Open Access Article
Open Access ArticleIdentification of pathogenic bacteria in complex samples using a smartphone based fluorescence microscope†
Vilhelm
Müller
 a,
José M.
Sousa
b,
Hatice
Ceylan Koydemir
cde,
Muhammed
Veli
cde,
Derek
Tseng
cde,
Laura
Cerqueira
bf,
Aydogan
Ozcan
a,
José M.
Sousa
b,
Hatice
Ceylan Koydemir
cde,
Muhammed
Veli
cde,
Derek
Tseng
cde,
Laura
Cerqueira
bf,
Aydogan
Ozcan
 *cde,
Nuno F.
Azevedo
*f and
Fredrik
Westerlund
*cde,
Nuno F.
Azevedo
*f and
Fredrik
Westerlund
 *a
*a
aDepartment of Biology and Biological Engineering, Chalmers University of Technology, Gothenburg, Sweden. E-mail: fredrikw@chalmers.se
bBiomode 2, S.A., Edifício GNRation, Praça Conde Agrolongo no. 123, 4700-312, Braga, Portugal
cElectrical and Computer Engineering Department, University of California, Los Angeles, 90095, CA, USA. E-mail: ozcan@ucla.edu
dBioengineering Department, University of California, Los Angeles, 90095, CA, USA
eCalifornia NanoSystems Institute (CNSI), University of California, Los Angeles, 90095, CA, USA
fLEPABE, Department of Chemical Engineering, Faculty of Engineering of the University of Porto, Rua Dr Roberto Frias, 4200-465 Porto, Portugal. E-mail: nazevedo@fe.up.pt
First published on 29th October 2018
Abstract
Diagnostics based on fluorescence imaging of biomolecules is typically performed in well-equipped laboratories and is in general not suitable for remote and resource limited settings. Here we demonstrate the development of a compact, lightweight and cost-effective smartphone-based fluorescence microscope, capable of detecting signals from fluorescently labeled bacteria. By optimizing a peptide nucleic acid (PNA) based fluorescence in situ hybridization (FISH) assay, we demonstrate the use of the smartphone-based microscope for rapid identification of pathogenic bacteria. We evaluated the use of both a general nucleic acid stain as well as species-specific PNA probes and demonstrated that the mobile platform can detect bacteria with a sensitivity comparable to that of a conventional fluorescence microscope. The PNA-based FISH assay, in combination with the smartphone-based fluorescence microscope, allowed us to qualitatively analyze pathogenic bacteria in contaminated powdered infant formula (PIF) at initial concentrations prior to cultivation as low as 10 CFU per 30 g of PIF. Importantly, the detection can be done directly on the smartphone screen, without the need for additional image analysis. The assay should be straightforward to adapt for bacterial identification also in clinical samples. The cost-effectiveness, field-portability and simplicity of this platform will create various opportunities for its use in resource limited settings and point-of-care offices, opening up a myriad of additional applications based on other fluorescence-based diagnostic assays.
Introduction
Optical imaging methods are essential tools for screening and diagnosing a variety of different diseases. However, a vast majority of the methods require advanced laboratory settings, using relatively costly and bulky imaging setups. Developing simple, cost-efficient and even field portable devices that allow for high throughput analysis would thus provide numerous opportunities for point of care (POC) diagnosis and an overall improvement of global health, especially in resource limited settings.1 Recently, smartphones have shown great potential as a platform for the development of cost-efficient and portable devices, with continuously improving optical hard-ware and increasing computational power. There are various applications that utilize smartphone-based technologies as part of field-portable analytical measurement platforms, including e.g., DNA detection,2 tissue cell monitoring,3 and environmental monitoring of water samples, among others.4–10One field that could benefit from simple and cost-efficient applications is the screening and detection of pathogenic bacteria in areas such as human infections, food safety and environmental control. The need for screening and detection of bacteria will increase even further with the rising threat to human health caused by bacteria that have acquired resistance to last-resort antibiotics.11,12 Using a fluorescence microscope, bacteria can be visualized by a variety of different fluorescent probes. However, a general probe will not allow for selective targeting of different bacterial species and may to some extent also non-specifically stain the surrounding sample matrix. Therefore, bacterial identification at the species level usually requires highly specific probes. The increasing specificity of the probe typically lowers the amount of target, such as RNA, rendering lower number of fluorophores in the target cells, and thus increasing the demand on the detection setup.
Fluorescence in situ hybridization (FISH) is a broadly used method that can be used for bacterial identification. It is based on binding of specifically designed nucleic acid probes to specific regions on DNA or RNA.13,14 The use of a synthetic DNA analogue, peptide nucleic acid (PNA), as the selective probe has been demonstrated to improve the FISH process significantly.15,16 By designing the PNA probes to target specific parts of the bacterial rRNA, it is possible to not only selectively target bacteria in a sample, but even target a specific bacterial species, which can be very useful in a variety of applications. Unlike PCR, FISH allows for direct visualization of microorganisms and, because it does not require nucleic acid amplification, is less prone to be affected by inhibitors that might be present in a sample.17
One example of an opportunistic pathogenic bacteria is Cronobacter spp. (originally described as Enterobacter sakazakii), commonly found in the environment as well as in various types of food, such as powdered infant formula (PIF). Cronobacter spp. is capable of causing severe infections associated with a high fatality rate in newborn infants.18–20 Hence, simple and efficient ways of detecting Cronobacter spp., and tracing potential sources of infections, are critical for limiting the impact of these pathogens on neonatal and pediatric health.
