C. N. Hunter, J. D. Olsen and V. Subramaniam, et al., Combined AFM and confocal fluorescence microscope for applications in bio-nanotechnology, J. Microsc., 2005, 217, 109–116 CrossRef CAS PubMed.
- Z. Deng, V. Lulevich, F. T. Liu and G. Y. Liu, Applications of Atomic Force Microscopy in Biophysical Chemistry of Cells, J. Phys. Chem. B, 2010, 114(18), 5971–5982 CrossRef CAS PubMed.
- M. S. Kuyukina, I. B. Ivshina, I. O. Korshunova and E. V. Rubtsova, Assessment of bacterial resistance to organic solvents using a combined confocal laser scanning and atomic force microscopy (CLSM/AFM), J. Microbiol. Methods, 2014, 107, 23–29 CrossRef CAS PubMed.
- A. R. Burns, D. J. Frankel and T. Buranda, Local mobility in lipid domains of supported bilayers characterized by atomic force microscopy and fluorescence correlation spectroscopy, Biophys. J., 2005, 89(2), 1081–1093 CrossRef CAS PubMed.
- I. W. Sutherland, Biofilm exopolysaccharides: a strong and sticky framework, Microbiology, 2001, 147, 3–9 CrossRef CAS PubMed.
- A. Dazzi and C. B. Prater, AFM-IR: Technology and Applications in Nanoscale Infrared Spectroscopy and Chemical Imaging, Chem. Rev., 2017, 117(7), 5146–5173 CrossRef CAS PubMed.
- A. Dazzi, C. B. Prater, Q. C. Hu, D. B. Chase, J. F. Rabolt and C. Marcott, AFM-IR: Combining Atomic Force Microscopy and Infrared Spectroscopy for Nanoscale Chemical Characterization, Appl. Spectrosc., 2012, 66(12), 1365–1384 CrossRef CAS PubMed.
- D. E. Barlow, J. C. Biffinger, A. L. Cockrell-Zugell, M. Lo, K. Kjoller and D. Cook, et al., The importance of correcting for variable probe-sample interactions in AFM-IR spectroscopy: AFM-IR of dried bacteria on a polyurethane film, Analyst, 2016, 141(16), 4848–4854 RSC.
- P. Vitry, R. Rebois, E. Bourillot, A. Deniset-Besseau, M. J. Virolle and E. Lesniewska, et al., Combining infrared and mode synthesizing atomic force microscopy: Application to the study of lipid vesicles inside Streptomyces bacteria, Nano Res., 2016, 9(6), 1674–1681 CrossRef CAS.
- D. Kurouski, Advances of tip-enhanced Raman spectroscopy (TERS) in electrochemistry, biochemistry, and surface science, Vib. Spectrosc., 2017, 91, 3–15 CrossRef CAS.
- R. M. Stockle, Y. D. Suh, V. Deckert and R. Zenobi, Nanoscale chemical analysis by tip-enhanced Raman spectroscopy, Chem. Phys. Lett., 2000, 318(1–3), 131–136 CrossRef CAS.
- N. Hayazawa, Y. Inouye, Z. Sekkat and S. Kawata, Metallized tip amplification of near-field Raman scattering, Opt. Commun., 2000, 183(1–4), 333–336 CrossRef CAS.
- M. S. Anderson, Locally enhanced Raman spectroscopy with an atomic force microscope, Appl. Phys. Lett., 2000, 76(21), 3130–3132 CrossRef CAS.
- U. Neugebauer, P. Rosch, M. Schmitt, J. Popp, C. Julien and A. Rasmussen, et al., On the way to nanometer-sized information of the bacterial surface by tip-enhanced Raman spectroscopy, ChemPhysChem, 2006, 7(7), 1428–1430 CrossRef CAS PubMed.
- U. Neugebauer, U. Schmid, K. Baumann, W. Ziebuhr, S. Kozitskaya and V. Deckert, et al., Towards a detailed understanding of bacterial metabolism - Spectroscopic characterization of Staphylococcus epidermidis, ChemPhysChem, 2007, 8(1), 124–137 CrossRef CAS PubMed.
- C. Budich, U. Neugebauer, J. Popp and V. Deckert, Cell wall investigations utilizing tip-enhanced Raman scattering, J. Microsc., 2008, 229(3), 533–539 CrossRef CAS PubMed.
- G. Rusciano, G. Zito, R. Isticato, T. Sirec, E. Ricca and E. Bailo, et al., Nanoscale Chemical Imaging of Bacillus subtilis Spores by Combining Tip-Enhanced Raman Scattering and Advanced Statistical Tools, ACS Nano, 2014, 8(12), 12300–12309 CrossRef CAS PubMed.
- D. Naumenko, V. Snitka, E. Serviene, I. Bruzaite and B. Snopok, In vivo characterization of protein uptake by yeast cell envelope: single cell AFM imaging and mu-tip-enhanced Raman scattering study, Analyst, 2013, 138(18), 5371–5383 RSC.
- W. Q. Wang, V. Hynninen, L. Qiu, A. W. Zhang, T. Lemma and N. N. Zhang, et al., Synergistic enhancement via plasmonic nanoplate-bacteria-nanorod supercrystals for highly efficient SERS sensing of food-borne bacteria, Sens. Actuators, B, 2017, 239, 515–525 CrossRef CAS.
- K. Jia, P. M. Adam, R. S. Marks and R. E. Ionescu, Fixed Escherichia coli bacterial templates enable the production of sensitive SERS-based gold nanostructures, Sens. Actuators, B, 2015, 211, 213–219 CrossRef CAS.
- D. T. Yang, H. B. Zhou, C. Haisch, R. Niessner and Y. B. Ying, Reproducible E. coli detection based on label-free SERS and mapping, Talanta, 2016, 146, 457–463 CrossRef CAS PubMed.
- R. Zhang, Y. Zhang, Z. C. Dong, S. Jiang, C. Zhang and L. G. Chen, et al., Chemical mapping of a single molecule by plasmon-enhanced Raman scattering, Nature, 2013, 498(7452), 82–86 CrossRef CAS PubMed.
- A. B. Zrimsek, N. H. Chiang, M. Mattei, S. Zaleski, M. O. McAnally and C. T. Chapman, et al., Single-Molecule Chemistry with Surface- and Tip-Enhanced Raman Spectroscopy, Chem. Rev., 2017, 117(11), 7583–7613 CrossRef CAS PubMed.
- G. Sharma, T. Deckert-Gaudig and V. Deckert, Tip-enhanced Raman scattering-Targeting structure-specific surface characterization for biomedical samples, Adv. Drug Delivery Rev., 2015, 89, 42–56 CrossRef CAS PubMed.
- M. McMahon, R. Lopez, H. M. Meyer, L. C. Feldman and R. F. Haglund, Rapid tarnishing of silver nanoparticles in ambient laboratory air, Appl. Phys. B: Lasers Opt., 2005, 80(7), 915–921 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2020 |