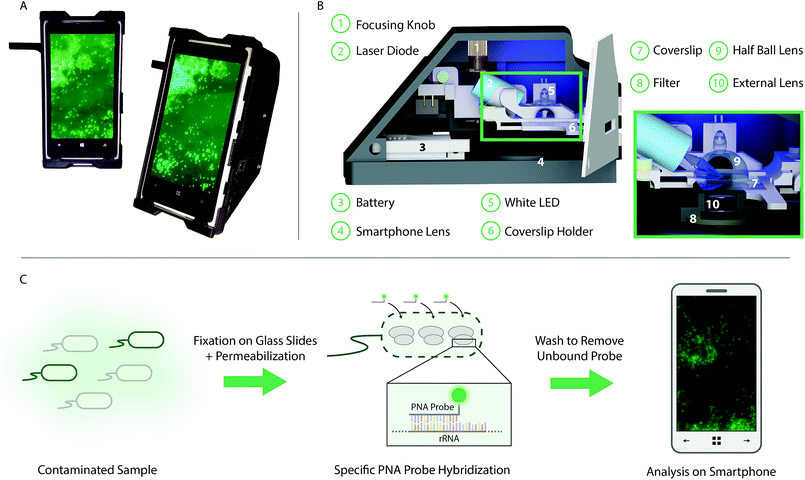
In this paper we present the development of a smartphone-based fluorescence microscope, used in combination with an optimized PNA-FISH assay, for selectively targeting bacteria in complex samples (Fig. 1). The bacterial samples are fixed and stained on conventional glass slides, using a FISH-based approach with designed selective rRNA-targeting PNA probes, before being qualitatively analyzed, directly on the smartphone screen, without any need for additional image analysis. The PNA-probes are designed to either target only a specific bacterial species or a broad range of bacteria. The smartphone is integrated with a 3D printed optomechanical attachment to create the smartphone-based fluorescence microscope. The microscopy unit, weighing less than 400 g (including 160 g smartphone), uses a standard blue laser-diode (488 nm, 60 mW) to excite the fluorescently labeled molecules in the sample at an experimentally optimized angle of incidence of 61°. The high illumination angle is combined with a long pass emission filter (>514 nm) in order to almost completely block the background noise created by the powerful excitation beam. In addition to the smartphone lens, an external lens (<$10) with a focal length of 2.6 mm is integrated into the design for further magnification of the sample onto the Complementary Metal-Oxide-Semiconductor (CMOS) sensor of the smartphone. A sample stage was designed to allow scanning of the entire sample while maintaining an oil film between the coverslip and the half ball lens, which in turn is connected to a miniature dover stage allowing for the adjustment of the sample focus. The field portable light weight microscope unit, which also includes a white light emitting diode (LED) for bright field imaging and focus adjustment purposes, costs approximately $400, excluding the application specific optical filter and the excitation laser diode, which can be customized based on the choice of the fluorophore (details in Methods). This level of cost for device parts is already more than one order of magnitude lower than any conventional benchtop fluorescence microscope, and it could be significantly reduced further by large volume manufacturing. In particular, the heat sink, which costs $230, could potentially be replaced with much cheaper options. The presented design adds to the previous set of smartphone-based fluorescence microscopes, further pushing the boundaries for what is possible to achieve in terms of signal to noise ratio and resolution.4,21–23
Using the smartphone-based microscope we demonstrate how we can selectively detect Cronobacter spp. from contaminated PIF samples. We also show that the setup works with a general PNA-probe targeting all prokaryotes and compare the performance of our mobile system against that of a conventional fluorescence microscope. The assay is general, and we foresee that it could be used to identify bacteria in a variety of applications, including clinical samples.
Experimental
Design of smartphone integrated fluorescence microscope
A smartphone based and handheld fluorescence microscope was created by integrating the rear camera module of a Nokia Lumia 1020 smartphone with a 3D-printed optomechanical attachment. A laser diode (PLT5 488 nm, 60 mW, Φ 5.6 mm, Osram), powered by a rechargeable battery (3.7 V, 1700 mA h, Vivitar) and a constant output driver, was mounted in a collimation tube (LTN330-A, Thorlabs Inc.), used as a heat sink as well as for focusing the laser beam onto the sample. The laser diode was integrated into the sample stage attaining an experimentally optimized illumination angle (angle of incidence) of 61°. The light from the laser diode was focused by a 20 mm half ball lens, separated from a sample coverslip (22 mm × 50 mm, Premium Cover Glass 12-548-5E, Fisher Scientific) only by a thin layer of immersion oil (n = 1.51). Magnets were used to keep the coverslip holder in place, while still allowing for x/y-movement. A miniature dovetail stage (DT12, Thorlabs) was used for focusing, moving both laser diode and coverslip to keep the illumination angle constant. The fluorescence emission was collected through an integrated external lens (f = 2.6 mm, LS-40166 – M12xP0.5 Camera Lens) and passed through a long pass emission filter (>514 nm, 12.5 mm Diameter Raman Edge Filter, Edmund Optics) before reaching the smartphone camera lens (f = 7.2 mm) and finally recorded by the smartphone CMOS sensor (7152 × 5368 pixels). The inner walls of the prototype were covered with a black aluminum foil (T205-1.0, Thorlabs Inc.) to reduce autofluorescence of the prototype. Additionally, a white LED (897-1183-ND, DigiKey) was integrated into the sample stage for focusing.Evaluation of smartphone microscope
The accuracy and dynamic range for qualitative detection using the smartphone-based fluorescence microscope were evaluated by preparing three separate dilution series (N = 3) of fluorescent beads (FluoSpheres™ Carboxylate-Modified Microspheres, 0.5 μm, yellow-green fluorescent (505/515), ThermoFisher) in ultra-pure water, with concentrations ranging from 102 to 108 beads per mL. Twenty μL were placed on a coverslip (22 mm × 50 mm, Premium Cover Glass 12-548-5E, Fisher Scientific) and the solution was allowed to evaporate at room temperature prior to imaging, leaving only the fluorescent beads on the glass coverslips.Bacterial growth and culture media
The bacterial strains and species used in this study – Cronobacter sakazakii ATCC 29544 and Staphylococcus aureus ATCC 25923 were maintained on tryptic soy agar (TSA) plates (Liofilchem) at 37 °C, and streaked onto fresh plates every 48 h.SYTO 9 staining
The comparison between the performances of our smartphone setup and a conventional benchtop microscope was performed by preparing a serial of ten-fold dilutions of a pure culture suspension of Cronobacter sakazakii ATCC 29544, from 104 to 107 CFU mL−1, in sterile distilled water. 20 μL sample of each dilution (104, 105, 106 and 107 CFU mL−1) was placed on the glass microscope slides. The samples were then dried at 61 °C in a conventional incubator for 10 minutes. After the drying step, a 30 μL aliquot (12 μM) of SYTO9 (ThermoFisher) was applied directly to the samples and the samples were incubated in low light conditions for 10 min at room temperature (25 ± 1 °C). The samples were then washed by immersing the slides in a washing solution containing 5 mM Tris base (Sigma), 15 mM NaCl (Sigma) and 1% (v/v) of Triton X-100 (Sigma-Aldrich) for 30 minutes.EUB hybridization
A 107 CFU mL−1C. sakazakii ATCC 25944 suspension was prepared and placed on a microscope slide as described above. The fluorescent in situ hybridization was performed in three main steps: (i) sample fixation/permeabilization, (ii) hybridization and (iii) washing of the unbound probe. The dried 20 μL samples were covered with 30 μL of 4% (wt/vol) paraformaldehyde (Sigma), followed by 50% (vol/vol) ethanol (Fisher Scientific) for 10 min each, and subsequently air dried. The samples were then covered with 30 μL of hybridization solution containing 10% (wt/vol) dextran sulphate (Sigma), 10 mM NaCl (Sigma), 30% (vol/vol) formamide (Sigma), 0.1% (wt/vol) sodium pyrophosphate (Sigma), 0.2% (wt/vol) polyvinylpyrrolidone (Sigma), 0.2% (wt/vol) ficoll (Sigma), 5 mM disodium EDTA (Sigma), 0.1% (vol/vol) Triton X-100 (Sigma), 50 mM Tris–HCl (pH 7.5; Sigma), and 200 nM of EUB338 probe (Panagene). Samples were covered with coverslips, placed in moist chambers, and incubated for 60 min at 61 °C. Subsequently, the coverslips were removed, and the slides were submerged in a prewarmed (61 °C) washing solution containing 15 mM NaCl (Sigma), 1% (vol/vol) Triton X-100 (Sigma), and 5 mM Tris base (pH 10; Sigma). Washing was performed at 61 °C for 30 min, and the slides were subsequently air dried. The samples could be stored (dark) for a minimum 24 h before observation, if needed.Mixed culture experiments
C. sakazakii ATCC 25944 and S. aureus ATCC 25923, both in 107 CFU mL−1 suspensions, were prepared as described above and intermixed in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. 20 μL of sample was placed on the slide followed by EUB hybridization and subsequent DAPI staining. The EUB hybridization was performed as described above, with two modifications: (i) the CRONO probe (SakPNA971 with Alexa Fluor 488, Panagene) was used instead of the EUB probe and (ii) the hybridization time and temperature were 30 min and 57 °C, respectively. The DAPI staining was performed immediately after the CRONO probe hybridization. A 30 μL aliquot (0.1% wt/vol) of DAPI (Merck) was applied to the slide following incubation in low light conditions for 10 min at room temperature (25 ± 1 °C). Finally, the slides were washed in the same way as described above. The samples could be stored (dark) for a minimum 24 h before observation, if needed.
1 ratio. 20 μL of sample was placed on the slide followed by EUB hybridization and subsequent DAPI staining. The EUB hybridization was performed as described above, with two modifications: (i) the CRONO probe (SakPNA971 with Alexa Fluor 488, Panagene) was used instead of the EUB probe and (ii) the hybridization time and temperature were 30 min and 57 °C, respectively. The DAPI staining was performed immediately after the CRONO probe hybridization. A 30 μL aliquot (0.1% wt/vol) of DAPI (Merck) was applied to the slide following incubation in low light conditions for 10 min at room temperature (25 ± 1 °C). Finally, the slides were washed in the same way as described above. The samples could be stored (dark) for a minimum 24 h before observation, if needed.
Powdered infant formula experiments
For spiked powdered infant formula samples, a pure culture suspension of C. sakazakii ATCC 29544 was prepared in a phosphate-buffered saline (PBS) solution and adjusted to a cell density corresponding to approximately 108 cells per mL. The suspension was further diluted in PBS to obtain a concentration of 101 CFU to be inoculated on retail available powdered infant formula (Nan 1, Nestlé). Cell concentrations were confirmed by plating on TSA plates. 30 g portions of PIF were mixed with 270 mL of sterile distilled water in sealed stomacher bags (with filters). The samples were then artificially contaminated with 101 CFU of C. sakazakii ATCC 29544 followed by homogenization with a stomacher (Seward 3500) and incubated for 24 ± 2 hours at 37 °C. A pure PIF sample was included for each experiment to check for any possible natural contamination with Cronobacter spp. The samples for the FISH assay were taken from the enrichment broth and diluted with sterile distilled water in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 to reduce the intense background in the PIF samples. The samples were then stained as described previously for the CRONO probe.
5 to reduce the intense background in the PIF samples. The samples were then stained as described previously for the CRONO probe.
Reference benchtop imaging
Prior to acquiring images using the smartphone-based microscope, the slides were imaged using a conventional benchtop inverted fluorescence microscope (Zeiss AxioObserver.Z1) with a 63× oil immersion objective (NA = 1.46) in combination with an EMCCD camera (Andor Xion). The images were acquired as tiles of 15 × 15 individual images and the exposure time was set to 20 ms. To set a reference point, the slides were marked with a small dot using a regular marker pen prior to imaging. This made it possible to find the same position on both the benchtop microscope and the smartphone-based microscope. For the dual staining experiments, a Nikon Eclipse 80i epifluorescence microscope, equipped with a 60× oil immersion objective and a Nikon DS-Fi1 camera, was used. All images were acquired using DAPI and FITC filter sets.Smartphone image acquisition
The coverslips with fixed labeled bacteria were placed on the 3D-printed holder face down and immersion oil was placed on the back of the coverslip. The holder and coverslip were secured in the sample stage using magnets, pressing the back of the coverslip against the half ball lens, separated by a thin layer of the applied immersion oil. The white LED of the smartphone was used to locate the reference dot used for the benchtop images, as well as for preliminary focusing. Next, the 488 nm laser diode was turned on, focus fine-tuned, and images acquired of the bacteria on the slides. The coverslip holder was moved to image multiple positions per slide. The images were captured using the built in Nokia Pro Cam application with an integration time of 1/25 s (Fluorospheres and SYTO 9) or 1 s (EUB and CRONO) with additional settings kept the same (disabled auto-focusing, white balance as daylight, ISO as 100). The images were saved in lossless digital negative (DNG) format (43 MB) as well as in compressed JPEG images (1 MB), displayed on the smartphone screen.Image analysis
The smartphone images presented in the paper have been converted from the raw dng format to 16 bit tiff images. The CMOS sensor of the Nokia Lumia has a Bayer-pattern (RGGB). Since the emission from the fluorospheres, SYTO9, and the Alexa Fluor 488 dye is mainly in the green channel, the red and blue pixels were discarded and the image rotated 45 degrees, as described elsewhere,21 leaving only the green pixels in the final image.The number of fluorescent beads per mm2 in both the smartphone and the benchtop images was obtained by selecting a region of interest in each image for which the beads were counted using a custom written ImageJ macro (ESI Methods M1†). For the images acquired with the smartphone-based microscope at the lowest concentrations (102 to 104), the thresholding parameters in the macro were manually adjusted due to the very low number of beads compared to background.
The average signal to noise ratio (SNR) for bacteria was calculated from 50 individual bacterial cells on a coverslip. The signal to noise ratio is defined as: 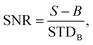 where S is the signal from the bacteria, B is the mean background intensity of a 9 × 9 pixel region of interest (ROI) adjacent to each bacterium, and STDB is the standard deviation of the background region.
where S is the signal from the bacteria, B is the mean background intensity of a 9 × 9 pixel region of interest (ROI) adjacent to each bacterium, and STDB is the standard deviation of the background region.
Results and discussion
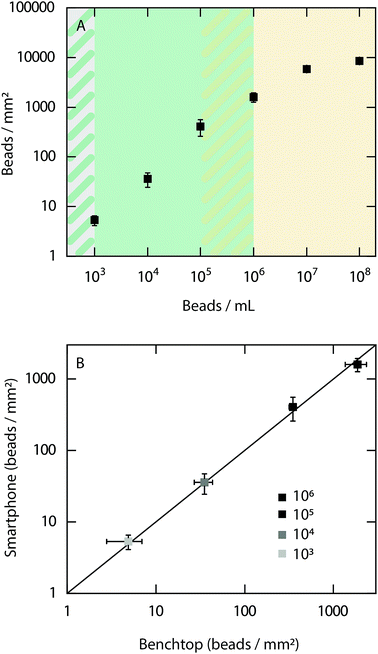
As an initial evaluation of the performance of the smartphone-based microscope, quantitative detection of 500 nm fluorescent beads (Ex/Em, 505/515 nm) at concentrations ranging from 102 to 108 beads per mL was performed (Fig. 2A).The results show that it is possible to quantitatively detect beads from a concentration of at least 105 beads per mL down to the limit of detection of 103 beads per mL. At concentrations above 106 beads per mL the bead density is too high to detect each bead individually. However, it should be noted that a qualitative read out is obtained at all concentrations investigated between 103 to 108 beads per mL. Example images of fluorescent beads visualized at different concentrations with the smartphone-based microscope can be found in ESI Fig. S1.† Next, the bead count within the dynamic range of the smartphone-based fluorescence microscope was compared with the detected number of beads when using a conventional benchtop microscope (Fig. 2B). The results show excellent agreement between both microscopes, demonstrating the precision and accuracy of the smartphone-based device.
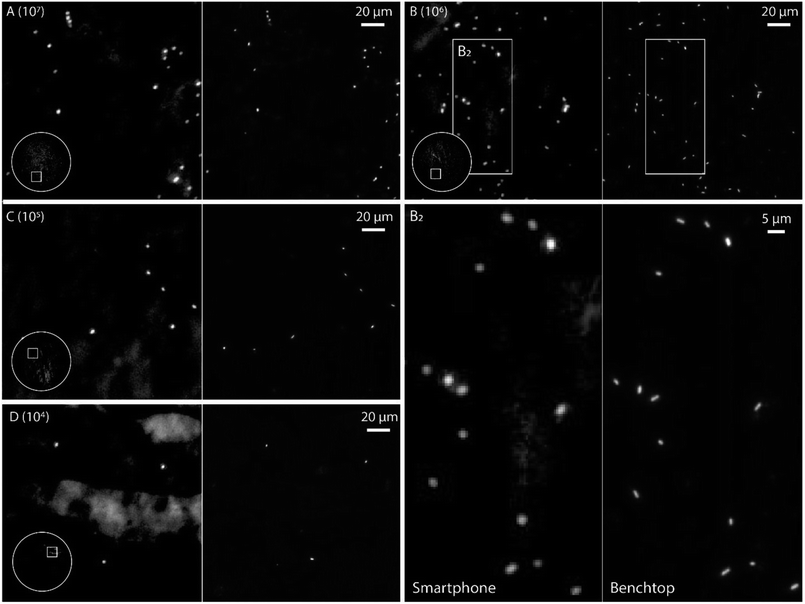
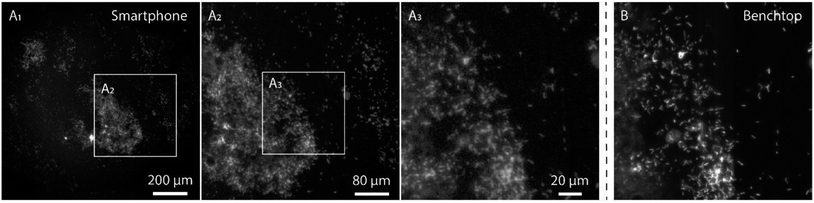
In order to evaluate the performance of the smartphone-based microscope on bacterial samples, Cronobacter spp. were stained with the general nucleic acid stain SYTO 9 (Ex/Em, 486/501 nm (RNA)). The Cronobacter spp. were cultured and diluted to obtain final concentrations of 104 to 107 colony forming units (CFUs) per mL and fixed on glass slides for subsequent imaging. Prior to each image acquired by the smartphone, a reference image of the same position on the glass slide was recorded using a conventional benchtop fluorescence microscope (Fig. 3, see Methods for details).
The images in Fig. 3 demonstrate that it is possible to detect fluorescently labeled bacteria using the smartphone-based microscope. Moreover, as predicted by the experiments on fluorescent beads, it is possible to detect bacteria down to concentrations of at least 104 CFUs per mL, which is comparable to the performance of a conventional fluorescence microscope.24 The extremely large field of view (FOV) of the smartphone (∼1 mm in diameter, i.e. ∼3.2 mm2, see inserts in Fig. 3) compared to the benchtop microscope (∼0.04 mm2 with a 63× objective-lens or ∼0.41 mm2 with a 20× objective-lens) is an advantage when quickly scanning an entire slide for bacteria and simplifies the detection process. The image quality of the smartphone microscope is, as expected, lower than that of the conventional microscope, however the results in Fig. 3 show that it is still more than enough for detecting the bacteria with a high degree of certainty.
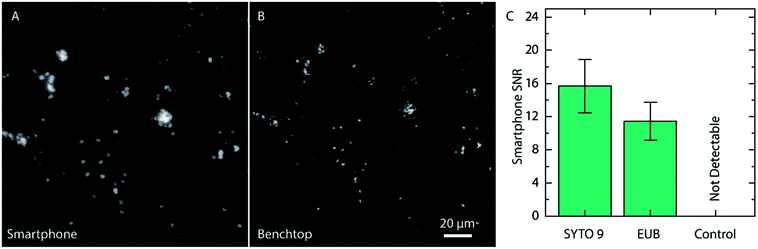
Even if the bacteria were straightforward to detect when stained with SYTO 9, the fluorophore is not specific to bacteria and targets all nucleic acids in the cell without any sequence specificity. Therefore, we evaluated the compatibility of the PNA-probe EUB338 (EUB) labeled with Alexa Fluor 488 (Ex/Em, 490/525 nm), which targets the common parts of the rRNA in all bacteria,25 making the probe universal for detection of bacteria. Since the PNA is designed to only bind to the rRNA of bacteria, the signal intensity is expected to be lower. Images of Cronobacter spp. stained with the EUB probe are shown in Fig. 4.
Once again, the same region on the slide was imaged using a conventional fluorescence microscope prior to acquiring the smartphone image. As for the samples stained with SYTO 9, the Cronobacter spp. were clearly visible also when using the EUB probe. Even if the signal from the EUB probe, as expected, was slightly lower than for SYTO 9, and provided, in the pure cultured samples, an SNR of 11.4 ± 2.3 (compared to 15.7 ± 3.2 for SYTO 9, Fig. 4C), the bacteria are still clearly visible. Moreover, negative controls were made with unlabeled bacteria to ensure that the autofluorescence of the bacteria does not provide a strong enough signal for detection (Fig. 4C and ESI Fig. S2†). When using the conventional microscope, the unlabeled bacteria provided a weak, but still measurable signal, approximately 47 times weaker than for Cronobacter spp. labeled with the EUB probe (ESI Fig. S2†). The drastically reduced emission intensity for the unlabeled bacteria rationalizes that they cannot be detected using the smartphone microscope and, in this way, do not cause false positives.
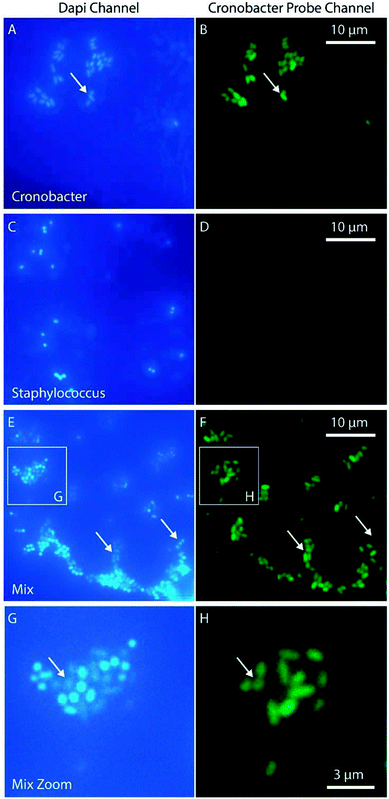
For some applications, such as qualitative detection of Cronobacter spp. in PIF, it is interesting to identify specific pathogenic bacteria in a complex matrix. With this aim, a PNA-probe designed to detect only Cronobacter spp. (CRONO Probe) was used. The probe is similar to the Cronobacter spp. specific probe SakPNA971,24 but with a different fluorophore attached (Alexa Fluor 488, Ex/Em 490/520 nm). In order to validate the specificity of the CRONO probe and provide an independent confirmation of our sample preparation method, a dual staining experiment was performed using a conventional microscope staining both Cronobacter spp. and Staphylococcus aureus with both the CRONO probe and the general DNA stain DAPI (Ex/Em, 350/470 nm) simultaneously (Fig. 5).
The results in Fig. 5 demonstrate how the Cronobacter spp. bacteria are clearly visible both in the DAPI and CRONO emission channels (Fig. 5A and B). For Staphylococcus, the bacteria are visible in the DAPI channel, but not in the CRONO channel (Fig. 5C and D), demonstrating the specificity of the CRONO probe to Cronobacter spp. Moreover, the lack of emission from the Staphylococcus bacteria in the CRONO channel shows that the unbound probe is efficiently removed during the washing step. A mix of Cronobacter spp. and Staphylococcus was also prepared (Fig. 5E, and 5F), demonstrating how the rod shaped Cronobacter spp. bacteria are visible in both channels, compared to the more spherically shaped Staphylococcus bacteria that are only visible in the DAPI channel. From the same figure it is also clear that the intensity in the DAPI channel is lower for Cronobacter spp. compared to Staphylococcus. This could be explained by resonance energy transfer between DAPI and the CRONO probe, were the emission spectrum of DAPI overlaps with the excitation spectrum of the CRONO probe. Extensive evaluation and verification of the specificity of the CRONO (SakPNA971) probe can be found elsewhere.24,26 The SNR from Cronobacter spp. stained with the CRONO probe was measured to 12.3 ± 2.1 (N = 50) using the smartphone microscope, thus providing a similar value to the other PNA probe (EUB).
Cronobacter spp., which are frequently found in PIF, are capable of causing severe infections with a high fatality rate in newborn infants.18–20 Therefore, as a final evaluation, we used the smartphone-based fluorescence microscope to detect Cronobacter spp. in contaminated PIF samples, which are expected to provide a higher background signal than pure bacterial cultures due to the autofluorescence of the formula matrix, increasing the difficulty of bacterial detection. A small sample of contaminated PIF, with a bacteria concentration of only 10 CFU per volume of 30 g of PIF (standard sampling size), was dissolved in distilled water prior to incubation and staining (details in Methods). In order to minimize any additional background from the formula matrix, the slides were washed, removing the unbound CRONO probe prior to imaging.
Example images from both the smartphone and benchtop microscopes can be seen in Fig. 6, showing that it is possible to qualitatively detect the bacteria also in the PIF samples. As discussed above, the large FOV of the smartphone proved to be an important advantage compared to a conventional microscope for effortlessly scanning large areas of the slide for bacteria. The reference image acquired with the benchtop microscope shows once again that the loss in quality using the smartphone microscope is not an issue for the final readout of the assay. Since the bacteria are visible directly on the smartphone screen (ESI Fig. S3†), there is no need for any additional data analysis that would require transmitting the data to distant computers, further promoting the use of the device at remote settings.
For the application of detecting fluorescently labeled bacteria, this study demonstrates a smartphone microscope design that allows for qualitative detection even in a single image. However, for future applications, when detection of even weaker signals might be required, there is still the possibility of averaging many frames in order to significantly increase the SNR.21 Moreover, deep learning approaches have recently shown promising results in transforming lower resolution and/or aberrated microscopic images into images that match the quality of high-end diffraction-limited microscopes and might be used to further improve the image quality.27,28 It should be noted that none of these approaches require any alterations or improvements to the design of the smartphone microscope presented in this study, since they are only used in the subsequent image processing.
Previous smartphone based approaches for detection of bacteria have been based on indirect methods,2,10 and more recently direct imaging of aptamer-functionalized fluorescent magnetic nanoparticles bound to the outer surface of bacteria was demonstrated.9 Compared to these approaches, the PNA-FISH assay allows us to directly target the nucleic acids within a bacterial cell. Hence, the assay has the potential to detect different mutations within a bacterial cell, as well as target specific genes, such as those encoding antibiotic resistance. Moreover, adding to previous designs, the advanced smartphone-based fluorescence microscope, provided at low cost, could open a myriad of additional applications in resource limited settings for POC applications, such as further advances in genetic-based detection, immunoassay quantification, sensing of viruses or detection of microorganisms and parasites. Also, the broad availability of smartphones globally makes them a perfect platform for portable, cost-efficient diagnostic tools.
As demonstrated here, the combination of smartphone-based microscopy with a well-designed assay, such as utilizing highly specific PNA probes, can be an invaluable tool for simple qualitative detection of pathogenic bacteria. As in the case of the PIF samples, the targeted Cronobacter spp. bacteria were clearly visible on the smartphone screen, even in the presence of the PIF matrix. The presented assay could improve food safety, which is crucial especially when the end users are as sensitive as infants, and where infections from contaminated PIF can have a fatal outcome. The time required to perform the PNA-based FISH assay is much less than culture based approaches and complementing biochemical tests commonly used to confirm the presence of an organism, which take up to a week for completion.29–31 Regarding novel molecular technologies, such as PCR-based and CRISPR assays,29,30,32–34 smartphone FISH does not require the use of enzymes, and it is hence less prone to inhibiting substances or the action of proteases.17 Furthermore, in contrast to PCR, smartphone FISH based on PNA probes is not sensitive to the presence of nucleases, reducing the requirements for sterile sample preparation. Finally, FISH has the potential to be carried out at room temperature,35 circumventing the need for the high temperature as in PCR based strategies, such as LAMP.36
Moreover, even if beyond the scope of this paper, we foresee that the PNA-based FISH assay should be straightforward to adapt for bacterial identification in other types of samples, such as cerebrospinal fluid and blood.37–39 Even if the PNA-FISH protocol in its current form does not require high-end lab-grade equipment, further simplifications of its protocol would increase the range of applications that it can be used for. As such, and particularly in resource limited settings, the smartphone-based microscope in combination with the PNA assay could be used in hospitals, as well as labs where a quick analysis of a few samples is needed each day, or even for control of food safety directly in a factory, due to its simplicity and cost-effectiveness. For applications such as the one presented here, only a qualitative read out is necessary for the end user since any initial amount of contaminating pathogenic bacteria is unacceptable and pre-enrichment is always performed. However, if needed in other applications, a calibration curve can be established similar to the one in Fig. 2, allowing for an additional quantitative readout of bacterial, or similar, samples. Moreover, the wireless link between a smartphone and computers could be very useful for quick transfer and documentation of the obtained results.
Conclusions
In conclusion, we demonstrate how a smartphone-based fluorescence microscope can be used in combination with highly specific PNA probes to detect pathogenic bacteria in complex samples. We have established that the smartphone setup can detect bacteria down to at least 104 CFUs per mL, comparable to a conventional microscope, and evaluated the compatibility with both a general nucleic acid stain and species-specific bacterial PNA probes. Even in a complex sample, such as powdered infant formula, we were able to acquire high quality images of pathogenic Cronobacter spp. bacteria using the smartphone-based microscope, demonstrating the great potential of the presented setup. Moreover, bacteria can be qualitatively detected directly on the smartphone screen, with no need for additional data analysis, further promoting the use in resource limited settings. We foresee that the assay presented here also could be used for bacterial detection in other samples, for example of clinical origin, and that the smartphone-based microscope in general could find a variety of additional applications in other fluorescence-based assays.Conflicts of interest
N. F. A. and L. C. are co-founders and shareholders of Biomode, a company that specializes in the development of PNA-FISH methods for the detection of microorganisms. A. O. has pending patent applications on mobile phone-based microscopy technologies.Acknowledgements
This project was funded by a grant to F. W. from the Bill and Melinda Gates Foundation (OPP1158530) and a grant to F. W. and N. F. A. from EuroNanoMedII (NanoDiaBac project). N. F. A. acknowledges funding from POCI-01-0145-FEDER-006939 (LEPABE-UID/EQU/00511/2013) funded by the European Regional Development Fund (ERDF), through COMPETE2020 and by national funds, through FCT – Fundação para a Ciência e a Tecnologia; and NORTE-01-0145-FEDER-000005 – LEPABE-2-ECO-INNOVATION, supported by the North Portugal Regional Operational Programme (NORTE 2020). The Ozcan Research Group at UCLA acknowledges the support of NSF Engineering Research Center (ERC, PATHS-UP) and the Howard Hughes Medical Institute (HHMI).References
- A. Ozcan, Lab Chip, 2014, 14, 3187–3194 RSC.
- C. F. Fronczek, T. S. Park, D. K. Harshman, A. M. Nicolini and J.-Y. Yoon, RSC Adv., 2014, 4, 11103–11110 RSC.
- S. Cho, A. Islas-Robles, A. M. Nicolini, T. J. Monks and J.-Y. Yoon, Biosens. Bioelectron., 2016, 86, 697–705 CrossRef CAS.
- Q. Wei, H. Qi, W. Luo, D. Tseng, S. J. Ki, Z. Wan, Z. Gorocs, L. A. Bentolila, T. T. Wu, R. Sun and A. Ozcan, ACS Nano, 2013, 7, 9147–9155 CrossRef CAS.
- S. K. Vashist, O. Mudanyali, E. M. Schneider, R. Zengerle and A. Ozcan, Anal. Bioanal. Chem., 2014, 406, 3263–3277 CrossRef CAS.
- O. Mudanyali, S. Dimitrov, U. Sikora, S. Padmanabhan, I. Navruz and A. Ozcan, Lab Chip, 2012, 12, 2678–2686 RSC.
- H. Zhu, U. Sikora and A. Ozcan, Analyst, 2012, 137, 2541–2544 RSC.
- A. Tapley, N. Switz, C. Reber, J. L. Davis, C. Miller, J. B. Matovu, W. Worodria, L. Huang, D. A. Fletcher and A. Cattamanchi, J. Clin. Microbiol., 2013, 51, 1774–1778 CrossRef.
- S. Shrivastava, W.-I. Lee and N.-E. Lee, Biosens. Bioelectron., 2018, 109, 90–97 CrossRef CAS.
- A. M. Nicolini, C. F. Fronczek and J.-Y. Yoon, Biosens. Bioelectron., 2015, 67, 560–569 CrossRef CAS.
- World Health Organization, Antimicrobial resistance: global report on surveillance, 2014 Search PubMed.
- The Review on Antimicrobial Resistance, Tackling drug-resistant infections globally: final report and recommendations, 2016 Search PubMed.
- R. Amann and B. M. Fuchs, Nat. Rev. Microbiol., 2008, 6, 339–348 CrossRef CAS.
- A. M. Costa, F. J. Mergulhao, R. Briandet and N. F. Azevedo, Future Microbiol., 2017, 12, 987–999 CrossRef CAS.
- H. Stender, M. Fiandaca, J. J. Hyldig-Nielsen and J. Coull, J. Microbiol. Methods, 2002, 48, 1–17 CrossRef CAS.
- L. Cerqueira, N. F. Azevedo, C. Almeida, T. Jardim, C. W. Keevil and M. J. Vieira, Int. J. Mol. Sci., 2008, 9, 1944–1960 CrossRef CAS.
- C. Schrader, A. Schielke, L. Ellerbroek and R. Johne, J. Appl. Microbiol., 2012, 113, 1014–1026 CrossRef CAS.
- D. Drudy, M. O'Rourke, M. Murphy, N. R. Mullane, R. O'Mahony, L. Kelly, M. Fischer, S. Sanjaq, P. Shannon, P. Wall, M. O'Mahony, P. Whyte and S. Fanning, Int. J. Food Microbiol., 2006, 110, 127–134 CrossRef CAS.
- J. Chap, P. Jackson, R. Siqueira, N. Gaspar, C. Quintas, J. Park, T. Osaili, R. Shaker, Z. Jaradat, S. H. Hartantyo, N. Abdullah Sani, S. Estuningsih and S. J. Forsythe, Int. J. Food Microbiol., 2009, 136, 185–188 CrossRef CAS.
- O. Holy and S. Forsythe, J. Hosp. Infect., 2014, 86, 169–177 CrossRef CAS PubMed.
- Q. Wei, W. Luo, S. Chiang, T. Kappel, C. Mejia, D. Tseng, R. Y. L. Chan, E. Yan, H. Qi, F. Shabbir, H. Ozkan, S. Feng and A. Ozcan, ACS Nano, 2014, 8, 12725–12733 CrossRef CAS.
- Q. Wei, G. Acuna, S. Kim, C. Vietz, D. Tseng, J. Chae, D. Shir, W. Luo, P. Tinnefeld and A. Ozcan, Sci. Rep., 2017, 7, 2124 CrossRef.
- M. Kühnemund, Q. Wei, E. Darai, Y. Wang, I. Hernández-Neuta, Z. Yang, D. Tseng, A. Ahlford, L. Mathot, T. Sjöblom, A. Ozcan and M. Nilsson, Nat. Commun., 2017, 8, 13913 CrossRef.
- C. Almeida, N. F. Azevedo, C. Iversen, S. Fanning, C. W. Keevil and M. J. Vieira, Appl. Environ. Microbiol., 2009, 75, 2925–2930 CrossRef CAS.
- R. S. Santos, N. Guimaraes, P. Madureira and N. F. Azevedo, J. Biotechnol., 2014, 187, 16–24 CrossRef CAS.
- R. Rocha, M. Sousa, L. Cerqueira and N. F. Azevedo, Comparative Evaluation of the Biomode 2, S.A. Probe4Cronobacter for the Identification of Cronobacter Species in Infant Formula, AOAC Performance Tested, Certification Number 081702, 2017 Search PubMed.
- Y. Rivenson, H. Ceylan Koydemir, H. Wang, Z. Wei, Z. Ren, H. Günaydın, Y. Zhang, Z. Göröcs, K. Liang, D. Tseng and A. Ozcan, ACS Photonics, 2018, 5, 2354–2364 CrossRef CAS.
- Y. Rivenson, Z. Göröcs, H. Günaydin, Y. Zhang, H. Wang and A. Ozcan, Optica, 2017, 4, 1437–1443 CrossRef.
- M. K. Mohan Nair and K. S. Venkitanarayanan, Appl. Environ. Microbiol., 2006, 72, 2539–2546 CrossRef.
- S. Derzelle and F. Dilasser, BMC Microbiol., 2006, 6, 100 CrossRef PubMed.
- X. Song, S. Shukla and M. Kim, Food Microbiol., 2018, 72, 23–30 CrossRef CAS.
- Y. Liu, Q. Gao, X. Zhang, Y. Hou, J. Yang and X. Huang, Mol. Cell. Probes, 2006, 20, 11–17 CrossRef CAS.
- Y. Liu, X. Cai, X. Zhang, Q. Gao, X. Yang, Z. Zheng, M. Luo and X. Huang, J. Microbiol. Methods, 2006, 65, 21–31 CrossRef CAS PubMed.
- J. S. Gootenberg, O. O. Abudayyeh, J. W. Lee, P. Essletzbichler, A. J. Dy, J. Joung, V. Verdine, N. Donghia, N. M. Daringer, C. A. Freije, C. Myhrvold, R. P. Bhattacharyya, J. Livny, A. Regev, E. V. Koonin, D. T. Hung, P. C. Sabeti, J. J. Collins and F. Zhang, Science, 2017, 356, 438–442 CrossRef CAS.
- S. Fontenete, N. Guimarães, M. Leite, C. Figueiredo, J. Wengel and N. Filipe Azevedo, PLoS One, 2013, 8, e81230 CrossRef.
- K. Yogesh, B. Sangita and J. Pranita, Compr. Rev. Food Sci. Food Saf., 2017, 16, 1359–1378 CrossRef.
- H. Hartmann, H. Stender, A. Schäfer, I. B. Autenrieth and V. A. J. Kempf, J. Clin. Microbiol., 2005, 43, 4855–4857 CrossRef CAS.
- M. Søgaard, H. Stender and H. C. Schønheyder, J. Clin. Microbiol., 2005, 43, 1947–1949 CrossRef.
- D. A. Wilson, M. J. Joyce, L. S. Hall, L. B. Reller, G. D. Roberts, G. S. Hall, B. D. Alexander and G. W. Procop, J. Clin. Microbiol., 2005, 43, 2909–2912 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra06473c |
| This journal is © The Royal Society of Chemistry 2018 |